ABSTRACT
BK polyomavirus (PyV) infects the genitourinary tract of >90% of the adult population. Immunosuppression increases the risk of viral reactivation, making BKPyV a leading cause of graft failure in kidney transplant recipients. Polyomaviruses have a small double-stranded DNA (dsDNA) genome that requires host replication machinery to amplify the viral genome. Specifically, polyomaviruses promote S phase entry and delay S phase exit by activating the DNA damage response (DDR) pathway via an uncharacterized mechanism requiring viral replication. BKPyV infection elevates expression of MutSα, a mismatch repair (MMR) pathway protein complex that senses and repairs DNA mismatches and can activate the DDR. Thus, we investigated the role of the MMR pathway by silencing the MutSα component, Msh6, in BKPyV-infected primary cells. This resulted in severe DNA damage that correlated with weak DNA damage response activation and a failure to arrest the cell cycle to prevent mitotic entry during infection. Furthermore, silencing Msh6 expression resulted in significantly fewer infectious viral particles due to significantly lower levels of VP2, a minor capsid protein important for trafficking during subsequent infections. Since viral assembly occurs in the nucleus, our findings are consistent with a model in which entry into mitosis disrupts viral assembly due to nuclear envelope breakdown, which disperses VP2 throughout the cell, reducing its availability for encapsidation into viral particles. Thus, the MMR pathway may be required to activate the ATR (ATM-Rad3-related) pathway during infection to maintain a favorable environment for both viral replication and assembly.
IMPORTANCE Since there are no therapeutics that target BKPyV reactivation in organ transplant patients, it is currently treated by decreasing immunosuppression to allow the natural immune system to fight the viral infection. Antivirals would significantly improve patient outcomes since reducing immunosuppression carries the risk of graft failure. PyVs activate the DDR, for which there are several promising inhibitors. However, a better understanding of how PyVs activate the DDR and what role the DDR plays during infection is needed. Here, we show that a component of the mismatch repair pathway is required for DDR activation during PyV infection. These findings show that the mismatch repair pathway is important for DDR activation during PyV infection and that inhibiting the DDR reduces viral titers by generating less infectious virions that lack the minor capsid protein VP2, which is important for viral trafficking.
KEYWORDS: BK polyomavirus, DNA damage response, MSH6, mismatch repair
INTRODUCTION
Polyomaviruses (PyVs) are small, double-stranded DNA, nonenveloped viruses. There are 13 known human PyVs, with BKPyV, JCPyV, and Merkel cell PyV (MCPyV) causing the majority of PyV-related maladies. Despite significant sequence and functional differences between PyV species, the PyV genome universally consists of three regions, early genes, late genes, and the noncoding control region, which contains the viral origin of replication (1). The BKPyV early genes are expressed by alternative splicing of a single transcript to generate large (TAg/LTAg), small (tAg/sTAg), and truncated (truncTAg) tumor antigens, which promote cell cycle entry into the S phase. The late genes encode agnoprotein and three structural proteins (VP1, VP2, and VP3). The BKPyV virion has an outer shell that consists of 72 pentamers of the major capsid protein, VP1, and an inner shell that is not solvent exposed, consisting of VP2 and VP3 monomers (2). The minor capsid proteins, VP2 and VP3, are translated from the same transcript, but VP2 has an N-terminal extension (3). VP1 is required for viral entry through endocytosis and microtubule transport to the endoplasmic reticulum (ER), at which point the capsid is uncoated, and VP2 and VP3 are required for ER exit and nuclear import by the importin pathway where the genome remains episomal (4, 5). TAg expression drives the cells into the S phase by sequestering retinoblastoma protein (pRb), resulting in derepression of E2F-dependent transcription, which leads to the expression of genes important for G1/S transition as well as DNA repair, among others (6). BKPyV does not encode a polymerase for replication; thus, the virus depends on cellular proteins for replication, such as replication protein A (RPA), DNA polymerase δ (Pol δ), and topoisomerase I.
In primary kidney cells, BKPyV induces a prolonged S phase by inducing cell cycle arrest wherein host and viral genomes are rereplicated. This cell cycle arrest is achieved by activation of the DNA damage response (DDR) (7) via an unknown mechanism (8, 9). Replication of the viral genome is linked to activation of ATM (ataxia telangiectasia mutated) and ATR (ATM-Rad3 related), which are DDR kinases that respond to dsDNA breaks and single-stranded DNA lesions, respectively (7, 9–13). Studies of simian virus 40 (SV40), a related PyV, suggest that activation of the DDR is important for resolving replication intermediates produced during amplification of the circular viral genome (12). Another meaningful role for DDR activation in PyV infection is to promote cell cycle arrest to prolong the S phase during BKPyV infection in primary kidney cells, which allows more time for viral replication (7, 9).
A proteomics analysis found that the most significantly upregulated repair pathway during BKPyV infection is the mismatch repair (MMR) pathway (14), which is a known activator of the DDR. MMR is an excision repair DDR pathway that targets DNA mismatches, insertion-deletion loops, and certain nucleotide base modifications (15). Two protein complexes, MutSα and MutSβ, serve as the DNA damage sensors that initiate this pathway. MutSα is an Msh2 and Msh6 heterodimeric complex that is primarily responsible for recognizing mismatched nucleotides, single base insertion or deletion loops, and DNA adducts (16). MutSβ is a heterodimer of Msh2 and Msh3 that recognizes larger insertion-deletion loops (15–17). Importantly, MutSα can bind both base-base mispairs and insertion-deletion mispairs, while MutSβ can only bind insertion-deletion mispairs and double-strand breaks (18). Once a mismatch is identified by MutSα or MutSβ, they are converted into closed clamp-like conformation that can translocate along the DNA. Then, they recruit the nicking endonuclease, MutLα complex, to initiate removal of the error-containing DNA strand. The single-stranded terminus that is generated allows entry of exonuclease I (ExoI) for strand excision (19). The resulting single-stranded DNA lesion is repaired by DNA polymerase δ and DNA ligase. Msh6 and Msh2 expression is promoted by E2F transcription factors (20–22), which are derepressed by TAg sequestration of pRB during PyV infection (23). However, the role that the MMR pathway plays during BKPyV infection has not been studied.
This study used primary kidney cells to determine whether the MMR pathway was required for productive BKPyV infection. In agreement with the proteomics data (14), Western blot analysis showed that BKPyV infection elevated Msh6 expression disproportionately to Msh2 and Msh3, suggesting that MutSα was the dominant MMR complex upregulated by BKPyV. BKPyV virions produced in Msh6-deficient cells were less infectious and contained smaller amounts of the minor capsid protein VP2 despite having no defect in packaging viral DNA, suggesting a role for viral assembly. Given that the MMR pathway is a known activator of the DDR (15, 24, 25), we determined if Msh6 was required for robust ATR activation and cell cycle arrest during a BKPyV infection. Msh6 depletion resulted in increased mitotic entry, nuclear fragmentation, and metaphase shattering, which recapitulates the phenotype when the DDR is inhibited during a BKPyV infection (7, 9). These findings suggest that Msh6 is necessary for DDR activation during a BKPyV infection and for production of infectious virions.
RESULTS
BKPyV infection upregulates MutSα expression.
