Abstract
Introduction:
Diagnostic and therapeutic strategies in eosinophilic esophagitis (EoE) are evolving. New knowledge regarding the pathophysiology of EoE has been the foundation for updated diagnostic recommendations and new therapeutic trials.
Areas Covered:
We performed structured literature searches in Medline and PubMed, Cochrane meta-analyses, and abstracts of international congresses to review therapeutic approaches for EoE in July 2018. Additional articles were obtained by perusing the references of articles identified in the original PubMed search. Articles were excluded if they did not focus on the mechanism of disease, diagnosis, or treatment of humans with eosinophilic esophagitis.
Expert Commentary:
Recent advances in the understanding of mechanisms underlying the pathology of eosinophilic esophagitis (EoE) have resulted in significant change in the diagnostic algorithm for EoE, and are identifying promising potential targets for personalized medicine. There is clinical need for improved targeted therapy for EoE, and better understanding the underlying pathophysiology of EoE will help to determine therapeutic targets. In this review, we highlight key mechanisms in the pathophysiology of EoE and how they are being utilized to change therapy in EoE.
Keywords: eosinophilic esophagitis, physiopathology, etiology, diagnosis, diagnostics, treatment, biologics, diet therapy
1. Introduction:
Eosinophilic esophagitis (EoE) is a chronic relapsing inflammatory disorder of the esophagus and is a common cause of esophageal dysfunction, fibrosis, stricture and food impaction in children and adults[1]. EoE incidence and prevalence is rapidly increasing in developed westernized countries and it is estimated to affect 1 in 1000 patients in the United States [2–9]. EoE is due to a T helper type 2 (Th2) atopic inflammation which, if left untreated, can cause irreversible fibrosis [10]. It can occur at any age and has heterogeneous clinical presentations depending on age at the time of presentation. The younger the patient, the more likely symptoms reflect acute inflammation, whereas the older the patient, the more likely it is that symptoms are due to fibrosis [10]. Therefore typically infants and young children present with non-specific symptoms of esophageal dysfunction such as vomiting, food refusal, and failure to thrive[11]; whereas adolescents and adults present with a spectrum of symptoms reflecting ongoing fibrosis like dysphagia, odynophagia, nausea, vomiting and esophageal strictures[12]. The long term consequences of EoE are fibrosis, stricture, and food impaction which can require esophageal dilation [13, 14].
EoE is diagnosed by demonstration of greater than 15 eosinophils per high powered field in the esophageal epithelium on biopsy [15, 16]. Despite recent research, there are no noninvasive biomarkers with sufficient sensitivity or specificity to replace examination of the esophageal biopsies by an experienced pathologist [10].
Like in many atopic diseases it has been demonstrated by multiple groups independently that in EoE there is Th2-predominant inflammation driven by chronic antigen exposure from food and possibly environmental allergens [17–25]. Treatment options for patients with EoE, as for many other atopic diseases, include antigen avoidance, i.e. dietary modification, and topical swallowed steroids [15, 16].Of note, no topical steroid preparation is FDA approved for EoE and asthma medications are currently used “off label” to treat the disease. However in contrast with typical allergic disorder, a portion of EoE patients will have histologic improvement while taking proton pump inhibitors PPI, suggesting a more complex and diverse mechanisms in EoE development [15].
Finally there is a substantial proportion of patients who have partial or no response to these therapies. To address the unmet needs in those patients many clinical trials are underway to develop new method of delivery for topical steroid or new more specific biological therapies for EoE aiming at controlling both eosinophilic inflammation and patients’ symptoms [26]. Better understanding of the pathophysiology of EoE is necessary to direct development of targeted therapies for this disorder. The aim of this article is to review the cellular and molecular mechanisms of EoE and how this knowledge influence present and future management of EoE.
2. Genetic susceptibility
Since the first few case descriptions in the mid-nineties, great progress has been made in understanding EoE pathophysiology in a relative short period of time and the advancement in genomic studies of the last decades have been pivotal in sustaining the rapid pace of this research[27]. Indeed since the first description of EoE, genetic predisposition has been clearly shown to be a critical factor in EoE as demonstrated by the increased risk of developing EoE in first degree relative or siblings of patients affected by the disease [28] [29].
Like every atopic disease the genetics of EoE appears to be the one typical of most of the complex diseases such as asthma and obesity: genetics are permissive of the disease, but the disease development happens only in the presence of the crucial environmental factors [27, 30] (Figure 1). Indeed if several inheritable gene loci have now been described linked to EoE risk, [31–39], a strong environmental component in EoE development is suggested by the rapid increase in EoE prevalence experienced in western countries, the fact that fraternal twins are more at risk for developing EoE than siblings and the possibility of identical twins not equally affected [3, 28].
Figure 1:
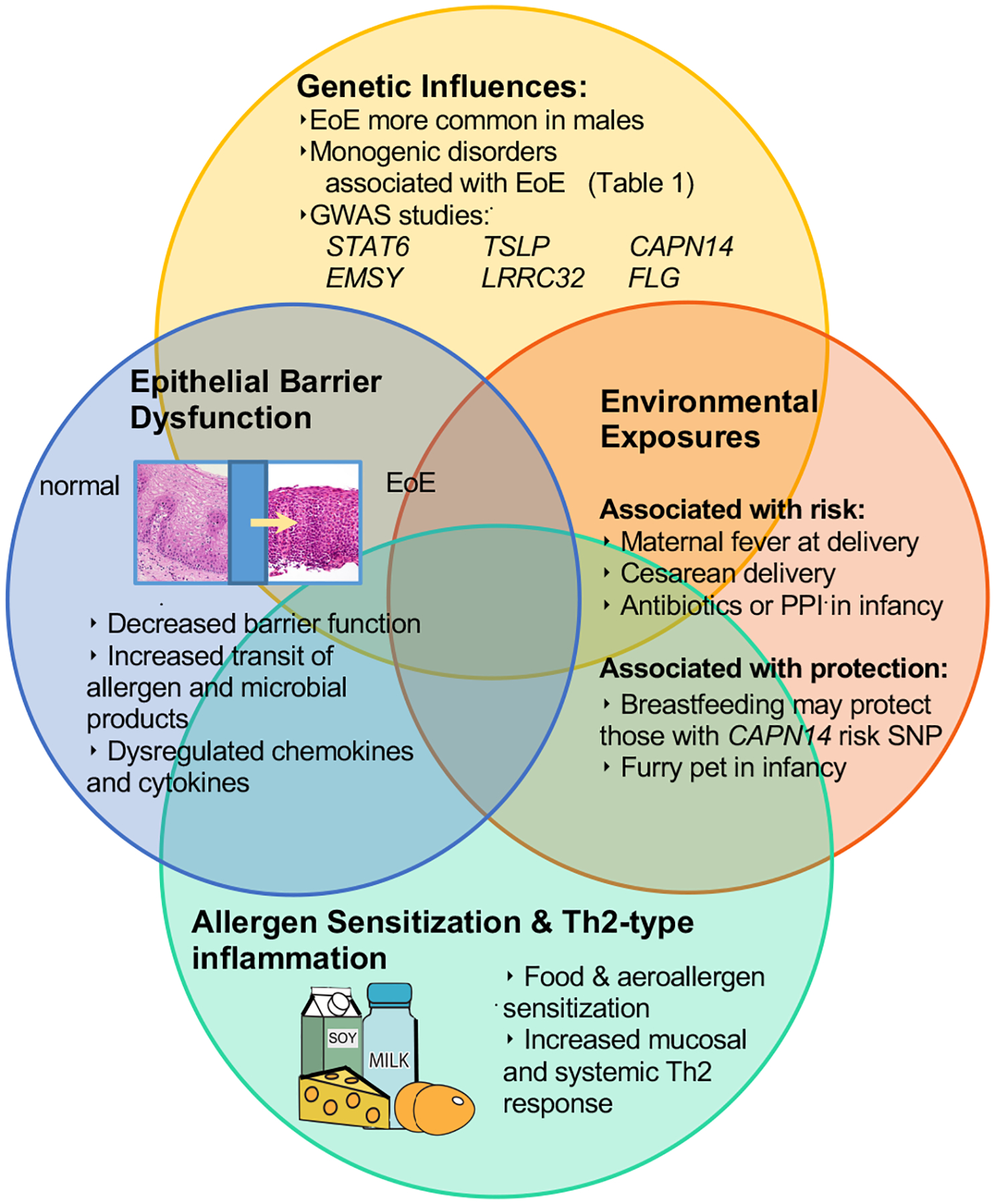
Pathogenesis of EoE: interaction between environment, genetics, epithelial barrier and immune response
GWAS= Genome Wide associated studies, TSLP=Thymic Stromal Lymphopoietin, CAPN14= Calpain 14, FLG= filaggerin
Small and large studies have shown that genetic determinants are far greater than other atopic diseases such as asthma[27]. In a study of 4,423 EoE cases and 24,322 controls from the Utah population database, it was demonstrated that risks of EoE were significantly increased among first-degree relatives (odds ratio [OR], 7.19; 95% CI, 5.65–9.14)[40], especially those diagnosed by 18 years of age (OR, 16.3; 95% CI, 9.4–28.3)[40]. Similar results have been shown in twin studies, in which monozygotic twins have 41% disease concordance while the non-twin siblings of patients with EoE have a 2.4% disease concordance and the non-related population of 0.05% [28]. Therefore the sibling risk ratio in EoE is estimated to be over 40 and only 2 for example for atopy[41]. However the aforementioned studies shows also a great influence of the environment. Interestingly the Utah study found an increased risk of EoE in the spouses of EoE probands, which suggests either nonrandom mating or potential shared environmental contribution to development of EoE [40]. Similarly in the twin study the dizygotic twins had a disease concordance of 22% much higher than the siblings’ concordance, indicating that both genetics and early life factors play important roles in the development of EoE [28].
In complex diseases typically multiple single nucleotide gene differences (polymorphism-SNP) can have a protective or causative effects for a certain disease in a specific subject depending on the environmental exposure [39, 42–45]. Two study designs are commonly used to determine the genetic contributions in complex diseases: candidate gene association studies and genome-wide association studies(GWAS)[45].
The candidate gene association studies compare the incidence of SNP in biologically plausible gene between a population affected by the disease (cases) and a group of controls[45]. The main limitations of such a design are its inability to identify novel genes and pathways contributing to the pathogenesis of a disorder [45].
