Abstract
Background
Colorectal cancer (CRC) is the third most common of cancer-related deaths. Nucleolar protein 14 (NOP14) is known to play different roles in diverse types of cancers. However, little is known about its roles in CRC. Here, we assessed the prognostic value and functions of NOP14 in CRC using the data from The Cancer Genome Atlas (TCGA) and validated them based on the data from Gene Expression Omnibus (GEO).
Methods
NOP14 mRNA and protein data in CRC was obtained from the TCGA, GEO, human protein atlas (HPA), and UALCAN databases. Survival and Cox regression analysis was performed to assess the prognostic value of NOP14 in CRC patients. Next, to evaluate the potential functions of NOP14, a protein–protein interaction (PPI) network was constructed and gene set enrichment analysis (GSEA) of differential expression genes (DEGs) associated with dysregulated NOP14 was performed. Finally, to investigate the immune response associated with NOP14 expression in CRC, we analyzed the correlations between immune cells infiltration and NOP14 expression level. Additionally, the correlations between immune molecule expression levels with NOP14 expression level were analyzed.
Results
High NOP14 mRNA expression was observed in CRC tissues based on the data from TCGA and GEO datasets. Similarly, high NOP14 protein levels were found in CRC tissues according to the immunohistochemical images from HPA. Interestingly, high NOP14 expression level was associated with an improved prognosis in CRC patients. Univariate and multivariate Cox regression analysis indicated that high NOP14 expression level was an independent protective factor for CRC patients. With the support of PPI network analysis, we found several risk genes interacted with NOP14. GSEA revealed that high NOP14 expression inhibited several signal pathways involved in tumor formation and development. Additionally, high NOP14 expression was positively associated with most kinds of immune cell infiltrations and the expression levels of some molecules related to immune activation.
Conclusion
Altogether, these results indicated that high NOP14 expression leads to improved prognosis in CRC patients by inhibiting the signaling pathways involved in tumor growth and promoting the immune responses.
Keywords: Colorectal cancer, NOP14, Prognostic marker, Immune infiltration, Nucleolar protein
Introduction
Colorectal cancer (CRC) is a very prevalent cancer worldwide and remains the third leading cause of cancer-related death [1]. The most effective screening test for the early diagnose of CRC is colonoscopy. However, the implementation of colonoscopies in a clinical setting is limited, resulting in approximately 60% of CRC patients being at an advanced stage at the time of diagnosed. Despite the recent progress of surgery, radiotherapy, chemotherapy, and targeted therapy have improved the prognosis of CRC patients, it still remains relatively poor due to the delayed diagnosis. According to the data of China National Cancer Center, the 5-year survival rate of colorectal cancer in China is about 50% [2]. The formation and development of CRC is a complex process associated with increased expression of oncogenes and reduced expression of tumor suppressor genes. Interestingly, CXCL11, encoding the chemokine ligand C-X-C motif chemokine ligand 11, has been reported to be significantly upregulated in colon cancer tissues and to be associated with a better prognosis. Additionally, high CXCL11 expression was reported to be positively correlated with immune infiltration and expression levels of cytotoxic genes, which can activate the antitumor immune response, thereby resulting in improved survival. This exemplifies the importance of studying the role of the genes upregulated in cancer, and as it may contribute to understand the tumorigenic process and reveal potential therapeutic targets [3].
NOP14 encodes Nucleolar Protein 14, an 875 amino protein, which is highly conserved in eukaryotes and participates in numerous biological processes, such as DNA repair and replication and cell cycle control. A recent study reported NOP14 regulates the expression of the methyltransferase Emg1, thereby being required for 18 s rRNA maturation [4]. Moreover, increasing studies supported that NOP14 is involved in cancer initiation and development. For example, NOP14 inhibited the Wnt/β-catenin signaling pathway, which reduced the proliferation and metastasis of melanoma [5] and breast cancer [6], thereby acting as a tumor suppressor gene. Similarly, in bladder cancer, NOP14 attenuated the miR-502-5p-mediated inhibition of migration and proliferation of tumor cells. Nonetheless, the roles of NOP14 varies depending on the specific types of cancer. In pancreatic cancer, NOP14 was highly expressed, and had been shown to promote the growth and invasion of tumor cells in vitro by stabilizing mutant P53, which suppressed the expression of P21 via induction of miR-17-5p [7]. Thereby, NOP14 acts as an oncogene in some tumors, and as tumor suppressor gene in others.
To date, the role of NOP14 in CRC has not been reported. In this study, we analyzed the differential expression of NOP14 between CRC tissues and adjacent normal specimens using the data from the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) database, and then evaluated its prognostic value in CRC patients. Next, a protein–protein interaction (PPI) network was construct and Gene Set Enrichment Analysis (GSEA) was performed to predict the potential functions of NOP14. Finally, we assessed the correlations between the NOP14 expression level and immune infiltration as well as immune molecules, to unravel the role of NOP14 in the prognosis of CRC patients.
Materials and methods
Data acquisition, preprocessing, and ethics statement
Pan-cancer RNA sequencing (RNA-seq) data was obtained from UCSC XENA database (https://xenabrowser.net/datapages/). Level 3 RNA RNA-seq expression data and clinical data for CRC (647 CRC tissues vs. 51 normal adjacent cancer tissues) were downloaded from TCGA portal (https://portal.gdc.cancer.gov/). Before analysis, transcripts per million reads (TPM) RNA-seq data was log transformed (Log2(TPM + 1)). ChIP data of expression profiles for CRC samples were downloaded from following GEO datasets (https://www.ncbi.nlm.nih.gov/geo/): GSE 20482 (65 pairs of CRC tumor and matched adjacent non-tumor tissue samples; platform GPL4133), GSE38839 (101 CRC samples and 35 normal tissues, platform GPL10558), GSE87211 (230 CRC samples and 133 normal colorectal mucosa tissues, platform GPL13497), and GSE161158 (250 CRC tissues and supplementary clinical information, platform GPL570). As TCGA and GEO are open publicly available databases, the data collection from the databases was compliant with all applicable laws, regulations, and policies for the protection of human subjects, and all written informed consents were obtained from all subjects involved.
Analysis of differential expression of NOP14
The R package edgeR was used to analyze differentially expressed genes (DEGs) between CRC and normal colon tissue within TCGA database using the threshold parameters |log2(FC)|> 1 and p adj value < 0.01. To compare the expression levels of NOP14 in pan-cancer and CRC, statistical analyses were performed using R (V3.6.3), considering a p value of < 0.05 significant. The CRC tissue samples were divided into two groups according to median NOP14 expression level (high- and low- NOP14 expression level), among which 3 cases were duplicated, which had been deleted and not included in the analysis. Finally, these samples were used to investigate the prognostic value, and immune infiltration associated with NOP14 expression levels.
Analysis of NOP14 protein expression in CRC tissue samples
Immunohistochemistry staining images of NOP14 in CRC and normal tissue sections were downloaded from HPA (https://www.proteinatlas.org/), in which these sections used the same antibodies and experimental methods. Next, the differential expression of NOP14 protein in CRC and normal adjacent tissues was obtained from UALCAN (http://ualcan.path.uab.edu/), which is a web resource including gene transcriptional data and clinical information of cancers from TCGA [8].