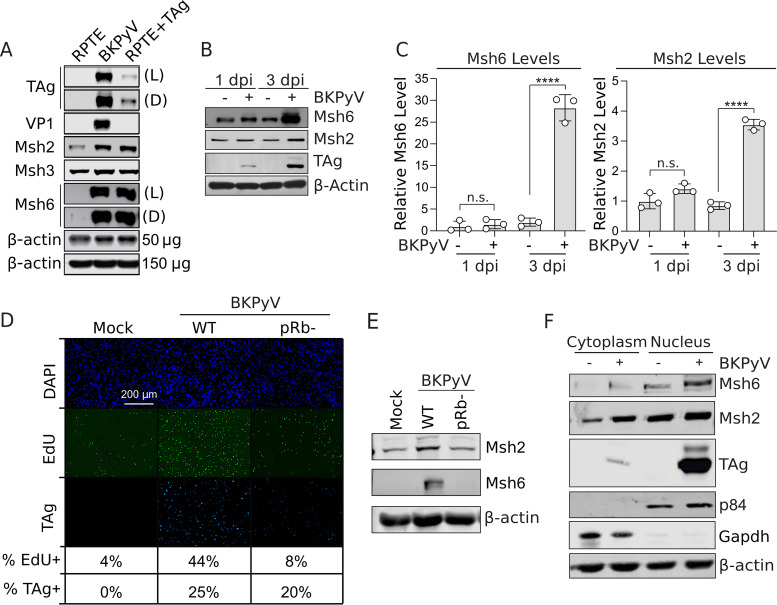
To study the role of the mismatch repair pathway during BKPyV infection, we used primary human renal proximal tubule epithelial (RPTE) cells, which are the predicted site of productive infection in patients with reactivated BKPyV (26). Furthermore, primary RPTE cells have an intact DNA damage response (DDR) and normal cell cycle regulation in contrast to transformed or immortalized cells that often have mutations in these pathways (27). First, the proteomic analysis (14) was confirmed by Western blot analysis, which showed that MutSα (Msh6 and Msh2) protein expression was increased in BKPyV-infected cells and in cells expressing TAg alone (Fig. 1A and B). Since Msh3 levels were not impacted by either infection or TAg expression, nor were they significantly enriched in either a proteomic analysis or a transcriptomic analysis of BKPyV infection (14, 23), this suggests that MutSα is preferentially dysregulated during infection in contrast to MutSβ (Msh3 and Msh2). Msh6 protein levels were not affected at early time points (1 day postinfection [dpi]) and subsequently increased (14-fold) late in infection (3 dpi) (Fig. 1B). In contrast, Msh2 levels only increased 4-fold at 3 dpi (Fig. 1A to C). Our findings demonstrated that TAg expression was sufficient to upregulate Msh6 protein levels. Msh6 is thought to be expressed dependent upon E2F activation (20–22). To test if E2F activation was required for Msh6 upregulation in BKPyV-infected cells, we infected RPTE cells with BKPyV that had mutations in the LxCxE domain of TAg (E109K and D110K; pRb−) to ablate pRb binding (28). Infection with the pRb− mutant failed to promote the S phase, consistent with the TAg pRb− mutant being unable to inhibit pRb and promote activation of the E2F S phase transcription factors. Importantly, the pRb− mutant BKPyV did not induce Msh6 upregulation (Fig. 1D and E) despite expressing a similar amount of TAg.
FIG 1.
BKPyV infection increases Msh6 levels. (A) Western blot analysis of Msh2, Msh6, Msh3 (MutSα and MutSβ), and TAg expression levels following BKPyV infection at an MOI of 0.5 focus-forming units (FFU)/cell of RPTE or RPTE TAg-transduced cells at 3 days post-BKPyV infection (dpi). TAg and VP1 levels represent viral protein expression, and β-actin level was used as a loading control. (B) Representative quantitative Western blot analysis of Msh6 and Msh2 at 1 dpi and 3 dpi of MOI of 0.5 FFU/cell. TAg levels represent viral protein expression, and β-actin levels are used as a loading control. (C) Msh6 and Msh2 protein levels normalized to β-actin levels from panel B and graphed relative to the mock-infected cells at day 1, which were set to 1. (D and E) RPTE cells were infected (MOI = 0.2) with wild-type (WT) BKPyV or a pRb binding mutant (pRb−). (D) Portion of cells expressing TAg and in S phase were determined by fluorescence microscopy at 3 dpi. (E) The impact of the pRb binding mutant on Msh6 and Msh2 expression was compared to mock- and WT BKPyV-infected RPTE cells by Western blot analysis (using β-actin as a loading control) at 3 dpi for n = 3 biological replicates. (F) Western blot analysis of nuclear and cytoplasmic fractions from mock- or BKPyV-infected RPTE cells 3 dpi. Western blots were probed for Msh6, Msh2, TAg, and p84 (nuclear markers), GAPDH (cytoplasmic marker), and β-actin (loading control). Shown are representative of n = 3 biological repeats.
Since Msh6 and Msh3 are only active when bound to Msh2, and Msh2 preferentially forms a heterodimer with Msh6 (29, 30), this suggests that the increase in Msh6 and Msh2 protein levels represents active MutSα complexes. A recent small interfering RNA (siRNA) screen for host factors that are important for BKPyV, using polyploidy as a measurement for productive BKPyV infection, revealed that Msh6 silencing, but not Msh2 or Msh3, significantly decreased polyploidy (indicating decreased BKPyV infection) in RPTE cells (31). However, this study did not confirm knockdown. Despite using multiple siRNAs, we have been unable to achieve knockdown of Msh2. Since our findings and others suggest that Msh6 was preferentially upregulated over Msh3 during BKPyV infection, we focused on Msh6 to specifically understand the role of the MutSα complex during infection.
Since MutSα translocates to the nucleus in response to genotoxic stress (32), we determined if BKPyV infection induced nuclear translocation of MutSα by performing a nuclear fractionation followed by Western blot analysis for Msh2, Msh6, and TAg. Msh6 levels accumulated in the nucleus following BKPyV infection accompanied by a partial shift in Msh2 into the nucleus (Fig. 1F). It is possible that Msh2 is retained in the cytoplasm due to binding with its alternative partner, Msh3. These data suggest that upon BKPyV infection, MutSα is upregulated and becomes localized in the nucleus, which is the site of viral replication.
MutSα is required for proper assembly of BKPyV particles but not for viral replication.
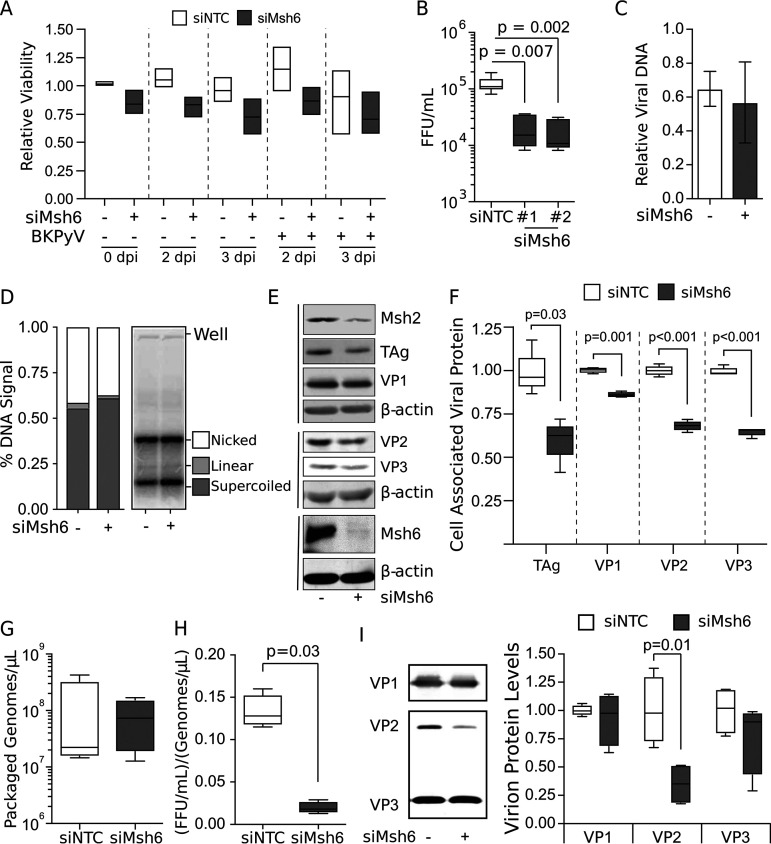
Msh6 binds to Msh2 to form MutSα, which is the predominant MMR complex. Alternatively, Msh2 can bind with Msh3 to form MutSβ (33). Msh6 and Msh3 are only active when bound to Msh2, and, in their monomeric form, Msh2, Msh6, and Msh3 are rapidly degraded (29, 30, 34–36). To assess the requirement of the MutSα complex for BKPyV, Msh6 was silenced in RPTE cells, cell viability was measured, and the titers of viral progeny titers were determined (Fig. 2A and B). Silencing Msh6 with either of two different siRNAs significantly reduced the infectious titers relative to the nontargeting siRNA control without impacting cell viability (Fig. 2A and B).
FIG 2.
