Abstract
In addition to a conventional relaxed state, a fraction of myosins in the cardiac muscle exists in a low-energy consuming super-relaxed (SRX) state, which is kept as a reserve pool that may be engaged under sustained increased cardiac demand. The conventional relaxed and the super-relaxed states are widely assumed to correspond to a structure where myosin heads are in an open configuration, free to interact with actin, and a closed configuration, inhibiting binding to actin, respectively. Disruption of the myosin SRX population is an emerging model in different heart diseases, such as hypertrophic cardiomyopathy, which results in excessive muscle contraction, and stabilizing them using myosin inhibitors is budding as an attractive therapeutic strategy. Here we examined the structure-function relationships of two myosin ATPase inhibitors, mavacamten and para-nitroblebbistatin, and found that binding of mavacamten at a site different than para-nitroblebbistatin populates myosin into the SRX state. Para-nitroblebbistatin, binding to a distal pocket to the myosin lever arm near the nucleotide-binding site, does not affect the usual myosin SRX state but instead appears to render myosin into a new, perhaps much more inhibited, ‘ultra-relaxed’ state. X-ray scattering-based rigid body modeling of human β-cardiac heavy meromyosin shows that both mavacamten and para-nitroblebbistatin induce novel conformations that diverge significantly from the hypothetical open and closed states, and furthermore, mavacamten treatment causes greater compaction than para-nitroblebbistatin. Taken together, we conclude that mavacamten and para-nitroblebbistatin stabilize myosin in different structural states, and such states may give rise to different functional energy-sparing states.
Keywords: Super-Relaxed State (SRX), Interacting Heads Motif (IHM), Small Angle-X-ray Scattering (SAXS), mavacamten, blebbistatin, human β-cardiac myosin, Synthetic thick filaments (STF)
INTRODUCTION
Myosin thick filaments constitute one of the central components in the sarcomeres of striated muscles. Under activating conditions, the binding of Ca2+ to troponin tightly regulates myosin interaction with actin on the thin filaments, which leads to sarcomere contraction (reviewed in [1,2]). However, the discovery that in relaxed muscle, myosin can exist in two distinct structural states—a folded-back closed state termed as the interacting heads-motif (IHM) [3–10] and a more conventional open state (reviewed in [11–16])—emphasizes another mode of regulation within thick filaments that determines the number of myosin heads available for interaction with actin. In the folded-back closed state, myosin heads are closely associated with the thick filament shaft, where one of the heads of a single myosin dimer, known as the ‘blocked head,’ has its actin-binding domain sequestered into the folded molecule, while the other head is ‘free’ (reviewed in [11–16]). Based on many different low-resolution electron microscopy structures of thick filaments and soluble myosins, it has been hypothesized that structurally, the ‘blocked head’ interacts with its subfragment-2 as well as the converter domain of the ‘free head’ to give rise to the IHM (reviewed in [11–16]). On the other hand, myosins in the conventional open state, which remain in equilibrium with the closed state, project out from the thick filament and are ready to interact with actin under activating conditions of the muscle. Recently, in two independent studies using cryo-electron microscopy, a 4–8 Å resolution IHM structure of smooth muscle myosin has been determined [17,18], but this is not yet available for cardiac myosin.
As these fascinating structural discoveries were being made, parallel investigations discovered that, biochemically, myosin heads in relaxed thick filaments exist in an equilibrium between two distinct energy-sparing states (reviewed in [19–22]): (1) the disordered relaxed state (DRX), in which myosin has an average ATP turnover time of <60 s; and (2) the super-relaxed (SRX) state, in which myosin has a prolonged ATP turnover time of >100 s. In conjunction with the structural results, these biochemical discoveries led to a hypothesis within the central protein structure-function dogma that the structurally closed and open states of myosin are responsible for the low-energy consuming SRX and the high-energy consuming DRX states, respectively.
Since these discoveries, altering the distribution of the myosin population between closed and open states using a small-molecule approach has emerged as an attractive therapeutic strategy to correct the aberrant cardiac function in certain cardiomyopathies because many hypertrophic cardiomyopathy-causing mutations in myosin have been shown to destabilize the IHM state of myosin, thereby increasing the number of myosin heads in the open state that results in hypercontractility [19–25]. A newly discovered small molecule cardiac myosin inhibitor, mavacamten [26], which recently concluded phase-III clinical trials to treat obstructive hypertrophic cardiomyopathy, effectively reduces the strength of muscle contraction. The ability of mavacamten to counter enhanced contractility has also been investigated in vitro using hypertrophic cardiomyopathy-causing mutations in myosin, myosin-binding protein C, troponin T, and troponin I [20,22,26,27]. Many elegant mechanistic studies showed that, functionally, mavacamten populates myosin in the low-energy consuming SRX state [20,28,29]. Using electron microscopy and muscle fiber X-ray diffraction studies, it has also been shown that mavacamten structurally stabilizes a closed state reminiscent of the IHM [20], suggesting a link between the structurally closed state and the SRX functionality. Whether this structure-function relationship holds, in general, is an unanswered question.
To address this question and interrogate the mechanism of mavacamten’s action on cardiac myosin, we compared its activity with para-nitroblebbistatin, a non-phototoxic and photostable analog of blebbistatin that belongs to a different class of myosin inhibitors. Like mavacamten, the blebbistatin family of molecules binds and inhibits phosphate release from myosin, leading to inhibition of myosin ATPase function [30–32]. In the presence of ADP, blebbistatin is also known to prime the myosin lever arm to the pre-stroke orientation [33]. Structural studies using low-resolution electron microscopy and X-ray diffraction in skeletal systems [34–38] and fluorescent polarization spectroscopy in the cardiac system [39] have shown that, like mavacamten, blebbistatin can promote a folded-back myosin structure mimicking a closed or IHM-like state. However, single ATP turnover kinetics assessed using a single high concentration of blebbistatin in rabbit fast and slow skeletal muscle fibers showed that, unlike mavacamten, blebbistatin does not populate more myosins in the SRX state [40]. Instead, it slows the ATP turnover rate of myosins already in the SRX state, leading to a super-high relaxed state with a 10-fold lower ATPase turnover rate than those in the conventional SRX state [40]. Also, a recent study demonstrated that para-nitroblebbistatin is more potent in inhibiting the myosin ATPase of skeletal myosin than human β-cardiac myosin, suggesting some key differences in the way blebbistatin affects different myosin isoforms [41]. Altogether, these pieces of evidence suggest that although blebbistatin- and mavacamten-treated myosins may be enzymatically inhibited, the mechanism of inhibition may have different functional outcomes, thereby challenging the link between the closed structural state and the functional SRX state. Moreover, in the absence of high-resolution atomic structures, it remains unresolved whether the closed structural state of myosin stabilized by blebbistatin differs from mavacamten and whether this could lead to distinct SRX functional outcomes.
In this study, using biochemical and structural approaches, we report that mavacamten and para-nitroblebbistatin binding to different pockets of myosin leads to distinct functional outcomes. Importantly, we show that these functional differences arise from different structural states that are preferentially stabilized by these two molecules. We present arguments suggesting that these two small-molecule myosin inhibitors act via different mechanisms distinguishable by dissimilar structural and functional transitions.
RESULTS
Mavacamten and Para-nitroblebbistatin Effectively Inhibit Myosin ATPase Activity.
We first evaluated the concentration-dependent effects of mavacamten and para-nitroblebbistatin on the basal, actin-activated, as well as the myofibrillar ATPase activity of myosin (Fig. 1). Consistent with their inhibitory action, both these molecules showed potent inhibition of the basal ATPase activity of myosin in bovine cardiac synthetic thick filaments (BcSTF) (Fig. 1A), with mavacamten being more potent (P<0.001; IC50 of 0.63±0.05 μM) than para-nitroblebbistatin (2.37±0.12 μM). A similar pattern of inhibition was also observed in the actin-activated (Fig. 1B and Table 1) and bovine cardiac myofibrillar (BcMF) ATPase activity at pCa 6 (Fig. 1C and Table 1). IC50 values reported for mavacamten and para-nitroblebbistatin in this study are consistent with those reported in the literature [26,28,29,31,32,42].
Figure 1. Mavacamten and Para-nitroblebbistatin inhibit Basal, Actin-Activated, and Myofibrillar Myosin ATPase activities.
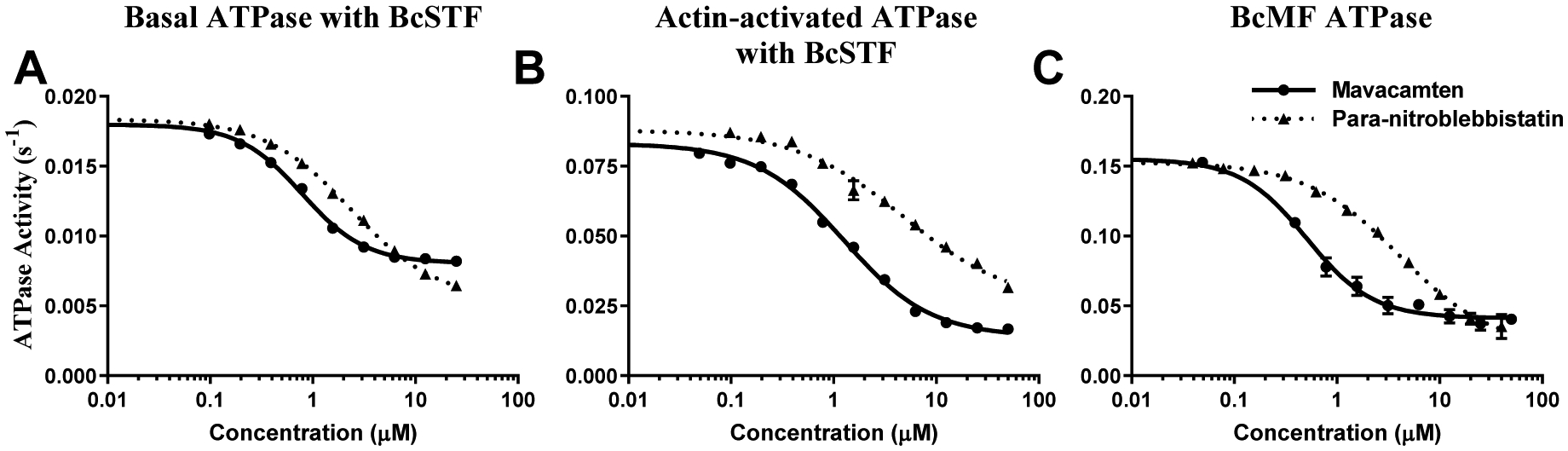
(A) Basal, (B) Actin-activated, and (C) Myofibrillar ATPase activity. All values are presented in the absolute scale (s−1). BcSTF refers to bovine cardiac synthetic thick filaments, and BcMF refers to bovine cardiac myofibrils. Circles and triangles correspond to mavacamten and para-nitroblebbistatin, respectively. Concentrations of these molecules required to attain half-maximal inhibition (IC50) in the ATPase activity are listed in Table 1. Data are expressed as mean ± SEM (n=8 from two experiments that used single preparation of the same reagent purification).
Table 1.
