Summary
Three principal methods are under discussion as possible pathways to “true” de-extinction; i.e., back-breeding, cloning, and genetic engineering.1,2 Of these, while the latter approach is most likely to apply to the largest number of extinct species, its potential is constrained by the degree to which the extinct species genome can be reconstructed. We explore this question using the extinct Christmas Island rat (Rattus macleari) as a model, an endemic rat species that was driven extinct between 1898 and 1908.3, 4, 5 We first re-sequenced its genome to an average of >60× coverage, then mapped it to the reference genomes of different Rattus species. We then explored how evolutionary divergence from the extant reference genome affected the fraction of the Christmas Island rat genome that could be recovered. Our analyses show that even when the extremely high-quality Norway brown rat (R. norvegicus) is used as a reference, nearly 5% of the genome sequence is unrecoverable, with 1,661 genes recovered at lower than 90% completeness, and 26 completely absent. Furthermore, we find the distribution of regions affected is not random, but for example, if 90% completeness is used as the cutoff, genes related to immune response and olfaction are excessively affected. Ultimately, our approach demonstrates the importance of applying similar analyses to candidates for de-extinction through genome editing in order to provide critical baseline information about how representative the edited form would be of the extinct species.
Keywords: Christmas Island rat, Rattus macleari, de-extinction, evolutionary divergence, ancient DNA, genomic sequencing, molecular dating
Highlights
-
•
Evolutionary divergence limits the completeness of extinct species genomes
-
•
The extinct Christmas Island rat was re-sequenced to ca. 60× coverage
-
•
Nevertheless, 4.85% of the Norway brown rat genome remains absent after mapping
-
•
Absences are not random; immune response and olfaction are excessively affected
Lin et al. explore how evolutionary divergence constrains the potential of de-extinction, using the Christmas Island rat as a model. Although 95% of its genome is recovered when re-sequenced at high depth, regions that remain unmapped to the reference likely have significant consequences for any attempt at de-extinction using genome editing.
Results and discussion
Unlike alternative potential de-extinction approaches such as targeted back-breeding and interspecies somatic cell nuclear transfer (iSCNT), genome reconstruction through genetic engineering approaches are not constrained by the requirement of working with still-living/viable material. In contrast, they propose to take advantage of recent advances in both ancient DNA (aDNA) and genome editing technology to potentially revive extinct species for which either no genomic tracts are preserved in living species (for back-breeding) or as viable frozen somatic cells (for iSCNT). Genetic engineering for de-extinction is conceptually based upon the idea of first describing the sequence of the extinct species, then editing the genomes of living cells from related species, for example using CRISPR-Cas9 technologies.1,2 However, currently this process is not straightforward. First, since DNA recovered from most historic/ancient samples is typically heavily fragmented,6,7 the extinct species' genome is unlikely to be reconstructed through de novo genome assembly.1,2,8,9 Rather, the extinct species’ genome sequence is obtained through mapping its DNA against the (ideally) de novo sequenced genome of a closely related living species in order to identify sequence differences7 for use in the subsequent editing. There are at least two key hurdles inherent in de-extinction through this route, both ultimately derived from the evolutionary divergence that separates the extinct from the extant species. First, as current gene editing technologies are typically limited to the range of introducing several tens to several hundreds of edits per cycle,10 multiple rounds of edits would be required to fully modify a genome that may differ at many thousands of positions (or even much more).1,2 However, even if the genome editing technology can be improved to efficiently edit every site required in a single generation, an additional possible challenge remains that may be far more problematic. Because ancient DNA molecules are typically very short as a result of post mortem diagenesis (most typically well under 50 bp in length),7,11 these map poorly and/or ambiguously (if at all) to any regions of the genome that are highly divergent from the reference, thus potentially rendering them unrecoverable.8,9 Although some computational solutions to this challenge have been proposed, such as reducing the evolutionary divergence through mapping to in silico predicted ancestral nodes on phylogenies,12 at best the effect is reduced, not eliminated. As such, given that the ultimate goal of at least some de-extinction projects may be the regeneration of species whose genomes are as representative as possible of the lost form (as opposed to the definition adopted by IUCN Species Survival Commission, that is the creation of “a proxy of an extinct species” that is “a functional equivalent able to restore ecological functions or processes that might have been lost as a result of the extinction of the original species”13), a key question is how exactly does evolutionary divergence affect genome reconstruction success? In particular, given that evolutionary rates can vary greatly across the genome,14,15 how might this information inform us about the biological reality of any resurrected species created in this way?7, 8, 9
We extracted and sequenced aDNA from two dry preserved skin samples of the Christmas Island rat (Rattus macleari), originally collected between 1900–1902 and held as part of the Oxford University Museum of Natural History collections. We assume that should gene editing be used to attempt resurrection, the Norway brown rat (Rattus norvegicus) would represent an ideal system for editing for several reasons. First, the relatively close estimated evolutionary divergence of the Christmas Island rat and Norway brown rat (previously estimated split at ca. 2.6 million years ago (mya) based on molecular phylogenetic analysis,5,16,17 assuming a mutation rate of 1.655 × 10−9 per generation per base pair and generation time of 0.5 years18). Second, the Norway brown rat is widely used as a laboratory model in both general genomic studies, but also those that require genome editing. Third, it has an excellent quality (i.e., highly complete and contiguous) reference genome that is more complete than that of another possibly relevant candidate, the black rat (Rattus rattus) with regards to contig N50 (29.20 Mb versus 1.64 Mb), and number of scaffolds (176 versus 2,173) and contigs (757 versus 1,635,336).1,19
Following sequencing using both Illumina and BGISeq technologies, the sequence data were trimmed and mapped to the Norway brown rat reference genome (mRatBN7.2, NCBI: GCA_015227675.2, male) using the Paleomix pipeline.20 The sequences displayed characteristic aDNA damage profiles such as misincorporations and fragmentation (Figure S1; Table S1). The amount of sequence data generated allowed us to map the Christmas Island rat’s genome sequence to an average depth of 60.81× once the data from the two samples was merged. Nevertheless, despite this high average depth of coverage, the data only spanned 95.15% of the Norway brown rat reference genome (Table 1), raising the question as to why. While this can be partly explained by the observation that 0.81% of the bases in the reference genome are undetermined (Ns), we hypothesized that the remaining 4.04% of the reference genome was unmappable because either (1) the short length of the ancient DNA templates (take the BGISeq data, for example; 48.27% of the reads are shorter than 50 bp; Figure S1A) reduces their mapping ability; (2) the AT richness of some genome regions introduces PCR amplification bias, thus sequencing bias; and/or (3) the missing regions are unmappable due to the evolutionary divergence of the two species.