MutSα is required for proper assembly of BKPyV particles, but not for viral replication. For all analyses, Msh6 was knocked down with siRNA in RPTE cells 3 days prior to mock or BKPyV infection (MOI = 0.5 FFU/cell). (A) Cell viability was monitored by cell titer glow at 0, 2, and 3 dpi. Box-and-whisker plots for n = 3 biological repeats. Two-tailed Student's t test indicated no significant change (P > 0.05). (B to E) Samples were collected from whole-cell lysates 2 dpi. (B) Msh6 was knocked down using two independent siRNAs. Viral titers were determined by focus-forming assay. A box-and-whisker plot is shown for n ≥ 5 biological repeats. P values were determined using the Kruskal-Wallis test. (C) Viral DNA levels were quantified by RT-qPCR of extrachromosomal DNA from cells using BKPyV-specific primers. Viral DNA levels from infected siNTC and siMsh6 knocked-down cells are shown relative to the no-siRNA viral DNA level set to 1 (not shown). SD is shown for n = 3 biological repeats. Unpaired Student's t test indicated no significant change (P = 0.62). (D) Southern blot analysis was performed on equivalent amounts of total DNA isolated from cells to detect BKPyV DNA. Southern blots were quantified by phosphorimager, and the fractions of total DNA in the nicked, linear, and supercoiled form are shown. There was no evidence of concatenated viral DNA species, which would have a higher molecular weight than the nicked DNA. Error bars indicate SD for n = 3 biological repeats. Unpaired Student's t test indicated no significant change between the siNTC and siMsh6 for any DNA species. (E) Western blot analyses for the indicated proteins were performed on whole-cell lysates. Representative of n = 3 biological replicates is shown. (F) Box-and-whisker plots are shown for the major and minor capsid proteins analyzed by Western blotting in panel E. Significant differences were determined by Student's t test. (G to I) Viral particles were purified from siNTC and siMsh6 cells 2 dpi, and box-and-whisker plots are shown for n = 4 biological repeats; P values were determined using the Mann-Whitney test. (G) Viral particles were purified from cellular lysates, and any unencapsidated DNA was degraded by DNase treatment. Encapsidated viral genomes were extracted, and BKPyV DNA levels were measured by RT-qPCR. (H) Viral titers from purified BKPyV particles were measured by focus-forming assay and normalized to the amount of encapsidated genome content determined in panel G. (I) VP1, VP2, and VP3 protein levels from purified viral particles were determined by Western blot analysis (representative of n = 4). Box-and-whisker plots showing protein levels normalized to the no siRNA control and are shown relative to the average siNTC set to 1.
MutSα is an important cofactor for high-fidelity homologous recombination—a DNA repair pathway whose proteins are important for repairing dsDNA breaks and protecting stalled replication forks (37–39). Furthermore, homologous recombination is thought to be important for efficient PyV replication (12, 40). Since Msh6 was needed to produce high viral titers, we determined the impact of Msh6 silencing on viral genomes isolated from intact cells. Msh6 knockdown had no effect on levels of intracellular viral genomes (Fig. 2C). Furthermore, Southern blot analysis of encapsidated viral DNA revealed that Msh6 knockdown did not alter the relative abundance of nicked, linear, or supercoiled viral DNA, nor was there evidence of alternative viral chromatin species that might indicate aberrant viral DNA replication (Fig. 2D). Because neither gross DNA levels nor viral DNA species were affected by Msh6 silencing, the decrease in viral titers was not due to defective DNA replication.
To determine if progeny fitness could be attributed to the relative abundance of the viral proteins, Msh6 was silenced, followed by Western blot analysis (Fig. 2E). As mentioned above, Msh2, Msh3, and Msh6 must form either MutSα or MutSβ or be rapidly degraded. Msh2 levels were decreased by Msh6 silencing, which is consistent with Msh6 being bound to Msh2 during infection to mediate their mutual stability by forming MutSα (Fig. 2E). Residual Msh2 is likely protected from proteolysis due to binding to either Msh3 or undetectable Msh6. The major capsid VP1 protein abundance was only subtly affected by Msh6 knockdown (Fig. 2E). While there was a decrease in TAg levels with Msh6 knockdown, this did not affect viral DNA levels, suggesting TAg is not limiting. However, Msh6 silencing resulted in a more pronounced decrease in the levels of the minor capsid proteins VP2 and VP3 (Fig. 2E and F), which are required for the assembly of infectious progeny virions (4, 5).
Since Msh6 knockdown had no significant effect on viral DNA replication in RPTE cells, yet the level of infectious virions and minor capsid proteins were decreased by Msh6 silencing, we examined viral assembly by quantifying packaged viral DNA and viral proteins from purified virions. Msh6 knockdown did not significantly impact the number of encapsidated viral genomes isolated from equal volumes of purified viral particles (Fig. 2G). Further analysis of the ratio of infectious virions per encapsidated genome revealed that Msh6 knockdown decreased viral infectivity of these purified particles 6.8-fold compared to the nontargeting siRNA control (Fig. 2H). We cannot rule out that there were inactivating mutations occurring during the amplification of viral DNA in the absence of MutSα; however, this is unlikely since the error rate of DNA polymerase δ is 1/106 nucleotides in the absence of MMR activity, meaning that MutSα knockdown would result in less than 1 mutation per 100 viral genomes (41, 42). Therefore, it is more likely that the reduction in viral infectivity is due to a defect in particle assembly rather than an increase in mutations in viral genomes.
Mature BKPyV virions have 72 pentamers of the VP1 that are solvent exposed, while VP2 and VP3 are monomers in the core. VP1 can form pseudovirions containing viral DNA in the absence of VP2 and VP3; however, the nuclear localization sequences of VP2 and VP3 are important for BKPyV trafficking from the endoplasmic reticulum to the nucleus (3, 4, 43, 44). Therefore, we examined whether there were alterations in the relative protein composition of the viral particle that could indicate viral assembly defects that would explain decreased viral infectivity. Consistent with our finding that Msh6 did not alter the number of encapsidated viral genomes, the relative VP1 levels between Msh6 knockdown and nontargeting control siRNA (siNTC) conditions were not significantly altered (Fig. 2I). Since encapsidated viral DNA and VP1 levels were not affected by Msh6 silencing, this suggests that the total number of virions produced during infection was not altered relative to the nontargeting siRNA control. However, virion-associated VP2 levels were significantly reduced when Msh6 was silenced, although VP3 levels were unaffected (Fig. 2I). These findings indicate that Msh6 knockdown may impair viral assembly by skewing the composition of the minor capsid VP2 protein in the cell and thus within viral progeny. Because VP2 is required for efficient particle transition from the ER to the nucleus to establish infection for PyVs (45), particles formed in the absence of Msh6 may be defective in trafficking, which would explain the apparent decrease in infectious particles.
MutSα is required for host genomic stability and ATR activation during BKPyV infection.
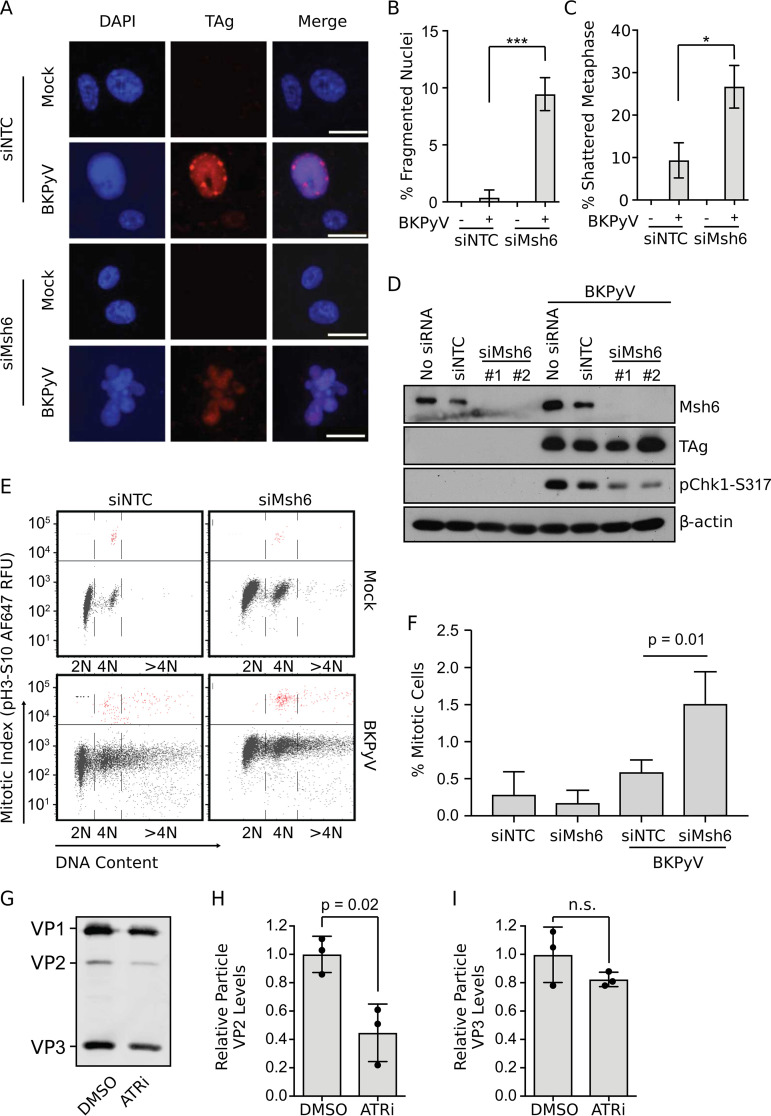
While investigating the impact of Msh6 on BKPyV infection, we observed a significant population of BKPyV-infected cells with fragmented nuclei, which is a sign of severe DNA damage (Fig. 3A and B). Previously, BKPyV infection was found to induce nuclear fragmentation only when the DNA damage response governed by the ATR kinases was inhibited (7, 9, 46). ATR inhibition during infection was also linked to metaphase shattering (an uncountable number of DNA breaks), which is indicative of catastrophic DNA damage occurring during mitosis (7, 9). We found that silencing Msh6 resulted in a significant increase in shattered metaphases when cells were infected with BKPyV (Fig. 3C). Our previous work identified that the ATR pathway was required during BKPyV infection in order to prevent DNA damage that occurs when S phase cells enter mitosis prematurely (7). MutSα is known to interact with various DDR proteins and pathways, including ATR, in response to DNA damage (37, 47, 48). Therefore, we examined Msh6-silenced cells for evidence of defective ATR pathway activation since the nuclear fragmentation and metaphase shattering were similar to ATR knockdown or inhibition during a BKPyV infection.
