ABSTRACT
In Pseudomonas aeruginosa, the complex multisensing regulatory networks RetS-GacS/GacA have been demonstrated to play key roles in controlling the switch between planktonic and sessile lifestyles. However, whether this multisensing system is involved in the regulation of phage infection has not been investigated. Here, we provide a link between the sensors RetS/GacS and infection of phages vB_Pae_QDWS and vB_Pae_W3. Our data suggest that the sensors kinases RetS and GacS in Pseudomonas aeruginosa play opposite regulatory functions on phage infection. Mutation in retS increased phage resistance. Cellular levels of RsmY and RsmZ increased in PaΔretS and were positively correlated with phage resistance. Further analysis demonstrated that RetS regulated phage infection by affecting the type IV pilus (T4P)-mediated adsorption. The regulation of RetS on phage infection depends on the GacS/GacA two-component system and is likely a dynamic process in response to environmental signals. The findings offer additional support for the rapid emergence of phage resistance.
IMPORTANCE Our knowledge on the molecular mechanisms behind bacterium-phage interactions remains limited. Our study reported that the complex multisensing regulatory networks RetS-GacS/GacA of Pseudomonas aeruginosa PAO1 play key roles in controlling phage infection. The main observation was that the mutation in RetS could result in increased phage resistance by reducing the type IV pilus-mediated phage adsorption. The bacterial defense strategy is generally applicable to various phages since many P. aeruginosa phages can use type IV pilus as their receptors. The results also suggest that the phage infection is likely to be regulated dynamically, which depends on the environmental stimuli. Reduction of the signals that RetS favors would increase phage resistance. Our study is particularly remarkable for uncovering a signal transduction system that was involved in phage infection, which may help in filling some knowledge gaps in this field.
KEYWORDS: GacS/GacA two-component system, Pseudomonas aeruginosa, RetS, adsorption, phage
INTRODUCTION
Phages are the most abundant entities in nature and highly effective bacterial predators that specifically infect and lyse bacterial cells. Phage therapy is gradually becoming a reality in clinical, veterinary, and agricultural settings for preventing and controlling bacterial infections, as antibiotic abuse has led to an explosion in bacterial resistance (1–3). Although phage biocontrol is a promising alternative to antibiotics, the emergence of phage resistance imposed difficulties in phage therapy.
A range of bacterial defense mechanisms have been reported, including CRISPR-based immunity (4, 5), bacteriophage exclusion (BREX) system (6), abortive infection (Abi) system (7), and toxin-antitoxin (TA) system (8). Besides these antiphage strategies, several host genes have also been shown to play roles in controlling phage infection. Mutations in genes associated with phage receptor synthesis, such as lipopolysaccharide (LPS), pili, and outer membrane proteins, may decrease phage adsorption and stop phage attacks (9, 10). Disruptions of the genes for Escherichia coli RNA polymerase (11), E. coli thioredoxin (12), Pseudomonas aeruginosa small regulatory protein (SrpA) (13), and Bacillus subtilis DNA polymerase (14) would affect the phage infection process. Recently, some metabolic pathways that were modulated by quorum sensing (QS) were shown to help bacteria to avoid phage infection (15–17). Mutations in QS-related genes may also lead to changes in phage resistance. Although a number of antiphage strategies have been shown, our knowledge on the mechanisms of phage resistance is still limited, considering the enormous genetic diversity of both phages and bacteria.
P. aeruginosa is a leading cause of health care-acquired infections, especially in cystic fibrosis patients (18). The two-component system GacS-GacA has been reported to play important roles in bacterial adaptability and infection (19). GacS is a membrane-associated sensor histidine kinase (SK) which senses environmental signals, working with its cognate response regulator (RR) GacA, which positively controls the expression of two central regulatory noncoding RNAs, namely, RsmY and RsmZ. RsmY and RsmZ have been proposed as key players in regulating genes required for infection (20, 21). However, GacS is reciprocally regulated by the sensor histidine kinase RetS. When RetS senses signals, it will interfere with the initial autophosphorylation of GacS and inhibit the GacS-GacA two-component system (22, 23). RetS and GacS signaling converge on the master virulence regulator GacA, influencing levels of RsmY and RsmZ. To date, the sensor kinases RetS and GacS have been studied widely and reported to control the transition between acute and chronic infection (24).
A wide range of virulence factors have been reported to be controlled by RetS and GacS regulons, including motility and biofilm production. Flagella and pili are closely related to bacterial motility, and they often act as receptors for phages (25, 26). The formation of biofilm can provide potential physical and chemical barriers that protect bacteria against phage infection (27). More importantly, bacteria commonly use two-component systems to sense the environmental changes for survival. Phage predation, as environmental stresses, can also affect microbial communities (15). Thus, we speculate that there is a close correlation between sensor kinases RetS and GacS and phage infection.
The aim of the current study was to clarify the effects of two sensor kinases from P. aeruginosa PAO1, namely, RetS and GacS, on phage infection. Our findings revealed that the multisensing regulatory networks RetS-GacS/GacA regulate phage infection processes, which offer additional support for the rapid emergence of phage resistance. This work will help in providing important implications for minimizing the development of phage resistance by changing the environmental factors.
RESULTS
Characterization of P. aeruginosa phages vB_Pae_QDWS and vB_Pae_W3.
