ABSTRACT
Sp100 (speckled protein 100 kDa) is a constituent component of nuclear structure PML (promyelocytic leukemia) bodies, playing important roles in mediating intrinsic and innate immunity. The Sp100 gene encodes four isoforms with distinct roles in the transcriptional regulation of both cellular and viral genes. Since Sp100 is a primary intranuclear target of infected-cell protein 0 (ICP0), an immediate early E3 ligase encoded by herpes simplex virus 1 (HSV-1), previous investigations attempting to analyze the functions of individual Sp100 variants during HSV-1 infection mostly avoided using a wild-type virus. Therefore, the role of Sp100 under natural infection by HSV-1 remains to be clarified. Here, we reappraised the antiviral capacity of four Sp100 isoforms during infection by a nonmutated HSV-1, examined the molecular behavior of the Sp100 protein in detail, and revealed the following intriguing observations. First, Sp100 isoform A (Sp100A) inhibited wild-type HSV-1 propagation in HEp-2, Sp100−/−, and PML−/− cells. Second, endogenous Sp100 is located in both the nucleus and the cytoplasm. During HSV-1 infection, the nuclear Sp100 level decreased drastically upon the detection of ICP0 in the same subcellular compartment, but cytosolic Sp100 remained stable. Third, transfected Sp100A showed subcellular localizations similar to those of endogenous Sp100 and matched the protein size of endogenous cytosolic Sp100. Fourth, HSV-1 infection induced increased secretion of endogenous Sp100 and ectopically expressed Sp100A, which copurified with extracellular vesicles (EVs) but not infectious virions. Fifth, the Sp100A level in secreting cells positively correlated with its level in EVs, and EV-associated Sp100A restricted HSV-1 in recipient cells.
IMPORTANCE Previous studies show that the PML body component Sp100 protein is immediately targeted by ICP0 of HSV-1 in the nucleus during productive infection. Therefore, extensive studies investigating the interplay of Sp100 isoforms with HSV-1 were conducted using a mutant virus lacking ICP0 or in the absence of infection. The role of Sp100 variants during natural HSV-1 infection remains blurry. Here, we report that Sp100A potently and independently inhibited wild-type HSV-1 and that during HSV-1 infection, cytosolic Sp100 remained stable and was increasingly secreted into the extracellular space, in association with EVs. Furthermore, the Sp100A level in secreting cells positively correlated with its level in EVs and the anti-HSV-1 potency of these EVs in recipient cells. In summary, this study implies an active antiviral role of Sp100A during wild-type HSV-1 infection and reveals a novel mechanism of Sp100A to restrict HSV-1 through extracellular communications.
KEYWORDS: PML, Sp100A, cytosolic Sp100, HSV-1, EVs, extracellular vesicle, subcellular localization
INTRODUCTION
Speckled protein 100 kDa (Sp100) is a constituent component of multifunctional promyelocytic leukemia protein (PML) nuclear bodies (NBs) (also known as ND10), playing important roles in mediating both intrinsic and innate immunity (1). The Sp100 gene, located on human chromosome 2, is a typical target of interferon (IFN), containing two IFN-responding elements in the promoter region: the IFN-stimulated response element (ISRE, -ACTTTCACTTCTCT-) and Gamma Interferon Activation Site, (GAS, -TTCCAGGAA-) domains (2, 3). Evidence from two aspects implies that Sp100 plays a critical antiviral role against infection. First, various RNA and DNA viruses employ diverse strategies to modify, target, or disperse Sp100 from its normal subcellular locations (4–18). Second, the depletion of Sp100 protein significantly promotes the progression of both wild-type and attenuated viruses (19–26). The Sp100 gene encodes a major isoform, Sp100A, and three longer isoforms, Sp100B, Sp100C, and Sp100HMG, through RNA splicing of a single primary transcript (27). Sp100 splice variants share an N terminus of 477 amino acids (aa) in length, comprising a PML NB-targeting signal (aa 29 to 152), a dimerization domain (DD) (aa 1 to 182), a heterochromatin protein 1 (HP1) binding domain (aa 287 to 334), a transcription activation domain (aa 333 to 407), and a nuclear localization signal (NLS) sequence (aa 444 to 450) (1, 28–31), and differ in their C termini (27, 32, 33). While Sp100B, Sp100C, and Sp100HMG harbor a SAND (Sp100, AIRE-1, NucP41/75, and DEAF-1) domain, which enables direct binding to DNA, the major isoform Sp100A contains no known DNA-interacting domain and is thus believed to be brought to DNA by association with other chromatin binding proteins such as HP1 and Bright, etc. (27, 34). In correlation with their various C-terminal domains, Sp100 isoforms have been reported to possess divergent spatiotemporal behaviors and similar or opposite transcriptional regulatory functions (27, 28, 32, 33, 35–40).
Nuclear Sp100 protein is a primary host target during productive infection by herpes simplex virus 1 (HSV-1), a highly prevalent human pathogen and prototype member of the subfamily Alphaherpesvirinae, family Herpesviridae (5, 41). HSV-1 encodes an immediate early protein, infected-cell protein 0 (ICP0), that dismantles PML NBs and degrades SUMOylated Sp100 through a UbcH5a-mediated proteasome-dependent pathway, leaving only a portion of unSUMOylated Sp100A untargeted (4–6, 8, 42). SUMOylation is an important mechanism to regulate the functions of the Sp100 protein and mediate its differential interaction with the SUMO-interacting motif (SIM)-containing proteins in the nucleus (12, 33, 43–48). For instance, SUMO modification of Sp100 has been reported to strongly correlate with the nuclear entry of the protein (28) and to enhance its in vitro interaction with HP1 and, hence, the protein’s ability to suppress transcription (33). Enthusiasm on Sp100 in the field of HSV-1 is fueled by accruing evidence implying that Sp100 protein affects productive infection of the virus with isoform-dependent differences. In brief, studies have shown that (i) IFN induces shifted splicing of Sp100 isoforms in a cell type-dependent manner (27, 49); (ii) the SAND domain-containing Sp100 isoforms Sp100B, Sp100C, and Sp100HMG effectively suppress transcription from the ICP0 promoter of HSV-1, while Sp100A promotes expression (49); (iii) Sp100A drives chromatin decondensation on a cytomegalovirus (CMV) promoter, while the SAND domain-containing Sp100 isoforms function opposingly (27, 36, 49); and (iv) Sp100A accumulates on viral DNA of HSV-1 in a SUMO-independent but SIM-dependent manner during infection (50, 51). Sp100 is suspected to participate in multiple stages of the lytic replication cycle of HSV-1 through distinct mechanisms (50, 52–57), and important observations argue for a beneficial role of unSUMOylated Sp100A during HSV-1 propagation (41). It should be noted that these studies were performed using HSV-1 mutant viruses lacking a functional ICP0 protein or conducted in the absence of infection.
Meanwhile, previous work by us and others shows that the depletion or knockdown of Sp100 significantly promotes wild-type HSV-1 infection (53, 58), implying that Sp100 isoforms in sum serve as host-restrictive factors of HSV-1. Whether Sp100 is a friend, an enemy, or a frenemy during natural HSV-1 infection needs to be clarified. We thus set out to investigate the roles of individual Sp100 variants during productive infection by wild-type HSV-1 and examine the molecular behavior of Sp100 in detail. By utilizing overexpression cell lines and recombinant HSV-1 carrying the cDNA of Sp100 isoform A, we made the following observations: (i) Sp100A significantly inhibited HSV-1 infection in HEp-2 and Sp100−/− cells; (ii) combined evidence that both endogenous Sp100 and transfected Sp100A were detected in the cytosol and that cytosolic Sp100 and Sp100A had similar electrophoretic mobilities in polyacrylamide gels indicated that the endogenous cytosolic Sp100 was Sp100 isoform A; (iii) during HSV-1 infection, nuclear Sp100 was degraded soon after the nuclear entry of ICP0, while the protein level of cytosolic Sp100 remained stable; (iv) HSV-1 infection induced the secretion of endogenous Sp100 and ectopically overexpressed Sp100A, in association with noninfectious extracellular vesicles (EVs); and (v) the Sp100A level in secreting cells positively correlated with its level in EVs, and EV-associated Sp100A restricted HSV-1 in recipient cells.
RESULTS
Sp100A localized differently from other Sp100 isoforms.
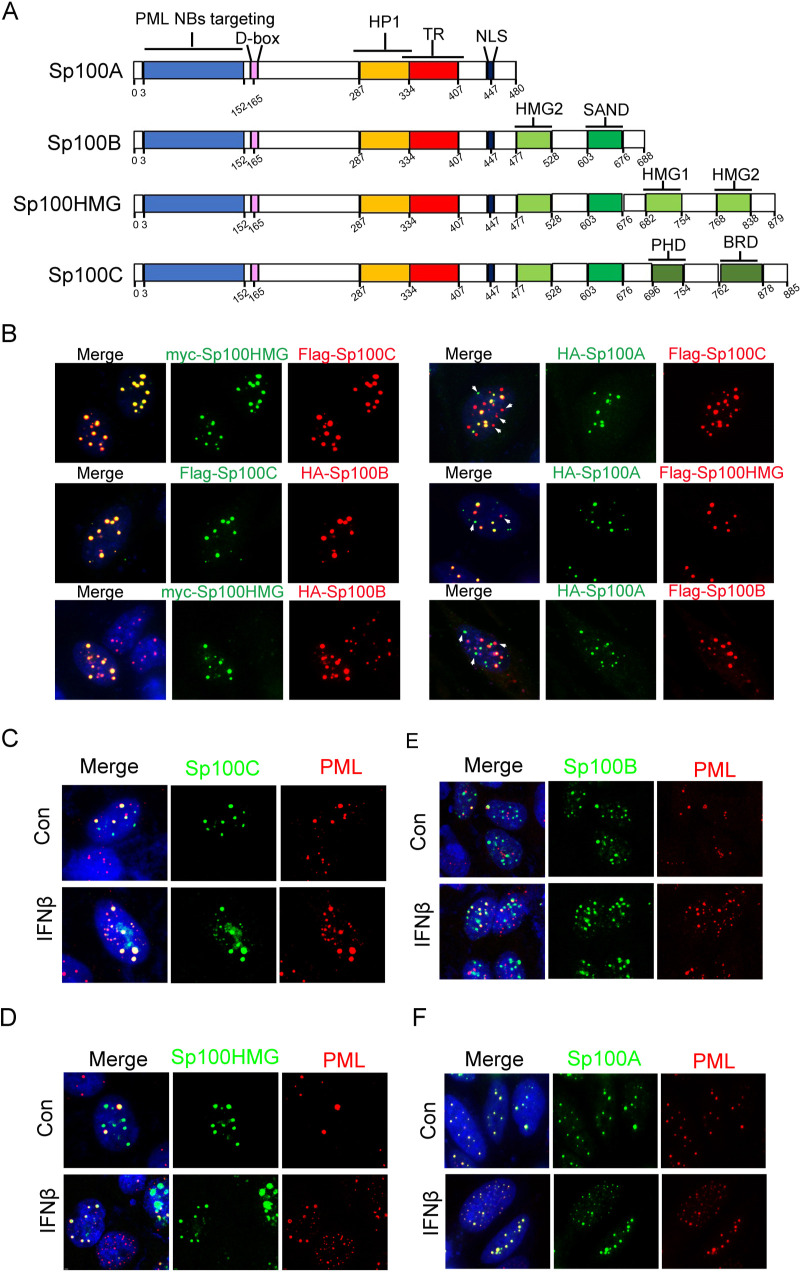
The Sp100 gene encodes multiple protein variants through alternative splicing (41). Four of them are experimentally detected in several cell lines and commonly investigated, namely, Sp100A, Sp100B, Sp100HMG, and Sp100C (Fig. 1A). Transient transfection of HEp-2 cells with plasmids coding for individual Sp100 isoforms, tagged with either a Flag, a myc, or a hemagglutinin (HA) epitope, in pairs revealed that although all Sp100 isoforms were stained as intranuclear speckles in the nuclei of cotransfected HEp-2 cells, Sp100A did not completely colocalize with the other isoforms (Fig. 1B, white arrowheads), and the other three isoforms overlapped each other well in those punctate structures (Fig. 1B). Furthermore, coimmunofluorescence staining of the ectopically expressed Sp100 isoforms and endogenous PML with or without IFN-β treatment showed that Sp100C (Fig. 1C), Sp100HMG (Fig. 1D), and Sp100B (Fig. 1E) only partially colocalized with PML NBs, delegated by the staining of endogenous PML proteins, while Sp100A shared a better colocalization with PML NBs in HEp-2 cells. Note that Sp100A appeared to be more stable than the other isoforms; these distinct intranuclear localization patterns may result from a combinational effect of the concentration-dependent detection efficiency in immunofluorescence (IF) and the intrinsically divergent biological functions of these isoforms (Fig. 1F).