Table 1.
Summary of mapping genomic sequencing data of five Rattus species to Norway brown rat reference genome, related to Table S1 and Figure S1
| Hit reads | Coverage | MaxDepth | 1× | 5× | 10× | ||
|---|---|---|---|---|---|---|---|
| Christmas Island rat/Maclear’s rat (Merged data) | Rattus macleari | 2892096135 | 60.80977094 | 287 | 0.9515 | 0.9142 | 0.8793 |
| Christmas Island rat/Maclear’s rat (BGISeq data) | Rattus macleari | 1095698394 | 22.59070271 | 97 | 0.9365 | 0.8601 | 0.766 |
| Christmas Island rat/Maclear’s rat (HiSeq data) | Rattus macleari | 1794361241 | 38.17020817 | 205 | 0.937 | 0.8771 | 0.816 |
| Christmas Island rat/Maclear’s rat (MiSeq data) | Rattus macleari | 2036500 | 0.048860062 | 3 | 0.0444 | 0.0001 | 0.0001 |
| Norway brown rat (Simulative ancient DNA) | Rattus norvegicus | 2921567669 | 60.77177418 | 114 | 0.9919 | 0.9919 | 0.9919 |
| Norway brown rat (Simulative modern DNA) | Rattus norvegicus | 1616683907 | 60.79049948 | 83 | 0.9919 | 0.9919 | 0.9919 |
| Norway brown rat (Real modern DNA, five samples)a | Rattus norvegicus | 1555330238 | 60.83049687 | 205 | 0.9914 | 0.987 | 0.9759 |
| Norway brown rat (Real modern DNA, four samples)b | Rattus norvegicus | 1591467983 | 60.79035188 | 184 | 0.9912 | 0.9845 | 0.9593 |
| Himalayan field rat (seven samples) | Rattus nitidus | 2132796965 | 120.4604476 | 447 | 0.9848 | 0.9748 | 0.9664 |
| Himalayan field rat (three samples)c | Rattus nitidus | 1015013828 | 57.69764693 | 208 | 0.9805 | 0.9647 | 0.9502 |
| Himalayan field rat (four samples)d | Rattus nitidus | 1117783137 | 62.7628007 | 246 | 0.9815 | 0.9657 | 0.9512 |
| Asian house rat | Rattus tanezumi | 587134689 | 29.87708106 | 101 | 0.9276 | 0.8634 | 0.8002 |
| Black rat | Rattus rattus | 1152792528 | 42.48457157 | 139 | 0.9481 | 0.9098 | 0.8815 |
Norway brown rat (Real modern DNA, five samples): China1+Mali+AH2+BJ+Cambodia5
Norway brown rat (Real modern DNA, four samples): Mali+AH1+SD+Cambodia5
Himalayan field rat (three samples): NZ1+NZ2+WH3
Himalayan field rat (four samples): SG1+SG2+WH1+WH2
To test these hypotheses, we undertook several different analyses. First we used gargammel21 to generate in silico simulative modern (60.79×) and ancient data (60.77×) of the Norway brown rat and mapped them back to the Norway brown rat reference genome. The results showed that when mapped back to the reference genomes, both the simulative modern and ancient datasets covered over 99.19% of the reference genome. Second, we mapped two sets of real Norway brown rat sequencing data of 60.83× and 60.79× coverage to its reference genome, and found that both of them covered 99.14% and 99.12% of the genome, respectively (Table 1). The slight difference between the real and simulative modern data in terms of recovering genomic regions was the result of the genetic variation between the sequenced individuals and the reference genome.
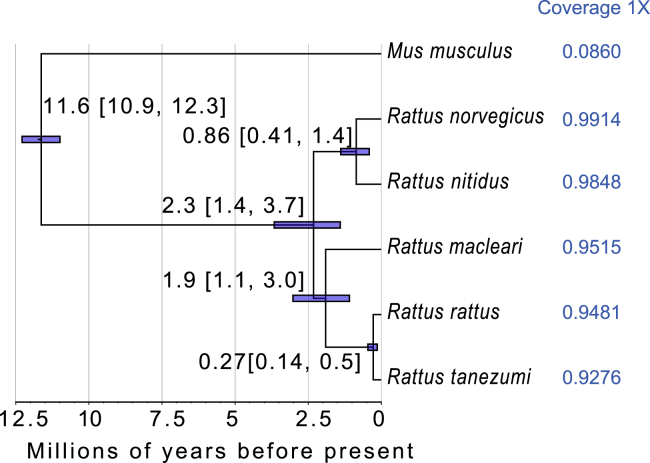
Third, we explored the relationship between depth of sequencing coverage and AT content and found that the regions in Norway rat genome with higher AT content did tend to have lower coverage (Figure S2), suggesting that PCR amplification bias may partly contribute to the problem, but not to the degree needed to explain the observations. Fourth, we explored the role of evolutionary divergence in reducing the mappability of sequence reads by obtaining the sequence datasets of three other Rattus species from public databases (the Himalayan field rat [Rattus nitidus], the Asian house rat [Rattus tanezumi], and the black rat),18,22,23 then mapping them to the Norway brown rat reference genome. Using this nuclear genome dataset, we both inferred the phylogenetic placement and evolutionary divergence times among the five Rattus species, and calculated the percent genome coverage for each species recovered after mapping to the Norway rat. Our results not only provide a new, nuclear genome-based estimate of the divergence times of the Christmas Island rat from other species, but more importantly show that although all five Rattus species share a last common ancestor only ca. 2.3 mya (Figure 1), the percentage genome coverage rapidly decreases to as low as 92.76% for R. tanezumi (Table 1).