Parameters derived from ATPase and single ATP turnover studies
| IC50/ EC50 (in μM) derived using data acquired in various bovine cardiac systems from ATPase and Single ATP turnover experiments | |||
|---|---|---|---|
| Para-nitroblebbistatin | Mavacamten | ||
| 0μM blebbistatin |
10μM blebbistatin |
||
| Basal ATPase (BcSTF) | 2.37±0.12* | 0.63±0.05 | |
| Actin-activated ATPase (BcSTF) | 4.81±0.16* | 1.25±0.11 | |
| Myofibrillar ATPase (BcMF) | 4.05±0.34* | 0.50±0.05 | |
| SRX Amplitude, Aslow (BcSTF) | >12.5* | 1.21±0.35 | 0.63±0.07* |
| DRX ATPase Rate, kfast (BcSTF) | >12.5* | 0.43±0.03 | 0.64±0.12 |
| SRX ATPase Rate, kslow (BcSTF) | >12.5 | >12.5 | >12.5 |
| SRX Amplitude, Aslow (BcMF) | >12.5* | 4.30±0.31 | |
| DRX ATPase Rate, kfast (BcMF) | >12.5* | 0.59±0.08 | |
| SRX ATPase Rate, kslow (BcMF) | >12.5 | >12.5 | |
Nomenclature is as follows: BcSTF refers to bovine β-cardiac synthetic thick filaments; BcMF, bovine cardiac myofibrils. Data are expressed as mean±SEM (n≥8 from two experiments that used single preparation of the same purification). The IC50/EC50 values of both blebbistatin and para-nitroblebbistain are statistically different from mavacamten based on a student’s t-test (*p<0.001). The Hill coefficient measured in these experiments is always between 2–4.
Binding of Mavacamten to a Different Site on Myosin than Para-nitroblebbistatin, Populates Myosin in the SRX State.
To understand if the ATPase inhibition of mavacamten and para-nitroblebbistatin arises from increasing the myosin population in the SRX state, we next studied the single ATP myosin turnover kinetics in reconstituted BcSTF in response to increasing concentrations of these molecules. Representative mant-ATP fluorescence decay profiles after chasing with non-fluorescent ATP in these experiments are shown in Fig. 2. A qualitative comparison showed that the addition of mavacamten (Fig. 2A) led to a concentration-dependent rightward shift in the fluorescence decay profile, suggesting a slower release of mant-nucleotides from myosin. Occasionally, but not consistently, fluorescence decay profiles in myosin samples at high concentrations (12.5 μM) of mavacamten presented a lag phase (up to 400 s) for reasons not well understood (compare blue trace with other traces in Fig. S1C in the Supporting Information). In contrast to the effect of mavacamten, para-nitroblebbistatin (Fig. 2B) did not show any noticeable change in the fluorescence decay up to a time point of ~ 200 sec. However, the baseline fluorescence gradually increased with the increasing concentration of para-nitroblebbistatin (compare grey shaded region in Fig. 2A with Fig. 2B). To quantitatively analyze the overall fluorescence decay kinetics, we assessed the area under these curves, which measures the total amount of the mant-ATP that remains bound to myosin and serves as a proxy for the total energy unused by the system. A comparison of the total area under these fluorescence decay curves shows a concentration-dependent increase of undissociated mant-nucleotides from myosin in the presence of mavacamten but not with para-nitroblebbistatin (Fig. 2C). However, the baseline fluorescence increase in response to para-nitroblebbistatin treatment suggests a considerable number of myosins with a prolonged nucleotide-bound state whose lifetime extends beyond the duration of the experiment (40 min) (Fig. 2B).
Figure 2. Mavacamten, but not Para-nitroblebbistatin, Slows the Release of Mant-Nucleotides from Myosin in a Concentration-dependent Manner.
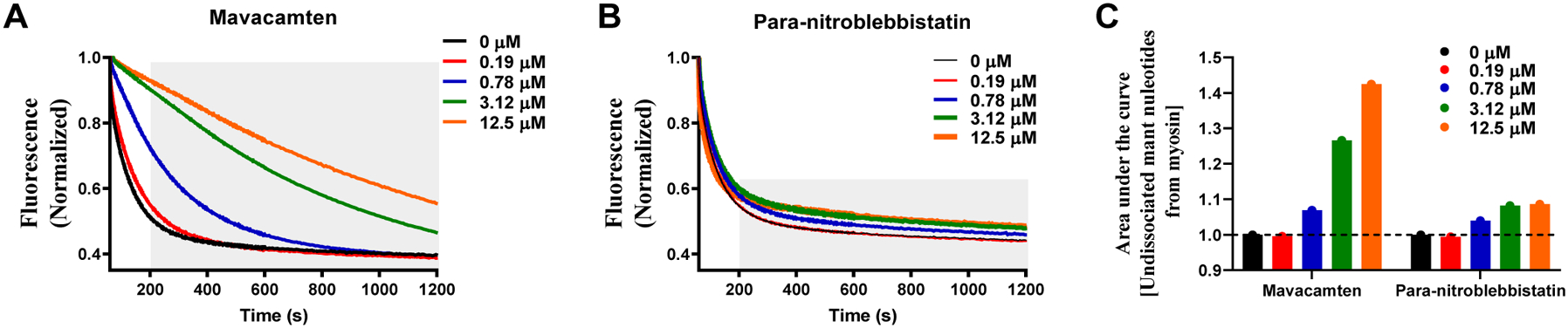
Representative single turnover fluorescence decay profiles for (A) Mavacamten and (B) Para-nitroblebbistatin. The grey shaded region in panels A & B highlights the differences in fluorescence decay profiles from 200 s onwards. The fluorescence decay profiles were acquired for 40 min but only the data corresponding to the first 20 minutes was shown in panels A & B to highlight important differences. (C) The area under the curve, which qualitatively describes the undissociated mant-nucleotides from myosin in a single turnover, is plotted for the traces shown in panels A & B. Only mavacamten, but not para-nitroblebbistatin, shows a concentration-dependent increase in the undissociated mant nucleotide.
The dose-dependent profiles of % amplitude of the slow (SRX) phase, Aslow, are derived by fitting a two-state exponential model to the fluorescence decay profiles (see Methods and as described in [43]), and these are shown in Fig. 3A. The SRX population increased cooperatively (with Hill coefficient ranging from 2 to 4) with increasing mavacamten concentration, reaching 100% at 3.125 μM, suggesting that mavacamten shifted the myosin DRX-SRX equilibrium more towards the SRX state, consistent with observations made in previous studies [20,28,29]. The concentration of mavacamten required to attain a half-maximal increase in Aslow was 1.21±0.35 μM, close to the IC50 of ATPase data measured in various systems (Table 1). In contrast, para-nitroblebbistatin did not affect Aslow up to a concentration of 12.5μM, indicating that it does not affect the normal myosin DRX-SRX equilibrium. Qualitatively similar observations were also made using skinned bovine cardiac myofibrils (Fig. 3D), verifying that the functional outcomes of these small molecules are not different between a reconstituted myosin bipolar thick filament system and a more intact myofibril system. However, the absolute magnitude of the SRX population in untreated BcMF (~ 35%) was higher than that in BcSTF (~15%). This may be due to the presence of other partner proteins, such as myosin binding protein-C, that stabilize nearly 60% of myosins in the SRX state compared to the rest of the thick filament that contributes to only ~10% [44]. More interestingly, at saturating concentrations of mavacamten, the magnitude of myosin SRX population was smaller in BcMF (~65%) than in BcSTF (~100%) (compare Fig. 2A and 2D), suggesting that some physiological constraints such as phosphorylation of other sarcomeric proteins may not fully favor the stabilization of myosins into the SRX state. This remains a topic of further investigation.
Figure 3. Mavacamten, but not Para-nitroblebbistatin, Populates Myosin in the SRX State.
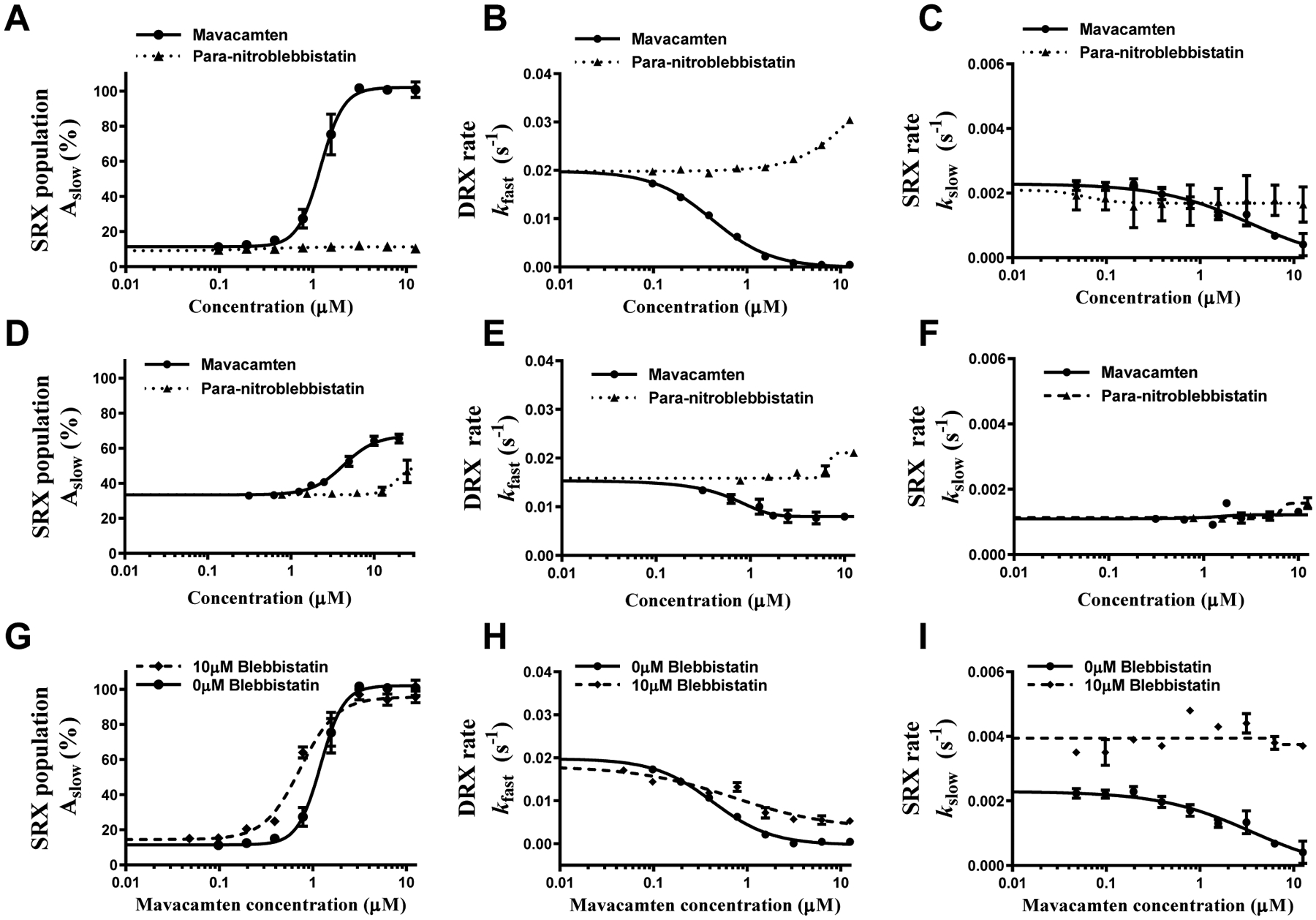
(A, B, and C) Show comparison of responses between mavacamten and para-nitroblebbistatin in BcSTF for SRX population (Aslow), DRX rate (kfast), and SRX rate (kslow), respectively, while (D, E, and F) show the respective comparisons in BcMF. (G, H, and I) Show a comparison of responses in BcSTF in the presence and absence of 10 μM blebbistatin for Aslow, kfast, and kslow, respectively. BcSTF refers to bovine cardiac synthetic thick filaments, and BcMF refers to bovine cardiac myofibrils. The data were presented on the absolute scale (% for amplitude and s−1 for rates). Concentrations of mavacamten required to attain half-maximal change (AC50/IC50) in parameters are listed in Table 1. Data are expressed as mean±SEM (n≥8 from two experiments that used single preparation of the same reagent purification).
