Abstract
Combinations of reverse transcriptase (RT) inhibitors are currently used in anti-human immunodeficiency virus therapy in order to prevent or delay the emergence of resistant virus and to improve the efficacy against viral enzymes carrying resistance mutations. Drug-drug interactions can result in either positive (additive or synergistic inhibition) or adverse (antagonistic interaction, synergistic toxicity) effects. Elucidation of the nature of drug interaction would help to rationalize the choice of antiretroviral agents to be used in combination. In this study, different combinations of nucleoside and nonnucleoside inhibitors, including d- and l-(β)-deoxy- and -dideoxynucleoside triphosphate analogues, have been tested in in vitro RT assays against either recombinant wild-type RT or RT bearing clinically relevant nonnucleoside inhibitor resistance mutations (L100I, K103N, Y181I), and the nature of the interaction (either synergistic or antagonistic) of these associations was evaluated. The results showed that (i) synergy of a combination was not always equally influenced by the individual agents utilized, (ii) a synergistic combination could improve the sensitivity profile of a drug-resistant mutant enzyme to the single agents utilized, (iii) l-(β)-enantiomers of nucleoside RT inhibitors were synergistic when combined with nonnucleoside RT inhibitors, and (iv) inter- and intracombination comparisons of the relative potencies of each drug could be used to highlight the different contributions of each drug to the observed synergy.
The majority of the drugs currently utilized for the clinical treatment of human immunodeficiency virus type 1 (HIV-1)-infected individuals are targeted against the viral reverse transcriptase (RT), the enzyme responsible for the conversion of viral genomic single-stranded RNA into double-stranded proviral DNA (2, 21, 22). These drugs can be divided into two broad classes: (i) dideoxynucleoside analogues (or nucleoside RT inhibitors [NRTIs]), such as zidovudine (3′-azido-2′,3′-dideoxythymidine) (AZT), zacitalbine (2′,3′-dideoxycytidine) (ddC), didanosine (2′,3′-dideoxyinosine) (ddI), stavudine (2′,3′-didehydro-2′,3′-dideoxythymidine) (d4T), lamivudine [(−)-β-l-2′-deoxy-3′-oxa-4′-thiocytidine] (3TC), and abacavir, which inhibit viral replication by acting in their triphosphate form as chain terminators of DNA synthesis, and (ii) nonnucleoside analogues (or nonucleoside RT inhibitors [NNRTIs]), including structurally different molecules, such as nevirapine, delavirdine, and efavirenz, which bind to a common allosteric site of RT distinct from the polymerase active site, thus inhibiting catalysis. Due to the emergence of drug resistance mutations in the RT gene, which is readily accomplished in vivo due to the low fidelity of RT and the massive viral turnover, all these drugs showed a significant but limited and transient beneficial effect on inhibition of viral replication when administered in monotherapy regimens. In addition, many of the selected mutations display cross-resistance to other NRTIs and NNRTIs (23, 29). Multiple drug combinations have been shown to suppress viral load for relatively longer periods of time compared to monotherapy (2, 10). Combination therapy could allow administration of lower dosages of individual drugs than monotherapy regimens, thus limiting the toxic side effects, and it could result in potentiation of their therapeutic efficacy due to synergism among the different compounds administered. Indeed, several studies reported synergistic activities for different combinations of NRTIs with NNRTIs and even of NNRTIs with NNRTIs (4, 9, 10, 19, 30, 31, 35, 37, 39). Combination of drugs does not always result in beneficial effects, and several examples have been reported of antagonistic activity, increased toxicity, and metabolic interference as well as increased drug resistance mutation rates for certain drug associations (2, 10). An additional important factor affecting the interaction between RT inhibitors in vivo is the heterogeneity of the viral population, where mutant virus strains which exhibit reduced susceptibility to RT inhibitors are present. Thus, expanding knowledge of the efficacy of RT inhibitor combinations against viral enzymes carrying resistance mutations to one or more of the utilized drugs would allow a better prediction of the in vivo outcome of the combinations. The evaluation of positive (synergistic) or negative (antagonistic) effects of combinations of RT inhibitors could be incorporated into screening programs for new drugs. Such approaches would allow the characterization of the efficacy of a new RT inhibitor in terms of its favorable (i.e., synergistic) action with other drugs in suppressing HIV-1 RT activity. Synergy assessment has been performed in the majority of investigations using infected-cell-based assays. However, in complex biological systems such as infected cells, it is very difficult to determine the precise mechanisms of any observed drug-drug interaction. Moreover, the impossibility of knowing the exact concentration of the drug within the cell prevents any detailed enzymological study of the interaction of different inhibitors at the level of their molecular target(s). Enzymatic assays employing purified enzymes are more suitable for kinetic studies, and indeed some detailed enzymological analyses of drug-drug interaction using such approaches have been published (5, 7, 15, 28, 36, 39). In many cases good correspondence between the results obtained with enzymatic assays and infected-cell-based assays has been found (7, 15, 39), even if the behavior of the drugs used in the combinations can be influenced by the reaction conditions (1, 6, 37). In the present work, different combinations of NRTIs and NNRTIs have been tested against recombinant RTs, either wild type (wt) or bearing clinically relevant NNRTI resistance mutations. It has been shown that the compound 3TC, with the unnatural l-(β)- conformation, selected for uncommon resistance mutations at Met184 of RT which were able to restore AZT sensitivity when expressed in a resistant genetic background (33). Since other l-(β)-NRTIs, such as l-(β)-ddC and -(−)-β-l-2′-deoxy-3′-oxa-4′-thiocytidine (dOTC), have been shown to potently inhibit as triphosphates HIV-1 RT as well as virus replication in infected cells (13, 16, 26, 27, 35), we wanted to further investigate the interaction of l-(β)-NRTIs with other NRTIs and NNRTIs. We have focused our attention on the clinically used NNRTIs nevirapine and efavirenz. In particular for efavirenz, the synergistic effects of its combination with d- and l-(β)-dideoxynucleoside triphosphate analogues were studied since, in spite of a large body of data about efavirenz's clinical use in combination with AZT and 3TC, there are few detailed studies on the nature of its interaction with NRTIs (38). Recently, our group identified a specific mechanism of action for efavirenz, which might suggest a possible synergistic action of this compound in combination with other NNRTIs (28). Thus, the effect of efavirenz association with nevirapine and nevirapine plus AZT was also studied. Our results highlight the different natures of the interactions among these drugs and suggest that the use of synergistic NRTI-NNRTI combinations could also be effective against NNRTI-resistant mutants and that l-(β)-dideoxynucleoside triphosphate analogues are synergistic when used in combination with NRTIs and NNRTIs in place of the corresponding d-enantiomers.
MATERIALS AND METHODS
Chemicals.
[3H]2′,3′-deoxythymidine triphosphate (dTTP) (40 Ci/mmol) was from Amersham, and unlabeled deoxynucleoside triphosphates (dNTPs) and ddNTPs were from Boehringer. Whatman was the supplier of the GF/C filters. All other reagents were of analytical grade and were purchased from Merck or Fluka. Efavirenz has been synthesized according to procedures of L. Tan et al. (34). The final preparation showed the following physicochemical properties. mp 133 to 136°C (hexane/toluene). High-performance liquid chromatography analysis: RT = 6.57 min. 1H NMR (CDCl3)_: 0.85, (m, 2H), 0.94 (m, 2H), 1.40 (m, 1H), 6.81 (d, J = 8.5 Hz, 1H), 7.37 (dd, J = 2.5, 8.5 Hz, 1H), 7.49 (d, J = 2.5 Hz, 1H), 8.71 (br s, 1H). 13C NMR 148.0, 133.2, 131.6, 129.0, 127.8, 127.7, 123.9, 120.1, 116.3, 115.7, 95.8, 77.3, 77.1, 76.9, 76.5, 55.0, 8.9, −0.7. MS m/z (ra%): 315 (M+, 30), 248 (23), 246 (100), 243 (33), 182 (13), 180 (36), 167 (12). Analysis calculated for: C14H9NO2CIF3; C, 53.27; H, 2.87; N, 4.44. Found: C, 52.90; H, 2.92; N, 4.77. l-(β)-dTTP and l-(β)-2′,3′-dideoxycytidine triphosphate (ddCTP) were synthetized as described previously (13, 16). 3′-Azido-2′,3′-dideoxythymidine triphosphate (AZTTP) was from USB. Nevirapine was a gift from M. Botta (University of Siena).