Figure 1.
The phylogenetic placement and evolutionary timescale of R. norvegicus, Rattus nitidus, R. macleari, Rattus rattus, and Rattus tanezumi
Numbers following the species names indicate the coverage of genomic sequencing data for the corresponding species when mapped to the Norway brown rat reference genome. Related to Table S6.
In summary, the above analyses provide clear evidence that a major part of the 4.04% of the Norway rat genome that is not covered by Christmas Island rat sequences derives from evolutionary divergence, as opposed to the quality of the reference genome itself or damage to the ancient DNA templates. In light of this, an interesting question is, how representative would a hypothetically re-generated Christmas Island rat be of the authentic extinct form?
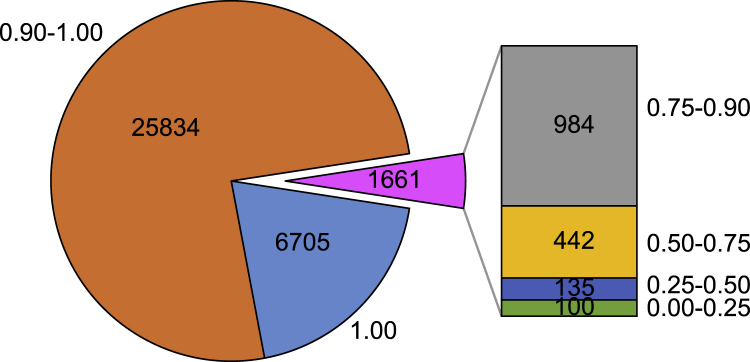
To answer this question, we explored the genomic distribution of the 128,423,913 bp of the Norway brown rat genome that was not covered by Christmas Island rat sequence data, and found that ca. one-quarter of it fell within gene regions (Table S2), thus implying that information is missing that would likely have functional consequences. We then calculated the coverage of each of the 34,200 genes annotated in the Norway rat reference genome, including 22,228 protein coding genes and 11,972 non-coding genes (Figure 2; Table S3). We found that 17,121 (50.19%) genes were covered at higher than 0.99 completeness. Almost all genes (83/86) encoding keratins or keratin-associated proteins, which are the key structural materials of hair and whiskers, have coverage higher than 90%. Additionally, all eight orthologs of the human round-ear phenotype-involved-genes (CEP57, ERF, MYH3, NALCN, PSMC3, TNNI2, TNNT3, and TPM2; https://hpo.jax.org/app/browse/term/HP:0100830) were found to be covered at higher than 97%. These results suggested that most of the long thick black hair, long dark whisker, and round ear phenotypes of the Christmas Island rat could likely be recreated if genome editing of a Norway rat was attempted. However, another 1,661, 677, 235, and 100 genes exhibited coverages lower than 0.90, 0.75, 0.50, and 0.25, respectively. And notably, 26 genes, including MAGEB18, PUF-like, five endogenous retrovirus group K members (ERVKs), eight snRNA, ten snoRNA, and one tRNA, were completely missed by Christmas Island rat data (Figure 2; Table S3). We speculate that the absent MAGEB18 and ERVKs genes may simply be due to the two species’ different evolutionary histories, while the 19 non-coding RNAs may be unrecoverable simply due to their very short lengths.
Figure 2.
Numbers of genes found at different coverage levels after mapping Christmas Island rat genomic sequencing data to the Norway brown rat reference genome
Coverage levels (on a scale of 0–1) are shown next to the figure. Related to Tables S2 and S3 and Figure S2.
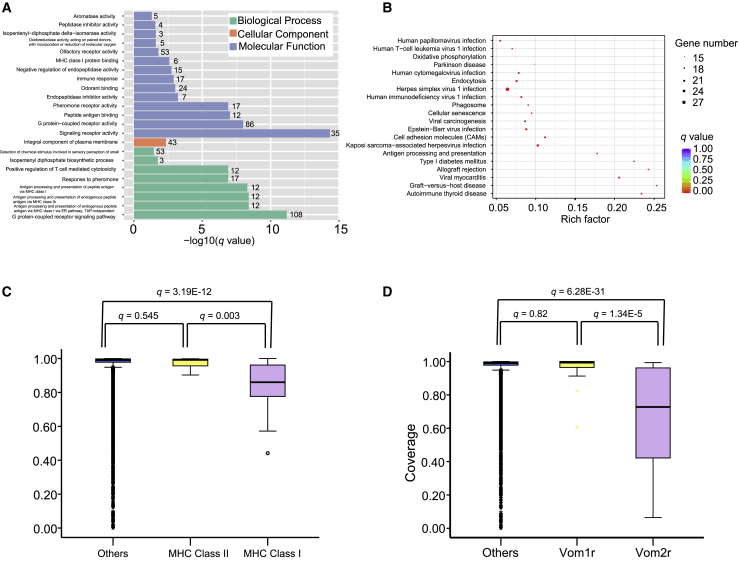
We furthermore found that these incompletely covered genes are not random representatives of the genome. Rather, genes that exhibit a coverage of lower than 0.9 are biased for GO/KEGG terms related to immune response (“autoimmune thyroid disease,” “antigen processing and presentation,” “herpes simplex virus 1 infection,” “MHC class I protein binding,” “immune response,” etc.) and olfaction (“olfactory receptor activity” and “odorant binding”) (Figures 3A and 3B; Tables S4 and S5). Additionally, even when ontology categories that appear at least superficially similar were compared, striking differences were observed. While the coverages of major histocompatibility complex (MHC) class I genes were significantly lower than other genes (q = 3.19E-12), MHC II yielded significantly higher coverage (higher than 0.9) (Figure S3A). The vomeronasal 2 receptor (Vom2r) genes, one of the olfactory receptor families associated with the detection of peptide pheromone,24 yielded significantly lower coverage than vomeronasal 1 receptor (Vom1r) genes, and indeed genes in other categories (Figures 3C and 3D).
