ABSTRACT
The high mutation rate of COVID-19 and the prevalence of multiple variants strongly support the need for pharmacological options to complement vaccine strategies. One region that appears highly conserved among different genera of coronaviruses is the substrate-binding site of the main protease (Mpro or 3CLpro), making it an attractive target for the development of broad-spectrum drugs for multiple coronaviruses. PF-07321332, developed by Pfizer, is the first orally administered inhibitor targeting the main protease of SARS-CoV-2, which also has shown potency against other coronaviruses. Here, we report three crystal structures of the main protease of SARS-CoV-2, SARS-CoV, and Middle East respiratory syndrome (MERS)-CoV bound to the inhibitor PF-07321332. The structures reveal a ligand-binding site that is conserved among SARS-CoV-2, SARS-CoV, and MERS-CoV, providing insights into the mechanism of inhibition of viral replication. The long and narrow cavity in the cleft between domains I and II of the main protease harbors multiple inhibitor-binding sites, where PF-07321332 occupies subsites S1, S2, and S4 and appears more restricted than other inhibitors. A detailed analysis of these structures illuminated key structural determinants essential for inhibition and elucidated the binding mode of action of the main proteases from different coronaviruses. Given the importance of the main protease for the treatment of SARS-CoV-2 infection, insights derived from this study should accelerate the design of safer and more effective antivirals.
IMPORTANCE The current pandemic of multiple variants has created an urgent need for effective inhibitors of SARS-CoV-2 to complement vaccine strategies. PF-07321332, developed by Pfizer, is the first orally administered coronavirus-specific main protease inhibitor approved by the FDA. We solved the crystal structures of the main protease of SARS-CoV-2, SARS-CoV, and MERS-CoV that bound to the PF-07321332, suggesting PF-07321332 is a broad-spectrum inhibitor for coronaviruses. Structures of the main protease inhibitor complexes present an opportunity to discover safer and more effective inhibitors for COVID-19.
KEYWORDS: COVID-19, PF-07321332, SARS-CoV-2, coronavirus, crystal structure, main protease
INTRODUCTION
Since its discovery in December 2019, cases of novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-infected pneumonia have rapidly continued to emerge, with the current case count at close to 438 million and a case mortality rate of ∼1.4% by the end of February 2022, causing huge economic and social loss to the world (1, 2). There have been several coronaviruses in human history that are pathogenic to humans, among which two were associated with severe respiratory disease outbreaks, SARS-CoV (severe acute respiratory syndrome coronavirus first emerged in Guangdong China in 2002) and MERS-CoV (Middle East respiratory syndrome coronavirus first detected in Saudi Arabia in 2012) (3–5). Genomic sequencing data showed that SARS-CoV-2 shares 79.6% sequence identity with SARS-CoV (1, 6–8). As SARS-CoV and SARS-CoV-2 successively emerged and COVID-19 continues spreading throughout the globe, developing broad-spectrum drugs remains an urgent and unmet clinical need in the treatment and prevention of COVID-19 infections.
Viral enzymes and proteins of CoVs that are involved in coronavirus replication are potential drug targets for COVID-19. In particular, the main protease (Mpro or 3CLpro), which cleaves the replicase polyproteins at 11 sites, is one of the most attractive targets for numerous classes of small molecule inhibitors for the development of drugs against coronavirus infections (9, 10). Mpro is highly conserved among coronaviruses, and the substrate-binding site in Mpro also shares several common features (6). Because Mpro has no human homolog, Mpro inhibitors should be highly specific to SARS-CoV-2 and have minimal side effects (11). Unfortunately, although several peptidomimetic covalent inhibitors of Mpro have been reported, few candidates have progressed into clinical trials (6, 12–14).
We previously screened an in-house small molecule library and identified shikonin as a noncovalent inhibitor for SARS-CoV-2 and SARS-CoV Mpro in vitro, with a half-maximum inhibitory concentration (IC50) of 1.57 μM and 7.89 μM, respectively (15, 16). In addition, crystal structures of SARS-CoV-2 Mpro with the antineoplastic drug carmofur and the natural products baicailin and baicalein have also been solved recently (17, 18). Despite the progress made in understanding the origin of human coronaviruses as well as the prediction and prevention of the emerging pandemics, we still lack effective and safe drugs and therapies to combat the global pandemic caused by coronavirus (5, 19, 20).
In 2021, Pfizer initiated a phase 1 clinical trial (NCT04756531) and later the phase 2/3 clinical trials (NCT04960202, NCT05011513) of a novel antiviral therapeutic agent against SARS-CoV-2. The clinical candidate, PF-07321332, is the first orally administered coronavirus-specific protease inhibitor, which has shown potent antiviral activity against SARS-CoV-2 in vitro, as well as activity against other coronaviruses (21). As a protease inhibitor, PF-07321332 binds to the viral enzyme and can block the activity of the protease that the coronavirus needs to reproduce itself. Ritonavir, an inhibitor of a key liver enzyme called CYP3A, can boost and maintain the plasma concentration of PF-07321332 when coadministered. This kind of strategy has been used to effectively treat other viral pathogens, such as HIV and hepatitis C virus. Recently, the emergency use of Paxlovid (PF-07321332 and ritonavir) has been authorized by the U.S. Food and Drug Administration (FDA). Thus, PF-07321332 could be an encouraging antiviral with potential for use in the treatment of COVID-19, as well as potential use to address future coronavirus threats.