Nucleic acid substrates.
The homopolymer poly(rA) (Pharmacia) was mixed at weight ratios in nucleotides of 10:1 with the oligomer oligo(dT)12–18 (Pharmacia) in 20 mM Tris-HCl (pH 8.0) containing 20 mM KCl and 1 mM EDTA, heated at 65°C for 5 min, and then slowly cooled at room temperature. Preparation of d24:d66-mer deoxyoligonucleotide was as previously described (26).
Expression and purification of recombinant HIV-1 RT forms.
Recombinant RT, either wt or mutated, was expressed and purified to >95% purity as described (25). Purified enzymes had the following specific activities on poly(rA) · oligo(dT) (see below): HIV-1 p66(His)/p51, 75,670 U/mg; p66(L100I)/p51, 56,690 U/mg; p66(K103N)/p51, 96,415 U/mg; p66(Y181I)/p51, 65,770 U/mg. One unit of DNA polymerase activity corresponds to the incorporation of 1 nmol of dNMP into acid-precipitable material in 60 min at 37°C.
HIV-1 RT RNA- or DNA-dependent DNA polymerase activity assay.
RNA-dependent DNA polymerase activity of RT was assayed as follows. A final volume of 25 μl contained buffer A (50 mM Tris-HCl [pH 7.5], 1 mM dithiothreitol, 0.2 mg of bovine serum albumin per ml, 4% glycerol), 10 mM MgCl2. 0.5 μg of poly(rA) · oligo(dT)10:1 (0.3 μM 3′-OH ends), 10 μM [3H]dTTP (1 Ci/mmol), and 2 to 4 nM RT. Reaction mixtures were incubated for 10 min at 37°C. Twenty-microliter aliquots were then spotted on glass fiber GF/C filters, which were immediately immersed in 5% ice-cold trichloroacetic acid (TCA). Filters were washed twice in 5% ice-cold TCA and once in ethanol for 5 min and then were dried, and acid-precipitable radioactivity was quantitated by scintillation counting.
DNA-dependent DNA synthesis activity of RT was assayed in buffer A in the presence of 0.3 μM (3′-OH ends) of a d66-mer oligodeoxynucleotide corresponding to nucleotides (nt) 1006 to 1071 of the sequence of the HIV-1 pol gene (codons 169 to 190) annealed to a d24-mer complementary primer, 10 μM concentrations each of dATP, dGTP, and dCTP, 10 μM [3H]dTTP (10 Ci/mmol), and 2 to 4 nM RT. Reaction mixtures were incubated for 10 min at 37°C, and reactions were stopped by addition of 5 μl of 0.4 M EDTA along with 200 μg of salmon sperm carrier DNA. Twenty-microliter aliquots were then spotted on glass fiber GF/C filters, which were immediately immersed in 5% ice-cold TCA. Filters were washed twice in 5% ice-cold TCA and once in ethanol for 5 min and then were dried, and acid-precipitable radioactivity was quantitated by scintillation counting
Inhibition assays.
Reactions were performed under the conditions described for the RNA- or DNA-dependent DNA synthesis activity of RT. Incorporation of radioactive dTTP into poly(rA) · oligo(dT) or d24:d66-mer at different concentrations of DNA or dNTPs was monitored in the presence of increasing amounts of inhibitors, either alone or in combination at fixed molar ratios. Drugs combinations were as follows (fixed molar ratios [M]/[M]): [NVP]/[ddTTP], 1.5:1; [EFV]/[AZTTP], 5:1; [NVP]/[L-(β)-dTTP], 1:140; [EFV]/[L-(β)-dTTP], 1:1,400; [EFV]/[L-(β)-ddCTP] and [EFV]/[D-(β)-ddCTP], 1:550; [NVP]/[EFV], 20:1 for wt RT (RTwt), 200:1 for L100I; [AZTTP]/[EFV]/[NVP], 1:4:12.5.
Determination of synergy.
The terms of agent interactions have been defined in different ways. In the present work, the consensus terminology established at the Fifth International Conference on the Combined Effects of Environmental Factors was used (17). Analysis of the interaction between two agents, both effective individually, has been performed according to the null reference mode of Loewe additivity (24). Inhibitors were combined at fixed molar ratios depending on their different potencies in order to ensure that all the compounds significantly contributed to the inhibition observed. Interaction indexes were derived according to earlier guidelines (3). The cases in which the observed effects were either significantly more or less than those predicted by the reference model for additivity were considered synergism or antagonism, respectively. These corresponded to interaction index (I) values of <1 for synergism or of >1 for antagonism.
The method was based on the additivity model originally developed by Loewe and Muischnek (24) and successively implemented by Greco et al. (18). Dose-response curves for drug action were assumed to follow the model originally developed by Hill (20), adapted following the guidelines of Greco et al. (18), and were generated by fitting the experimental data to the equation
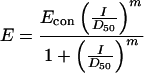 |
1 |
where E is the observed effect (% of activity), Econ is the control effect (activity in the absence of the inhibitor), and D50 is the concentration of inhibitor giving 50% inhibition.
The parameter m is the sigmoidicity term. The validity of the assumption of the Hill model for dose-response curves was tested by calculating the Ki values for each inhibitor according to a fully competitive (NRTI) or noncompetitive (NNRTI) mechanism (11). The Ki values were then compared with the respective D50 values calculated by equation 1, according to the relationships Ki = D50 for the noncompetitive cases and Ki = D50/(1+[S]/Km) for the competitive cases. In all cases, optimal correlation was found between D50 and Ki values (not shown).
For the combination of two drugs at a fixed molar ratio (R = [drug1]/[drug2]), D1 and D2 values were calculated from the D50 value derived from equation 1, with (D1 + D2) = D50 and D1 = RD2.
Expected D1, D2, and Di values for the combination of i drugs under the null reference hypothesis of no interaction were derived by inserting estimated D50 and m values for each drug in the combination in the specific form of the Loewe additivity equation, which assumes that equation 1 is appropriate for each drug individually (18).
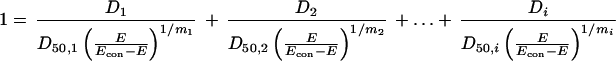 |
2 |
The null reference hypothesis of no interaction (equation 2) corresponded to I = 1. It must be noted that inhibitory doses calculated according to equations 1 and 2 were designed with the symbol D, whereas the correspondent values derived from equation 3 were indicated with the symbol ID. This was in order to be consistent with the different method used for calculations. In all cases, D50 = ID50.
Expected inhibitor concentrations at different fractional inhibitions were calculated from the parameters D50, Econ, and m according to the equation
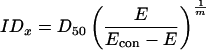 |
3 |
where IDx is the dose of drug giving x% of inhibition. I was then calculated according to Berenbaum (3) by the equation
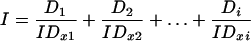 |
4 |
where D1, D2, and Di were the concentrations of the drugs in combination, and IDx1, IDx2, and IDxi are the predicted inhibitory concentrations of each drug individually giving the observed effect of the combination. An I value of <1 indicates synergy, >1 indicates antagonism, and 1 indicates additivity, according to the Loewe additivity model.