The availability of microarray technology and the sequencing of the whole human genome has made possible to study hundreds of thousands SNPs on the entire genome and comparing those in cases and controls like it happens in genome wide associated studies (GWAS) [45]. GWAS allows performance of hypothesis-free search for gene variants associated with a certain diseases and has been proven to be a powerful tool to unveil new replicable gene targets for researchers, when adequately powered[45]. Indeed SNPs are typically frequent in the general healthy population. To establish their role in GWAS, the larger the number of cases and controls for analysis, the better is the statistical power[45].
Using a candidate gene approach, 3 genes have been identified as important in EoE pathogenesis TGF-β [46–48], CCL-26 (Eotaxin 3) [49] and Toll-like receptor 3 (TLR3) [50].
Aceves et al showed that patients who responded to swallowed steroid therapy are more likely to have a CC genotype at the −509 position in the TGF-β promoter than were non-responders. These data suggest that TGF- β, that is known to induce fibrosis in atopic disease, could be a target for therapeutic intervention and that response to therapy maybe influenced by the genetic background [46–48].
Blanchard et al, found that CCL-26 is overexpressed by 50-fold in EoE patients compared with normal controls or patients with GERD due to a SNP present in about 13% of patients with EoE, suggesting for the first time how a dysregulated epithelium may play a central role in favoring eosinophil chemotaxis in certain individuals [49].
Most recently Avila-Castellano R et al. demonstrated that patients with EoE express more often the CC or CG SNP rs3775292 on TLR3 gene on Chromosome 4. Patient with TLR SNP tended also to be more frequently sensitized to both environmental and food allergens [50].
More recently, GWAS analysis done by several independent groups have identified 3 more epithelial genes as important candidate gene in the pathogenesis of EoE: the thymic stromal lymphopoietin (TSLP) gene, at 5q22, the Calpain 14 (CAPN14) on chr2p23.1 and the c11orf30/EMSY on chr11q13.5 [37, 39, 42, 51].
The first EoE GWAS was published in 2010 and included 351 total cases resulting in the identification of multiple genome wide significant variants on chromosome 5q22 at a locus that contained [36]. The TSLP risk allele (AA) is correlated with increased epithelial TSLP expression, increased mucosal basophil recruitment. The importance of TSLP in EoE pathophysiology is not surprising as TSLP plays a crucial role in many other atopic diseases such as asthma or phenotype as eczema [52]. Its central role in EoE has been confirmed in animal studies, where TSLP inhibition leads to a prevention in EoE development in an animal model [53]. More recently our group for the first time demonstrated that TSLP risk allele (AA) could be used clinically as EoE pediatric patients with this genotype have less chances to respond to a diet that eliminates only one food and have increased food allergen triggers, regardless of the degree of background atopic disease [35, 53]. Sherrill et al demonstrated a nonsynonymous polymorphism in the TSLP receptor (TSLPR) gene on Xp22.3 and Yp11.3 [37]. This SNP was significantly associated with EoE only in male patients [37], and could be responsible for the preponderance of EoE in male patients [37], who represent approximately 70% of EoE patients [3].
Two additional expanded GWAS were published in 2014. Both of these confirmed the association of TSLP with EoE and reported a novel locus including the Calpain 14 (CAPN14) gene. Kottyan et al [51, 54] demonstrated association of SNP at the TSLP and CAPN14 loci as well as food hypersensitivity loci LRRC32, IL33 and LPP with the development of EoE. In a separate cohort, Sleiman et al replicated association of TSLP and CAPN14 SNPs with EoE, and demonstrated novel association of c11orf30, ANKRD27, and STAT6[39]. CAPN14 appears to be specifically expressed in the upper GI tract at particularly high levels in the esophageal epithelium [51]. It is a protease that belongs to the Calpain family, which is believed to be important in protein turnover and, if dysregulated, can lead to loss of barrier function [40]. Atopic inflammation, IL-13 and the risk SNP allele favor CAPN14 overexpression in EoE, resulting in loss of critical barrier proteins such as Desmoglein 1, which are hydrolyzed by CAPN14, and consequent increased epithelial permeability[54].
C11orf30 and STAT6 have been found to be associated with allergic and inflammatory disease [55–57]. C11orf30, encodes EMSY, and an epithelium loss of EMSY has been associated with increased levels of the proinflammatory TSLP and CCL5, suggesting how small risk in multiple genes can amplify immune dysregulation[58]. GWAS studies have therefore confirmed how the epithelium is central in EoE pathogenesis and have identified several candidate targets for therapy.
Other studies that point to the importance of genetics in EoE are several monogenic inherited disorders associated with an increased risk of EoE (Table 1). Examination of the various mechanisms implicated by these associations provides insight into the diversity of EoE pathogenesis. Connective tissue disorders are thought to share a common pathogenic mechanism in the development of EoE through dysregulated TGF-β signaling. The risk of connective tissue disorders such as Marfan’s syndrome, hypermobile Ehrlos Danlos’ Syndrome and joint hypermobility syndrome is increased 8-fold in patients with EoE[59]. Recent studies have shown that STAT3 negatively regulates TGF-β signaling via ERBB2-interacting protein (ERBIN) which is a SMAD anchor for receptor activation [60]. Individuals with dominant-negative STAT3 mutations (autosomal dominant Hyper-IgE Syndrome, AD-HIES) have significantly increased incidence of EoE [61]. EoE has also been associated with defects in PTEN, the tumor suppressor lipid phosphatase and tensin homolog as well as with Netherton’s syndrome which is caused by a defect in the epithelial protease inhibitor SPINK5[62, 63]. In a 2018 study Sherrill et al used trio whole-exome sequencing of 63 EoE patients and 60 unaffected family members [38]. This study identified 5 rare, damaging variants in dehydrogenase E1 and transketolase domain–containing 1 (DHTKD1) as well as 7 variants in the DHTKD1 homolog oxoglutarate dehydrogenase-like (OGDHL) which seem to play a role in normal mitochondrial function in esophageal epithelium[38]. As these studies demonstrate, the genetic etiologies of EoE are heterogeneous and range from non-Mendelian to rare, EoE-associated variants to patients with EoE as a feature of a systemic syndrome such as connective tissue disease or AD-HIES. In the future, it will be necessary to determine how prevalent these different mechanisms of inheritance are within the population of EoE. Further, the impact of mechanism of inheritance on disease course and response to therapy needs to be determined.
TABLE 1:
MONOGENETIC DISEASES AS A RISK FACTOR FOR EOE.
| Name | Gene | Pathophysiology | References |
|---|---|---|---|
| Loeys-Dietz syndrome | TGFBR1, TGFBR2, SMAD3, TGFB2, TGFB3 | TGF-β1 promotes fibrosis and is a cytokine that modulates T cell function ・In Loeys-Dietz syndrome, TGF-β signaling is overactive which disrupts the extracellular matrix (ECM) ・In Marfan’s syndrome, deficiencies in fibrillin-1 alter ECM sequestration of the large complex of TGF-β |
[59] [60] |
| Marfan syndrome Type II | Fibrillin: FBN | [59] [60] | |
| Ehlers-Danlos syndrome | Collagen: COL5A1, COL3A1 | Mutated collagen pathways alter ECM; TGF-β signaling is enhanced due to decreased interactions with ECM | [59] |
| PTEN hamartoma tumor syndromes | PTEN gain of function | Tumor suppressor protein encodes phosphatase that negatively regulates MAPK and antagonizes PI3K | [63] |
| Autosomal dominant hyper-IgE syndrome | Autosomal dominant STAT3 deficiency | Patients have abnormal connective tissue remodeling, abnormal levels of matric metalloproteinases, impaired mucosal healing in addition to combined primary immunodeficiency with altered T cell function | [60, 61] |
| Severe dermatitis, multiple allergies, and metabolic wasting (SAM) syndrome | DSG1 or DSP loss of function | Loss of skin and esophageal epithelial integrity due to breakdown of desmosome integrity | [129] |
| Netherton Syndrome | SPINK5 loss of function | SPINK5 encodes LEKT1, a type of serine protease inhibitor on the epithelial surface of skin and esophagus | [62] |
| α-ketoglutarate dehydrogenase complex defects | DHTKD1 OGDHL | Dysregulation of gene expression secondary to mitochondrial dysfunction | [38] |
TGF-β= Transforming Growth Factor-Beta; TGFBR1= TGF-β Receptor; ECM= extracellular matrix; FBN= Fibrillin; DSG1= Desmoglein; COL= Collagen; SAM Severe dermatitis, multiple allergies, and metabolic wasting syndrome; DSP=Desmoplakin; SPINK5=Serine Peptidase Inhibitor Kazal Type 5; LEKT1=Lympho Epithelial Kazal Type Related Inhibitor; DHTKD1= α-ketoglutarate dehydrogenase; OGDHL=Oxoglutarate Dehydrogenase Like
3. Early life exposures
Early-life exposures have been demonstrated to confer additional risk in the development of atopic disease. There is evidence that neonatal microbiome composition modulates asthma risk, and maternal acute atopic symptoms during pregnancy have been associated with risk of early atopic dermatitis in offspring [64, 65] Several studies have also shown a possible link between specific exposures and an increased risk of EoE. These studies have mainly been performed in American populations, and if confirmed more broadly they may provide insight into potentially modifiable risk factors to prevent the development of EoE (Figure 1).
Specifically, a recent retrospective case control study of an American pediatric EoE cohort by Jensen et al [66] examined association between the development of EoE and exposures during the neonatal period. Development of EoE was positively associated with maternal fever (adjusted odds ratio, aOR 3.18, 95% CI, 1.27–7.98) preterm labor (aOR, 2.18 95% CI [1.06–4.48]), cesarean delivery (aOR 1.77, 95% CI [1.01, 3.09]), and antibiotic use during infancy (aOR, 2.30 [95% CI, 1.21–4.38]) as well as the use of an acid suppressant during infancy (aOR 6.05, 95% CI [2.55–14.40]).Notably, an inverse relationship was noted between having a furry pet in infancy and the development of EoE (aOR 0.58; 95% CI [0.34–0.97]. Interestingly, no associations were noted for breast feeding or maternal multi-vitamin use [66].
Additional work by Jensen et al[67] has investigated how early-life environmental exposures may interact with genetic susceptibility loci defined on EoE GWAS studies. Specific SNPs associated with EoE were evaluated and included TSLP at 5q22, the LOC283710 and KLF13 region at 15q13, CAPN14, chemokine (C-C motif) ligand (CCL) 26, and transforming growth factor β (TGF-β). Case control analysis suggested that breast feeding infants with the pathogenic variant CAPN14 may have a reduced risk of EoE (aOR .08, 95% CI [0.01–0.59])[67]. Those with admission to the NICU as well as the variant LOC283710/KLF13 may have an increased risk of EoE (aOR 4.83, 95% CI [1.49–15.66]). These results provide additional evidence into modifiable risk factors that may contribute to development of EoE, and provide insight into how these environmental risks may interact with inherited pathogenic variants. This may ultimately provide information regarding individual patient or population-based risks to implement risk modification strategies.