In this study, we aim to explore the molecular basis for the small molecule inhibitor PF-07321332 targeting Mpro of coronaviruses. We found that PF-07321332 potently inhibits the enzymatic activity of SARS-CoV-2 Mpro. We then determined the crystal structures of complexes of the main protease of SARS-CoV-2, SARS-CoV, and MERS-CoV bound to the inhibitor PF-07321332, revealing a novel binding mode of Mpro. Structural comparison with reported Mpro-inhibitor complex structures provides insight into the mechanism of Mpro inhibition by a small molecule inhibitor and a framework for small molecule drug discovery.
RESULTS
Inhibitory activity of PF-07321332 against SARS-CoV-2 Mpro.
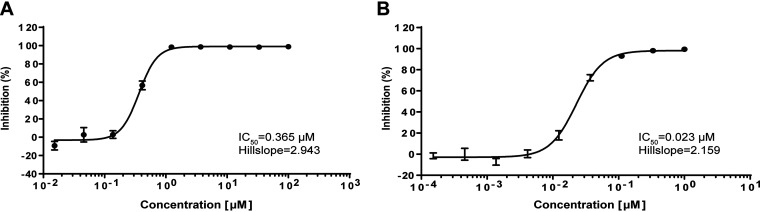
We purified the Mpro of SARS-CoV-2 as previously reported (15). A fluorescence resonance energy transfer (FRET) assay was employed to determine the inhibitory activity of PF-07321332 against SARS-CoV-2 Mpro. Ebselen was used as a control with an IC50 value of 0.365 μM, which is similar to that determined in previous studies (12, 22). The results showed that PF-07321332 has a potent inhibition, with the IC50 value against the Mpro of SARS-CoV-2 being 0.023 μM (Fig. 1), which is much lower than that of recently reported covalent inhibitors such as boceprevir, leupeptin, and carmofur (17, 23, 24).
FIG 1.
Enzymatic inhibition of SARS-CoV-2 Mpro. (A) Inhibition of ebselen against SARS-CoV-2 Mpro. (B) Inhibition of PF-07321332 against SARS-CoV-2 Mpro. SARS-CoV-2 Mpro was preincubated in the reaction buffer with various concentrations of PF-07321332 at room temperature for 30 min before reacting with the FRET substrate. Ebselen was used as a control. The IC50 was calculated using GraphPad Prism software.
Inhibitory mechanisms of PF-07321332 against SARS-CoV-2 Mpro.
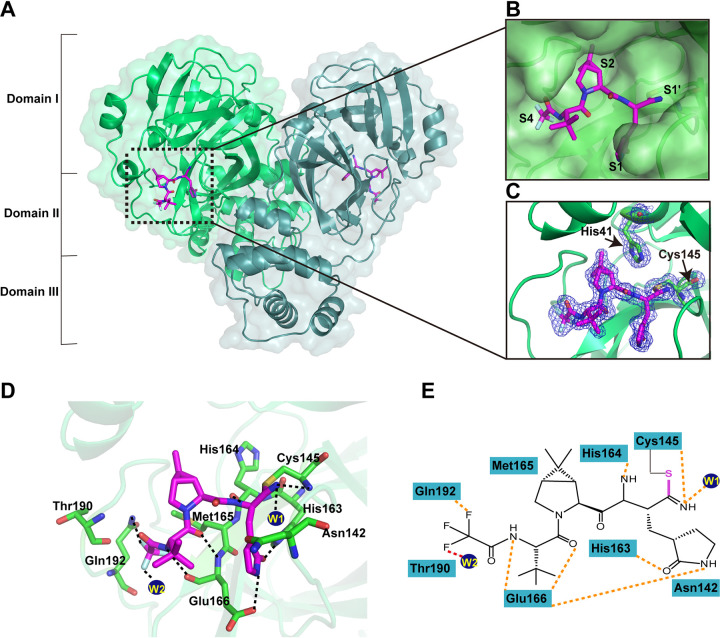
In order to figure out the inhibitory mechanisms of PF-07321332, we determined the crystal structure of SARS-CoV-2 Mpro in complex with PF-07321332 (PFMpro-Co) at 1.5-Å resolution using the cocrystallization method (see Fig. S1 in the supplemental material, Table 1). By comparison with the apo structure of SARS-CoV-2 Mpro at pH 7.5, the root mean square deviation (RMSD) of equivalent Cα positions between apo Mpro and PFMpro-Co is ∼0.985 Å (Fig. S2). As shown in Fig. 2, the Mpro molecule in the complex structure forms a homodimer and contains three domains, namely, domain I (residues 10 to 99), domain II (residues 100 to 184), and domain III (residues 201 to 303). PF-07321332 can be found in both protomer A and protomer B (Fig. 2A). Specifically, PF-07321332 binds to the active site situated in the cleft between domains I and II of Mpro in an extended conformation and occupies subsites S1, S2, and S4 of SARS-CoV-2 Mpro (Fig. 2B). The electron density map unambiguously shows that nitrile carbon of PF-07321332 forms a C-S covalent bond with the sulfur atom of catalytic residue C145 (Fig. 2C). The imine nitrogen of the thioimidate moiety occupies the oxyanion hole and forms hydrogen bonds with the backbone NH of C145 and the oxygen from a water molecule for stabilization. Besides the typical covalent interaction, PF-07321332 forms multiple noncovalent interactions with the active site. According to the Berger and Schechter nomenclature, PF-07321332 consists of five moieties, namely, P1 to P4 and P1′. As shown in Fig. 2D and E, PF-07321332 contains a γ-lactam ring at the P1 position just before the warhead nitrile group. The lactam ring inserts into the S1 subsite, with the oxygen and nitrogen atoms of the lactam ring forming hydrogen bonds with the Nε2 of H163 and carboxy group of E166, respectively. Further, a hydrogen bond is also found between the amide nitrogen at the P1 moiety and the main-chain carbonyl oxygen of H164. The P2 position contains a dimethyl cyclopropyl proline (DMCP) moiety which inserts into the S2 subsite and mainly forms hydrophobic interactions, similar to the previously reported compound boceprevir (24). PF-07321332 presents a tert-leucine residue at the P3 position. However, limited interactions are observed between the P3 moiety of PF-07321332 and the S3 subsite of Mpro. PF-07321332 displays a trifluoromethyl group at the P4 position. The amide nitrogen at the P4 position forms a hydrogen bond with the main chain carbonyl oxygen of E166, while the trifluoromethyl group forms additional hydrogen bonds by interacting with the nitrogen atom of Q192 and a water molecule. Thus, PF-07321332 occupies the active site of SARS-CoV Mpro by covalently binding to C145 and noncovalently interacting with conserved residues, including Cy145, H163, H164, E166, and Q192.