FIG 3.
MutSα is required for host genomic stability and ATR activation during BKPyV infection. Msh6 was knocked down in RPTE cells 3 days prior to mock or BKPyV infection (MOI = 0.5 FFU/cell). (A) Cells were fixed at 3 dpi, and an immunofluorescent micoscopy analysis (IFA) was performed to detect TAg (red) and DNA (DAPI). Nuclear morphology was scored for 100 TAg-positive nuclei per biological repeat. Scale bar, 20 μm. Representative images are shown. (B) Quantification of nuclei with fragmented morphology from panel A. (C) Metaphase spread analysis was performed, and 50 metaphase nuclei were scored per biological replicate. Error bars indicate SD for n = 3 biological repeats. Two-tailed Student's t test was used to determine significance (*, P < 0.05; ***, P < 0.001). (D) Western blot analyses of lysates collected 2 dpi to measure ATR activation (pChk1-S317) TAg, Msh6, and β-actin levels. Shown is representative of n = 4. (E) Cells were fixed at 3 dpi and stained for mitotic index (pH 3, Ser10) and DNA content (FxCycle Violet) and analyzed by flow cytometry (top). Mitotic cells are indicated in red (above the line). (F) Quantification of the percentage of cells in mitosis for n = 3 biological repeats for the indicated conditions is shown. Fluorescence-activated cell sorter (FACS) analyses are representative of n = 3 biological repeats. Error bars indicate SD for n = 3, and P values were determined using a two-tailed Student's t test. (G to I) BKPyV-infected cells (0.5 FFU/cell) were treated with ATRi (5 μM VE-821) or DMSO (vehicle control) at 1 dpi and then samples were harvested at 2 dpi for particle purification. (G) Equal volumes of purified particles were subjected for quantitative Western blot analysis for the viral structural proteins VP1, VP2, and VP3. The ratio of VP2 (H) or VP3 (I) to VP1 ± SD was calculated and then normalized to the DMSO control for n = 3 biological replicates.
BKPyV infection activates the ATR pathway (7–9), which can be evaluated by measuring Chk1 phosphorylation at serine-317 (pChk1-S317), a downstream marker of ATR activation (Fig. 3D, compare mock versus BKPyV). Notably, Chk1 phosphorylation (ATR activation) was severely impaired when Msh6 was knocked down (Fig. 3D), suggesting that Msh6 is required for BKPyV-driven ATR activation. Since the trigger for ATR activation by BKPyV is viral DNA replication (8), which was not affected by Msh6 knockdown, this suggests that Msh6 may be downstream of DNA replication for DDR activation.
Since Msh6 was required to prevent host DNA damage and for robust ATR activation during BKPyV infection, which is important for preventing host DNA damage by blocking mitosis, we determined whether MutSα is required for cell cycle arrest during a BKPyV infection. BKPyV infection leads to an accumulation of cells with a ≥4N DNA content compared to the uninfected control (Fig. 3E). This is due to viral induction of the S phase coupled to G2/M arrest in a phenomenon called endoreduplication that results in polyploidy (7). Msh6 silencing significantly increased mitotic entry (pH 3, Ser10) (Fig. 3E and F) in BKPyV-infected cells, but not mock-infected cells, as expected since the ATR pathway (pChk1) was not activated in the mock samples (Fig. 3D). Thus, our findings suggest that Msh6 is required for activation of the ATR pathway during BKPyV infection, which, in turn, arrests the cell cycle at the G2/M checkpoint and prevents DNA damage (7, 49).
Given that Msh6 was required for ATR activation and for proper assembly of infectious virions, we next sought to determine if ATR is required for BKPyV assembly. The ratio of the minor capsid proteins, VP2 and VP3, compared to the major capsid protein, VP1, in purified particles produced in cells treated with a potent ATR inhibitor revealed a significant decrease in the level of VP2 but not VP3 relative to VP1 (Fig. 3G to I), which is similar to what we observed when Msh6 was silenced during infection (Fig. 3H). Given that our data showed that Msh6 was required to activate the ATR pathway and both ATRi and Msh6 silencing decreased the abundance of VP2 in viral particles, our data suggest that Msh6 activation of the ATR pathway during BKPyV infection is required for viral assembly. Since PyVs assemble in the nucleus (50) and both Msh6 and ATR were required to prevent mitosis, which leads to disruption of the nuclear envelope, this suggests that mitotic entry disrupts BKPyV particle assembly by disrupting the viral assembly compartment.
DISCUSSION
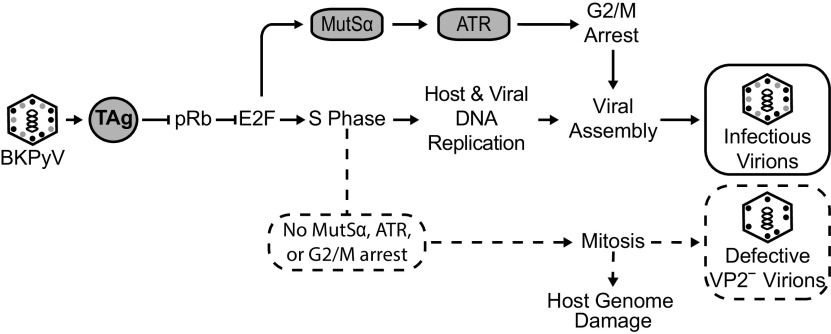
DDR activation is important during a BKPyV infection to prolong the S phase and prevent massive host DNA damage via cell cycle arrest (7). In particular, activation of the ATR kinase by the virus prevents unscheduled mitotic entry, while viral and host DNA is undergoing replication, termed premature mitosis, which can result in cellular DNA damage. Although DDR activation is critical for viral titers, the mechanisms that underlie DDR activation during BKPyV infection are not understood. Since the MMR pathway recruits ATR to damaged DNA, which leads to DDR activation in other contexts, we investigated whether the upregulation of Msh6 during a BKPyV infection was important for viral production and DDR activation. Here, we showed that Msh6, a component of the MMR pathway, is important for robust ATR activation during a BKPyV infection in primary kidney cells. BKPyV infection or TAg expression resulted in an increase in Msh6 protein levels, suggesting an increase in the MutSα complex. Msh6 was also localized to the nucleus where MutSα is known to recruit ATR to sites of DNA damage. Knockdown of Msh6 protein levels decreased ATR activation (Fig. 3D), which is required for cell cycle arrest to prevent premature mitosis during a BKPyV infection (7). In fact, inhibiting ATR or knockdown of Msh6 resulted in increased mitosis and massive DNA damage (Fig. 3) (7). Importantly, knocking down Msh6 blunted ATR activation, suggesting that it lies upstream of ATR activation. Silencing Msh6 revealed that MutSα was dispensable for viral genome amplification but was required for high viral titers. Viral particles that were formed in the absence of Msh6 contained less VP2, the minor capsid protein, relative to VP1, the major capsid protein. Although equal levels of viral DNA were encapsidated by particles assembled in Msh6 knockdown cells, their infectivity was decreased, demonstrating a defect in particle assembly important for generating infectious virions. Similarly, the decrease in VP2 levels in viral particles was also observed when the ATR kinase was inhibited during infection. Our findings are consistent with a model in which MutSα is required for efficient ATR activation, leading to G2/M arrest in BKPyV-infected primary RPTE cells (Fig. 4). Taken together with our previous finding that ATR is necessary to prevent mitotic entry during BKPyV infection to prolong the S phase, our data suggest that blocking mitosis is also important for maintaining the integrity of the nucleus for virion assembly.
FIG 4.
