ABSTRACT
The highly contagious and fast-spreading omicron variant of SARS-CoV-2 infects the respiratory tracts efficiently. The receptor-binding domain (RBD) of the omicron spike protein recognizes human angiotensin-converting enzyme 2 (ACE2) as its receptor and plays a critical role in the tissue tropism of SARS-CoV-2. Here, we showed that the omicron RBD (strain BA.1) binds to ACE2 more strongly than does the prototypic RBD from the original Wuhan strain. We also measured how individual omicron mutations affect ACE2 binding. We further determined the crystal structure of the omicron RBD (engineered to facilitate crystallization) complexed with ACE2 at 2.6 Å. The structure shows that omicron mutations caused significant structural rearrangements of two mutational hot spots at the RBD/ACE2 interface, elucidating how each omicron mutation affects ACE2 binding. The enhanced ACE2 binding by the omicron RBD may facilitate the omicron variant’s infection of the respiratory tracts where ACE2 expression level is low. Our study provides insights into the receptor recognition and tissue tropism of the omicron variant.
IMPORTANCE Despite the scarcity of the SARS-CoV-2 receptor—human angiotensin-converting enzyme 2 (ACE2)—in the respiratory tract, the omicron variant efficiently infects the respiratory tract, causing rapid and widespread infections of COVID-19. The omicron variant contains extensive mutations in the receptor-binding domain (RBD) of its spike protein that recognizes human ACE2. Here, using a combination of biochemical and X-ray crystallographic approaches, we showed that the omicron RBD binds to ACE2 with enhanced affinity and also elucidated the role of each of the omicron mutations in ACE2 binding. The enhanced ACE2 binding by the omicron RBD may contribute to the omicron variant’s new viral tropism in the respiratory tract despite the low level of ACE2 expression in the tissue. These findings help us to understand tissue tropism of the omicron variant and shed light on the molecular evolution of SARS-CoV-2.
KEYWORDS: COVID-19, omicron variant, receptor-binding domain (RBD), receptor-binding motif (RBM), mutational hotspots, angiotensin-converting enzyme 2, X-ray crystallography
INTRODUCTION
The omicron variant marks a new phase of the COVID-19 pandemic (1–4). Whereas previous SARS-CoV-2 strains mainly infect the lungs to cause severe illness, the omicron variant mainly infects the respiratory tracts and causes milder symptoms (5, 6). This new viral tropism speeds up the spread of the omicron variant, causing as many as 1 million new daily infections in the United States (7). Yet the respiratory tract tropism of the omicron variant is puzzling. The virus-surface spike protein mediates coronavirus entry into host cells (8, 9). A critical determinant of the species and tissue tropisms and infectivity of coronaviruses is the binding interactions between the receptor-binding domain (RBD) of the spike protein and its host receptor (10, 11). SARS-CoV-2 RBD recognizes human angiotensin-converting enzyme 2 (ACE2) as its receptor (12, 13). ACE2 is expressed in the lungs but is poorly expressed in the respiratory tracts (14, 15). Elucidating the binding interactions between the omicron RBD and ACE2 is important for understanding the tissue tropism and infectivity of the omicron variant.
Structural studies have delineated detailed interactions between SARS-CoV-2 RBD and human ACE2 (16, 17). The RBD of SARS-CoV-2 and that of closely related SARS-CoV-1 both contain the following two subdomains: a core structure and a receptor-binding motif (RBM) (16–18). The RBMs of these coronaviruses mediate the viral interactions with ACE2. Three mutational hot spots have been identified at both the SARS-CoV-1 RBD/ACE2 interface and the SARS-CoV-2 RBD/ACE2 interface as follows: two of them center around Lys353 and Lys31 on ACE2 (i.e., hot spot-353 and hot spot-31, respectively), and the third involves a receptor-binding ridge in the RBM (i.e., hot spot-ridge) (17, 19) (Fig. 1A). Compared with SARS-CoV-1 RBD, the RBD from the original SARS-CoV-2 Wuhan strain (i.e., prototypic RBD) binds to human ACE2 more tightly because its RBM contains a more compact receptor-binding ridge (which allows for more extensive interactions with ACE2) and several residue changes at hot spot-353 and hot spot-31 (which cause small structural adjustments of the two hot spots) (17, 19) (Fig. 1A). All of the RBM mutations in the omicron variant occurred around the three hot spots (Fig. 1A). To understand the molecular evolution of SARS-CoV-2 and future trajectory of the pandemic, it is critical to study how each of these RBM mutations in the omicron variant affects ACE2 binding.