Figure 3.
Annotation of genes with unrecoverable regions in of Christmas Island rat genome
(A) GO enrichment (q < 0.05) of Christmas Island rat genes obtained at coverage lower than 0.9. Numbers following bars: the number of genes; x axes: −log10(q value); y axes: the GO terms enriched in genes with coverage lower than 0.9.
(B) KEGG enrichment (q < 0.05) in genes with coverage lower than 0.9; x axes: rich factor, number of genes with coverage lower than 0.9/total genes in KEGG terms; y axes: the KEGG pathways enriched in genes with coverage lower than 0.9.
(C and D) The coverage of genomic sequencing data on major histocompatibility complex (MHC) and vomeronasal receptor (VoR) genes in the Norway brown rat reference genome.
Conclusion
Our results clearly demonstrate that, should genome editing (ignoring current technical limitations) be applied to the Norway brown rat in order to recreate the Christmas Island rat through editing every identifiable difference, a remarkable number of genes would either only partially resemble the extinct form, or in the worst case, remain 100% Norway rat-like. Naturally given that ultimately, evolutionary divergence is driving this phenomenon, the use of a more closely related species (e.g., the black rat) would lead to some improvements in the amount of the genome reconstructed, although any gains are likely to be small. For example, mapping of our Christmas Island rat data to the black rat allows recovery of a maximum of 96.56% of its genome, compared to 95.15% of the Norway rat genome. Furthermore, it is clear that the non-random distribution of these genes would have consequences for the resulting biology of the reconstructed animals, potentially precluding reintroduction of the species to its original environment. For example, given the role of olfaction in many critical behaviors; such as foraging and food selection, detecting predators,25 and mate choice;24 a reconstructed Christmas Island rat would lack attributes likely critical to surviving in its natural or natural-like environment. In contrast, however, in light of the hypothesis that the Christmas Island rat was driven extinct due to an infectious disease introduced from black rats,5,26 one might hypothesize that maintaining the immune genes of the Norway rat could even have some potential benefit.
Current de-extinction work is focused on species such as Mammuthus primigenius, the woolly mammoth (μ = 3.83 × 10−8, estimated generation time = 31 years),27 and Ectopistes migratorius, the passenger pigeon (μ = 5.68 × 10−9, estimated generation time = 4 years) (Table S6),28 and we suspect it is unfortunately unlikely that serious efforts will ever be attempted to bring back a rat species. Nevertheless, by using, as an example here, closely related extinct and extant rat species, we highlight that the scale of the challenge will only multiply as the evolutionary divergence between extinct and living species increases.9 In this context we note that the genomic divergence between the woolly mammoth and Asian elephant (Elephas maximus) is similar to that between the Christmas Island rat and the Norway brown rat, while the genomic divergence between the passenger pigeon and band-tailed pigeons (Patagioenas fasciata) is much larger (2.24 times) (Table S6). As divergence relates not only to absolute time, but generation time, ideally analyses such as ours should be done on a case-by-case basis. Therefore, we hope that the approach demonstrated here may offer a framework that others can consider when exploring the viability of other proposed de-extinction projects.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Dry preserved skin samples of the Christmas Island rat (Rattus macleari) | Oxford University Museum of Natural History | 18844 |
| Dry preserved skin samples of the Christmas Island rat (Rattus macleari) | Oxford University Museum of Natural History | 18845 |
| Deposited data | ||
| Christmas Island rat (Rattus macleari) resequencing datasets | This study; ENA (https://www.ebi.ac.uk/) | SAMEA12813846 (18844) SAMEA12813847 (18845) |
| Reference genome of Norway brown rat | NCBI website (https://www.ncbi.nlm.nih.gov/assembly/) | GCF_015227675.2 |
| Reference genome of black rat | NCBI website (https://www.ncbi.nlm.nih.gov/assembly/) | GCF_011064425.1 |
| Reference genome of house mouse | NCBI website (https://www.ncbi.nlm.nih.gov/assembly/) | GCF_000001635.27 |
| Norway brown rat resequencing datasets | CNCB-NGDC (http://gsa.big.ac.cn/) | CRX019583 (Mali), CRX019522 (Cambodia5), CRX019633 (China1), CRX019515 (AH1), CRX019516 (AH2), CRX019517 (BJ1) and CRX019639 (SD1) |
| Black rat resequencing dataset #1 | CNCB-NGDC (http://gsa.big.ac.cn/) | CRX019632 |
| Black rat resequencing dataset #2 | ENA (https://www.ebi.ac.uk/) | SAMEA2051945 |
| Black rat resequencing dataset #3 | NCBI (https://www.ncbi.nlm.nih.gov/sra/) | SRX9009079 |
| Himalayan field rat resequencing datasets | NCBI (https://www.ncbi.nlm.nih.gov/sra/) | SAMN05425704 (NZ2), SAMN05425705 (SG1) SAMN05425706 (SG2), SAMN05425709 (NZ1), SAMN05425641 (WH1), SAMN05425642 (WH2), and SAMN05425643 (WH3). |
| Asian house rat resequencing dataset | NCBI (https://www.ncbi.nlm.nih.gov/sra/) | SAMN05425710 |
| House mouse resequencing dataset | NCBI (https://www.ncbi.nlm.nih.gov/sra/) | SRX10650663 |
| Software and Algorithms | ||
| AdapterRemoval v2.3.1 | Schubert et al., 201629 | https://adapterremoval.readthedocs.io/en/stable/ |
| bwa | Li and Durbin30 | http://bio-bwa.sourceforge.net/ |