FIG 1.
Distinct intranuclear localizations of ectopically expressed Sp100 isoforms. (A) Schematics of protein domains of SP100 splice variants. Destruction-box (D-box), trans-activating region (TR), high mobility group (HMG), bromodomain (BRD) and plant homeodomain (PHD). (B) HEp-2 cells were transfected with plasmids coding for tagged Sp100 isoforms A, B, C, and HMG in pairs. At 24 h posttransfection, cells were fixed, permeabilized, and reacted with antibodies labeled with fluorophores, as indicated. White arrowheads indicate differential intranuclear localizations of Sp100A from the other three Sp100 isoforms. (C to F) HEp-2 cells were transfected with individual plasmids encoding Sp100C (C), Sp100HMG (D), Sp100B (E), or Sp100A (F). At 24 h posttransfection, cells were mock treated (control [Con]) or treated with IFN-β at 1,000 U/mL for 2 h, fixed, permeabilized, and reacted with anti-Flag and anti-PML antibodies. Representative images are shown.
Sp100A restricted HSV-1 infection in HEp-2 and Sp100−/− cells.
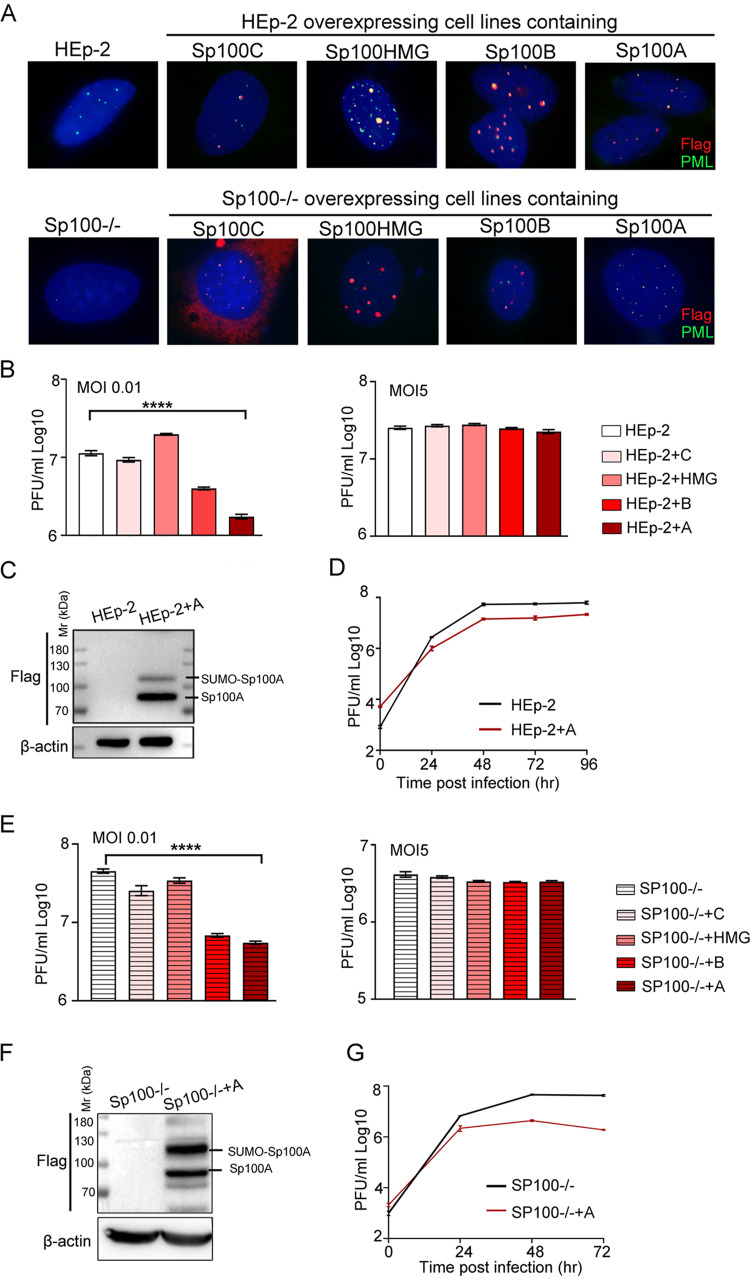
To analyze the role of individual Sp100 isoforms during HSV-1 infection, we constructed Sp100 isoform-overexpressing cell lines on the background of HEp-2 cells or Sp100 protein knockout (Sp100−/−) cells (58), termed HEp-2+A, HEp-2+B, HEp-2+HMG, HEp-2+C, Sp100−/−+A, Sp100−/−+B, Sp100−/−+HMG, and Sp100−/−+C cells. All isoforms were tagged with a Flag epitope at the N terminus. The expression and PML NB association of these isoforms were examined by immunofluorescence staining (Fig. 2A). Cells expressing Sp100C, Sp100HMG, and Sp100B were not stable, even under stringent drug selection, and tended to lose Sp100 expression over time, so only early passages of the cells were used in this study. Attempts to confirm the expression of Sp100 isoforms by immunoblot analysis consistently succeeded for HEp-2+A and Sp100+A cells but frequently failed for cells expressing the other three Sp100 isoforms, implying very low basal expression levels of these exogenously introduced isoforms in the stable cell lines (data not shown). HSV-1 replication in HEp-2+A, HEp-2+B, HEp-2+HMG, and HEp-2+C cells was examined in parallel with that in HEp-2 cells at multiplicities of infection (MOIs) of 0.01 and 5. Titration of the cell-associated viruses at 48 h postinfection (hpi) revealed that Sp100A and Sp100B significantly restricted HSV-1 lytic replication in HEp-2 cells at a low MOI (Fig. 2B). Confirmation of the expression of Sp100A in HEp-2+A cells by an immunoblot assay is shown in Fig. 2C, and the growth kinetics of HSV-1 in HEp-2+A cells at an MOI of 0.01 revealed sustained inhibited growth of the virus in the presence of overexpressed Sp100A (Fig. 2D). Given that Sp100A lacks a direct DNA binding domain and that other Sp100 isoforms have been previously shown to restrict transcription from the ICP0 promoter, it was important to examine the dependency of this antiviral capability of Sp100A on the presence of other Sp100 variants. To this end, similar experiments were conducted in Sp100−/− cells and the four Sp100−/−-derived cell lines expressing individual Sp100 isoforms (Fig. 2E to G). The ectopic expression of Sp100A in Sp100−/− cells was confirmed by immunoblot analysis (Fig. 2F). The results showed that Sp100−/−+A cells were the most resistant to HSV-1 replication (Fig. 2E) and restricted virus propagation at all time points postinfection (Fig. 2G). Note that the expression of Flag-Sp100A in Sp100−/− and HEp-2 cells led to distinct modification patterns of the protein. In HEp-2+A cells, Flag antibody recognized one major band between 70 and 100 kDa, corresponding to unSUMOylated Sp100, and a minor band above 100 kDa, corresponding to SUMOylated Sp100A, according to previous studies, while in Sp100−/−+A whole-cell lysates, there were multiple bands of different intensities reactive to Flag antibody besides the major band between 70 and 100 kDa and the SUMOylated Sp100A band. Since the expression levels of the other three Sp100 isoforms were frequently below the detection limit of our immunoblot analysis, likely due to cytotoxicity, no reliable conclusions regarding the anti-HSV-1 potency of these proteins could be made in this report. We thus conclude that Sp100A is an active host factor that restricts wild-type HSV-1 replication independently of other Sp100 isoforms.
FIG 2.
Sp100A inhibited HSV-1 lytic infection at a low multiplicity of infection. (A) Overexpression cell lines of individual Sp100 isoforms (Flag tagged) were established on the background of HEp-2 and Sp100−/− cells by a lentiviral transduction system and confirmed by immunofluorescence staining with antibodies against Flag (red) and PML (green). Nuclei were stained with DAPI. (B) HEp-2 cells and four Sp100 isoform-overexpressing HEp-2 cell lines were infected with HSV-1 at an MOI of 0.01 (left) or an MOI of 5 (right). Cells were collected at 48 hpi, and cell-associated viruses were titrated by a plaque assay. (C) Protein expression of Sp100A in HEp-2+A cells by immunoblotting with anti-Flag antibody. (D) Multicycle growth kinetics of HSV-1 in HEp-2 and HEp-2+A cells at an MOI of 0.01 by plaque titration of the cell-associated viruses. (E to G) Experiment conducted as described above for panels B to D but in Sp100−/− cells and the four Sp100 isoform-overexpressing Sp100−/− cell lines.
Cytosolic Sp100A remained stable during HSV-1 infection.