Association of NOP14 expression with clinical factors in CRC patients
To determine the diagnostic value of NOP14 expression levels, a receiver operating characteristics (ROC) curve was built using R package pROC (v1.17.0.1), and the area under cure (AUC) with 95% confidence interval (CI) was calculated to confirm the diagnostic efficacy. Next, to assess the potential value of NOP14 expression on prognostic prediction for pan-cancer and CRC patients, Kaplan–Meier (K—M) survival curves were generated according to NOP14 expression level and survival status data from TCGA and GSE161158 and analyzed using the R packages survival (v3.2–10) and survminer (v0.4.9). The difference between high- and low- NOP14 expression groups was analyzed using the log-rank test, using a p value < 0.05 as the significance threshold. In addition, 1-, 3- 5-year aspects of time dependent ROC were constructed using the R package timeROC. Similarly, to identify the independent survival risk factors for CRC patients, among gender, age, T and N stages, and NOP14 expression level, univariate and multivariate Cox regression analysis were performed using the R package survival (v3.2–10). The hazard rate (HR) with 95% confidence interval (CI) and p value were presented with forest plots using R package ggplot2 (v3.3.3).Next, a nomogram and a calibration curve were constructed based on the results of univariate and multivariate Cox regression analysis of NOP14 expression level and clinical feature using the rms package.
PPI network construction
To explore the proteins interacted with NOP14, a PPI network was constructed using STRING 11.5 ((www.string-db.org) [9]. The interacting proteins were selected based on the original criteria of STRING 11.5. The linkage scores were used to identify the interaction pairs.
Functional enrichment analysis of DEGs
GSEA associated with DEGs between the high- and low- NOP14 expression level groups was performed using the clusterProfiler package, with c2.cp.v7.2.symbols.gmtcurated gene sets from MSigDB Collections as reference gene sets [10, 11]. Clusters with a p value < 0.05 and a false discovery rate (FDR) < 0.25 were considered significant.
Immune infiltration analysis
Single-sample GSEA (ssGSEA) was used to assess the infiltration of individual immune cell populations, e.g., T helper 2 cells (Th2 cells), CD8 T cells, NK CD56dim cell, and others. The analysis was performed using GSVA package [12]. Next, the correlations between NOP14 expression level and tumor purity, B cell, CD8 + cell, CD4 + cell, macrophage, neutrophil and dendritic cell infiltration were calculated and plotted using TIMER2.0 (http://timer.cistrome.org/) [13, 14].
Analysis of immune-related molecules levels
To investigate the association between the expression levels of NOP14 with immune-related molecules, a Pearson’s correlation analysis was performed using a p-value < 0.05 as the threshold for significance. Heat maps of the significant immune-related molecules based on Pearson coefficients were generated using the R package ggplot2.
Statistical Analyses
Statistical analyses were performed using R (v.3.6.3). The ggplot2 package was used for data visualization. Data are presented as means ± standard deviation (SD). All tests were two-sided, and a p < 0.05 was defined as statistically significant.
Results
NOP14 expression in pan-cancer and CRC patients
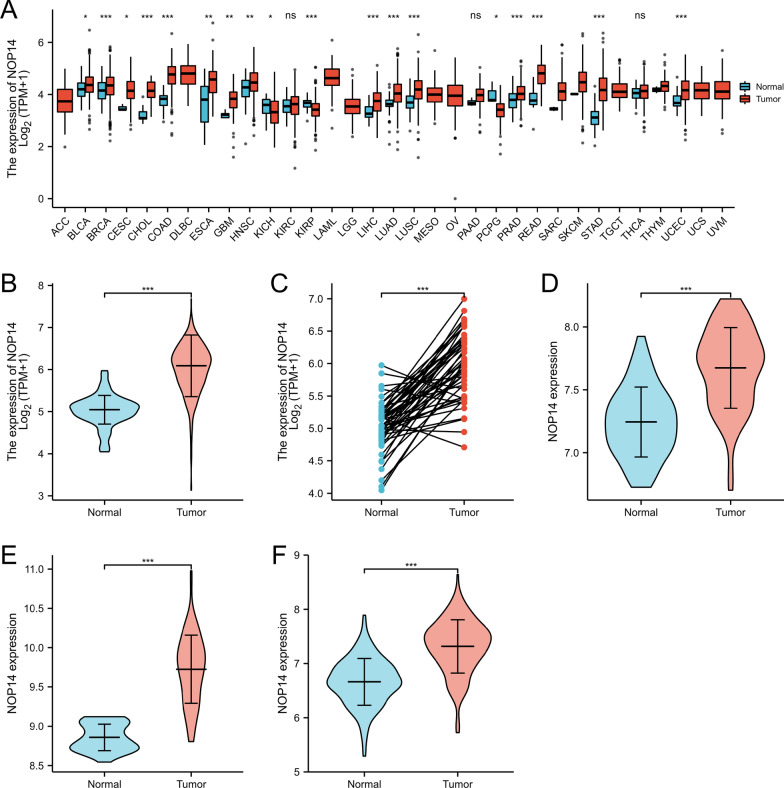
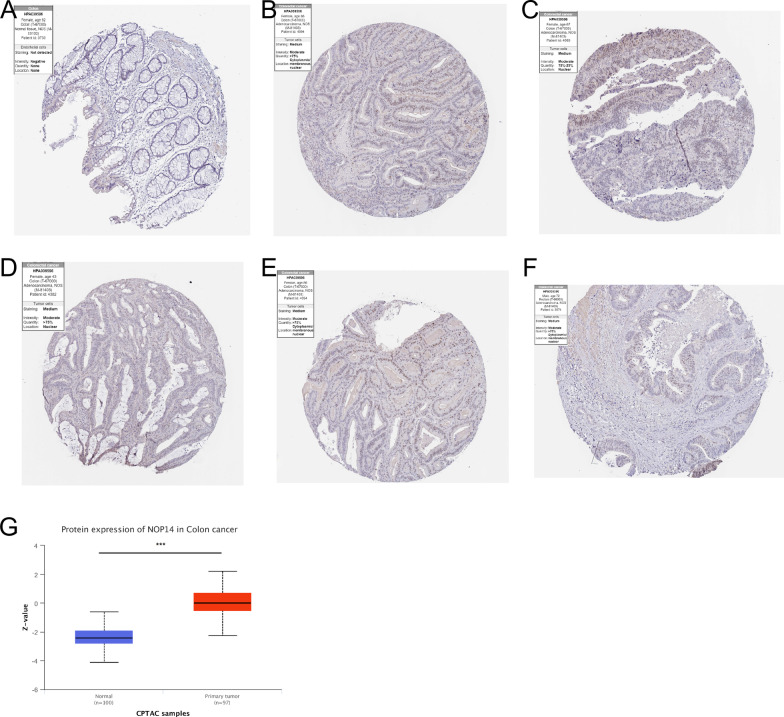
The clinical features of the 644 CRC patients included in this study are shown in Table 1. Using the data from TCGA, we found that NOP14 expression was significantly elevated in 19 of 33 cancer types, including bladder urothelial carcinoma (BLCA), colon adenocarcinoma (COAD), cholangiocarcinoma (CHOL), liver hepatocellular carcinoma (LIHC), lung squamous carcinoma (LUSC), stomach adenocarcinoma (STAD), among others (Fig. 1A). Similarly, NOP14 expression was significantly increased in CRC tissues compared with normal samples (Fig. 1B). For the 51 pared samples, NOP14 expression was also significantly higher in cancer tissues than in matched normal adjacent tissue samples (Fig. 1C). These results were confirmed in three GEO datasets: GSE 20482, GSE38839, and GSE87211 (Fig. 1D–F). Next, NOP14 protein expression levels were assessed using immunohistochemistry images obtained from HPA. Whereas NOP14 protein was not detected in normal tissue (Fig. 2A), but NOP14 protein positive cells were moderate in tumor tissues (Fig. 2B–F). Similarly, NOP14 protein expression level was significantly higher in cancer tissues than in normal tissue samples according to the data from UALCAN (Fig. 2G).