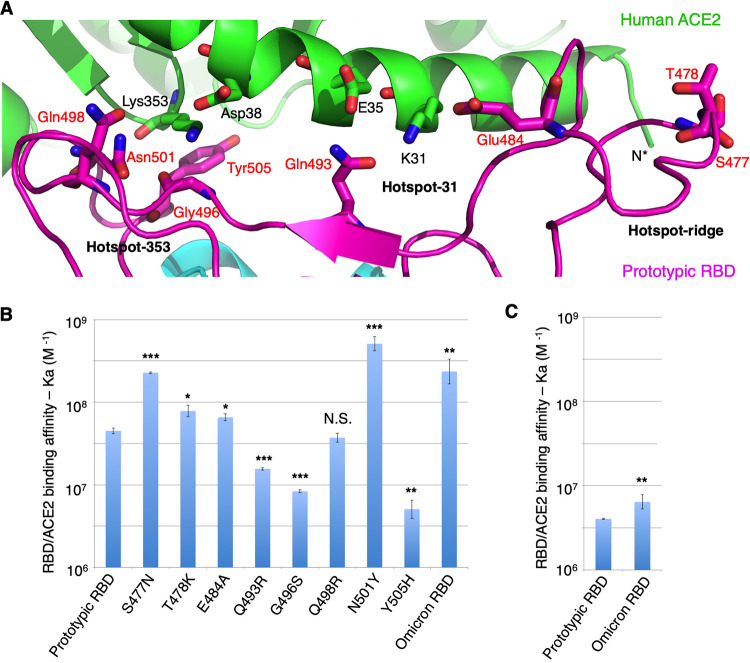
FIG 1.
Mutations in the receptor-binding motif (RBM) of the omicron variant. (A) Interface between prototypic RBM and ACE2. ACE2 is in green. RBD core structure is in cyan. RBM is in magenta. RBD residues that have undergone mutations from the prototypic strain to the omicron variant are labeled in red. Three mutational hot spots are highlighted as follows: hot spot-353 centers on Lys353 in ACE2, hot spot-31 centers on Lys31 in ACE2, and hot spot-ridge centers on the receptor-binding ridge in RBD. (B) Surface plasmon resonance (SPR) assay for ACE2/RBD binding. ACE2-Fc was coated to a protein A chip in a fixed direction, and individual RBDs flowed through. The data are presented as mean ± standard error of the mean (SEM) (n = 3) on a log scale. A Student’s two-tailed t test was performed to analyze the statistical difference between the prototypic RBD and each of the other RBDs. ***, P < 0.001; **, P < 0.01; *, P < 0.05. N.S., statistically not significant. See Table S1 in the supplemental material for detailed binding kinetics. (C) Reverse coating SPR assay for ACE2/RBD binding. Prototypic RBD-His or omicron RBD-His was coated to a CM5 chip in random orientations through chemical cross-linking, and His-tagged monomeric ACE2 flowed through. The data are presented as mean ± SEM (n = 3) on a log scale. A Student’s two-tailed t test was performed to analyze the statistical difference between the prototypic RBD and the omicron RBD. **, P < 0.01. See Table S1 in the supplemental material for detailed binding kinetics.
In this study, we measured the binding affinity between the omicron RBD and human ACE2 using the prototypic RBD as a comparison. We also measured how each of the RBM mutations in the omicron variant affected ACE2 binding. We further determined the crystal structure of the omicron RBD complexed with ACE2, elucidating the structural role for individual RBM mutations in ACE2 binding. Our findings have implications for tissue tropism, infectivity, and evolution of the omicron variant.
RESULTS
To evaluate the binding interactions between the omicron RBD and human ACE2, we prepared recombinant omicron RBD and human ACE2 ectodomain and measured their binding affinity using surface plasmon resonance (SPR). To this end, we coated Fc-tagged human ACE2 (ACE2-Fc) on a protein A chip, allowed His-tagged omicron RBD (RBD-His) to flow through, and calculated the binding kinetics (Fig. 1B; see also Table S1 in the supplemental material). Prototypic RBD-His was used as a comparison to omicron RBD-His. The result showed that the omicron RBD-His binds to ACE2-Fc 3.9 times as tightly as the prototypic RBD-His does. To confirm the above result, we coated omicron RBD-His and prototypic RBD-His on two separate chemical cross-linking chips, allowed His-tagged ACE2 (ACE2-His) to flow through, and calculated the binding kinetics separately. The result showed that omicron RBD-His binds to ACE2-His 1.7 times as tightly as the prototypic RBD-His does (Fig. 1C; Table S1). Overall, the omicron RBD binds to ACE2 more tightly than does the prototypic RBD.