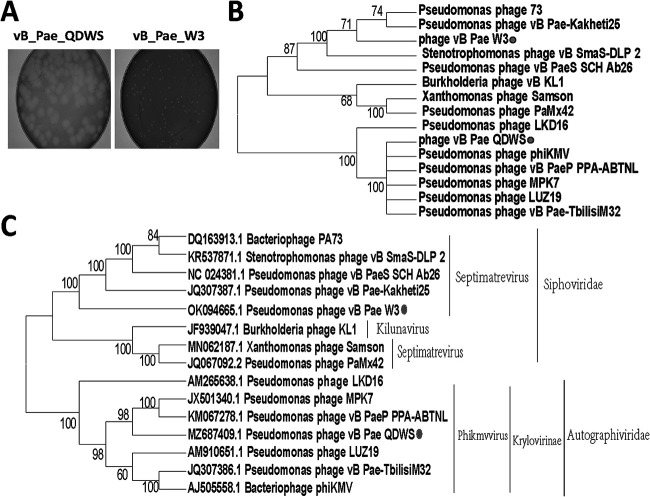
Two Pseudomonas phages were isolated from sewage using double-layer agar plates and named vB_Pae_QDWS and vB_Pae_W3. The phage vB_Pae_QDWS formed large plaques of approximately 6 mm in diameter, while vB_Pae_W3 produced small plaques (Fig. 1A). General genome characteristics of the phage isolates are summarized in Table 1. ORF39 (holin) and ORF40 (endolysin) were identified as the host lysis system of phage vB_Pae_W3. And phage vB_Pae_QDWS contained ORF49 (holin) and ORF50 (endolysin) for lysis. Since no lysogeny module or lysogens were found based on gene functional annotation analysis, the two phages vB_Pae_QDWS and vB_Pae_W3 are lytic. Both phage vB_Pae_QDWS and vB_Pae_W3 did not have antibiotic resistance genes and virulence genes. Phage vB_Pae_W3 shared the auxiliary metabolic genes (AMGs) that encode FAD/FMN-containing dehydrogenase (ORF4), pyrophosphatase (mazG, ORF16), and dCMP deaminase (DCD, ORF34). No AMGs were found in phage vB_Pae_QDWS. Phylogenetic analysis based on the capsid protein amino acid sequences revealed that vB_Pae_QDWS is most closely related to Phikmvvirus, Krylovirinae, and Autographiviridae and phage vB_Pae_W3 is most closely related to Septimatrevirus and Siphoviridae (Fig. 1B). The same result was also seen in the phylogenetic tree based on the whole-genome sequence, although the closest phage was different (Fig. 1C).
FIG 1.
Characterization of phages vB_Pae_QDWS and vB_Pae_W3. (A) Plaque morphology of phages vB_Pae_QDWS and vB_Pae_W3. (B) A phylogenetic tree was constructed based on the amino acid sequence of the capsid protein. (C) The phylogenetic analysis was based on genome sequences. ClustalW alignments were used to generate neighbor-joining trees with 1,000 bootstrap replicates.
TABLE 1.
General characteristics of Pseudomonas phages
| Family | Phage | Genome size (bp) | No. of ORFsa | GC content (%) | % Nucleotide identity (% coverage) |
|---|---|---|---|---|---|
| Autographiviridae | vB_Pae_QDWS | 43,170 | 53 | 62.3 | 93.94 (97) with vB_PaeP_PAO1_1-15pyo |
| Siphoviridae | vB_Pae_W3 | 43,014 | 59 | 53.8 | 98.78 (95) with TehO |
ORFs, open reading frames.
Effects of RetS and GacS on phage resistance.
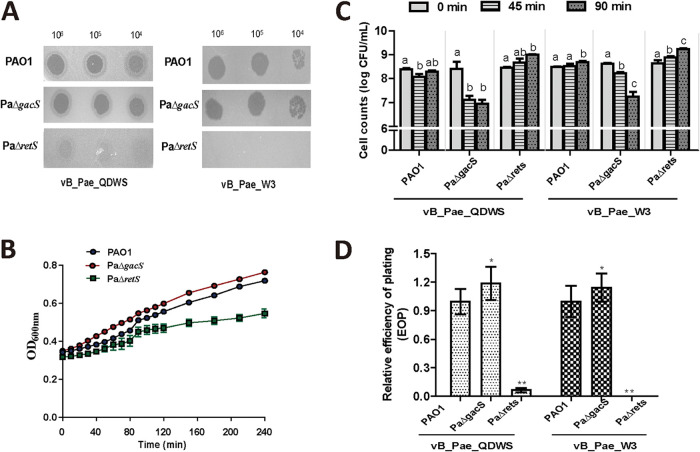
The deletion of retS genes increased resistance against phages vB_Pae_QDWS and vB_Pae_W3 compared with the wild-type strain PAO1. The deletion of gacS genes, however, decreased phage resistance as the transparency of plaques increased (Fig. 2A). When there were no phages, a similar growth rate was observed in PAO1 and PaΔgacS, but a slight growth delay occurred in PaΔretS (Fig. 2B). Phages vB_Pae_QDWS and vB_Pae_W3 significantly inhibited the growth of PaΔgacS, as the cell counts reduced significantly compared with PAO1. However, an increase in bacterial count was observed in the PaΔretS group following phage vB_Pae_QDWS or vB_Pae_W3 treatment (Fig. 2C), which indicated PaΔretS is more resistant to phages. By testing the efficiency of plating (EOP), we found the EOP of phage vB_Pae_QDWS on PaΔgacS was higher than that on PAO1, and the same result was also found for phage vB_Pae_W3. Phages vB_Pae_QDWS and vB_Pae_W3, however, were largely unsuccessful in infecting PaΔretS, as the EOP significantly decreased (Fig. 2D). Therefore, our data indicate that sensor kinases RetS and GacS had an opposite effect on phage resistance.
