ABSTRACT
Epstein-Barr virus (EBV) is an oncogenic herpesvirus that is associated with 200,000 new cases of cancer and 140,000 deaths annually. To date, there are no available vaccines or therapeutics for clinical usage. Recently, the viral heterodimer glycoprotein gH/gL has become a promising target for the development of prophylactic vaccines against EBV. Here, we developed the anti-gH antibody 6H2 and its chimeric version C6H2, which had full neutralizing activity in epithelial cells and partial neutralizing activity in B cells. C6H2 exhibited potent protection against lethal EBV challenge in a humanized mouse model. The cryo-electron microscopy (cryo-EM) structure further revealed that 6H2 recognized a previously unidentified epitope on gH/gL D-IV that is critical for viral attachment and subsequent membrane fusion with epithelial cells. Our results suggest that C6H2 is a promising candidate in the prevention of EBV-induced lymphoproliferative diseases (LPDs) and may inform the design of an EBV vaccine.
IMPORTANCE Epstein-Barr virus (EBV) is a ubiquitous gammaherpesvirus that establishes lifelong persistence and is related to multiple diseases, including cancers. Neutralizing antibodies (NAbs) have proven to be highly effective in preventing EBV infection and subsequent diseases. Here, we developed an anti-EBV-gH NAb, 6H2, which blocked EBV infection in vitro and in vivo. This 6H2 neutralizing epitope should be helpful to understand EBV infection mechanisms and guide the development of vaccines and therapeutics against EBV infection.
KEYWORDS: Epstein-Barr virus, glycoprotein H, neutralizing antibody, humanized mouse model, cryo-electron microscopy
INTRODUCTION
Epstein-Barr virus (EBV) is a prototypical member of human gammaherpesvirus (1, 2), which establishes lifelong latency in more than 90% of the world’s population and threatens global health as an oncogenic virus (3, 4). EBV is a causative agent of infectious mononucleosis (IM) and multiple malignancies, including Hodgkin’s lymphoma, diffuse large B-cell lymphoma, Burkitt’s lymphoma, T/NK cell lymphoma, nasopharyngeal carcinoma, and 10% of gastric carcinomas that are EBV positive (5–7). In addition, EBV is associated with lymphoproliferative diseases (LPDs) in immunodeficient individuals, including AIDS patients and organ transplant recipients (5). Prophylactic vaccines against human oncogenic pathogens such as hepatitis B virus (HBV) and human papillomavirus (HPV) have proven to be efficient in reducing tumorigenesis, indicating that vaccine strategies would be viable for cancer prevention (8). However, no prophylactic or therapeutic vaccine has been approved against EBV infection or EBV-associated diseases to date (9).
The close association of EBV with lymphomas and epithelial carcinomas suggests that EBV infection is a crucial event in these two types of cancers (10–12). B cells are the major reservoir for EBV, and EBV exhibits high tumorigenic potential, with the ability to transform human B cells into growing lymphoblastoid cell lines (LCLs) (13–15). Epithelial cell infection of EBV is essential for viral genome replication and production of infectious viral particles for transmission through saliva (16). During its life cycle, EBV shuttles between B cells and epithelial cells, which facilitates its persistence and transmission in infected hosts. Hence, EBV-related work has focused on these two major target cells. EBV infects B cells and epithelial cells through different mechanisms involving the participation of various envelope glycoproteins (1, 9, 17). At least five envelope glycoproteins, gp350, gp42, gH, gL, and gB, have been reported to participate in B-cell infection, while EBV entry into epithelial cells involves BMRF2, gH, gL, and gB (18). gH/gL and gB fusogen comprise the core fusion machinery for membrane fusion and virus entry (19). The gH/gL or gH/gL/gp42 entry complex is an essential component to trigger fusogen gB-mediated membrane fusion (20). Heterodimer gH/gL, which consists of soluble gL and transmembrane gH, is important for the infection of both B cells and epithelial cells. When infecting B cells, gp42 is required to form a stable gH/gL/gp42 complex to engage human leukocyte antigen (HLA) class II (21, 22). gH/gL directly binds to receptors such as Ephrin type-A receptor 2 (EphA2) and integrins αvβ5, αvβ6, and/or αvβ8 during entry into epithelial cells (23–26).
The crystal structure of the ectodomain of EBV gH in complex with gL (gH/gL) has been resolved and exhibits an elongated rod-like conformation, while HSV-2 and vesicular stomatitis virus (VSV) gH/gL adopt a boot-like shape (27–29). gH/gL comprises four major domains (D-I to D-IV), and D-I is associated with soluble gL (amino acids [aa] 24 to 131) and the N terminus (aa 20 to 65) of gH, while the remaining region of gH comprises D-II to D-IV (D-II, aa 66 to 344; D-III, aa 345 to 529; D-IV, aa 530 to 672) (27). Multiple regions of gH/gL are important for EBV entry. D-I is responsible for binding to the recently discovered epithelial cell receptor of EphA2, regulating gB activation and potentially defining a gB binding site (25, 26, 30, 31). A KGD (lysine-glycine-aspartic acid) motif in D-II was implicated in binding to integrins (28). The gp42 N-terminal binding site crosses through gH/gL D-II to D-IV (32, 33). Structural and mutagenicity analysis demonstrated that D-I, the D-I/D-II interface, and D-IV participate in the membrane fusion process (32, 34–38). Overall, different gH/gL domains display variable and indispensable functions, which makes gH/gL a promising target for the design of vaccines and therapeutics.
Recently, antibodies against gH/gL in human plasma were found to be indispensable to block EBV infection of both B cells and epithelial cells, indicating that gH/gL is a principal target of the humoral immune response (39). Furthermore, gH/gL-based immunogens elicited high levels of neutralizing antibodies (NAbs) in multiple animal models, including mice, rabbits, and nonhuman primates (39–41). NAbs have become an increasingly important tool for investigating viral infection mechanisms, and a series of anti-EBV gH/gL NAbs have been generated and characterized. Most of the anti-gH/gL NAbs, such as E1D1, CL40, and CL59, are of murine origin and efficiently block epithelial cell infection but not B-cell infection (23, 42–44). Two human NAbs, AMMO1 and 769B10, were proven to effectively neutralize EBV infection in both cell types (39, 45). AMMO1 defines a binding site at the D-I/D-II interface (45). Structural studies demonstrated that E1D1 binds exclusively to gL (32). CL40 binds to the region at the D-II/D-III interface, which would compete with AMMO1 and 769B10 (39, 46). Among the NAbs mentioned above, only CL59 recognized gH/gL D-IV, and mutations within gH/gL D-IV affected fusion function (38, 46). Epitope determination of these NAbs together revealed that multiple regions of gH/gL are critical for EBV infection.
Here, we obtained and characterized an EBV gH/gL-specific murine NAb 6H2 targeting gH/gL D-IV, which could block EBV infection of epithelial cells and partially prevent B-cell infection. The chimeric form of 6H2, C6H2, showed potent protection against EBV-induced LPD and reduced viral load in peripheral blood in a humanized mouse model. Cryo-electron microscopy (cryo-EM) revealed that 6H2 recognized a novel epitope referring to three loops on D-IV of gH/gL. 6H2 prevented gH/gL or gH/gL/gp42 complex binding to the cell surface and significantly inhibited subsequent membrane fusion with epithelial cell. Our study highlights the importance of D-IV as an alternative site for the rational design of gH/gL-based therapeutics and subunit vaccines.
RESULTS
Characterization of EBV gH/gL-specific NAb 6H2.
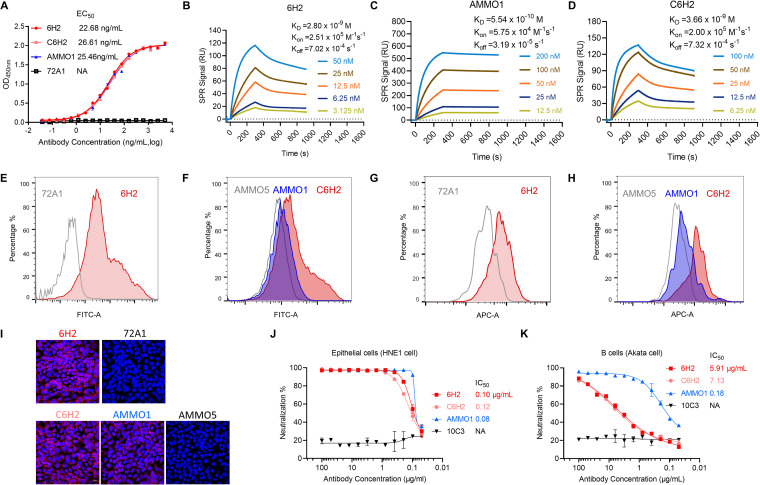
Recombinant EBV gH/gL derived from 293F cells was used for mouse immunization and subsequent NAb screening. One MAb, 6H2, that can neutralize EBV infection of epithelial cells and partially neutralize B-cell infection was obtained. We then evaluated the binding ability of 6H2 by both enzyme-linked immunosorbent assay (ELISA) and surface plasmon resonance (SPR) assay. To further evaluate the potential clinical application of 6H2, we also constructed a chimeric 6H2 (C6H2), which contained a human IgG1 Fc. Both 6H2 and C6H2 effectively bound to purified gH/gL protein with half-maximal effective concentrations (EC50) of 22.68 ng/mL and 26.61 ng/mL, respectively, which were comparable to that of AMMO1 (25.46 ng/mL) (Fig. 1A). For SPR analysis, 6H2 followed fast-on (association rate constant [Kon] of 2.51 × 105 M-1s-1) and slow-off (dissociation rate constant [Koff] of 7.02 × 10−4 s−1) binding kinetics, resulting in an equilibrium dissociation constant (KD) of 2.80 nM (Fig. 1B), which was ∼5-fold lower than that of AMMO1 (0.55 nM) (Fig. 1C). C6H2 showed a similar affinity of KD (3.66 nM) with that of murine 6H2 (2.80 nM) (Fig. 1D).
FIG 1.