All the analyses were based on the results of three independent experiments for each drug combination, and the standard deviation values for each parameter estimate are indicated. Values were calculated by non-least squares computer fitting of the data to the appropriate equations. It has been shown that an experimental factor affecting the relative potency (and also any observed synergistic effect) of an NRTI is the number of possible incorporation sites along the template to be replicated (37). For this reason, in order to study the effect of NNRTIs in combination with AZTTP, a synthetic homopolymeric template, poly(rA) · oligo(dT), in which the number of possible termination sites by AZTTP incorporation was not limiting, was used. This approach allowed the determination of the eventual synergism of AZTTP-NNRTI association without any template-dependent effect. Synergistic combinations were then tested on a more natural template, a heteropolymeric deoxyoligonucleotide substrate corresponding to nt 1061 to 1071 of the HIV-1 pol gene, which is a sequence which is physiologically replicated by HIV-1 RT. This template also possessed a nonlimiting number of possible incorporation sites for dideoxythymidine and -cytidine nucleoside triphosphate analogs. In order to directly verify that the nature of the substrate and/or the assay conditions did not affect the observed synergy, we have tested a double combination of ddCTP and AZTTP against RTwt on the heteropolymeric template. As expected, this combination showed only an additive effect, with I = 0.97 (data not shown).
Statistical analysis.
Equation 2 was used to estimate the expected values for each drug in combination under the hypothesis of additivity. Mean values and standard deviations for observed and expected value data sets were calculated, and a Student's t test was then conducted under the hypothesis that both mean values were equal. Successively, a modification of the method of Drewinko et al. (12) was used as an additional approach. Difference scores were calculated by subtracting expected from observed values for each drug. Mean difference score values and standard deviations for each drug were calculated, and a Student's t test was performed to test the hypothesis that the true mean value of the differences between observed and expected data sets was zero. As an example, observed and expected values for the efavirenz-AZTTP combination against RTwt, along with statistics, are listed in Table 2. Both approaches were consistent with significant synergy (P < 0.05). All the combinations which showed I values of <1 were analyzed in the same way, and significant synergy (P < 0.05) was found in all cases.
TABLE 2.
Comparison of observed and expected values and statistical analysis of the synergistic inhibition of the synthetic activity of wt HIV-1 RT by the efavirenz-AZTTP combination
| Ea (%) and statistics | Mean (nM) ± SDb
|
Δobs-exp[EFV] | Δobs-exp[AZTTP] | |||
|---|---|---|---|---|---|---|
| [EFV]obs for D1 | [EFV]exp for D1 | [AZTTP]obs for D2 | [AZTTP]exp for D2 | |||
| 10 | 1.4 ± 0.2 | 0.48 ± 0.01 | 0.28 ± 0.01 | 0.094 ± 0.002 | 0.92 | 0.18 |
| 20 | 3.1 ± 0.02 | 2.5 ± 0.01 | 0.62 ± 0.05 | 0.5 ± 0.02 | 0.6 | 0.12 |
| 30 | 5.36 ± 0.01 | 7.83 ± 0.01 | 1.07 ± 0.01 | 1.56 ± 0.01 | −2.47 | −0.49 |
| 40 | 8.33 ± 0.01 | 17.22 ± 0.2 | 1.67 ± 0.02 | 3.44 ± 0.02 | −8.89 | −1.77 |
| 50 | 12.5 ± 0.02 | 32 ± 0.3 | 2.5 ± 0.05 | 6.4 ± 0.02 | −19.5 | −3.9 |
| 60 | 18.75 ± 0.05 | 51.6 ± 0.2 | 3.75 ± 0.01 | 10.32 ± 0.5 | −32.85 | −6.57 |
| 70 | 29.1 ± 0.02 | 80.8 ± 0.1 | 5.8 ± 0.02 | 16.16 ± 0.1 | −51 | −10.36 |
| 80 | 50 ± 0.5 | 136.5 ± 0.6 | 10 ± 0.1 | 27.3 ± 0.2 | −86.5 | −17.3 |
| 90 | 112.5 ± 0.2 | 254 ± 0.8 | 22.5 ± 0.2 | 50.8 ± 0.6 | −141.5 | −28.3 |
| Statisticsc | μ1 = 26.78 ± 35.6 | μ2 = 64.77 ± 83.6∗ | μ1 = 5.35 ± 7.12 | μ2 = 12.95 ± 16.7∗ | μ = −37.9 ± 48.3 | μ = −7.59 ± 8.6∗ |
E, observed inhibition expressed as percent of control activity without drugs (Econ).
EFV, efavirenz; obs, observed effective concentration, calculated according to equation 3; exp, expected effective concentration under the null reference hypothesis I = 1, calculated according to equation 2. Δobs-exp, change between observed and expected values. D1 and D2, dose of drug 1 and drug 2 giving the indicated percentage of inhibition of RT activity when tested in the combination (D1 + D2).
μ, mean value; ∗, P < 0.05 compared to the observed value. This P value indicates that the probability that the observed value differ from the expected ones is ≥95%. Thus, the observed synergy can be considered highly significant. For details see the text.
RESULTS
Dose-response curve determination and calculation of effective inhibitory doses of individual drugs for RTwt and mutants.
In order to analyze the effects of multiple drug combinations on the catalytic activity of HIV-1 RT, each drug was first tested individually against either RTwt or the three mutant forms L100I, K103N, and Y181I, containing known NNRTI resistance mutations (described in Materials and Methods). The corresponding inhibitory doses for E = 10, 50, and 90% of the control activity (ID90, ID50, and ID10, respectively) were calculated according to equation 3. The computed ID50 values are listed in Table 1. The selected mutants showed, as expected, significant resistance to NNRTIs, but the dose-response behavior was different. For example, the mutant L100I showed maximal resistance to nevirapine at low inhibitory doses (ID10 of L100I [ID10L100I]/ID10 of wt [ID10wt] = 85), which, however, dropped at high inhibitory doses (ID90L100I/ID90wt = 9), while all the mutants tested were increasingly resistant to efavirenz as the inhibition increased, with K103N showing the lowest sensitivity to the drug compared to RTwt (ID10K103N/ID10wt = 9.8; ID90K103N/ID90wt = 59). On the other hand, none of the mutants tested showed significant resistance to the triphosphate forms of d- or l-(β)-NRTIs, in agreement with previous observations (25, 26).
TABLE 1.
Determined parameters (ID10, ID50, ID90, m) for inhibition of the synthetic activity of wt and mutant HIV-1 RT by NNRTIs and d- and l-(β)-NRTIsa
| RT |
ID50 and mb
|
||||||
|---|---|---|---|---|---|---|---|
| NVP (μM) | EFV (nM) | ddTTP (nM) | l-dTTP (μM) | AZTTP (nM) | ddCTP (μM) | l-ddCTP (μM) | |
| wt | 0.5 ± 0.1, −0.73 ± 0.01 | 47 ± 1, −1.3 ± 0.1 | 220 ± 15, −0.7 ± 0.1 | 75 ± 1, −1.1 ± 0.1 | 19 ± 5, −0.4 ± 0.1c | 9.8 ± 1, −0.6 ± 0.1 | 14 ± 1, −0.6 ± 0.1 |
| L100I | 10 ± 1, −1.2 ± 0.1 | 70 ± 1, −0.9 ± 0.1 | 18 ± 1, −0.4 ± 0.1 | 54 ± 5, −0.5 ± 0.1 | |||
| K103N | 1100 ± 10, −0.9 ± 0.1 | 40 ± 1, −0.3 ± 0.05 | |||||
| Y181I | 237 ± 15, −1 ± 0.1 | 25 ± 1, −0.4 ± 0.05 | |||||
ID50, dose of inhibitors giving 50% inhibition of RT activity, m, slope parameter. For details see Materials and Methods.