In addition to the myosin SRX population, we also analyzed the ATP turnover rates of the myosin population in both the DRX state (kfast) and the SRX state (kslow), both of which were similar in BcSTF and BcMF. kfast in the untreated (DMSO) myosin systems was in the range of 0.015–0.02 s−1, while kslow was roughly 10-fold slower (0.0012–0.002 s−1). In both BcSTF (Fig. 3B) and BcMF (Fig. 3E), mavacamten, but not para-nitroblebbistatin, decreased kfast in a concentration-dependent manner approaching a value of kslow at 3.125 μM, suggesting that mavacamten gradually sequesters more myosin from the fast DRX state (with a rate of ~0.02 s−1) to the slow SRX state (with a rate of ~0.002 s−1) without affecting kslow up to this point (Fig. 3C and Fig. 3F). The concentration of mavacamten required to attain a half-maximal decrease (IC50) in kfast was 0.43±0.03 μM, which is in good agreement with the IC50 values of basal, actin-activated, and myofibrillar ATPase activity (Table 1). Interestingly, an additional increase in the mavacamten concentration to 12.5 μM further slowed the fluorescence decay profile (compare orange trace relative to the green trace in Fig. 2A), which a single exponential function could well describe, but with an observed rate that is lower than that of the myosin heads in the conventional SRX state (kslow in Fig. 3C). This suggested that additional myosin conformational changes occur at high concentrations of mavacamten, an effect that was not seen in the myofibril system (kslow in Fig. 3F). Unraveling this mystery would require simultaneous structural and enzymatic determination of the SRX population in thick filaments, which is beyond the scope of this study.
In another experiment, to determine whether the ability of mavacamten to promote myosin heads into the SRX state is affected by the presence of blebbistatin, we performed single turnover experiments by pre-incubating BcSTF with a near-saturating concentration (10 μM) of blebbistatin and then titrating it with increasing concentrations of mavacamten. Because the competition experiments can be better performed using two molecules of similar potency, we used blebbistatin instead of para-nitro blebbistatin in these experiments because it has nearly the same potency as mavacamten [29,31,32,42]. Even in the presence of 10 μM blebbistatin, mavacamten effectively increased the myosin population in the SRX state in a concentration-dependent manner. The concentration of mavacamten required to attain a half-maximal increase in Aslow was 0.63±0.06 μM (Fig. 3G), while that required for the half-maximal decrease in kfast was 0.64±0.12 μM (Fig. 3H). These results compare very well with those measured in the absence of blebbistatin (1.21±0.35 and 0.43±0.03 μM, respectively). Similar to the observation made in the blebbistatin-free system (Fig. 3C), increasing concentration of mavacamten showed no discernible effect on kslow in the presence of 10 μM blebbistatin (Fig. 3I). In a separate experiment, we also confirmed that the binding of blebbistatin is not compromised in the presence of mavacamten (Fig. S2 in the Supporting Information), suggesting that these two molecules do not compete for the same site and that the presence of blebbistatin does not affect the ability of mavacamten to populate more myosins in the SRX state.
Mavacamten Stabilizes a More Compact Structure of Myosin Than Does Para-nitroblebbistatin
To evaluate the effect of mavacamten and para-nitroblebbistatin on myosin structure, we used Small-Angle X-ray Scattering (SAXS) (Fig. S3 in the Supporting Information) combined with Multi-Angle Light Scattering (MALS) and Dynamic Light Scattering (DLS) with purified human β-cardiac 25-hep heavy meromyosin (HMM) subfragment of myosin. The 25-hep HMM domain of the myosin used in this study consists of two S1 domains and 25 heptads (25 repeated patterns of seven amino acids) of the S2 domain. Molecular weights estimated by MALS ensured the mono-dispersity of the sample. They were estimated to be 365±18, 363±14, and 366±18 kDa for DMSO-treated, para-nitroblebbistatin-treated, and mavacamten-treated 25-hep HMM, respectively (Table 2), which compares well to the theoretical molecular weight of ~380 kDa. The hydrodynamic radius estimated by DLS for the 25-hep HMM was 84±2 Å, with no substantial changes observed upon treatment with either mavacamten or para-nitroblebbistatin (Table 2), suggesting that the apparent size adopted by the solvated, tumbling HMM does not change with either treatment. These studies were done with either 20 μM mavacamten or 50 μM para-nitroblebbistatin to ensure that we study their structural properties at complete biochemical inhibition levels (Fig. 1).
Table 2.
Parameters derived from SAXS studies
| Light scattering structural parameters for human β-cardiac 25-hep HMM (at 20 μM mavacamten and 50 μM para-nitroblebbistatin) | |||
|---|---|---|---|
| DMSO | Para-nitroblebbistatin | Mavacamten | |
| Molecular Mass (kDa) | 365±18 | 363±14 | 366±18 |
| Hydrodynamic radius (Å) | 84.1±2.1 | 84.1±2.2 | 84.2±2.2 |
| SAXS structural parameters of human β-cardiac 25-hep HMM based on Guinier Analysis (at 20 μM mavacamten and 50 μM para-nitroblebbistatin) | |||
| DMSO | Para-nitroblebbistatin | Mavacamten | |
| I(0) (cm−1) | 0.093±0.003 | 0.04±0.01 | 0.055±0.015 |
| Rg (Å) | 119±1 | 102.5±1.5 | 102±5 |
| q range (Å−1) | 0.0048–0.0110 | 0.0048–0.0122 | 0.0059–0.0122 |
| SAXS structural parameters of human β-cardiac 25-hep HMM based on P(r) Analysis (at 20 μM mavacamten and 50 μM para-nitroblebbistatin) | |||
| DMSO | Para-nitroblebbistatin | Mavacamten | |
| I(0) (cm−1) | 0.09±0 | 0.04±0 | 0.06±0.02 |
| Rg (Å) | 139±4 | 124±10 | 114±8 |
| Dmax(Å) | 541±36 | 486±56 | 412±32 |
| Porod volume (Å−3) | 1.22e6 | 1.42e6 | 1.49e6 |
| q range (Å−1) | 0.0043–0.3522 | 0.0043–0.3522 | 0.0043–0.3522 |
25-hep HMM refers to heavy meromyosin containing the subfragment S1 and 25 heptad repeats of the S2-subfragment; I(0), zero-angle scattering intensity; Rg, radius of gyration; q, , 2θ is the scattering angle, and λ is the wavelength of the radiation; Dmax, maximum dimension of the molecule; Porod volume, particle volume calculated as = 2π2I(0)/Qi, where Qi is the total scattered intensity [71]. Data reported are the mean±SEM from two experiments using two different protein preparations.
For isotropic solution scattering, scattering data reduction refers to the process of converting counts on the SAXS detector to the one-dimensional scattered intensity profile arising from the sample, with associated errors, as I(q) vs. q (where ; 2θ is the scattering angle, and λ is the radiation wavelength [45]). Guinier analysis (ln I(q) vs. q2) of the I(q) vs. q curves showed a significantly reduced radius of gyration (Rg) for both mavacamten-treated HMM (14%; P<0.01) and para-nitroblebbistatin-treated HMM (14%; P<0.05) when compared to untreated 25-hep HMM (Fig. S4 in the Supporting Information and Table 2). The Rg values for the untreated, para-nitroblebbistatin-treated, and mavacamten-treated myosins were 119±1 Å, 102±1.5 Å, and 102±5 Å, respectively. The dimensionless Kratky plot ((qRg)2I(q)/I(0) versus qRg) is commonly used for qualitative assessment of flexibility and compactness of macromolecules. The relative downward shift of the Kratky plot for para-nitroblebbistatin-treated and mavacamten-treated HMM compared to DMSO-treated HMM indicated that the HMM molecule becomes more compact in the presence of either para-nitroblebbistatin or mavacamten, as compared to the untreated HMM (Fig. 4A).
Figure 4. Mavacamten Stabilizes a More Compact Structure of Myosin as Compared to Para-niotroblebbistatin.
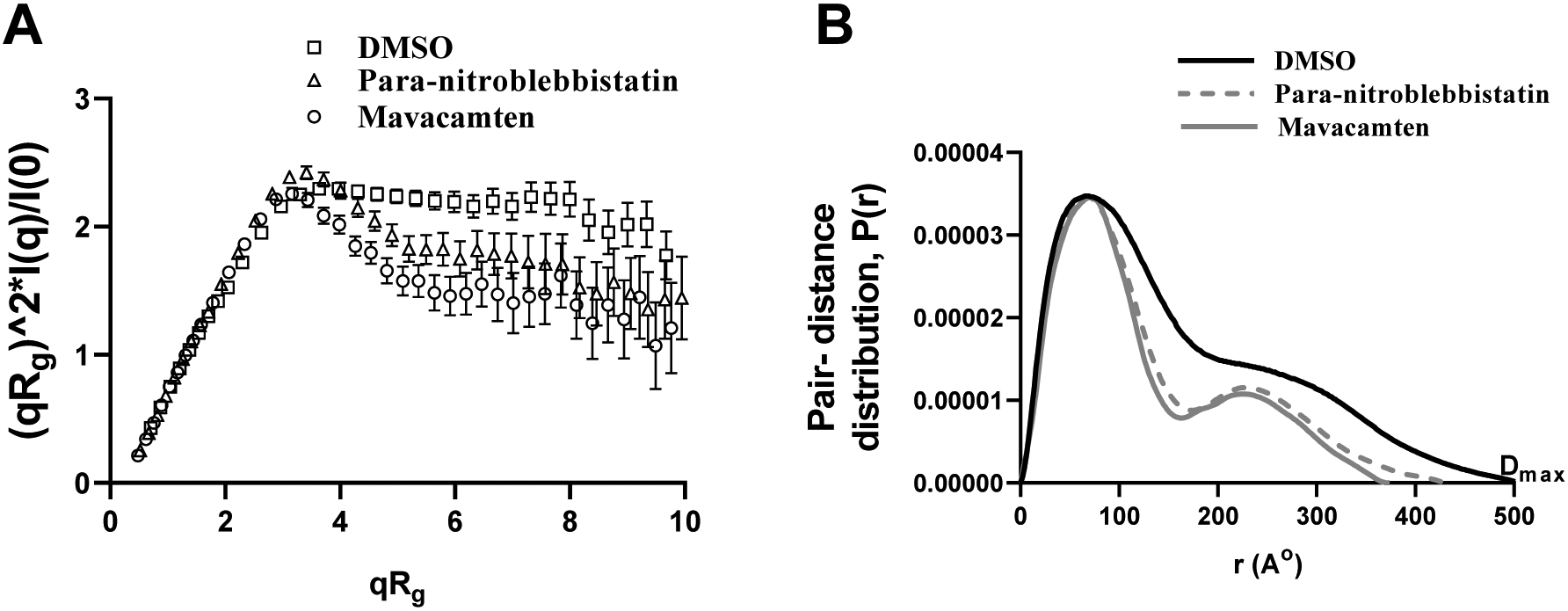
(A) Comparison of dimensionless Kratky plots from 25-hep HMM treated with DMSO, para-nitroblebbistatin, and mavacamten. Kratky plots are commonly used for qualitative assessment of flexibility and compactness of biological molecules and are independent of the molecular size. The downward shift of the Kratky plot indicates that 25-hep HMM becomes less flexible, probably more compacted, in the presence of either mavacamten or para-nitroblebbistatin compared to control (DMSO). (B) Comparison of the pair-distance distribution function, P(r), of 25-hep HMM treated with DMSO, para-nitroblebbistatin, and mavacamten. P(r) functions of 25-hep HMM indicate an elongated structure with two globular domains consistent with the classic HMM structure. HMM treated with mavacamten and para-nitroblebbistatin elicited a decrease in the maximum dimension (Dmax) by different amounts than the untreated (DMSO) control. Dmax is the maximum dimension of the molecule. At least three separate experiments were performed on different batches of samples for each treatment to confirm the reproducibility of the trend. Presented here are single representatives, which also produced the best fits during rigid body modeling shown in Fig. 5.