4. Allergen sensitization
The atopic march refers to the phenomenon of progression of allergic disease from infancy through childhood. Specifically this refers to the development of atopic dermatitis, food allergy, asthma and allergic rhinitis[68]. In several retrospective studies it has been shown that the risk of EoE is increased in pediatric patients with atopic comorbidities including asthma, allergic rhinitis and food allergy. For example patients with chronic rhinosinusitis and their first degree relatives have respectively a 3.4 fold and 1.5 fold increased risk of developing EoE [69]. A prospective, EMR-based primary care birth cohort, has also confirmed the association of atopic disorders with EoE[70]. The presence of atopic dermatitis (HR 3.2, 95% CI [2.2–4.6]), IgE mediated food allergy (HR 9.1, 95% CI [6.5–12.6]) and asthma (HR 1.9, 95% CI [1.3–2.7]) were independently and cumulatively associated with a later diagnosis of EoE[70]. These studies suggest that EoE is a member of the atopic march and raises questions about shared mechanisms of pathogenesis as well as potential targeted therapy (Figure 1).
Food allergy has been shown in numerous studies to be a common trigger of EoE, because dietary restriction therapies have been successfully used globally as effective treatment options [23, 24, 71–77]. Elemental diets have been shown in several reports to induce histological remission in both children and adults with EoE [24, 75, 78]. This strategy employs an amino acid based formula and is difficult to adhere to, especially for older children and adult patients. As such, multi-food elimination diets have come into play as more realistic options for patients that have also been shown to improve symptoms [23, 24, 71–77]. Studies have shown clinical improvement with both 6-food (milk, wheat, egg, soy, peanut/treenut, fish/shellfish) and comparatively 4-food (milk, wheat, egg, soy) elimination diets with a recent prospective study showing benefit to 2-food elimination (milk, wheat) with step-wise additional restriction if clinically necessary [23, 24, 71–77]. Sequential single food reintroduction once remission is achieved in order to identify the trigger is necessary. In EoE like in food allergic disease, immunological response to food is very reproducible [23, 24, 71–77] and food allergens have been demonstrated to have a causative role in EoE following Koch’s postulate as demonstrated by clinical and endoscopic resolution of EoE once the food is removed and exacerbation when the same food is reintroduced[11, 23, 72, 73].
Several investigators have examined methods to identify causative allergens in EoE, and this has been recently been reviewed in detail elsewhere[79] In general, conventional allergy testing methods that rely on food specific IgE measurement, have had low sensitivity and specificity to guide food elimination in EoE and therefore clinical response to food elimination on the endoscopic biopsy has remained the standard of care in the clinical guidelines[15]. This is probably due to the fact that the role of IgE in food induced EoE is probably minimal if any [71]. Indeed in animal models the lack of IgE didn’t prevent EoE development, omalizumab (anti-IgE) monoclonal antibody has not shown to be an effective treatment [71, 80, 81]. Similarly with previous IgE mediated allergy to a specific food that reintroduced the food in the diet either because they have spontaneously outgrown the FA or have undergone an oral immunotherapy protocol (OIT) are at risk of developing EoE [71]. A large meta-analysis concluded that new onset EoE occurs in 2.7% (95% confidence interval 1.7%–4.0%, I(2) = 0%) of patients following food OIT given for IgE mediated food allergy, most commonly to milk, egg or peanut[82]. Another two-year prospective study following children given milk and/or egg OIT showed a rate of subsequent diagnosis of eosinophilic gastroenterological disorders of 6.25% [83]. Additional investigation is needed to determine which patients would be at highest risk after receiving OIT for developing EoE.
Like measurement of specific IgE to food, also patch testing that look at delayed food reactions have lead to disappointing results[79]. Therefore diagnostic testing to guide food elimination in EoE remains a significant clinical problem due to the lack of effective clinical allergen testing in this disorder [84]. As a result, patients undergo multiple endoscopies and the development of allergen testing for food allergens with high specificity and sensitivity for use in EoE would have a significant quality of life impact for patients with this disorder. Recently we have described a specific CD154 induction in vitro of peripheral blood T cells to milk in patients with milk induced EoE, suggesting that this test could be used in the future to predict specific FA in patients with EoE, however further prospective study will be needed to confirm the clinical applicability of the test[20]
In addition to food allergens, some speculate that inhaled aeroallergens may play a role in exacerbating EoE for a subset of patients. Aeroallergen exposure and sensitization has been shown to induce EoE in mouse models[85]. Some studies have suggested that aeroallergen can cause seasonal flares in EoE disease activity. Ram et al performed a retrospective study of a pediatric EoE cohort and determined that 14% of 1,180 patients had seasonal flares in EoE symptoms and 20% of this subset of patients had biopsy-confirmed EoE exacerbations attributable to aeroallergen alone[21]. This suggests that in aeroallergen sensitization can be a clinically significant trigger of EoE flares and can be an issue in patients receiving oral immunotherapy (OIT) for environmental allergens. Several studies have unfortunately reported cases of newly diagnosed or relapsed EoE in patients receiving sublingual immunotherapy for pollen and grass [86, 87]. However a metanalysis by Lucendo et al. has failed to demonstrate a seasonal distribution of initial diagnosis and clinical recrudescence of EoE[88].
5. Immune response
The immune response responsible for the chronic-relapsing inflammation in EoE is to be attributed to an inter-talk between innate and adaptive immune response [10]. It is now believed that a dysfunctional esophageal epithelium in genetically predisposed individuals, when exposed to the right environmental condition, promotes a sensitization to food and a Th2 response [10]. The continual exposure to the antigen favors a chronic inflammation that ultimately leads to esophageal dysfunction and fibrosis [10].
5.1. Epithelial barrier dysfunction
The human esophagus is lined by a nonkeratinized stratified squamous epithelium [89]. The function of the epithelium is to serve as a barrier against microorganisms, acid, food antigens, and mechanical trauma [89]. The integrity of the epithelium is maintained by the intracellular connections provided by tight junctions, adherence junctions, and desmosomes [89].
Impaired epithelial barrier function is a hallmark of the allergic inflammation that characterizes EoE [10] (Figure 1). Abnormal epithelial features can be identified in the histological analysis of esophageal biopsies in patients with EoE [90]. The loss of cell-cell adhesion structures like tight junctions and Desmoglein have been demonstrated using in vitro assays such as transepithelial electrical resistance and FITC-Dextran assays and increased epithelial permeability correlated EoE disease activity [91]. An increased exposure to the Candida albicans via the impaired esophageal barrier can explain why patients with active EoE have higher rates of sensitization to such pathogen [92].
Barrier dysfunction in EoE is also characterized by a significantly altered mucosal transcriptome studied using RNA derived from patient biopsies, that is significantly different from the one seen in GERD patients or healthy controls [19]. As many as 1607 significantly altered genes in EoE have been identified, with approximately two-thirds upregulated [93]. In vitro studies have confirmed many of these findings. Studies using an air-liquid interface of human epithelial esophageal cells demonstrate that IL-13 stimulation induces gene expression changes in patterns that mimic the in vivo EoE transcriptome [93].
These studies have revealed complex, multiple level epithelial dysfunction in EoE involving cytokine production, downregulation of structural barrier genes, and increase in damaging proteases production. As previously discussed there are ample evidence of altered cytokine and chemokine signaling from the epithelium, including highly overexpressed levels of CCL26, IL-33, IL-25, Arachidonate 15-lipoxygenase (ALOX15) and TSLP[42, 49, 94, 95]. Next, there is significant downregulation of multiple structural genes important to maintain barrier integrity, including filaggrin (FLG), desmoglein 1 (DSG1), claudins (CLDN1 and CLDN7)[19, 93, 96, 97]. Finally, dysregulated expression of proteases and their inhibitors most likely result in further degradation of structurally important epithelial proteins, including DSG1[54]. For example upregulation of the protease CAPN14 and altered expression of protease inhibitors including SERPINS and SPINKs have been shown in EoE biopsy tissue [33]. SPINK7 depletion in the epithelium has been tied to increased secretion of the TH2 cytokine TSLP as well as eosinophil activation [98]. These effects are reduced in vitro via treatment with α1-antitrypsin [98]. Further exploration of how to revers epithelial barrier dysfunction in this disorder may result in potential future therapeutics.
There has also been significant effort to determine if the gene expression pattern seen in the esophagus can be harnessed as a diagnostic tool to distinguish EoE from other disorders. Wen et al developed a 96-gene quantitative PCR array with associated diagnostic algorithms to distinguish biopsies derived from EoE patients from those with reflux esophagitis [99]. This EoE diagnostic panel (EDP) has been validated in several cohorts, including adult and pediatric patients with EoE and can be used with RNA isolated from freshly isolated or formalin-fixed biopsy tissue to differentiate patients with EoE from those with GERD or without disease [100–102]. Gene expression signatures within this panel have also been correlated with response to topical steroid therapy, suggesting that this approach holds promise as an adjunct diagnostic tool [103].
Recently, there is growing evidence that proton pump inhibitors may have a second, anti-inflammatory mechanism of action by targeting the esophageal epithelium. This was of significant interest because there is a substantial subset of patients with esophageal eosinophilia who seem to respond to PPI monotherapy and Current guidelines now recommend that this subset should be treated indefinitely with a PPI as mono-therapy [15]. Therefore given the fact that PPI are easy to administer and have a very favorable risk benefit ratio, the new guidelines suggest to start treatment with PPI and only if there is a treatment failure with PPI it is recommended to start a therapy with dietary exclusion or swallowed steroid[15]. In a meta-analysis of 32 studies, 50.5% (95% CI 42.2%–58.7%) patients had histologic improvement (to <15 eosinophils/hpf) with PPI treatment with significant differences among different studies [104]. Interestingly, many of the same transcriptional changes were noted in patients whose EoE was responsive to PPI alone [105]. These included characteristic changes in genes for eosinophil chemotaxis (Eotaxin 3, CCL26), barrier molecules (Desmoglein 1, DSG1), tissue remodeling (periostea, POSTN), and mast cells (carboxypeptidase A, CPA3) [105]. PPI monotherapy resolved these changes. Further examination of PPI mechanism has revealed anti-cytokine effects at the level of the esophageal epithelium in addition to their well-established role as an inhibitor of the gastric parietal H+/K+-ATPase [105]. Evidence suggests that omeprazole may directly inhibit epithelial STAT6, downregulating the secretion of pro-inflammatory chemokines and cytokines [84]. In vitro studies of epithelial cells subjected to conventional doses of omeprazole have shown that omeprazole blocks eotaxin-3 secretion stimulated by Th2 cytokines in esophageal epithelial cells from patients with EoE [84]. Based on what we know of the epithelium dysregulation and PPI effectiveness other drugs that act on the epithelial barrier maybe developed in the future.