| Paleomix v1.3.2 | Schubert et al., 201420 | https://paleomix.readthedocs.io/en/stable/ |
| mapDamage v2.2.1 | Jónsson et al.31 | https://github.com/ginolhac/mapDamage/ |
| MEGA X | Kumar et al.32 | https://www.megasoftware.net/ |
| samtools v1.9 | Li et al.33 | https://github.com/samtools/samtools |
| bedtools v2.29.0 | Quinlan34 | |
| bcftools v1.9 | Li et al.33 | https://github.com/samtools/bcftools |
| seqkit v0.16.1 | Shen et al.35 | https://github.com/shenwei356/seqkit/tree/v0.16.1 |
| gargammel | Renaud et al.21 | https://grenaud.github.io/gargammel/ |
| KOBAS 3.0 | Bu et al.36 | http://bioinfo.org/kobas |
| MAFFT v7.4 | Katoh et al.37 | https://mafft.cbrc.jp/alignment/software/ |
| IQ-TREE v1.6 | Minh et al.38 | http://www.iqtree.org/ |
| PAML v4.8 | Yang39 | https://github.com/abacus-gene/paml |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jianqing Lin (linjianqing@stu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Historic specimens
The specimens, ID numbers 18844 and 18845 from the collections of the Oxford University Museum of Natural History, are dried skin samples collected on Christmas Island between 1900-1902, and originally sampled for genetic analyses for a prior study that explored whether the species’ extinction could be ascribed to introduced pathogens.5
Method details
Ancient DNA methods
DNA was extracted from ca 1x1 cm2 of the skin samples using the digestion buffer of Gilbert et al.40 combined with a silica-based purification following Dabney et al.41
Library construction and sequencing
The specimen 18844 was used for Illumina (HiSeq/Miseq) and 18845 for BGISEQ-500 library construction. Extracted DNA was converted into both Illumina and BGISEQ-500 compatible libraries using blunt-end protocols, with both sequenced with 100 bp SE chemistry. Illumina library construction used the NEBNext 6070L kit, following Wales et al. (2015),42 while BGISEQ library construction followed Mak et al. (2017).43 In total 2,694,229,632 reads of Illumina and 2,754,802,455 reads of BGISeq data were generated from these libraries.
Mapping and calculation of coverage/depth
Before mapping, the last 10 bases of each read from the BGISEQ-500 sequencing perform were removed because they represent the index. Subsequently the sequence data was trimmed and mapped against the reference genomes of the Norway brown rat (mRatBN7.2, GCA_015227675.2) and black rat, using Paleomix v1.3.2.20 Specifically, the adapters in BGISEQ-500 data (–adapter1: AAGTCGGAGGCCAAGCGGTCTTAGGAAGACAA;–adapter2: GAACGACATGGCTACGATCCGACTT) and Illumina data (–adapter1: AGATCGGAAGAGCACACGTCTGAACTCCAGTCACNNNNNNATCTCGTATGCCGTCTTCTGCTTG;–adapter2: AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTATCATT) were trimmed by AdapterRemoval v2.3.1 with default setting.29 bwa v0.7.17 (the backtrack algorithm) was used to map the reads to the genome with options “MinQuality: 0; FilterUnmappedReads: yes; UsedSeed: no.”30 In the mapping step, we used the same setting for modern and ancient samples to avoid introducing any biases. For the ancient samples, mapDamage v2.2.131 was used to estimate the ancient DNA damage parameters, to both validate that the data is truly ancient and to provide input values for the gargammel simulations. We recovered the mtDNA consensus sequences (see below) from each of the Christmas Island specimens, and compared them using MEGA X,32 and found that they exhibited very little genetic distance (0.002890), thus we elected to merge the two sequence datasets to obtain the final high coverage dataset. The bam files generated from each species were merged into one bam file using samtools v1.9.33 We used paleomix coverage and Paleomix depths to calculate the coverage and depth histogram for a bam file. The “bedtools coverage” command in bedtools v2.29.0 was used to calculate the coverage rate of each gene in the Norway rat genome.34 The “bedtools nuc command in bedtools v2.29.0 was used to calculate the base composition of each 100-bp window across chromosome NC_051336.1 of the Norway rat genome.34
Consensus genome
We identified the SNPs in each of the rat/mouse species’ genomes, and replaced them in the Norway brown rat reference genome to create the consensus genomes using the bam file from one sample of each species and samtools v1.9 and bcftools v1.9.33 The “seqkit fq2fa” command in seqkit v0.16.1 was used to converse fastq file into fasta format.35
Quantification and statistical analysis
Sequence simulation
We used gargammel,21 a DNA sequence simulator, to generate simulative modern and simulative ancient Norway brown rat data. The simulated reads were set to be single end and 100 bp in length (consistent with the BGISEQ-500 data). We set the overall raw data coverage of the modern data 62.63 × and that of ancient data 63.59 × to ensure that the coverage of both the modern and ancient data to be about 60.81 × , consistent with that of the Christmas Island rat. For the modern data, the fixed fragment length (-l) is 100 bp. For the ancient data, the size frequency file (f) and the miscorporation file (-mapdamage) input values were taken from the estimates made by mapDamage on the BGISEQ-500 Christmas Island rat data.
GO and KEGG annotation and enrichment
GO term and KEGG pathway enrichment analyses were carried out using the KOBAS 3.0 web server.36 The Statistical test method was Fisher’s Exact Test and the false discovery rate (FDR) correction method was Benjamini and Hochberg. GO terms and KEGG pathways with an FDR (q value) < 0.05 were regarded as significantly enriched.