NVP, nevirapine; EFV, efavirenz.
Determined on poly(dA) · oligo(dT). When tested on the d24:d66-mer, the ID50 was 50 nM.
Statistical analysis.
According to equation 4, a combination was considered synergistic when the observed effective inhibiting concentrations for each drug were significantly lower than the corresponding values predicted by equation 2 under the null reference hypothesis of additivity. Thus, a critical point to address was whether the observed differences were statistically significant. As an example, observed and expected values for the efavirenz-AZTTP combination against RTwt, along with statistics, are listed in Table 2. Two different approaches were used, as described in Materials and Methods, and both were found to be consistent with significant synergy (P < 0.05). All the combinations which showed I values of <1 were analyzed in the same way, and significant synergy (P < 0.05) was found in all cases.
Double and triple NNRTI–d-(β)-NRTI combinations show synergism in the inhibition of RTwt.
The effects of the nevirapine-ddTTP, efavirenz-AZTTP, and nevirapine-efavirenz-AZTTP combinations were tested on RTwt, and the interaction parameters at 10, 50, and 90% inhibition were determined as outlined in Materials and Methods. The calculated values are listed in Table 3. All the combinations were found to be significantly synergistic at inhibition doses of ≥50%; however, the efavirenz-AZTTP combination displayed an already-high degree of synergy at 50% inhibition. According to the Loewe additivity model, the expected D1 and D2 values for two drugs acting independently can be calculated from equation 2 and corresponded to I = 1. Under the experimental conditions used for efavirenz and AZTTP (molar ratio of 5:1), the expected values in the case of additivity at 50% inhibition would be 33 nM for efavirenz and 6.6 nM for AZTTP (see Materials and Methods). Comparison with the actual values derived from the experimental data showed that both drugs had comparable reductions of their effective concentrations, thus equally contributing to the observed synergism. A similar analysis for the nevirapine-ddTTP combination (molar ratio of 1.5:1) gave expected D1 and D2 values of 0.25 μM for nevirapine and 0.1 μM for ddTTP. In this case, the contribution of nevirapine to the observed synergy was more significant than that of ddTTP, since the observed value was 2.5-fold lower than that expected in the case of nevirapine but only 1.3-fold in the case of ddTTP. For the triple combination, expected D1, D2, and D3 values under the hypothesis I = 1 were 6 nM for AZTTP, 24 nM for efavirenz, and 75.1 nM for nevirapine. Thus, as for the efavirenz-AZTTP combination, the three drugs equally contributed to the observed synergy. The triple AZTTP-efavirenz-AZTTP combination was also tested on a heteropolymeric deoxyoligonucleotide substrate corresponding to nt 1061 to 1071 of the HIV-1 pol gene, which is a sequence which is physiologically replicated by HIV-1 RT. This was done in order to directly verify that the nature of the substrate and/or the assay conditions did not affect the observed synergy. As reported in Table 3, the combination also showed comparable synergy (I = 0.68) on this template, indicating that neither the RNA-DNA versus DNA-DNA structure nor the homopolymeric versus the heteropolymeric nature of the template affected the observed synergy.
TABLE 3.
Interaction parameters (I10, I50, I90) for 10, 50, and 90% inhibition of DNA synthesis of wt and mutant HIV-1 RT by combinations of NNRTIs and d-(β)-NRTIsa
| RT | Drug (d1-d2-d3)b | Mean ± SD
|
||||||
|---|---|---|---|---|---|---|---|---|
| Econ (%) | D1 (nM) | D2 (nM) | D3 (nM) |
I
|
||||
| 10 | 50 | 90 | ||||||
| wt | EFV-AZTTP | 83 ± 5 | 12.5 ± 0.1 | 2.5 ± 0.1 | 0.39 ± 0.01 | |||
| wt | NVP-ddTTP | 100 ± 7 | 100 ± 20 | 75 ± 1 | 1.2 ± 0.1 | 0.54 ± 0.02 | 0.45 ± 0.01 | |
| wt | EFV-NVP-AZTTPc | 101 ± 5 | 15 ± 2 | 50 ± 5 | 4 ± 1 | 0.7 ± 0.1 | 0.62 ± 0.02 | 0.46 ± 0.02 |
| 91 ± 7 | 60 ± 5 | 180 ± 7 | 15 ± 1 | 0.68 ± 0.05 | ||||
| L100I | EFV-AZTTP | 93 ± 5 | 18 ± 0.1 | 4.7 ± 0.1 | 0.9 ± 0.1 | 0.54 ± 0.02 | 0.45 ± 0.01 | |
| K103N | EFV-AZTTP | 94 ± 5 | 118 ± 2 | 34 ± 0.5 | 1.5 ± 0.1 | 0.97 ± 0.02 | 0.81 ± 0.01 | |
| Y181I | EFV-AZTTP | 90 ± 5 | 73 ± 5 | 18.7 ± 0.1 | 1.2 ± 0.1 | 0.74 ± 0.02 | 0.65 ± 0.01 | |
I10, I50, and I90, interaction index at 10, 50, and 90% inhibition of RT activity, respectively; Econ, estimated parameter for the control effect in the absence of the drug; D1, D2, and D3, dose of inhibitors giving 50% inhibition of RT activity when tested in the combination (D1 + D2 + D3). For details see Materials and Methods.
EFV, efavirenz; NVP, nevirapine. d1, d2, and d3, drugs 1, 2, and 3 in the combination.
Data on the first line are for the drug combination on poly(rA) · oligo(dT), and data on the second line are for the combination on d24:d66-mer oligodeoxynucleotide.
l-(β)-dideoxy but not l-(β)-deoxy-nucleoside triphosphate analogs show synergistic effects in double NNRTI-NRTI combinations against RTwt and the NNRTI-resistant mutant forms L100I, K103N, and Y1811.
The combination efavirenz-AZTTP was also tested against NNRTI-resistant mutants of HIV-1 RT. The calculated interaction indexes are listed in Table 3. The combination showed synergism towards all the mutants tested, even though in the case of the K103N and Y181I mutants significant synergism was observed only at inhibition doses of ≥90%. Expected values under the null reference model of Loewe additivity for the absence of interaction at 50% inhibition were AZTTP = 8.3 nM and efavirenz = 41 nM for L100I, AZTTP = 33 nM and efavirenz = 169 nM for K103N, and AZTTP = 16 nM and efavirenz = 82 nM for Y181I. Comparison of the expected values with the observed ones listed in Table 3 indicated that in all cases both drugs were contributing equally to the observed synergy, with the exception of K103N, where the potentiation was at the level of efavirenz inhibition. The efficacy of the nevirapine–l-(β)-dTTP, efavirenz–l-(β)-dTTP, efavirenz–d-(β)-ddCTP, and efavirenz–l-(β)-ddCTP combinations was tested against HIV-1 RTwt. Calculated values are listed in Table 4. Only the combinations with either d-(β)- or l-(β)-ddCTP proved to be significantly synergistic, even though at inhibition doses of ≥90%. The l-(β)-ddCTP combination was also synergistic towards the mutant L100I.
TABLE 4.