FIG 2.
Phage resistance was regulated by RetS and GacS. (A) A total of 3 μL of serial dilutions of vB_Pae_QDWS and vB_Pae_W3 were spotted onto PAO1, PaΔgacS, and PaΔretS for lytic activity assays. (B) Growth curves of PAO1, PaΔgacS, and PaΔretS. (C) Cell counts of PAO1, PaΔgacS, and PaΔretS infected with phage vB_Pae_QDWS or vB_Pae_W3 at an MOI of 1 were detected at different time points. Different lowercase letters in column mean significant differences (P < 0.05) among different time points. (D) Relative efficiency of plating (EOP) of phages vB_Pae_QDWS and vB_Pae_W3 on P. aeruginosa strains. The values were the averages of three measures with standard deviation. Symbol * indicates the sample is different (0.01 < P < 0.05) from the control PAO1, and symbol ** indicates the sample is significantly different (P < 0.01) from PAO1 (Student’s paired t test).
Effects of the two small RNAs RsmY and RsmZ on phage resistance.
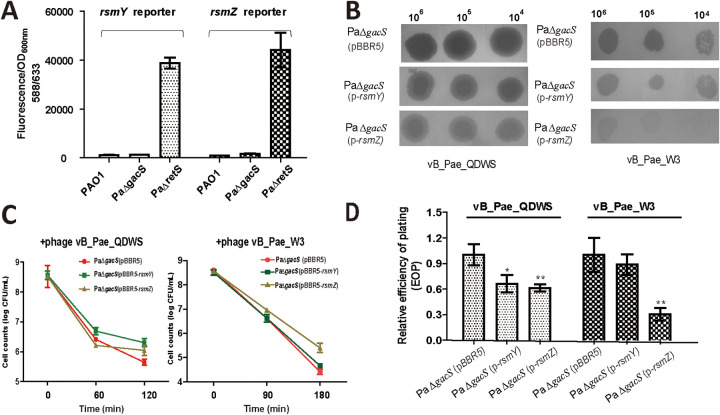
The mkate expression was significantly increased in PaΔretS compared with that in PAO1 and PaΔgacS, which indicated higher rsmY and rsmZ expression in retS mutant (Fig. 3A). To investigate whether PaΔretS promoted phage resistance through RsmY and RsmZ, the complementation plasmids pBBR5-rsmY and pBBR5-rsmZ were generated based on the PaΔgacS mutant. As expected, results of a spot assay showed that the complementation strains PaΔgacS (p-rsmY) and PaΔgacS (p-rsmZ) were more resistant to both phage vB_Pae_QDWS and vB_Pae_W3 than the control PaΔgacS with the empty plasmid (Fig. 3B). In the CFU reduction assay, we found that the maximum reductions of PaΔgacS (p-rsmY) and PaΔgacS (p-rsmZ) after a 120-min infection were 2.26 ± 0.14 (mean ± SEM) and 2.48 ± 0.17 log10 CFU/mL, respectively, while the control PaΔgacS (pBBR5) was decreased by 2.88 ± 0.27 after treatment with phage vB_Pae_QDWS. When infected with phage vB_Pae_W3 for 180 min, bacterial counts of PaΔgacS (p-rsmY) and PaΔgacS (p-rsmZ) reduced by 3.84 ± 0.11 and 3.16 ± 0.19 log10 CFU/mL, respectively, while the control PaΔgacS (pBBR5) was decreased by 4.20 ± 0.11 (Fig. 3C). Results of the EOP assay also suggested a reduction in EOP of the two phages on the complementation strains, especially for PaΔgacS (p-rsmZ) compared with the control group PaΔgacS (pBBR5) (Fig. 3D). Thus, our results suggest that these two sRNAs, namely, RsmY and RsmZ, play a role in the pathway by which RetS regulates the phage resistance.
FIG 3.
RsmY/Z levels were closely associated with phage resistance. (A) P. aeruginosa PAO1, PaΔgacS, and PaΔretS cells containing pBBR-PrsmY-mkate or pBBR-PrsmZ-mkate grew in LB medium until an OD600 of 1 and were collected for fluorescence assays. (B) A total of 3 μL of serial dilutions of vB_Pae_QDWS and vB_Pae_W3 were spotted onto PAO1 (pBBR5), PaΔgacS (p-rsmY), and PaΔgacs (pBBR5-rsmZ) for lytic activity assays. (C) Cell counts of PaΔgacS (pBBR5)(●, red mark), PaΔgacS (p-rsmY)(■, green mark), and PaΔgacS (p-rsmZ)(▲, yellow mark) strains infected with phage vB_Pae_QDWS or vB_Pae_W3 at an MOI of 1 were detected at different time points. (D) Relative efficiency of plating (EOP) of phage vB_Pae_QDWS and vB_Pae_W3 on P. aeruginosa strains. The values were the averages of three measures with standard deviation. Symbol * indicates the sample is different (0.01 < P < 0.05) from the control PaΔgacS (pBBR5), and symbol ** indicates the sample is significantly different (P < 0.01) from PaΔgacS (pBBR5) (Student’s paired t test).