5.2. Immune dysregulation
As the name of the disorder implies, the esophageal mucosa in EoE is infiltrated by large numbers of eosinophils, as well as a diverse Th2-type inflammatory infiltrate (Figure 1). Eosinophils are attracted by migratory factors such as chemokine (C-C motif) ligand (CCL) 26 (eotaxin 3) and TSLP which are overexpressed in the esophageal epithelium [19]. TSLP is a basophil chemoattractant and has been demonstrated with EoE in humans [53]. Similarly IL-33 production, a cytokine with many redundant function compared to TSLP, is increased in the epithelium of EoE patients[79], and induces local production of IL-4, IL-5, IL-9, and IL-13[19]. This local environment most likely promotes the well documented eosinophil, mast cell, innate lymphocyte type 2 (ILC2), invariant natural killer T cell, T and B lymphocyte and basophil migration and Th2 cytokine secretion[53, 106–110].
Th2-skewing in EoE is not limited to the mucosal environment, but can also be observed systemically. Th2 cytokines like IL-5 and IL-13 can be found in the circulating Th2 type CD4+CD154+ cells [20]. Further, IL-5 and IL-13 are upregulated in CD4+ T cells from EoE patients following stimulation by protein milk antigens in patients with milk-sensitive EoE but not in control subjects [20]. Similarly milk derived lipids induced Th2 cytokine secretion in iNKT cells derived patients with EoE [108]. These studies indicate that EoE is a systemic Th2 antigen–driven disease.
Additional targeted therapy for EoE has been an area of active research because there is a subset of patients with EoE who do not respond well to treatment with dietary therapy or topical steroid, and there is clinical need to address this unmet therapeutic need. Several of the biologic agents tried to date have been those which share common mechanisms of atopy and relative Th2 inflammation, such as anti-IgE, anti-IL5, and anti-IL-13 monoclonal antibodies (Table 2) [80, 110–113]. While these molecular targets have been demonstrated to play a role in EoE, the trials of monoclonal antibodies targeting the Th2 pathway in EoE failed to induce clinical remission, despite the success in other Th2-predominant allergic disorders such as asthma and atopic dermatitis [111, 113–115]. There were several challenging limitations to the patient selection in these studies, including enrollment of patients with severe EoE with steroid-resistant disease. Additionally, the lack of clinically-validated measures to uniformly assess disease outcome in EoE has hindered comparison of results between trials. On the other hand antibodies which were able to block Th2 cell and other atopic cells like basophils and eosinophils recruitment and activation like anti Chemoattractant receptor-homologous molecule on Th2 cells (CRTH2) (a prostaglandin D (2) (PGD (2)) receptor) [114] or antibodies that block multiple Th2 pathways like the anti-IL-4 anti-IL-13 (dupilumab) have been shown to be more promising in small clinical trials. In an 8-week treatment with in 26 patients with in adult patients with active, corticosteroid-dependent or corticosteroid-refractory EoE the CRTH2-antagonist, OC000459, had a modest, but significant, anti-eosinophil and beneficial clinical effects [114]. Similarly a phase II placebo controlled randomized trial on 47 adult with EoE have shown that dupilumab significantly improved dysphagia, esophageal eosinophil counts, endoscopic features, histology, and esophageal distensibility in adults with active EoE compared with placebo[116].
TABLE 2:
BOLOGICS FOR TREATMENT OF EOE.
| Target | Mechanism | Medications | Study Design & Outcome | References |
|---|---|---|---|---|
| IgE | IgE bound to mast cells & basophils cause degranulation on antigen recognition. Role of IgE in EoE is unclear. | Omalizumab | ◦ Open label, 24 pts (age 14–71 yr). 33% with improved histology but no improvement in symptoms ◦ RDBCT, 27 adult & 3 children. No improvement in histology |
[80, 130, 131] |
| IL-5 | Elevated IL-5 has been described in numerous EoE studies | Mepolizumab | ◦ RDBCT, 11 adults. No patients with patients with <5 eos/hpf but some with significant improvement in histology ◦ RDBCT, 59 children (age 2–17 yr). 8.8% of patients with <5 eos/hpf; significant improvement in histology but symptoms not improved |
[110, 111, 132] |
| Reslizumab | RDBCT, 226 children (age 5–18 yr). Significant improvement in pathology with reslizumab (59%–67%) compared with placebo (24%); No significant improvement in Children’s Health Questionnaire or physician assessment of symptoms | [113] | ||
| IL-13 | IL-13 is increased in EoE, induces eotaxin-1, eotaxin-2, and eotaxin-3 expression via STAT6, and causes epithelial barrier dysfunction | QAX576 | RDBCT, 25 adults. After 12 weeks, 40% of QAX576 group had 75% reduction in peak esophageal count on biopsy (13% in placebo group). No difference in Mayo dysphagia symptom questionnaires. | [115] |
| IL-4rα | IL-4 is increased in EoE. IL-4 and IL-13 receptor for heterodimer with IL-4rα subunit: therapy blocking IL-4 signaling through IL4Ralpha will block both IL-4 and IL-13 pathways. | Dupilumab | RDBCT with 47 adult patients. With Dupilumab, histologic score, distensibility, and Straumann dysphagia score were improved at week 12 | [116] |
| TNF-α | TNF-α is increased in epithelium of EoE patients [4–6] | Infliximab | Open label, nonrandomized study. No significant difference in histology, however, two of three adult patients had improved symptoms[133]. | [133] |
| CRTH2 | Chemoattractant receptor-homologous molecule on TH2 cells (CRTH2) is the receptor for prostaglandin D2. ↑ prostaglandin D2 has been seen in plasma from EoE patients | OC000459 | RDBPCT of 26 adults with active EoE randomized to OC000459 or placebo. Esophageal eosinophil load decreased significantly, from 114.83 to 73.26 eos/hpf in OC000459 group with significant improvement in physician assessed disease activity[114][115][115][115][114][114](114). | [114][7] |
RDBPCT= Randomized Double Blind Placebo Controlled Trial; IL= interleukin; Ig+ Immunoglobulin; TNF= Tumor Mecrosis Factor; CRTH2= Chemoattractant receptor-homologous molecule on TH2 cells
These data suggest that possibly redundant Th2 pathway may need to be inhibited. Antibodies targeting Th2 pathway are very desirable as many patients with EoE have a number of atopic disorders including asthma or atopic dermatitis which are amenable to management with these biologic therapies and therefore it remains important to understand their effects on the disease process in EoE.
Recent data suggest that IgG4 is elevated in EoE [80, 117]. Clayton et al demonstrated that adult EoE patients have increased levels of IgG4 in esophageal tissue and that removal of dietary triggers decreased the levels of IgG4. The positive association between EoE and esophageal IgG4 production has also been demonstrated in the pediatric population [117]. It is suspected that this IgG4 is produced locally. Recent studies have correlated to the overall levels local IgG4 production to the esophageal eosinophil count as well as classical alterations seen with EoE on the mucosal transcriptome signature using the EDP 96-gene PCR array [117].
6. Fibrosis
Fibrosis begins early in EoE disease course, and has been documented in children with EoE [118, 119]. It is estimated that 57% to 88% of children have evidence of fibrosis, and that 89% of adults with EoE have some degree of fibrosis on biopsy tissue[118, 120]. These estimates can be difficult to assess on biopsy tissue as the pieces of tissue to not always include the lamina propria, which is where fibrotic tissue can initially be the most prominent. Broadly, fibrosis is correlated with increased esophageal stiffness, muscular hypertrophy, risk of food impaction as well as endoscopic appearance of tissue pallor [118]. On a molecular level, pro-fibrotic and pro-angiogenic factors such as TGFβ1, IL-5, CCL-18, VCAM-1 and VEGF are elevated in EoE[119].
The reversibility of tissue fibrosis in EoE is complex and can be difficult to assess. One study demonstrated that pediatric patients on steroid therapy with decrease in mucosal eosinophilia below 7 eos/hpf had reduced fibrosis scores [121, 122]. Other studies suggest that dietary restriction reduced fibrosis in 17.6% of patients, whereas 56% of steroid-treated patients had reduced fibrosis scores[123]. However, in adult patients evidence suggests that there may be discordance between the epithelial and subepithelial response to therapy, with evidence of persistent submucosal fibrosis despite resolution of eosinophilic inflammation [123]. More recently a study showed an association between refractory EoE and the Resisting-like-β (RTNLB) gene, which has been associated with asthma remodeling suggesting that genes important in fibrosis may play a role in therapy resistance [124]
As a result, there is interest in agents which specifically target fibrosis in EoE. Clinical trials of losartan are currently underway to evaluate its anti-fibrotic effects in EoE[125]. Losartan is an angiotensin II receptor blocker that is FDA-approved to treat hypertension. There is evidence that it may reduce signaling of TGFβ1, and may potentially be a therapy for EoE[126].
7. Expert commentary
EoE continues to present unique challenges for caregivers and patients alike.
Currently there are several limitations in the management of patients with EoE
Lack of not invasive diagnostic tool for diagnosis and monitoring of the disease
Lack of diagnostic test to predict specific food allergy
Lack of early diagnostic predictor factor
Inability to predict who will develop fibrosis
Inability to predict which therapeutic intervention will work better for the patients
Inability to induce remission or to improve at all the inflammation in subset of patients with EoE who do not respond to PPI dietary or steroid therapy
Research has helped us to better understand EoE and continue to innovate therapy. The understanding garnered from translational investigation in EoE has recently changed the treatment paradigm in this disorder in unexpected ways. Following the observation that omeprazole blocks STAT6 activity and secretion of CCL26 from esophageal epithelial cells and that the transcriptome of patients with PPI-responsive esophageal eosinophilia and EoE shared many dysregulated features, changes have been made to EoE treatment guidelines. PPI-responsive esophageal eosinophilia has therefore been reclassified and PPI does not play a required role in diagnosis of EoE any longer. This illustrates the utility of the gene expression signature from the esophageal biopsies in identifying features of this disorder. Similarly the demonstration that EOE is a Th2 mediated disease led to the development of trials with biological therapeutic agents that block Th2 cytokines.