Interaction parameters (I10, I50, I90) for 10, 50 and 90% inhibition of DNA synthesis of wt and mutant HIV-1 RT by combinations of NNRTIs and l-(β)-NRTIsa
| RT | Drug (d1-d2)b | Mean ± SD
|
|||||
|---|---|---|---|---|---|---|---|
| Econ (%) | D1 (μM) | D2 (μM) |
I
|
||||
| 10 | 50 | 90 | |||||
| wt | NVP–l-dTTP | 95 ± 7 | 0.25 ± 0.02 | 34.5 ± 0.1 | 0.94 ± 0.02 | ||
| wt | EFV–l-dTTP | 103 ± 5 | 0.03 ± 0.005 | 44.5 ± 0.5 | 1.1 ± 0.01 | ||
| wt | EFV–l-ddCTP | 91 ± 0.5 | 0.013 ± 0.001 | 7 ± 0.5 | 0.96 ± 0.02 | 0.62 ± 0.01 | |
| wt | EFV–l-ddCTP | 101 ± 5 | 0.016 ± 0.001 | 8.95 ± 0.05 | 1.2 ± 0.1 | 0.97 ± 0.02 | 0.66 ± 0.01 |
| L100I | EFV–l-ddCTP | 93 ± 5 | 0.05 ± 0.001 | 26.3 ± 0.1 | 1.1 ± 0.1 | 0.88 ± 0.02 | 0.75 ± 0.01 |
I10, I50, and I90, interaction index at 10, 50, and 90% inhibition of RT activity, respectively; Econ, estimated parameter for the control effect in the absence of the drug; D1 and D2, dose of inhibitors giving 50% inhibiton of RT activity when tested in the combination (D1 + D2). For details see Materials and Methods.
NVP, nevirapine; EFV, efavirenz; d1 and d2, drugs 1 and 2 in the combination.
Double NNRTI–NNRTI combinations show different synergistic effects against wt and L100I mutant RT.
The effects of a combination of nevirapine and efavirenz on HIV-1 RTwt and the mutant L100I were assayed. Calculated values are listed in Table 5. The combination proved to be synergistic for both enzymes at inhibition doses of ≥50%. Under the hypothesis of no interaction (I = 1), expected values at 50% inhibition according to the Loewe additivity model were 16.3 nM for efavirenz and 0.33 μM for nevirapine in the case of RTwt and 29 nM for efavirenz and 5.8 μM for nevirapine in the case of the L100I mutant. Comparison with the observed values reported in Table 5 showed that in the case of RTwt, the observed synergy was almost exclusively due to a twofold potentiation of the effect of nevirapine, whereas in the case of the mutant L100I both drugs contributed equally.
TABLE 5.
Interaction parameters (I10, I50, I90) for 10, 50, and 90% inhibition of DNA synthesis of wt and L100I mutant HIV-1 RT by combinations of different NNRTIsa
| RT | Drug (d1-d2)b | Mean ± SD
|
|||||
|---|---|---|---|---|---|---|---|
| Econ (%) | D1 (μM) | D2 (μM) |
I
|
||||
| 10 | 50 | 90 | |||||
| wt | NVP-EFV | 103 ± 5 | 0.16 ± 0.02 | 0.02 ± 0.005 | 1.2 ± 0.1 | 0.72 ± 0.02 | 0.26 ± 0.02 |
| L100I | NVP-EFV | 93 ± 5 | 4.2 ± 0.1 | 0.023 ± 0.002 | 0.9 ± 0.1 | 0.72 ± 0.02 | 0.15 ± 0.01 |
I10, I50, and I90, interaction index at 10, 50, and 90% inhibition of RT activity, respectively; Econ, estimated parameter for the control effect in the absence of the drug; D1 and D2, dose of inhibitors giving 50% inhibition of RT activity when tested in the combination (D1 + D2). For details see Materials and Methods.
NVP, nevirapine; EFV, efavirenz. d1 and d2, drugs 1 and 2 in the combination.
RTwt and NNRTI-resistant mutant RT have different sensitivities to double NRTI-NNRTI combinations.
One of the disadvantages in using Berenbaum's interaction index for determining synergy is the difficulty of deriving a quantitative measure of the intensity of the interaction from the different calculated values of I (18). This is, however, a crucial point for the evaluation of the relative efficacy of multiple drug combinations against mutant forms of RT. Thus, in order to compare the efficacy of different combinations, a plot of the uninhibited fraction (fu = % of the control activity Econ) versus the calculated inhibitory doses (IDx) derived from experimental data for each combination according to equation 3 was constructed. In Fig. 1A the plot relative to the AZTTP-efavirenz combination is shown. It was evident that different mutations conferred increasing degrees of resistance to the synergistic combination in all cases. When the effects of the efavirenz–l-(β)-ddCTP combination against RTwt and mutant L100I were compared, again the mutant displayed a degree of resistance which was proportional to its resistance to efavirenz alone, even if the combination proved to be synergistic (not shown).
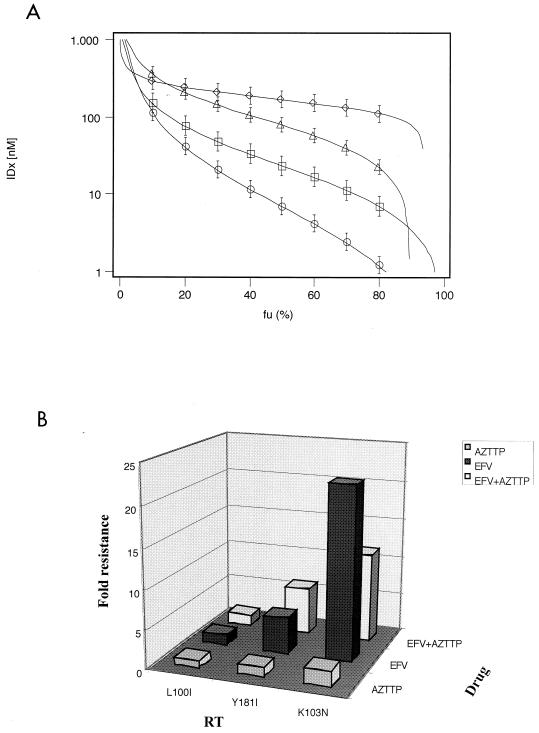
FIG. 1.
Resistance of wt and three mutant HIV-1 RTs to different combinations of NNRTIs and NRTIs. Dose-response curves for RTwt and mutants have been generated, as outlined in Materials and Methods, by fitting experimental data to equation 2 and then calculating the respective inhibition doses (IDx) at different x (%) of inhibition according to equation 3. (A) Dose-response curves for the combination of efavirenz (EFV) and AZTTP. The fraction of uninhibited activity (fu = %Econ) has been plotted versus the calculated IDx for RTwt (circles), L100I (squares), Y181I (triangles), and K103N (rhombics). Calculated parameters: (D1 + D2)wt = 15 nM; mwt = −0.79; (D1 + D2)L100I = 22.7 nM, mL100It = −1.17; (D1 + D2)Y1811 = 91.7 nM, mY1811 = −1.49; (D1 + D2)K103N = 172 nM, mK103N = −3.95. (B) Comparison of the relative resistance values (D50mut/D50wt) of different mutant RT enzymes towards efavirenz and AZTTP either individually or in combination. D50 values for the combination were calculated from the dose-response curves shown in panel A, whereas the correspondent values for the single drugs were derived from Table 1.
AZTTP can reduce the level of resistance to efavirenz of the K103N mutant RT.
In Fig. 1B, the relative resistance values (D50mut/D50wt) of each mutant to the combination were compared with the corresponding values for AZTTP and efavirenz alone (Table 1). The sensitivity of each enzyme to the combination correlated very closely with its resistance to efavirenz. A relevant exception was the mutant K103N, which was twofold more sensitive to the combination than to efavirenz alone with respect to RTwt. As shown in Table 1, none of the mutants was significantly resistant to AZTTP inhibition.