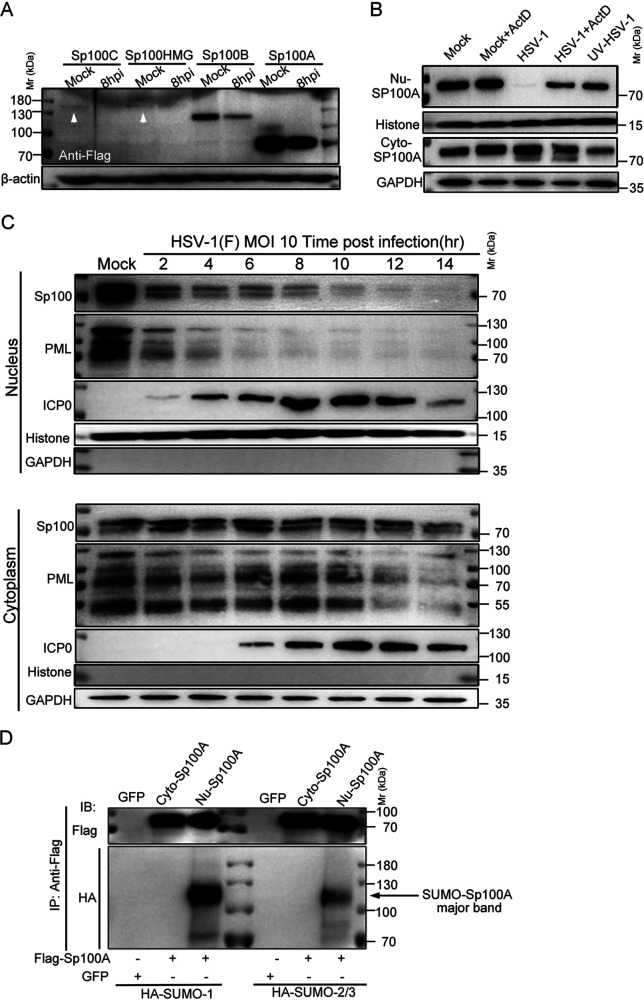
It has long been noticed that HSV-1 dismantles nuclear structure PML NBs early in infection (59–62). Subsequent studies confirmed that the immediate early protein ICP0 of the virus targets PML protein (the scaffold protein of PML NBs) and all SUMOylated forms of Sp100 protein but not unSUMOylated Sp100A (41, 52). Studies by others and our group have shown that nuclear Sp100 was immediately degraded during productive HSV-1 infection, and there was barely any detectable Sp100 left in the nuclei of the HSV-1-infected cells (6, 42, 63). The subsequent logical inquiries are where the escaped Sp100A is located in the infected cells and whether unSUMOylated Sp100A plays a role during HSV-1 replication. To address these questions, we conducted a set of experiments. First, HEp-2 cells were transfected with plasmids coding for individual Sp100 isoforms and superinfected with HSV-1 at an MOI of 5. HSV-1 infection led to a significant decrease in the total protein amount for all isoforms, which was consistent with previous reports (Fig. 3A). We then infected HEp-2+A cell lines (Sp100A-overexpressing cells) with HSV-1, HSV-1 with actinomycin D (ActD) (a protein synthesis blocker), or the same amount of HSV-1 but pretreated with UV and divided the infected cells into the cytosolic and the nuclear compartments to examine the subcellular protein levels of Sp100A. While nuclear Sp100A disappeared quickly upon HSV-1 infection when new proteins were allowed to be synthesized, the protein level of cytosolic Sp100A in HSV-1-infected cells remained unchanged (Fig. 2B). Note that in cells inoculated with UV-treated HSV-1, the cytosolic Sp100A level was lower than that in the HSV-1-infected group (Fig. 3B). To trace the protein level of endogenous Sp100 during HSV-1 infection, HEp-2 cells were infected with HSV-1 at an MOI of 10. Proteins from subcellular compartments (cytoplasm and nucleus) were collected at different time points postinfection and examined by immunoblotting with antibodies against PML, Sp100, and ICP0. As shown in Fig. 3C and in accordance with previous reports, nuclear PML and Sp100 levels dramatically decreased upon nuclear detection of the viral E3 ligase ICP0 by as soon as 2 hpi and were no longer detectable after 4 hpi and 10 hpi, respectively, whereas the protein in the cytoplasm reactive to Sp100 polyclonal antibody was stable in the presence of massive cytosolic ICP0. Cytosolic Sp100, loaded at a portion and under conditions similar to those of nuclear Sp100 (Fig. 3C), is present at a much lower level than nuclear Sp100 and accounts for a minor portion of the whole-cell Sp100. To test whether cytosolic Sp100A was subjected to SUMO modification, HEp-2 cells were transfected with plasmids coding for Flag-Sp100A and HA-SUMO1 or HA-SUMO2/3. Immunoblot analysis of Sp100A isolated from different subcellular compartments with anti-HA antibody revealed that only nuclear Sp100A was heavily modified by SUMO1 and SUMO2/3 and that there was no detectable SUMO modification of cytosolic Sp100A (Fig. 3D). We thus conclude that during HSV-1 infection, cytosolic Sp100A escaped targeting by the viral protein ICP0.
FIG 3.
Cytosolic Sp100 remained stable during HSV-1 infection. (A) HEp-2 cells were transfected with plasmids coding for individual Sp100 isoforms (Flag tagged) for 24 h and mock infected or infected with HSV-1 at an MOI of 5. At 8 hpi, cell lysates were collected and immunoblotted with antibodies against Flag and β-actin. (B) HEp-2+A cells were nontreated, treated with 5 μg/mL actinomycin D (ActD) for 2 h, infected with HSV-1 at an MOI of 10 in the presence or absence of ActD, or inoculated with UV-inactivated HSV-1 of the same amount. Subcellular fractions of different groups of cells were collected at 6 hpi, and the protein levels of Sp100A in the nucleus (Nu-Sp100A) and the cytoplasm (Cyto-Sp100A) were analyzed by immunoblotting with polyclonal anti-Sp100 antibody. Histone and GAPDH served as loading controls. (C) HEp-2 cells were infected with HSV-1 at an MOI of 10. Subcellular fractions were collected at the indicated time points postinfection, and protein levels of Sp100, PML, and ICP0 were analyzed by immunoblotting with polyclonal anti-Sp100 antibody and anti-PML and anti-ICP0 antibodies. Histone and GAPDH served as markers for subcellular compartments. Cytosolic Sp100A was loaded at portions and exposed under conditions similar to those of nuclear Sp100. (D) HEp-2 cells were cotransfected with plasmids coding for Flag-Sp100A or GFP and plasmids coding for HA-SUMO1 or HA-SUMO2/3, as indicated. At 24 h posttransfection, Sp100A was immunoprecipitated (IP) down from the cytosolic and nuclear compartments by anti-Flag antibody and immunoblotted (IB) with antibodies against the HA tag and Flag tag. Fivefold more total cytosolic immunoprecipitated lysate was loaded so that cytosolic Sp100A was at an amount similar to that of nuclear Sp100A. The arrow points to the SUMOylated Sp100A band.
HSV-1 infection in HEp-2 cells promoted the secretion of EV-associated Sp100.
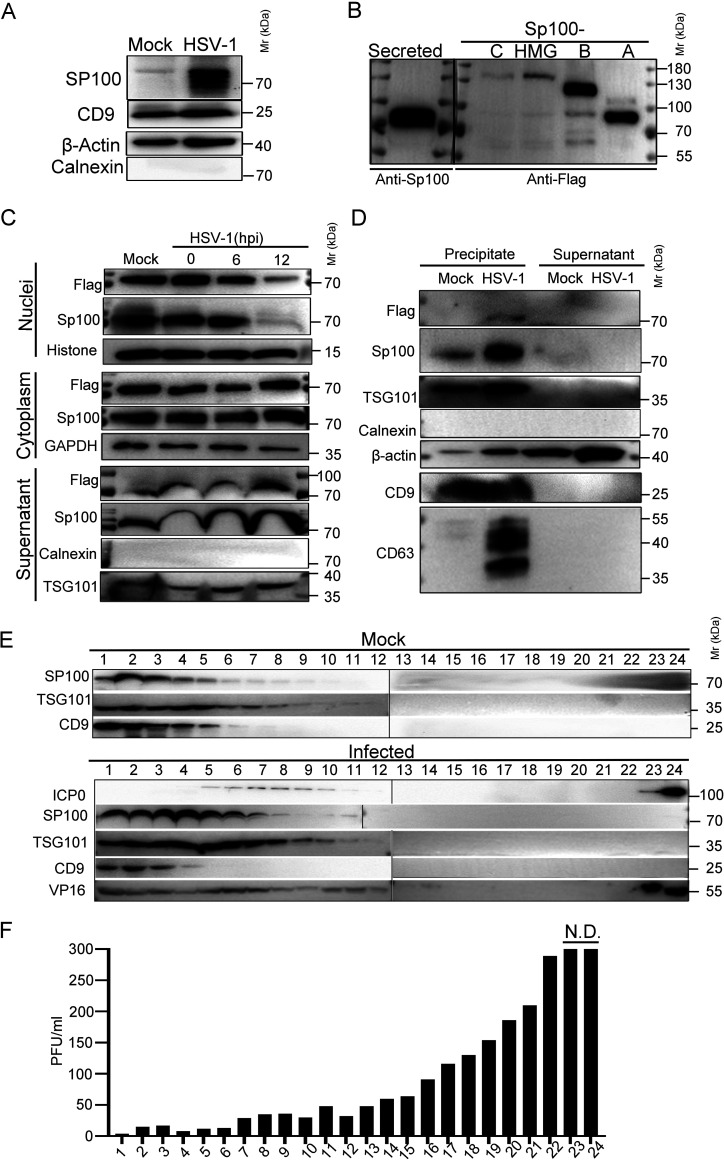
Recent studies reveal that EVs are an important means of communication between infected and uninfected cells during herpesvirus replication, and host cells were reported to secrete restrictive factors, such as STING and CD63, to block HSV-1 infection in neighboring cells (64–70). We wondered if the cytosolic Sp100 protein could be secreted during infection to exert antiviral functions. First, to examine the extracellular Sp100 protein level in HSV-1-infected HEp-2 cells, HEp-2 cells were mock infected or infected with HSV-1 at an MOI of 0.01, and the culture medium was collected at 50 hpi and processed as described in Materials and Methods to remove cell debris and to pellet down all extracellular vesicles, including HSV-1 virions. The pellet was positive for CD9 and β-actin and negative for calnexin, and its associated Sp100 level was significantly higher in the HSV-1-infected group (Fig. 4A). Second, the EV-associated Sp100 was loaded in parallel with the whole-cell lysates of HEp-2 cells transfected with individual Sp100 isoforms. As shown in Fig. 4B, the electrophoretic mobility of secreted Sp100 in polyacrylamide gels matched that of the Sp100A isoform. To further confirm that Sp100A could be secreted by HEp-2 cells during infection, HEp-2+A cells were infected with HSV-1, and proteins from subcellular compartments (cytoplasm and nucleus) and the extracellular space (cell culture medium) were collected and examined by immunoblotting with antibodies against PML, Sp100, and marker proteins of cellular compartments. Interestingly, proteins reactive to polyclonal Sp100 antibody and proteins reactive to Flag antibody were accumulating in the extracellular samples upon HSV-1 infection, implying that HSV-1 infection induced the increased secretion of Sp100A.