Phylogenetics and molecular dating of Rattus
We inferred the phylogenetic placement and evolutionary timescale of five rat species using a set of 3,095 loci regions with 1000 nucleotides each (3,095,000 nucleotides). Loci were first identified by mapping the genomic data for five species of rats (R. macleari, R. rattus, R. tanezumi, R. norvegicus, and R. nitidus) and the mouse (M. musculus) to the reference genome of R. norvegicus. While it is ideal to use outgroup species as reference, we found that using the mouse as a reference led to a near-complete lack of phylogenetic signal among rat species. This can be explained by the substantial distance of rat species to the mouse, relative to the distances within rat species. This difference leads to a bias toward excessively slowly-evolving nucleotide substitutions that lack information about very recent divergences. Therefore, we focused on the data using the Norway rat genome as a reference. Future research using a close outgroup relative as reference would be a valuable contribution. We randomly extracted contiguous windows with 100 Kb each from 1 Mb window in the genomes. We then performed automated multiple sequence alignment using MAFFT v7.4,37 and randomly extracted 1 Kb windows from alignments, in order to minimize the impact of recombination breakpoints on the data.44 Maximum likelihood phylogenetic searches were performed for each locus under a GTR+R4 substitution model45 as implemented in IQ-TREE v1.6.38
Data selection and individual locus tree inference was followed by two methods of species tree inference. First, we concatenated our loci and performed maximum likelihood phylogenetic inference on the concatenated dataset under the GTR+R10 substitution model using IQ-TREE. Approximate likelihood ratio tests (aLRT) per branch were used as branch supports with 1,000 bootstrap replicates.46 Second, we inferred the species tree under the multi-species coalescent, used the individual locus trees as input to the summary coalescent method implemented in ASTRAL-III.47 Local posterior probabilities were taken as branch supports following the multispecies coalescent analysis.48 Both methods of species tree inference led to identical tree topologies and maximal statistical supports for all branches. Most branches were supported by nearly the whole set of loci and nucleotides. The single exception was the placement of the Christmas Island rat, which was supported by 60.3% of gene trees and 53.8% of nucleotide sites. This low concordance across the data is likely driven by short times between divergence events involving these taxa, and therefore large amounts of incomplete lineage sorting in the data.
Molecular dating was performed by assuming our inferred species tree topology and a further subset of 1,407 loci (1,407,000 nucleotides). Loci were retained for molecular dating to minimize rate variation among lineages,49 and gene tree discordance.50 The loci included were those that led to locus trees with coefficients of variation in root-to-tip lengths (non-clocklikeness) < 0.1, and Robinson-Foulds distances to our inferred species tree £2. Selected loci were then concatenated for molecular dating. A single time-calibration was used at the root of the tree, taking the mouse-rat divergence to have occurred between 11 and 12.3 Mya following evidence from palaeontology (genus Prognomys)51 and phylogenetics.52 The prior distribution for this root calibration was a uniform with soft maximum and minimum bounds, with a 2.5% prior probability of the age occurring beyond each bound. Bayesian molecular dating was performed using a GTR+Γ substitution model and an uncorrelated-gamma relaxed clock model as implemented in MCMCtree, using PAML v4.8.39 Approximate Bayesian computation was implemented to improve the efficiency of the analysis.53 The posterior distribution was approximated using Markov chain Monte Carlo (MCMC), starting with a burn-in phase of 105 MCMC steps, and then drawing samples every 103 MCMC steps over a total of 107 steps. Convergence to the stationary distribution was verified by comparing parameter estimates from four independent runs, and confirming that effective sample sizes were above 200 for sampled parameters.
Additional resources
The reference genome of Norway brown rat (R. norvegicus), black rat (Rattus rattus) and house mouse (Mus musculus) was downloaded from the NCBI website (assembly accession: GCA_015227675.2, GCF_011064425.1 and GCF_000001635.27). Additional Norway brown rat resequencing datasets were downloaded from CNCB-NGDC (http://gsa.big.ac.cn/) under accession IDs CRX019583 (Mali), CRX019522 (Cambodia5), CRX019633 (China1), CRX019515 (AH1), CRX019516 (AH2), CRX019517 (BJ1) and CRX019639 (SD1).18 The black rat resequencing datasets were downloaded from CNCB-NGDC under Accession ID CRX019632,18 from EBI under accession ID SAMEA205194522 and from NCBI under accession ID SRX9009079. The Himalayan field rat (R. nitidus) sequence data were downloaded from NCBI under accession ID SAMN05425704 (NZ2), SAMN05425705 (SG1), SAMN05425706 (SG2), SAMN05425709 (NZ1), SAMN05425641 (WH1), SAMN05425642 (WH2), and SAMN05425643 (WH3).23 The Asian house rat (R. tanezumi) sequence data were downloaded from NCBI under accession ID SAMN05425710.23 The house mouse sequence data were downloaded from NCBI under accession ID SRX10650663.
Acknowledgments
The authors acknowledge the Oxford University Museum of Natural History for originally providing the loan of the materials to ADG, and Mikkel-Holger Strander Sinding for ongoing discussion about the potential and challenges of de-extinction. We also acknowledge ERC Consolidator Award 681396 “Extinction Genomics,” Danish National Research Foundation Award DNRF143, Program of Study Abroad for Postdoctoral Researcher of College of Life Sciences, ZJU, and Scientific Research Foundation for Talents, STU (NTF21026) for funding this research.
Author contributions
Designed the study: M.T.P.G. Generated palaeogenomic data: C.C., O.S., and M.M.C. Analyzed data: J.L., D.D., J.N., and D.R. Provided additional computational and laboratory guidance/support: S.G. and G.Z. Provided samples and context: A.D.G. and R.M. Provided modern reference datasets and wrote the paper: J.L. and M.T.P.G. with input from all other authors.
Declaration of interests
M.T.P.G. is on the advisory board of Current Biology.
Published: March 9, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2022.02.027.