Binding and neutralizing efficacies of neutralizing antibody (NAb) 6H2. (A) The binding abilities of 6H2, C6H2 (chimeric version of 6H2), and AMMO1 to purified gH/gL protein were determined by indirect enzyme-limited immunosorbent assay (ELISA), and the half-maximal effective concentration (EC50) values are shown. (B, C, and D) The binding affinity of (B) 6H2, (C) AMMO1, and (D) C6H2 to gH/gL was measured by surface plasmon resonance. (E to H) 6H2, C6H2, AMMO1, and isotype antibody controls (72A1 and AMMO5) were used to test their binding abilities to cell surface-expressed gH/gL in transfected 293T cells (E and F) and EBV-positive CNE2 cells (G and H) using flow cytometry. (I) Detection of the antibody binding abilities with gH/gL-expressed Cos7 cells using immunofluorescence. (J and K) Serial dilutions of the indicated antibodies were evaluated for their abilities to neutralize EBV infection in HNE1 cells (E) and Akata cells (F). 6H2, C6H2, AMMO1, and 10C3 (negative control) are shown as red, light red, blue, and black lines, respectively. Data are presented as the mean ± standard error of the mean (SEM).
Next, we assessed whether 6H2 can recognize native gH/gL. 293T cells were cotransfected with plasmids encoding the full-length gH and gL. The cells were then incubated with 6H2, C6H2, and AMMO1. 72A1 (47) and AMMO5 (45) were used as negative controls. Flow cytometric analysis demonstrated that 6H2, C6H2, and AMMO1 could bind to the heterodimeric gH/gL complex expressed on the cell surface (Fig. 1E and F). We also used EBV-positive CNE2 cells to test the binding properties of 6H2, C6H2, and AMMO1. We found that three antibodies could specifically bind to the virus-encoded native form of gH/gL embedded on the surface of the tested cells (Fig. 1G and H). The binding activities of 6H2 were further confirmed by immunofluorescence (IF) assay (Fig. 1I). Heterodimer gH/gL is a trigger factor for gB activation and promotes gB-induced membrane fusion. (19, 48). We next evaluated the abilities of 6H2 and C6H2 to neutralize EBV infection in both epithelial and B cells. In the HNE1 cell-based neutralization assay, 6H2 and C6H2 showed potent neutralizing activity with half-maximal inhibitory concentration (IC50) values of 0.10 μg/mL and 0.12 μg/mL, comparable to that of AMMO1 (0.08 μg/mL) (Fig. 1J). The neutralizing efficacy of 6H2 against B-cell infection was less potent than that in epithelial cells. Even though it was unable to completely block B-cell infection, 6H2 was able to greatly reduce it (Fig. 1K). AMMO1 showed complete neutralizing ability in B-cell infection, with an IC50 value of 0.18 μg/mL (Fig. 1K). We identified that NAb 6H2 could prevent EBV infection of epithelial cells and greatly reduce EBV infection of B cells, indicating that multiple mechanisms are involved in the neutralization of EBV infection by 6H2 in different cell types.
6H2 conferred protection against lethal EBV challenge in humanized mice.
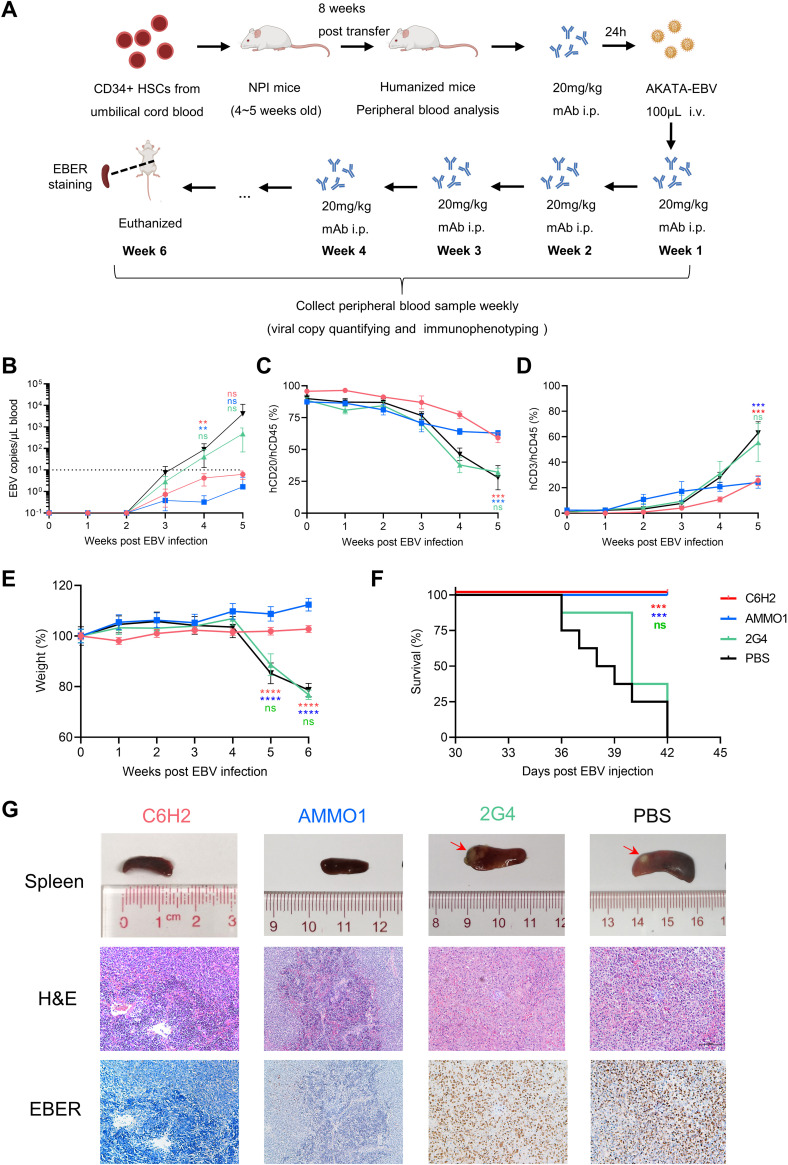
Several antibodies have been reported previously to prevent EBV-driven tumor formation in humanized mice (49, 50). To examine whether 6H2 can protect mice against EBV-related disease, we carried out a similar in vivo assessment in a humanized mouse model susceptible to EBV infection (51–54). NOD-Prkdcnull IL2Rγnull (NPI) mice were chosen and inoculated with CD34-positive (CD34+) hematopoietic stem cells from umbilical cord blood. The humanized immune system was reconstructed after 8 weeks (Fig. 2A). Next, we assessed the protection of C6H2 in vivo. AMMO1 (49) and the anti-Ebola antibody 2G4 (55, 56) were used as positive and negative controls, respectively. Animal physical condition was monitored by documenting body weight. All animals were euthanized 6 weeks after EBV challenge. EBV DNA copies in peripheral blood became detectable on week 3 post EBV challenge. C6H2-treated and AMMO1-treated mice remained aviremic, with viral DNA copy numbers under 10 copies/μL (Fig. 2B), although antibody was likely still present in the blood in C6H2-treated animals at the time the blood was drawn. Mice in the 2G4 and phosphate-buffered saline (PBS) groups showed viremia in which EBV duplicated rapidly, and the DNA copy numbers were approximately 100-fold higher than those of the C6H2 and AMMO1 groups at week 5 postchallenge (Fig. 2B).
FIG 2.
6H2 provides potent protection against EBV challenge in humanized mice. (A) Experimental timeline for CD34+ hematopoietic stem cell (HSC) engraftment, antibody injection, virus challenge, and monitoring for various biological and clinical features. Humanized mice received 20 mg/kg C6H2, AMMO1 positive control, 2G4 negative control, or an equal volume of phosphate-buffered saline (PBS) (infected control) via intraperitoneal (i.p.) injection 24 h prior to intravenous (i.v.) challenge with Akata-EBV. After virus challenge, antibodies or PBS were reinjected weekly 4 additional times. Schematic diagram of mouse was created with BioRender.com. (B) Viral DNA copies in the peripheral blood of infected mice were evaluated by real-time PCR (RT-PCR). The dashed line indicated the limit of detection (10 copies/μL). (C and D) Blood samples from antibody-treated or PBS-treated humanized mice were collected, and peripheral blood mononuclear cells (PBMCs) were isolated weekly. The frequencies of hCD45+ hCD20+ cells (C) and hCD45+ hCD3+ cells (D) in PBMCs were analyzed by flow cytometry. (E) Body weights of infected mice were monitored weekly. (F) The survival curves of mice treated with C6H2, AMMO1, 2G4, or PBS. (G) Pathological analyses of EBV-infected mice. Photograph of spleens and photomicrographs of hematoxylin and eosin (H&E) and EBV-encoded small RNA (EBER) in situ hybridization of spleen sections from infected mice treated with C6H2, AMMO1, 2G4, and PBS. All data are presented as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001, ****; P < 0.0001; ns, not significant.
During the animal trials, peripheral blood mononuclear cells (PBMCs) were isolated and analyzed each week. hCD20+ B cells in mice from the 2G4 and PBS groups had a significant decrease that was concurrent with a marked increase in the frequency of hCD3+ T cells compared to that of the C6H2 and AMMO1 groups (Fig. 2C and D). This was likely because human B cells from the 2G4 and PBS groups were infected by EBV, and infected B cells were effectively recognized and removed by EBV-specific cytotoxic T cells, which was consistent with previous reports (49, 54). C6H2 and AMMO1 successfully blocked in vivo EBV infection without a decline in B cells or an increase in T cells (Fig. 2C and D).
Animals treated with 2G4 and PBS exhibited significant body weight loss from week 4 to week 6 that was concurrent with increasing levels of EBV DNA (Fig. 2B and E). In contrast, the mouse body weights of the C6H2 and AMMO1 groups were relatively stable (Fig. 2E). By the end of the study, all of the mice treated with PBS and 2G4 were euthanized due to substantial body weight loss, while all mice from the C6H2 and AMMO1 groups survived the challenge (Fig. 2F). Furthermore, spleens from different groups were collected and photographed after euthanasia. The spleens from C6H2-treated and AMMO1-treated mice were normal and of equal size (Fig. 2G). Animals from the 2G4 and PBS groups showed extreme splenomegaly with overt and pale tumor formation on the spleen surface, which should be a sign of lymphoproliferative disorder (Fig. 2G). We further performed histopathology analysis on paraffin sections of spleens using hematoxylin and eosin (H&E) staining and in situ hybridization for Epstein-Barr virus-encoded RNAs (EBERs). The spleens of mice treated with 2G4 and PBS presented with typical lymphoproliferative disease (LPD) due to EBV infection, which consisted of large and atypical lymphoid cells that were positive for EBER staining (Fig. 2G). In contrast, C6H2 and AMMO1 protected mice from LPD without obvious EBER staining (Fig. 2G).
Structural insight into the interaction of NAb 6H2 and gH.