However there are still many roadblock that we need to clear to provide optimal care to the patients
The lack of reliable biomarkers and in vitro testing to predict food allergies continue to present a unique challenge to treat the ever increasing EoE population. At the present time diagnosis and follow after each therapeutic intervention require to perform endoscopies, as no biomarkers of disease or disease activity have been found to have enough sensitivity and specificity. As there are no in vitro testing to predict food allergen triggers, patients who chose to do a dietary management of the disease typically undergo several biopsies before reaching the least restrictive and effective diet. This medical approach can be prohibitive in countries where availability of endoscopies and health care resources are limited. In other setting this approach maybe cost prohibitive for the patients who don’t have medical insurance or have insurances with high deductibles.
Also the current diagnostic criteria based solely on the numbers of eosinophils infiltrating the esophageal mucosa maybe not able to capture the full spectrum of the disease as many patient do well despite persistent esophageal eosinophilia and other continue to manifest symptoms after eosinophilia resolves.
Therefore research is needed to understand pathogenesis the disease and food allergy mechanism to develop noninvasive diagnostics tools for disease diagnosis, monitoring and food allergy evaluation.
Genetic testing has been helping in finding molecular targets in EoE pathogenesis and may provide in the future guidance for deep phenotyping of the disease and therefore open the door to personalized medicine.
Ideally in the future we will have a noninvasive test to diagnose and monitor the disease, a genetic/epigenetic profile of the patient will guide us to know the patient specific long term risk of fibrosis, the chances of being allergic to multiple foods and the mechanism causing its disease so the doctor can adequately counsel the patient to obtain the most appropriate treatment based on his specific long term risks
8. Five year view
Even the research that has been conducted to date has altered our understanding of this disorder.
In the next five years the use of biologicals, deep phenotyping and noninvasive tools to diagnose and monitor the disease, will revolutionize the way we manage patients today
A focus of next five year research will be to determine of gene expression features can be used to risk stratify groups of EoE patients in a clinically meaningful way
The use of less invasive esophageal sampling methods such as the esophageal string test or cytosponge timely monitoring of disease activity and quick adjustment of the therapeutic intervention. It will also help to establish minimum duration of dosage of therapeutic agents’ uses [127, 128].
Studies on T cell reactive to specific foods via Th2 response may provide a much needed in vitro testing for Food allergens that trigger EoE
Lastly, there is significant interest in developing a biologic-based agent for the treatment of EoE, these product will certainly help to provide personalized and multisystem treatments, but also will shed light on pathogenesis and phenotyping of patient population
9. Key issues
Eosinophilic esophagitis (EoE) is common and occurs in approximately 1 in 2000 persons. It is more common in males, whites, and in persons with a history of atopy or connective tissue disorder.
Several GWAS studies have established risk loci near TSLP, CAPN14, LRRC32, IL33, STAT6, c11orf30 and ANKRD27. Mechanistic studies have established roles for TSLP, CAPN14, IL33, and STAT6 in the esophageal epithelium highlighting the importance of the epithelium in this disorder.
The risk of EoE is increased in several monogenic disorders as outlined in Table 1, and these studies demonstrate that the genetic contributions to the pathogenesis of EoE are diverse
The role of environmental exposures in the etiopathogenesis of EoE is just beginning to be understood, however, the role of food and aeroallergen sensitization is well-established as a major trigger for ongoing inflammation in the disorder.
The mainstays of therapy for EoE are PPI, avoidance of dietary triggers, topical steroids and dilation therapy as needed for strictures.
There have been a number of trials of novel biologic therapies for EoE, but in general these trials were less successful than anticipated. This raises questions about which mechanisms involved in EoE represent the most promising targets for development of novel therapy.
Funding
M Ruffner was supported by a grant NIH KL2TR001879. A Cianferono is supported by CHOP Food allergy fund.
Footnotes
Declaration of interest
A Cianferoni has received speaker fees from DBV and consulting fees from Sanofi. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
- 1.Dellon ES, Jensen ET, Martin CF, Shaheen NJ, Kappelman MD. Prevalence of eosinophilic esophagitis in the United States. Clin Gastroenterol Hepatol. 2014;12(4):589–96 e1. doi: 10.1016/j.cgh.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dellon ES, Erichsen R, Baron JA, Shaheen NJ, Vyberg M, Sorensen HT, et al. The increasing incidence and prevalence of eosinophilic oesophagitis outpaces changes in endoscopic and biopsy practice: national population-based estimates from Denmark. Aliment Pharmacol Ther. 2015;41(7):662–70. doi: 10.1111/apt.13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dellon ES, Hirano I. Epidemiology and Natural History of Eosinophilic Esophagitis. Gastroenterology. 2018;154(2):319–32 e3. doi: 10.1053/j.gastro.2017.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hruz P, Straumann A, Bussmann C, Heer P, Simon HU, Zwahlen M, et al. Escalating incidence of eosinophilic esophagitis: a 20-year prospective, population-based study in Olten County, Switzerland. J Allergy Clin Immunol. 2011;128(6):1349–50 e5. doi: 10.1016/j.jaci.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 5.van Rhijn BD, Verheij J, Smout AJ, Bredenoord AJ. Rapidly increasing incidence of eosinophilic esophagitis in a large cohort. Neurogastroenterol Motil. 2013;25(1):47–52 e5. doi: 10.1111/nmo.12009. [DOI] [PubMed] [Google Scholar]
- 6.Hommeida S, Grothe RM, Hafed Y, Lennon RJ, Schleck CD, Alexander JA, et al. Assessing the incidence trend and characteristics of eosinophilic esophagitis in children in Olmsted County, Minnesota. Dis Esophagus. 2018. doi: 10.1093/dote/doy062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robson J, O’Gorman M, McClain A, Mutyala K, Davis C, Barbagelata C, et al. Incidence and Prevalence of Pediatric Eosinophilic Esophagitis in Utah Based on a 5-Year Population-Based Study. Clin Gastroenterol Hepatol. 2018. doi: 10.1016/j.cgh.2018.06.028. [DOI] [PubMed] [Google Scholar]
- 8.Arias A, Lucendo AJ. Incidence and prevalence of eosinophilic oesophagitis increase continiously in adults and children in Central Spain: A 12-year population-based study. Dig Liver Dis. 2018. doi: 10.1016/j.dld.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 9.Molina-Infante J, Gonzalez-Cordero PL, Ferreira-Nossa HC, Mata-Romero P, Lucendo AJ, Arias A. Rising incidence and prevalence of adult eosinophilic esophagitis in midwestern Spain (2007–2016). United European Gastroenterol J. 2018;6(1):29–37. doi: 10.1177/2050640617705913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spergel JM, Aceves SS, Kliewer K, Gonsalves N, Chehade M, Wechsler JB, et al. New developments in patients with eosinophilic gastrointestinal diseases presented at the CEGIR/TIGERS Symposium at the 2018 American Academy of Allergy, Asthma & Immunology Meeting. J Allergy Clin Immunol. 2018;142(1):48–53. doi: 10.1016/j.jaci.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spergel JM, Brown-Whitehorn TF, Beausoleil JL, Franciosi J, Shuker M, Verma R, et al. 14 years of eosinophilic esophagitis: clinical features and prognosis. J Pediatr Gastroenterol Nutr. 2009;48(1):30–6. doi: 10.1097/MPG.0b013e3181788282. [DOI] [PubMed] [Google Scholar]
- 12.Noel RJ, Putnam PE, Rothenberg ME. Eosinophilic esophagitis. N Engl J Med. 2004;351(9):940–1. [DOI] [PubMed] [Google Scholar]
- 13.Straumann A The natural history and complications of eosinophilic esophagitis. Thorac Surg Clin. 2011;21(4):575–87. doi: 10.1016/j.thorsurg.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Straumann A, Spichtin HP, Grize L, Bucher KA, Beglinger C, Simon HU. Natural history of primary eosinophilic esophagitis: a follow-up of 30 adult patients for up to 11.5 years. Gastroenterology. 2003;125(6):1660–9. [DOI] [PubMed] [Google Scholar]
- 15.Dellon ES, Liacouras CA, Molina-Infante J, Furuta GT, Spergel JM, Zevit N, et al. Updated international consensus diagnostic criteria for eosinophilic esophagitis: Proceedings of the AGREE conference. Gastroenterology. 2018. doi: 10.1053/j.gastro.2018.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucendo AJ, Molina-Infante J, Arias A, von Arnim U, Bredenoord AJ, Bussmann C, et al. Guidelines on eosinophilic esophagitis: evidence-based statements and recommendations for diagnosis and management in children and adults. United European Gastroenterol J. 2017;5(3):335–58. doi: 10.1177/2050640616689525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sayej WN, Menoret A, Maharjan AS, Fernandez M, Wang Z, Balarezo F, et al. Characterizing the inflammatory response in esophageal mucosal biopsies in children with eosinophilic esophagitis. Clin Transl Immunology. 2016;5(7):e88. doi: 10.1038/cti.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. IL-5 promotes eosinophil trafficking to the esophagus. J Immunol. 2002;168(5):2464–9. [DOI] [PubMed] [Google Scholar]