The sensitivity to double NNRTI-NNRTI combinations is different between wt and L100I mutant RT.
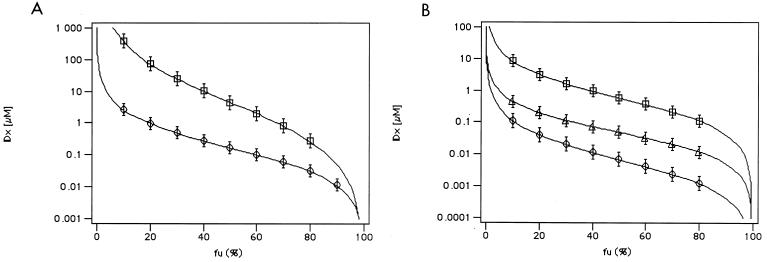
In the above discussed cases, each enzyme was resistant to only one of the two drugs used for the combination. A case in which the mutant enzyme was resistant to both drugs in the combination was also analyzed. Figure 2A shows the fu versus IDx plot for the inhibition of RTwt and mutant L100I by the efavirenz-nevirapine combination. This association proved to be synergistic with both enzymes; however, the mutant L100I displayed a significant resistance. The relative resistance of the L100I mutant at 50% inhibition was 20-fold to nevirapine and 1.5-fold to efavirenz when the inhibitors were tested individually and 26-fold to their combination. However, at 90% inhibition, the relative resistance values to nevirapine and efavirenz individually were 6-fold and 3.5-fold. respectively, whereas for the combination the resistance increased up to >100-fold.
FIG. 2.
(A) Resistance of wt and mutant L100I HIV-1 RT to combinations of two different NNRTIs. Dose-response curves for RTwt and mutants have been generated, as outlined in Materials and Methods, by fitting experimental data to equation 2 and then calculating the respective inhibition doses (IDx) at different x (%) of inhibition according to equation 3. The fraction of uninhibited activity (fu = %Econ) has been plotted versus the calculated IDx for the combination of efavirenz (EFV) and nevirapine (NVP) against RTwt (circles) and L100I (squares). Calculated parameters: (D1 + D2)wt = 0.18 μM, mwt = −0.79; (D1 + D2)L100I = 4.2 μM, mL100It = −0.5. (B) Efficacy of triple versus double RT inhibitor combinations on wt HIV-1 RT. Dose-response curves for RTwt and mutants have been generated, as outlined in Materials and Methods, by fitting experimental data to equation 1 or 2 and then calculating the respective inhibition doses (IDx) at different x (%) of inhibition according to equation 3. The fraction of uninhibited activity (fu = %Econ) has been plotted versus the calculated IDx for RTwt (circles) for the combinations EFV-NVP (squares), EFV-AZTTP (circles), and EFV-NVP-AZTTP (triangles). Calculated parameters: (D1 + D2)EFV-AZTTP = 15 nM, mEFV-AZTTP = −1; (D1 + D2)EFV-NVP = 0.18 μM, mEFV-NVP = −0.8; (D1 + D2 + D3)EFV-NVP-AZTTP = 69 nM, mEFV-NVP-AZTTP = −0.9.
Comparison of double versus triple NRTI-NNRTI combinations.
In Fig. 2B, the double combinations efavirenz-AZTTP and nevirapine-efavirenz were compared to the triple nevirapine-efavirenz-AZTTP combination. The plot indicated that the triple combination was more advantageous with respect to the nevirapine-efavirenz association than the efavirenz-AZTTP association. Indeed, when the D1 and D2 values listed in Table 3 and Table 5 for each double combination are compared to the D1, D2, and D3 values listed in Table 3 for the triple combination, it can be seen that efavirenz and AZTTP doses required to achieve 50% inhibition in all the combinations were similar, whereas the nevirapine dose was decreased by threefold in the triple versus the double combination.
DISCUSSION
As a first step towards the elucidation of the nature of the interactions between different anti-HIV RT drugs, associations between NRTIs and NNRTIs have been tested in in vitro assays against RTwt and compared to three RT forms containing clinically relevant NNRTI resistance mutations, namely L100I, K103N, and Y181I. We tested combinations of clinically used NNRTIs (nevirapine and efavirenz) with either AZTTP (the most widely used NRTI) or other model nucleoside analogs (ddTTP and ddCTP). In particular, both d-(β)- and l-(β)-dCTPs were compared for their ability to act synergistically in combination with efavirenz. In this study, the inhibitory activity of individual doses of the combined drugs was found to differ significantly in additive versus synergistic associations. Synergisms among inhibitors could be due to an increase of the affinity of the enzyme for one or more of the components of the associations with respect to the case of simple additivity. Synergism can also occur among drugs acting on the same binding site but specific for different enzyme-substrate intermediates (10, 14, 28). When the median effective doses (D50) for the different combinations tested were dissected into the individual contributions of each drug and these latter values were compared with the expected corresponding values for the single agents in the combination under the hypothesis of no interaction, the results showed that synergy of a combination was not always equally influenced by the individual agents. For example, the synergy of combinations including nevirapine with either ddTTP or efavirenz resulted almost exclusively from potentiation of the effects of nevirapine with respect to the ones expected under the hypothesis of additivity. Thus, optimizing synergistic interactions would also require the rational choice of agents to be associated. Moreover, the same drug could behave differently when included in double versus triple combinations. The relative contribution of nevirapine to the observed synergy of the triple efavirenz-nevirapine-AZTTP combination was comparable to those of the other two drugs, but as shown in Fig. 2B, inclusion of AZTTP in the combination was clearly advantageous over the double efavirenz-nevirapine association, since the absolute potency (i.e., effective inhibiting dose) of nevirapine was potentiated with respect to its association with efavirenz only. In agreement with published data, nevirapine was not found to inhibit HIV-1 RT synergistically in combination with AZTTP only (36, 38), and AZTTP only showed an additive effect when combined with ddCTP, as expected (38) (data not shown). By comparison of the potencies of different drug associations against drug-resistant viral isolates in infected-cell-based assays, it has been shown that the reduced susceptibility to a drug may affect the synergistic effect of combinations containing that drug (8). The same effect was also evident in our experimental approach. In fact, it appeared that resistance mutations towards one or more of the agents utilized can influence the efficacy of the combination. As shown in Fig. 1, in general the absolute degree of resistance of each mutant enzyme to the different double drug combinations correlated with their relative resistance to the individual agents in the combination. However, for the AZTTP-efavirenz combination, the mutant K103N showed a level of resistance to the double combination which was twofold lower than its resistance to efavirenz alone, suggesting that in the presence of AZTTP, the sensitivity of the K103N mutant to efavirenz was increased. These data indicated that a synergistic combination could improve the sensitivity profile to the single drugs utilized. In all cases when the RT mutant was resistant to only one of the drugs utilized, the relative contribution of each individual agent to the observed synergy with the mutant enzymes was comparable to that observed with the wild-type enzyme (Table 3 and 4). On the other hand, for the efavirenz-nevirapine combination against the mutant L100I, which was resistant to both drugs, contribution of efavirenz to the observed synergy was also evident, contrary to the case of RTwt, where the major contribution was attributable to nevirapine only (Table 5). The recently reported inhibition of different enzyme-substrate complexes by efavirenz (28) provides a molecular explanation for the observed synergy between this compound and nevirapine. Moreover, the fact that additional potentiation of nevirapine inhibition was gained by adding AZTTP in the combination provided direct biochemical evidence of the advantage of using triple versus double drug combinations. The l-(β)-cytidine analog 3TC has become an important component of combined anti-HIV drug therapy in association with efavirenz. Other l-(β)-enantiomers of NRTIs are currently being evaluated as potential antiviral agents; however, little is known about the influence of the l-(β)- configuration in determining the kind of interaction (whether synergistic, additive, or antagonistic) with currently used antiretroviral agents. The data presented showed that l-(β)-enantiomers of NRTIs were synergistic when combined with NNRTIs, as in the case of the combination of d-(β)- or l-(β)-ddCTP with efavirenz, which showed comparable synergy against RTwt. l-(β)-ddCTP was also synergistic in combination with efavirenz towards the L100I mutant. The results of a clinical study showed an advantage of the efavirenz-AZT-3TC combination therapy over an indinavir-AZT-3TC combination in suppressing viral replication in infected patients (32). The observed synergy of efavirenz in inhibiting HIV-1 RT when combined with d- and l-(β)-NRTIs might provide an explanation for such a difference. Synergy, however, was observed in the case of l-(β)-dideoxy- but not with l-(β)-deoxynucleoside triphosphate analogs [compare the I values for l-(β)-dTTP with the ones for l-(β)-ddCTP], confirming the advantage of using dideoxy- versus deoxy-l-(β)-NRTIs (26, 27). In conclusion, the presented approach was found to reliably reflect previous observations made with infected-cell-based assays. Moreover, the use of purified enzymes and defined in vitro systems proved to be suitable for detailed kinetic studies of drug-drug interactions. The data indicated that (i) the synergy of a combination was not always equally influenced by the individual agents utilized, (ii) a synergistic combination could improve the sensitivity profile of a drug-resistant mutant enzyme towards the single agents utilized, (iii) l-(β)-enantiomers of NRTIs were synergistic when combined with NNRTIs, and (iv) inter- and intracombination comparisons of the relative potencies of each drug could be used to highlight the different contributions of each drug to the observed synergy.