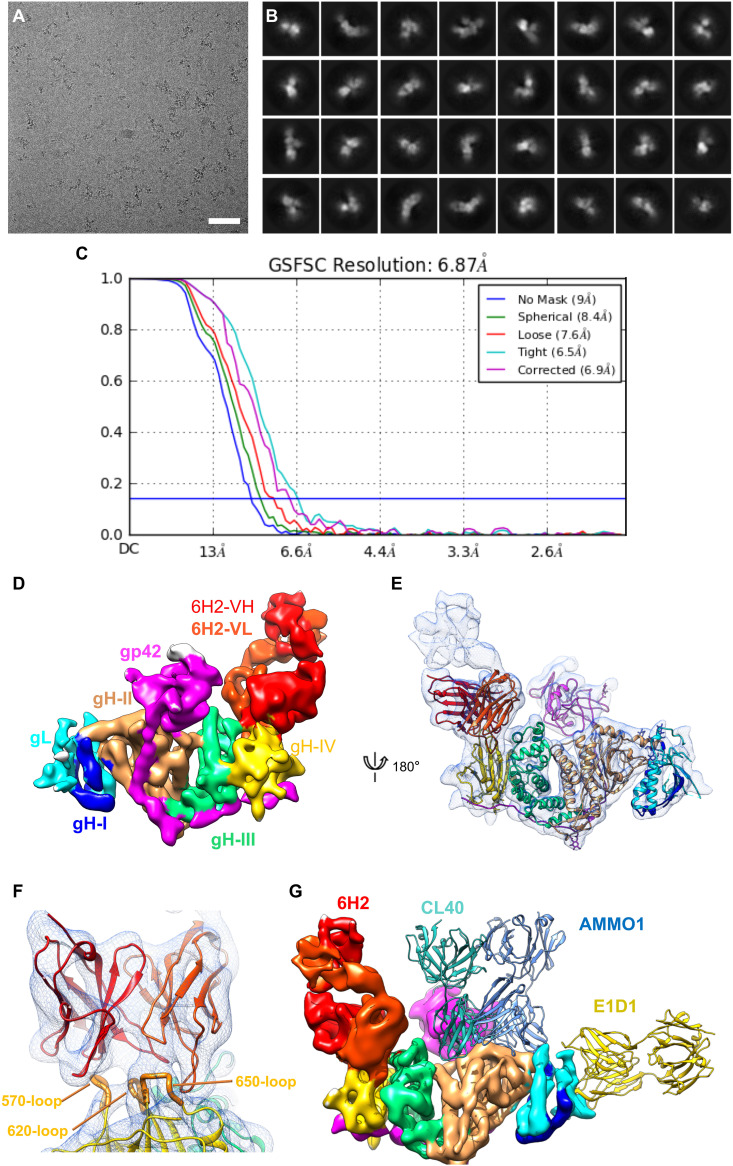
To characterize the structural basis for the interaction between NAb 6H2 and gH/gL, we determined the structure of the 6H2 Fab in complex with gH/gL/gp42 using single particle cryo-EM and obtained a three-dimensional (3D) structure of the complex at an overall resolution of ∼6.9 Å (Fig. 3A to C). In the moderate resolution density map of the complex, most of the α-helixes and β-strands were separated, and several bulky side chains could be visualized (Fig. 3D and E). The previously reported crystal structures of EBV gH/gL/gp42 (PDB no. 5T1D) could be well fitted to our density map with distinguishable D-I to D-IV of the gH/gL and gp42 subunits (Fig. 3E). The 6H2 Fab bound mainly to the D-IV of gH/gL, without extra interaction with gL or gp42 (Fig. 3E).
FIG 3.
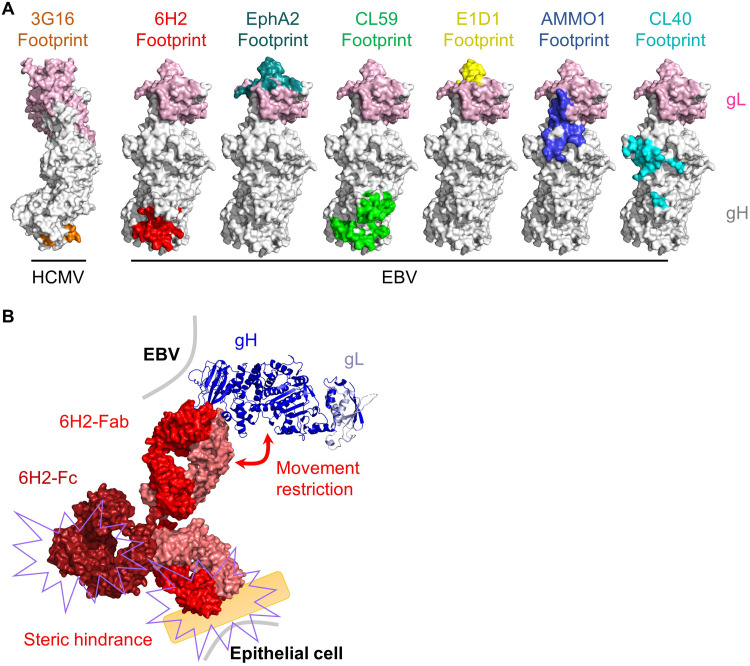
6H2 targets a novel epitope on gH/gL D-IV. (A) Motion-corrected micrograph of frozen-hydrated gH/gL/gp42/6H2 complex; bar, 50 nm. (B) Representative two-dimensional (2D) class averages of gH/gL/gp42/6H2. (C) The gold-standard Fourier shell correlation (FSC) curve of the 3D reconstruction of gH/gL/gp42/6H2. (D) Cryo-electron microscopy (cryo-EM) structure of gH/gL/gp42/6H2. gL is colored in cyan. gH is colored blue in D-I, orange in D-II, green in D-III, and yellow in D-IV. gp42 is colored in pink. 6H2 is colored in red. (E) Ribbon diagram of the gH/gL/gp42/6H2 complex structure. gH/gL, gp42 and 6H2 are colored as in panel D. (F) Close-up view of 6H2 CDR interfaces with gH. 6H2 contacts three loops on gH/gL D-IV (570, 620, and 650 loops). (G) Structure comparison of the binding modes of gH/gL NAbs. E1D1 (PDB no. 5T1D), CL40 (PDB no. 5W0K), and AMMO1 (PDB no. 6C5V) are fitted in a density map of gH/gL/gp42/6H2 and shown in ribbon style and colored yellow, cyan, and light blue, respectively.
To further inspect the 6H2 binding interface with gH, we also built a homology model of 6H2 Fab using Discovery Studio 2.0. The Fab density in contact with the D-IV of gH/gL was well resolved and allowed us to fit the 6H2 Fab model into a density map (Fig. 3F). All six complementarity-determining region (CDR) loops fit into the density unambiguously and therefore revealed a solid piece of density connection between 6H2 Fab and gH/gL. The 6H2 Fab interacted with gH mainly by the contact of its HCDR3 to the gH 570 loop and that of LCDR2 and LCDR3 to the gH 620 and 650 loops. We then fitted the complex structures of AMMO1, E1D1, and CL40 in our density map to compare the binding sites of these NAbs (Fig. 3G). D-IV was excluded from the binding sites of AMMO1, E1D1, and CL40, and the location of 6H2 on gH/gL represented a novel binding mode without any overlap with other reported NAbs. Taken together, our structural analysis combining the cryo-EM map and homology model information revealed a unique neutralizing epitope located on D-IV of gH/gL.
Residues involved in 6H2 binding are critical for correct folding and fusion function of gH/gL.
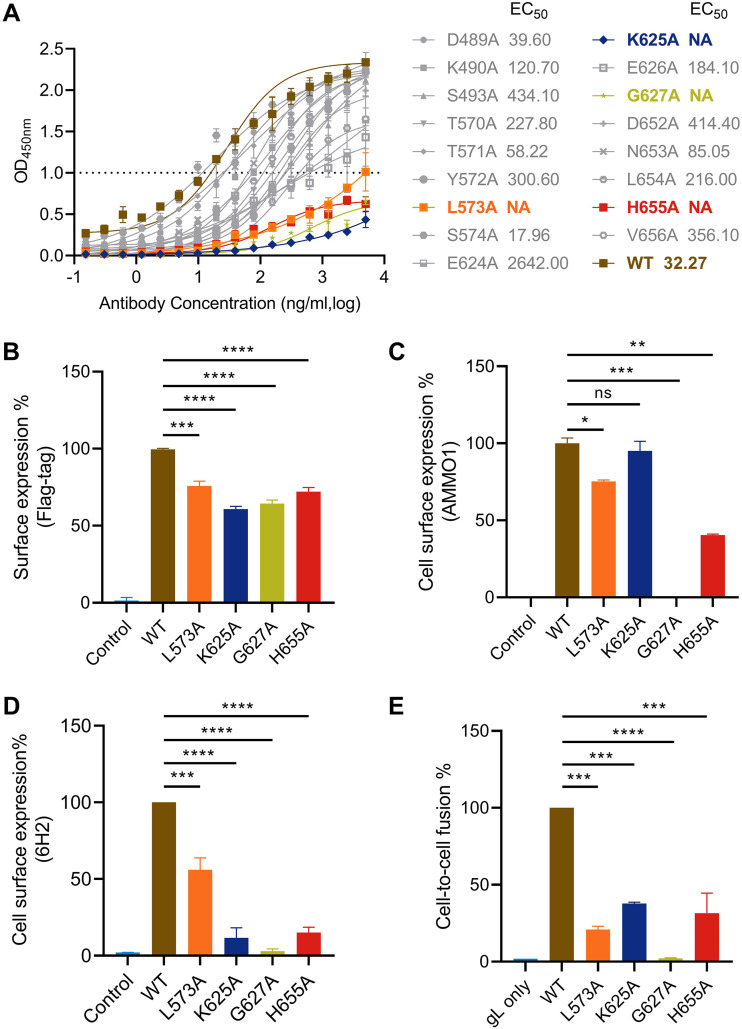
To further identify the key residues located on the 6H2-gH/gL interface, we chose a series of residues based on structural analysis for single alanine substitution. We found that 6H2 Fab interacted with three loops (the 570, 620, and 650 loops) on gH D-IV. Hence, we selected the amino acids on these three loops for further alanine-scanning mutagenesis (T570/T571/Y572/L573/S574 of the 570 loop, E624/K625/E626/G627 of the 620 loop, and D652/N653/L654/H655/V656 of the 650 loop). The adjacent amino acids, including D489, K490, and S493, were included as well. The soluble mutant gH/gL proteins were constructed, recombinantly expressed, and purified. The binding ability of 6H2 to mutants was assessed by indirect ELISA. Soluble gH/gL mutants showed similar expression efficiency to that of wild-type (WT) recombinant gH/gL but resulted in different impacts on 6H2 binding (Fig. 4A). Among all 17 residues, most of the mutants retained their binding with 6H2, while four mutated residues (L573A, K625A, G627A, and H655A) significantly decreased the 6H2 binding capacity (optical density [OD] value below 1.0 at 5 μg/mL) (Fig. 4A). These four residues were located on the 570, 620, and 650loops, which was consistent with the structural analysis.