FIG 4.
Sp100A was secreted into the extracellular space during HSV-1 infection. (A) HEp-2 cells were mock infected or infected with HSV-1 at an MOI of 0.1. At 50 hpi, culture medium was collected, and cell debris was removed as described in Materials and Methods. Proteins secreted into the extracellular space were concentrated and examined by immunoblotting with polyclonal anti-Sp100 antibody. β-Actin, CD9, and calnexin served as controls. (B) HEp-2 cells were transfected with plasmids coding for individual Flag-tagged Sp100 isoforms for 24 h. Cell lysates were collected; separated in a polyacrylamide gel side by side with the concentrated, cell debris-free extracellular proteins from panel A; and immunoblotted with anti-Flag or anti-Sp100 antibodies, as indicated. (C) HEp-2+A cells were infected with HSV-1 at an MOI of 10. Subcellular fractions and culture medium were collected at the indicated time points and analyzed by immunoblotting with anti-Flag and anti-Sp100 antibodies. Histone, GAPDH, calnexin, and TSG101 served as markers for the origin and purity of the samples. (D) HEp-2+A cells were infected with HSV-1 at an MOI of 10 for 24 h. Extracellular vesicles ranging in sizes between30 and 200 nm in diameter were isolated using a commercial exosome extraction kit according to the manufacturer’s protocol. The protein contents were analyzed by immunoblotting with the indicated antibodies. (E) HEp-2 cells were mock infected or infected with HSV-1 at an MOI of 0.1 for 50 h. Culture medium was collected and processed as described in Materials and Methods. Extracellular vesicles and HSV-1 virions were separated by iodixanol-sucrose gradient-based ultracentrifugation. Twenty-four fractions, each in 500 μL, were collected from top to bottom from the density gradient, labeled 1 to 24. The proteins present in each fraction were analyzed by immunoblotting with the indicated antibodies. (F) Infectious HSV-1 virions in the 24 fractions from panel E were titrated by a plaque assay on Vero cells. The cells exposed to the bottom 2 fractions of the gradient (fractions 23 and 24) were fully infected, so plaques could not be determined. N.D., not determined.
Next, we examined the biological properties of the EVs that contained extracellular Sp100. Utilizing a size-based exosome purification kit, we precipitated all vesicles within a diameter of 30 nm to 200 nm from the culture medium of HSV-1-infected HEp-2+A cells. These vesicles were positive for β-actin and the exosomal markers TSG101 and CD9, and the ones isolated from HSV-1-infected cells contained significantly more Sp100 protein (Fig. 4D). Note that EVs purified from HSV-1-infected HEp-2+A cells contained a significantly larger amount of CD63, which is in accordance with previous observations (71). As HSV-1 is known to pack a variety of both viral and cellular factors into the tegument structure of virion to participate in the next round of infection in vitro and in vivo (72) and the size of viral virions coincides with the size of exosomes, we set out to investigate if Sp100A was packed into virions or into noninfectious extracellular microvesicles. We adopted an iodixanol-sucrose gradient-based ultracentrifugation method to separate all extracellular vesicles, including HSV-1 virions, into 24 portions numbered in sequence from top to bottom on the iodixanol-sucrose gradient after 16 h of ultracentrifugation (64). Analysis of the protein contents by immunoblotting with antibodies against Sp100, ICP0, VP16, TSG101, and CD9 revealed that Sp100 protein cosedimented with EVs that were positive for TSG101 and CD9 in the top lanes but not with fractions 23 and 24 in the bottom lanes containing virion-associated ICP0 protein (Fig. 4E). Titration of the infectious viral particles in the 24 portions confirmed the successful separation of EVs (Fig. 4E, top lanes 1 to 6) from infectious HSV-1 virions (Fig. 4E, bottom lanes, and Fig. 4F).
Taken together, these results indicate that endogenous Sp100 and ectopically expressed Sp100A are increasingly secreted into the extracellular space during HSV-1 infection, and they are associated with the non-virion-containing EV portion that matches the size, biological membrane markers, and sedimentation velocity in the iodixanol-sucrose gradient during the ultracentrifugation of exosomes.
EV-associated Sp100A restricted HSV-1 in the recipient cells.
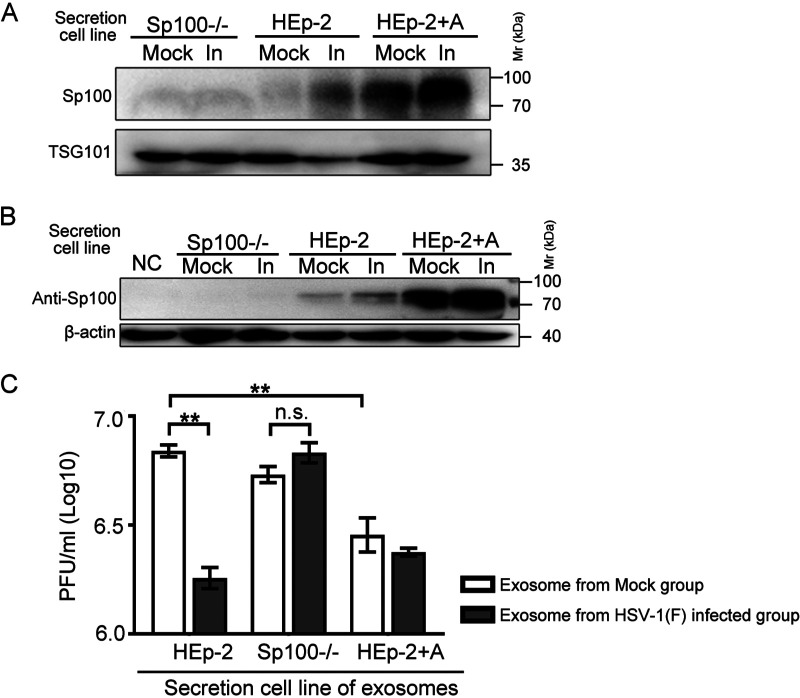
In the series of experiments to address the antiviral role of Sp100A against HSV-1, it was of importance to test if the secreted Sp100A had an impact on HSV-1 replication. We first separated EVs from HSV-1 virions by the iodixanol-sucrose gradient ultracentrifugation method and collected the top 6 portions, designated EVs for the following studies in this section (Fig. 4E). Analysis of the protein levels of Sp100 and TSG101 in EVs from mock- or HSV-1-infected Sp100−/−, HEp-2, and HEp-2+A cells showed elevated Sp100 protein levels in the EVs from HSV-1-infected HEp-2 and HEp-2+A cells, while no Sp100 antibody-reactive proteins were detected in the EVs from Sp100−/− cells, confirming the specificity of the anti-Sp100 antibody (Fig. 5A). When Sp100−/− cells were incubated with equal volumes of EVs from mock- or HSV-1-infected Sp100−/−, HEp-2, and HEp-2+A cells, a significantly higher Sp100 protein level was detected in the cells incubated with the EVs from HSV-1-infected HEp-2 and HEp-2+A cells than in the ones that received the EVs from the mock-infected HEp-2 group or from either Sp100−/− group (Fig. 5B).
FIG 5.
Effect of EVs from Sp100−/−, HEp-2, and HEp-2+A cells on HSV-1 replication in the recipient HEp-2 cells. (A) Sp100−/−, HEp-2, and HEp-2+A cells were mock infected or infected (In) with HSV-1(F) at an MOI of 0.1 for 50 h. Exosome-enriched fractions (fractions 1 to 6 in Fig. 4E) were used. The Sp100 protein level in an equal volume of EVs was examined by immunoblotting with anti-Sp100 antibody. (B and C) A total of 4 × 105 Sp100−/− cells were incubated with an equal volume (500 μL) of EVs from mock-infected or HSV-1-infected Sp100−/−, HEp-2, and HEp-2+A cells without infection (B) or with 4,000 PFU of HSV-1 (MOI of 0.01) (C) at 37°C for 2 h. NC represents negative control. Five hundred microliters of EVs from different infected groups was contaminated with on average 7 to 173 PFU (EVs from infected HEp-2 cells, 172 to 173 PFU/500 μL; EVs from infected SP100−/− cells, 7 to 8 PFU/500 μL; EVs from infected HEp-2+A cells, not detectable). (B) Cells were extensively washed and collected for immunoblotting with antibodies against Sp100 and β-actin. (C) At 24 hpi, cell-associated viruses were titrated by a plaque assay.
To test the antiviral ability of the EVs from different experimental groups against HSV-1, cells treated as described in the legend of Fig. 5B were exposed to HSV-1 at an MOI of 0.01, and cell-associated viruses were titrated at 24 hpi (Fig. 5C). There were four interesting observations: (i) in accordance with a previous report (65), the EVs from HSV-1-infected HEp-2 cells showed a strong antiviral effect; (ii) the depletion of Sp100 in the host cells abolished the antiviral difference between the EVs from HSV-1-infected cells and those from noninfected cells; and (iii) in the absence of infection, the EVs from HEp-2+A cells, containing higher Sp100A protein levels, significantly inhibited HSV-1 growth in the recipient cells compared with the EVs from mock-infected HEp-2 cells. We thus conclude that the level of Sp100A in HSV-1-infected host cells strongly correlates with its level in the secreted EVs and that EV-associated Sp100A protein has antiviral potency in HSV-1 infection in the recipient cells. Note that the increase of the Sp100A level in the EVs from HSV-1-infected HEp-2+A cells led to no further inhibition of HSV-1 replication in the recipient cells compared to the EVs from mock-infected HEp-2+A cells, implying that the antiviral effect of Sp100A was saturated or that Sp100A may not be the sole player in this process.
Recombinant HSV-1 expressing Sp100A was attenuated.