Table 1.
Relationship between NOP14 expression levels and clinical features of colorectal cancer patients based on data from TCGA
| Characteristic | Low NOP14 expression | High NOP14 expression | p |
|---|---|---|---|
| n | 322 | 322 | |
| Gender, n (%) | 0.527 | ||
| Female | 155 (24.1%) | 146 (22.7%) | |
| Male | 167 (25.9%) | 176 (27.3%) | |
| Age, n (%) | 0.032 | ||
| ≤ 65 | 124 (19.3%) | 152 (23.6%) | |
| > 65 | 198 (30.7%) | 170 (26.4%) | |
| T stage, n (%) | 0.890 | ||
| T1 | 10 (1.6%) | 10 (1.6%) | |
| T2 | 55 (8.6%) | 56 (8.7%) | |
| T3 | 214 (33.4%) | 222 (34.6%) | |
| T4 | 40 (6.2%) | 34 (5.3%) | |
| N stage, n (%) | 0.209 | ||
| N0 | 176 (27.5%) | 192 (30%) | |
| N1 | 76 (11.9%) | 77 (12%) | |
| N2 | 68 (10.6%) | 51 (8%) | |
| M stage, n (%) | 0.395 | ||
| M0 | 235 (41.7%) | 240 (42.6%) | |
| M1 | 49 (8.7%) | 40 (7.1%) |
Fig. 1.
NOP14 expression pattern in pan-cancer and colorectal cancer (CRC) based on data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO). A NOP14 was highly expressed in 18 of 33 cancers compared with its expression in normal tissues. ***p < 0.001, **p < 0.01, *p < 0.05; n.s., no significance. B NOP14 expression is significantly higher in tumor tissues than in normal tissues. ***p < 0.01. C NOP14 expression in 51 CRC tissues and their paired adjacent para-carcinomatous tissues. *** p < 0.001. D–F Differential NOP14 expression between cancer and normal tissues according to the datasets GSE 20,482 (D), GSE38839 (E), and GSE87211 (F). ***p < 0.01
Fig. 2.
NOP14 protein expression pattern in colorectal cancer (CRC) tissues. NOP14 protein was not detected in normal colon tissues (A), and was detected in CRC tissues with moderate signal intensity (B–F) based on the immunohistochemical images from Human Protein Atlas (HPA). (G) NOP14 protein expression level was significantly higher in CRC tissues than in normal tissues based on the data from UALCAN, ***p < 0.01
Clinical significance associated with NOP14 expression in CRC patients
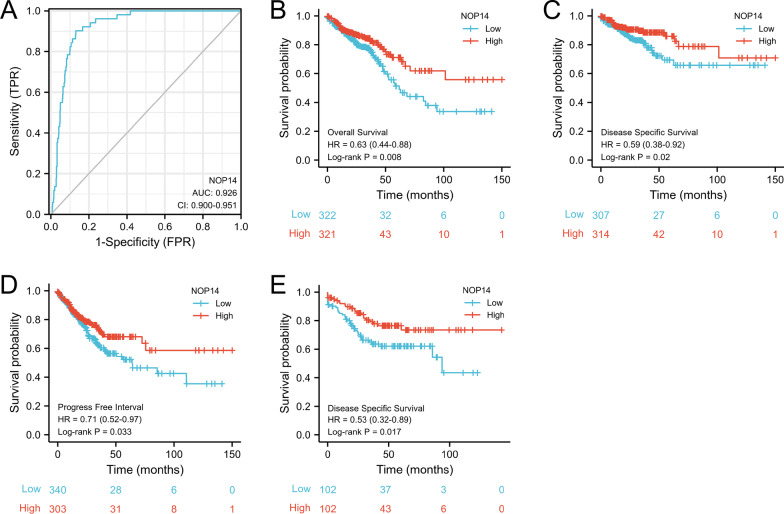
To evaluate the diagnostic value of NOP14 expression level, a ROC curve was constructed. As shown in Fig. 3A, the NOP14 expression had a high sensitivity and specificity for CRC diagnosis. The area under the curve (AUC) was 0.926, and Youden index was 0.771. Next, the prognostic value of NOP14 expression was evaluated using a K–M analysis. According to the data from TCGA, the overall survival (OS), disease specific survival (DSS), and progress free interval (PFI) were significantly better in high NOP14-expression group than in low NOP14-expression group (Fig. 3B–D). Additionally, we validated the DSS results in the cohort from the GEO dataset GSE161158 (HR:0.53, p < 0.05, Fig. 3E). Interestingly, as shown in Table 2, high NOP14 expression levels were associated with different prognostic impacts in different types of cancers. For example, in colon adenocarcinoma (COAD), stomach adenocarcinoma (STAD), rectum adenocarcinoma (READ), and kidney renal clear cell carcinoma (KIRC), high NOP14 expression levels were associated with a good prognosis (HR < 1, p < 0.05). Whereas in brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), sarcoma (SARC), adrenocortical carcinoma (ACC), high expression levels of NOP14 were associated with a poor prognosis (HR > 1, p < 0.05).
Fig. 3.
NOP14 expression level has potential diagnostic and prognostic values for patients with colorectal cancer (CRC). A Receiver operating characteristic (ROC) curve for NOP14 (area under the curve [AUC]: 0.926, P < 0.001). Kaplan–Meier (K–M) survival curves for overall survival (OS) (B), disease specific survival (DSS) (C), and disease free interval (DFI) (D) constructed based on data from The Cancer Genome Atlas (TCGA). E K–M survival curves for DSS constructed based on data from the Gene Expression Omnibus (GEO) dataset GSE161158
Table 2.