Model for how Msh6 promotes G2/M arrest and maintains host genome stability during BKPyV infection. Expression of TAg following BKPyV infection relieves repression of pRb, which induces a burst of E2F transcription to initiate S phase entry whereupon host and viral replication occur. TAg also upregulates MutSα (Msh6 and Msh2) through derepression of pRb. MutSα and viral replication activate ATR to arrest the cell cycle at the G2/M checkpoint (solid arrows). Viral assembly occurs in the nucleus. Here, we showed that MUTSα is upregulated by TAg and is required for cell cycle arrest in order to prevent host DNA damage stemming from premature mitosis (dotted lines/arrows). Without MUTSα, ATR activation, and cell cycle arrest, viral assembly is defective, resulting in virions deficient in VP2 (dotted lines). Thus, G2/M arrest is necessary for production of infectious virions. Solid lines and arrows represent the normal viral infection pathway, whereas the dotted lines emphasize what happens when MUTSα is not upregulated during a BKPyV infection.
We observed that Msh6 levels increased with either BKPyV infection or TAg expression in RPTE cells, suggesting that Msh6 expression may be regulated by TAg. TAg is a viral protein that is expressed early in infection. While TAg has many functions, one of its main functions is to inactivate pRb, the transcriptional repressor of S phase promoting transcription factors (E2Fs). Indeed, the Msh6 promoter has predicted E2F binding sites, and it is known to be activated by Sp1, which is upregulated by E2F at the G1/S transition (51–53). Msh6 protein levels are regulated by expression levels of the Sp1 activator relative to the Sp3 repressor; thus, an increase in abundance of the Sp1 transcription factor (52), which occurs at the G1/S phase boundary, could explain the increased levels of Msh6 that we and others observed (14, 51). Indeed, infection with a pRb binding mutant BKPyV failed to upregulate Msh6 (Fig. 1), indicating that its upregulation is dependent on E2F activation. It is notable that while some MMR factors are upregulated upon growth stimulation, such as Msh2 and Msh6, other transcripts are constitutively transcribed, such as Msh3 (54). In addition, SV40-transformed cells had increased mRNA and protein levels of Msh6 and Msh2 compared to untransformed cells (33). This agrees with our findings showing that BKPyV infection or TAg expression increased Msh2 and Msh6 levels but not Msh3 levels (Fig. 1). These findings suggest that the major MMR complex during BKPyV infection is MutSα.
Our studies found that Msh6 levels increased to a greater extent than Msh2 (14-fold versus 4-fold, respectively). Msh6 and Msh2 are degraded through the ubiquitin-proteasome pathway, but phosphorylation of MutSα by PKCζ interferes with ubiquitination and degradation (34–36). In relation to our studies showing that Msh6 levels increased more than Msh2, another study provides a possible explanation for this, as they found that the direct phosphorylation of MutSα by PKCζ revealed that Msh6 is phosphorylated to a greater extent than Msh2 (36). Given that small TAg expression activates PKCζ (55), it is possible that Msh6 is stabilized in BKPyV- or Tag-expressing cells. While understanding differences in these protein levels is beyond the scope of our studies here, the increased expression and stabilization of Msh6 and, to a lesser extent, Msh2, are consistent with what is known about how the protein levels are regulated. Specifically, they are transcriptionally upregulated during the cell cycle (14, 51), and their protein levels are stabilized by PKCζ (34–36), which is known to be activated by small TAg (55). Nonetheless, important for our conclusions, knockdown of Msh6 demonstrated an important role for Msh6 in DDR activation through ATR. MutSα recognizes DNA damage and recruits ATR and ATR-interacting protein (ATRIP) to regions of single-stranded DNA (ssDNA) bound to RPA, which leads to activation of the DNA damage response and cell cycle arrest (25). Thus, the DNA damage we observed with Msh6 knockdown is likely due to a failure to robustly activate ATR to arrest the cell cycle rather than a failure to repair DNA mismatches.
Virions formed in the absence of Msh6 or ATR activity were less infectious and had decreased levels of the minor capsid protein VP2 relative to VP1. The nuclear localization signals of VP2 and VP3 are important for trafficking of the BKPyV genome from the ER to the nucleus (4, 43), but little is known about the function of VP2 independent of VP3 for infection. VP2 is expressed at a lower level than VP3 during infection and contains a putative myristylation site that is absent in VP3 and is required for infection of a related polyomavirus, MCPyV, in many cell types (3, 5). Mutation of VP2 separate from VP3 results in decreased infectivity of particles produced during infection (44). Thus, it is reasonable to assume that the decrease of VP2 levels in particles produced in Msh6-deficient cells affects viral infectivity at the trafficking step for subsequent infections.
It is still unclear how MutSα contributes to virion assembly. One possibility is that MutSα prevents nuclear envelope disassembly by arresting the cell cycle through ATR activation, which prevents mitosis (7). As we have previously shown, blocking mitosis by inhibiting Cdk1 rescues BKPyV titers when ATR is inactivated (7). We noted that both VP2 and VP3 expression in cells was decreased, but only VP2 was dramatically deficient in viral particles. As the nucleus is the major site of virion assembly (50), it is possible that nuclear breakdown during mitosis may affect the assembly of viral particles by mislocalizing VP2. As VP2 is expressed at lower levels than VP3, viral assembly may be more sensitive to a decrease in VP2 levels locally when the nucleus breaks down, which redistributes nuclear proteins throughout the cell. In addition, myristoylation of VP2, which is important for VP2 interactions with membranes, may be more affected by breakdown of the membrane structures during mitosis. Another possibility is that VP2 is targeted by an E3-ubiquitin ligase that is activated during mitotic entry, such as APC/CCdc20, which may explain why silencing Msh6 decreased cellular viral protein levels generally (Fig. 2F). More detailed studies of viral assembly during PyV infection are required.
MATERIALS AND METHODS
Cell culture, viruses, infections, and transfections.
Renal proximal tubule epithelial (RPTE) cells (Lonza) were expanded for 2 passages, and freezer stocks were prepared in renal epithelial cell growth medium (REGM; Lonza) with 10% dimethyl sulfoxide (DMSO) at 2 × 106 cells/mL (56). TAg-expressing RPTE cells were generated as previously described (8). For experiments, RPTEs were maintained at 37°C in 5% CO2. Stocks of infectious BKPyV were prepared from the pBR322-Dunlop genomic clone (ATCC 45025) or, for the Rb binding mutant, from pBR322-Dunlop plasmid bearing the E109K and D110K mutations to disrupt the LxCxE domain. Four micrograms of the plasmids were digested with BamHI (Promega) and ligated with T4 ligase (Promega) followed by phenol-chloroform extraction and ethanol precipitation. The purified viral genomes were transfected using Lipofectamine 2000 (Thermo Fisher) into 5 × 106 RPTE cells that were 60% confluent for BKDunlop or into TAg-expressing RPTE cells for the Rb binding mutant to rescue viral replication. Cells were collected in media 21 days later and subjected to 3 freeze-thaw cycles. The titer of virus was determined by focus-forming assay using pAb416 (57). Subsequent viral stocks were prepared by inoculating 70% confluent Vero cells (ATCC) maintained in Dulbecco’s modified eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Altanta Biologicals) at a multiplicity of infection (MOI) of 0.1 and harvested and their titer determined as described above. For BKPyV infections, RPTE cells were chilled at 4°C for 15 min; then, BKPyV in REGM was added to cells at 4°C for 1 hour before replacing the media with REGM at 37°C, and cells were placed in 5% CO2. TAg-expressing lentivirus was prepared, and transductions were performed as previously described (8).
siRNA knockdown.
Msh6 siRNAs (Ambion; Silencer Select; catalog nos. s6286 and s6288) and nontargeting siRNA (Silencer Select; negative control no. 1) were reverse transfected into RPTE cells adapted from reference 9. Briefly, 10 nM siRNAs were combined with 2.7 μL/mL Lipofectamine RNAiMax (Thermo Fisher) in REGM, and then, 1.25 × 105 cells/mL RPTE cells were added to a plate to achieve 50% confluence. The optimal Msh6 siRNA concentration was determined to be 10 nM, which yielded >90% knockdown efficiency (determined by Western blot analysis) with no effect on cell viability (Fig. S1 in the supplemental material).
BKPyV particle purification.