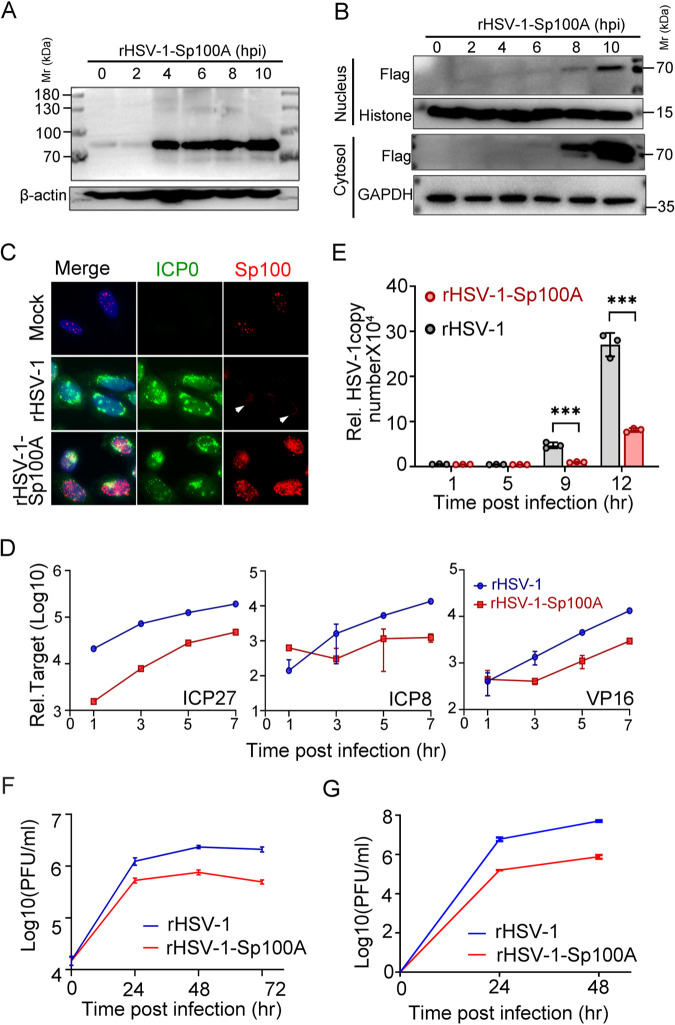
The reduced HSV-1 replication in Sp100A-overexpressing cell lines could be due to the elevated basal expression level of Sp100A in every single cell and/or due to an increased secretion of Sp100A, which helped to mount an anti-HSV-1 status in the recipient cells. To express Sp100A only in the infected cells, we constructed a recombinant HSV-1 expressing a Flag-tagged Sp100A protein and rescued and single-plaque purified the virus in Vero cells (rHSV-1-Sp100A) (73). A wild-type HSV-1 was rescued and repaired in parallel to serve as a control virus in this section (rHSV-1). The successful expression of Flag-Sp100A by rHSV-1-Sp100A during infection was confirmed by immunoblotting (Fig. 6A and B). Tracking Sp100A expression and subcellular localization during rHSV-1-Sp100A infection showed that the protein accumulated to a detectable level on the whole-cell level by as early as 4 hpi and was simultaneously detected in the cytoplasm and the nucleus of infected cells at between 6 and 8 hpi, with a significantly higher level in the cytosol (Fig. 6B). Immunofluorescence staining revealed the coexpression of Sp100A and the viral protein ICP0 in rHSV-1-Sp100A-infected cells only (Fig. 6C). Very faint cytosolic staining around the nucleus of rHSV-1-infected HEp-2 cells by anti-Sp100 antibody could sometimes be visualized under an immunofluorescence microscope (Fig. 6C, white arrowheads). Whether this was cytosolic Sp100 needs further investigation. Both viral gene expression and viral DNA multiplication rates were thwarted during rHSV-1-Sp100A infection (Fig. 6D and E), and the virus yield of rHSV-1-Sp100A in HEp-2 cells was significantly lower than that of rHSV-1 at 24, 48, and 72 hpi during low-MOI infection (Fig. 6F). An important question was whether Sp100A suppressed HSV-1 replication through PML bodies. To address this inquiry, the growth of rHSV-1-Sp100A was compared to that of rHSV-1 in PML−/− cells (a PML knockout cell line previously established by our group) (63). As shown in Fig. 6G, the overexpression of Sp100A significantly inhibited HSV-1 replication in the absence of PML protein and intact PML bodies, indicating that Sp100A restricted HSV-1 independently of PML.
FIG 6.
rHSV-1-Sp100A was attenuated compared to rHSV-1. (A and B) Protein levels of Sp100A in rHSV-1-Sp100A-infected HEp-2 cells (MOI of 10) at the indicated time points postinfection were examined by immunoblotting with anti-Flag antibody, in whole-cell lysates (A) or subcellular fractions (B). (C) HEp-2 cells were mock infected or infected with rHSV-1 or rHSV-1-Sp100A at an MOI of 10. At 12 hpi, cells were fixed and stained with anti-Flag (red) and anti-ICP0 (green) antibodies. White arrowheads mark the faint cytosolic staining of proteins reactive to anti-Sp100 antibody. (D to F) HEp-2 cells were infected with rHSV-1-Sp100A at an MOI of 0.01. (D) Expression levels of the representative α (ICP27), β (ICP8), and γ (VP16) genes at the indicated time points were quantified by quantitative PCR (qPCR). (E) Viral DNA levels at the indicated time points postinfection were quantified by qPCR using primers targeting the ICP27 gene region. (F) Growth kinetics of rHSV-1 and rHSV-1-Sp100A in HEp-2 cells at an MOI of 0.01 by a plaque assay. (G) Growth kinetics of rHSV-1 and rHSV-1-Sp100A in PML−/− cells at an MOI of 0.01 by a plaque assay.
DISCUSSION
Previous investigations identified distinct functional domains in individual Sp100 isoforms and revealed that they play independent or opposing roles in regulating the transcription of endogenous genes and viral genes through interacting with various partners (36, 41). Due to the rapid degradation of PML bodies and nuclear Sp100 by the HSV-1-encoded protein ICP0 early in infection, extensive investigations of the relationship between Sp100 and HSV-1 were conducted using mutated HSV-1 lacking a functional ICP0 (41). Here, in this study, we examined the fate of Sp100 protein during wild-type HSV-1 infection in detail and reported the following findings.
First, overexpressing Sp100 isoforms in pairs by transient transfection revealed an interesting phenomenon that while all Sp100 isoforms formed punctate structures in the nucleus, there were speckles positive only for Sp100A or positive only for one of the other Sp100 isoforms, which may imply disparate functions among Sp100 variants. To clarify the role of individual Sp100 isoforms in HSV-1 replication, we established cell lines in the background of HEp-2 and Sp100−/− cells that expressed individual Sp100 isoforms and tested their susceptibility to a wild-type HSV-1. The Sp100C, Sp100HMG, and Sp100B isoforms were expressed at low levels in the transduced cells and frequently lost upon passaging of the cells, which was in accordance with the observed low endogenous levels of these isoforms and implied that their overexpression may be adverse for cell survival or proliferation (52). With regard to the major isoform Sp100A, although it has been previously shown to activate viral promoters (15, 36, 49), the overexpression of Sp100A in HEp-2 and Sp100−/− cells seemed to potently inhibit HSV-1 lytic replication at a low multiplicity of infection (Fig. 2B to G and Fig. 6D to F), implying that Sp100A functions as an active restrictive factor on HSV-1 independently of other Sp100 isoforms and PML.
Second, and importantly, it has long been noticed that a portion of unSUMOylated Sp100A escapes targeting during HSV-1 infection. Recent studies on Sp100A, SUMO modification of the protein, and HSV-1 infection suggest that the virus may degrade SUMOylated Sp100 and take advantage of the unSUMOylated form of Sp100A to activate the transcription of viral genes. An inquiry into the subcellular localization of untargeted Sp100A during HSV-1 infection in this report revealed that nuclear Sp100, SUMOylated or not, decreased dramatically upon the detection of the viral protein ICP0 in the nucleus at 4 hpi and was below the detection level at 10 to 12 hpi (MOI = 10), while the protein level of cytosolic Sp100 remained stable and coexisted with the massive ICP0 that translocated into the cytoplasm during late infection (8 to 12 hpi) (MOI = 10), as was ectopically expressed Sp100A (Fig. 4B to D). These results imply that instead of the previously believed SUMOylation, subcellular localization is a more important factor influencing the targeting efficiency of ICP0 on endogenous Sp100, and the molecular mechanisms responsible for this spatial bias of ICP0 need to be further elucidated.
Third, EV secretion is an important means of communication between infected and uninfected cells during herpesvirus replication (64–67). Valuable research results have shown that HSV-1-, Epstein-Barr virus-, and Kaposi’s sarcoma-associated herpesvirus (KSHV)-infected cells incorporate viral RNA, viral proteins, host factors, and/or virions into EVs to achieve either an anti- or a proinfection role (64–68, 74–78). Interestingly, in this report, we found that endogenous Sp100 was actively secreted into the extracellular space during HSV-1 infection, supported by three pieces of evidence: (i) HSV-1 infection in HEp-2 cells induced increased secretion of endogenous Sp100 that showed electrophoretic mobility similar to that of Sp100A during SDS-PAGE (Fig. 4B); (ii) in HEp-2+A cells, Flag-tagged Sp100A was located in both the nuclear and cytosolic compartments, and cytosolic Sp100A remained stable during HSV-1 infection (Fig. 4C); and (iii) HSV-1 infection in HEp-2+A cells induced increased secretion of Sp100 to the extracellular space recognized by both anti-Flag and anti-Sp100 antibodies. It should be noted that in our experimental settings, Sp100 in the culture medium of HEp-2-A cells was constantly more reactive to anti-Sp100 polyclonal antibody than to anti-Flag antibody, and it was not a phenomenon observed in the immunoblot assays of the lysates of the cytoplasm or the nuclei of HEp-2+A cells (Fig. 4C and D). Whether Sp100A is the only Sp100 isoform secreted and whether Flag-Sp100A is processed in the extracellular secretion pathway remain to be clarified.
Fourth, the Sp100 protein was likely to be exported with exosomes during HSV-1 infection in HEp-2 cells. Evidence that agreed with this assumption was that (i) Sp100 protein was coprecipitated with CD9-, CD63-, and TSG101-positive extracellular vesicles with a size of 30 to 200 nm in diameter by a commercial exosome extraction kit (Fig. 4D) and (ii) Sp100 protein cosedimented with infectious virion-free extracellular vesicles positive for the exosome markers CD9 and TSG101 during iodixanol-sucrose-based density ultracentrifugation (Fig. 4E and F). Importantly, the intracellular Sp100A level positively correlated with the protein levels in EVs in an infection-independent manner, and EV-associated Sp100A restricted HSV-1 in the recipient cells (Fig. 5C). One key question is how the Sp100 delivered by EVs helps to mount an anti-HSV-1 effect on uninfected recipient cells. The facts that Sp100 protein is a constituent component of PML bodies, HSV-1 immediately targeted Sp100 in the nucleus upon infection, and Sp100 family proteins are reported to be active transcriptional regulators strongly suggest that EV-delivered Sp100 may also carry out its functions as a host-restrictive factor in the nucleus of recipient cells. Given this assumption being true, it is intriguing to investigate the molecular mechanisms that indicate this cytosol-nuclear transportation of Sp100A since it may be an important part of the host defense system against viruses. Another related question concerns the potential function of EV-carried Sp100 during infection by HSV-1 in vivo. Accruing evidence reveals that there is active communication among periphery neurons, their innervating epithelial cells, and local immune cells (79–81). Whether HSV-1-infected epithelial cells serve as a source of increased Sp100 delivery not only to adjacent epithelial cells but also to local immune cells and/or sensory neurons remains an unaddressed question in the full picture of the HSV-1 and Sp100 interplay.