NOP14 expression levels significantly related to overall survival of patients with 8 eight types of cancer according to the data from TCGA
| Cancer types | HR | 95% CI | P value |
|---|---|---|---|
| STAD | 0.69 | 0.50–0.96 | 0.028 |
| READ | 0.41 | 0.19–0.89 | 0.029 |
| COAD | 0.59 | 0.40–0.88 | 0.015 |
| KIRC | 0.62 | 0.45–0.84 | 0.002 |
| LGG | 1.75 | 1.25–2.45 | 0.001 |
| LIHC | 1.49 | 1.06–2.11 | 0.021 |
| SARC | 1.59 | 1.07–2.37 | 0.02 |
| ACC | 2.24 | 1.06–4.73 | 0.035 |
Stomach adenocarcinoma (STAD), rectum adenocarcinoma (READ), colon adenocarcinoma (COAD), kidney renal clear cell carcinoma (KIRC), lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), sarcoma (SARC), adrenocortical carcinoma (ACC)
Prognostic efficacy of NOP14 expression levels
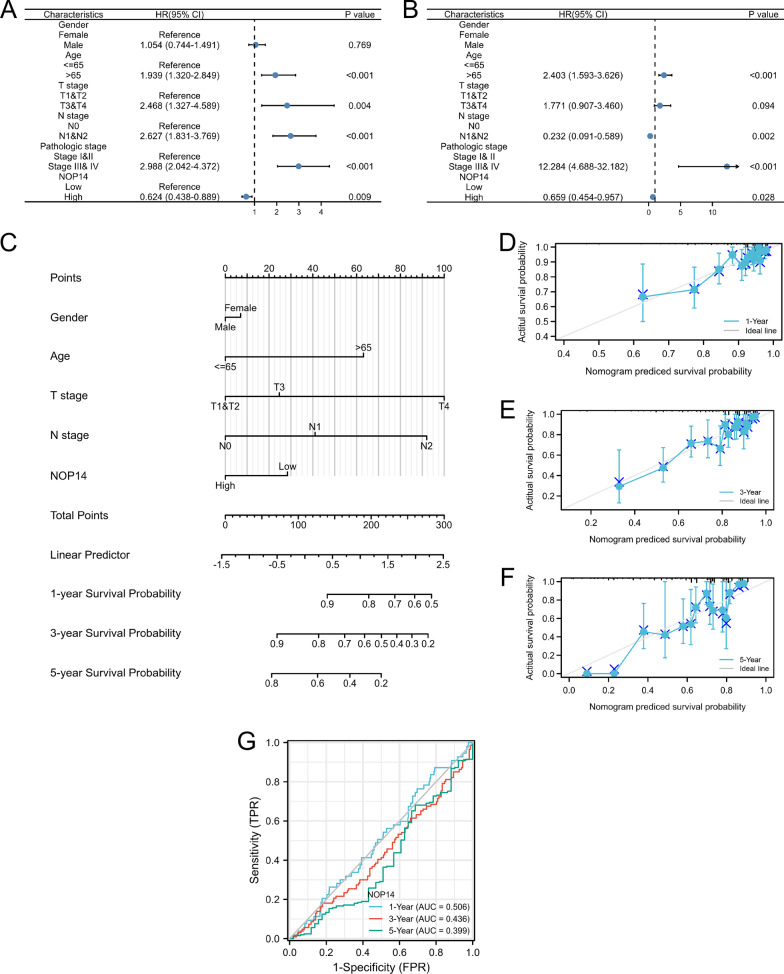
We then performed univariate and multivariate Cox regression analyses using several clinical features and NOP14 expression level. As shown in Fig. 4A, B, high NOP14 expression was an independent protective factor for OS in CRC patients. In contrast, advanced age (> 65 years old) and clinical stage (T3-4 stage and N1 stage) predicted poor clinical outcomes. Then, we generated a simple-to-use nomogram based on clinical features, including gender, age, T and N stage, as well as NOP14 expression level. As shown in Fig. 4C, NOP14 expression level had a good performance in predicting 1-, 3- and 5-years survival rate in the patients with CRC. The concordance index (C-index) was 0.726 (95% CI: 0.701–0.751). Similarly, the calibration plots ran very close to the diagonals, which showed a good performance in calibrating (Fig. 4D–F). Time dependent ROC revealed similar results for 1-, 3- 5-year follow-up, the AUCs were < 0.5, indicating that high NOP14 expression might be a protective factor for CRC patients (Fig. 4G).
Fig. 4.
Prognostic efficacy of NOP14 expression levels in patients with colorectal cancer (CRC). Results of univariate (A) and multivariate (B) Cox regression analyses displayed using forest plots. C Nomogram for predicting clinical outcomes associated with NOP14 expression in CRC patients. D–F Calibration plots validating 1-, 3-, and 5-year clinical outcomes for CRC patients. G Time-dependent receiver operating characteristic (ROC) analysis of 1-, 3-, and 5-year overall survival based on NOP14 expression levels in CRC patients
Proteins interacting with NOP14
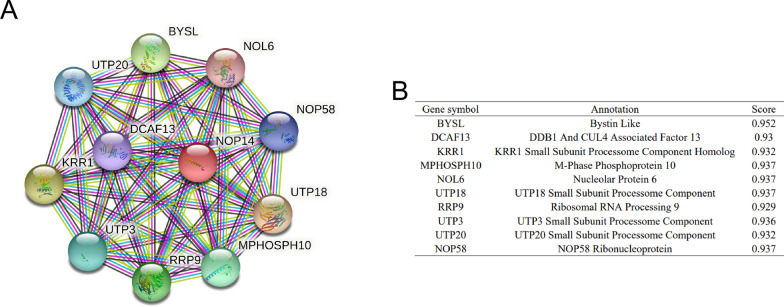
To explore the proteins interacted with NOP14, we generated a PPI network of NOP14 protein using the tool STRING (Fig. 5A). The top 10 proteins and their gene symbol, annotation, and scores are listed in Fig. 5B. The proteins interacted with NOP14 include BYSL, DCAF13, KRR1, MPHOSPH10, NOL6, UPT20, RRP9, UPT3, UPT18, and NOP58, some of which closely related to carcinogenesis.
Fig. 5.
Proteins interacting with NOP14. A Protein–protein interaction (PPI) network of NOP14. B Annotation of proteins that interact with NOP14 and their co-expression scores
Functional enrichment analysis of the genes associated with NOP14 expression level
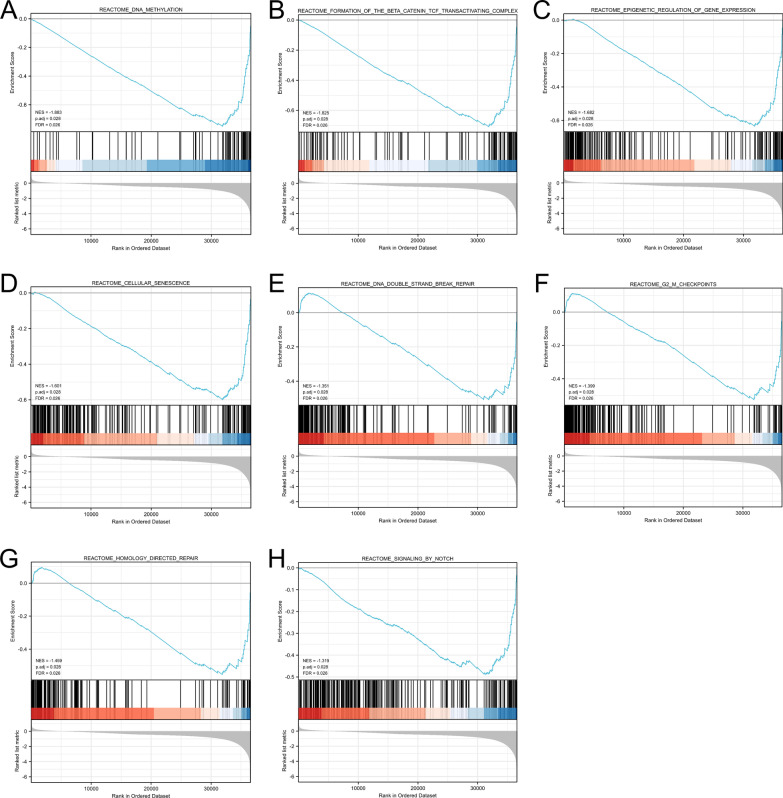
To further investigate the potential function of NOP14 in CRC, CRC samples were divided into NOP14 low- and high- expression groups, and DEGs were identify, and GSEA was performed. The results of GSEA revealed that some pathways associated with tumor cell growth were significantly enriched in NOP14 low- expression group (p < 0.05, FDR < 0.025), e.g., FORMATION_OF_THE_BETA_CATENIN_TCF_TRANSACTIVATING_COMPLEX, DNA_METHYLATION, SIGNALING_BY_NOTCH (Fig. 6A-H).