TABLE 1.
Statistics for data processing and model refinement of Mpro-PF-07321332
| Parameter | SARS-CoV-2 Mpro-PF-07321332 (cocrystal) | SARS-CoV-2 Mpro-PF-07321332 (soaking) | SARS-CoV Mpro-PF-07321332 (soaking) | MERS-CoV Mpro-PF-07321332 (soaking) |
|---|---|---|---|---|
| PDB code | 7VLP | 7VLQ | 7VLO | 7VTC |
| Data collection | ||||
| Synchrotron | SSRF | SSRF | SSRF | SSRF |
| Beam line | BL17U1 | BL17U1 | BL17U1 | BL17U1 |
| Wavelength (Å) | 0.97918 | 0.97918 | 0.97918 | 0.97918 |
| Space group | P1211 | P212121 | P1 | P212121 |
| a,b,c (Å) | 55.48, 98.70, 59.42 | 67.85, 102.02, 103.27 | 55.46, 60.46, 68.14 | 83.42, 93.24, 97.58 |
| α,β,γ (°) | 90.00, 108.72, 90.00 | 90.00, 90.00, 90.00 | 91.92, 102.42, 108.77 | 91.00, 90.00, 90.00 |
| Total reflections | 612,117 | 399,180 | 157,101 | 136,239 |
| Unique reflections | 94,054 | 51,490 | 50,908 | 23,783 |
| Resolution (Å) | 1.50 (1.58–1.50) | 1.94 (2.04–1.94) | 2.02 (2.13–2.02) | 2.54 (2.68–2.54) |
| R-merge (%) | 3.5 (55.6) | 3.4 (68.1) | 2.6 (24.3) | 4.8 (122.8) |
| Mean I/σ (I) | 15.0/2.7 | 15.3/2.7 | 9.8/2.7 | 11.6/2.1 |
| Completeness (%) | 97.7 (100.0) | 95.5 (100.0) | 95.7 (96.1) | 92.0 (99.5) |
| Redundancy | 6.5 (5.8) | 7.8 (5.3) | 3.1 (2.8) | 5.7 (5.7) |
| Refinement | ||||
| Resolution (Å) | 33.43–1.50 | 51.64–1.94 | 30.08–2.02 | 48.79–2.54 |
| Rwork/Rfree (%) | 19.47/21.92 | 20.15/23.07 | 20.37/23.52 | 19.62/24.62 |
| Atoms | 5175 | 4,724 | 4,565 | 4,587 |
| Mean temp factor (Å2) | 23.3 | 26.8 | 43.0 | 47.7 |
| Bond lengths (Å) | 0.006 | 0.007 | 0.008 | 0.009 |
| Bond angles (°) | 0.95 | 0.95 | 1.09 | 1.15 |
| Preferred | 97.83 | 97.47 | 96.12 | 96.28 |
| Allowed | 2.17 | 2.53 | 3.54 | 3.55 |
| Outliers | 0 | 0 | 0.34 | 0.17 |
FIG 2.
Crystal structure of SARS-CoV-2 Mpro in complex with PF-07321332. (A) Overall structure of SARS-CoV-2 Mpro in complex with PF-07321332. The three domains and two protomers of Mpro are labeled. The substrate-binding pocket is located within the black dotted box. PF-07321332 is shown as sticks with the carbon atoms in magentas, oxygen atoms in bright red, nitrogen atoms in blue, and fluorine atom in pale cyan. (B) An enlarged view of the substrate-binding pocket. PF-07321332 forms a covalent bond with C145. The substrate-binding subsites (S1′, S1, S2, and S4) are labeled. (C) A C-S covalent bond forms between the Sγ atom of C145 and the nitrile carbon of PF-07321332. The 2Fo-Fc density map contoured at 1.0σ is shown as a blue mesh. (D) The detailed interaction in the complex structure is shown with the residues involved in inhibitor binding (within 3.5 Å) displayed as sticks. W1 and W2 represent the water molecules. Hydrogen bond interactions are shown as black dashed lines. (E) Schematic interaction between PF-07321332 and Mpro. Hydrogen bond interactions are shown as orange dashed lines.