The mechanism of RetS and GacS regulating phage resistance.
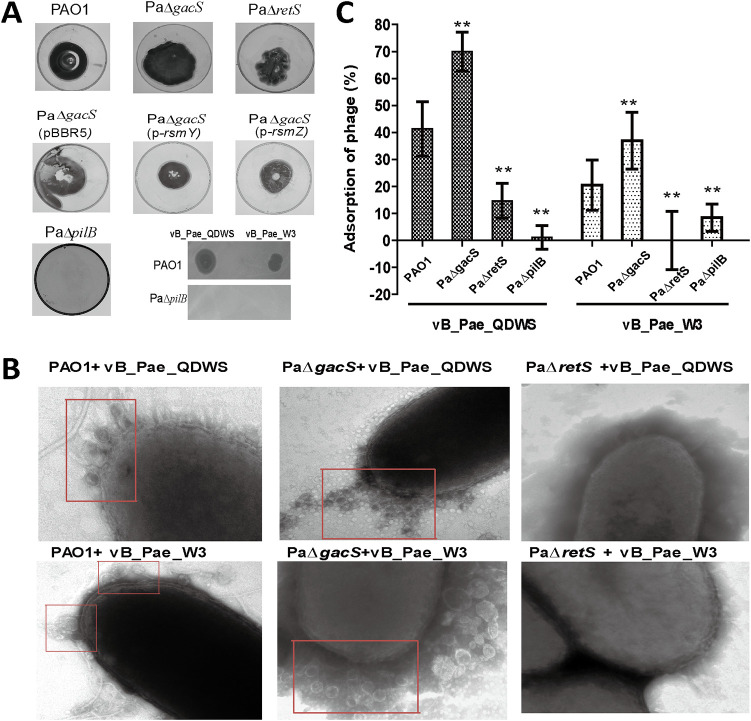
Compared with PAO1, PaΔretS decreased twitching motility, but PaΔgacS increased. When detecting the levels of RsmY and RsmZ on twitching motility, we found that higher levels of RsmY and RsmZ lead to a decrease in the twitching motility (Fig. 4A). Since type IV pilus (T4P) are located on the surface of a wide variety of bacteria and are always involved in mediating twitching motility (28), we speculated that RetS and GacS regulate phage resistance via affecting the function of T4P. Furthermore, we deleted the pilB gene that is essential for T4P (29) and found both vB_Pae_QDWS and vB_Pae_W3 could not lyse PaΔpilB, but it could lyse PAO1 (Fig. 4A). A transmission electron microscopy (TEM) analysis showed that more phages, either vB_Pae_QDWS or vB_Pae_W3, were distributed around the PaΔgacS and PAO1 and almost no phages attached to the surface of the host PaΔretS (Fig. 4B). The reduction in adsorption rate was found in PaΔretS and PaΔpilB compared with PAO1, and PaΔgacS showed the increased adsorption rate (Fig. 4C). Based on these results, we proposed that the impairment of T4P mediated phage adsorption that posed the phage-resistant phenotypes of PaΔretS. However, results of quantitative reverse transcription-PCR (RT-qPCR) demonstrated that no significant changes in the mRNA level of T4P-related genes were found (data not shown), although the significant enhancement of T4P-mediated twitching motility was found in PaΔgacS, in addition to a reduction by PaΔretS (Fig. 4A). Our finding is consistent with a previous report that no entire or partial T4P-related gene clusters were negatively regulated by the Gac system, which indicated that RetS and GacS controlled the function of T4P most likely through a series of indirect and complex pathways or mechanisms (30).
FIG 4.
RetS and GacS affected phage resistance by regulating phage adsorption. (A) The T4P-mediated twitching motility was assayed in different P. aeruginosa strains. A total of 3 μL of serial dilutions of vB_Pae_QDWS and vB_Pae_W3 were spotted onto PAO1 and PaΔpilB strains for lytic activity assays. (B) Different P. aeruginosa strains were mixed with phages vB_Pae_QDWS and vB_Pae_W3 at an MOI of 100 for 5 min and subjected to TEM analysis. Phages adsorbed on the host surface were labeled (red boxes). (C) Adsorption assays of phages vB_Pae_QDWS and vB_Pae_W3 to different P. aeruginosa strains. The values were the averages of three measures with standard deviation. Symbol ** indicates the sample is significantly different (P < 0.01) from the wide-type PAO1 sample (Student’s paired t test).