Contributor Information
Jianqing Lin, Email: linjianqing@stu.edu.cn.
M. Thomas P. Gilbert, Email: tgilbert@sund.ku.dk.
Supplemental information
Data and code availability
Raw sequencing reads from whole genome sequencing of the two historic Christmas Island rat samples have been deposited at the European Nucleotide Archive (ENA; study accession number PRJEB50610).
References
- 1.Richmond D.J., Sinding M.H.S., Gilbert M.T.P. The potential and pitfalls of de-extinction. Zool. Scr. 2016;45:22–36. [Google Scholar]
- 2.Shapiro B., Seddon P. Pathways to de-extinction: how close can we get to resurrection of an extinct species? Funct. Ecol. 2016;31:996–1002. [Google Scholar]
- 3.Andrews C.W. Mammalia. A monograph of Christmas Island (Indian Ocean) London: British Museum. 1900 [Google Scholar]
- 4.Pickering J., Norris C.A. New evidence concerning the extinction of the endemic murid Rattus macleari from Christmas Island, Indian Ocean. Australian Mammalogy. 1996;19:19–25. [Google Scholar]
- 5.Wyatt K.B., Campos P.F., Gilbert M.T., Kolokotronis S.O., Hynes W.H., DeSalle R., Ball S.J., Daszak P., MacPhee R.D., Greenwood A.D. Historical mammal extinction on Christmas Island (Indian Ocean) correlates with introduced infectious disease. PLoS ONE. 2008;3:e3602. doi: 10.1371/journal.pone.0003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pääbo S. Ancient DNA: extraction, characterization, molecular cloning, and enzymatic amplification. Proc. Natl. Acad. Sci. USA. 1989;86:1939–1943. doi: 10.1073/pnas.86.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poinar H.N., Schwarz C., Qi J., Shapiro B., Macphee R.D., Buigues B., Tikhonov A., Huson D.H., Tomsho L.P., Auch A., et al. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 8.Kircher M. Analysis of high-throughput ancient DNA sequencing data. Methods Mol. Biol. 2012;840:197–228. doi: 10.1007/978-1-61779-516-9_23. [DOI] [PubMed] [Google Scholar]
- 9.Shapiro B., Hofreiter M. A paleogenomic perspective on evolution and gene function: new insights from ancient DNA. Science. 2014;343:1236573. doi: 10.1126/science.1236573. [DOI] [PubMed] [Google Scholar]
- 10.Campa C.C., Weisbach N.R., Santinha A.J., Incarnato D., Platt R.J. Multiplexed genome engineering by Cas12a and CRISPR arrays encoded on single transcripts. Nat. Methods. 2019;16:887–893. doi: 10.1038/s41592-019-0508-6. [DOI] [PubMed] [Google Scholar]
- 11.Leonardi M., Librado P., Der Sarkissian C., Schubert M., Alfarhan A.H., Alquraishi S.A., Al-Rasheid K.A., Gamba C., Willerslev E., Orlando L. Evolutionary patterns and processes: Lessons from ancient DNA. Syst. Biol. 2017;66:e1–e29. doi: 10.1093/sysbio/syw059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrett Vieira F., Samaniego Castruita J.A., Gilbert M.T.P. Using in silico predicted ancestral genomes to improve the efficiency of paleogenome reconstruction. Ecol. Evol. 2020;10:12700–12709. doi: 10.1002/ece3.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.SSC I . IUCN Species Survival Commission; 2016. IUCN guiding principles on creating proxies of extinct species for conservation benefit. Version 1.0. [Google Scholar]
- 14.Duchêne D.A., Tong K.J., Foster C.S.P., Duchêne S., Lanfear R., Ho S.Y.W. Linking branch lengths across sets of loci provides the highest statistical support for phylogenetic inference. Mol. Biol. Evol. 2020;37:1202–1210. doi: 10.1093/molbev/msz291. [DOI] [PubMed] [Google Scholar]
- 15.Prendergast J.G., Campbell H., Gilbert N., Dunlop M.G., Bickmore W.A., Semple C.A. Chromatin structure and evolution in the human genome. BMC Evol. Biol. 2007;7:72. doi: 10.1186/1471-2148-7-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rowe K.C., Aplin K.P., Baverstock P.R., Moritz C. Recent and rapid speciation with limited morphological disparity in the genus Rattus. Syst. Biol. 2011;60:188–203. doi: 10.1093/sysbio/syq092. [DOI] [PubMed] [Google Scholar]
- 17.Robins J.H., McLenachan P.A., Phillips M.J., McComish B.J., Matisoo-Smith E., Ross H.A. Evolutionary relationships and divergence times among the native rats of Australia. BMC Evol. Biol. 2010;10:375. doi: 10.1186/1471-2148-10-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng L., Ming C., Li Y., Su L.Y., Su Y.H., Otecko N.O., Dalecky A., Donnellan S., Aplin K., Liu X.H., et al. Out of southern east Asia of the brown rat revealed by large-scale genome sequencing. Mol. Biol. Evol. 2018;35:149–158. doi: 10.1093/molbev/msx276. [DOI] [PubMed] [Google Scholar]
- 19.Howe K., Dwinell M., Shimoyama M., Corton C., Betteridge E., Dove A., Quail M.A., Smith M., Saba L., Williams R.W., et al. The genome sequence of the Norway rat, Rattus norvegicus Berkenhout 1769. Wellcome Open Res. 2021;6:118. doi: 10.12688/wellcomeopenres.16854.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schubert M., Ermini L., Der Sarkissian C., Jónsson H., Ginolhac A., Schaefer R., Martin M.D., Fernández R., Kircher M., McCue M., et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 2014;9:1056–1082. doi: 10.1038/nprot.2014.063. [DOI] [PubMed] [Google Scholar]