To evaluate how individual RBM mutations in the omicron variant affect ACE2 binding, we prepared prototypic RBD-His containing one of the RBM mutations from the omicron variant and measured their ACE2 binding using SPR. ACE2-Fc was coated on the protein A chip and mutant RBD-His flowed through. The results showed that, compared with the prototypic RBD, mutations S477N and N501Y significantly enhanced ACE2 binding, mutations Q493R, G496S, and Y505H significantly reduced ACE2 binding, mutation T478K slightly enhanced ACE2 binding, and mutations E484A and Q498R had less significant impacts on ACE2 binding (Fig. 1B; Table S1). Among these mutations, it is known that the S477N and T478K mutations (parts of hot spot-ridge) enhance ACE2 binding by introducing new interactions with the N terminus of ACE2 (20). However, the structural bases for the other mutations (parts of hot spot-353 and hot spot-31) are not known.
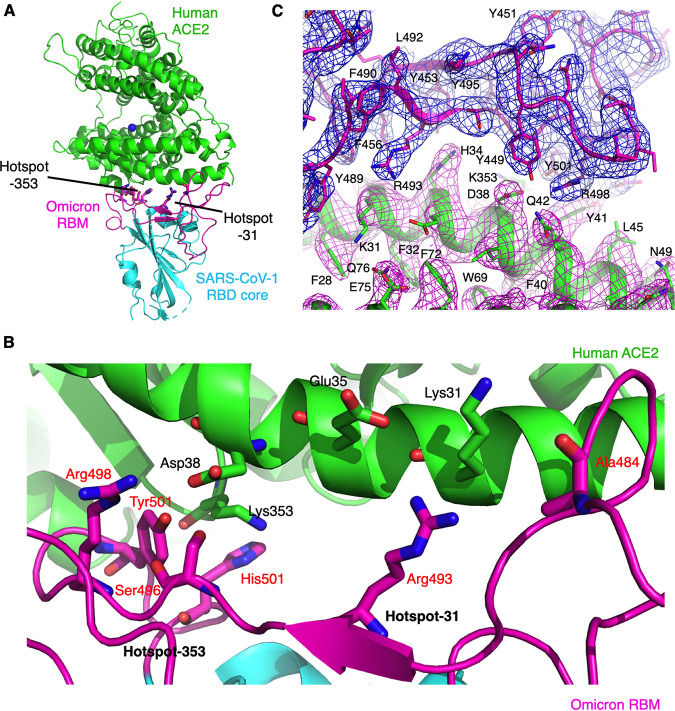
To provide a structural understanding for each of the omicron mutations (parts of hot spot-353 and hot spot-31) on ACE2 binding, we determined the structure of the interface between the omicron RBD and human ACE2. We previously established a structural platform for studying the interactions between the RBD from SARS-CoV-1 or SARS-CoV-2 and ACE2. More specifically, we constructed a chimeric RBD containing the core structure from SARS-CoV-1 RBD and the RBM region from SARS-CoV-1-like coronaviruses (e.g., SARS-CoV-2). The complex of this chimeric RBD and ACE2 can be crystallized reliably under the same condition as the complex of SARS-CoV-1 RBD and ACE2. In other words, the chimeric RBD provides a crystallization scaffold for studying the interactions between the RBMs of SARS-CoV-1-like coronaviruses (e.g., SARS-CoV-2) and ACE2 (17–19, 21). Indeed, structural comparison between the complex of the chimeric SARS-CoV-2 prototypic RBD and ACE2 and the complex of the wild-type SARS-CoV-2 prototypic RBD and ACE2 showed that the two structures are highly similar to each other in both the overall structure and structural details at the RBD/ACE2 interface (16, 17), confirming the success of the design of the chimeric SARS-CoV-2 prototypic RBD. In this study, we redesigned the chimeric SARS-CoV-2 RBD by incorporating the six mutations from the omicron RBM (E484A, Q493R, G496S, Q498R, N501Y, Y505H), which are parts of hot spot-353 and hot spot-31. We successfully crystallized the complex of this new chimeric RBD (i.e., omicron chimeric RBD) and human ACE2 and determined its structure at 2.6 Å resolution (Fig. 2; Table 1). The overall structure of the omicron RBD/ACE2 complex is highly similar to that of the prototypic RBD/ACE2 complex, but structural details at the hot spot-353 and hot spot-31 differ significantly, revealing the structural role of each of these mutations in ACE2 binding.
FIG 2.