- 19.Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127(1):208–17, 17 e1–7. Epub 2011/01/08. doi: S0091–6749(10)01654–4 [pii] 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cianferoni A, Ruffner MA, Guzek R, Guan S, Brown-Whitehorn T, Muir A, et al. Elevated expression of activated TH2 cells and milk-specific TH2 cells in milk-induced eosinophilic esophagitis. Ann Allergy Asthma Immunol. 2018;120(2):177–83 e2. doi: 10.1016/j.anai.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ram G, Lee J, Ott M, Brown-Whitehorn TF, Cianferoni A, Shuker M, et al. Seasonal exacerbation of esophageal eosinophilia in children with eosinophilic esophagitis and allergic rhinitis. Ann Allergy Asthma Immunol. 2015;115(3):224–8 e1. doi: 10.1016/j.anai.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Lucendo AJ, Molina-Infante J, Arias A, Gonzalez-Cervera J. Seasonal Variation in the Diagnosis of Eosinophilic Esophagitis: There and Back Again. J Pediatr Gastroenterol Nutr. 2017;64(1):e25. doi: 10.1097/MPG.0000000000001417. [DOI] [PubMed] [Google Scholar]
- 23.Molina-Infante J, Arias A, Alcedo J, Garcia-Romero R, Casabona-Frances S, Prieto-Garcia A, et al. Step-up empiric elimination diet for pediatric and adult eosinophilic esophagitis: The 2-4-6 study. J Allergy Clin Immunol. 2018;141(4):1365–72. doi: 10.1016/j.jaci.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 24.Lucendo AJ. Meta-Analysis-Based Guidance for Dietary Management in Eosinophilic Esophagitis. Curr Gastroenterol Rep. 2015;17(10):464. doi: 10.1007/s11894-015-0464-y. [DOI] [PubMed] [Google Scholar]
- 25.Arias A, Gonzalez-Cervera J, Tenias JM, Lucendo AJ. Efficacy of dietary interventions for inducing histologic remission in patients with eosinophilic esophagitis: a systematic review and meta-analysis. Gastroenterology. 2014;146(7):1639–48. doi: 10.1053/j.gastro.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Wechsler JB, Hirano I. Biological therapies for eosinophilic gastrointestinal diseases. J Allergy Clin Immunol. 2018;142(1):24–31 e2. doi: 10.1016/j.jaci.2018.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cianferoni A, Spergel JM. From genetics to treatment of eosinophilic esophagitis. Curr Opin Allergy Clin Immunol. 2015;15(5):417–25. doi: 10.1097/ACI.0000000000000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexander ES, Martin LJ, Collins MH, Kottyan LC, Sucharew H, He H, et al. Twin and family studies reveal strong environmental and weaker genetic cues explaining heritability of eosinophilic esophagitis. J Allergy Clin Immunol. 2014;134(5):1084–92 e1. doi: 10.1016/j.jaci.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Straumann A [Eosinophilic esophagitis: a novel entity?]. Praxis (Bern 1994). 2006;95(6):191–5. doi: 10.1024/0369-8394.95.6.191. [DOI] [PubMed] [Google Scholar]
- 30.Cianferoni A, Spergel JM, Muir A. Recent advances in the pathological understanding of eosinophilic esophagitis. Expert Rev Gastroenterol Hepatol. 2015;9(12):1501–10. doi: 10.1586/17474124.2015.1094372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kottyan LC, Maddox A, Braxton JR, Stucke EM, Mukkada V, Putnam PE, et al. Genetic variants at the 16p13 locus confer risk for eosinophilic esophagitis. Genes Immun. 2018. doi: 10.1038/s41435-018-0034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin LJ, He H, Collins MH, Abonia JP, Biagini Myers JM, Eby M, et al. Eosinophilic esophagitis (EoE) genetic susceptibility is mediated by synergistic interactions between EoE-specific and general atopic disease loci. J Allergy Clin Immunol. 2018;141(5):1690–8. doi: 10.1016/j.jaci.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothenberg ME. Molecular, genetic, and cellular bases for treating eosinophilic esophagitis. Gastroenterology. 2015;148(6):1143–57. doi: 10.1053/j.gastro.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherrill JD, Rothenberg ME. Genetic and epigenetic underpinnings of eosinophilic esophagitis. Gastroenterol Clin North Am. 2014;43(2):269–80. doi: 10.1016/j.gtc.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fahey LM, Chandramouleeswaran PM, Guan S, Benitez AJ, Furuta GT, Aceves SS, et al. Food allergen triggers are increased in children with the TSLP risk allele and eosinophilic esophagitis. Clin Transl Gastroenterol. 2018;9(3):139. doi: 10.1038/s41424-018-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nature Genetics. 2010;42(4):289–91. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherrill JD, Gao PS, Stucke EM, Blanchard C, Collins MH, Putnam PE, et al. Variants of thymic stromal lymphopoietin and its receptor associate with eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126(1):160–5 e3. doi: 10.1016/j.jaci.2010.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sherrill JD, Kc K, Wang X, Wen T, Chamberlin A, Stucke EM, et al. Whole-exome sequencing uncovers oxidoreductases DHTKD1 and OGDHL as linkers between mitochondrial dysfunction and eosinophilic esophagitis. JCI Insight. 2018;3(8). doi: 10.1172/jci.insight.99922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sleiman PM, Wang ML, Cianferoni A, Aceves S, Gonsalves N, Nadeau K, et al. GWAS identifies four novel eosinophilic esophagitis loci. Nature communications. 2014;5:5593. doi: 10.1038/ncomms6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allen-Brady K, Firszt R, Fang JC, Wong J, Smith KR, Peterson KA. Population-based familial aggregation of eosinophilic esophagitis suggests a genetic contribution. J Allergy Clin Immunol. 2017;140(4):1138–43. doi: 10.1016/j.jaci.2016.12.979. [DOI] [PubMed] [Google Scholar]
- 41.Lebowitz MD, Barbee R, Burrows B. Family concordance of IgE, atopy, and disease. J Allergy Clin Immunol. 1984;73(2):259–64. [DOI] [PubMed] [Google Scholar]
- 42.Rothenberg ME, Spergel JM, Sherrill JD, Annaiah K, Martin LJ, Cianferoni A, et al. Common variants at 5q22 associate with pediatric eosinophilic esophagitis. Nat Genet. 2010;42(4):289–91. doi: 10.1038/ng.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spergel JM. New genetic links in eosinophilic esophagitis. Genome medicine. 2010;2(9):60. doi: 10.1186/gm181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanchard C, Wang N, Rothenberg ME. Eosinophilic esophagitis: pathogenesis, genetics, and therapy. J Allergy Clin Immunol. 2006;118(5):1054–9. [DOI] [PubMed] [Google Scholar]
- 45.March ME, Sleiman PM, Hakonarson H. The genetics of asthma and allergic disorders. Discovery medicine. 2011;11(56):35–45. [PubMed] [Google Scholar]
- 46.Aceves S, Hirano I, Furuta GT, Collins MH. Eosinophilic gastrointestinal diseases--clinically diverse and histopathologically confounding. Semin Immunopathol. 2012;34(5):715–31. doi: 10.1007/s00281-012-0324-x. [DOI] [PubMed] [Google Scholar]
- 47.Aceves SS, Ackerman SJ. Relationships between eosinophilic inflammation, tissue remodeling, and fibrosis in eosinophilic esophagitis. Immunol Allergy Clin North Am. 2009;29(1):197–211, xiii–xiv. Epub 2009/01/15. doi: S0889–8561(08)00107–0 [pii] 10.1016/j.iac.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aceves SS, Broide DH. Airway fibrosis and angiogenesis due to eosinophil trafficking in chronic asthma. Current molecular medicine. 2008;8(5):350–8. [DOI] [PubMed] [Google Scholar]
- 49.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116(2):536–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Avila-Castellano R, Garcia-Lozano JR, Cimbollek S, Lucendo AJ, Bozada JM, Quiralte J. Genetic variations in the TLR3 locus are associated with eosinophilic esophagitis. United European Gastroenterol J. 2018;6(3):349–57. doi: 10.1177/2050640617732643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kottyan LC, Davis BP, Sherrill JD, Liu K, Rochman M, Kaufman K, et al. Genome-wide association analysis of eosinophilic esophagitis provides insight into the tissue specificity of this allergic disease. Nat Genet. 2014;46(8):895–900. doi: 10.1038/ng.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cianferoni A, Spergel J. The importance of TSLP in allergic disease and its role as a potential therapeutic target. Expert Rev Clin Immunol. 2014;10(11):1463–74. doi: 10.1586/1744666X.2014.967684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noti M, Wojno ED, Kim BS, Siracusa MC, Giacomin PR, Nair MG, et al. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nat Med. 2013. Epub 2013/07/23. doi: nm.3281 [pii] 10.1038/nm.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis BP, Stucke EM, Khorki ME, Litosh VA, Rymer JK, Rochman M, et al. Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment. JCI Insight. 2016;1(4):e86355. doi: 10.1172/jci.insight.86355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Asai Y, Eslami A, van Ginkel CD, Akhabir L, Wan M, Ellis G, et al. Genome-wide association study and meta-analysis in multiple populations identifies new loci for peanut allergy and establishes C11orf30/EMSY as a genetic risk factor for food allergy. J Allergy Clin Immunol. 2018;141(3):991–1001. doi: 10.1016/j.jaci.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 56.Amaral AF, Minelli C, Guerra S, Wjst M, Probst-Hensch N, Pin I, et al. The locus C11orf30 increases susceptibility to poly-sensitization. Allergy. 2015;70(3):328–33. doi: 10.1111/all.12557. [DOI] [PubMed] [Google Scholar]
- 57.Li X, Ampleford EJ, Howard TD, Moore WC, Li H, Busse WW, et al. The C11orf30-LRRC32 region is associated with total serum IgE levels in asthmatic patients. J Allergy Clin Immunol. 2012;129(2):575–8, 8 e1–9. doi: 10.1016/j.jaci.2011.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fahey LM, Guzek R, Ruffner MA, Sullivan KE, Spergel J, Cianferoni A. EMSY is increased and activates TSLP & CCL5 expression in eosinophilic esophagitis. Pediatr Allergy Immunol. 2018;29(5):565–8. doi: 10.1111/pai.12907. [DOI] [PubMed] [Google Scholar]