ACKNOWLEDGMENTS
We thank S. H. Hughes for kindly providing us with the coexpression vectors pUC12N/p66(His)/p51 with the wild-type or the mutant forms of HIV-1 RT p66.
This work was supported by TMR grant ERBMRXCT 970125 to S.S. and U.H., by an ISS-AIDS fellowship to G.M., by the ISS II AIDS Research National Program, Project 2.1.3, Research proposal no. 133, by the Kanton of Zürich to U.H., and by the CNR-Target Project on Biotechnology to S.S.
REFERENCES
- 1.Althaus I W, Chou J J, Gonzales A J, Deibel M R, Chou K C, Kezdy F J, Romero D L, Palmer J R, Thomas R C, Aristoff P A. Kinetic studies with the non-nucleoside HIV-1 reverse transcriptase inhibitor U-88204E. Biochemistry. 1993;32:6548–6554. doi: 10.1021/bi00077a008. [DOI] [PubMed] [Google Scholar]
- 2.Balzarini J. Suppression of resistance to drugs targeted to human immunodeficiency virus reverse transcriptase by combination therapy. Biochem Pharm. 1999;58:1–27. doi: 10.1016/s0006-2952(99)00029-5. [DOI] [PubMed] [Google Scholar]
- 3.Berenbaum M C. A method for testing for synergy with any number of agents. J Infect Dis. 1978;137:122–130. doi: 10.1093/infdis/137.2.122. [DOI] [PubMed] [Google Scholar]
- 4.Brennan T M, Taylor D L, Bridges C G, Leyda J P, Tymsa A S. The inhibition of human immunodeficiency virus type 1 in vitro by a non-nucleoside reverse transcriptase inhibitor MKC-442, alone and in combination with other anti-HIV compounds. Antivir Res. 1995;26:173–187. doi: 10.1016/0166-3542(94)00074-i. [DOI] [PubMed] [Google Scholar]
- 5.Byrnes W W, Sardana V V, Schleif W A, Condra J H, Waterbury J A, Wolfgang J A, Long W J, Schneider C L, Schlabach A J, Wolanski B S. Comprehensive mutant enzyme and viral variant assessment of human immunodeficiency virus type 1 reverse transcriptase resistance to nonnucleoside inhibitors. Antimicrob Agents Chemother. 1993;37:1576–1579. doi: 10.1128/aac.37.8.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll S S, Olsen D B, Bennett C D, Gotlib L, Graham D J, Condra J H, Stern A M, Shafer J A, Kuo L C. Inhibition of HIV-1 reverse transcriptase by pyridinone derivatives. Potency, binding characteristics and effect of template sequence. J Biol Chem. 1993;268:276–281. [PubMed] [Google Scholar]
- 7.Carroll S S, Stahlhut M, Geib J, Olsen D B. Inhibition of HIV-1 reverse transcriptase by a quinazolinone and comparison with inhibition by pyridinones. J Biol Chem. 1994;269:32351–32357. [PubMed] [Google Scholar]
- 8.Cox S W, Albert J, Ljungdahl-Stahle E, Wahren B. Effect of resistance on combination chemotherapy for human immunodeficiency virus infection. Adv Enzyme Regul. 1993;33:27–36. doi: 10.1016/0065-2571(93)90007-z. [DOI] [PubMed] [Google Scholar]
- 9.Cushman M, Golebiewski W M, Graham L, Turpin J A, Rice W G, Fliakas-Boltz V, Buckheit R W., Jr Synthesis and biological evaluation of certain alkenyldiarylmethanes as anti-HIV-1 agents which act as non-nucleoside reverse transcriptase inhibitors. J Med Chem. 1996;39:3217–3227. doi: 10.1021/jm960082v. [DOI] [PubMed] [Google Scholar]
- 10.De Clercq E. Perspectives of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the therapy of HIV-1 infection. Farmaco. 1999;54:26–45. doi: 10.1016/s0014-827x(98)00103-7. [DOI] [PubMed] [Google Scholar]
- 11.Dixon M, Webb E C. Enzymes. London, United Kingdom: Longman; 1979. [Google Scholar]
- 12.Drewinko B, Loo T L, Brown B, Gottlieb J A, Freireich E J. Combination chemotherapy in vitro with adriamycin. Observations of additive, antagonistic and synergistic effects when used in two-drug combinations on cultured human lymphoma cells. Cancer Biochem Biophys. 1976;1:187–195. [PubMed] [Google Scholar]
- 13.Faraj A, Agrofoglio L, Wakefield J K, McPherson S, Morrow C D, Gosselin G, Mathe C, Imbach J-L, Schinazi R F, Sommadossi J-P. Inhibition of human immunodeficiency virus type 1 reverse transcriptase by the 5′-triphosphate β enantiomers of cytidine analogs. Antimicrob Agents Chemother. 1994;38:2300–2305. doi: 10.1128/aac.38.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fletcher R S, Syed K, Mithani S, Dimitrienko G I, Parniak M A. Carboxanilide derivative non-nucleoside inhibitors of HIV-1 reverse transcriptase interact with different mechanistic forms of the enzyme. Biochemistry. 1995;34:4346–4353. doi: 10.1021/bi00013a025. [DOI] [PubMed] [Google Scholar]
- 15.Fletcher R S, Arion D, Borkow G, Wainberg M, Dmitrienko G I, Parniak M A. Synergistic inhibition of HIV-1 reverse transcriptase DNA polymerase activity and virus replication in vitro by combinations of carboxanilide nonnucleoside compounds. Biochemistry. 1995;34:10106–10112. doi: 10.1021/bi00032a002. [DOI] [PubMed] [Google Scholar]
- 16.Gosselin G, Schinazi R F, Sommadossi J-P, Mathé C, Bergogne M-C, Aubertine A-M, Imbach J-L. Anti-human immunodeficiency virus activity of the β-l-enantiomer of 2′,3′-dideoxycytidine and its 5-fluoro derivative in vitro. Antimicrob Agents Chemother. 1994;38:1292–1297. doi: 10.1128/aac.38.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greco W R, Dembinski W E. Fundamental concepts in the assessment of the joint interaction of biological response modifiers with other agents. Can J Infect Dis. 1992;3:60B–68B. [Google Scholar]
- 18.Greco W R, Bravo G, Parsons J C. The search for synergy: a critical review from a response surface perspective. Pharm Rev. 1995;47:331–385. [PubMed] [Google Scholar]
- 19.Gu Z, Wainberg M A, Nguyen-Ba N, L'Heureux L, deMuys J M, Bowlin T L, Rando R F. Mechanism of action and in vitro activity of 1′,3′-dioxolanylpurine nucleoside analogues against sensitive and drug-resistant human immunodeficiency virus type 1 variants. Antimicrob Agents Chemother. 1999;43:2376–2382. doi: 10.1128/aac.43.10.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill A V. The possible effects of the aggregation of the molecules of haemoglobin on its dissociation curves. J Physiol. 1910;40:iv–vii. [Google Scholar]