FIG 4.
Identification of key residues involved in the gH/gL-6H2 interaction and their roles in virus-triggered cell fusion. (A) Binding activities of 6H2 to mutant gH/gL proteins were assessed by ELISA. (B) Detection of surface expression of Flag-tagged gH cotransfected with wild-type gL (WT-gL) in 293T cells using anti-Flag antibody. (C and D) Expression amount of WT-gH or mutated gH cotransfected with WT-gL in 293T cells. Surface expression amount of gH/gL evaluated by AMMO1 (C) and 6H2 (D). (E) The influence of gH mutants on gH/gL-mediated cell-to-cell fusion. All values are normalized as a percentage to WT gH/gL. All data are presented as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
We chose these four residues for further evaluation to identify their roles in the 6H2-gH/gL interaction. We individually introduced the mutations into full-length gH carrying a Flag tag in the N terminus and transfected mutated gH together with WT-gL into 293T cells. As previously reported, AMMO1 binding is dependent on the coexpression of both gH and gL and recognizes a conformational epitope across D-I and D-II and distant from D-IV (45). Then, we used AMMO1 to examine whether the mutated gH/gL correctly folded on the cell surface. All four mutants retained the capacity to traffic to cell surfaces as detected by anti-Flag antibody (Fig. 4B). The binding efficiencies of AMMO1 against L573A, K625A, and H665A mutants were lower for L573A (75%) and H655A (40%) than for the WT (100%) and K625A (95%) (Fig. 4C). The G627A mutant was undetectable on the cell surface for AMMO1 (Fig. 4C). Residues L573, K625, and H655 disrupted 6H2 binding when mutated to alanine (Fig. 4D). Among the four mutants, G627A was undetectable for 6H2 (Fig. 4D).
Subsequently, the functional effects of gH mutants were evaluated in a gB-gH/gL-mediated epithelial cell-membrane fusion assay. 293T cells cotransfected with plasmids encoding gB, gH (WT-gH or mutated gH), WT-gL, and T7 RNA polymerase were used as effector cells. 293T cells transfected with plasmid expressing luciferase under the T7 promoter control were selected as target cells. The coincubation of effector cells and target cells mimicked the process of membrane fusion between virus and epithelial cells (57). In contrast to WT-gH (100%), gH mutants were markedly impaired for membrane fusion (L573A, 21%; K625A, 37%; H655A, 31%), and no fusion activity was observed for the gH mutant G627A, as expected (Fig. 4E). The evaluation of gH/gL mutants identified that four residues (L573A, K625A, G627A, and H655A) largely contribute to the 6H2 epitope. In addition, we illustrated that key residues recognized by 6H2 on gH/gL D-IV were critical for correct protein folding and membrane fusion with epithelial cells.
6H2 effectively blocks gH-mediated viral attachment and subsequent membrane fusion.
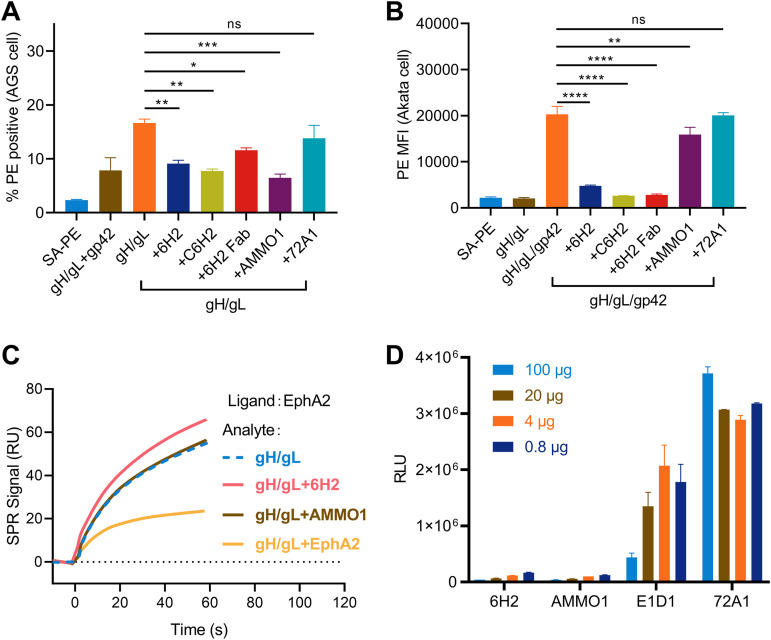
During epithelial cell infection, gH/gL binds directly to receptors on the cell surface and triggers gB-driven membrane fusion between the viral envelope and cell membrane (19). We found that 6H2 was capable of reducing gH/gL binding to the surface of AGS epithelial cells (Fig. 5A). AMMO1 binding to gH/gL also affected gH/gL binding to epithelial cells, whereas the anti-gp350 MAb 72A1 had no such effect (Fig. 5A). For B-cell infection, EBV entry into B cells is initiated with gp42 binding to HLA class II, and gB-mediated fusion is then triggered through gH/gL/gp42 complex formation (58). We found that preincubation of 6H2 with the gH/gL/gp42 complex also significantly inhibited the binding of the latter to the Akata B-cell surface (Fig. 5B). In contrast, AMMO1 only slightly reduced the binding of the gH/gL/gp42 complex to B cells and failed to completely block this binding (Fig. 5B). We further evaluated whether 6H2 could inhibit gH/gL binding to the reported cell receptor EphA2, which has been proven to be essential for EBV infection (25, 26, 30). However, we found that the interaction between EphA2 and gH/gL was not influenced by preincubation with 6H2 and AMMO1, while soluble EphA2 inhibited the interaction (Fig. 5C), which was consistent with a previous study (45). To further examine whether 6H2 could inhibit EBV-initiated membrane fusion, a virus-free cell fusion assay was performed as described above. Preincubation with 6H2 and AMMO1 significantly blocked membrane fusion with epithelial cells by the addition of as little as 0.8 μg of antibody, whereas E1D1 only partially reduced membrane fusion even at the highest dose (100 μg), and the control antibody 72A1 failed to do so (Fig. 5D). Taken together, 6H2 was able to block membrane fusion with epithelial cells by restricting gH/gL binding to the cell surface.
FIG 5.
6H2 interferes with gH/gL-mediated cell attachment and subsequent cell fusion. (A and B) Inhibition of gH/gL or gH/gL/gp42 binding to cell surfaces by MAbs. (A) Epithelial cells (AGS cells) were stained with phycoerythrin (PE)-conjugated gH/gL complex with or without MAbs. (B) B cells (Akata cells) were stained with PE-conjugated gH/gL/gp42 complex with or without MAbs. (C) Surface plasmon resonance was used to detect the influence of EphA2 binding efficiency to gH/gL when premixed with 6H2, AMMO1, and EphA2. (D) 6H2, AMMO1, E1D1, and the negative control 72A1 were evaluated in a virus-free fusion assay. All data are presented as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
DISCUSSION
EBV entry into host cells is a complex process that requires the coordinated action of multiple viral envelope glycoproteins (1, 18). Heterodimer gH/gL, an essential component of the core fusion machinery, is indispensable for EBV infection of both epithelial cells and B cells (27). Previous studies have proven that antibodies against gH/gL display potent neutralization effects and that gH/gL-based immunogens elicit a robust humoral immune response against EBV infection (39–41). gH/gL is therefore becoming an attractive target for the development of therapeutic drugs and promising prophylactic vaccine candidates. In this study, we isolated one anti-gH antibody, 6H2, that showed neutralizing ability in vitro and displayed potent protective activity against EBV-induced LPD in vivo. The cryo-EM structure of 6H2 complexed with gH/gL/gp42 mapped a new epitope at D-IV of gH/gL.
Recently, the in vivo protective effect of EBV NAbs was evaluated in humanized mouse models (49, 50). Accordingly, we carried out a similar study to evaluate the potential clinical use of 6H2. We found that C6H2 provided efficient and comparable protection with AMMO1 by preventing viremia and EBV-induced LPD (Fig. 2B and G). The antibody levels in the plasma are important for controlling EBV replication. In our study, the last dose of antibody was given at the 4th week and the last detection for viremia was at the 5th week post EBV challenge. Even though the peripheral EBV DNA copy numbers of C6H2 and AMMO1 were still within the threshold at the 5th week, both of them exhibited weak growth trends. Due to the short time (1 week) between the last dose of antibody and when the last blood was obtained, it is not known whether viremia, weight loss, or LPD would have occurred when antibody was no longer present in the blood. Considering that C6H2 is a chimeric antibody containing a murine component in variable regions, C6H2 may induce a potentially undiscovered human anti-mouse antibody (HAMA) response and prevent C6H2 from working more effectively. Hence, construction of highly humanized 6H2 and further antibody affinity maturation are required for future clinical evaluation. In addition, antibody cocktails have proven to be a promising approach for preventing viral infection and treating disease (59–61). 6H2 and AMMO1 could simultaneously bind to nonoverlapping epitopes and exert their functions through different mechanisms. An antibody cocktail combining 6H2 and AMMO1 would be an ideal antibody-based therapy and prophylactic for EBV infection.
Several anti-gH/gL NAbs have been characterized and define multiple regions on gH/gL with different neutralizing mechanisms. NAb E1D1 overlaps with the binding site of the identified epithelial cell receptor EphA2 and possibly neutralizes epithelial cell infection by directly blocking gH/gL binding to EphA2 (Fig. 6A) (30, 32). The remaining anti-gH/gL NAbs do not bind close to known receptor-binding sites of gH/gL, for which the neutralizing mechanisms are not explained clearly (45, 46). 6H2 displayed potent neutralizing ability against epithelial cell infection and efficiently blocked cell surface binding and membrane fusion with epithelial cells (Fig. 1J and Fig. 5A and D). Our structural study revealed that the 6H2 epitope was distant from the known receptor-binding site (αVβ5, αVβ6, αVβ8, and EphA2) of gH/gL (23–26) (Fig. 3 and Fig. 6A) and that 6H2 could not prevent the gH/gL-EphA2 interaction (Fig. 5C). Considering the complexity of herpesvirus infection, 6H2 restricted gH/gL access to the epithelial cell surface, possibly by physically obstructing gH/gL binding to potential receptors or cofactors (Fig. 5A and 6A). Similarly to the anti-HCMV gH/gL MAb 3G16 (62), 6H2 bound to the virus membrane-proximal domain of gH and possibly restricted the conformational change of gH/gL required for subsequent receptor binding or gB binding and activation to trigger membrane fusion with epithelial cells (Fig. 6A and B).