The pair-distance distribution function, P(r), representing the frequency histogram of all the interatomic vector lengths within the sample particle, can provide structural information concerning the underlying electron density distribution in the molecules. The P(r) of para-nitroblebbistatin-treated and mavacamten-treated 25-hep HMM showed similar features as the DMSO-treated HMM control (Fig. 4B). However, the maximum dimension (Dmax) of 25-hep HMM was significantly lower in mavacamten-treated HMM (24%; P<0.001) but not in para-nitroblebbistatin-treated HMM (11%; P=0.07). The Dmax values for the untreated, para-nitroblebbistatin-treated, and mavacamten-treated HMM were 541±36 Å, 486±56 Å, 412±32 Å, respectively (Table 2). Similarly, when compared to the data in the untreated sample, Rg values of 25-hep HMM from the P(r) analysis were significantly lower in mavacamten-treated samples (18%; P<0.01) but not in the para-nitroblebbistatin-treated sample (11%; P=0.07). The Rg values of untreated, para-nitroblebbistatin-treated, and mavacamten-treated 25-hep HMM were 139±4 Å, 124±10 Å, and 114±8 Å, respectively (Table 2). These results collectively demonstrate that mavacamten induces greater compaction of myosin than does para-nitroblebbistatin, suggesting that these two molecules affect the structure of myosin differently.
Mavacamten Populates a More Compact Closed State of Myosin than Para-nitroblebbistatin.
To reconcile the SAXS data with known structural conformations, we attempted to fit the two starting human β-cardiac myosin homology models representing the hypothetical open and closed states as described in Nag et al. [46] (MS03, open-source material downloaded from http://spudlab.stanford.edu/homology-models) against SAXS data obtained from DMSO, para-nitroblebbistatin, and mavacamten-treated HMM using CRYSOL [47]. Open and closed head conformations were modified by extending the tail and adding Green Fluorescent Protein (GFP, PDB-ID 4KW9) to the tail to match the 25-hep HMM construct used for the SAXS studies (Fig. 5A and B). SAXS curves representing untreated, para-nitroblebbistatin- and mavacamten-treated HMM fitted relatively well with the closed head conformation, when compared to the open head conformation (indicated by the χ2 values of 3.34, 3.17, and 2.61 respectively, as opposed to 20.56, 22.39, and 20.29, respectively; Fig 5A and B). Previous EM studies [20] and the failure to fit the open configuration suggest that DMSO-treated HMM, in solution, may be very flexible and represented by either multiple conformations or a single alternate conformation that differs from the conventional open state. While the HMM treated with para-nitroblebbistatin or mavacamten seemed to have an obvious preference for the closed head conformation, the differences in Rg and Dmax values from the Guinier and pair distance distribution analyses strongly indicated conformational differences, which suggested the need to optimize further the structural models resulting from small molecule treatment of HMM.
Figure 5. Rigid-body Modelling Using the SAXS data Suggests Conformational Heterogeneity and a Higher Propensity for Conformations Closer To the Closed Head Configuration in Mavacamten-treated HMM.
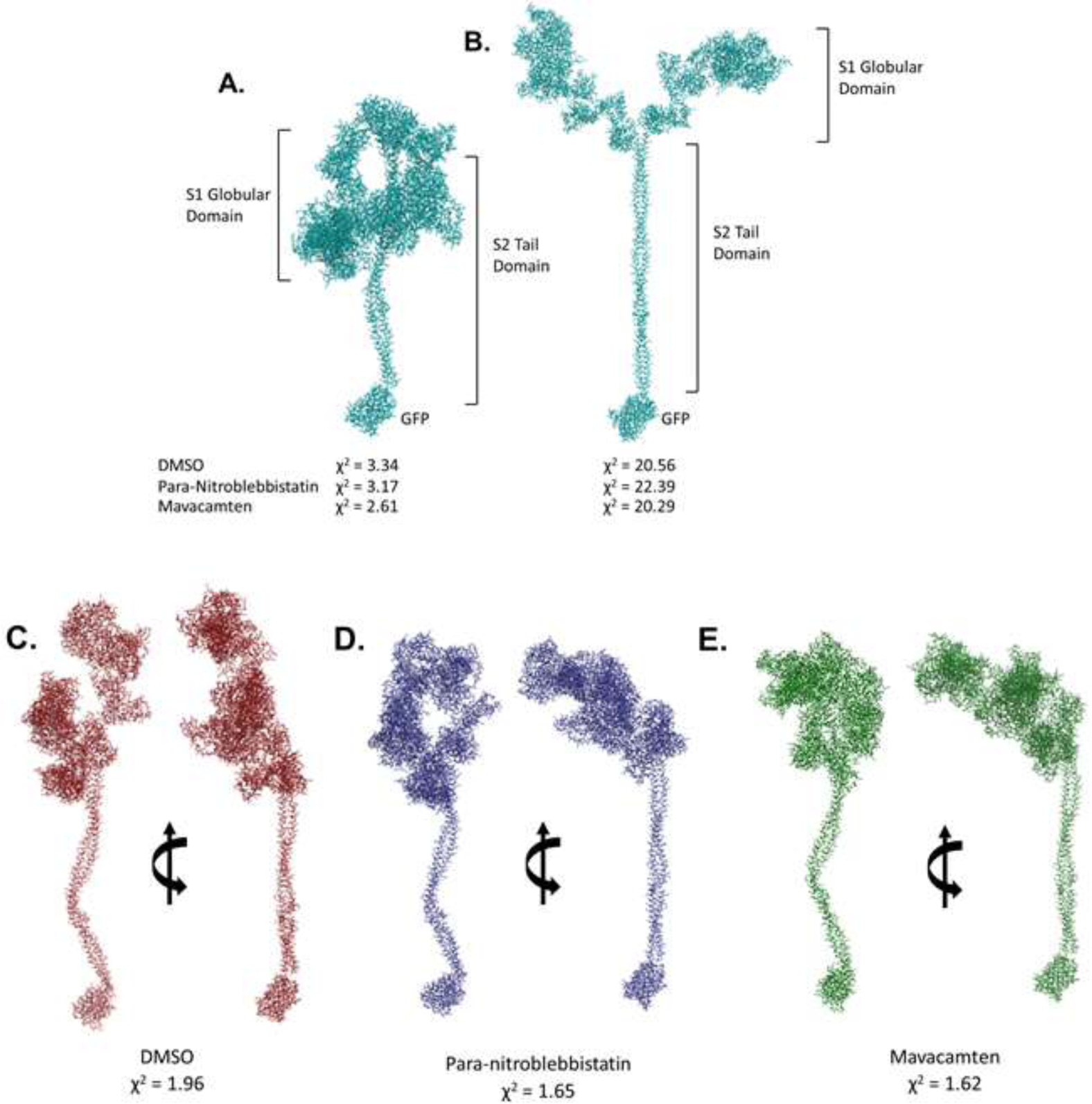
Closed head (A) open head (B) conformation with the 25-hep tail and the GFP used for fitting against SAXS data obtained on untreated (DMSO), para-nitroblebbistatin, and mavacamten treated HMM using CRYSOL [47]. χ2 values from CRYSOL indicated. Rigid body models obtained using SASREF [48] performed using open and closed head conformations, respectively, as starting models for HMM treated with (C) DMSO, (D) Para-nitroblebbistatin, and (E) Mavacamten. χ2 values from SASREF analysis are presented under the corresponding models.
We next used the rigid-body modeling software SASREF [48] to explore the possibility of a single conformation other than the hypothetical open and closed head starting models. We used SASREF to sample different relative orientations between the lever arm in the S1-subfragment and the proximal S2-subfragment domain in the hypothetical closed state because the initial CRYSOL fit was better for the two preparations compared to the open state. The S1–S2 disordered hinge is known to have flexibility in all myosins providing the S1 heads the needed rotational freedom to make productive interactions with actin (recently illustrated in the review [49]). Multiple SAXS experiments were performed using different preparations of HMM, and numerous iterations of SASREF were performed with the closed conformation as a starting model. DMSO-treated HMM converged to a more extended conformation than the theoretical open conformation, consistent with the higher Dmax and Rg estimates (Fig. 5C). Para-nitroblebbistatin-treated and mavacamten-treated HMM consistently reached a conformation best described as an intermediate between the open and closed states (Fig. 5D and E). While the P(r) analysis showed a greater reduction in Dmax in mavacamten-treated HMM than in para-nitroblebbistatin-treated HMM, SAXS-based rigid body modeling proved to be inadequate for distinguishing such structural features between the two. Based on these data, we posit that while para-nitroblebbistatin and mavacamten both induce conformations between the hypothetical open and closed states, the average myosin structure stabilized by mavacamten is more compact than by para-nitroblebbistatin. We also note that it is quite likely that the ensemble structure that myosin adapts in the presence of these agents may not be well-represented by any linear interpolation of the “canonical” structures. Notably, the open conformation is probably not a single configuration, but instead, myosin can undergo many structural reconfigurations as it advances through the chemo-mechanical cycle.