Viral particles were purified using a modified procedure adapted from reference 12. Briefly, 2 days postinfection (dpi), 1.6 × 107 RPTE cells in REGM were harvested by centrifugation at 8,000 × g for 30 min at 4°C. The supernatant (S1) containing free virus was stored at 4°C, and the pellet (P1) containing cell-associated virus was resuspended in 5 mL buffer A (10 mM HEPES, pH 7.9, 1 mM CaCl2, and 5 mM KCl) and incubated in a sonicating water bath for 5 min at 4°C. Then, it was adjusted to pH 5.4 with 0.5 M HEPES (pH 5.0) and treated with 1 U/mL type V neuraminidase (Sigma) at 25°C for 2 h. Then, the lysate was adjusted to pH 7.4 with 0.5 M HEPES, pH 8.0, incubated at 42°C for 5 min, and centrifuged at 16,000 × g for 5 min at 4°C in a swinging bucket rotor. The resulting supernatant (S2) was combined with S1, and the remaining pellet (P2) was suspended in 1 mL buffer A with 0.1% (wt/vol) sodium deoxycholate for 15 min at 25°C with occasional agitation. P2 was pelleted at 16,000 × g for 15 min at 4°C, and the final supernatant (S3) was combined with S1 and S2. The combined supernatants were layered carefully over a 4-mL 20% sucrose cushion (in buffer A), and the viral particles were pelleted at 50,000 × g for 3 h at 4°C in a SW 28 Ti rotor. The pellet containing the purified virions was resuspended in 1 mL buffer A and stored at −80°C.
Total and particle DNA extraction.
Intracellular viral DNA was isolated from 8 × 106 RPTE cells infected with BKPyV by pelleting at 300 × g for 5 min and incubating overnight at 37°C in 500 μL of 0.4% sodium dodecyl sulfate, 0.2 mg/mL RNase A, 50 μg/mL proteinase K, and 100 mM Tris-HCl, pH 7.5 (12). Total DNA was phenol/chloroform/isoamyl alcohol (25:24:1) extracted 3× and precipitated with 2.5 volumes 100% ethanol and 0.3 M sodium acetate, pH 5.2. The DNA was resuspended in 50 μL TE buffer (10 mM Tris, pH 8.0, 1 mM EDTA) and then digested using 40 units of EcoRV and XmaI (NEB) to selectively digest cellular DNA and not the viral DNA. Encapsidated viral DNA was prepared from 50 μL of purified BKPyV particles by adding 100 μL 10× DNase buffer and 4 U RQ1 DNase I (Promega) for 1 h at 37°C. The reaction was stopped with 10 μL RQ1 Stop and incubated for 1 h at 37°C. Proteinase K buffer (10 mM Tris-HCl pH 8.0, 10 mM EDTA, and 0.25% SDS), 0.2 μg/mL RNase A, and 50 μg/mL proteinase K (Roche) were added and incubated for 1 h at 37°C. The DNA was extracted and precipitated as above and resuspended in 50 μL TE buffer. DNA was analyzed by Southern blot analysis using a radiolabeled BKPyV fragment generated by digestion of pBR322-Dunlop with PvuII (3.2-kbp fragment) as described in reference 58 and quantified using Storm 820 phosphorimager from Amersham Biosciences.
Western blot analysis and quantification.
Cellular lysates were harvested in E1A lysis buffer (50 mM HEPES, pH 7.0, 250 mM NaCl, and 0.1% NP-40) with protease inhibitors (5 μg/mL phenylmethylsulfonyl fluoride [PMSF], 5 μg/mL aprotinin, 5 μg/mL leupeptin, 50 mM NaF, and 0.2 mM sodium orthovanadate) and frozen at −20°C (22). Whole-cell lysates were pelleted at 13,000 × g for 10 min at 4°C. Protein concentrations were determined by Bradford assay, and equal amounts of lysate (20 to 40 μg) were loaded. For Western blotting of purified viral particles, 50 μL was analyzed per lane. Proteins were separated by 8% or 10% SDS-PAGE and transferred to Immobilon-FL polyvinylidene difluoride membrane (EMD Millipore) and blocked in 2% fat-free dry milk in PBST (1 mM KH2PO4, 155 mM NaCl, 3 mM Na2HPO4-7H2O, and 0.1% Tween 20). The following primary antibodies were diluted 1:1,000 unless otherwise specified: Msh6 (BD; catalog no. 610918; RRID AB_398233), Msh2 (Cell Signaling; catalog no. 2017; RRID AB_2235387), Msh3 (Novus Biologicals; catalog no. NBP2-19417), pChk1-S317 (Cell Signaling; catalog no. 2344; RRID AB_331488), Chk1 (Santa Cruz; catalog no. 8408; RRID AB_627257), pATM-S1981 (Abcam; catalog no. ab81282; RRID AB_1640207), ATM (Cell Signaling; catalog no. 2873; RRID AB_2062659), β-actin (1:2,000; Cell Signaling; catalog no. 4967; RRID AB_330288), T antigen (1:3,000; pAb416), VP1 (1:5,000; pAb597), and VP2 and VP3 (1:5,000; recombinant rabbit antibody). Quantitative Western blot analysis of β-actin, TAg, VP1, VP2, and VP3 was performed using IRDye 800-conjugated goat anti-mouse IgG (Li-Cor; catalog no. 926-32210; RRID AB_621842) or IRDye 680-conjugated goat anti-rabbit IgG (LiCor; catalog no. 926-68071; RRID AB_10956166) at 1:20,000 and quantified with Li-Cor Odyssey. pATM-S1981, ATM, pChk1-S317, Chk1, Msh2, Msh6, and β-actin Western blotting was visualized by horseradish peroxidase-conjugated donkey anti-rabbit IgG (GE; catalog no. NA934; RRID AB_772206) and sheep anti-mouse IgG (GE; catalog no. NA931; RRID AB_772210) that were diluted 1:3,000 to 1:5,000 and visualized by autoradiography.
Cell fractionation.
Cellular fractionation was performed as previously described (14). Briefly, 8 × 106 RPTE cells were scraped and lysed in hypotonic solution (20 mM HEPES, pH 7.0, 10 mM KCl, 2 mM MgCl2, and 0.5% Nonidet P-40) by dounce homogenization for 30 strokes. Nuclei were pelleted at 1,500 × g for 5 min. The supernatant (cytoplasmic fraction) was placed at 4°C. The nuclei pellet was resuspended in the hypotonic solution with 0.5 M NaCl for 30 min on ice. Antibodies against GAPDH (glyceraldehyde-3-phosphate dehydrogenase; Abcam; catalog no. 9484; RRID AB_307274) and p84 (Genetex; catalog no. GTX70220; RRID AB_372637) were used as markers for cytoplasmic and nuclear proteins, respectively.
Cell cycle analysis.
Cells were harvested by trypsinization, pelleted, and resuspended in phosphate-buffered saline (PBS; 1 mM KH2PO4, 155 mM NaCl, and 3 mM Na2HPO4-7H2O). RPTE cells were fixed in 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) for 20 min, washed in 2% FBS in PBS, and then permeabilized with 0.25% Triton X-100 in 2% FBS in PBS. Mitotic indexing was performed by staining for phosphohistone H3 (Ser10) (Cell Signaling; catalog no. 3458; RRID AB_10694086) and DNA staining with FxCycle Violet (Thermo Fisher) according to the manufacturer’s protocol. Samples were analyzed on an LSR-II flow cytometer, and cell cycle status was measured using FlowJo software. To monitor cells in the S phase, newly synthesized DNA was labeled with ethynyl-2′-deoxyuridine (EdU) for 3 h and then conjugated by click chemistry to azide-Alexa Fluor 488 as previous described (14).
Immunofluorescence microscopy.
Immunofluorescence staining was performed as described in reference 9. Anti-Msh6 rabbit monoclonal antibody (Cell Signaling; catalog no. 3996; RRID AB_1904050) or Msh2 rabbit polyclonal antibody (Cell Signaling; catalog no. 2017; RRID AB_2235387) and TAg antibody (pAb416; Abcam; catalog no. ab16879; RRID AB_302561) were used at 1:200 in PBS. Secondary antibodies were Alexa Fluor 594-conjugated goat anti-mouse IgG (Invitrogen; catalog no. A11005; RRID AB_2534073) or Alexa Fluor 488-conjugated goat anti-rabbit IgG (Invitrogen; catalog no. A11008; RRID AB_143165) used at 1:250 in 5% goat serum in PBS. Coverslips were mounted in ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole) (Thermo Fisher). To quantify nuclear fragmentation, at least 100 nuclei were scored under each condition using an Olympus IX81 microscope with a Zeiss Plan 40×/0.75 objective. For S phase quantification, the Keyence BZ-X810 all-in-one fluorescence microscope using a Nikon Pan Fluor 10×/0.30 objective and the following Chroma filters: enhanced green fluorescent protein (EGFP) for EdU conjugated to Alexa Fluor 488 with an exposure time of 1/3.5 s, ET-DAPI for FxCycle Violet with an exposure time of 1/6 s with black balance, and ET-Cy3/tetramethyl rhodamine isocyanate (TRITC) for the goat anti-mouse secondary conjugated to DyLight 594 (Novus; catalog no. NBP1-75957) with an exposure time of 1/3 s. Ten random images per sample (three replicates using RPTE cells from two donors, average of >8,000 cells per sample/replicate) were taken using the BZ-X800 Viewer software. Region of interest (ROI) data were extracted using ImageJ 1.53f51 (Fiji) and analyzed using R version 4.1.2 on RStudio 2021.09.2+382.