FIG 6.
Possible mechanisms of 6H2-mediated neutralization. (A) Surface mapping of the anti-HCMV gH/gL 3G16 epitope, anti-EBV gH/gL NAb epitopes, and EphA2 epitope. The structures were drawn using PyMOL. (B) For epithelial cells, 6H2 could possibly restrict gH/gL binding to epithelial cell receptors by steric hindrance through the Fab and Fc regions of the antibody. 6H2 may also restrict the movement of gH/gL required for subsequent receptor binding or gB binding.
We determined the structure of gH/gL/gp42/6H2 using cryo-EM and identified the key residues involved in the contact interface by mutagenesis studies. 6H2 uncovered a new epitope at D-IV (Fig. 3). Key residues recognized by 6H2 are directly involved in membrane fusion with epithelial cells (Fig. 4E). Alanine mutation in G627 resulted in undetectable cell surface gH/gL expression by 6H2 and AMMO1 (Fig. 4C and D), suggesting that the mutation to G627 may greatly impair gH/gL folding. Similar to the G627A mutant, insertion mutations into gH 564AL565 and 621SY622 abolished the cell surface and total expression of gH/gL (38). L573, G627, and H655 are conserved in gammaherpesviruses, including rhesus lymphocryptovirus (rhLCV) and Kaposi’s sarcoma-associated herpesvirus (KSHV), suggesting a conserved function of gH/gL D-IV. Identification of functional residues on EBV gH/gL D-IV provides compelling evidence that the virus membrane-proximal domain is important in gH/gL expression, heterodimer formation, and membrane fusion.
Collectively, we isolated the anti-gH NAb 6H2, which can neutralize EBV infection of epithelial cells and partially neutralize B cell infection. 6H2 recognizes a previously undetected epitope located on D-IV and exerts neutralizing effects by restricting gH/gL and gH/gL/gp42 complex binding to the cell surface and therefore prevents subsequent membrane fusion with epithelial cells. The chimeric form of 6H2 provides potent protection against lethal EBV challenge in vivo. This study provides new insight into rational therapeutics or vaccine design based on gH/gL.
MATERIALS AND METHODS
Plasmid construction.
The coding sequences of the gH ectodomain (residues 19 to 678), gL (residues 24 to 137) and the gp42 ectodomain (residues 34 to 223) were obtained from the M81 strain (GenBank accession no. KF373730.1). The plasmid containing EphA2 (GenBank accession no. NP_004422.2) sequence was provided by Mu-Sheng Zeng. The gL and the gH were connected by a (GGGGS)3 linker. gH/gL, gp42, and the ectodomain of EphA2 (residues 28 to 530) fragments were cloned into pcDNA3.1(+) by homologous recombination with an N-terminal CD5 signal peptide and a C-terminal 6×His tag. pCAGGS-Flag-gH was constructed by introducing a DYKDDDDK sequence between residues 22 and 23 of the EBV gH sequence (38). The mutant gH/gL-His was constructed by introducing alanine mutations into pcDNA3.1-gH/gL-His using a Mut Express fast mutagenesis kit (Vazyme) and confirmed by Sanger sequencing. pCAGGS-gH, pCAGGS-gL, pCAGGS-gB, pCAG-T7, and pT7EMCLuc (which carries a luciferase-containing reporter plasmid under the control of the T7 promoter) were provided by Richard Longnecker. The mutant pCAGGS-gH was constructed by introducing alanine mutations into pCAGGS-gH using a Mut Express fast mutagenesis kit (Vazyme) and confirmed by Sanger sequencing.
Recombinant expression.
Plasmids encoding wild-type (WT) gH/gL-His, gH/gL-His mutants, gp42-His, and recombinant antibodies were transiently transfected into 293F cells using polyetherimide (PEI) at a mass ratio of 1:3 (plasmids:PEI). The culture was collected 7 days after transfection. The cells and cell debris were removed by centrifugation at 10,000 rpm for 2 h, after which the supernatant was filtered through a 0.22-μm filter. The WT gH/gL-His, mutant gH/gL-His, and gp42-His were further purified with Ni2+ Sepharose 6 Fast Flow resin (GE Healthcare). The target proteins were eluted by elution buffer (PBS with 250 mM imidazole; pH 7.4). Purified proteins were then dialyzed in PBS overnight and further identified by SDS-PAGE. The antibody was purified by protein A affinity chromatography (GE Healthcare). Purified antibodies were then dialyzed in PBS overnight and further identified by SDS-PAGE.
Antibody and Fab preparation.
Eight-week-old female BALB/c mice were immunized subcutaneously three times at 2-week intervals. At 2 weeks after the final immunization, mice were boosted by administering soluble recombinant proteins. Three days later, spleen cells were collected from immunized mice and fused with mouse myeloma Sp2/0 cells. The hybridomas were sequentially screened for the secretion of gH/gL-specific MAbs by ELISA and neutralization assays. The hybridomas were cloned three times by limiting dilution and then purified from mouse ascites using protein A affinity chromatography (GE Healthcare). Fab fragments were prepared using papain cleavage. Antibodies were mixed with papain in reducing l-cysteine buffer and digested at 37°C for 12 h. The reaction was stopped by adding iodoacetamide, and the product was analyzed by SDS-PAGE. Fab fragments were separated from Fc fragments and undigested MAbs with protein A affinity chromatography (GE Healthcare).
Cell lines.
All cell lines were cultured and routinely maintained at 37°C in humidified air containing 5% CO2. 293T cells (ATCC), AGS cells (ATCC), and Cos7 cells (ATCC) were maintained in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS). Akata cells and HNE1 cells (63) were grown in RMPI 1640 plus 10% FBS. EBV-positive Akata cells (44) (used for producing Akata-EBV-free fluorescent protein [GFP] virus) were grown in RMPI containing 5% FBS. EBV-positive CNE2 cells (64) (used for producing CNE2-EBV-GFP virus) were maintained in DMEM plus 10% FBS. 293F cells (Thermo Fisher) were maintained in FreeStyle medium (Union) with gentle shaking.
Sequence analysis and chimeric antibody construction.
Total RNA from 107 hybridoma cells was extracted using a MiniBest Universal RNA extraction kit (TaKaRa). The extracted RNA was then subjected to a reverse transcription reaction with the following primers: 5′-CCCAAGCTTCCAGGGRCCARKGGATARACIGRTGG-3′ for reverse transcription of the heavy (H) chain variable region gene and 5′-CCCAAGCTTACTGGATGGTGGGAAGATGGA-3′ for reverse transcription of the light (L) chain variable region gene. The coding regions of the H and L chains of the antibodies were amplified by PCR using the following primers: 5′-ACTAGTCGACATGGATTTTGGGCTGATTTTTTTTATTG-3′ (H-chain-forward) and 5′-CCCAAGCTTCCAGGGRCCARKGGATARACIGRTGG-3′ (H-chain-reverse); and 5′-ACTAGTCGACATGAKGTHCYCIGCTCAGYTYCTIRG-3′ (L-chain-forward) and 5′-CCCAAGCTTACTGGATGGTGGGAAGATGGA-3′ (L-chain-reverse). The PCR products were cloned into a pMD18-T vector and sent to Sangon Biotech Company (Shanghai, China) for sequencing. The sequences of the antibody variable regions were confirmed using the VBASE2 database (http://www.vbase2.org/), and the corresponding amino acid sequences were determined. To construct the chimeric version of 6H2 (C6H2), the coding sequences for the variable heavy (VH) chain and variable light (VL) were subsequently cloned into the pTT5 vector containing the constant region of human IgG1 gamma heavy chain (HC) and kappa light chain (LC). The recombinant antibodies were expressed in 293F cells through transient transfection and purified from culture media by protein A affinity chromatography (GE Healthcare).
Enzyme-linked immunosorbent assay.
Then, 100 ng/well gH/gL-His or gH/gL-His mutant was added to microplates at 37°C for 2 h. The plates were washed once with PBS containing 0.1% vol/vol Tween 20 (PBST) and blocked in PBS containing 2% wt/vol nonfat milk (blocking buffer) at 37°C for 2 h. After blocking, plates were washed once with phosphate-buffered saline with Tween 20 (PBST). Serial 2-fold dilutions of purified antibody were added to the wells and incubated at 37°C for 30 min. After five washes, 100 μL of horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-human IgG buffer was added to each well and incubated at 37°C for 30 min. After five washes, 100 μL of tetramethylbenzidine (TMB) substrate was added and incubated at 37°C for 15 min. The reaction was stopped with a 2 M H2SO4 solution, and the absorbance was measured at 450 nm. All samples were run in triplicate. EC50 was calculated using the Prism 8 package (GraphPad Software).
Surface plasmon resonance assays.
Equilibrium dissociation constants (Kd values) for antibodies were determined on a Biacore 8K system (GE Healthcare). The sensor chip NTA was used to capture gH/gL-His. gH/gL-His was diluted with PBS to a final concentration of 5 μg/mL. Serially diluted Fab was injected for 300 s at 30 μL/min, followed by dissociation for 600 s at 30 μL/min. The Series S sensor chip CM-5 has a carboxylated dextran polymer matrix covalently coupled to a surface coating on a gold film. EphA2 were attached via covalent coupling. The carboxyl groups on the dextran surface were activated with 70 µL of a 1:1 (vol/vol) mixed solution of N-ethyl-N-(3-diethylarninopropyl) carbodiimide (EDC) and N-hydroxysuccinimide (NHS) for 7 min. EphA2 was diluted with 10 mM sodium acetate (pH 5.0) to a final concentration of 75 µg/mL for coupling. In each channel, flow cell 2 of the chip was coated with ligand, whereas flow cell 1 was left uncoated and blocked with ethanolamine as a control. For the EphA2 blocking assay, 500 nM gH/gL and 500 nM gH/gL mixed with equal molar amounts of 6H2, AMMO1, and EphA2-His were injected onto sensors for 60 s at 30 μL/min. The results were analyzed by Biacore Insight Evaluation software.