In sum, this study examined the fate and role of Sp100 isoforms during HSV-1 infection by utilizing Sp100 overexpression cell lines and recombinant HSV-1 expressing the Sp100A isoform and shows that Sp100A is an active restrictive factor of wild-type HSV-1. During natural HSV-1 infection, cytosolic Sp100A remained stable and was increasingly secreted into the extracellular space in EVs, which restricted HSV-1 replication in the recipient cells. These results indicate a novel antiviral mechanism of Sp100 protein through intercellular communications of EVs.
MATERIALS AND METHODS
Cell lines and viruses.
HEp-2, HEK293T, and Vero cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (catalog number 10-013-CVRC; Corning) supplemented with 10% fetal bovine serum (FBS) (catalog number 10270-106; Gibco). 143TK−/− cells were cultured in minimum essential medium (MEM) (catalog number 12561-056; Gibco) supplemented with 10% FBS and 1% nonessential amino acid solution (catalog number 25-025-CI; Corning). Sp100A/B/C/HMG overexpression cell lines were constructed by lentiviral transduction of HEp-2 or Sp100−/− cells (58). HSV-1(F), abbreviated HSV-1 in this study, is the prototype strain used in the laboratory. Recombinant viruses rHSV-1 and rHSV-1-Sp100A were constructed using a bacterial artificial chromosome (BAC)-based system for the generation of recombinant herpes simplex virus, rescued, and plaque purified in Vero cells as previously described (82). All HSV-1 viruses in this study were amplified in HEp-2 cells and titrated in Vero cells by a plaque assay.
Plaque assay.
All infected cells were collected, frozen-thawed three times, and sonicated briefly to fully release and separate all cell-associated viruses. Titration was conducted in Vero cells in triplicates of 10-fold serial dilutions of the virus stocks. The error bars were calculated using Student’s t test.
Antibodies and drugs.
The antibodies and drugs used in this study included rabbit polyclonal anti-Flag antibody (catalog number 80010; Proteintech), rabbit polyclonal anti-HA antibody (catalog number 3724s; CST), mouse anti-HA antibody (catalog number 26186; Invitrogen), rabbit anti-myc polyclonal antibody (catalog number 16286-1-AP; Proteintech), rabbit polyclonal anti-Sp100 antibody (catalog number GTX131569; GeneTex), mouse monoclonal anti-Flag antibody (catalog number AB0008; Abways), mouse monoclonal anti-β-actin antibody (catalog number 1000166; Sino Biological), rabbit monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (catalog number AB0037; Abways Technology), rabbit polyclonal anti-PML antibody (catalog number ab179466; Abcam), mouse monoclonal anti-PML antibody (catalog number sc966; Santa Cruz), rabbit polyclonal antibody anti-TSG101 (catalog number 28283-I-AP; Proteintech), mouse monoclonal anti-VP16 antibody (catalog number sc7545; Santa Cruz), mouse monoclonal anti-CD63 antibody (catalog number sc5275; Santa Cruz), mouse monoclonal anti-CD9 antibody (catalog number 13118; Santa Cruz), mouse monoclonal anticalnexin antibody (catalog number sc23954; Santa Cruz), mouse monoclonal anti-ICP0 (catalog number sc53070; Santa Cruz), Alexa Fluor 594-labeled goat anti-rabbit IgG(H+L) (catalog number 2165334; Invitrogen), Alexa Fluor plus 488-labeled goat anti-mouse IgG(H+L) (catalog number A32723; Invitrogen), goat anti-mouse IgG-horseradish peroxidase (HRP) (catalog number 31430; Invitrogen), goat anti-rabbit IgG(H+L)-HRP (catalog number 32460; Invitrogen), anti-Flag magnetic beads (catalog number L-1011; Biolinkedin), a protease inhibitor cocktail (catalog number EO0492; Thermo Scientific), and actinomycin D (catalog number A9415-10MG; Sigma).
Plasmids and transient transfection.
PCR-amplified Flag-tagged human Sp100A/B/C/HMG was cloned into the pcDNA3.1 or PLVX vector and verified by sequencing. The PCR primers for cloning of the four Sp100 isoforms are Flag-Sp100 F (AACTTAAGCTTGCCACCATGGACTACAAAGACGATGACGACAAGGCAGGTGGGGGCGGCGACCTGAGCAC), Sp100A R (TGTGCTGGATATCCGCCTAATCTTCTTTACCTGACCCTCTTC), Sp100B R (GCGTTAGATATCCGCTCACTTGATCATCACCTT), Sp100C R (CTAGGCGATATCCCGCTAAATAAACATTATAATG), and Sp100HMG R (TTAGGCGATATCCCGTTATTTATCATCATCTTCTTCA).
For HEp-2 cells and other mammalian cell lines, JetPRIME (catalog number 114-15; Polyplus) was used for transfection according to the manufacturer’s protocol.
Lentiviral transduction.
HEK293T cells were transfected with PLVX-Flag-SP100A/B/C/HMG or PLVX-GFP along with the packaging plasmids VSV-G and Δ8.9 using standard polyethylenimine (PEI) (catalog number 408727; Sigma) at appropriate ratios. At 48 h posttransfection, culture medium containing lentivirus was collected and passed through a 0.45-μm filter. HEp-2 or Sp100−/− cells were inoculated with the lentiviruses for 4 h. Forty-eight hours after lentivirus infection, cells continued to be cultured under hygromycin B (200 μg/mL) (catalog number V900372; Sigma) or puromycin (1 μg/mL) (catalog number A1113803; Gibco) selection pressure for another 4 to 7 days. All cell lines routinely tested negative for mycoplasma contamination.
Collection of total EVs and HSV-1 virions by ultracentrifugation.
As shown in Fig. 4A, 1 × 107 cells at 90% confluence were infected with HSV-1 at 0.1 PFU per cell. At 50 h postinfection (hpi), culture medium was collected and centrifuged at 300 × g for 5 min, 2,000 × g for 20 min, and 10,000 × g for 30 min. The supernatant was collected and ultracentrifuged in an SW40 Ti rotor for 90 min at 100,000 × g at 4°C (Optima XE-100 ultracentrifuge; Beckman) to pellet down all EVs and HSV-1 virions (83). The collected EVs and HSV-1 virions were washed with phosphate-buffered saline (PBS), repelleted by ultracentrifugation, and eventually collected in a total volume of 200 μL.
EV purification by density gradient ultracentrifugation.
The experiments for EV purification were conducted as described previously (64). FBS used for cell culture in this set of experiments was depleted of EVs by ultracentrifugation. Briefly, 5 × 107 cells at 90% confluence were infected with HSV-1 at a multiplicity of infection (MOI) of 0.1. At 50 hpi, culture medium was collected and centrifuged at 300 × g for 5 min and then at 2,000 × g for 20 min prior to filtration through a 0.45-μm-pore-size filter. The flowthrough was then concentrated to 1,000 μL using an Amicon Ultra 15-mL, 30-kDa centrifugal filter unit (catalog number UFC903024; Merck) and loaded on top of an iodixanol-sucrose gradient ranging from 6 to 18%, with 1.2% increments. Samples were centrifuged in an SW40 Ti rotor for 16 h at 160,000 × g at 4°C. Fractions in 500 μL were collected from top to bottom, numbered 1 to 24, and analyzed.
Nanoparticle tracking analysis.
Isolated EVs were quantified by nanoparticle tracking analysis (NTA) by applying a monochromatic 488-nm laser to the diluted samples and measuring the Brownian movements of each particle with a NanoSight NS300 instrument (Malvern, United Kingdom). NanoSight NTA 3.3 was used to analyze the vesicle size and concentration.
Subcellular fractionation.
Subcellular fractionation was performed as previously described (58). In short, 1 × 106 cells were collected, pelleted by centrifugation at 1,000 × g for 5 min, resuspended in 0.3 mL of ice-cold buffer 1 (150 mM NaCl, 50 mM HEPES [pH 7.4], 25 μg/mL digitonin, 10 μL/mL protease inhibitor), incubated for 30 min at 4°C, and then centrifuged at 4,600 rpm for 5 min. The supernatants represented the cytosol-enriched fraction. The pellets were then washed and resuspended in 0.3 mL of ice-cold buffer 2 (150 mM NaCl, 50 mM HEPES [pH 7.4], 1% [vol/vol] NP-40, 10 μL/mL protease inhibitor) and incubated for 30 min on ice. The samples were centrifuged at 8,700 rpm for 5 min to pellet nuclei, and the supernatants were collected as cytosolic membrane-associated components. The pellets were washed and resuspended in 0.2 mL of ice-cold buffer 3 (150 mM NaCl, 50 mM HEPES [pH 7.4], 0.5% [wt/vol] sodium deoxycholate, 0.5% [wt/vol] SDS, 1 mM dithiothreitol [DTT], 10 μL/mL protease inhibitor) on ice for 30 min, followed by sonication (10 s, 20% amplification). The final solution contained the extract comprising nuclear membranes and nuclear proteins.
Immunofluorescence staining, immunoprecipitation, and immunoblot analyses.
For immunofluorescence staining, in short, cells cultured on slides were washed with PBS, fixed, and permeabilized in methanol at −80°C overnight. Cells were then blocked with PBS-TBH (10% FBS, 1% bovine serum albumin [BSA], 1× PBS, 0.1% Triton X-100) at room temperature for 30 min and incubated with primary antibodies at appropriate dilutions in PBS-TBH overnight at 4°C or for 1 h at 37°C and then with fluorophore-conjugated secondary antibodies at appropriate dilutions in PBS-TBH for 30 min at 37°C in dark, with extensive washes between the steps. The slides were mounted using mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (catalog number 104139; Abcam).
For immunoprecipitation, nuclear and cytosolic fractions of 2 × 106 HEp-2 cells transfected with Flag-Sp100A or green fluorescent protein (GFP) and HA-SUMO1 or HA-SUMO2/3 were extracted and then incubated with anti-Flag antibody-conjugated magnetic beads overnight at 4°C. The resin was washed 3 times with PBS-Tween (PBST) and denatured in 1× SDS-PAGE loading buffer.
For immunoblotting, whole-cell extracts or subcellular fractions were subjected to SDS-PAGE separation, transferred onto polyvinylidene difluoride (PVDF) membranes, blocked with 5% BSA in PBS with 1% Tween 20, incubated with primary and secondary antibodies, and developed using the SuperSignal West Pico Plus chemiluminescent substrate (catalog number 34580; Thermo Fisher). Subcellular Sp100 was loaded with a larger amount or exposed for a longer time to increase the visibility of the protein if not otherwise noted.
RNA extraction and qRT-PCR.