Cell viability.
Cell viability was measured using CellTiter-Glo (Promega) according to the manufacturer’s protocol and measured on a Cytation3 (BioTek) plate reader and analyzed with Gen5 software. Technical triplicates were performed for 3 biological repeats.
Real-time quantitative PCR.
DNA was quantified by real-time quantitative PCR (RT-qPCR) using TAg-Forward (5′-AAGGAAAGGCTGGATTCTGA-3′) and TAg-Reverse (5′-TGTGATTGGGATTCAGTGCT-3′) or mitochondrial 16S rRNA-Forward (5′-GAGGAACAGCTCTTTGGACA-3′) and mitochondrial 16S rRNA-Reverse (5′-CAATTGGGTGTGAGGAGTTC-3′) primers (59), which all had efficiencies of >80%. Absolute viral copy number was determined using a standard curve of pBR322-Dunlop plasmid and analyzed by the threshold cycle (ΔΔCT) method against relative mitochondrial DNA levels to normalize for any potential unequal cell input. RT-qPCR was performed in 25 μL total volume with 12.5 μL SYBR green PCR master mix (Applied Biosystems), 2.5 μL of DNA template diluted 1:500, and 300 nM each primer. Amplification was performed and analyzed as described in reference 59 on a Bio-Rad C1000 thermocycler with the following reaction conditions: 2 min at 50°C, 10 min at 95°C, and then 40 cycles of 15 s at 95°C and 30 s at 58°C.
Metaphase chromosome preparations.
At 2 dpi, BKPyV-infected (MOI = 0.5 IU/cell) RPTE cells (9) were treated with 50 ng/mL Colcemid (Thermo Fisher) for 1 h to arrest cells in metaphase. Cells were trypsinized and incubated with 0.8% sodium citrate for 1 h and then fixed with Carnoy’s fixative (3:1 methanol/acetic acid). Cells were dropped onto slides and incubated at 55°C overnight prior to staining with Giemsa (Sigma) to visualize chromatin. Metaphase chromosomes were visualized using Olympus IX81 microscope with a plan 40×/0.75 objective under ×100 magnification. Greater than seventy-five metaphases per condition per biological replicate were counted and classified as either normal or shattered chromatin.
Inhibitor treatment.
Fresh REGM was supplemented with 5 μM VE-821 (ATRi) (46). DMSO was added to control and single knockdown (KD) wells at equal concentrations to offset any off-target effects of the vehicle. All inhibitors were added to RPTE cells at 24 hpi, which is after TAg is expressed in the nucleus.
ACKNOWLEDGMENTS
This work was supported by NIH (R01AI123162 to M.J. and S.R.T.) and the Training Program in Cell, Molecular, and Developmental Biology T32 (T32 GM008111 to J.L.J. and J.M.N.) led by Bradley Yoder at the University of Alabama at Birmingham.
We thank Marla I. Hertz for careful reading of the manuscript.
Contributor Information
Sunnie R. Thompson, Email: sunnie@uab.edu.
Felicia Goodrum, University of Arizona.
REFERENCES
- 1.Bennett SM, Broekema NM, Imperiale MJ. 2012. BK polyomavirus: emerging pathogen. Microbes Infect 14:672–683. 10.1016/j.micinf.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hurdiss DL, Morgan EL, Thompson RF, Prescott EL, Panou MM, Macdonald A, Ranson NA. 2016. New structural insights into the genome and minor capsid proteins of BK polyomavirus using cryo-electron microscopy. Structure 24:528–536. 10.1016/j.str.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krauzewicz N, Streuli CH, Stuart-Smith N, Jones MD, Wallace S, Griffin BE. 1990. Myristylated polyomavirus VP2: role in the life cycle of the virus. J Virol 64:4414–4420. 10.1128/JVI.64.9.4414-4420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakanishi A, Itoh N, Li PP, Handa H, Liddington RC, Kasamatsu H. 2007. Minor capsid proteins of simian virus 40 are dispensable for nucleocapsid assembly and cell entry but are required for nuclear entry of the viral genome. J Virol 81:3778–3785. 10.1128/JVI.02664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambalathingal GR, Francis RS, Smyth MJ, Smith C, Khanna R. 2017. BK polyomavirus: clinical aspects, immune regulation, and emerging therapies. Clin Microbiol Rev 30:503–528. 10.1128/CMR.00074-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An P, Sáenz Robles MT, Pipas JM. 2012. Large T antigens of polyomaviruses: amazing molecular machines. Annu Rev Microbiol 66:213–236. 10.1146/annurev-micro-092611-150154. [DOI] [PubMed] [Google Scholar]
- 7.Justice JL, Needham JM, Thompson SR. 2019. BK polyomavirus activates the DNA damage response to prolong S phase. J Virol 93:e00130-19. 10.1128/JVI.00130-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verhalen B, Justice JL, Imperiale MJ, Jiang M. 2015. Viral DNA replication-dependent DNA damage response activation during BK polyomavirus infection. J Virol 89:5032–5039. 10.1128/JVI.03650-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang M, Zhao L, Gamez M, Imperiale MJ. 2012. Roles of ATM and ATR-mediated DNA damage responses during lytic BK polyomavirus infection. PLoS Pathog 8:e1002898. 10.1371/journal.ppat.1002898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Justice JL, Verhalen B, Jiang M. 2015. Polyomavirus interaction with the DNA damage response. Virol Sin 30:122–129. 10.1007/s12250-015-3583-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orba Y, Suzuki T, Makino Y, Kubota K, Tanaka S, Kimura T, Sawa H. 2010. Large T antigen promotes JC virus replication in G2-arrested cells by inducing ATM- and ATR-mediated G2 checkpoint signaling. J Biol Chem 285:1544–1554. 10.1074/jbc.M109.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sowd GA, Li NY, Fanning E. 2013. ATM and ATR activities maintain replication fork integrity during SV40 chromatin replication. PLoS Pathog 9:e1003283. 10.1371/journal.ppat.1003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahl J, You J, Benjamin TL. 2005. Induction and utilization of an ATM signaling pathway by polyomavirus. J Virol 79:13007–13017. 10.1128/JVI.79.20.13007-13017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Justice JL, Verhalen B, Kumar R, Lefkowitz EJ, Imperiale MJ, Jiang M. 2015. Quantitative proteomic analysis of enriched nuclear fractions from BK polyomavirus-infected primary renal proximal tubule epithelial cells. J Proteome Res 14:4413–4424. 10.1021/acs.jproteome.5b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G-M. 2008. Mechanisms and functions of DNA mismatch repair. Cell Res 18:85–98. 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 16.Jiricny J. 2006. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol 7:335–346. 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 17.Kunkel TA, Erie DA. 2005. DNA mismatch repair. Annu Rev Biochem 74:681–710. 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 18.van Oers JMM, Edwards Y, Chahwan R, Zhang W, Smith C, Pechuan X, Schaetzlein S, Jin B, Wang Y, Bergman A, Scharff MD, Edelmann W. 2014. The MutSβ complex is a modulator of p53-driven tumorigenesis through its functions in both DNA double-strand break repair and mismatch repair. Oncogene 33:3939–3946. 10.1038/onc.2013.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Pearlman AH, Hsieh P. 2016. DNA mismatch repair and the DNA damage response. DNA Repair (Amst) 38:94–101. 10.1016/j.dnarep.2015.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polager S, Kalma Y, Berkovich E, Ginsberg D. 2002. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene 21:437–446. 10.1038/sj.onc.1205102. [DOI] [PubMed] [Google Scholar]
- 21.Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev 16:245–256. 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lario LD, Ramirez-Parra E, Gutierrez C, Casati P, Spampinato CP. 2011. Regulation of plant MSH2 and MSH6 genes in the UV-B-induced DNA damage response. J Exp Bot 62:2925–2937. 10.1093/jxb/err001. [DOI] [PubMed] [Google Scholar]
- 23.Abend JR, Low JA, Imperiale MJ. 2010. Global effects of BKV infection on gene expression in human primary kidney epithelial cells. Virology 397:73–79. 10.1016/j.virol.2009.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burdova K, Mihaljevic B, Sturzenegger A, Chappidi N, Janscak P. 2015. The mismatch-binding factor MutSβ can mediate ATR activation in response to DNA double-strand breaks. Mol Cell 59:603–614. 10.1016/j.molcel.2015.06.026. [DOI] [PubMed] [Google Scholar]