DISCUSSION
The use of small-molecule agents that target cardiac myosin and inhibit its ATPase activity is emerging as an attractive therapeutic strategy to normalize excessive contractility in hypertrophic cardiomyopathy. In this regard, although several clinical and non-clinical small molecule inhibitors have been identified and characterized, the underlying mechanism of their inhibitory actions remains unclear. This study aimed at filling some of this gap by investigating the structure-function relationships of cardiac myosin using two different classes of small-molecule inhibitors—mavacamten and blebbistatin. It has been known that mavacamten and blebbistatin are myosin inhibitors, although much of the data on the inhibitory action of blebbistatin comes from studies using skeletal muscle, smooth muscle, and non-muscle myosin systems, and a few in cardiac systems [30–32,41]. Here, we found that, in the presence of mavacamten and para-nitroblebbistatin, a non-phototoxic and photostable analog of blebbistatin, the inhibition of basal myosin ATPase activity was comparable to the inhibition observed in the cardiac actomyosin or myofibrillar system (Fig. 1 and Table 1), suggesting that these two molecules directly act upon myosin to inhibit its enzymatic activity and that this effect does not involve any other sarcomere proteins, including the regulatory components of the thin filaments. This leads to an immediate question “how do these small molecules inhibit cardiac myosin function?”
Analysis from single turnover experiments showed that mavacamten shifts the myosin equilibrium between DRX and SRX states more towards the SRX state, making them unavailable for interaction with actin, thereby suggesting a unified mechanism of both basal and actin-activated ATPase inhibition. This hypothesis is supported by the linear regression analysis, which demonstrates that the inhibition in either the basal or the actin-activated myosin ATPase activity by mavacamten linearly correlates to the increase in myosin SRX population (Fig. 6). This mechanism may also explain the observed reduction in cardiac contractility in pre-clinical and clinical models by mavacamten treatment [26,50–56]. In contrast, at inhibiting concentrations, para-nitroblebbistatin did not show any effect on the myosin DRX state (Fig. 3 and 6). This finding agrees with another study using rabbit slow skeletal muscle fibers that expresses the same myosin isoform as cardiac muscle in which the authors show that, at a saturating concentration of 40 μM, blebbistatin does not affect the early (fast) fluorescence decay phase in the single turnover experiments [40]. Instead, it considerably slows the late (slow) phase corresponding to the myosin activity in the SRX state, which can be explained by blebbistatin stabilizing a small but significant fraction of SRX heads into a third ‘ultra-relaxed’ state in which myosins have greater than 10-fold lower ATP turnover rate than those in the usual SRX state, thereby practically manifesting these myosins to have negligible ATPase activity (Fig. 1). Our data at a high para-nitroblebbistatin concentration of 50 μM (Fig. S5 in the Supporting Information; close to 40 μM used in the previous study) supports this possibility. For example, a progressive increase of mavacamten concentration resulted in slower fluorescence decay from 200 s to the end of the experimental period (2400 s), suggesting a slower release of mant-nucleotides (grey shaded region Fig. 2A). This contrasts with the treatment of para-nitroblebbistatin (grey shaded region Fig. 2B and Fig. S5 in the Supporting Information), which does not result in further fluorescence decay during this period, suggesting that the lifetime of mant-nucleotides for para-nitroblebbistatin-bound myosin is greater than the duration of the experiment (i.e., >2400 s), consistent with a previous study [44]. The notion that mavacamten and blebbistatin populate myosin in different relaxed states is further supported by our observation that mavacamten can populate myosin heads into the normal SRX state with similar effectiveness even in the presence of blebbistatin bound to myosin, suggesting that blebbistatin binding to myosin does not alter the usual DRX-SRX equilibrium of myosin. Although speculative at this moment, these observations suggest the possibility that, unlike mavacamten, binding of blebbistatin near the nucleotide-binding site of myosin [57], but far from the myosin lever arm, may make myosin unable to prime the lever arm more towards the pre-stroke direction in the ATP-bound state, which is proposed to be a pre-requisite for populating myosin in the normal SRX state [15,58].
Figure 6. Inhibition of Myosin ATPase Caused by Mavacamten, but not Para - nitroblebbistatin, Linearly Correlates with Increase in Myosin SRX Population.
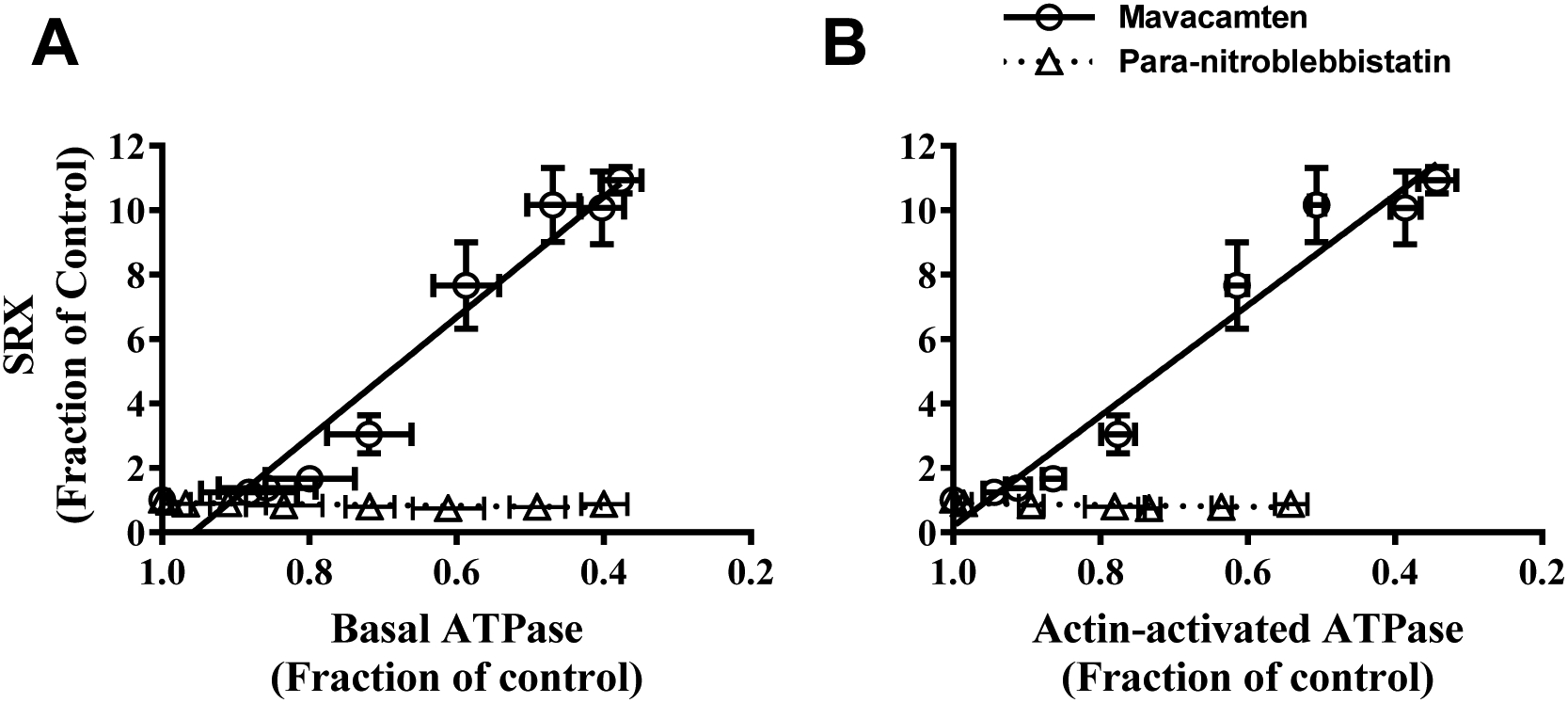
Correlation plot between myosin SRX population and (A) basal myosin ATPase activity and (B) actin-activated ATPase activity for mavacamten (circles) and para-nitroblebbistatin (triangles). Regression analysis showed a strong linear correlation between myosin population in SRX state and ATPase activity for mavacamten (R2≥0.94) but not for para-nitroblebbistatin-treated samples. All data were normalized by the DMSO-treated sample data and presented as mean±SEM (n≥8 from two experiments that used single preparation of the same reagent purification).
Although our results at low concentrations (up to 3.125 μM) of mavacamten provide simple insight into its inhibitory mechanism, this is not the case at higher concentrations. For example, while our data show that, in reconstituted myosin synthetic thick filaments, mavacamten stabilize 100% of myosin heads in the SRX state at 3.125 μM and beyond (Fig. 2A and Fig. 3A), the ATP turnover rate of these myosin heads progressively dips below (~5-fold lower) that of the conventional SRX state. We postulate that mavacamten at low concentration (up to 3.125 μM) cooperatively (the Hill coefficient is between 2–4; Table 1) populate myosin from the DRX state to the SRX state by priming the lever arm more towards the pre-stroke direction [15,20,58], while still leaving some heads unoccupied by the drug. At concentrations above 3.125 μM, the rest of the unbound myosin heads are occupied by mavacamten that could potentially prime the lever arm even more towards the pre-stroke direction, possibly stabilizing myosin in a third relaxed state, similar to that observed in tarantula skeletal muscles [59–62]. However, in the absence of atomic-level structural data, this description remains speculative and awaits the results of future structural studies.
Additional evidence that para-nitroblebbistatin structurally affects myosin differently from mavacamten comes from structural parameters derived using SAXS/MALS/DLS measurements. For example, while the Kratky plots suggested that both mavacamten and para-nitroblebbistatin qualitatively decrease the flexibility and increase the compactness of the 25-hep HMM, parameters derived from pair-distance distribution function (P(r)) quantified the differences among treatments. In the P(r) analysis, the DMSO-treated HMM showed a bell-shaped curve with a peak at low r values (0–170Å), a shoulder at intermediate r values (170–340Å), and an extended tail at high r values (>340Å), indicating an extended structure with Dmax of ~ 505 Å (Fig. 4B). Intuitively, this kind of P(r) functions for proteins represents a long-tailed structure with globular domains, consistent with the classic HMM structure in which the S2-subfragment of two myosin molecules form a coiled-coil rod on which the flexible globular S1 heads rests. The bell-shaped curve in P(r) at low r values presumably comes from the rigid, well-defined intra-head vector length, while the broad shoulder in P(r) for intermediate r values comes mostly from the inter-head vector lengths indicating a flexible inter-head spacing. Interestingly, compared to untreated (DMSO) and para-nitroblebbistatin-treated samples, the shoulder region of P(r) for mavacamten-treated HMM elicited a more prominent peak with higher probability, indicating a qualitatively well-defined, less scattered inter-head spacing. A greater reduction in physical parameters such as Rg and Dmax in mavacamten-treated HMM than in para-nitroblebbistatin-treated HMM, when compared to DMSO-treated control (Table 2), strongly suggest that mavacamten induces a much more compact and rigid form of myosin than para-nitroblebbistatin, supporting the idea that mavacamten structurally populates myosin in a different closed state when compared to para-nitroblebbistatin.