Overall structure of omicron chimeric RBD complexed with human ACE2. (A) Overall structure of omicron chimeric RBD complexed with ACE2. The omicron chimeric RBD contains the core structures (in cyan) from SARS-CoV-1 RBD and receptor-binding motif (RBM) (in magenta) from the prototypic RBD. ACE2 is in green. Two mutational hot spots in the RBM are shown. (B) Interface between omicron RBM and ACE2. Coloring and labeling are the same as Fig. 1A. (C) Unbiased composite omit map of the interface between the omicron RBD and ACE2. Contour level is 1σ.
TABLE 1.
Crystallization data collection and refinement statistics
| Data collection | Value(s)a |
|---|---|
| Space group | P1211 |
| Unit cell dimensions | |
| a, b, c (Å) | 81.130, 120.179, 113.325 |
| α, β, γ (°) | 90, 92.443, 90 |
| Resolution (Å) | 113.22–2.61 (2.92–2.61) |
| Rsym or Rmerge | 0.042 (0.647) |
| I/σ | 15.5 (1.9) |
| Completeness (%) | 92.6 (95.6) |
| Redundancy | 4.0 (4.1) |
| Refinement | |
| Resolution (Å) | 30.11–2.61 (2.68–2.61) |
| No. reflections | 36,475 (1,734) |
| Rwork/Rfree | 0.183/0.238 |
| No. atoms | 13,148 |
| Protein | 12,741 |
| Ligand/ion | 394 |
| Water | 13 |
| B-factor | 95.30 |
| Protein | 94.46 |
| Ligand/ion | 123.25 |
| Water | 69.35 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.44 |
Statistics for the highest-resolution shell are shown in parentheses.
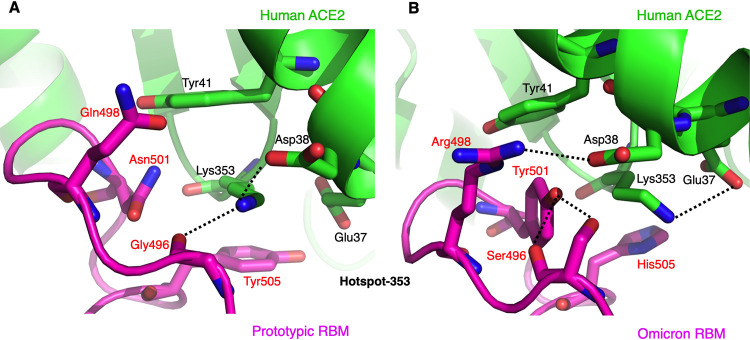
We compared the structural details of hot spot-353 between the prototypic RBD/ACE2 interface and the omicron RBD/ACE2 interface. At the interface between the prototypic RBD and ACE2, Lys353 in ACE2 is buried in a hydrophobic tunnel with four walls as follows: Tyr41 and Asp38 in ACE2 and Tyr505 and Asn501 in the RBD (Fig. 3A). At the end of the tunnel, Lys353 forms a salt bridge and a hydrogen bond with Asp38 in ACE2 and the main chain of Gly496 in the RBD, respectively. At the interface between the omicron RBD and ACE2, four omicron mutations have occurred around hot spot-353 (Fig. 3B). The previous Asn501 in the RBD, one of the tunnel walls, has become a tyrosine, which forms two hydrogen bonds with the main chain and side chain of the glycine-turned Ser496 in the RBD. The previous Gln498 in the RBD has become an arginine, which forms a salt bridge with Asp38 in ACE2. Since Asp38 in ACE2 has found a new salt bridge partner, Lys353 in ACE2 points to a different direction and forms a salt bridge with Glu37 in ACE2. The previous Tyr505 in the RBD, another one of the tunnel walls, has become a histidine, which forms weaker hydrophobic stacking with Lys353. Instead, the new Tyr501 and Arg498 in the RBD and Tyr41 and Lys353 in ACE2 form hydrophobic and aromatic stacking interactions. Overall, hot spot-353 at the RBD/ACE2 interface has undergone significant structural rearrangements in the omicron RBD compared to the prototypic RBD.
FIG 3.
Structural details at the mutational hot spot-353. (A) Interface between the prototypic RBM and ACE2. (B) Interface between the omicron RBM and ACE2. RBM residues that have undergone mutations from the prototypic strain to the omicron variant are labeled in red. Dotted lines indicate hydrogen bonds or salt bridges.