Fig. 6.
Biological pathways enriched in NOP14 low-expression group (A–H) determined using Gene Set Enrichment Analysis (GSEA). NES, normalized enrichment score
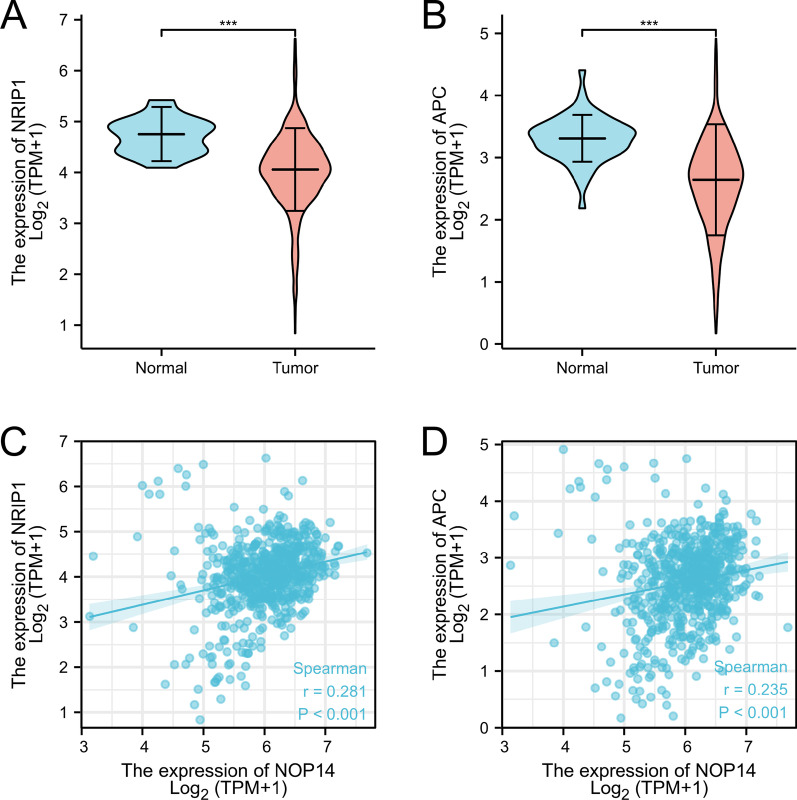
Next, we analyzed the expression levels of the inhibitors of the Wnt/β-catenin signaling pathway, Nuclear Receptor Interacting Protein 1 (NRIP1) and Adenomatous Polyposis Coli (APC) in CRC tissues in CRC tissues. The expression levels of NRIP1 and APC were significantly lower in tumor tissues than normal samples (Fig. 7A, B). Additionally, NOP14 expression levels were positive correlated to NRIP1 and APC expression levels (p < 0.01, Fig. 7C, D). Therefore, NOP14 might act as a tumor suppressor gene through inhibiting some tumor growth-related signal pathways in CRC cells, such as Wnt/β-catenin pathway.
Fig. 7.
NOP14 expression pattern associated with inhibitors of the Wnt/catenin pathway. NRIP1 (A) and APC (B) expression is significantly lower in cancer tissues than in normal tissues. *** p < 0.001. Positive correlations between NOP14 and NRIP1 (C) and NOP14 and APC (D) expression are shown. p < 0.001
Association between NOP14 expression and immune cell infiltration and immune molecule expression levels
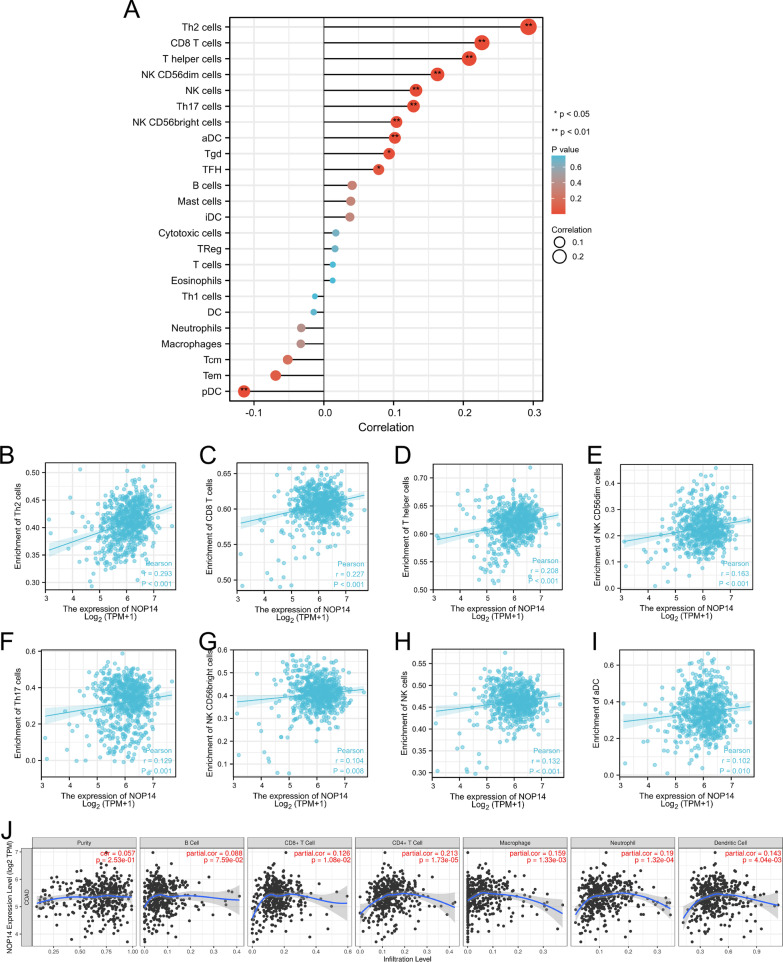
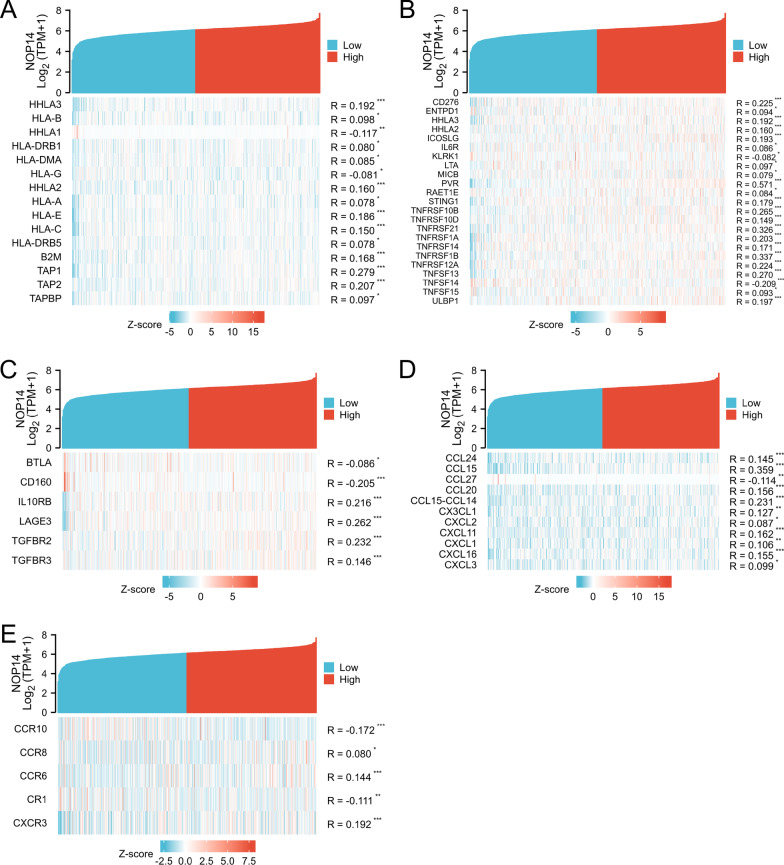
To understand the effects of NOP14 expression on tumor microenvironment, immune infiltration analysis was performed using ssGSEA method. The correlations between enrichments of immune cell in CRC tissues and NOP14 expression levels were calculated using Spearman correlation analyses. As shown in Fig. 8A–I, NOP14 expression levels were positively correlated with numerous types of immune cells, e.g., Th2 cells, CD8 T cells, T helper cells, NKCD56 dim cells, NK cells, and aDC cells, all of which play an important role in anti-tumor immunity. Next, to observe the infiltrations of other immune cells in CRC tissues, TIMER2.0 was used to analyze the correlations between NOP14 expression levels and the infiltrations of CD8 + cells, CD4 + cells, B cells, macrophages, neutrophils, and dendritic cells. As shown in Fig. 8J, NOP14 expression levels were significantly positively correlated with the infiltrations of those immune cells (p < 0.01). Similarly, the correlations between NOP14 expression levels and the expression levels of several immune-related molecules, such as MHC genes (Fig. 9A), immune activation genes (Fig. 9B), immunosuppressive genes (Fig. 9C), chemokine receptors (Fig. 9D) and chemokines (Fig. 9E) were also statistically significant.
Fig. 8.
Relationship between NOP14 expression and immune cell infiltration in the tumor microenvironment. A Correlations between NOP14 expression and 24 types of immune cell infiltrations. B–I Correlations between T helper (h) 2, cluster of differentiation (CD)8 + , Th, NK CD56dim, Th17, NK CD56bright, NK, and B cell and aDC enrichment levels and NOP14 expression. J Correlations between NOP14 expression and infiltrations of CD8+, CD4+, B, and dendritic cells as well as macrophages and neutrophils determined using TIMER2.0 tool
Fig. 9.
Coexpression heatmaps for correlations between expression of NOP14 and immune-related factors. The heatmaps display the expression levels of MHC genes, immune activation genes (A), immunosuppressive genes (B), chemokine receptors (C), and chemokines (D) significant correlated with NOP14 expression
Discussion