The crystal structure of the Mpro-PF-07321332 complex (PFMpro-So) has also been solved at 1.9-Å resolution by soaking (Table 1). By superimposition of PFMpro-Co and PFMpro-So, we found that the binding mode of PF-07321332 with SARS-CoV-2 Mpro was highly similar to the RMSD, being 0.781 Å over the 523 best-aligned Cα atoms (Fig. S3).
Crystal structures of PF-07321332 in complex with SARS-CoV and MERS-CoV Mpro.
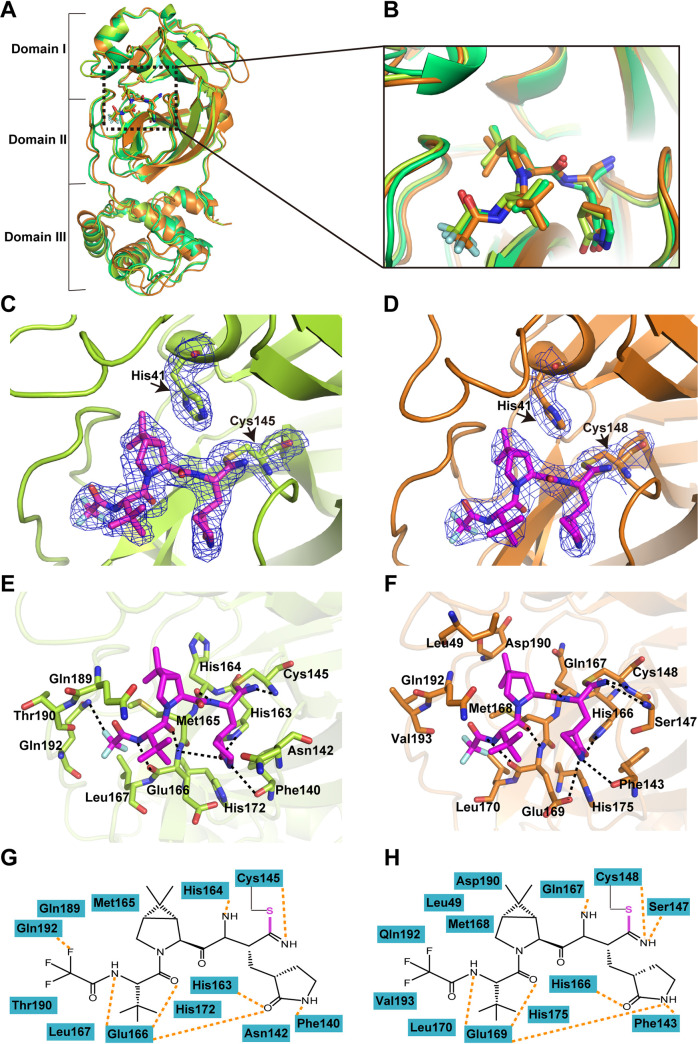
We also determined the crystal structure of PF-07321332 in complex with SARS-CoV and MERS-CoV Mpro at 2.0-Å and 2.5-Å resolutions (Fig. S1, Table 1), respectively. PF-07321332 displays a highly similar conformation in the substrate-binding site of SARS-CoV, MERS-CoV, and SARS-CoV-2 Mpro, even though the orientation of each moiety of PF-07321332 has slight differences (Fig. 3A and B). Like the case with SARS-CoV-2 Mpro, PF-07321332 fits into the S1, S2, and S4 subsites and forms a covalent bond with the catalytic residue cysteine as expected (Fig. 3C and D). In addition, several amino acid residues, including F140, C145, H163, H164, E166, and Q192, in the protease form hydrogen bond interactions with the inhibitor in the SARS-CoV Mpro-PF-07321332 complex. The key residues of MERS-CoV Mpro interacting with PF-07321332 are highly conserved (Fig. 3E to H). These observations are consistent with the fact that the structure of Mpro in the coronaviruses is highly conserved and that PF-07321332 may be a potent inhibitor with broad-spectrum potential to defeat diseases caused by various coronaviruses.
FIG 3.
Crystal structures of SARS-CoV and MERS-CoV Mpros in complex with PF-07321332. (A) Structural alignment of CoV Mpros complexed with PF-07321332 with SARS-CoV-2 Mpro-inhibitor complex in lime green, SARS-CoV Mpro-inhibitor complex in yellow, and MERS-CoV Mpro-inhibitor complex in orange. (B) An enlarged view of the substrate-binding pocket. (C and D) A C-S covalent bond forms between C145 of SARS-CoV Mpro (C) or MERS-CoV Mpro (D) and the nitrile group of PF-07321332. The 2Fo-Fc density map contoured at 1.0σ is shown as a blue mesh. (E and F) The detailed interaction in the complex structure is shown with the residues of SARS-CoV Mpro (E) or MERS-CoV Mpro (F) involved in inhibitor binding (within 3.5 Å) displayed as sticks. Hydrogen bond interactions are shown as black dashed lines. (G and H) Schematic interaction between PF-07321332 and SARS-CoV Mpro (G) or MERS-CoV Mpro (H). Hydrogen bonds interactions are shown as orange dashed lines.
Structural comparison of PF-07321332 with other covalent inhibitors in complex with SARS-CoV-2 Mpro.