- 59.Abonia JP, Wen T, Stucke EM, Grotjan T, Griffith MS, Kemme KA, et al. High prevalence of eosinophilic esophagitis in patients with inherited connective tissue disorders. J Allergy Clin Immunol. 2013. Epub 2013/04/24. doi: S0091–6749(13)00361–8 [pii] 10.1016/j.jaci.2013.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lyons JJ, Liu Y, Ma CA, Yu X, O’Connell MP, Lawrence MG, et al. ERBIN deficiency links STAT3 and TGF-beta pathway defects with atopy in humans. J Exp Med. 2017;214(3):669–80. doi: 10.1084/jem.20161435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arora M, Bagi P, Strongin A, Heimall J, Zhao X, Lawrence MG, et al. Gastrointestinal Manifestations of STAT3-Deficient Hyper-IgE Syndrome. J Clin Immunol. 2017;37(7):695–700. doi: 10.1007/s10875-017-0429-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paluel-Marmont C, Bellon N, Barbet P, Leclerc-Mercier S, Hadj-Rabia S, Dupont C, et al. Eosinophilic esophagitis and colonic mucosal eosinophilia in Netherton syndrome. J Allergy Clin Immunol. 2017;139(6):2003–5 e1. doi: 10.1016/j.jaci.2016.10.045. [DOI] [PubMed] [Google Scholar]
- 63.Henderson CJ, Ngeow J, Collins MH, Martin LJ, Putnam PE, Abonia JP, et al. Increased prevalence of eosinophilic gastrointestinal disorders in pediatric PTEN hamartoma tumor syndromes. J Pediatr Gastroenterol Nutr. 2014;58(5):553–60. doi: 10.1097/MPG.0000000000000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karvonen AM, Hyvarinen A, Rintala H, Korppi M, Taubel M, Doekes G, et al. Quantity and diversity of environmental microbial exposure and development of asthma: a birth cohort study. Allergy. 2014;69(8):1092–101. doi: 10.1111/all.12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Illi S, Weber J, Zutavern A, Genuneit J, Schierl R, Strunz-Lehner C, et al. Perinatal influences on the development of asthma and atopy in childhood. Ann Allergy Asthma Immunol. 2014;112(2):132–9 e1. doi: 10.1016/j.anai.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 66.Jensen ET, Kuhl JT, Martin LJ, Rothenberg ME, Dellon ES. Prenatal, intrapartum, and postnatal factors are associated with pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2018;141(1):214–22. doi: 10.1016/j.jaci.2017.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jensen ET, Kuhl JT, Martin LJ, Langefeld CD, Dellon ES, Rothenberg ME. Early-life environmental exposures interact with genetic susceptibility variants in pediatric patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2018;141(2):632–7 e5. doi: 10.1016/j.jaci.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hill DA, Spergel JM. The atopic march: Critical evidence and clinical relevance. Ann Allergy Asthma Immunol. 2018;120(2):131–7. doi: 10.1016/j.anai.2017.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Padia R, Curtin K, Peterson K, Orlandi RR, Alt J. Eosinophilic esophagitis strongly linked to chronic rhinosinusitis. Laryngoscope. 2016;126(6):1279–83. doi: 10.1002/lary.25798. [DOI] [PubMed] [Google Scholar]
- 70.Hill DA, Grundmeier RW, Ramos M, Spergel JM. Eosinophilic Esophagitis Is a Late Manifestation of the Allergic March. J Allergy Clin Immunol Pract. 2018. doi: 10.1016/j.jaip.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simon D, Cianferoni A, Spergel JM, Aceves S, Holbreich M, Venter C, et al. Eosinophilic esophagitis is characterized by a non-IgE-mediated food hypersensitivity. Allergy. 2016;71(5):611–20. doi: 10.1111/all.12846. [DOI] [PubMed] [Google Scholar]
- 72.Spergel JM, Brown-Whitehorn TF, Cianferoni A, Shuker M, Wang ML, Verma R, et al. Identification of causative foods in children with eosinophilic esophagitis treated with an elimination diet. J Allergy Clin Immunol. 2012;130(2):461–7 e5. Epub 2012/06/30. doi: S0091–6749(12)00859–7 [pii] 10.1016/j.jaci.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 73.Gonsalves N, Yang GY, Doerfler B, Ritz S, Ditto AM, Hirano I. Elimination diet effectively treats eosinophilic esophagitis in adults; food reintroduction identifies causative factors. Gastroenterology. 2012;142(7):1451–9 e1; quiz e14–5. Epub 2012/03/07. doi: S0016–5085(12)00309–5 [pii] 10.1053/j.gastro.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 74.Kagalwalla AF, Sentongo TA, Ritz S, Hess T, Nelson SP, Emerick KM, et al. Effect of six-food elimination diet on clinical and histologic outcomes in eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2006;4(9):1097–102. [DOI] [PubMed] [Google Scholar]
- 75.Liacouras CA, Spergel JM, Ruchelli E, Verma R, Mascarenhas M, Semeao E, et al. Eosinophilic esophagitis: a 10-year experience in 381 children. Clin Gastroenterol Hepatol. 2005;3(12):1198–206. [DOI] [PubMed] [Google Scholar]
- 76.Lucendo AJ, Arias A, Gonzalez-Cervera J, Yague-Compadre JL, Guagnozzi D, Angueira T, et al. Empiric 6-food elimination diet induced and maintained prolonged remission in patients with adult eosinophilic esophagitis: a prospective study on the food cause of the disease. J Allergy Clin Immunol. 2013;131(3):797–804. Epub 2013/02/05. doi: S0091–6749(12)02644–9 [pii] 10.1016/j.jaci.2012.12.664. [DOI] [PubMed] [Google Scholar]
- 77.Molina-Infante J, Arias A, Barrio J, Rodriguez-Sanchez J, Sanchez-Cazalilla M, Lucendo AJ. Four-food group elimination diet for adult eosinophilic esophagitis: A prospective multicenter study. J Allergy Clin Immunol. 2014;134(5):1093–9 e1. doi: 10.1016/j.jaci.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 78.Kelly KJ, Lazenby AJ, Rowe PC, Yardley JH, Perman JA, Sampson HA. Eosinophilic esophagitis attributed to gastroesophageal reflux: improvement with an amino acid-based formula. Gastroenterology. 1995;109(5):1503–12. [DOI] [PubMed] [Google Scholar]
- 79.Spergel J, Aceves SS. Allergic components of eosinophilic esophagitis. J Allergy Clin Immunol. 2018;142(1):1–8. doi: 10.1016/j.jaci.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clayton F, Fang JC, Gleich GJ, Lucendo AJ, Olalla JM, Vinson LA, et al. Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology. 2014;147(3):602–9. doi: 10.1053/j.gastro.2014.05.036. [DOI] [PubMed] [Google Scholar]
- 81.Noti M, Wojno ED, Kim BS, Siracusa MC, Giacomin PR, Nair MG, et al. Thymic stromal lymphopoietin-elicited basophil responses promote eosinophilic esophagitis. Nat Med. 2013;19(8):1005–13. doi: 10.1038/nm.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lucendo AJ, Arias A, Tenias JM. Relation between eosinophilic esophagitis and oral immunotherapy for food allergy: a systematic review with meta-analysis. Ann Allergy Asthma Immunol. 2014;113(6):624–9. doi: 10.1016/j.anai.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 83.Echeverria-Zudaire LA, Fernandez-Fernandez S, Rayo-Fernandez A, Munoz-Archidona C, Checa-Rodriguez R. Primary eosinophilic gastrointestinal disorders in children who have received food oral immunotherapy. Allergol Immunopathol (Madr). 2016;44(6):531–6. doi: 10.1016/j.aller.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 84.Zhang X, Cheng E, Huo X, Yu C, Zhang Q, Pham TH, et al. Omeprazole blocks STAT6 binding to the eotaxin-3 promoter in eosinophilic esophagitis cells. PLoS One. 2012;7(11):e50037. doi: 10.1371/journal.pone.0050037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest. 2001;107(1):83–90. doi: 10.1172/JCI10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miehlke S, Alpan O, Schroder S, Straumann A. Induction of eosinophilic esophagitis by sublingual pollen immunotherapy. Case Rep Gastroenterol. 2013;7(3):363–8. doi: 10.1159/000355161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wells R, Fox AT, Furman M. Recurrence of eosinophilic oesophagitis with subcutaneous grass pollen immunotherapy. BMJ Case Rep. 2018;2018. doi: 10.1136/bcr-2017-223465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lucendo AJ, Arias A, Redondo-Gonzalez O, Gonzalez-Cervera J. Seasonal distribution of initial diagnosis and clinical recrudescence of eosinophilic esophagitis: a systematic review and meta-analysis. Allergy. 2015;70(12):1640–50. doi: 10.1111/all.12767. [DOI] [PubMed] [Google Scholar]
- 89.Rosekrans SL, Baan B, Muncan V, van den Brink GR. Esophageal development and epithelial homeostasis. Am J Physiol Gastrointest Liver Physiol. 2015;309(4):G216–28. doi: 10.1152/ajpgi.00088.2015. [DOI] [PubMed] [Google Scholar]
- 90.Collins MH, Martin LJ, Alexander ES, Boyd JT, Sheridan R, He H, et al. Newly developed and validated eosinophilic esophagitis histology scoring system and evidence that it outperforms peak eosinophil count for disease diagnosis and monitoring. Dis Esophagus. 2017;30(3):1–8. doi: 10.1111/dote.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Warners MJ, van Rhijn BD, Verheij J, Smout A, Bredenoord AJ. Disease activity in eosinophilic esophagitis is associated with impaired esophageal barrier integrity. Am J Physiol Gastrointest Liver Physiol. 2017;313(3):G230–G8. doi: 10.1152/ajpgi.00058.2017. [DOI] [PubMed] [Google Scholar]
- 92.Simon D, Straumann A, Dahinden C, Simon HU. Frequent sensitization to Candida albicans and profilins in adult eosinophilic esophagitis. Allergy. 2013;68(7):945–8. doi: 10.1111/all.12157. [DOI] [PubMed] [Google Scholar]
- 93.Sherrill JD, Kiran KC, Blanchard C, Stucke EM, Kemme KA, Collins MH, et al. Analysis and expansion of the eosinophilic esophagitis transcriptome by RNA sequencing. Genes Immun. 2014;15(6):361–9. doi: 10.1038/gene.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Matoso A, Resnick MB. Immunohistochemical analysis of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2015;13(6):1209–10. doi: 10.1016/j.cgh.2014.12.024. [DOI] [PubMed] [Google Scholar]
- 95.Hui Y, Chen S, Lombardo KA, Resnick MB, Mangray S, Matoso A. ALOX15 Immunohistochemistry Aids in the Diagnosis of Eosinophilic Esophagitis on Pauci-eosinophilic Biopsies in Children. Pediatr Dev Pathol. 2017;20(5):375–80. doi: 10.1177/1093526617693106. [DOI] [PubMed] [Google Scholar]
- 96.Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 2010;184(7):4033–41. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Matoso A, Mukkada VA, Lu S, Monahan R, Cleveland K, Noble L, et al. Expression microarray analysis identifies novel epithelial-derived protein markers in eosinophilic esophagitis. Mod Pathol. 2013;26(5):665–76. doi: 10.1038/modpathol.2013.41. [DOI] [PubMed] [Google Scholar]