- 21.Hottiger M, Hübscher U. Human immunodeficiency virus type 1 reverse transcriptase. Biol Chem. 1996;377:97–120. [PubMed] [Google Scholar]
- 22.Hübscher U, Spadari S. DNA replication and chemotherapy. Physiol Rev. 1994;74:259–304. doi: 10.1152/physrev.1994.74.2.259. [DOI] [PubMed] [Google Scholar]
- 23.Larder B A. Interactions between drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase. J Gen Virol. 1994;75:951–957. doi: 10.1099/0022-1317-75-5-951. [DOI] [PubMed] [Google Scholar]
- 24.Loewe S, Muischnek H. Effect of combinations: mathematical basis of problem. Arch Exp Pathol Pharmakol. 1926;114:313–326. [Google Scholar]
- 25.Maga G, Amacker M, Hübscher U, Gosselin G, Imbach J L, Mathé C, Faraj A, Sommadossi J P, Spadari S. Structural determinants of HIV-1 reverse transcriptase stereoselectivity towards (β)-L-deoxy- and dideoxy-pyrimidine nucleoside triphosphates: molecular basis for the combination of L-dideoxynucleoside analogs with non-nucleoside inhibitors in anti-HIV chemotherapy. Nucleosides Nucleotides. 1999;18:795–805. doi: 10.1080/15257779908041566. [DOI] [PubMed] [Google Scholar]
- 26.Maga G, Amacker M, Hübscher U, Gosselin G, Imbach J-L, Mathé C, Faraj A, Sommadossi J-P, Spadari S. Molecular basis for the enantioselectivity of HIV-1 reverse transcriptase: role of the 3′-hydroxyl group of the L-(β)-ribose in chiral discrimination between D- and L-enantiomers of deoxy- and dideoxy-nucleoside triphosphate analogs. Nucleic Acids Res. 1999;27:972–978. doi: 10.1093/nar/27.4.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maga G, Amacker M, Ruel N, Hübscher U, Spadari S. Resistance to nevirapine of HIV-1 reverse transcriptase mutants: loss of stabilizing interactions and thermodynamic or steric barriers are induced by different single amino acid substitutions. J Mol Biol. 1997;274:738–747. doi: 10.1006/jmbi.1997.1427. [DOI] [PubMed] [Google Scholar]
- 28.Maga G, Ubiali D, Salvetti R, Pregnolato M, Spadari S. Selective interaction of the HIV-1 reverse transcriptase non-nucleoside inhibitor efavirenz and its thio-analog with different enzyme-substrate complexes. Antimicrob Agents Chemother. 2000;44:1186–1194. doi: 10.1128/aac.44.5.1186-1194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moyle G J. Current knowledge of HIV-1 reverse transcriptase mutations selected during nucleoside analogue therapy: the potential to use resistance data to guide clinical decisions. J Antimicrob Chemother. 1997;40:765–777. doi: 10.1093/jac/40.6.765. [DOI] [PubMed] [Google Scholar]
- 30.Quann Y, Motakis D, Buckheit R, Jr, Xu Z Q, Flavin M T, Parniak M A, Wainberg M A. Sensitivity and resistance to (+)-calanolide A of wild type and mutated forms of HIV-1 reverse transcriptase. Antivir Ther. 1999;4:203–209. [PubMed] [Google Scholar]
- 31.Richman D, Rosenthal A S, Skoog M, Eckner R J, Chou T C, Sabo J P, Merluzzi V J. BI-RG-587 is active against zidovudine-resistant human immunodeficiency virus type 1 and synergistic with zidovudine. Antimicrob Agents Chemother. 1991;35:305–308. doi: 10.1128/aac.35.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staszewski S, Morales-Ramirez J, Tashima K T, Rachlis A, Skiest D, Stanford J, Stryker R, Johnson P, Labriola D F, Farina D, Manion D J, Ruiz N M. Efavirenz plus zidovudine and lamivudine, efavirenz plus indinavir and indinavir plus zidovudine and lamivudine in the treatment of HIV-1 infection in adults. Study 006 Team. N Engl J Med. 1999;341:1865–1873. doi: 10.1056/NEJM199912163412501. [DOI] [PubMed] [Google Scholar]
- 33.Stuyver L, Wyseur A, Rombout A, Louwagie J, Scarcez T, Verhofstede V, Rimland D, Schinazi R F, Rossau R. Line probe assay for rapid detection of drug-selected mutations in the human immunodeficiency virus type 1 reverse transcriptase gene. Antimicrob Agents Chemother. 1997;41:284–291. doi: 10.1128/aac.41.2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan L, Chen C-Y, Tyller R D, Grabowsky E J J, Reider P J. A novel, highly enantioselective ketone alkynylation reaction mediated by chiral zinc aminoalkoxides. Angew Chem Int Ed. 1999;38:711–713. doi: 10.1002/(SICI)1521-3773(19990301)38:5<711::AID-ANIE711>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 35.Taylor D L, Ahmed P S, Wood L J, Kelly L A, Chambers P, Clarke J, Bedard J, Bowlin T L, Rando R F. Drug resistance and drug combination features of the human immunodeficiency virus inhibitor, BCH-10652 [(+/−)-2′-deoxy-3′-oxa-4′-thiocytidine, dOTC] Antivir Chem Chemother. 2000;11:291–301. doi: 10.1177/095632020001100405. [DOI] [PubMed] [Google Scholar]
- 36.Tramontano E, Cheng Y C. HIV-1 reverse transcriptase inhibition by a dipyridodiazepinone derivative: BI-RG-587. Biochem Pharmacol. 1992;17:1371–1376. doi: 10.1016/0006-2952(92)90515-k. [DOI] [PubMed] [Google Scholar]
- 37.Villahermosa M L, Martinez-Irujo J J, Cabodevilla F, Santiago E. Synergistic inhibition of HIV-1 reverse transcriptase by combinations of chain-terminating nucleotides. Biochemistry. 1997;36:13223–13231. doi: 10.1021/bi970852k. [DOI] [PubMed] [Google Scholar]
- 38.Young S D, Britcher S F, Tran L O, Payne L S, Lumma W C, Lyle T A, Huff J R, Anderson P S, Olsen D B, Carroll S S, et al. L-743,726 (DMP-266): a novel, highly potent nonnucleoside reverse transcriptase inhibitor of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1995;39:2602–2605. doi: 10.1128/aac.39.12.2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuasa S, Sadakata Y, Takashima H, Sekiya K, Inouye N, Ubasawa M, Baba M. Selective and synergistic inhibition of human immunodeficiency virus type 1 reverse transcriptase by a non-nucleoside inhibitor, MKC-442. Mol Pharmacol. 1993;44:895–900. [PubMed] [Google Scholar]