We then correlated the structural details of hot spot-353 with the RBD/ACE2 binding data (Fig. 1B). First, the N501Y mutation in the omicron RBD enhances ACE2 binding because Tyr501 has formed new favorable interaction network around hot spot-353. The new interactions include two new hydrogen bonds that stabilize the RBM, as well as several hydrophobic and aromatic stacking interactions that enhance RBM/ACE2 binding (Fig. 3B). Second, the Q498R mutation in the omicron RBD has insignificant net impact on ACE2 binding because it forms a new favorable salt bridge with Asp38 in ACE2 while also breaking a favorable salt bridge between Lys353 and Asp38 in ACE2 (Fig. 3B). Third, the G496S mutation significantly reduces ACE2 binding because Ser496 has steric interference with Lys353 in ACE2 (Fig. 3B). Last, the Y505H mutation reduces ACE2 binding because His505 forms a weaker stacking interaction with Lys353 in ACE2 (Fig. 3B). Therefore, the structural arrangements of hot spot-353 are consistent with the RBD/ACE2 binding data in explaining the role of individual omicron mutations in ACE2 binding.
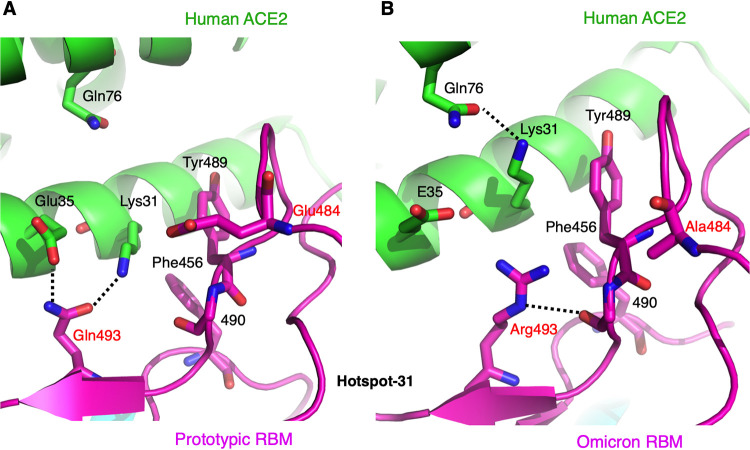
We also analyzed the structural details of hot spot-31 at both the prototypic RBD/ACE2 interface and the omicron RBD/ACE2 interface. At the prototypic RBD/ACE2 interface, Lys31 and Glu35 in ACE2 both form a hydrogen bond with Gln493 in the RBD, while Glu484 in the RBD is slightly outside the salt bridge range from Lys31 (Fig. 4A). At the omicron RBD/ACE2 interface, the previous Gln493 in the RBD has become an arginine, which has charge repulsion with Lys31 and hence forces Lys31 to point to a different direction (Fig. 4B). Instead, Lys31 forms a hydrogen bond with Gln76 in ACE2. The newly appeared Arg493 in the RBD forms a hydrogen bond with the main chain of residue 490 in the RBD. The E484A mutation in the RBD did not form any new interaction or change the structure of the residing loop. Therefore, the Q493R mutation reduces ACE2 binding by disrupting two favorable interactions at hot spot-31 (those between Gln493 and Lys31 and between Gln493 and Glu35), while the E484A mutation has little impact on ACE2 binding.
FIG 4.
Structural details at the mutational hot spot-31. (A) Interface between the prototypic RBM and ACE2. (B) Interface between the omicron RBM and ACE2. Coloring and labeling are the same as Fig. 3.
DISCUSSION
The recent tropism change of the omicron variant from the lungs to respiratory tracts marks an important phase of the COVID-19 pandemic because the newly gained respiratory tract tropism allows the omicron variant to be more transmissible while causing milder symptoms and lower fatality rate (5, 6). Two important determinants of the species and tissue tropisms of coronaviruses are the binding affinity between the spike protein and its host receptor and the cleavage of the spike protein by host proteases (10, 11, 22–24). While it remains to be seen whether the omicron spike protein has altered protease susceptibility compared to previous strains, the current study examined whether the binding affinity between the spike RBD and human ACE2 has altered for the omicron variants.
Our study demonstrated that the omicron RBD binds to Fc-tagged dimeric ACE2 3.9 times as strongly as the prototypic RBD does. This is in contrast to a recently published study showing that the omicron RBD and prototypic RBD bind to Fc-tagged ACE2 with similar affinity (25). However, there was a key difference in the SPR methodology between the current study and the published study. In the current study, Fc-tagged ACE2 was coated to the protein A chip in a fixed orientation, such that the virus-binding surface of ACE2 was facing up and fully accessible to the flown-though His-tagged RBD; in the published study, Fc-tagged ACE2 was coated to the CM5 chip in random orientations (or certain unknown preferred orientation) through chemical cross-linking. It is worth noting that on cell surfaces, ACE2 is a dimer with the virus-binding surface facing outwards. Hence, our SPR measurement is more physiologically relevant. We confirmed our result using the reverse coating SPR: His-tagged RBD was coated to the CM5 chip in random orientations through chemical cross-linking, and His-tagged monomeric ACE2 flowed through. The result confirmed that omicron RBD binds to ACE2 more strongly than does the prototypic RBD, although the difference was only 1.7-fold. Again, our first SPR approach with Fc-tagged ACE2 coated to protein A chips was the most physiologically relevant, while our second SPR approach confirmed the conclusion.