- 21.Renaud G., Hanghøj K., Willerslev E., Orlando L. gargammel: a sequence simulator for ancient DNA. Bioinformatics. 2017;33:577–579. doi: 10.1093/bioinformatics/btw670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deinum E.E., Halligan D.L., Ness R.W., Zhang Y.H., Cong L., Zhang J.X., Keightley P.D. Recent evolution in Rattus norvegicus is shaped by declining effective population size. Mol. Biol. Evol. 2015;32:2547–2558. doi: 10.1093/molbev/msv126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teng H., Zhang Y., Shi C., Mao F., Cai W., Lu L., Zhao F., Sun Z., Zhang J. Population genomics reveals speciation and introgression between Brown Norway Rats and their sibling species. Mol. Biol. Evol. 2017;34:2214–2228. doi: 10.1093/molbev/msx157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brennan P.A., Zufall F. Pheromonal communication in vertebrates. Nature. 2006;444:308–315. doi: 10.1038/nature05404. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi L.K., Nakashima B.R., Hong H., Watanabe K. The smell of danger: a behavioral and neural analysis of predator odor-induced fear. Neurosci. Biobehav. Rev. 2005;29:1157–1167. doi: 10.1016/j.neubiorev.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 26.Green P. Mammal extinction by introduced infectious disease on Christmas Island (Indian Ocean): the historical context. Australian Zoologist. 2014;37:1–14. [Google Scholar]
- 27.Palkopoulou E., Mallick S., Skoglund P., Enk J., Rohland N., Li H., Omrak A., Vartanyan S., Poinar H., Götherström A., et al. Complete genomes reveal signatures of demographic and genetic declines in the woolly mammoth. Curr. Biol. 2015;25:1395–1400. doi: 10.1016/j.cub.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray G.G.R., Soares A.E.R., Novak B.J., Schaefer N.K., Cahill J.A., Baker A.J., Demboski J.R., Doll A., Da Fonseca R.R., Fulton T.L., et al. Natural selection shaped the rise and fall of passenger pigeon genomic diversity. Science. 2017;358:951–954. doi: 10.1126/science.aao0960. [DOI] [PubMed] [Google Scholar]
- 29.Schubert M., Lindgreen S., Orlando L. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes. 2016;9:88. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jónsson H., Ginolhac A., Schubert M., Johnson P.L., Orlando L. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29:1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinlan A.R. BEDTools: The Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47 doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen W., Le S., Li Y., Hu F. SeqKit: A cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE. 2016;11:e0163962. doi: 10.1371/journal.pone.0163962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bu D., Luo H., Huo P., Wang Z., Zhang S., He Z., Wu Y., Zhao L., Liu J., Guo J., et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021;49(W1):W317–W325. doi: 10.1093/nar/gkab447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., von Haeseler A., Lanfear R. IQ-TREE 2: New Models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert M.T., Tomsho L.P., Rendulic S., Packard M., Drautz D.I., Sher A., Tikhonov A., Dalén L., Kuznetsova T., Kosintsev P., et al. Whole-genome shotgun sequencing of mitochondria from ancient hair shafts. Science. 2007;317:1927–1930. doi: 10.1126/science.1146971. [DOI] [PubMed] [Google Scholar]
- 41.Dabney J., Knapp M., Glocke I., Gansauge M.T., Weihmann A., Nickel B., Valdiosera C., García N., Pääbo S., Arsuaga J.L., Meyer M. Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragments. Proc. Natl. Acad. Sci. USA. 2013;110:15758–15763. doi: 10.1073/pnas.1314445110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wales N., Carøe C., Sandoval-Velasco M., Gamba C., Barnett R., Samaniego J.A., Madrigal J.R., Orlando L., Gilbert M.T. New insights on single-stranded versus double-stranded DNA library preparation for ancient DNA. Biotechniques. 2015;59:368–371. doi: 10.2144/000114364. [DOI] [PubMed] [Google Scholar]
- 43.Mak S.S.T., Gopalakrishnan S., Carøe C., Geng C., Liu S., Sinding M.S., Kuderna L.F.K., Zhang W., Fu S., Vieira F.G., et al. Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience. 2017;6:1–13. doi: 10.1093/gigascience/gix049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendes F.K., Livera A.P., Hahn M.W. The perils of intralocus recombination for inferences of molecular convergence. Philos Trans R Soc Lond B Biol Sci. 2019;374 doi: 10.1098/rstb.2018.0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anisimova M., Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 47.Zhang C., Rabiee M., Sayyari E., Mirarab S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinformatics. 2018;19(Suppl 6):153. doi: 10.1186/s12859-018-2129-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sayyari E., Mirarab S. Fast coalescent-based computation of local branch support from quartet frequencies. Mol. Biol. Evol. 2016;33:1654–1668. doi: 10.1093/molbev/msw079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doyle V.P., Young R.E., Naylor G.J., Brown J.M. Can we identify genes with increased phylogenetic reliability? Syst. Biol. 2015;64:824–837. doi: 10.1093/sysbio/syv041. [DOI] [PubMed] [Google Scholar]
- 50.Mendes F.K., Hahn M.W. Gene tree discordance causes apparent substitution rate variation. Syst. Biol. 2016;65:711–721. doi: 10.1093/sysbio/syw018. [DOI] [PubMed] [Google Scholar]
- 51.Benton M.J., Donoghue P.C. Paleontological evidence to date the tree of life. Mol. Biol. Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- 52.Robins J.H., McLenachan P.A., Phillips M.J., Craig L., Ross H.A., Matisoo-Smith E. Dating of divergences within the Rattus genus phylogeny using whole mitochondrial genomes. Mol. Phylogenet. Evol. 2008;49:460–466. doi: 10.1016/j.ympev.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 53.Thorne J.L., Kishino H., Painter I.S. Estimating the rate of evolution of the rate of molecular evolution. Mol. Biol. Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing reads from whole genome sequencing of the two historic Christmas Island rat samples have been deposited at the European Nucleotide Archive (ENA; study accession number PRJEB50610).