DISCUSSION
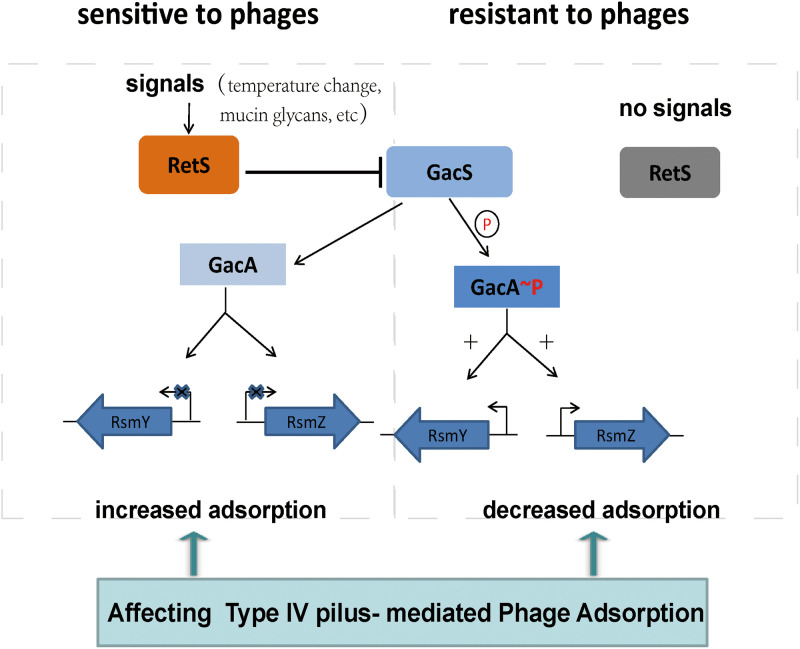
Here, we present evidence that the sensor kinase RetS of P. aeruginosa PAO1 plays an important role in regulating the resistance of both phages vB_Pae_QDWS and vB_Pae_W3. RetS inhibits GacS activity, which controls the expression of the regulatory RNAs RsmY and RsmZ (31). When introducing the mutation in the key genes of RetS or reducing the amounts of signals that RetS favors, the GacS/GacA signaling pathway can be activated, which would result in an enhanced production of RsmY and RsmZ and promoting phage resistance. This increase in phage resistance is due largely to a reduction in T4P-mediated adsorption (Fig. 4). A model whereby phage infection is regulated by RetS-GacS/GacA pathway is proposed (Fig. 5).
FIG 5.
Schematic representation of RetS involved in the regulation of phage infection via modulating the GacS/GacA two-component systems.
The expression levels of the regulatory RNAs RsmY and RsmZ, which are controlled by RetS/GacS, corresponded positively with phage resistance. The significant elevation in RsmY and RsmZ levels was seen in the ΔretS mutant (Fig. 3A). Overexpression of either rsmY or rsmZ could override the negative effects of gacS mutation and increase phage resistance (Fig. 3B). However, rsmY and rsmZ expression is essentially dependent on environmental stimuli. The signals that favor the activation of RetS function to repress the expression of rsmY and rsmZ. Under normal growth conditions, the GacS-GacA signal pathway is inhibited by RetS, as the levels of RsmY and RsmZ were very low (Fig. 3A), which indicated that the RetS is active and RetS-dependent signals are ubiquitous. RetS is reported to sense temperature changes (32). Mucin glycans, also act as signals through RetS that promote the ability of RetS to directly inhibit GacS-GacA activity (33). Thus, it is possible for RetS to regulate phage infection dynamically in response to these environmental stimuli.
Mutation of retS decreased the T4P-mediated twitching motility of P. aeruginosa PAO1 (Fig. 4A). The results are consistent with an retS mutant of P. aeruginosa PA103 that decreased the expression level of pili, which is associated with acute cytotoxicity (34). PaΔretS reduced the virulence and pathogenicity of P. aeruginosa but increased phage resistance. P. aeruginosa is the dominant pathogen in chronic lung infections of cystic fibrosis patients, and phages have also been abundant in cystic fibrosis lungs (35, 36). For P. aeruginosa, it is a relative cost-saving strategy to balance virulence and survival in the microecosystem of the cystic fibrosis lungs.
Changes in the T4P-mediated phage adsorption is the main mechanism of RetS and GacS regulating phage infection. Besides the phages vB_Pae_QDWS and vB_Pae_W3 that we reported, many other P. aeruginosa phages can also recognize and use T4P as their receptors (25, 37). The mechanism we proposed whereby RetS regulates T4P-mediated adsorption is likely a common strategy for P. aeruginosa PAO1 to defend against different phages. Our findings will provide new ideas for phage therapy against P. aeruginosa infection.
MATERIALS AND METHODS
Bacteria and phages.
P. aeruginosa strains were cultured in lysogeny broth (LB) medium at 37°C and stored in 20% glycerol at −80°C until use. Tetracycline (50 μg/mL) and gentamicin (50 μg/mL) were added as required. Phages vB_Pae_QDWS and vB_Pae_W3 were isolated against the host bacterial strain P. aeruginosa PAO1, from water samples collected from Qingdao, China. The phage suspension titer was determined by the double-layer agar method using LB as the culture medium. After an incubation step at 37°C for 12 h, the number of lysis plaques was counted.
Gene knockout and complementation.
The deletions in P. aeruginosa PAO1 were determined according to a published method (38). For complementation, the target genes were amplified by PCR and cloned into linearized pBBR1MCS5 by using an In-Fusion cloning kit (Clontech, Mountain View, CA). The resulting plasmids were then transferred into the PAO1 strain via electroporation.
Reporter plasmid construction and fluorescence assays.
The reporter plasmids pBBR5-PrsmY-mKate and pBBR5-Prsmz-mKate were constructed by placing the mkate gene after the rsmY/Z promoter and inserting them into pBBR1MCS5 plasmid. The constructed plasmids were then transformed into PAO1, PaΔgacS, and PaΔretS mutants for fluorescence assays.