- 25.Yoshioka K, Yoshioka Y, Hsieh P. 2006. ATR kinase activation mediated by MutSα and MutLα in response to cytotoxic O6-methylguanine adducts. Mol Cell 22:501–510. 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Low J, Humes HD, Szczypka M, Imperiale M. 2004. BKV and SV40 infection of human kidney tubular epithelial cells in vitro. Virology 323:182–188. 10.1016/j.virol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 27.Baudot A, de la Torre V, Valencia A. 2010. Mutated genes, pathways and processes in tumours. EMBO Rep 11:805–810. 10.1038/embor.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harris KF, Christensen JB, Radany EH, Imperiale MJ. 1998. Novel mechanisms of E2F induction by BK virus large-T antigen: requirement of both the pRb-binding and the J domains. Mol Cell Biol 18:1746–1756. 10.1128/MCB.18.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marra G, Iaccarino I, Lettieri T, Roscilli G, Delmastro P, Jiricny J. 1998. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc Natl Acad Sci USA 95:8568–8573. 10.1073/pnas.95.15.8568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drummond JT, Genschel J, Wolf E, Modrich P. 1997. DHFR/MSH3 amplification in methotrexate-resistant cells alters the hMutSalpha/hMutSbeta ratio and reduces the efficiency of base-base mismatch repair. Proc Natl Acad Sci USA 94:10144–10149. 10.1073/pnas.94.19.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao L, Imperiale MJ. 2017. Identification of Rab18 as an essential host factor for BK polyomavirus infection using a whole-genome RNA interference screen. mSphere 2:e00291-17. 10.1128/mSphereDirect.00291-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christmann M, Kaina B. 2000. Nuclear translocation of mismatch repair proteins MSH2 and MSH6 as a response of cells to alkylating agents. J Biol Chem 275:36256–36262. 10.1074/jbc.M005377200. [DOI] [PubMed] [Google Scholar]
- 33.Chang DK, Ricciardiello L, Goel A, Chang CL, Boland CR. 2000. Steady-state regulation of the human DNA mismatch repair system. J Biol Chem 275:18424–18431. 10.1074/jbc.M001140200. [DOI] [PubMed] [Google Scholar]
- 34.Arlow T, Kim J, Haye-Bertolozzi JE, Martínez CB, Fay C, Zorensky E, Rose MD, Gammie AE. 2021. MutSα mismatch repair protein stability is governed by subunit interaction, acetylation, and ubiquitination. G3 (Bethesda) 11:jkaa065. 10.1093/g3journal/jkaa065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez-Pigeon H, Laurent G, Humbert O, Salles B, Lautier D. 2004. Degadration of mismatch repair hMutSα heterodimer by the ubiquitin-proteasome pathway. FEBS Lett 562:40–44. 10.1016/S0014-5793(04)00181-4. [DOI] [PubMed] [Google Scholar]
- 36.Hernandez-Pigeon H, Quillet-Mary A, Louat T, Schambourg A, Humbert O, Selves J, Salles B, Laurent G, Lautier D. 2005. hMutSα is protected from ubiquitin-proteasome-dependent degradation by atypical protein kinase Cζ phosphorylation. J Mol Biol 348:63–74. 10.1016/j.jmb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Smith JA, Bannister LA, Bhattacharjee V, Wang Y, Waldman BC, Waldman AS. 2007. Accurate homologous recombination is a prominent double-strand break repair pathway in mammalian chromosomes and is modulated by mismatch repair protein Msh2. Mol Cell Biol 27:7816–7827. 10.1128/MCB.00455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costes A, Lambert SAE. 2012. Homologous recombination as a replication fork escort: fork-protection and recovery. Biomolecules 3:39–71. 10.3390/biom3010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schofield MJ, Hsieh P. 2003. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol 57:579–608. 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 40.Sowd GA, Mody D, Eggold J, Cortez D, Friedman KL, Fanning E. 2014. SV40 utilizes ATM kinase activity to prevent non-homologous end joining of broken viral DNA replication products. PLoS Pathog 10:e1004536. 10.1371/journal.ppat.1004536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PMJ, Kunkel TA. 2005. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem 280:29980–29987. 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- 42.Albertson TM, Ogawa M, Bugni JM, Hays LE, Chen Y, Wang Y, Treuting PM, Heddle JA, Goldsby RE, Preston BD. 2009. DNA polymerase epsilon and delta proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci USA 106:17101–17104. 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schowalter RM, Buck CB. 2013. The Merkel cell polyomavirus minor capsid protein. PLoS Pathog 9:e1003558. 10.1371/journal.ppat.1003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henriksen S, Hansen T, Bruun J-A, Rinaldo CH. 2016. The presumed polyomavirus viroporin VP4 of simian virus 40 or human BK polyomavirus is not required for viral progeny release. J Virol 90:10398–10413. 10.1128/JVI.01326-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett SM, Zhao L, Bosard C, Imperiale MJ. 2015. Role of a nuclear localization signal on the minor capsid proteins VP2 and VP3 in BKPyV nuclear entry. Virology 474:110–116. 10.1016/j.virol.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charrier J-D, Durrant SJ, Golec JMC, Kay DP, Knegtel RMA, MacCormick S, Mortimore M, O'Donnell ME, Pinder JL, Reaper PM, Rutherford AP, Wang PSH, Young SC, Pollard JR. 2011. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem 54:2320–2330. 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- 47.Wang Y, Qin J. 2003. MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation. Proc Natl Acad Sci USA 100:15387–15392. 10.1073/pnas.2536810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Fang Y, Shao H, Lindsey-Boltz L, Sancar A, Modrich P. 2010. Interactions of human mismatch repair proteins MutSalpha and MutLalpha with proteins of the ATR-Chk1 pathway. J Biol Chem 285:5974–5982. 10.1074/jbc.M109.076109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamane K, Taylor K, Kinsella TJ. 2004. Mismatch repair-mediated G2/M arrest by 6-thioguanine involves the ATR–Chk1 pathway. Biochem Biophys Res Commun 318:297–302. 10.1016/j.bbrc.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 50.Erickson KD, Bouchet-Marquis C, Heiser K, Szomolanyi-Tsuda E, Mishra R, Lamothe B, Hoenger A, Garcea RL. 2012. Virion assembly factories in the nucleus of polyomavirus-infected cells. PLoS Pathog 8:e1002630. 10.1371/journal.ppat.1002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tapias A, Ciudad CJ, Roninson IB, Noé V. 2008. Regulation of Sp1 by cell cycle related proteins. Cell Cycle 7:2856–2867. 10.4161/cc.7.18.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gazzoli I, Kolodner RD. 2003. Regulation of the human MSH6 gene by the Sp1 transcription factor and alteration of promoter activity and expression by polymorphisms. Mol Cell Biol 23:7992–8007. 10.1128/MCB.23.22.7992-8007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicolás M, Noé V, Ciudad CJ. 2003. Transcriptional regulation of the human Sp1 gene promoter by the specificity protein (Sp) family members nuclear factor Y (NF-Y) and E2F. Biochem J 371:265–275. 10.1042/BJ20021166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwanaga R, Komori H, Ohtani K. 2004. Differential regulation of expression of the mammalian DNA repair genes by growth stimulation. Oncogene 23:8581–8590. 10.1038/sj.onc.1207976. [DOI] [PubMed] [Google Scholar]
- 55.Sontag E, Fedorov S, Kamibayashi C, Robbins D, Cobb M, Mumby M. 1993. The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates the map kinase pathway and induces cell proliferation. Cell 75:887–897. 10.1016/0092-8674(93)90533-v. [DOI] [PubMed] [Google Scholar]
- 56.Abend JR, Low JA, Imperiale MJ. 2007. Inhibitory effect of gamma interferon on BK virus gene expression and replication. J Virol 81:272–279. 10.1128/JVI.01571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jiang M, Abend JR, Tsai B, Imperiale MJ. 2009. Early events during BK virus entry and disassembly. J Virol 83:1350–1358. 10.1128/JVI.02169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Broekema NM, Imperiale MJ. 2013. miRNA regulation of BK polyomavirus replication during early infection. Proc Natl Acad Sci USA 110:8200–8205. 10.1073/pnas.1301907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang M, Entezami P, Gamez M, Stamminger T, Imperiale MJ. 2011. Functional reorganization of promyelocytic leukemia nuclear bodies during BK virus infection. mBio 2:e00281-10. 10.1128/mBio.00281-10. [DOI] [PMC free article] [PubMed] [Google Scholar]