To strengthen our assertion, rigid body modeling using SASREF [48] converged to a myosin conformation that was more extended than the theoretical open conformation (Fig. 5C), which when paired with a broader shoulder region in the P(r) distribution (Fig. 4) and higher ensemble averages of Rg and Dmax (Table 2) suggest that there is a preferential shift in the distribution of myosin more towards the open state in the DMSO-treated HMM. However, in the HMM samples treated with either mavacamten or para-nitroblebbistatin, the SASREF modeling converged to a single homogeneous conformation that may be best described as a structural intermediate between the closed and open states (Fig. 5D–E) but proved to be inadequate to distinguish the structural features between treatments. However, these modeling-based solutions, when combined with other structural parameters such as a narrow shoulder region in the P(r) distribution indicating less inter-head flexibility, paired with lower Rg and Dmax, indicated that the degree of compactness in myosin is greater in the presence of mavacamten than with para-nitroblebbistatin and that mavacamten shifted the myosin distribution more towards a compact closed state.
It is worth noting that, in all HMM samples, including the control (DMSO), neither a classic closed myosin conformation mimicking the actual IHM state (i.e., two heads in the dimeric myosin folding back and interacting with the S2-subfragment tail) nor a genuine open conformation best described the model predicted by the SASREF modeling. While these findings differ from previous studies, which show that both mavacamten and blebbistatin can promote myosin into a fully compact folded-state [20,36], they are consistent with other findings from earlier reports. For example, findings from an X-ray diffraction study show that mavacamten treatment increases myosin density along thick filaments in cardiac muscle fibers, reflecting a compact arrangement [20]. Likewise, electron microscopy and structural studies using fluorescent polarization show that blebbistatin induces compactness by aligning myosin heads closer and parallel to the thick filament backbone [39] and improves the helical ordering of myosin along thick filaments in skeletal systems [34,35]. The discrepancy between findings from this study and previous studies may be likely attributed to the differences in the techniques used (e.g., solution-based SAXS structure vs. static electron microscopy images). Also, hydrolysis of ATP to different nucleotide states in solution during the chemo-mechanical cycle can render myosin into different conformations, and the modeling of SAXS data represents an ensemble average of these structures. Also, mavacamten and blebbistatin may not bind to myosin equally in all these nucleotide states, thereby giving dissimilar weights to different structures in the ensemble averaging.
Collectively, these studies lead us to posit that para-nitroblebbistatin traps myosin in a structural intermediate between closed and open states that prevents myosin from undergoing a productive ATPase cycle by probably populating an ‘ultra-relaxed’ state, which dramatically inhibits nucleotide release. This mechanism needs to be fully uncovered using better functional and structural elucidations in future investigations. In contrast, in the therapeutic range of concentrations, mavacamten shifts the open-closed equilibrium more towards the closed state and inhibits myosin ATPase function by increasing the energy-sparing myosin SRX population, most likely by promoting myosin into more compact conformational states in which the lever arm may be primed towards the pre-stroke direction [15,20]. Inferences drawn from our study support the idea that different interventions may structurally stabilize myosin in many other closed states in addition to the classic IHM state, and not all such closed states may necessarily correlate to the functional SRX state. Our study provides novel insights into the mechanistic actions of small molecules by connecting structural properties to biological functions, which is fundamental to designing better molecules for improved therapies.
MATERIALS AND METHODS
All materials and methods are briefly described here, but more details can be found in the Supporting Information (SI).
Preparation of Protein Reagents
Bovine cardiac actin, bovine cardiac full-length myosin, and bovine cardiac myofibrils were prepared following the methods described previously [63–65]. Human β-cardiac 25-hep heavy meromyosin (HMM) was purified using methods described elsewhere [46]. The human β-cardiac 25-hep HMM cDNA consists of a truncated version of MYH7 (residues 1–855), corresponding to S1-subfragment and the first 25 heptad repeats (175 amino acids) of S2-subfragment, followed by a GCN4 leucine zipper to ensure dimerization. This is further linked to a flexible GSG (Gly-Ser-Gly) linker, then a GFP moiety followed by another GSG linker, and finally ending with an 8-residue (RGSIDTWV) PDZ binding peptide.
Reconstitution of Myosin Thick Filaments
Starting from a buffer of high ionic strength (300 mM KCl or higher) in which full-length myosin remains completely soluble, the buffer’s ionic strength was reduced to 30 mM to allow spontaneous self-assembly of bi-polar thick filaments as described previously [66,67]. This study uses bovine β-cardiac full-length myosin to form reconstituted myosin filaments, referred to as bovine cardiac synthetic thick filaments (BcSTF).
Steady-state ATPase Measurements
Measurements of basal (pCa 10), actin-activated, and myofibrillar (pCa 6) myosin ATPase activity were performed at 23°C on a plate-based reader (SpectraMax 96-well) using an enzymatically coupled assay described elsewhere [26,42]. The buffer conditions used were 12 mM Pipes (pH 6.8), 2 mM MgCl2, 10 mM KCl, and 1 mM DTT. All basal and actin-activated ATPase experiments were done with myosin in the form of STFs. A final myosin concentration of 1 μM was attained in all cases. The concentration of actin in the actin-activated myosin ATPase experiments was 14 μM. When using myofibrils, approximately 40% of the total weight was assumed to be from myosin, and accordingly, the myofibril amount was loaded to attain a final myosin concentration of 1 μM.
Preparation of Myosin-based Small Molecule Inhibitors
Three different small molecule myosin inhibitors, blebbistatin, para-nitroblebbistatin, and mavacamten, were used in this study. Mavacamten was synthesized by MyoKardia, Inc., whereas blebbistatin and para-nitroblebbistatin were purchased from external sources (blebbistatin from Fisher Scientific, IL, and para-nitroblebbistatin from Cayman Chemical Company, MI). Concentrated stocks (20 mM) of each molecule were first prepared using dimethyl sulfoxide (DMSO). These are used to achieve a final concentration ranging from 0 to 50 μM in the experimental buffer samples. The final concentration of DMSO was 2% in all cases. Structural studies involving Small-angle X-ray scattering (SAXS), Multi-Angle Light Scattering (MALS), and Dynamic Light Scattering (DLS) measurements were carried out using a saturating concentration of each compound (20 μM for mavacamten and 50 μM for para-nitroblebbistatin).
Single Nucleotide Turnover Experiments
Single ATP turnover kinetic experiments using a fluorescent 2′/3′-O-(N-Methylanthraniloyl) (mant)-ATP were conducted in a 96-well plate fluorescence plate reader, as described in detail in previous work [29]. Briefly, in the first step, 50 μL of the experimental buffer containing 3.2 μM mant-ATP is combined with 100 μL of the same buffer containing 0.8 μM myosin in a UV plate. The reaction was aged 60 s to allow binding and hydrolysis of mant-ATP to inorganic phosphate (Pi) and mant-ADP. In the second step, mant-nucleotides were chased with non-fluorescent ATP by adding 50 μL of 16 mM non-fluorescent ATP to the above mixture. The resulting fluorescence decay was due to mant-nucleotide dissociation from myosin was monitored over time. The experimental buffer contained the following : 20 mM Tris-HCl (pH 7.4), 30 mM KCl, 1 mM EGTA, 3 mM MgCl2, and 1 mM DTT. The concentrations of myosin, mant-ATP and non-fluorescent ATP attained in the final mixture were 0.4 μM, 0.8 μM, and 4 mM, respectively. The excitation wavelength for mant-nucleotides was 385 nm. The emission was monitored using a long-pass filter with a cutoff wavelength of 465 nm. All experiments were performed at 25°C, and the fluorescence decay traces were collected for 2400 s.
As described in other studies [20,28,43], the fluorescence decay profile during the chase phase characteristically depicted two phases, a fast phase followed by a slow phase. Therefore, a bi-exponential function was fitted to each trace to estimate four parameters corresponding to fast and slow phases — Afast, kfast, Aslow, and kslow — where A represents the % amplitude, and k represents the observed ATP turnover rate of each phase. The fast and slow phases in this study correspond to myosin activity in the DRX and SRX states, respectively.
SEC-MALS-DLS-SAXS Studies
Small-angle X-ray scattering (SAXS) was performed with in-line size-exclusion chromatography (SEC), multi-angle light scattering (MALS), and dynamic light scattering (DLS) at the BioCAT beamline 18ID (Advanced Photon Source, Argonne National Laboratory, Lemont IL). The 25-hep HMM was loaded on a superose-6 increase 10/300 column (GE Lifesciences), which was run at 0.7 ml/min on an Agilent Infinity II HPLC system. The column eluant passed through a UV detector, MALS and DLS detectors (Wyatt DAWN HELEOS II), and a differential refractive index detector (Wyatt Optilab t-rEX) before reaching a co-flow SAXS cell. Samples and experimental buffers were treated with DMSO, 20 μM mavacamten, or 50 μM para-nitroblebbistatin. Because these experiments were run for a longer period (~2 hours), we avoided using blebbistatin in these experiments due to its unstable nature in the aqueous medium [68]. Instead, we used para-nitroblebbistatin, which is a non-cytotoxic and photostable derivative of blebbistatin [69]. All such experiments were done at 25°C, and the buffer used for these experiments contained 20 mM Tris-HCl (pH 7.4), 30 mM KCl, 1 mM EGTA, 3 mM MgCl2, 1 mM ATP, and 1 mM DTT.
Scattering intensity was recorded using a Pilatus3 1M detector, which was placed ~3.5m from the sample giving access to a q range of ~0.004Å−1 to 0.4 Å−1. 0.5-second exposures were acquired every second during elution, and data were reduced using BioXTAS RAW 1.6.0 [70]. Buffer blanks were created by averaging regions flanking the elution peak and subtracted from exposures selected from the elution peak to create the I(q) vs. q curves used for all subsequent analyses, which included Guinier approximation (radius of gyration, Rg and zero-angle scattering, I0), pair-distance distribution function (Rg and the maximum dimension – Dmax) and calculation of structural models. ASTRA (Wyatt Inc.) was used to calculate the molecular weight and hydrodynamic radii based on the MALS and DLS data, respectively. Four repeat experiments were performed on four different preparations of HMM to confirm the trends in the calculated parameters, and the results presented here are from the experiments with the best signal to noise.
Data Analysis
For biochemical studies, each assay was repeated at least twice with a minimum of two replicates per experiment. Sample number n refers to the total number of measurements made (number of experiments*number of replicates), and these data were presented as mean±SEM. The dose-response curve in each assay was used to estimate the concentration (in μM) required to attain half-maximal decrease (IC50) or half-maximal increase (EC50) in a given parameter for each small-molecule treatment. Linear regression analysis was performed between data from various experiments to determine the strength of such correlations. For SAXS studies, experiments were repeated at least thrice for each treatment (DMSO, para-nitroblebbistatin, mavacamten) and representative data (lowest chi-squared values for fits) are presented hereafter, confirming the reproducibility of the fundamental trend.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank the protein production team and the medicinal chemistry team of MyoKardia Inc. for producing biological reagents and mavacamten. The authors thank Dr. Leslie Leinwand at the University of Colorado, Boulder, for supervising producing cell pellets and purifying the human β-cardiac 25-hep HMM. The authors thank Dr. Daniel Chin at MyoKardia Inc. for helping develop a custom-built program to analyze the semi-high throughput single nucleotide turnover data. The authors also thank Dr. Roger Cooke, Professor Emeritus at UCSF; Dr. Robert McDowell, Chief Scientific Officer of MyoKardia Inc.; and Robert Anderson, Biology leader at MyoKardia Inc., for providing critical inputs to the manuscript. Finally, the authors thank Dr. Raul Padron and Dr. Roger Craig from the University of Massachusetts for all the critical discussions about myosin SRX and IHM states. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. This project was supported by grant P41 GM103622 from the National Institute of General Medical Sciences of the National Institutes of Health. The content is solely the authors’ responsibility and does not necessarily reflect the official views of the National Institute of General Medical Sciences or the National Institutes of Health.