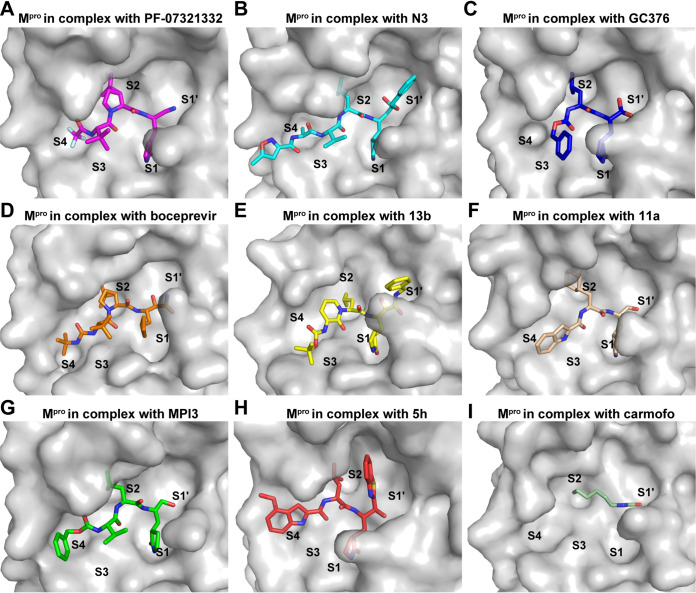
Several covalent inhibitors that show good inhibitory activity of the SARS-CoV-2 main protease have recently been reported (Table S1). Among these, the structures of 13b and N3 complexed with Mpro have been determined (12, 14). Based on the complex structure, the covalent inhibitor 11a was designed with better inhibitory activities (6). The clinically approved drugs carmofur and boceprevir were repurposed, and they displayed low micromolar inhibitory activity against SARS-CoV-2 Mpro (17, 24), while the other three preclinical inhibitors, GC376 (24), 5h (25), and MPI3 (26), showed inhibitory activities (Ki or IC50) in the nanomolar range. We compared the structures of these recently identified covalent inhibitors complexed with SARS-CoV-2 Mpro. All these small molecules form a C-S covalent bond with the catalytic residue cysteine (Fig. 4) and display a lactam ring at the P1 position which fits well with the S1 subsite except for carmofur and boceprevir. The amide nitrogen of the lactam ring can form hydrogen-bond interactions with the ketonic oxygen of Glu166 or Phe140, while the oxygen atom forms hydrogen-bond interactions with the backbone NH of Glu166 or Nε2 of His163. Subsite S2 of SARS-CoV-2 Mpro appears to prefer hydrophobic interactions, and all these inhibitors display a hydrophobic group at the P2 position, such as isopropyl and the dimethyl cyclopropyl proline (DMCP) group. However, the P3 position of these inhibitors does not fit well with the S3 pocket. Indeed, structural optimization may be conducted in the future to investigate more suitable groups for this subsite. The binding pattern of PF-07321332 is similar to that of boceprevir at the P4 position but differs at substitution of F atoms and formation of hydrogen bonds with Q192. Therefore, structures of inhibitors in complex with Mpro reported in this study and other studies, will provide the structural basis for the development and optimization of more potent drugs against SARS-CoV-2 infection.
FIG 4.
Comparison of the binding modes of different inhibitors targeting SARS-CoV-2 Mpro. (A to I) The binding pockets of PF-07321332 (A) (PDB ID 7VLQ), N3 (B) (PDB ID 6LU7), GC376 (C) (PDB ID 7D1M), boceprevir (D) (PDB ID 7BRP), 13b (E) (PDB ID 6Y2F), 11a (F) (PDB ID 6LZE), MPI3 (G) (PDB ID 7JQO), 5h (H) (PDB ID 7JKV), and carmofur (I) (PDB ID 7BUY) bound to SARS-CoV-2 Mpro are shown. Mpros are shown as the gray surface, and the inhibitors are shown as sticks.
DISCUSSION
There is an urgent need to develop effective drugs as the novel coronavirus pandemic continues to wreak havoc on human society. Mpro is a promising drug target for its vital role in viral replication and high conservation among all coronaviruses and because it has no homolog in humans. Although a great number of inhibitors have shown inhibitory activity against Mpro, most of these are not potent enough and not highly bioavailable following oral administration, which limits its clinical application to hospitalized patients with relatively advanced disease. Thus, potent orally bioavailable antiviral drugs for treatment of SARS-CoV-2 infection are urgently needed. PF-07321332 is the first orally available antiviral drug developed by Pfizer to tackle the SARS-CoV-2 virus by targeting its Mpro (21, 27). In this study, we found that PF-07321332 is a potent inhibitor with an IC50 of 0.023 μM. As shown in Table S1, this value is a little higher than that of MPI3 (IC50, 8.5 nM) but lower than that of most reported inhibitors, including 11a (IC50, 0.053 μM) (6), 13b (IC50, 0.67 μM) (14), boceprevir (IC50, 8.0 μM) (24), and GC376 (IC50, 0.15 μM) (24), which suggests its huge potential to treat COVID-19 clinically. In addition, the main advantage of PF-07321332 with respect to other inhibitors of SARS-CoV-2 Mpro is the possibility of oral administration, a feature that could dramatically facilitate the treatment of COVID-19.