Our study also measured how each of the RBM mutations in the omicron variant affected ACE2 binding. We showed that S477N, N501Y, and T478K enhanced ACE2 binding; Q493R, G496S, and Y505H reduced ACE2 binding; and E484A and Q498R had less significant impact on ACE2 binding. These mutations were distributed around the following three previously identified mutational hot spots: hot spot-353 (centering on ACE2 residue Lys353), hot spot-31 (centering on ACE2 residue Lys31), and hot spot-ridge (centering on the receptor-binding ridge in RBD). Because the structural basis for the mutations in hot spot-ridge has been elucidated, the current study focused on the mutations in hot spot-353 and hot spot-31. To this end, we determined the structural interface between the omicron RBM and human ACE2 using a well-established crystallization platform involving a chimeric RBD. We previously showed that both SARS-CoV-2 and SARS-CoV-1 RBDs have evolved to accommodate Lys353 and Lys31 at the RBD/ACE2 interface that is rich in hydrophobic residues, particularly tyrosines (17–19, 21). The current study revealed that at both hot spot-353 and hot spot-31, significant structural arrangements have taken place for the omicron RBD compared to the prototypic RBD. Particularly, the previous salt bridge between Lys353 and Asp38 and hydrogen bonds between Lys31 and Gln493 and between Glu35 and Gln493 have been broken by new mutations; instead, both Lys353 and Lys31 point to a different direction and form a new hydrogen bond with different residues. Each of the new RBM mutations in the omicron RBD (except E484A) plays a role in the structural arrangements of the two hot spots, explaining our RBD/ACE2 binding data. These significant structural arrangements at the hot spots mark a significant evolutionary twist of SARS-CoV-2 RBD.
Our finding about enhanced ACE2 binding by the omicron RBD has important implications for understanding the COVID-19 pandemic. Human ACE2 is expressed at lower levels in the respiratory tracts than in the lungs (14, 15). Enhanced ACE2 binding by the RBD may facilitate the omicron variant’s infection of the respiratory tracts. Moreover, we recently showed that whereas the RBD in the prototypic spike protein takes a mixture of standing-up position for receptor binding and lying-down position for immune evasion, the RBD in the omicron spike protein predominantly takes a standing-up conformation (26), which should further enhance ACE2 binding. Thus, the omicron variant appears to have evolved the following two strategies for enhanced ACE2 binding: evolution of the RBD and opening up of the spike. Both of these strategies may have contributed to the respiratory tract tropism of the omicron variant. Our findings shed light on the tissue tropism and infectivity of the omicron variant and provide insights into the molecular evolution of SARS-CoV-2 and future trajectory of the COVID-19 pandemic.
MATERIALS AND METHODS
Plasmids.
The genes encoding the receptor-binding domain (RBD) of SARS-CoV-2 prototypic strain (GenBank accession number QHD43416.1) and human ACE2 (GenBank accession number NM_021804) were both synthesized (GenScript Biotech). The gene encoding the RBD from SARS-CoV-2 omicron strain (B.1.1.529; GISAID EPI_ISL_6647960) was constructed by site-directed mutagenesis of the prototypic RBD (residues 319 to 535). The gene encoding the chimeric omicron RBD was constructed by site-directed mutagenesis of the chimeric prototypic RBD (17). The prototypic RBD-His, prototypic RBD-His mutants, and omicron RBD-His were subcloned into pLenti-transfer vector (Addgene) with an N-terminal tissue plasminogen activator (tPA) signal peptide and a C-terminal His tag. Human ACE2-Fc (residues 1 to 615) was subcloned into the same vector except that a C-terminal human IgG4 Fc region replaced the His tag. Chimeric omicron RBD-His and human ACE2-His (residues 1 to 615) were subcloned into pFastBac I vector (Life Technologies) with an N-terminal honeybee melittin signal peptide and a C-terminal His tag.
Protein expression and purification.