The reporter strains were grown at 37°C with shaking in LB medium to an optical density at 600 nm (OD600) of 1. Then, the cultures were transferred to a 96-well plate for mKate fluorescence measurement by using the SynergyH1 microplate reader. The excitation wavelength was set at 588 nm, and the emission wavelength was set at 633 nm.
Transmission electron microscopy (TEM).
P. aeruginosa cells were grown in LB broth to an OD600 of 2.5, harvested, and washed with phosphate-buffered saline (PBS). Then, high titers (1 × 1011 PFU/mL) of phages were added to the P. aeruginosa cells with a multiplicity of infection (MOI) of 100. After 5 min of adsorption, the bacterial particles and phage pellet were deposited on carbon-coated copper grids and negatively stained with 2% phosphotungstic acid (pH 6.8). The samples were observed under a JEM-1200EX transmission electron microscope (JEOL) at 100 kV.
Twitching motility assay.
Twitching motility was tested as described previously (39). In brief, cells from an overnight culture were stab-inoculated through the 1% LB agar layer. The plates were inoculated at 37°C for 30 h. To measure the twitching motility zone, the agar was carefully removed, and the motility zone was measured by staining the petri dish with 2% crystal violet for 1 h.
Adsorption rate assay.
P. aeruginosa strains were inoculated in LB medium until they reached an optical density at 600 nm (OD600) of about 1. Then, the cells were diluted 10-fold in LB and mixed with the phage vB_Pae_QDWS and vB_Pae_W3 solutions at a multiplicity of infection (MOI) of 0.0025. The phage adsorptions for vB_Pae_QDWS and vB_Pae_W3 were performed at 37°C for 5 min and 10 min, respectively. Finally, the cells were centrifuged at 10,000 rpm for 2 min at 4°C to obtain the free phages, which were detected using a double-layer agar method. The adsorption rate was calculated by adsorption rate (%) = [(initial phage titer − phage titer in the supernatant)/(initial phage titer)] × 100.
Phage resistance assays.
The phage resistance of P. aeruginosa strains was detected by spotting 3 μL of the phage suspension with serial dilutions into the lawn of each strain using the double-layer technique. Then, the samples were incubated at 37°C without shaking before examination.
The efficiency of plating (EOP) assay was performed according to a published method with some modifications (40). In brief, 3 μL of the phage suspension with serial dilutions were spotted into the lawn of each P. aeruginosa strain. The plates were incubated for 7 h (for vB_Pae_QDWS) or 12 h (for vB_Pae_W3) at 37°C, and the number of PFUs was counted. Finally, the relative EOP was calculated (average PFU on target bacteria/average PFU on the control PAO1 bacteria). Each experiment was performed at least three independent times.
The CFU reduction assay was determined as follows. Briefly, P. aeruginosa strains were inoculated in LB medium until an OD600 of about 1 was reached, and then they were cocultivated with the phage vB_Pae_QDWS or vB_Pae_W3 at MOI of 1 at 37°C. The bacterial counts were determined at different time points.
Real-time quantitative reverse transcription-PCR (RT-qPCR).
For RT-qPCR, strains were grown in LB medium until the OD600 reached 1, and then cells were collected for RNA extraction. RNA samples were prepared by using the TRIzol RNA purification kit (12183555; Invitrogen). Total cDNA was synthesized using the HiScript II reverse transcriptase (Vazyme) reagent. RT-qPCR was performed using the SYBR green real-time PCR master mix and the StepOnePlus real-time PCR system (ABI). rplS was selected as the reference gene for normalization.
Genome sequencing and bioinformatics analysis.
DNA sequencing of phages was performed by Shanghai Biozeron Biotechnology Co., Ltd. (Shanghai, China). The genomes were submitted to the online annotation tool RAST (http://rast.nmpdr.org) for genome-wide alignment quick annotation. BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi), Gene Ontology (GO) (http://geneontology.org/), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.kegg.jp/) annotations were performed for the prediction of gene and protein functions. The bioinformatics analysis was done as reported previously, with minor modifications (41). Briefly, the alignment of phage capsid protein sequences from different phages were carried out using ClustalW in MEGA 7.0 software (42), and then a phylogenetic analysis was performed by using neighbor-joining with a pairwise deletion, p-distance distribution, and bootstrap analysis of 1,000 repeats as the parameters.
Data availability.
The complete genome sequence of phage vB_Pae_QDWS is available in GenBank under accession number MZ687409, and the phage vB_Pae_W3 is under OK094665. All data within the paper are available from the authors upon request.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program (2017YFC1600703) and National Nature Science Foundation of China (31870166).
G.X. acquired and analyzed the data, X.L. and J.K. performed TEM assays, H.L. supervised the research, and J.W. designed the study and wrote the manuscript.
We declare no conflict of interest.
Contributor Information
Jingxue Wang, Email: snow@ouc.edu.cn.
Rebecca Ellis Dutch, University of Kentucky College of Medicine.