We also solved the crystal structure of PF-07321332 in complex with Mpro of three deadly coronoviruses (SARS-CoV-2, SARS-CoV, and MERS-CoV), which is of great importance for structure-based drug development. The complex structures indicate that PF-07321332 covalently bound to the catalytic cysteine of Mpro and formed multiple hydrogen bonds with conserved residues within the active site. Owen et al. and Zhao et al. also reported the structure of PF-07321332 in complex with SARS-CoV-2 Mpro (21, 28). Superimpositions of the complex structure reported in this study (PDB ID 7VLQ) with the complex structure reported by Owen et al. (PDB ID 7RSF) and Zhao et al. (PDB ID 7VH8) show root mean square deviation (RMSD) values of 1.329 and 1.225 Å over the 292 and 288 best-aligned Cα atoms, respectively. Compared with other covalent inhibitors, PF-07321332 employs a unique binding pattern to SARS-CoV-2 Mpro. Further structure-based optimization of such covalent inhibitors will help generate drugs against the current COVID-19 pandemic with high efficiency and a broad spectrum.
Another orally bioavailable drug candidate, masitinib, has been demonstrated to show an inhibitory effect against SAR-CoV-2 Mpro. Until now, masitinib has been approved for treatment of mast cell tumors in dogs and evaluated in phase 2 and 3 clinical trials in humans for the treatment of cancer, asthma, Alzheimer’s disease, multiple sclerosis, and amyotrophic lateral sclerosis (29). However, unlike PF-07321332 as a covalent inhibitor, masitinib is a noncovalent inhibitor, which could complement the design of covalent inhibitors against the SARS-CoV-2 main protease. Both covalent and noncovalent Mpro inhibitor will contribute to increased public health preparedness for potential future pandemics.
MATERIALS AND METHODS
Expression and purification of human CoVs.
The codon-optimized cDNAs for Mpro of SARS-CoV-2, SARS-CoV, and MERS-CoV were synthesized fused with 6× His at the N terminus. Synthesized genes were subcloned into the pET-28a vector. The expression and purification of each main protease were performed by a standard method described previously by our lab (15).
Enzymatic assays.
PF-07321332 was purchased from MedChemExpress (MCE; Chemical Abstracts Service [CAS] no. 2628280-40-8). Fluorogenic substrates as a donor and quencher pair were synthesized. The IC50 values of PF-07321332 against the SARS-CoV-2 main protease were measured with a common protocol as the following: first, 1 μL of main protease (200 nM) was incubated with various concentrations of testing inhibitors at room temperature for 30 min in its reaction buffer (50 mM Tris 7.3,150 mM NaCl, 1 mM EDTA) in a 384-well plate, and then fluorescence resonance energy transfer (FRET) substrate was added to the reaction system. The reaction was monitored for 20 min, and the data were calculated at 10 min intervals by linear regression. Ebselen was used as a positive control. The IC50 was determined by plotting the initial velocity against various concentrations of testing inhibitor by using the dose-response curve in GraphPad Prism software.
Crystallization.
Cocrystallization of SARS-CoV-2 Mpro with PF-07321332 (0.1 M HEPES, pH 7.5, 20% wt/vol PEG 10,000), SARS-CoV Mpro with PF-07321332 (0.1 M HEPES, pH 7.5, 10% PEG 8,000, 8% ethylene glycol), and MERS-CoV Mpro with PF-07321332 (10% PEG 200, 0.1 M bis-Tris-propane, pH 9.0, 18% PEG 8,000) was carried out at 20°C using the hanging drop vapor-diffusion method. PF-07321332 was added to Mpros according to a 3:1 molar ratio, and the mixture was incubated for 30 min on ice. After 3 to 5 days, the complex crystals of Mpros with PF-07321332 were obtained. For the soaking method, SARS-CoV-2 Mpro crystals were obtained first using the sitting-drop vapor diffusion method at 20°C. PF-07321332 was then soaked with crystals of SARS-CoV-2 Mpro (0.1 M HEPES sodium, pH 7.5, 10% vol/vol 2-propanol, 20% wt/vol PEG 4,000) within 24 h.
Data collection, structure determination, and refinement.
The crystals were tailored with cryo-loop and then flash-frozen in liquid nitrogen to collect better X-ray data. All data sets were collected at 100 K on a macromolecular crystallography beamline 17U1 (BL17U1) at the Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China). All collected data were handled with the HKL 2000 software package. The structures were determined by molecular replacement with Phenix software. The program Coot was used to rebuild the initial model. The complete data collection and statistics of refinement are shown in Table 1.
Data availability.
Coordinates and structure factors for the SARS-CoV Mpro-PF-07321332, SARS-CoV-2 Mpro-PF-07321332 (soaking and cocrystallization), and MERS-CoV Mpro-PF-07321332 complexes have been deposited in the Protein Data Bank (PDB) under accession numbers 7VLO, 7VLP, 7VLQ, and 7VTC, respectively.
ACKNOWLEDGMENTS
We thank the cryo-electron microscopy (cryo-EM) center of the Southern University of Science and Technology for our cryo-EM work and for their help with cryo-EM data collection.
J.L. was supported by the Open Project of Key Laboratory of Prevention and Treatment of Cardiovascular and Cerebrovascular Diseases, Ministry of Education (no. XN201904), Gannan Medical University (QD201910), Jiangxi Key Research and Development Program (20203BBG73063), and Jiangxi “Double Thousand Plan.” J.Z. was supported by the Thousand Young Talents Program of China, the National Natural Science Foundation of China (grant no. 31770795 and 81974514), and the Jiangxi Province Natural Science Foundation (grant no. 20181ACB20014). P.J.M was supported by the Foreign Talent project of Jiangxi Province. Y.F. was supported by the Shenzhen Science and Technology Program (JCYJ20210324115611032 and KQTD20200909113758004). This work was also supported by the Ganzhou COVID-19 Emergency Research Project (2020.17), major science and technology programs of Ganzhou City (2020.67), and the Ganzhou Zhanggong District COVID-19 prevention and control key research projects (2020.67).