CRC is a prevalent malignant tumor of the digestive tract. Despite the prognosis of CRC has remarkably improved in the last years due to the progress in the fields of diagnosis and therapy, the prognosis of advanced CRC patients remains unsatisfactory. Therefore, it is urgent to find new molecular markers to improve the ability of predicting the prognosis of CRC. In this study, we assessed the genes that highly expressed in tumor tissues and was associated with a good prognosis in CRC patients. According the data from TCGA and GEO, we found that NOP14 expression was significantly higher than normal samples. Interestingly, high NOP14 expression was associated with improved prognosis for CRC patients, and acted as an independent protective factor. The underlying mechanism might be related to the inhibition of signaling pathways associated with tumor growth, and the activation of immune cell infiltration and immune response.
NOP14, which is an evolutionally conserved protein in eukaryotes, mainly locates in nucleus and nucleus membrane and plays an important role in pre-18 s rRNA processing and small ribosomal subunit assembly [15]. It has been recently reported that NOP14 can be considered as an immune regulator factor, and that is involved in various diseases [16, 17]. In chronic inflammatory disease, NOP14 activates NF-κB signaling pathway, and upregulates proinflammatory gene expression, and increases the expression of adhesion molecules in endothelial cells, thereby promoting the infiltrations of immune cells [18]. This highlights that NOP14 is closely related with immune response and that it plays an important role in biological processes of various cells.
The involvement of NOP14 in cancer initiation and development is still unclear. Some studies have reported that NOP14 expression levels are associated with prognosis of various types of cancer [19–21]. In our study, we showed that NOP14 expression was significantly elevated in many kinds of cancer, such as bladder urothelial carcinoma, cholangiocarcinoma, and liver hepatocellular carcinoma. Because colon adenocarcinoma and rectum adenocarcinoma share similar biological features, we integrated them as CRC during the analysis processes. DEGs analysis showed that NOP14 expression level was also significantly higher in CRC tissues than in normal tissues, suggesting in may play an important role in carcinogenesis and development of CRC, among other cancers. So far, only a few studies have reported the prognostic value of NOP14 expression levels in cancer. Isaksson et al. reported that low expression of NOP14 associated with poor prognosis in ovarian cancer [21]. Similarly, Chang et al. found that NOP14 highly expressed in colon cancer tissues, and that high expression predicted a good prognosis [20].We further investigated this relationship, and found that the high expression of NOP14 had different effects on OS for different cancers (Table 2). In COAD, STAD, READ, and KIRC, high NOP14 expression level was associated with an improved prognosis. However, in LGG, LIHC, SARC, and ACC, high NOP14 expression level was associated with a poor prognosis. These results suggest that NOP14 plays different roles in different cancers. For CRC patients, according to the results of K–M survival analyses based the data from TCGA and GEO, we found that high NOP14 expression level was associated with an improved prognosis. Furthermore, Univariate and multivariate Cox regression analysis identified that high NOP14 expression level as an independent protective factor for CRC patients. Our nomogram and calibration plots confirmed that NOP14 expression level was a good predictor for 1-, 3, 5-year OS in CRC patients. Therefore, NOP14 had a good performance in prognostic prediction, illustrating that NOP14 could be used as a biomarker of diagnosis and therapeutic target.