The prototypic RBD-His, prototypic RBD-His mutants, omicron RBD-His, and ACE2-Fc were prepared from 293F mammalian cells as previously described (27). Briefly, lentiviral particles were packaged for construction of stable cell lines expressing prototypic RBD-His, prototypic RBD-His mutants, omicron RBD-His, or ACE2-Fc. Puromycin (Gibco) was used for the selection of stable cell lines. The proteins were collected from the cell culture medium, purified on a nickel-nitrilotriacetic acid (Ni-NTA) column for His-tagged proteins or on a protein A column for Fc-tagged protein, and purified further on a Superdex 200 gel filtration column (Cytiva).
Chimeric omicron RBD-His and human ACE2-His were prepared from sf9 insect cells using the Bac-to-Bac system (Life Technologies) as previously described (17). Briefly, the His-tagged proteins were harvested from cell culture medium, purified on an Ni-NTA column, and purified further on a Superdex 200 gel filtration column (Cytiva).
Crystallization and structure determination.
The complex of the omicron chimeric RBD and human ACE2 was purified on gel filtration chromatography. Crystals of the complex were grown at room temperature over wells containing 100 mM Tris (pH 8.5), 18 to 22% polyethylene glycol 6000 (PEG 6000), and 100 mM NaCl. Crystals were flash-frozen in liquid nitrogen. X-ray diffraction data were collected at beamline 12-1 of the Stanford Synchrotron Radiation Lightsource (SSRL). Data processing was done using HKL2000 (28). The structure was determined by molecular replacement using the structure of prototypic chimeric RBD complexed with ACE2 as the search template (Protein Data Bank accession number 6VW1). Molecular replacement and model refinement were done using PHENIX and CCP4 (29, 30). Model building was done using COOT (31). Structural figures were made using PYMOL (The PyMOL Molecular Graphics System, Version 2.0; Schrödinger, LLC.). Structure data and refinement statistics are shown in Table 1.
Protein-protein binding assay.
Surface plasmon resonance assay using a Biacore S200 system (Cytiva) were carried out as described previously (17). Briefly, ACE2-Fc was immobilized to a protein A chip (Cytiva). The running buffer composed of 10 mM HEPES pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% Tween 20. Serial dilutions of purified recombinant RBD-His were injected ranging in concentration from 20 to 320 nM. In a different approach, RBD-His was immobilized to a CM5 sensor chip through chemical cross-linking (Cytiva). Serial dilutions of purified recombinant ACE2-His were injected ranging in concentration from 100 to 1,600 nM. Binding kinetics were calculated using Biacore evaluation software (Cytiva).
Data availability.
Coordinates and structure factors have been submitted to the Protein Data Bank under accession number 7U0N.
ACKNOWLEDGMENTS
This work was supported by NIH grants R01AI089728 and R01AI110700 (to F.L.) and R35GM118047 (to H.A.).
We thank staff (Silvia Russi and Aina E. Cohen) at beamline 12-1 of the Stanford Synchrotron Radiation Lightsource (SSRL) (contract number DE-AC02-76SF00515) for assistance in data collection.
The Biacore S200 system was supported by NIH ORIP grant 1S10OD021539.
Footnotes
[This article was published on 28 March 2022 with “SARS-CoV” instead of “SARS-CoV-2” in the title. The title was updated in the current version, posted on 5 April 2022.]
Supplemental material is available online only.
Contributor Information
Hideki Aihara, Email: aihar001@umn.edu.
Fang Li, Email: lifang@umn.edu.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Karim SSA, Karim QA. 2021. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet 398:2126–2128. 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maslo C, Friedland R, Toubkin M, Laubscher A, Akaloo T, Kama B. 2022. Characteristics and outcomes of hospitalized patients in South Africa during the COVID-19 omicron wave compared with previous waves. JAMA 327:583–584. 10.1001/jama.2021.24868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxena SK, Kumar S, Ansari S, Paweska JT, Maurya VK, Tripathi AK, Abdel-Moneim AS. 2022. Characterization of the novel SARS-CoV-2 omicron (B.1.1.529) variant of concern and its global perspective. J Med Virol 94:1738–1744. 10.1002/jmv.27524. [DOI] [PubMed] [Google Scholar]
- 4.Gu H, Krishnan P, Ng DYM, Chang LDJ, Liu GYZ, Cheng SSM, Hui MMY, Fan MCY, Wan JHL, Lau LHK, Cowling BJ, Peiris M, Poon LLM. 2022. Probable transmission of SARS-CoV-2 omicron variant in quarantine hotel, Hong Kong, China, November 2021. Emerg Infect Dis 28:460–462. 10.3201/eid2802.212422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolter N, Jassat W, Walaza S, Welch R, Moultrie H, Groome M, Amoako DG, Everatt J, Bhiman JN, Scheepers C, Tebeila N, Chiwandire N, Du Plessis M, Govender N, Ismail A, Glass A, Mlisana K, Stevens W, Treurnicht FK, Makatini Z, Hsiao N-Y, Parboosing R, Wadula J, Hussey H, Davies M-A, Boulle A, von Gottberg A, Cohen C. 2022. Early assessment of the clinical severity of the SARS-CoV-2 omicron variant in South Africa: a data linkage study. Lancet 399:437–446. 10.1016/S0140-6736(22)00017-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peacock TP, Brown JC, Zhou J, Thakur N, Newman J, Kugathasan R, Sukhova K, Kaforou M, Bailey D, Barclay WS. 2022. The SARS-CoV-2 variant, omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry. bioRxiv. 10.1101/2021.12.31.474653. [DOI]
- 7.COVID Data Tracker, February . 2022. https://covidcdcgov/covid-data-tracker/#variant-proportions.