J.L. and J.Z. initiated and supervised the project. C.L., X.Z., F.Z., and P.Z. crystallized the protein complexes and performed the soaking experiments. J.L., C.L., X.Z., F.Z., P.Z., and J.Z. collected X-ray data and solved and refined structures. B.Y., Y.Z., and H.J. performed docking and identified compounds to be tested in the initial screens and assisted with the enzymatic assay by FRET assay. H.J., P.J.M., Y.Y., X.F., Y.F., and R.F. assisted with the design of experiments, project management, and interpretation of results.
Footnotes
Supplemental material is available online only.
Contributor Information
Jin Zhang, Email: zhangxiaokong@hotmail.com.
Rozanne M. Sandri-Goldin, University of California, Irvine
REFERENCE
- 1.Chan JF-W, Yuan S, Kok K-H, To KK-W, Chu H, Yang J, Xing F, Liu J, Yip CC-Y, Poon RW-S, Tsoi H-W, Lo SK-F, Chan K-H, Poon VK-M, Chan W-M, Ip JD, Cai J-P, Cheng VC-C, Chen H, Hui CK-M, Yuen K-Y. 2020. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 395:514–523. 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO. WHO COVID-19 dashboard. https://covid19.who.int/.
- 3.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, Xing X, Xiang N, Wu Y, Li C, Chen Q, Li D, Liu T, Zhao J, Liu M, Tu W, Chen C, Jin L, Yang R, Wang Q, Zhou S, Wang R, Liu H, Luo Y, Liu Y, Shao G, Li H, Tao Z, Yang Y, Deng Z, Liu B, Ma Z, Zhang Y, Shi G, Lam TTY, Wu JT, Gao GF, Cowling BJ, Yang B, Leung GM, Feng Z. 2020. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382:1199–1207. 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, Nagel J, Johnson JB, Agnihothram S, Gates JE, Frieman MB, Baric RS, Donaldson EF. 2012. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol 86:12816–12825. 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai W, Zhang B, Jiang X-M, Su H, Li J, Zhao Y, Xie X, Jin Z, Peng J, Liu F, Li C, Li Y, Bai F, Wang H, Cheng X, Cen X, Hu S, Yang X, Wang J, Liu X, Xiao G, Jiang H, Rao Z, Zhang L-K, Xu Y, Yang H, Liu H. 2020. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368:1331–1335. 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zumla A, Chan JFW, Azhar EI, Hui DSC, Yuen K-Y. 2016. Coronaviruses: drug discovery and therapeutic options. Nat Rev Drug Discov 15:327–347. 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anand K, Ziebuhr J, Wadhwani P, Mesters JR, Hilgenfeld R. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763–1767. 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 11.Ren Z, Yan L, Zhang N, Guo Y, Yang C, Lou Z, Rao Z. 2013. The newly emerged SARS-like coronavirus HCoV-EMC also has an “Achilles’ heel”: current effective inhibitor targeting a 3C-like protease. Protein Cell 4:248–250. 10.1007/s13238-013-2841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Peng C, Duan Y, Yu J, Wang L, Yang K, Liu F, Jiang R, Yang X, You T, Liu X, Yang X, Bai F, Liu H, Liu X, Guddat LW, Xu W, Xiao G, Qin C, Shi Z, Jiang H, Rao Z, Yang H. 2020. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 582:289–293. 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Lin D, Kusov Y, Nian Y, Ma Q, Wang J, von Brunn A, Leyssen P, Lanko K, Neyts J, de Wilde A, Snijder EJ, Liu H, Hilgenfeld R. 2020. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J Med Chem 63:4562–4578. 10.1021/acs.jmedchem.9b01828. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. 2020. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 368:409–412. 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Zhou X, Zhang Y, Zhong F, Lin C, McCormick PJ, Jiang F, Luo J, Zhou H, Wang Q, Fu Y, Duan J, Zhang J. 2021. Crystal structure of SARS-CoV-2 main protease in complex with the natural product inhibitor shikonin illuminates a unique binding mode. Sci Bull (Beijing) 66:661–663. 10.1016/j.scib.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Gao H, Hu X, Wang Q, Zhong F, Zhou X, Lin C, Yang Y, Wei J, Du W, Huang H, Zhou H, He W, Zhang H, Zhang Y, McCormick PJ, Fu J, Wang D, Fu Y, Lu X, Zhang T, Duan J, Qin B, Jiang H, Luo J, Zhang Y, Chen Q, Luo Q, Cheng L, Zhang Z, Zhang J, Li J. 2022. Structure-based discovery and structural basis of a novel broad-spectrum natural product against the main protease of coronavirus. J Virol 96:e0125321. 10.1128/JVI.01253-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin Z, Zhao Y, Sun Y, Zhang B, Wang H, Wu Y, Zhu Y, Zhu C, Hu T, Du X, Duan Y, Yu J, Yang X, Yang X, Yang K, Liu X, Guddat LW, Xiao G, Zhang L, Yang H, Rao Z. 2020. Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol 27:529–532. 