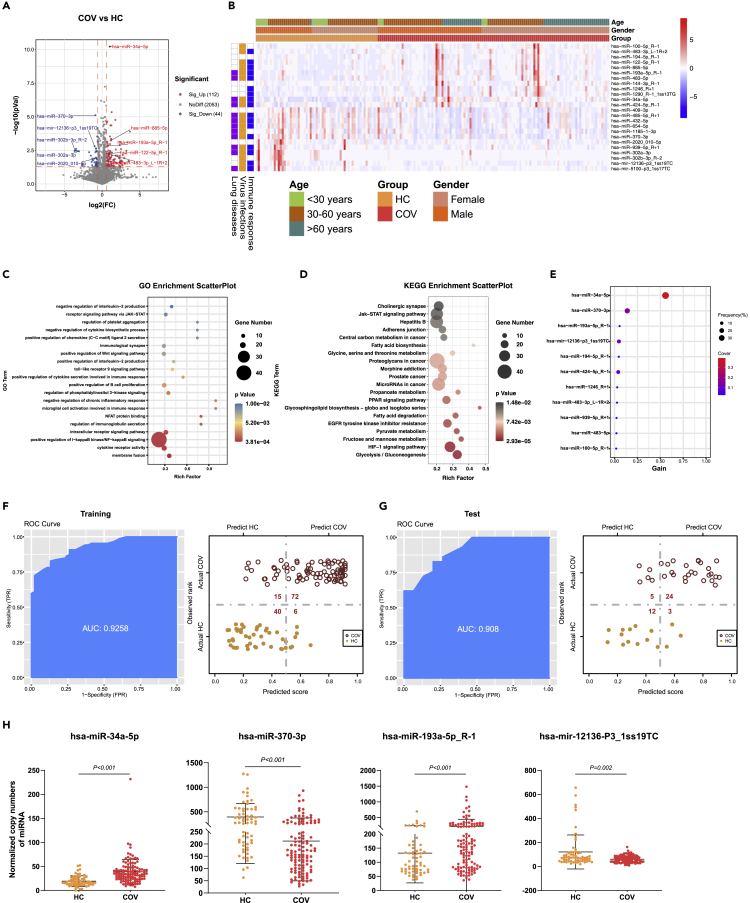
Figure 2.
DE-miRNAs associated with SARS-COV-2 infections
(A), The volcano plot of COV vs HC. Red dots represent the upregulated DE-miRNAs (fold change>1.5, p ≤ 0.05); blue dots represent the downregulated DE-miRNAs (fold change<0.67, p ≤ 0.05); gray dots represent miRNAs without significant changes (0.67 ≤ fold change≤1.5, p > 0.05).
(B), The heatmap of 24 DE-miRNAs in COV vs HC. In the left panel, blue, orange, and purple squares represent the function of miRNAs associated with immune responses, virus infections, and lung diseases, respectively.
(C), The GO analysis of the targeted genes in COV vs HC. Top 20 GO terms associated with immune responses, virus infections, and lung diseases were shown.
(D), The KEGG analysis of the targeted genes in COV vs HC. Top 20 KEGG pathways associated with immune responses, virus infections, and lung diseases were shown.
(E), The important DE-miRNAs prioritized by XGBoost analysis.
(F), Receiver operating characteristic (ROC) and performance of the model in the training set.
(G), ROC and performance of the model in the test set.
(H), Normalized copy numbers of four representative DE-miRNAs in the COV vs HC group, the bars showing the mean value ± standard deviation (sd) (COV n = 116, HC n = 61).