Based on previous literature, we hypothesized that the effects mediated by NOP14 in tumor formation and metastasis were mediated by the Wnt/β-catenin pathway. In melanoma and breast cancer, NOP14 inhibited the proliferation and metastasis of tumor cells by down regulating the Wnt/β-catenin signal pathway [5, 6]. In this study, we found that Wnt/β-catenin signal pathway was enriched in NOP14 low expression group, indicating that high NOP14 expression might downregulate the pathway, acting as a tumor suppressor gene. Nonetheless, some other pathways associated with tumor growth were also enriched in the low NOP14 expression group, e.g., Notch signaling, DNA methylation, G2/M checkpoints. Next, we analyzed the expression levels of the key proteins in Wnt/β-catenin pathway. NRIP1, a transcription co-regulatory factor, negatively regulated the Wnt/β-catenin pathway by increasing APC expression level and degrading β-catenin [22, 23]. Consistent with previous studies, we found that NRIP1 and APC expression were significantly lower in CRC tissues than in normal tissues. However, their expression levels were significantly positively correlated with NOP14 expression levels, suggesting that NOP14 might upregulate NRIP1 and APC expression in CRC tissues. To further unravel the biological function of NOP14 in CRC, we generated a PPI network using the tool STRING. As shown in Fig. 5, we found that NOP14 interacted with some proteins closely related to tumor initiation and development, such as BYSL or NOL6. Mechanistically, BYSL activated the AKT pathway by regulating RIOK2 and mTOR, acting as an oncogene in gliomas [24]. NOL6 promoted the proliferation and migration of various kinds of tumor cells, including endometrial cancer, hepatocellular carcinoma, and prostate cancer [25–27]. Therefore, the biological functions of NOP14 are closely related to carcinogenesis and development of CRC.
Many reports supported that immune cell infiltration in tumor tissues associate with tumor formation, growth, and metastasis [28, 29]. A high concentration of immune-promoting cells in tumor tissue improved patient prognosis, whereas a low concentration of immune-promoting cells may lead to immune escape of cancer cells, resulting in a poor prognosis [30, 31]. In this study, we found that NOP14 expression levels were positively correlated to the infiltration levels of numerous immune cells, such as CD8 cells, CD4 cells, NK cells, and dendritic cells, suggesting that NOP14 might promote the anti-tumor immune response in tumor tissues. Additionally, NOP14 expression levels were significantly correlated with the expression levels of many immune molecules, like MHC genes, immune activation genes, chemokine receptor, and chemokines, which might recruit immune cell infiltration, thereby exerting an anti-tumor effect. Altogether, these results suggest that NOP14 may exert an anti- oncogenic function by triggering anti-tumor immune responses in CRC tissues.
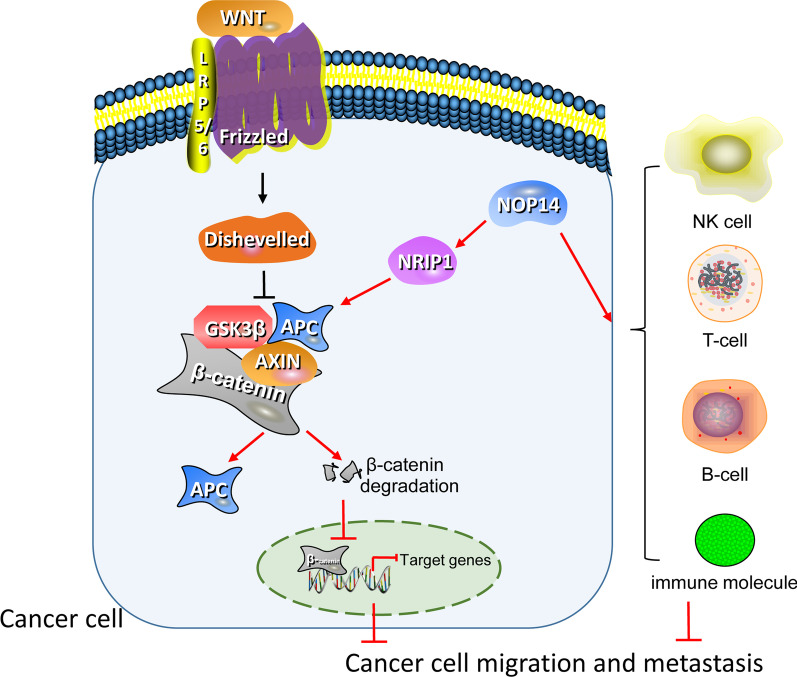
In conclusion, we preliminarily conformed that NOP14 is as a prognostic biomarker in CRC patients. Additionally, we suggested the potential mechanism relying on inhibiting the pathways of tumor growth and promoting anti-tumor immune response (Fig. 10). Nonetheless, further research is necessary to confirm this mechanism of action.
Fig. 10.
Schematic diagram of a possible mechanism of action of NOP14 in colorectal cancer (CRC) formation and metastasis, that is inhibiting the Wnt/β-catenin pathway and promoting anti-tumor response
Acknowledgements
Not applicable.
Author contributions
Conceptualization and Funding acquisition, YL; Data curation, CL; Formal analysis; W H, WHand YW; Methodology, YH; Writing-original draft, LC, WH, and YL; Writing-review and editing, CL and YL. All authors read and approved the final manuscript.
Funding
This study was supported by the Key Medical Disciplines Building Project of Shenzhen Longhua District, the Guangzhou Medical and Health Scientific Research Project (No. 20211A010004), and the Science and Technology Planning Project of Guangzhou (No. 202102080680).
Availability of data and materials
The data used for analysis in this study was obtained from public available databases, which were UCSC XENA (https://xenabrowser.net/datapages/), TCGA data portal (https://portal.gdc.cancer.gov/), and GEO database (https://www.ncbi.nlm.nih.gov/geo/).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yaoxing Huang, Email: huangyaoxing@sina.com.
Yuqi Luo, Email: luoyuqi2004@tom.com.