Surface staining of heterodimer gH/gL by flow cytometry.
293T cells were cotransfected with full-length gH and gL at a mass ratio of 1:1. Following 48 h of culture, the cells were trypsinized and pelleted by centrifugation at 1,500 rpm for 5 min. Trypsinized cells were resuspended in ice-cold 2% fetal bovine serum (FBS) and prepared at 106 cells per sample. The cells were stained with 6H2, C6H2, AMMO1, isotype control antibodies (72A1 and AMMO5), or anti-Flag antibody (Sigma-Aldrich) at 4°C for 30 min to detect the surface expression of gH/gL, followed by goat anti-mouse IgG fluorescein isothiocyanate (FITC; Sigma-Aldrich) or anti-human IgG Allophycocyanin (APC) (BD Biosciences) incubation at 4°C for another 30 min. The number of fluorescently stained cells was analyzed by flow cytometry on an LSRFortessa X-20 instrument (BD Biosciences). The data were collected and analyzed using FlowJo 10.0.7 software (Tree Star). CNE2-EBV-GFP cells were induced by 12-O-tetradecanoylphorbol 13-acetate (TPA) and sodium butyrate and stained as described above. The number of stained cells was analyzed by flow cytometry as described above.
Surface staining of heterodimer gH/gL by immunofluorescence.
COS7 cells were seeded on glass coverslips at a density of 3 × 105 cells per coverslip and transfected with 1 μg each of pCDH–full-length gL and pCDH–full-length gH. At 48 h after transfection, the cells were washed twice with PBS and fixed with 4% paraformaldehyde at room temperature for 10 min. Then, the cells were incubated in blocking solution (1.5% bovine serum albumin [BSA] and 0.25% Triton X-100 in PBS) for 1 h at room temperature and washed with PBS. Cells were incubated with primary antibodies against 6H2 (1:100), c6H2 (1:100), AMMO1 (1:100), 72A1 (1:100), and AMMO5 (1:100) at 4°C overnight. After washing three times with PBS, cells were incubated with secondary goat anti-rabbit IgG Alexa Fluor 647 (1:500; Invitrogen) or goat anti-human IgG Alexa Fluor 647 (1:500; Invitrogen) for 1 h at room temperature. Cells were washed three times with PBS and incubated with 4′,6-diamidino-2-phenylindole (DAPI; 1:1,000) for 10 min at room temperature. Cells incubated without primary antibody were used as a negative control. Images were captured by LSM 980 confocal microscope.
EBV production.
To produce epithelial tropic EBV virus, Akata-EBV-GFP cells were resuspended in RPMI 1640 to a density of 2 × 106 cells/mL, and then goat anti-human IgG (Promega) was added to the cell suspension at a concentration of 0.8% (vol/vol) to induce EBV production. After 6 h of induction, the medium was changed to RPMI 1640 medium supplemented with 10% FBS. After 72 h, the supernatant was clarified by centrifugation at 200 × g for 5 min and passed through a 0.45-μm filter. Viruses were concentrated 100× by centrifugation at 50,000 × g for 2.5 h and resuspended in RPMI 1640. The viruses were stored at −80°C. To produce B-cell tropic EBV virus, CNE2-EBV-GFP cells were induced by 12-O-tetradecanoylphorbol 13-acetate (TPA) (20 ng/mL) and sodium butyrate (2.5 mM) for 12 h, and the medium was changed to RPMI 1640 with 3% FBS. The viruses were collected and stored as described above.
B-cell infection neutralization assay.
Twofold serially diluted antibodies (starting from 100 μg/mL) were mixed with 100 μL CNE2-EBV-GFP and incubated at room temperature for 2 h. EBV-negative Akata cells (1 × 104 cells/well) were resuspended in the mixture and incubated in a 1.5-mL Eppendorf tube at 37°C for 3 h, after which the cells were pelleted by centrifugation at 200 × g for 5 min and washed once with PBS. The mixture was cultured in RPM1640 with 10% FBS in 96-well plates for 48 h and then collected and washed once with PBS. The infection rate was determined by the numbers of GFP-positive cells using a CytoFlex flow cytometer (Beckman Coulter) and analyzed using FlowJo 10.0.7 software (Tree Star). Uninfected cells were used as negative controls, and Akata-negative cells incubated with EBV in the absence of antibodies were used as positive controls. The neutralizing efficiency of antibodies was calculated using the following equation: neutralization % = [(% of GFP-positive cells in infected control) – (% of GFP-positive cells in antibody-treated group)]/% GFP-positive cells in infected control.
Epithelial cell infection neutralization assay.
Twofold serially diluted purified antibodies (starting from 100 μg/mL) were mixed and incubated with 50 μL Akata-EBV-GFP at room temperature for 2 h. The mixture was added to 5 × 103 HNE1 cells/well in 96-well plates and then incubated at 37°C for 3 h, after which the wells were washed once with PBS and cultured in RPM 1640 with 10% FBS for 48 h. After trypsinization with 0.25% trypsin-EDTA, the numbers of GFP-positive cells were analyzed by flow cytometry, and the infection rate was determined. The percent neutralization of antibody was determined as described in the B-cell neutralization assay.
EBV infection in humanized mice.
Approximately 4- to 5-week-old NOD Cg-Prkdcem1IDMOIl2rgem2IDMO (NOD-Prkdcnull IL2Rγnull, NPI) mice (Beijing Idmo Co., Ltd.) were chosen for human CD34+ stem cell (HSC) transfer through the intravenous (i.v.) route. At 8 weeks post HSC transfer, the efficiency of human cell engraftment was evaluated by detecting the percentage of human CD45+ cells in peripheral blood using flow cytometry. Experimental or control antibodies (20 mg/kg) were intraperitoneally (i.p.) injected in each humanized mouse. At 24 h later, the mice received a dose of Akata-EBV equivalent to 1,000 50% transforming dose (TD50) via i.v. injection. The same dose (20 mg/kg) of antibody was administered weekly for 4 weeks. Peripheral blood samples were collected, and mouse body weight was monitored weekly. DNA was extracted from whole blood samples using a tissue DNA kit (Omega), and then EBV copy number was quantified by real-time PCR (RT-PCR) using EBV BALF5 gene-specific primers (F: 5′-GGTCACAATCTCCACGCTGA-3′; R: 5′-CAACGAGGCTGACCTGATCC-3′). PBMCs isolated from peripheral blood were used to evaluate the immunophenotypes of circulating lymphocytes by staining with anti-human CD45-phycoerythrin (PE) (BD Biosciences), CD3-PerCP-Cy5.5 (BD Biosciences), and CD20-FITC (BD Biosciences).
Cryo-electron microscopy sample preparation and data collection.
Complex of gH/gL/gp42 (molar ratio of 1:1:1) was prepared and purified by TSK-Gel 3000PWXL (TOSOH) and then concentrated to 2 mg/mL. Aliquots (3 μL) of purified immune complex were deposited onto fresh glow-discharged holey carbon Quantifoil Cu grids (R1.2/1.3, 200 mesh; Quantifoil Micro Tools). Grids were blotted for 6 s at 100% humidity and 8°C for plunge freezing (Vitrobot Mark IV; FEI) in liquid ethane cooled by liquid nitrogen. The data set of gH/gL/gp42/6H2 was recorded on an FEI Titan Krios equipped with a K2 Summit (Gatan) direct electron detector at a nominal magnification of 21,000×, corresponding to a pixel size of 1.1 Å. The total electron dose was approximately 60 e−/Å2, which was fractionated into 40 frames (K2 Summit) with an exposure time of 8 s (K2 Summit). Data were automatically collected using FEI EPU or SerialEM software. The EMDB code for this deposition (gH/gL/gp42/6H2 complex) is EMD-33102.
Virus-free fusion assay.
293T cells were seeded in 10-cm dishes in DMEM with 10% FBS at a density of 2 × 106 cells/dish. After reaching 80% confluence, cells were transfected with 2.5 μg each of pCAGGS-gB, pCAGGS-gH, pCAGGS-gL, and pCAG-T7 polymerase, and cells in another dish were transfected with 10 μg pT7EMCLuc expressing luciferase under the control of the T7 promoter. At 24 h posttransfection, 2 × 105 cells transfected with plasmids encoding gB, gH, gL, and T7 polymerase were trypsinized and incubated with serially diluted antibodies (starting from 100 μg) at 37°C for 30 min. Then, 2 × 105 cells transfected with pT7EMCLuc were added to the mixture described above in a 24-well plate and cultured at 37°C for 24 h. Cells were lysed using a Dual-Glo luciferase assay (Promega). Cell lysate was transferred to a white-bottomed assay plate, and luciferase activity was quantified on a GloMax 96 microplate luminometer (Promega). To evaluate the epithelial cell-membrane fusion inhibition effect of gH mutants L573A, K625A, G627A, and H655A, 293T cells were seeded in 10-cm dishes in DMEM with 10% FBS at a density of 2 × 106 cells/dish. After reaching 80% confluence, cells were transfected with 2.5 μg each of pCAGGS-WT gH or pCAGGS-gH mutants, pCAGGS-gB, pCAGGS-gL, and pCAG-T7 polymerase, and cells in another dish were transfected with 10 μg pT7EMCLuc. At 24 h after transfection, cells were trypsinized, and 2 × 105 cells were mixed with each other in a 48-well plate and cultured at 37°C for 24 h in DMEM with 10% FBS. Cells were lysed, and luciferase activity was quantified using a Dual-Glo luciferase assay (Promega).
Cell surface binding assay.
gH/gL was biotinylated with the EZ-Link sulfo-NHS-biotinylation kit (Pierce). Streptavidin (SA)-PE-conjugated gH/gL-biotin (0.5 μg/μL) was added to a 96-well plate in a volume of 50 μL per well. An equimolar amount of gp42 was added to selected wells that contained gH/gL-PE. Antibody (35 μg) was added to selected wells containing gH/gL-PE with or without gp42 and incubated at room temperature for 1.5 h. Adherent epithelial cells (AGS cells) were trypsinized and recovered in RPMI 1640 with 10% FBS at 37°C for 1 h. Suspended B cells (Akata cells) were pelleted by centrifugation at 300 × g for 5 min, and then resuspended at a density of 1 × 106 cells/mL in ice-cold 0.5% BSA in PBS. Then, 100 μL of epithelial or B cells was added to wells containing SA-PE, gH/gL-PE, gH/gL-PE plus gp42, and with or without antibodies. The mixture described above was incubated on ice for 1 h. The cells were pelleted by centrifugation at 300 × g for 5 min, washed with 200 μL of ice-cold 0.5% BSA in PBS, pelleted again, and resuspended in 4% paraformaldehyde. The number of PE-positive cells was quantified using a flow cytometer on an LSRFortessa X-20 instrument (BD Biosciences).