Footnotes
DISCLOSURES
SKG, NS, and SN are all employees of MyoKardia, Inc, a wholly-owned subsidiary of Bristol Myers Squibb (TM), and hold company shares through their employment. The authors declare that they have no conflicts of interest with the contents of this article.
DATA AVAILABILITY
All data presented in this manuscript is either included in the main article or the Supporting Information.
REFERENCES
- [1].Gordon AM, Homsher E, Regnier M, Regulation of contraction in striated muscle, Physiol Rev. 80 (2000) 853–924. 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- [2].Kobayashi T, Solaro RJ, Calcium, thin filaments, and the integrative biology of cardiac contractility, Physiology. 67 (2005) 39–67. 10.1146/annurev.physiol.67.040403.114025. [DOI] [PubMed] [Google Scholar]
- [3].Woodhead JL, Zhao F-Q, Craig R, Egelman EH, Alamo L, Padrón R, Atomic model of a myosin filament in the relaxed state, Nature. 436 (2005) 1195–1199. 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- [4].Zoghbi ME, Woodhead JL, Moss RL, Craig R, Three-dimensional structure of vertebrate cardiac muscle myosin filaments, Proc National Acad Sci. 105 (2008) 2386–2390. 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wendt T, Taylor D, Trybus KM, Taylor K, Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2, Proc National Acad Sci. 98 (2001) 4361–4366. 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wendt T, Taylor D, Messier T, Trybus KM, Taylor KA, Visualization of head–head interactions in the inhibited state of smooth muscle myosin, J Cell Biology. 147 (1999) 1385–1390. 10.1083/jcb.147.7.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Burgess SA, Yu S, Walker ML, Hawkins RJ, Chalovich JM, Knight PJ, Structures of smooth muscle myosin and heavy meromyosin in the folded, shutdown state, J Mol Biol. 372 (2007) 1165–1178. 10.1016/j.jmb.2007.07.014. [DOI] [PubMed] [Google Scholar]
- [8].Jung HS, Burgess SA, Billington N, Colegrave M, Patel H, Chalovich JM, Chantler PD, Knight PJ, Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution, Proc National Acad Sci. 105 (2008) 6022–6026. 10.1073/pnas.0707846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hu Z, Taylor DW, Reedy MK, Edwards RJ, Taylor KA, Structure of myosin filaments from relaxed Lethocerus flight muscle by cryo-EM at 6 Å resolution, Sci Adv. 2 (2016) e1600058. 10.1126/sciadv.1600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Gillilan RE, Kumar VSS, O’Neall-Hennessey E, Cohen C, Brown JH, X-ray solution scattering of squid heavy meromyosin: strengthening the evidence for an ancient compact off state, Plos One. 8 (2013) e81994. 10.1371/journal.pone.0081994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Alamo L, Koubassova N, Pinto A, Gillilan R, Tsaturyan A, Padrón R, Lessons from a tarantula: new insights into muscle thick filament and myosin interacting-heads motif structure and function., Biophysical Rev. 9 (2017) 461–480. 10.1007/s12551-017-0295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Alamo L, Pinto A, Sulbarán G, Mavárez J, Padrón R, Lessons from a tarantula: new insights into myosin interacting-heads motif evolution and its implications on disease, Biophysical Rev. 10 (2017) 1465–1477. 10.1007/s12551-017-0292-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Trivedi DV, Adhikari AS, Sarkar SS, Ruppel KM, Spudich JA, Hypertrophic cardiomyopathy and the myosin mesa: viewing an old disease in a new light., Biophysical Rev. 10 (2017) 27–48. 10.1007/s12551-017-0274-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Spudich JA, Hypertrophic and dilated cardiomyopathy: four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases., Biophys J. 106 (2014) 1236–49. 10.1016/j.bpj.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Spudich JA, Three perspectives on the molecular basis of hypercontractility caused by hypertrophic cardiomyopathy mutations., Pflugers Archiv European J Physiology. 471 (2019) 701–717. 10.1007/s00424-019-02259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Spudich JA, The myosin mesa and a possible unifying hypothesis for the molecular basis of human hypertrophic cardiomyopathy., Biochem Soc T. 43 (2015) 64–72. 10.1042/bst20140324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Scarff CA, Carrington G, Casas-Mao D, Chalovich JM, Knight PJ, Ranson NA, Peckham M, Structure of the shutdown state of myosin-2, Nature. (2020) 1–6. 10.1038/s41586-020-2990-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang S, Tiwari P, Lee KH, Sato O, Ikebe M, Padrón R, Craig R, Cryo-EM structure of the inhibited (10S) form of myosin II, Nature. (2020) 1–5. 10.1038/s41586-020-3007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Robert-Paganin J, Auguin D, Houdusse A, Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition., Nat Commun. 9 (2018) 4019. 10.1038/s41467-018-06191-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Anderson RL, Trivedi DV, Sarkar SS, Henze M, Ma W, Gong H, Rogers CS, Gorham JM, Wong FL, Morck MM, Seidman JG, Ruppel KM, Irving TC, Cooke R, Green EM, Spudich JA, Deciphering the super relaxed state of human β-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers., P Natl Acad Sci Usa. 115 (2018) E8143–E8152. 10.1073/pnas.1809540115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sarkar SS, Trivedi DV, Morck MM, Adhikari AS, Pasha SN, Ruppel KM, Spudich JA, The hypertrophic cardiomyopathy mutations R403Q and R663H increase the number of myosin heads available to interact with actin, Sci Adv. 6 (2020) eaax0069. 10.1126/sciadv.aax0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Toepfer CN, Wakimoto H, Garfinkel AC, McDonough B, Liao D, Jiang J, Tai AC, Gorham JM, Lunde IG, Lun M, Lynch TL, McNamara JW, Sadayappan S, Redwood CS, Watkins HC, Seidman JG, Seidman CE, Hypertrophic cardiomyopathy mutations in MYBPC3 dysregulate myosin, Sci Transl Med. 11 (2019) eaat1199. 10.1126/scitranslmed.aat1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].McNamara JW, Li A, Lal S, Bos JM, Harris SP, van der Velden J, Ackerman MJ, Cooke R, Remedios CGD, MYBPC3 mutations are associated with a reduced super-relaxed state in patients with hypertrophic cardiomyopathy., Plos One. 12 (2017) e0180064. 10.1371/journal.pone.0180064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Adhikari AS, Kooiker KB, Sarkar SS, Liu C, Bernstein D, Spudich JA, Ruppel KM, Early-Onset Hypertrophic Cardiomyopathy Mutations Significantly Increase the Velocity, Force, and Actin-Activated ATPase Activity of Human β-Cardiac Myosin, Cell Reports. 17 (2016) 2857–2864. 10.1016/j.celrep.2016.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Adhikari AS, Trivedi DV, Sarkar SS, Song D, Kooiker KB, Bernstein D, Spudich JA, Ruppel KM, β-Cardiac myosin hypertrophic cardiomyopathy mutations release sequestered heads and increase enzymatic activity., Nat Commun. 10 (2019) 2685. 10.1038/s41467-019-10555-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Green EM, Wakimoto H, Anderson RL, Evanchik MJ, Gorham JM, Harrison BC, Henze M, Kawas R, Oslob JD, Rodriguez HM, Song Y, Wan W, Leinwand LA, Spudich JA, McDowell RS, Seidman JG, Seidman CE, A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice., Science. 351 (2016) 617–21. 10.1126/science.aad3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sparrow AJ, Watkins H, Daniels MJ, Redwood C, Robinson P, Mavacamten rescues increased myofilament calcium sensitivity and dysregulation of Ca2+ flux caused by thin filament hypertrophic cardiomyopathy mutations, Am J Physiol-Heart C. 318 (2020) H715–H722. 10.1152/ajpheart.00023.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Rohde JA, Roopnarine O, Thomas DD, Muretta JM, Mavacamten stabilizes an autoinhibited state of two-headed cardiac myosin., P Natl Acad Sci Usa. 115 (2018) E7486–E7494. 10.1073/pnas.1720342115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gollapudi SK, Yu M, Gan Q-F, Nag S, Synthetic thick filaments: A new avenue for better understanding the myosin super-relaxed state in healthy, disease, and mavacamten-treated cardiac systems, J Biol Chem. 296 (2020) 100114. 10.1074/jbc.ra120.016506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ramamurthy B, Yengo CM, Straight AF, Mitchison TJ, Sweeney HL, Kinetic mechanism of blebbistatin inhibition of nonmuscle myosin IIB, Biochemistry-Us. 43 (2004) 14832–14839. 10.1021/bi0490284. [DOI] [PubMed] [Google Scholar]
- [31].Kovács M, Tóth J, Hetényi C, Málnási-Csizmadia A, Sellers JR, Mechanism of blebbistatin inhibition of myosin II, J Biol Chem. 279 (2004) 35557–35563. 10.1074/jbc.m405319200. [DOI] [PubMed] [Google Scholar]
- [32].Limouze J, Straight AF, Mitchison T, Sellers JR, Specificity of blebbistatin, an inhibitor of myosin II, J Muscle Res Cell Motil. 25 (2004) 337–341. 10.1007/s10974-004-6060-7. [DOI] [PubMed] [Google Scholar]