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 18.Su HX, Yao S, Zhao WF, Li MJ, Liu J, Shang WJ, Xie H, Ke CQ, Hu HC, Gao MN, Yu KQ, Liu H, Shen JS, Tang W, Zhang LK, Xiao GF, Ni L, Wang DW, Zuo JP, Jiang HL, Bai F, Wu Y, Ye Y, Xu YC. 2020. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol Sin 41:1167–1177. 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu B, Ge X, Wang LF, Shi Z. 2015. Bat origin of human coronaviruses. Virol J 12:221. 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anthony SJ, Gilardi K, Menachery VD, Goldstein T, Ssebide B, Mbabazi R, Navarrete-Macias I, Liang E, Wells H, Hicks A, Petrosov A, Byarugaba DK, Debbink K, Dinnon KH, Scobey T, Randell SH, Yount BL, Cranfield M, Johnson CK, Baric RS, Lipkin WI, Mazet JAK. 2017. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. mBio 8:e00373-17. 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Owen DR, Allerton CMN, Anderson AS, Aschenbrenner L, Avery M, Berritt S, Boras B, Cardin RD, Carlo A, Coffman KJ, Dantonio A, Di L, Eng H, Ferre R, Gajiwala KS, Gibson SA, Greasley SE, Hurst BL, Kadar EP, Kalgutkar AS, Lee JC, Lee J, Liu W, Mason SW, Noell S, Novak JJ, Obach RS, Ogilvie K, Patel NC, Pettersson M, Rai DK, Reese MR, Sammons MF, Sathish JG, Singh RSP, Steppan CM, Stewart AE, Tuttle JB, Updyke L, Verhoest PR, Wei L, Yang Q, Zhu Y. 2021. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. Science 374:1586–1593. 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 22.Amporndanai K, Meng X, Shang W, Jin Z, Rogers M, Zhao Y, Rao Z, Liu ZJ, Yang H, Zhang L, O’Neill PM, Samar Hasnain S. 2021. Inhibition mechanism of SARS-CoV-2 main protease by ebselen and its derivatives. Nat Commun 12:3061. 10.1038/s41467-021-23313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu L, Shao S, Feng Y, Ye F, Sun X, Wang Q, Yu F, Wang Q, Huang B, Niu P, Li X, Wong CCL, Qi J, Tan W, Gao GF. 2021. Mechanism of microbial metabolite leupeptin in the treatment of COVID-19 by traditional Chinese medicine herbs. mBio 12:e0222021. 10.1128/mBio.02220-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu L, Ye F, Feng Y, Yu F, Wang Q, Wu Y, Zhao C, Sun H, Huang B, Niu P, Song H, Shi Y, Li X, Tan W, Qi J, Gao GF. 2020. Both boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat Commun 11:4417. 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hattori SI, Higashi-Kuwata N, Hayashi H, Allu SR, Raghavaiah J, Bulut H, Das D, Anson BJ, Lendy EK, Takamatsu Y, Takamune N, Kishimoto N, Murayama K, Hasegawa K, Li M, Davis DA, Kodama EN, Yarchoan R, Wlodawer A, Misumi S, Mesecar AD, Ghosh AK, Mitsuya H. 2021. A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat Commun 12:668. 10.1038/s41467-021-20900-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang KS, Ma XR, Ma Y, Alugubelli YR, Scott DA, Vatansever EC, Drelich AK, Sankaran B, Geng ZZ, Blankenship LR, Ward HE, Sheng YJ, Hsu JC, Kratch KC, Zhao B, Hayatshahi HS, Liu J, Li P, Fierke CA, Tseng CK, Xu S, Liu WR. 2021. A quick route to multiple highly potent SARS-CoV-2 main protease inhibitors. ChemMedChem 16:942–948. 10.1002/cmdc.202000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramos-Guzmán CA, Ruiz-Pernía JJ, Tuñón I. 2021. Computational simulations on the binding and reactivity of a nitrile inhibitor of the SARS-CoV-2 main protease. Chem Commun (Camb) 57:9096–9099. 10.1039/d1cc03953a. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y, Fang C, Zhang Q, Zhang R, Zhao X, Duan Y, Wang H, Zhu Y, Feng L, Zhao J, Shao M, Yang X, Zhang L, Peng C, Yang K, Ma D, Rao Z, Yang H. 2021. Crystal structure of SARS-CoV-2 main protease in complex with protease inhibitor PF-07321332. Protein Cell. 10.1007/s13238-021-00883-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drayman N, DeMarco JK, Jones KA, Azizi SA, Froggatt HM, Tan K, Maltseva NI, Chen S, Nicolaescu V, Dvorkin S, Furlong K, Kathayat RS, Firpo MR, Mastrodomenico V, Bruce EA, Schmidt MM, Jedrzejczak R, Muñoz-Alía M, Schuster B, Nair V, Han KY, O'Brien A, Tomatsidou A, Meyer B, Vignuzzi M, Missiakas D, Botten JW, Brooke CB, Lee H, Baker SC, Mounce BC, Heaton NS, Severson WE, Palmer KE, Dickinson BC, Joachimiak A, Randall G, Tay S. 2021. Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2. Science 373:931–936. 10.1126/science.abg5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S3 and Table S1 . Download jvi.02013-21-s0001.pdf, PDF file, 0.5 MB (549.9KB, pdf)
Data Availability Statement
Coordinates and structure factors for the SARS-CoV Mpro-PF-07321332, SARS-CoV-2 Mpro-PF-07321332 (soaking and cocrystallization), and MERS-CoV Mpro-PF-07321332 complexes have been deposited in the Protein Data Bank (PDB) under accession numbers 7VLO, 7VLP, 7VLQ, and 7VTC, respectively.