Quantification of the cell surface and total expression of gH/gL mutants.
293T cells were seeded in 10-cm dishes in DMEM with 10% FBS at a density of 2 × 106 cells/dish. When reaching 80% confluence, cells were cotransfected with 5 μg of gL with 5 μg gH mutants L573A, K625A, G627A, H655A, and WT gH. Cells were collected at 24 h posttransfection and incubated with 5 μg of AMMO1 with or without permeabilization at room temperature for 1 h. After washing with PBS, a 1:500 diluted goat anti-human IgG Alexa Fluor 647 antibody (Invitrogen) was applied, followed by incubation at 4°C for 30 min. Data were collected with a CytoFlex instrument (Beckman Coulter) and analyzed using FlowJo 10.0.7 software (Tree Star).
Statistics.
All statistical calculations were conducted with Prism 8 (GraphPad Software, Inc.). P values were generated by one-way analysis of variance (ANOVA) unless noted otherwise. P values of ≤0.05 were considered statistically significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
ACKNOWLEDGMENTS
We thank Richard Longnecker for kindly providing the plasmids pCAG-T7, pCAGGS-gH, pCAGGS-gL, pCAGGS-gB, and pT7EMCLuc.
The study was supported by grants from the National Natural Science Foundation of China (grants 82073756 and 81872228), the Guangdong Basic and Applied Basic Research Foundation (grant 2020B1515020002), and Science and Technology Major Projects of Xiamen (grant 3502Z20203023).
J.H., M.-S.Z., Y.Z., N.X., X.Z., M.X., and Y.C. designed the studies; J.H., L.Z., Q.Z., Q.W., Z.Z., D.W., H.C., W.Z., S.Z., Y.H., K.C., J.C., and X.Z. performed experiments; J.H., L.Z., Q.Z., X.Z., M.X., and Y. C analyzed the data; J.H., Q.Z., X.Z., and Y. C. wrote the paper.
We declare that we have no conflicts of interest.
Contributor Information
Xiao Zhang, Email: zhangxiao1@sysucc.org.cn.
Miao Xu, Email: xumiao@sysucc.org.cn.
Yixin Chen, Email: yxchen2008@xmu.edu.cn.
Lori Frappier, University of Toronto.
REFERENCES
- 1.Connolly SA, Jardetzky TS, Longnecker R. 2021. The structural basis of herpesvirus entry. Nat Rev Microbiol 19:110–121. 10.1038/s41579-020-00448-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young LS, Yap LF, Murray PG. 2016. Epstein-Barr virus: more than 50 years old and still providing surprises. Nat Rev Cancer 16:789–802. 10.1038/nrc.2016.92. [DOI] [PubMed] [Google Scholar]
- 3.Cohen JI, Fauci AS, Varmus H, Nabel GJ. 2011. Epstein-Barr virus: an important vaccine target for cancer prevention. Sci Transl Med 3:107fs107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat Rev Cancer 4:757–768. 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 5.Taylor GS, Long HM, Brooks JM, Rickinson AB, Hislop AD. 2015. The immunology of Epstein-Barr virus-induced disease. Annu Rev Immunol 33:787–821. 10.1146/annurev-immunol-032414-112326. [DOI] [PubMed] [Google Scholar]
- 6.Khan G, Fitzmaurice C, Naghavi M, Ahmed LA. 2020. Global and regional incidence, mortality and disability-adjusted life-years for Epstein-Barr virus-attributable malignancies, 1990–2017. BMJ Open 10:e037505. 10.1136/bmjopen-2020-037505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kutok JL, Wang F. 2006. Spectrum of Epstein-Barr virus-associated diseases. Annu Rev Pathol 1:375–404. 10.1146/annurev.pathol.1.110304.100209. [DOI] [PubMed] [Google Scholar]
- 8.Crews DW, Dombroski JA, King MR. 2021. Prophylactic cancer vaccines engineered to elicit specific adaptive immune response. Front Oncol 11:626463. 10.3389/fonc.2021.626463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rühl J, Leung CS, Münz C. 2020. Vaccination against the Epstein-Barr virus. Cell Mol Life Sci 77:4315–4324. 10.1007/s00018-020-03538-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsao SW, Tsang CM, Lo KW. 2017. Epstein-Barr virus infection and nasopharyngeal carcinoma. Philos Trans R Soc Lond B Biol Sci 372:20160270. 10.1098/rstb.2016.0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsao SW, Tsang CM, To KF, Lo KW. 2015. The role of Epstein-Barr virus in epithelial malignancies. J Pathol 235:323–333. 10.1002/path.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vockerodt M, Yap L-F, Shannon-Lowe C, Curley H, Wei W, Vrzalikova K, Murray PG. 2015. The Epstein-Barr virus and the pathogenesis of lymphoma. J Pathol 235:312–322. 10.1002/path.4459. [DOI] [PubMed] [Google Scholar]
- 13.Miller G, Lipman M. 1973. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci U S A 70:190–194. 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller G, Lipman M. 1973. Comparison of the yield of infectious virus from clones of human and simian lymphoblastoid lines transformed by Epstein-Barr virus. J Exp Med 138:1398–1412. 10.1084/jem.138.6.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minamitani T, Ma Y, Zhou H, Kida H, Tsai C-Y, Obana M, Okuzaki D, Fujio Y, Kumanogoh A, Zhao B, Kikutani H, Kieff E, Gewurz BE, Yasui T. 2017. Mouse model of Epstein-Barr virus LMP1- and LMP2A-driven germinal center B-cell lymphoproliferative disease. Proc Natl Acad Sci U S A 114:4751–4756. 10.1073/pnas.1701836114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutt-Fletcher LM. 2017. The long and complicated relationship between Epstein-Barr virus and epithelial cells. J Virol 91:e01677-16. 10.1128/JVI.01677-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sathiyamoorthy K, Chen J, Longnecker R, Jardetzky TS. 2017. The COMPLEXity in herpesvirus entry. Curr Opin Virol 24:97–104. 10.1016/j.coviro.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hutt-Fletcher LM. 2015. EBV glycoproteins: where are we now? Future Virol 10:1155–1162. 10.2217/fvl.15.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Möhl BS, Chen J, Sathiyamoorthy K, Jardetzky TS, Longnecker R. 2016. Structural and mechanistic insights into the tropism of Epstein-Barr virus. Mol Cells 39:286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connolly SA, Jackson JO, Jardetzky TS, Longnecker R. 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery. Nat Rev Microbiol 9:369–381. 10.1038/nrmicro2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirschner AN, Omerovic J, Popov B, Longnecker R, Jardetzky TS. 2006. Soluble Epstein-Barr virus glycoproteins gH, gL, and gp42 form a 1:1:1 stable complex that acts like soluble gp42 in B-cell fusion but not in epithelial cell fusion. J Virol 80:9444–9454. 10.1128/JVI.00572-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sathiyamoorthy K, Jiang J, Hu YX, Rowe CL, Möhl BS, Chen J, Jiang W, Mellins ED, Longnecker R, Zhou ZH, Jardetzky TS. 2014. Assembly and architecture of the EBV B cell entry triggering complex. PLoS Pathog 10:e1004309. 10.1371/journal.ppat.1004309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chesnokova LS, Hutt-Fletcher LM. 2011. Fusion of Epstein-Barr virus with epithelial cells can be triggered by αvβ5 in addition to αvβ6 and αvβ8, and integrin binding triggers a conformational change in glycoproteins gHgL. J Virol 85:13214–13223. 10.1128/JVI.05580-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chesnokova LS, Nishimura SL, Hutt-Fletcher LM. 2009. Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins αvβ6 or αvβ8. Proc Natl Acad Sci U S A 106:20464–20469. 10.1073/pnas.0907508106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J, Sathiyamoorthy K, Zhang X, Schaller S, Perez White BE, Jardetzky TS, Longnecker R. 2018. Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat Microbiol 3:172–180. 10.1038/s41564-017-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Li Y, Wang H-B, Zhang A, Chen M-L, Fang Z-X, Dong X-D, Li S-B, Du Y, Xiong D, He J-Y, Li M-Z, Liu Y-M, Zhou A-J, Zhong Q, Zeng Y-X, Kieff E, Zhang Z, Gewurz BE, Zhao B, Zeng M-S. 2018. Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat Microbiol 3:1–8. 10.1038/s41564-017-0080-8. [DOI] [PubMed] [Google Scholar]
- 27.Matsuura H, Kirschner AN, Longnecker R, Jardetzky TS. 2010. Crystal structure of the Epstein-Barr virus (EBV) glycoprotein H/glycoprotein L (gH/gL) complex. Proc Natl Acad Sci U S A 107:22641–22646. 10.1073/pnas.1011806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chowdary TK, Cairns TM, Atanasiu D, Cohen GH, Eisenberg RJ, Heldwein EE. 2010. Crystal structure of the conserved herpesvirus fusion regulator complex gH-gL. Nat Struct Mol Biol 17:882–888. 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xing Y, Oliver SL, Nguyen TVi, Ciferri C, Nandi A, Hickman J, Giovani C, Yang E, Palladino G, Grose C, Uematsu Y, Lilja AE, Arvin AM, Carfí A. 2015. A site of varicella-zoster virus vulnerability identified by structural studies of neutralizing antibodies bound to the glycoprotein complex gHgL. Proc Natl Acad Sci U S A 112:6056–6061. 10.1073/pnas.1501176112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su C, Wu L, Chai Y, Qi J, Tan S, Gao GF, Song H, Yan J. 2020. Molecular basis of EphA2 recognition by gHgL from gammaherpesviruses. Nat Commun 11:5964. 10.1038/s41467-020-19617-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Möhl BS, Chen J, Park SJ, Jardetzky TS, Longnecker R. 2017. Epstein-Barr virus fusion with epithelial cells triggered by gB is restricted by a gL glycosylation site. J Virol 91:e01255-17. 10.1128/JVI.01255-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sathiyamoorthy K, Hu YX, Möhl BS, Chen J, Longnecker R, Jardetzky TS. 2016. Structural basis for Epstein-Barr virus host cell tropism mediated by gp42 and gHgL entry glycoproteins. Nat Commun 7:13557. 