Total RNA was extracted using an Omega RNA extraction kit (catalog number R6834-01) according to the manufacturer’s protocol. The gene expression level was quantified by quantitative real-time PCR (qRT-PCR) (StepOnePlus; Thermo Fisher) using a SYBR green detection system (catalog number AG11701; Accurate Biology). The following primers were used: ICP27-F (CGGGCCTGATCGAAATCCTA), ICP27-R (GACACGACTCGAACACTCCT), ICP8-F (CATCAGCTGCTCCACCTCGCG), ICP8-R (GCAGTACGTGGACCAGGCGGT), VP16-F (CCATTCCACCACATCGCT), VP16-R (GAGGATTTGTTTTCGGCGTT), GAPDH-F (GAAGGTGAAGGTCGGAGTC), and GAPDH-R (GAAGATGGTGATGGGATTTC).
Quantification of HSV-1 genome copy numbers.
Total DNA was extracted using an Omega DNA extraction kit (catalog number D3396-02) according to the manufacturer’s protocol. Relative DNA expression was quantified by quantitative real-time PCR using a SYBR green detection system. Viral DNA was measured using primers targeting ICP27, and host DNA was quantified using GAPDH primers.
Virus Infection.
Cells at 80 to 90% confluence were inoculated with viruses diluted in DMEM at the desired MOI at 37°C for 2 h, washed with PBS, and cultured in 1% FBS–DMEM.
Statistical analysis.
Data are presented as the means ± standard deviations (SD), calculated by GraphPad Prism 6.0 software. Two-tailed unpaired Student’s t test was used to calculate P values. In the figures, P values of <0.05 are marked as “*,” P values of <0.01 are marked as “**,” P values of <0.001 are marked as “***,” and P values of <0.0001 are marked as “****.”
ACKNOWLEDGMENTS
This project is supported by the National Natural Science Foundation of China (numbers 31870157 and 81802006) and the Shenzhen Science and Technology Program (JCYJ20180307151536743 and KQTD20180411143323605).
We declare no conflict of interest.
Contributor Information
Pei Xu, Email: xupei3@mail.sysu.edu.cn.
Jae U. Jung, Lerner Research Institute, Cleveland Clinic
REFERENCES
- 1.Szostecki C, Guldner HH, Netter HJ, Will H. 1990. Isolation and characterization of cDNA encoding a human nuclear antigen predominantly recognized by autoantibodies from patients with primary biliary cirrhosis. J Immunol 145:4338–4347. [PubMed] [Google Scholar]
- 2.Guldner HH, Szostecki C, Grotzinger T, Will H. 1992. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J Immunol 149:4067–4073. [PubMed] [Google Scholar]
- 3.Grotzinger T, Jensen K, Will H. 1996. The interferon (IFN)-stimulated gene Sp100 promoter contains an IFN-gamma activation site and an imperfect IFN-stimulated response element which mediate type I IFN inducibility. J Biol Chem 271:25253–25260. 10.1074/jbc.271.41.25253. [DOI] [PubMed] [Google Scholar]
- 4.Puvion-Dutilleul F, Venturini L, Guillemin MC, de The H, Puvion E. 1995. Sequestration of PML and Sp100 proteins in an intranuclear viral structure during herpes simplex virus type 1 infection. Exp Cell Res 221:448–461. 10.1006/excr.1995.1396. [DOI] [PubMed] [Google Scholar]
- 5.Chelbi-Alix MK, de The H. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935–941. 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 6.Gu H, Roizman B. 2003. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci USA 100:8963–8968. 10.1073/pnas.1533420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walters MS, Kyratsous CA, Silverstein SJ. 2010. The RING finger domain of varicella-zoster virus ORF61p has E3 ubiquitin ligase activity that is essential for efficient autoubiquitination and dispersion of Sp100-containing nuclear bodies. J Virol 84:6861–6865. 10.1128/JVI.00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perusina Lanfranca M, Mostafa HH, Davido DJ. 2013. Two overlapping regions within the N-terminal half of the herpes simplex virus 1 E3 ubiquitin ligase ICP0 facilitate the degradation and dissociation of PML and dissociation of Sp100 from ND10. J Virol 87:13287–13296. 10.1128/JVI.02304-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mullen MA, Gerstberger S, Ciufo DM, Mosca JD, Hayward GS. 1995. Evaluation of colocalization interactions between the IE110, IE175, and IE63 transactivator proteins of herpes simplex virus within subcellular punctate structures. J Virol 69:476–491. 10.1128/JVI.69.1.476-491.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doucas V, Ishov AM, Romo A, Juguilon H, Weitzman MD, Evans RM, Maul GG. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev 10:196–207. 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 11.Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp Cell Res 229:155–158. 10.1006/excr.1996.0353. [DOI] [PubMed] [Google Scholar]
- 12.Muller S, Dejean A. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J Virol 73:5137–5143. 10.1128/JVI.73.6.5137-5143.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Echendu CW, Ling PD. 2008. Regulation of Sp100A subnuclear localization and transcriptional function by EBNA-LP and interferon. J Interferon Cytokine Res 28:667–678. 10.1089/jir.2008.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kyratsous CA, Walters MS, Panagiotidis CA, Silverstein SJ. 2009. Complementation of a herpes simplex virus ICP0 null mutant by varicella-zoster virus ORF61p. J Virol 83:10637–10643. 10.1128/JVI.01144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berscheminski J, Wimmer P, Brun J, Ip WH, Groitl P, Horlacher T, Jaffray E, Hay RT, Dobner T, Schreiner S. 2014. Sp100 isoform-specific regulation of human adenovirus 5 gene expression. J Virol 88:6076–6092. 10.1128/JVI.00469-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porter SS, Stepp WH, Stamos JD, McBride AA. 2017. Host cell restriction factors that limit transcription and replication of human papillomavirus. Virus Res 231:10–20. 10.1016/j.virusres.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao Y, Lupiani B, Reddy SM. 2021. Manipulation of promyelocytic leukemia protein nuclear bodies by Marek’s disease virus encoded US3 protein kinase. Microorganisms 9:685. 10.3390/microorganisms9040685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim YE, Lee JH, Kim ET, Shin HJ, Gu SY, Seol HS, Ling PD, Lee CH, Ahn JH. 2011. Human cytomegalovirus infection causes degradation of Sp100 proteins that suppress viral gene expression. J Virol 85:11928–11937. 10.1128/JVI.00758-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stepp WH, Meyers JM, McBride AA. 2013. Sp100 provides intrinsic immunity against human papillomavirus infection. mBio 4:e00845-13. 10.1128/mBio.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brasier AR, Spratt H, Wu Z, Boldogh I, Zhang Y, Garofalo RP, Casola A, Pashmi J, Haag A, Luxon B, Kurosky A. 2004. Nuclear heat shock response and novel nuclear domain 10 reorganization in respiratory syncytial virus-infected A549 cells identified by high-resolution two-dimensional gel electrophoresis. J Virol 78:11461–11476. 10.1128/JVI.78.21.11461-11476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Habiger C, Jager G, Walter M, Iftner T, Stubenrauch F. 2016. Interferon kappa inhibits human papillomavirus 31 transcription by inducing Sp100 proteins. J Virol 90:694–704. 10.1128/JVI.02137-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stepp WH, Stamos JD, Khurana S, Warburton A, McBride AA. 2017. Sp100 colocalizes with HPV replication foci and restricts the productive stage of the infectious cycle. PLoS Pathog 13:e1006660. 10.1371/journal.ppat.1006660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shishido-Hara Y, Higuchi K, Ohara S, Duyckaerts C, Hauw JJ, Uchihara T. 2008. Promyelocytic leukemia nuclear bodies provide a scaffold for human polyomavirus JC replication and are disrupted after development of viral inclusions in progressive multifocal leukoencephalopathy. J Neuropathol Exp Neurol 67:299–308. 10.1097/NEN.0b013e31816a1dd3. [DOI] [PubMed] [Google Scholar]
- 24.Khaiboullina SF, Morzunov SP, Boichuk SV, Palotas A, St Jeor S, Lombardi VC, Rizvanov AA. 2013. Death-domain associated protein-6 (DAXX) mediated apoptosis in hantavirus infection is counter-balanced by activation of interferon-stimulated nuclear transcription factors. Virology 443:338–348. 10.1016/j.virol.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 25.Neumann F, Czech-Sioli M, Dobner T, Grundhoff A, Schreiner S, Fischer N. 2016. Replication of Merkel cell polyomavirus induces reorganization of promyelocytic leukemia nuclear bodies. J Gen Virol 97:2926–2938. 10.1099/jgv.0.000593. [DOI] [PubMed] [Google Scholar]
- 26.Hahn AS, Großkopf AK, Jungnickl D, Scholz B, Ensser A. 2016. Viral FGARAT homolog ORF75 of rhesus monkey rhadinovirus effects proteasomal degradation of the ND10 components SP100 and PML. J Virol 90:8013–8028. 10.1128/JVI.01181-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negorev DG, Vladimirova OV, Maul GG. 2009. Differential functions of interferon-upregulated Sp100 isoforms: herpes simplex virus type 1 promoter-based immediate-early gene suppression and PML protection from ICP0-mediated degradation. J Virol 83:5168–5180. 10.1128/JVI.02083-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sternsdorf T, Jensen K, Reich B, Will H. 1999. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J Biol Chem 274:12555–12566. 10.1074/jbc.274.18.12555. [DOI] [PubMed] [Google Scholar]
- 29.Bloch DB, de la Monte SM, Guigaouri P, Filippov A, Bloch KD. 1996. Identification and characterization of a leukocyte-specific component of the nuclear body. J Biol Chem 271:29198–29204. 10.1074/jbc.271.46.29198. [DOI] [PubMed] [Google Scholar]
- 30.Xie K, Lambie EJ, Snyder M. 1993. Nuclear dot antigens may specify transcriptional domains in the nucleus. Mol Cell Biol 13:6170–6179. 10.1128/mcb.13.10.6170-6179.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Negorev D, Ishov AM, Maul GG. 2001. Evidence for separate ND10-binding and homo-oligomerization domains of Sp100. J Cell Sci 114:59–68. 10.1242/jcs.114.1.59. [DOI] [PubMed] [Google Scholar]
- 32.Seeler JS, Marchio A, Sitterlin D, Transy C, Dejean A. 1998. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc Natl Acad Sci USA 95:7316–7321. 10.1073/pnas.95.13.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seeler JS, Marchio A, Losson R, Desterro JM, Hay RT, Chambon P, Dejean A. 2001. Common properties of nuclear body protein SP100 and TIF1alpha chromatin factor: role of SUMO modification. Mol Cell Biol 21:3314–3324. 10.1128/MCB.21.10.3314-3324.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Surdo PL, Bottomley MJ, Sattler M, Scheffzek K. 2003. Crystal structure and nuclear magnetic resonance analyses of the SAND domain from glucocorticoid modulatory element binding protein-1 reveals deoxyribonucleic acid and zinc binding regions. Mol Endocrinol 17:1283–1295. 10.1210/me.2002-0409. [DOI] [PubMed] [Google Scholar]
- 35.Guldner HH, Szostecki C, Schroder P, Matschl U, Jensen K, Luders C, Will H, Sternsdorf T. 1999. Splice variants of the nuclear dot-associated Sp100 protein contain homologies to HMG-1 and a human nuclear phosphoprotein-box motif. J Cell Sci 112:733–747. 10.1242/jcs.112.5.733. [DOI] [PubMed] [Google Scholar]