References
- 1.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 2.Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: Good or bad news from the 2018 Global Cancer Statistics? Cancer Commun. 2019;39(1):22. doi: 10.1186/s40880-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao Y, Jiao N, Sun T, Ma Y, Zhang X, Chen H, Hong J, Zhang Y. CXCL11 correlates with antitumor immunity and an improved prognosis in colon cancer. Front Cell Dev Biol. 2021;9:646252. doi: 10.3389/fcell.2021.646252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas SR, Keller CA, Szyk A, Cannon JR, Laronde-Leblanc NA. Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic Acids Res. 2011;39(6):2445–2457. doi: 10.1093/nar/gkq1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Fang R, Wang J, Deng L. NOP14 inhibits melanoma proliferation and metastasis by regulating Wnt/beta-catenin signaling pathway. Braz J Med Biol Res. 2018;52(1):e7952. doi: 10.1590/1414-431X20187952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lei JJ, Peng RJ, Kuang BH, Yuan ZY, Qin T, Liu WS, Guo YM, Han HQ, Lian YF, Deng CC, Zhang HJ, Chen LZ, Feng QS, Xu M, Feng L, Bei JX, Zeng YX. NOP14 suppresses breast cancer progression by inhibiting NRIP1/Wnt/beta-catenin pathway. Oncotarget. 2015;6(28):25701–14. doi: 10.18632/oncotarget.4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Y, Liu Z, You L, Hou P, Ren X, Jiao T, Zhao W, Li Z, Shu H, Liu C, Zhao Y. pancreatic cancer progression relies upon mutant p53-induced oncogenic signaling mediated by NOP14. Cancer Res. 2017;77(10):2661–2673. doi: 10.1158/0008-5472.CAN-16-2339. [DOI] [PubMed] [Google Scholar]
- 8.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, Bork P, von Mering C. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database issue):D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;16(14):7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–14. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu PC, Thiele DJ. Novel stress-responsive genes EMG1 and NOP14 encode conserved, interacting proteins required for 40S ribosome biogenesis. Mol Biol Cell. 2001;12(11):3644–3657. doi: 10.1091/mbc.12.11.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirsch VC, Orgler C, Braig S, Jeremias I, Auerbach D, Müller R, Vollmar AM, Sieber SA. The cytotoxic natural product vioprolide A targets nucleolar protein 14, which is essential for ribosome biogenesis. Angew Chem Int Ed Engl. 2020;59(4):1595–1600. doi: 10.1002/anie.201911158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgers LD, Luong B, Li Y, Fabritius MP, Michalakis S, Reichel CA, Müller R, Fürst R. The natural product vioprolide A exerts anti-inflammatory actions through inhibition of its cellular target NOP14 and downregulation of importin-dependent NF-ĸB p65 nuclear translocation. Biomed Pharmacother. 2021;144:112255. doi: 10.1016/j.biopha.2021.112255. [DOI] [PubMed] [Google Scholar]
- 18.Burgers LD, Luong B, Li Y, Fabritius MP, Michalakis S, Reichel CA, Muller R, Furst R. The natural product vioprolide A exerts anti-inflammatory actions through inhibition of its cellular target NOP14 and downregulation of importin-dependent NF-kB p65 nuclear translocation. Biomed Pharmacother. 2021;144:112255. doi: 10.1016/j.biopha.2021.112255. [DOI] [PubMed] [Google Scholar]
- 19.Fan X, Liu L, Shi Y, Guo F, Wang H, Zhao X, Zhong D, Li G. Integrated analysis of RNA-binding proteins in human colorectal cancer. World J Surg Oncol. 2020;18(1):222. doi: 10.1186/s12957-020-01995-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang K, Yuan C, Liu X. A new RBPs-related signature predicts the prognosis of colon adenocarcinoma patients. Front Oncol. 2021;11:627504. doi: 10.3389/fonc.2021.627504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isaksson HS, Sorbe B, Nilsson TK. Whole genome expression profiling of blood cells in ovarian cancer patients -prognostic impact of the CYP1B1, MTSS1, NCALD, and NOP14. Oncotarget. 2014;5(12):4040–9. doi: 10.18632/oncotarget.193810.18632/oncotarget.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Triki M, Lapierre M, Cavailles V, Mokdad-Gargouri R. Expression and role of nuclear receptor coregulators in colorectal cancer. World J Gastroenterol. 2017;23(25):4480–4490. doi: 10.3748/wjg.v23.i25.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lapierre M, Bonnet S, Bascoul-Mollevi C, Ait-Arsa I, Jalaguier S, Del Rio M, Plateroti M, Roepman P, Ychou M, Pannequin J, Hollande F, Parker M, Cavailles V. RIP140 increases APC expression and controls intestinal homeostasis and tumorigenesis. J Clin Invest. 2014;124(5):1899–1913. doi: 10.1172/JCI65178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao S, Sha Z, Zhou J, Wu Y, Song Y, Li C, Liu X, Zhang T, Yu R. BYSL contributes to tumor growth by cooperating with the mTORC2 complex in gliomas. Cancer Biol Med. 2021;18(1):88–104. doi: 10.20892/j.issn.2095-3941.2020.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong D, Song M, Wu X, Wang W. NOL6, a new founding oncogene in human prostate cancer and targeted by miR-590-3p. Cytotechnology. 2020;72(3):469–478. doi: 10.1007/s10616-020-00394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng L, Xu KX, Zhao MX, Li K, Zhu K, Yuan DW, Wang HN, Dai PG, Yan R. Nucleolar protein 6 promotes cell proliferation and acts as a potential novel prognostic marker for hepatocellular carcinoma. Chin Med J. 2021;134(21):2611–2618. doi: 10.1097/CM9.0000000000001655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang J, Sun W, Song H, Wang C, Li Q, Li C, Wei D, Zhao Y, Li C, Zhang H. NOL6 promotes the proliferation and migration of endometrial cancer cells by regulating TWIST1 expression. Epigenomics. 2021;13(19):1571–1585. doi: 10.2217/epi-2021-0218. [DOI] [PubMed] [Google Scholar]
- 28.Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, Hess JM, Uzunangelov V, Walter V, Danilova L, Lichtenberg TM, Kucherlapati M, Kimes PK, Tang M, Penson A, Babur O, Akbani R, Bristow CA, Hoadley KA, Iype L, Chang MT, Network TR, Cherniack AD, Benz C, Mills GB, Verhaak RGW, Griewank KG, Felau I, Zenklusen JC, Gershenwald JE, Schoenfield L, Lazar AJ, Abdel-Rahman MH, Roman-Roman S, Stern MH, Cebulla CM, Williams MD, Jager MJ, Coupland SE, Esmaeli B, Kandoth C, Woodman SE. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell. 2017;32(2):204–220. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ringgaard L, Melander F, Eliasen R, Henriksen JR, Jolck RI, Engel TB, Bak M, Fliedner FP, Kristensen K, Elema DR, Kjaer A, Hansen AE, Andresen TL. Tumor repolarization by an advanced liposomal drug delivery system provides a potent new approach for chemo-immunotherapy. Sci Adv. 2020 doi: 10.1126/sciadv.aba5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buisseret L, Pommey S, Allard B, Garaud S, Bergeron M, Cousineau I, Ameye L, Bareche Y, Paesmans M, Crown JPA, Di Leo A, Loi S, Piccart-Gebhart M, Willard-Gallo K, Sotiriou C, Stagg J. Clinical significance of CD73 in triple-negative breast cancer: multiplex analysis of a phase III clinical trial. Ann Oncol. 2018;29(4):1056–1062. doi: 10.1093/annonc/mdx730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H, Xie J, Jin P. Assessment of hazard immune-related genes and tumor immune infiltrations in renal cell carcinoma. Am J Transl Res. 2020;12(11):7096–7113. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used for analysis in this study was obtained from public available databases, which were UCSC XENA (https://xenabrowser.net/datapages/), TCGA data portal (https://portal.gdc.cancer.gov/), and GEO database (https://www.ncbi.nlm.nih.gov/geo/).