- [33].Takács B, Billington N, Gyimesi M, Kintses B, Málnási-Csizmadia A, Knight PJ, Kovács M, Myosin complexed with ADP and blebbistatin reversibly adopts a conformation resembling the start point of the working stroke, Proc National Acad Sci. 107 (2010) 6799–6804. 10.1073/pnas.0907585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhao F-Q, Padrón R, Craig R, Blebbistatin stabilizes the helical order of myosin filaments by promoting the switch 2 closed state, Biophys J. 95 (2008) 3322–3329. 10.1529/biophysj.108.137067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Xu S, White HD, Offer GW, Yu LC, Stabilization of helical order in the thick filaments by blebbistatin: further evidence of coexisting multiple conformations of myosin, Biophys J. 96 (2009) 3673–3681. 10.1016/j.bpj.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jung HS, Komatsu S, Ikebe M, Craig R, Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells., Mol Biol Cell. 19 (2008) 3234–42. 10.1091/mbc.e08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Iwamoto H, Effects of myosin inhibitors on the X-ray diffraction patterns of relaxed and calcium-activated rabbit skeletal muscle fibers, Biophysics Physicobiology. 15 (2018) 111–120. 10.2142/biophysico.15.0_111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ma W, Gong H, Irving T, Myosin head configurations in resting and contracting murine skeletal muscle, Int J Mol Sci. 19 (2018) 2643. 10.3390/ijms19092643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kampourakis T, Zhang X, Sun Y, Irving M, Omecamtiv mercabil and blebbistatin modulate cardiac contractility by perturbing the regulatory state of the myosin filament, J Physiology. 596 (2018) 31–46. 10.1113/jp275050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wilson C, Naber N, Pate E, Cooke R, The myosin inhibitor blebbistatin stabilizes the super-relaxed state in skeletal muscle., Biophys J. 107 (2014) 1637–46. 10.1016/j.bpj.2014.07.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Tang W, Blair CA, Walton SD, Málnási-Csizmadia A, Campbell KS, Yengo CM, Modulating Beta-Cardiac Myosin Function at the Molecular and Tissue Levels, Front Physiol. 7 (2017) 659. 10.3389/fphys.2016.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kawas RF, Anderson RL, Ingle SRB, Song Y, Sran AS, Rodriguez HM, A small-molecule modulator of cardiac myosin acts on multiple stages of the myosin chemomechanical cycle, J Biol Chem. 292 (2017) 16571–16577. 10.1074/jbc.m117.776815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Stewart MA, Franks-Skiba K, Chen S, Cooke R, Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers., P Natl Acad Sci Usa. 107 (2009) 430–5. 10.1073/pnas.0909468107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nelson SR, Li A, Beck-Previs S, Kennedy GG, Warshaw DM, Imaging ATP Consumption in Resting Skeletal Muscle: One Molecule at a Time, Biophys J. 119 (2020) 1050–1055. 10.1016/j.bpj.2020.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Faruqi AR, Cross RA, Kendrick-Jones J, Small angle X-ray scattering studies on myosin, J Cell Sci. 1991 (1991) 23–26. 10.1242/jcs.1991.supplement_14.5. [DOI] [PubMed] [Google Scholar]
- [46].Nag S, Trivedi DV, Sarkar SS, Adhikari AS, Sunitha MS, Sutton S, Ruppel KM, Spudich JA, The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations., Nat Struct Mol Biol. 24 (2017) 525–533. 10.1038/nsmb.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Franke D, Petoukhov MV, Konarev PV, Panjkovich A, Tuukkanen A, Mertens HDT, Kikhney AG, Hajizadeh NR, Franklin JM, Jeffries CM, Svergun DI, ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions, J Appl Crystallogr. 50 (2017) 1212–1225. 10.1107/s1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Petoukhov MV, Svergun DI, Global rigid body modeling of macromolecular complexes against small-angle scattering data, Biophys J. 89 (2005) 1237–1250. 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Trivedi DV, Nag S, Spudich A, Ruppel KM, Spudich JA, The myosin family of mechanoenzymes: from mechanisms to therapeutic approaches, Annu Rev Biochem. 89 (2020) 1–27. 10.1146/annurev-biochem-011520-105234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang A, Heitner SB, Jacoby D, Lester S, Fang L, Balaratnam G, Sehnert AJ, 228Long-term safety and effectiveness of mavacamten in symptomatic obstructive hypertrophic cardiomyopathy (oHCM) patients (pts): update from PIONEER open-label extension (PIONEER-OLE) study, Eur Heart J. 40 (2019). 10.1093/eurheartj/ehz747.0063. [DOI] [Google Scholar]
- [51].Ho CY, Mealiffe ME, Bach RG, Bhattacharya M, Choudhury L, Edelberg JM, Hegde SM, Jacoby D, Lakdawala NK, Lester SJ, Ma Y, Marian AJ, Nagueh SF, Owens A, Rader F, Saberi S, Sehnert AJ, Sherrid MV, Solomon SD, Wang A, Wever-Pinzon O, Wong TC, Heitner SB, Mavacamten treatment for obstructive hypertrophic cardiomyopathy: a clinical trial, J Am Coll Cardiol. 75 (2020) 2649–2660. 10.1016/j.jacc.2020.03.064. [DOI] [PubMed] [Google Scholar]
- [52].Olivotto I, Oreziak A, Barriales-Villa R, Abraham TP, Masri A, Garcia-Pavia P, Saberi S, Lakdawala NK, Wheeler MT, Owens A, Kubanek M, Wojakowski W, Jensen MK, Gimeno-Blanes J, Afshar K, Myers J, Hegde SM, Solomon SD, Sehnert AJ, Zhang D, Li W, Bhattacharya M, Edelberg JM, Waldman CB, Lester SJ, Wang A, Ho CY, Jacoby D, E.-H. study investigators, Bartunek J, Bondue A, Craenenbroeck EV, Kubanek M, Zemanek D, Jensen M, Mogensen J, Thune JJ, Charron P, Hagege A, Lairez O, Trochu J-N, Axthelm C, Duengen H-D, Frey N, Mitrovic V, Preusch M, Schulz-Menger J, Seidler T, Arad M, Halabi M, Katz A, Monakier D, Paz O, Viskin S, Zwas D, Olivotto I, Rocca HPB-L, Michels M, Dudek D, Oko-Sarnowska Z, Oreziak A, Wojakowski W, Cardim N, Pereira H, Barriales-Villa R, Pavia PG, Blanes JG, Urbano RH, Diaz LMR, Elliott P, Yousef Z, Abraham T, Afshar K, Alvarez P, Bach R, Becker R, Choudhury L, Fermin D, Jacoby D, Jefferies J, Kramer C, Lakdawala N, Lester S, Marian A, Masri A, Maurer M, Nagueh S, Owens A, Owens D, Rader F, Saberi S, Sherrid M, Shirani J, Symanski J, Turer A, Wang A, Wever-Pinzon O, Wheeler M, Wong T, Yamani M, Mavacamten for treatment of symptomatic obstructive hypertrophic cardiomyopathy (EXPLORER-HCM): a randomised, double-blind, placebo-controlled, phase 3 trial, Lancet. 396 (2020) 759–769. 10.1016/s0140-6736(20)31792-x. [DOI] [PubMed] [Google Scholar]
- [53].Heitner SB, Jacoby D, Lester SJ, Owens A, Wang A, Zhang D, Lambing J, Lee J, Semigran M, Sehnert AJ, Mavacamten treatment for obstructive hypertrophic cardiomyopathy: a clinical trial, Ann Intern Med. 170 (2019) 741. 10.7326/m18-3016. [DOI] [PubMed] [Google Scholar]
- [54].Mamidi R, Li J, Doh CY, Verma S, Stelzer JE, Impact of the myosin modulator mavacamten on force generation and cross-bridge behavior in a murine model of hypercontractility, J Am Heart Assoc. 7 (2018) e009627. 10.1161/jaha.118.009627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Stern JA, Markova S, Ueda Y, Kim JB, Pascoe PJ, Evanchik MJ, Green EM, Harris SP, A small molecule inhibitor of sarcomere contractility acutely relieves left ventricular outflow tract obstruction in feline hypertrophic cardiomyopathy, PLOS ONE. 11 (2016) e0168407. 10.1371/journal.pone.0168407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Saberi S, Cardim N, Yamani MH, Schulz-Menger J, Li W, Florea V, Sehnert AJ, Kwong RY, Jerosch-Herold M, Masri A, Owens A, Lakdawala NK, Kramer CM, Sherrid M, Seidler T, Wang A, Sedaghat-Hamedani F, Meder B, Havakuk O, Jacoby D, Mavacamten favorably impacts cardiac structure in obstructive hypertrophic cardiomyopathy: EXPLORER-HCM CMR substudy analysis, Circulation. (2020). 10.1161/circulationaha.120.052359. [DOI] [PubMed] [Google Scholar]
- [57].Allingham JS, Smith R, Rayment I, The structural basis of blebbistatin inhibition and specificity for myosin II, Nature Structural and Molecular Biology. 12 (2005) 378–379. 10.1038/nsmb908. [DOI] [PubMed] [Google Scholar]
- [58].Nag S, Trivedi DV, To lie or not to lie: Super-relaxing with myosins, Elife. 10 (2021) e63703. 10.7554/elife.63703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ma W, Duno-Miranda S, Irving T, Craig R, Padrón R, Relaxed tarantula skeletal muscle has two ATP energy-saving mechanisms, J Gen Physiol. 153 (2021) e202012780. 10.1085/jgp.202012780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Naber N, Cooke R, Pate E, Slow myosin ATP turnover in the super-relaxed state in tarantula muscle., J Mol Biol. 411 (2011) 943–50. 10.1016/j.jmb.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Alamo L, Qi D, Wriggers W, Pinto A, Zhu J, Bilbao A, Gillilan RE, Hu S, Padrón R, Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis., J Mol Biol. 428 (2016) 1142–64. 10.1016/j.jmb.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Padrón R, Ma W, Duno-Miranda S, Koubassova N, Lee KH, Pinto A, Alamo L, Bolaños P, Tsaturyan A, Irving T, Craig R, The myosin interacting-heads motif present in live tarantula muscle explains tetanic and posttetanic phosphorylation mechanisms, Proc National Acad Sci. 117 (2020) 11865–11874. 10.1073/pnas.1921312117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Spudich JA, Watt S, The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin., J Biological Chem. 246 (1971) 4866–71. [PubMed] [Google Scholar]
- [64].Margossian SS, Lowey S, Methods in enzymology, Methods Enzymol. 85 (1982) 55–71. 10.1016/0076-6879(82)85009-x. [DOI] [PubMed] [Google Scholar]
- [65].Solaro RJ, Pang DC, Briggs FN, The purification of cardiac myofibrils with Triton X-100, Biochimica Et Biophysica Acta Bba - Bioenergetics. 245 (1971) 259–262. 10.1016/0005-2728(71)90033-8. [DOI] [PubMed] [Google Scholar]
- [66].Huxley HE, Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle, J Mol Biol. 7 (1963) 281–IN30. 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- [67].Davis JS, Assembly processes in vertebrate skeletal thick filament formation, Annu Rev Biophys Bio. 17 (1988) 217–239. 10.1146/annurev.bb.17.060188.001245. [DOI] [PubMed] [Google Scholar]
- [68].Rauscher AÁ, Gyimesi M, Kovács M, Málnási-Csizmadia A, Myosin by blebbistatin derivatives: optimization and pharmacological potential., Trends in Biochemical Sciences. (2018). 10.1016/j.tibs.2018.06.006. [DOI] [PubMed] [Google Scholar]
- [69].Képiró M, Várkuti BH, Végner L, Vörös G, Hegyi G, Varga M, Málnási-Csizmadia A, para-Nitroblebbistatin, the non-cytotoxic and photostable myosin II inhibitor, Angewandte Chemie Int Ed. 53 (2014) 8211–8215. 10.1002/anie.201403540. [DOI] [PubMed] [Google Scholar]
- [70].Hopkins JB, Gillilan RE, Skou S, BioXTAS RAW: improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis, J Appl Crystallogr. 50 (2017) 1545–1553. 10.1107/s1600576717011438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Trewhella J, Duff AP, Durand D, Gabel F, Guss JM, Hendrickson WA, Hura GL, Jacques DA, Kirby NM, Kwan AH, Pérez J, Pollack L, Ryan TM, Sali A, Schneidman-Duhovny D, Schwede T, Svergun DI, Sugiyama M, Tainer JA, Vachette P, Westbrook J, Whitten AE, 2017 publication guidelines for structural modelling of small-angle scattering data from biomolecules in solution: an update, Acta Crystallogr Sect D Struct Biology. 73 (2017) 710–728. 10.1107/s2059798317011597. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in this manuscript is either included in the main article or the Supporting Information.