- 36.Newhart A, Negorev DG, Rafalska-Metcalf IU, Yang T, Maul GG, Janicki SM. 2013. Sp100A promotes chromatin decondensation at a cytomegalovirus-promoter-regulated transcription site. Mol Biol Cell 24:1454–1468. 10.1091/mbc.E12-09-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Zhao D, Xiong X, He Z, Li H. 2016. Multifaceted histone H3 methylation and phosphorylation readout by the plant homeodomain finger of human nuclear antigen Sp100C. J Biol Chem 291:12786–12798. 10.1074/jbc.M116.721159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yordy JS, Li RZ, Sementchenko VI, Pei HP, Muise-Helmericks RC, Watson DK. 2004. SP100 expression modulates ETS1 transcriptional activity and inhibits cell invasion. Oncogene 23:6654–6665. 10.1038/sj.onc.1207891. [DOI] [PubMed] [Google Scholar]
- 39.Isaac A, Wilcox KW, Taylor JL. 2006. SP100B, a repressor of gene expression preferentially binds to DNA with unmethylated CpGs. J Cell Biochem 98:1106–1122. 10.1002/jcb.20841. [DOI] [PubMed] [Google Scholar]
- 40.Taylor JL, Isaac A, Yang M, Wilcox K. 2005. SP100B as a repressor of viral gene expression. Invest Ophthalmol Vis Sci 46:1022. [Google Scholar]
- 41.Collados Rodriguez M. 2021. The fate of speckled protein 100 (Sp100) during herpesviruses infection. Front Cell Infect Microbiol 10:607526. 10.3389/fcimb.2020.607526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Everett RD, Parsy ML, Orr A. 2009. Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J Virol 83:4963–4977. 10.1128/JVI.02593-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maul GG, Negorev D, Bell P, Ishov AM. 2000. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol 129:278–287. 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- 44.Seeler JS, Dejean A. 2001. SUMO: of branched proteins and nuclear bodies. Oncogene 20:7243–7249. 10.1038/sj.onc.1204758. [DOI] [PubMed] [Google Scholar]
- 45.Lang M, Jegou T, Chung I, Richter K, Munch S, Udvarhelyi A, Cremer C, Hemmerich P, Engelhardt J, Hell SW, Rippe K. 2010. Three-dimensional organization of promyelocytic leukemia nuclear bodies. J Cell Sci 123:392–400. 10.1242/jcs.053496. [DOI] [PubMed] [Google Scholar]
- 46.Sternsdorf T, Jensen K, Will H. 1997. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol 139:1621–1634. 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maarifi G, Maroui MA, Dutrieux J, Dianoux L, Nisole S, Chelbi-Alix MK. 2015. Small ubiquitin-like modifier alters IFN response. J Immunol 195:2312–2324. 10.4049/jimmunol.1500035. [DOI] [PubMed] [Google Scholar]
- 48.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. 2006. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281:16117–16127. 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 49.Negorev DG, Vladimirova OV, Ivanov A, Rauscher F, III, Maul GG. 2006. Differential role of Sp100 isoforms in interferon-mediated repression of herpes simplex virus type 1 immediate-early protein expression. J Virol 80:8019–8029. 10.1128/JVI.02164-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cuchet D, Sykes A, Nicolas A, Orr A, Murray J, Sirma H, Heeren J, Bartelt A, Everett RD. 2011. PML isoforms I and II participate in PML-dependent restriction of HSV-1 replication. J Cell Sci 124:280–291. 10.1242/jcs.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cuchet-Lourenco D, Boutell C, Lukashchuk V, Grant K, Sykes A, Murray J, Orr A, Everett RD. 2011. SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog 7:e1002123. 10.1371/journal.ppat.1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol 80:7995–8005. 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Everett RD, Parada C, Gripon P, Sirma H, Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol 82:2661–2672. 10.1128/JVI.02308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lukashchuk V, Everett RD. 2010. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J Virol 84:4026–4040. 10.1128/JVI.02597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sloan E, Orr A, Everett RD. 2016. MORC3, a component of PML nuclear bodies, has a role in restricting herpes simplex virus 1 and human cytomegalovirus. J Virol 90:8621–8633. 10.1128/JVI.00621-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McFarlane S, Orr A, Roberts APE, Conn KL, Iliev V, Loney C, da Silva Filipe A, Smollett K, Gu Q, Robertson N, Adams PD, Rai TS, Boutell C. 2019. The histone chaperone HIRA promotes the induction of host innate immune defences in response to HSV-1 infection. PLoS Pathog 15:e1007667. 10.1371/journal.ppat.1007667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alandijany T, Roberts APE, Conn KL, Loney C, McFarlane S, Orr A, Boutell C. 2018. Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection. PLoS Pathog 14:e1006769. 10.1371/journal.ppat.1006769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu P, Roizman B. 2017. The SP100 component of ND10 enhances accumulation of PML and suppresses replication and the assembly of HSV replication compartments. Proc Natl Acad Sci USA 114:E3823–E3829. 10.1073/pnas.1703395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maul GG, Guldner HH, Spivack JG. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J Gen Virol 74:2679–2690. 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 60.Maul GG, Everett RD. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol 75:1223–1233. 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 61.Merkl PE, Orzalli MH, Knipe DM. 2018. Mechanisms of host IFI16, PML, and Daxx protein restriction of herpes simplex virus 1 replication. J Virol 92:e00057-18. 10.1128/JVI.00057-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jan Fada B, Kaadi E, Samrat SK, Zheng Y, Gu H. 2020. Effect of SUMO-SIM interaction on the ICP0-mediated degradation of PML isoform II and its associated proteins in herpes simplex virus 1 infection. J Virol 94:e00470-20. 10.1128/JVI.00470-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu P, Mallon S, Roizman B. 2016. PML plays both inimical and beneficial roles in HSV-1 replication. Proc Natl Acad Sci USA 113:E3022–E3028. 10.1073/pnas.1605513113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deschamps T, Kalamvoki M. 2018. Extracellular vesicles released by herpes simplex virus 1-infected cells block virus replication in recipient cells in a STING-dependent manner. J Virol 92:e01102-18. 10.1128/JVI.01102-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dogrammatzis C, Saleh S, Deighan C, Kalamvoki M. 2021. Diverse populations of extracellular vesicles with opposite functions during herpes simplex virus 1 infection. J Virol 95:e02357-20. 10.1128/JVI.02357-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kalamvoki M, Deschamps T. 2016. Extracellular vesicles during herpes simplex virus type 1 infection: an inquire. Virol J 13:63. 10.1186/s12985-016-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bello-Morales R, Lopez-Guerrero JA. 2018. Extracellular vesicles in herpes viral spread and immune evasion. Front Microbiol 9:2572. 10.3389/fmicb.2018.02572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalamvoki M, Du T, Roizman B. 2014. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc Natl Acad Sci USA 111:E4991–E4996. 10.1073/pnas.1419338111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bello-Morales R, Ripa I, Lopez-Guerrero JA. 2020. Extracellular vesicles in viral spread and antiviral response. Viruses 12:623. 10.3390/v12060623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bello-Morales R, Praena B, de la Nuez C, Rejas MT, Guerra M, Galan-Ganga M, Izquierdo M, Calvo V, Krummenacher C, Lopez-Guerrero JA. 2018. Role of microvesicles in the spread of herpes simplex virus 1 in oligodendrocytic cells. J Virol 92:e00088-18. 10.1128/JVI.00088-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dogrammatzis C, Deschamps T, Kalamvoki M. 2019. Biogenesis of extracellular vesicles during herpes simplex virus 1 infection: role of the CD63 tetraspanin. J Virol 93:e01850-18. 10.1128/JVI.01850-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roizman B, Knipe DM. 2001. Herpes simplex viruses and their replication, p 2399–2459. In Knipe DM, Howley P, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 73.Saeki Y, Ichikawa T, Saeki A, Chiocca EA, Tobler K, Ackermann M, Breakefield XO, Fraefel C. 1998. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum Gene Ther 9:2787–2794. 10.1089/hum.1998.9.18-2787. [DOI] [PubMed] [Google Scholar]
- 74.Nkosi D, Howell LA, Cheerathodi MR, Hurwitz SN, Tremblay DC, Liu X, Meckes DG, Jr.. 2018. Transmembrane domains mediate intra- and extracellular trafficking of Epstein-Barr virus latent membrane protein 1. J Virol 92:e00280-18. 10.1128/JVI.00280-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao M, Nanbo A, Sun L, Lin Z. 2019. Extracellular vesicles in Epstein-Barr virus’ life cycle and pathogenesis. Microorganisms 7:48. 10.3390/microorganisms7020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nkosi D, Sun L, Duke LC, Patel N, Surapaneni SK, Singh M, Meckes DG, Jr.. 2020. Epstein-Barr virus LMP1 promotes syntenin-1- and Hrs-induced extracellular vesicle formation for its own secretion to increase cell proliferation and migration. mBio 11:e00589-20. 10.1128/mBio.00589-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jeon H, Yoo S-M, Choi HS, Mun JY, Kang H-G, Lee J, Park J, Gao S-J, Lee M-S. 2017. Extracellular vesicles from KSHV-infected endothelial cells activate the complement system. Oncotarget 8:99841–99860. 10.18632/oncotarget.21668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeon H, Lee J, Lee S, Kang SK, Park SJ, Yoo SM, Lee MS. 2019. Extracellular vesicles from KSHV-infected cells stimulate antiviral immune response through mitochondrial DNA. Front Immunol 10:876. 10.3389/fimmu.2019.00876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Najjar SA, Davis BM, Albers KM. 2020. Epithelial-neuronal communication in the colon: implications for visceral pain. Trends Neurosci 43:170–181. 10.1016/j.tins.2019.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kowtharapu BS, Stahnke T, Wree A, Guthoff RF, Stachs O. 2014. Corneal epithelial and neuronal interactions: role in wound healing. Exp Eye Res 125:53–61. 10.1016/j.exer.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 81.Kabata H, Artis D. 2019. Neuro-immune crosstalk and allergic inflammation. J Clin Invest 129:1475–1482. 10.1172/JCI124609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J Virol 77:1382–1391. 10.1128/jvi.77.2.1382-1391.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thery C, Amigorena S, Raposo G, Clayton A. 2006. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3:Unit 3.22. 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]