- 98.Azouz NP, Ynga-Durand MA, Caldwell JM, Jain A, Rochman M, Fischesser DM, et al. The antiprotease SPINK7 serves as an inhibitory checkpoint for esophageal epithelial inflammatory responses. Sci Transl Med. 2018;10(444). doi: 10.1126/scitranslmed.aap9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wen T, Stucke EM, Grotjan TM, Kemme KA, Abonia JP, Putnam PE, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology. 2013;145(6):1289–99. doi: 10.1053/j.gastro.2013.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wen T, Rothenberg ME. Clinical Applications of the Eosinophilic Esophagitis Diagnostic Panel. Front Med (Lausanne). 2017;4:108. doi: 10.3389/fmed.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dellon ES, Veerappan R, Selitsky SR, Parker JS, Higgins LL, Beitia R, et al. A Gene Expression Panel is Accurate for Diagnosis and Monitoring Treatment of Eosinophilic Esophagitis in Adults. Clin Transl Gastroenterol. 2017;8(2):e74. doi: 10.1038/ctg.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dellon ES, Rusin S, Gebhart JH, Covey S, Higgins LL, Beitia R, et al. Utility of a Noninvasive Serum Biomarker Panel for Diagnosis and Monitoring of Eosinophilic Esophagitis: A Prospective Study. Am J Gastroenterol. 2015;110(6):821–7. doi: 10.1038/ajg.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Butz BK, Wen T, Gleich GJ, Furuta GT, Spergel J, King E, et al. Efficacy, dose reduction, and resistance to high-dose fluticasone in patients with eosinophilic esophagitis. Gastroenterology. 2014;147(2):324–33 e5. doi: 10.1053/j.gastro.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lucendo AJ, Arias A, Molina-Infante J. Efficacy of Proton Pump Inhibitor Drugs for Inducing Clinical and Histologic Remission in Patients With Symptomatic Esophageal Eosinophilia: A Systematic Review and Meta-Analysis. Clin Gastroenterol Hepatol. 2016;14(1):13–22 e1. doi: 10.1016/j.cgh.2015.07.041. [DOI] [PubMed] [Google Scholar]
- 105.Wen T, Dellon ES, Moawad FJ, Furuta GT, Aceves SS, Rothenberg ME. Transcriptome analysis of proton pump inhibitor-responsive esophageal eosinophilia reveals proton pump inhibitor-reversible allergic inflammation. J Allergy Clin Immunol. 2015;135(1):187–97. doi: 10.1016/j.jaci.2014.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Doherty TA, Baum R, Newbury RO, Yang T, Dohil R, Aquino M, et al. Group 2 innate lymphocytes (ILC2) are enriched in active eosinophilic esophagitis. J Allergy Clin Immunol. 2015;136(3):792–4 e3. doi: 10.1016/j.jaci.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Strasser DS, Seger S, Bussmann C, Pierlot GM, Groenen PMA, Stalder AK, et al. Eosinophilic oesophagitis: relevance of mast cell infiltration. Histopathology. 2018;73(3):454–63. doi: 10.1111/his.13653. [DOI] [PubMed] [Google Scholar]
- 108.Jyonouchi S, Smith CL, Saretta F, Abraham V, Ruymann KR, Modayur-Chandramouleeswaran P, et al. Invariant natural killer T cells in children with eosinophilic esophagitis. Clin Exp Allergy. 2014;44(1):58–68. doi: 10.1111/cea.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Stuck MC, Straumann A, Simon HU. Relative lack of T regulatory cells in adult eosinophilic esophagitis - no normalization after corticosteroid therapy. Allergy. 2011;66(5):705–7. doi: 10.1111/j.1398-9995.2010.02525.x. [DOI] [PubMed] [Google Scholar]
- 110.Conus S, Straumann A, Bettler E, Simon HU. Mepolizumab does not alter levels of eosinophils, T cells, and mast cells in the duodenal mucosa in eosinophilic esophagitis. J Allergy Clin Immunol. 2010;126(1):175–7. doi: 10.1016/j.jaci.2010.04.029. [DOI] [PubMed] [Google Scholar]
- 111.Straumann A, Conus S, Grzonka P, Kita H, Kephart G, Bussmann C, et al. Anti-interleukin-5 antibody treatment (mepolizumab) in active eosinophilic oesophagitis: a randomised, placebo-controlled, double-blind trial. Gut. 2010;59(1):21–30. doi: 10.1136/gut.2009.178558. [DOI] [PubMed] [Google Scholar]
- 112.Conus S, Straumann A, Simon HU. Anti-IL-5 (mepolizumab) therapy does not alter IL-5 receptor alpha levels in patients with eosinophilic esophagitis. J Allergy Clin Immunol. 2009;123(1):269; author reply −70. doi: 10.1016/j.jaci.2008.09.031. [DOI] [PubMed] [Google Scholar]
- 113.Spergel JM, Rothenberg ME, Collins MH, Furuta GT, Markowitz JE, Fuchs G 3rd, et al. Reslizumab in children and adolescents with eosinophilic esophagitis: results of a double-blind, randomized, placebo-controlled trial. J Allergy Clin Immunol. 2012;129(2):456–63, 63 e1–3. doi: 10.1016/j.jaci.2011.11.044. [DOI] [PubMed] [Google Scholar]
- 114.Straumann A, Hoesli S, Bussmann C, Stuck M, Perkins M, Collins LP, et al. Anti-eosinophil activity and clinical efficacy of the CRTH2 antagonist OC000459 in eosinophilic esophagitis. Allergy. 2013;68(3):375–85. doi: 10.1111/all.12096. [DOI] [PubMed] [Google Scholar]
- 115.Rothenberg ME, Wen T, Greenberg A, Alpan O, Enav B, Hirano I, et al. Intravenous anti-IL-13 mAb QAX576 for the treatment of eosinophilic esophagitis. J Allergy Clin Immunol. 2015;135(2):500–7. doi: 10.1016/j.jaci.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 116.Sastre J, Davila I. Dupilumab: A New Paradigm for the Treatment of Allergic Diseases. J Investig Allergol Clin Immunol. 2018;28(3):139–50. doi: 10.18176/jiaci.0254. [DOI] [PubMed] [Google Scholar]
- 117.Rosenberg CE, Mingler MK, Caldwell JM, Collins MH, Fulkerson PC, Morris DW, et al. Esophageal IgG4 levels correlate with histopathologic and transcriptomic features in eosinophilic esophagitis. Allergy. 2018;73(9):1892–901. doi: 10.1111/all.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chehade M, Sampson HA, Morotti RA, Magid MS. Esophageal subepithelial fibrosis in children with eosinophilic esophagitis. J Pediatr Gastroenterol Nutr. 2007;45(3):319–28. doi: 10.1097/MPG.0b013e31806ab384. [DOI] [PubMed] [Google Scholar]
- 119.Aceves SS, Newbury RO, Dohil R, Bastian JF, Broide DH. Esophageal remodeling in pediatric eosinophilic esophagitis. J Allergy Clin Immunol. 2007;119(1):206–12. doi: 10.1016/j.jaci.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 120.Schoepfer AM, Simko A, Bussmann C, Safroneeva E, Zwahlen M, Greuter T, et al. Eosinophilic Esophagitis: Relationship of Subepithelial Eosinophilic Inflammation With Epithelial Histology, Endoscopy, Blood Eosinophils, and Symptoms. Am J Gastroenterol. 2018;113(3):348–57. doi: 10.1038/ajg.2017.493. [DOI] [PubMed] [Google Scholar]
- 121.Aceves SS, Newbury RO, Chen D, Mueller J, Dohil R, Hoffman H, et al. Resolution of remodeling in eosinophilic esophagitis correlates with epithelial response to topical corticosteroids. Allergy. 2010;65(1):109–16. doi: 10.1111/j.1398-9995.2009.02142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Menard-Katcher C, Benitez AJ, Pan Z, Ahmed FN, Wilkins BJ, Capocelli KE, et al. Influence of Age and Eosinophilic Esophagitis on Esophageal Distensibility in a Pediatric Cohort. Am J Gastroenterol. 2017;112(9):1466–73. doi: 10.1038/ajg.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lieberman JA, Morotti RA, Konstantinou GN, Yershov O, Chehade M. Dietary therapy can reverse esophageal subepithelial fibrosis in patients with eosinophilic esophagitis: a historical cohort. Allergy. 2012;67(10):1299–307. doi: 10.1111/j.1398-9995.2012.02881.x. [DOI] [PubMed] [Google Scholar]
- 124.Siddique AS, Corney DC, Mangray S, Lombardo KA, Chen S, Marwaha AS, et al. Clinicopathologic and gene expression analysis of initial biopsies from patients with eosinophilic esophagitis refractory to therapy. Hum Pathol. 2017;68:79–86. doi: 10.1016/j.humpath.2017.08.027. [DOI] [PubMed] [Google Scholar]
- 125.C. CsHMC. Testing Effectiveness of Losartan in Patients With EoE With or Without a CTD. In: C CsHMC, editor.: ClinicalTrials.gov [Internet].; 2018.
- 126.Nataatmadja M, West J, Prabowo S, West M. Angiotensin II Receptor Antagonism Reduces Transforming Growth Factor Beta and Smad Signaling in Thoracic Aortic Aneurysm. Ochsner J. 2013;13(1):42–8. [PMC free article] [PubMed] [Google Scholar]
- 127.Katzka DA, Geno DM, Ravi A, Smyrk TC, Lao-Sirieix P, Miremadi A, et al. Accuracy, safety, and tolerability of tissue collection by Cytosponge vs endoscopy for evaluation of eosinophilic esophagitis. Clin Gastroenterol Hepatol. 2015;13(1):77–83 e2. doi: 10.1016/j.cgh.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 128.Furuta GT, Kagalwalla AF, Lee JJ, Alumkal P, Maybruck BT, Fillon S, et al. The oesophageal string test: a novel, minimally invasive method measures mucosal inflammation in eosinophilic oesophagitis. Gut. 2013;62(10):1395–405. doi: 10.1136/gutjnl-2012-303171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McAleer MA, Pohler E, Smith FJ, Wilson NJ, Cole C, MacGowan S, et al. Severe dermatitis, multiple allergies, and metabolic wasting syndrome caused by a novel mutation in the N-terminal plakin domain of desmoplakin. J Allergy Clin Immunol. 2015;136(5):1268–76. doi: 10.1016/j.jaci.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Foroughi S, Foster B, Kim N, Bernardino LB, Scott LM, Hamilton RG, et al. Anti-IgE treatment of eosinophil-associated gastrointestinal disorders. J Allergy Clin Immunol. 2007;120(3):594–601. doi: 10.1016/j.jaci.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Loizou D, Enav B, Komlodi-Pasztor E, Hider P, Kim-Chang J, Noonan L, et al. A pilot study of omalizumab in eosinophilic esophagitis. PLoS One. 2015;10(3):e0113483. doi: 10.1371/journal.pone.0113483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Assa’ad AH, Gupta SK, Collins MH, Thomson M, Heath AT, Smith DA, et al. An antibody against IL-5 reduces numbers of esophageal intraepithelial eosinophils in children with eosinophilic esophagitis. Gastroenterology. 2011;141(5):1593–604. doi: 10.1053/j.gastro.2011.07.044. [DOI] [PubMed] [Google Scholar]
- 133.Straumann A, Bussmann C, Conus S, Beglinger C, Simon HU. Anti-TNF-alpha (infliximab) therapy for severe adult eosinophilic esophagitis. J Allergy Clin Immunol. 2008;122(2):425–7. doi: 10.1016/j.jaci.2008.06.012. [DOI] [PubMed] [Google Scholar]