REFERENCES
- 1.Silva YJ, Costa L, Pereira C, Mateus C, Cunha A, Calado R, Gomes NC, Pardo MA, Hernandez I, Almeida A. 2014. Phage therapy as an approach to prevent Vibrio anguillarum infections in fish larvae production. PLoS One 9:e114197. 10.1371/journal.pone.0114197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harada LK, Silva EC, Campos WF, Del Fiol FS, Vila M, Dąbrowska K, Krylov VN, Balcão VM. 2018. Biotechnological applications of bacteriophages: state of the art. Microbiol Res 212–213:38–58. 10.1016/j.micres.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Pollock J, Low AS, McHugh RE, Muwonge A, Stevens MP, Corbishley A, Gally DL. 2020. Alternatives to antibiotics in a One Health context and the role genomics can play in reducing antimicrobial use. Clin Microbiol Infect 26:1617–1621. 10.1016/j.cmi.2020.02.028. [DOI] [PubMed] [Google Scholar]
- 4.Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, Moineau S, Mojica FJM, Scott D, Shah SA, Siksnys V, Terns MP, Venclovas C, White MF, Yakunin AF, Yan W, Zhang F, Garrett RA, Backofen R, van der Oost J, Barrangou R, Koonin EV. 2020. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18:67–83. 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rostol JT, Marraffini L. 2019. (Ph)ighting phages: how bacteria resist their parasites. Cell Host Microbe 25:184–194. 10.1016/j.chom.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldfarb T, Sberro H, Weinstock E, Cohen O, Doron S, Charpak-Amikam Y, Afik S, Ofir G, Sorek R. 2015. BREX is a novel phage resistance system widespread in microbial genomes. EMBO J 34:169–183. 10.15252/embj.201489455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azam AH, Tanji Y. 2019. Bacteriophage-host arm race: an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl Microbiol Biotechnol 103:2121–2131. 10.1007/s00253-019-09629-x. [DOI] [PubMed] [Google Scholar]
- 8.Wei Y, Gao ZQ, Otsuka Y, Naka K, Yonesaki T, Zhang H, Dong YH. 2013. Structure-function studies of Escherichia coli RnlA reveal a novel toxin structure involved in bacteriophage resistance. Mol Microbiol 90:956–965. 10.1111/mmi.12409. [DOI] [PubMed] [Google Scholar]
- 9.Cui XL, You JJ, Sun L, Yang XJ, Zhang T, Huang KC, Pan XW, Zhang FJ, He Y, Yang HJ. 2016. Characterization of Pseudomonas aeruginosa phage C11 and identification of host genes required for virion maturation. Sci Rep 6:39130. 10.1038/srep39130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan FX, Li X, Pang B, Zhang C, Li Z, Zhang LJ, Li J, Zhang JY, Yan MY, Liang WL, Kan B. 2018. The outer-membrane protein TolC of Vibrio cholerae serves as a second cell-surface receptor for the VP3 phage. J Biol Chem 293:4000–4013. 10.1074/jbc.M117.805689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kruger DH, Schroeder C. 1981. Bacteriophage T3 and bacteriophage T7 virus-host cell interactions. Microbiol Rev 45:9–51. 10.1128/mr.45.1.9-51.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber HE, Tabor S, Richardson CC. 1987. Escherichia coli thioredoxin stabilizes complexes of bacteriophage T7 DNA polymerase and primed templates. J Biol Chem 262:16224–16232. 10.1016/S0021-9258(18)47719-8. [DOI] [PubMed] [Google Scholar]
- 13.You JJ, Sun L, Yang XJ, Pan XW, Huang ZW, Zhang XX, Gong MX, Fan Z, Li LY, Cui XL, Jing ZY, Jin SG, Rao ZM, Wu WH, Yang HJ. 2018. Regulatory protein SrpA controls phage infection and core cellular processes in Pseudomonas aeruginosa. Nat Commun 9:1846. 10.1038/s41467-018-04232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seco EM, Ayora S. 2017. Bacillus subtilis DNA polymerases, PolC and DnaE, are required for both leading and lagging strand synthesis in SPP1 origin-dependent DNA replication. Nucleic Acids Res 45:8302–8313. 10.1093/nar/gkx493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyland-Kroghsbo NM, Maerkedahl RB, Svenningsen SL. 2013. A quorum-sensing-induced bacteriophage defense mechanism. mBio 4:e00362-12. 10.1128/mBio.00362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan D, Svenningsen SL, Middelboe M. 2015. Quorum sensing determines the choice of antiphage defense strategy in Vibrio anguillarum. mBio 6:e00627. 10.1128/mBio.00627-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin XY, Sun QH, Yang BX, Pan XW, He Y, Yang HJ. 2017. Quorum sensing influences phage infection efficiency via affecting cell population and physiological state. J Basic Microbiol 57:162–170. 10.1002/jobm.201600510. [DOI] [PubMed] [Google Scholar]
- 18.Pang Z, Raudonis R, Glick BR, Lin TJ, Cheng Z. 2019. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and alternative therapeutic strategies. Biotechnol Adv 37:177–192. 10.1016/j.biotechadv.2018.11.013. [DOI] [PubMed] [Google Scholar]
- 19.Brencic A, McFarland KA, McManus HR, Castang S, Mogno I, Dove SL, Lory S. 2009. Th GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol Microbiol 73:434–445. 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7:745–754. 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 21.Heeb S, Blumer C, Haas D. 2002. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol 184:1046–1056. 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan Kaler KM, Nix JC, Schubot FD. 2021. RetS inhibits Pseudomonas aeruginosa biofilm formation by disrupting the canonical histidine kinase dimerization interface of GacS. J Biol Chem 297:101193. 10.1016/j.jbc.2021.101193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis VI, Waters EM, Finton-James SE, Gori A, Kadioglu A, Brown AR, Porter SL. 2018. Multiple communication mechanisms between sensor kinases are crucial for virulence in Pseudomonas aeruginosa. Nat Commun 9:2219. 10.1038/s41467-018-04640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coggan KA, Wolfgang MC. 2012. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr Issues Mol Biol 14:47–70. [PubMed] [Google Scholar]