10.1038/ncomms13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong J, Wei D, Wu Q, Zhong L, Chen K, Huang Y, Zhang W, Chen J, Xia N, Zhang X, Chen Y. 2021. Antibody generation and immunogenicity analysis of EBV gp42 N-terminal region. Viruses 13:2380. 10.3390/v13122380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen J, Jardetzky TS, Longnecker R. 2013. The large groove found in the gH/gL structure is an important functional domain for Epstein-Barr virus fusion. J Virol 87:3620–3627. 10.1128/JVI.03245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Möhl BS, Sathiyamoorthy K, Jardetzky TS, Longnecker R. 2014. The conserved disulfide bond within domain II of Epstein-Barr virus gH has divergent roles in membrane fusion with epithelial cells and B cells. J Virol 88:13570–13579. 10.1128/JVI.02272-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Omerović J, Lev L, Longnecker R. 2005. The amino terminus of Epstein-Barr virus glycoprotein gH is important for fusion with epithelial and B cells. J Virol 79:12408–12415. 10.1128/JVI.79.19.12408-12415.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plate AE, Reimer JJ, Jardetzky TS, Longnecker R. 2011. Mapping regions of Epstein-Barr virus (EBV) glycoprotein B (gB) important for fusion function with gH/gL. Virology 413:26–38. 10.1016/j.virol.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu L, Borza CM, Hutt-Fletcher LM. 2005. Mutations of Epstein-Barr virus gH that are differentially able to support fusion with B cells or epithelial cells. J Virol 79:10923–10930. 10.1128/JVI.79.17.10923-10930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bu W, Joyce MG, Nguyen H, Banh DV, Aguilar F, Tariq Z, Yap ML, Tsujimura Y, Gillespie RA, Tsybovsky Y, Andrews SF, Narpala SR, McDermott AB, Rossmann MG, Yasutomi Y, Nabel GJ, Kanekiyo M, Cohen JI. 2019. Immunization with components of the viral fusion apparatus elicits antibodies that neutralize Epstein-Barr virus in B cells and epithelial cells. Immunity 50:1305–1316.e1306. 10.1016/j.immuni.2019.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui X, Cao Z, Chen Q, Arjunaraja S, Snow AL, Snapper CM. 2016. Rabbits immunized with Epstein-Barr virus gH/gL or gB recombinant proteins elicit higher serum virus neutralizing activity than gp350. Vaccine 34:4050–4055. 10.1016/j.vaccine.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 41.Cui X, Cao Z, Ishikawa Y, Cui S, Imadome K-I, Snapper CM. 2021. Immunization with Epstein-Barr virus core fusion machinery envelope proteins elicit high titers of neutralizing activities and protect humanized mice from lethal dose EBV challenge. Vaccines 9:285. 10.3390/vaccines9030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balachandran N, Oba DE, Hutt-Fletcher LM. 1987. Antigenic cross-reactions among herpes simplex virus types 1 and 2, Epstein-Barr virus, and cytomegalovirus. J Virol 61:1125–1135. 10.1128/JVI.61.4.1125-1135.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Q, Turk SM, Hutt-Fletcher LM. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol 69:3987–3994. 10.1128/JVI.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molesworth SJ, Lake CM, Borza CM, Turk SM, Hutt-Fletcher LM. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J Virol 74:6324–6332. 10.1128/jvi.74.14.6324-6332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Snijder J, Ortego MS, Weidle C, Stuart AB, Gray MD, McElrath MJ, Pancera M, Veesler D, McGuire AT. 2018. An antibody targeting the fusion machinery neutralizes dual-tropic infection and defines a site of vulnerability on Epstein-Barr virus. Immunity 48:799–811.e799. 10.1016/j.immuni.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sathiyamoorthy K, Jiang J, Möhl BS, Chen J, Zhou ZH, Longnecker R, Jardetzky TS. 2017. Inhibition of EBV-mediated membrane fusion by anti-gHgL antibodies. Proc Natl Acad Sci U S A 114:E8703–E8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoffman GJ, Lazarowitz SG, Hayward SD. 1980. Monoclonal antibody against a 250,000-dalton glycoprotein of Epstein-Barr virus identifies a membrane antigen and a neutralizing antigen. Proc Natl Acad Sci U S A 77:2979–2983. 10.1073/pnas.77.5.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stampfer SD, Heldwein EE. 2013. Stuck in the middle: structural insights into the role of the gH/gL heterodimer in herpesvirus entry. Curr Opin Virol 3:13–19. 10.1016/j.coviro.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh S, Homad LJ, Akins NR, Stoffers CM, Lackhar S, Malhi H, Wan Y-H, Rawlings DJ, McGuire AT. 2020. Neutralizing antibodies protect against oral transmission of lymphocryptovirus. Cell Rep Med 1:100033. 10.1016/j.xcrm.2020.100033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haque T, Johannessen I, Dombagoda D, Sengupta C, Burns DM, Bird P, Hale G, Mieli-Vergani G, Crawford DH. 2006. A mouse monoclonal antibody against Epstein-Barr virus envelope glycoprotein 350 prevents infection both in vitro and in vivo. J Infect Dis 194:584–587. 10.1086/505912. [DOI] [PubMed] [Google Scholar]
- 51.Fujiwara S, Imadome K, Takei M. 2015. Modeling EBV infection and pathogenesis in new-generation humanized mice. Exp Mol Med 47:e135. 10.1038/emm.2014.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Münz C. 2017. Humanized mouse models for Epstein Barr virus infection. Curr Opin Virol 25:113–118. 10.1016/j.coviro.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 53.Lee EK, Joo EH, Song K-A, Choi B, Kim M, Kim S-H, Kim SJ, Kang M-S. 2015. Effects of lymphocyte profile on development of EBV-induced lymphoma subtypes in humanized mice. Proc Natl Acad Sci U S A 112:13081–13086. 10.1073/pnas.1407075112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yajima M, Imadome K-I, Nakagawa A, Watanabe S, Terashima K, Nakamura H, Ito M, Shimizu N, Honda M, Yamamoto N, Fujiwara S. 2008. A new humanized mouse model of Epstein-Barr virus infection that reproduces persistent infection, lymphoproliferative disorder, and cell-mediated and humoral immune responses. J Infect Dis 198:673–682. 10.1086/590502. [DOI] [PubMed] [Google Scholar]
- 55.Saphire EO, Schendel SL, Gunn BM, Milligan JC, Alter G. 2018. Antibody-mediated protection against Ebola virus. Nat Immunol 19:1169–1178. 10.1038/s41590-018-0233-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loh HS, Green BJ, Yusibov V. 2017. Using transgenic plants and modified plant viruses for the development of treatments for human diseases. Curr Opin Virol 26:81–89. 10.1016/j.coviro.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McShane MP, Longnecker R. 2005. Analysis of fusion using a virus-free cell fusion assay. Methods Mol Biol 292:187–196. [DOI] [PubMed] [Google Scholar]
- 58.Kirschner AN, Sorem J, Longnecker R, Jardetzky TS. 2009. Structure of Epstein-Barr virus glycoprotein 42 suggests a mechanism for triggering receptor-activated virus entry. Structure 17:223–233. 10.1016/j.str.2008.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yao H, Sun Y, Deng Y-Q, Wang N, Tan Y, Zhang N-N, Li X-F, Kong C, Xu Y-P, Chen Q, Cao T-S, Zhao H, Yan X, Cao L, Lv Z, Zhu D, Feng R, Wu N, Zhang W, Hu Y, Chen K, Zhang R-R, Lv Q, Sun S, Zhou Y, Yan R, Yang G, Sun X, Liu C, Lu X, Cheng L, Qiu H, Huang X-Y, Weng T, Shi D, Jiang W, Shao J, Wang L, Zhang J, Jiang T, Lang G, Qin C-F, Li L, Wang X. 2021. Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection. Cell Res 31:25–36. 10.1038/s41422-020-00444-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gilchuk P, Murin CD, Milligan JC, Cross RW, Mire CE, Ilinykh PA, Huang K, Kuzmina N, Altman PX, Hui S, Gunn BM, Bryan AL, Davidson E, Doranz BJ, Turner HL, Alkutkar T, Flinko R, Orlandi C, Carnahan R, Nargi R, Bombardi RG, Vodzak ME, Li S, Okoli A, Ibeawuchi M, Ohiaeri B, Lewis GK, Alter G, Bukreyev A, Saphire EO, Geisbert TW, Ward AB, Crowe JE. 2020. Analysis of a therapeutic antibody cocktail reveals determinants for cooperative and broad ebolavirus neutralization. Immunity 52:388–403.e312. 10.1016/j.immuni.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weinreich DM, Sivapalasingam S, Norton T, Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, Perry C, Pan C, Hosain R, Mahmood A, Davis JD, Turner KC, Hooper AT, Hamilton JD, Baum A, Kyratsous CA, Kim Y, Cook A, Kampman W, Kohli A, Sachdeva Y, Graber X, Kowal B, DiCioccio T, Stahl N, Lipsich L, Braunstein N, Herman G, Yancopoulos GD, Trial Investigators. 2021. REGN-COV2, a neutralizing antibody cocktail, in outpatients with Covid-19. N Engl J Med 384:238–251. 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ciferri C, Chandramouli S, Leitner A, Donnarumma D, Cianfrocco MA, Gerrein R, Friedrich K, Aggarwal Y, Palladino G, Aebersold R, Norais N, Settembre EC, Carfi A. 2015. Antigenic characterization of the HCMV gH/gL/gO and pentamer cell entry complexes reveals binding sites for potently neutralizing human antibodies. PLoS Pathog 11:e1005230. 10.1371/journal.ppat.1005230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhan F, Jiang N, Cao L, Deng L, Tan G, Zhou M, Xie Y, Li G. 1998. Primary study of differentially expressed cDNA sequences in cell line HNE1 of human nasopharyngeal carcinoma by cDNA representational difference analysis. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 15:341–344. [PubMed] [Google Scholar]
- 64.Zhang H-J, Tian J, Qi X-K, Xiang T, He G-P, Zhang H, Yu X, Zhang X, Zhao B, Feng Q-S, Chen M-Y, Zeng M-S, Zeng Y-X, Feng L. 2018. Epstein-Barr virus activates F-box protein FBXO2 to limit viral infectivity by targeting glycoprotein B for degradation. PLoS Pathog 14:e1007208. 10.1371/journal.ppat.1007208. [DOI] [PMC free article] [PubMed] [Google Scholar]