- 25.Shah MG, Taylor VL, Bona D, Tsao Y, Stanley SY, Pimentel-Elardo SM, McCallum M, Bondy-Denomy J, Howell PL, Nodwell JR, Davidson AR, Moraes TF, Maxwell KL. 2021. A phage-encoded anti-activator inhibits quorum sensing in Pseudomonas aeruginosa. Mol Cell 81:571–583.e6. 10.1016/j.molcel.2020.12.011. [DOI] [PubMed] [Google Scholar]
- 26.Bai J, Jeon B, Ryu S. 2019. Effective inhibition of Salmonella Typhimurium in fresh produce by a phage cocktail targeting multiple host receptors. Food Microbiol 77:52–60. 10.1016/j.fm.2018.08.011. [DOI] [PubMed] [Google Scholar]
- 27.Abedon ST. 2017. Phage “delay” towards enhancing bacterial escape from biofilms: a more comprehensive way of viewing resistance to bacteriophages. AIMS Microbiol 3:186–226. 10.3934/microbiol.2017.2.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craig L, Forest KT, Maier B. 2019. Type IV pili: dynamics, biophysics and functional consequences. Nat Rev Microbiol 17:429–440. 10.1038/s41579-019-0195-4. [DOI] [PubMed] [Google Scholar]
- 29.Chiang P, Sampaleanu LM, Ayers M, Pahuta M, Howell PL, Burrows LL. 2008. Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology (Reading) 154:114–126. 10.1099/mic.0.2007/011320-0. [DOI] [PubMed] [Google Scholar]
- 30.Wei X, Huang XQ, Tang LL, Wu DQ, Xu YQ. 2013. Global control of GacA in secondary metabolism, primary metabolism, secretion systems, and motility in the rhizobacterium Pseudomonas aeruginosa M18. J Bacteriol 195:3387–3400. 10.1128/JB.00214-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ventre I, Goodman AL, Vallet-Gely I, Vasseur P, Soscia C, Molin S, Bleves S, Lazdunski A, Lory S, Filloux A. 2006. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci USA 103:171–176. 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humair B, Gonzalez N, Mossialos D, Reimmann C, Haas D. 2009. Temperature-responsive sensing regulates biocontrol factor expression in Pseudomonas fluorescens CHA0. ISME J 3:955–965. 10.1038/ismej.2009.42. [DOI] [PubMed] [Google Scholar]
- 33.Wang BX, Wheeler KM, Cady KC, Lehoux S, Cummings RD, Laub MT, Ribbeck K. 2021. Mucin glycans signal through the sensor kinase RetS to inhibit virulence-associated traits in Pseudomonas aeruginosa. Curr Biol 31:90–102.e7. 10.1016/j.cub.2020.09.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zolfaghar I, Angus AA, Kang PJ, To A, Evans DJ, Fleiszig SM. 2005. Mutation of retS, encoding a putative hybrid two-component regulatory protein in Pseudomonas aeruginosa, attenuates multiple virulence mechanisms. Microbes and Infection 7:1305–1316. 10.1016/j.micinf.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 35.Willner D, Furlan M, Haynes M, Schmieder R, Angly FE, Silva J, Tammadoni S, Nosrat B, Conrad D, Rohwer F. 2009. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One 4:e7370. 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim YW, Evangelista JS, Schmieder R, Bailey B, Haynes M, Furlan M, Maughan H, Edwards R, Rohwer F, Conrad D. 2014. Clinical insights from metagenomic analysis of sputum samples from patients with cystic fibrosis. J Clin Microbiol 52:425–437. 10.1128/JCM.02204-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bae HW, Cho YH. 2013. Complete genome sequence of Pseudomonas aeruginosa podophage MPK7, which requires type IV pili for infection. Genome Announc 1:e00744-13. 10.1128/genomeA.00744-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu CJ, Xia YZ, Liu DX, Zhao R, Gao R, Liu HL, Xun LY. 2017. Cupriavidus necator H16 uses flavocytochrome c sulfide dehydrogenase to oxidize self-produced and added sulfide. Appl Environ Microbiol 83:e01610-17. 10.1128/AEM.01610-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitchurch CB, Erova TE, Emery JA, Sargent JL, Harris JM, Semmler ABT, Young MD, Mattick JS, Wozniak DJ. 2002. Phosphorylation of the Pseudomonas aeruginosa response regulator AlgR is essential for type IV fimbria-mediated twitching motility. J Bacteriol 184:4544–4554. 10.1128/JB.184.16.4544-4554.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mirzaei MK, Nilsson AS. 2015. Isolation of phages for phage therapy: a comparison of spot tests and efficiency of plating analyses for determination of host range and efficacy. PLoS One 10:e0118557. 10.1371/journal.pone.0118557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li P, Lin H, Mi ZQ, Xing SZ, Tong YG, Wang JX. 2019. Screening of polyvalent phage-resistant Escherichia coli strains based on phage receptor analysis. Front Microbiol 10:850. 10.3389/fmicb.2019.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of phage vB_Pae_QDWS is available in GenBank under accession number MZ687409, and the phage vB_Pae_W3 is under OK094665. All data within the paper are available from the authors upon request.