- 8.Li F. 2016. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3:237–261. 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Peacock SJ, Robertson DL, COVID-19 Genomics UK (COG-UK) Consortium. 2021. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 19:409–424. 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F. 2015. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol 89:1954–1964. 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li WH, Wong SK, Li F, Kuhn JH, Huang IC, Choe H, Farzan M. 2006. Animal origins of the severe acute respiratory syndrome coronavirus: insight from ACE2-S-protein interactions. J Virol 80:4211–4219. 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. Addendum: a pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 588:E6. 10.1038/s41586-020-2951-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan Y, Shang J, Graham R, Baric RS, Li F. 2020. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 94:e00127-20. 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salamanna F, Maglio M, Landini MP, Fini M. 2020. Body localization of ACE-2: on the trail of the keyhole of SARS-CoV-2. Front Med (Lausanne) 7:594495. 10.3389/fmed.2020.594495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hikmet F, Méar L, Edvinsson Å, Micke P, Uhlén M, Lindskog C. 2020. The protein expression profile of ACE2 in human tissues. Mol Syst Biol 16:e9610. 10.15252/msb.20209610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 17.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F, Li WH, Farzan M, Harrison SC. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868. 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 19.Wu KL, Peng GQ, Wilken M, Geraghty RJ, Li F. 2012. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J Biol Chem 287:8904–8911. 10.1074/jbc.M111.325803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zahradník J, Marciano S, Shemesh M, Zoler E, Harari D, Chiaravalli J, Meyer B, Rudich Y, Li C, Marton I, Dym O, Elad N, Lewis MG, Andersen H, Gagne M, Seder RA, Douek DC, Schreiber G. 2021. SARS-CoV-2 variant prediction and antiviral drug design are enabled by RBD in vitro evolution. Nat Microbiol 6:1188–1198. 10.1038/s41564-021-00954-4. [DOI] [PubMed] [Google Scholar]
- 21.Li F. 2008. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J Virol 82:6984–6991. 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Y, Shang J, Yang Y, Liu C, Wan Y, Geng Q, Wang M, Baric R, Li F. 2018. Lysosomal proteases are a determinant of coronavirus tropism. J Virol 92:e01504-18. 10.1128/JVI.01504-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heald-Sargent T, Gallagher T. 2012. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses 4:557–580. 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Millet JK, Whittaker GR. 2015. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202:120–134. 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han P, Li L, Liu S, Wang Q, Zhang D, Xu Z, Han P, Li X, Peng Q, Su C, Huang B, Li D, Zhang R, Tian M, Fu L, Gao Y, Zhao X, Liu K, Qi J, Gao GF, Wang L. 2022. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell 185:630–640. 10.1016/j.cell.2022.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye G, Liu B, Li F. 2022. Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain. Nat Commun 13:1214. 10.1038/s41467-022-28882-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geng Q, Tai W, Baxter VK, Shi J, Wan Y, Zhang X, Montgomery SA, Taft-Benz SA, Anderson EJ, Knight AC, Dinnon KH, III, Leist SR, Baric RS, Shang J, Hong SW, Drelich A, Tseng CK, Jenkins M, Heise M, Du L, Li F. 2021. Novel virus-like nanoparticle vaccine effectively protects animal model from SARS-CoV-2 infection. PLoS Pathog 17:e1009897. 10.1371/journal.ppat.1009897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode, p 307–326. In Abelson JN, Simon MI, Carter CW, Jr, Sweet RM, Macromolecular crystallography, part A, vol 276. Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 29.Liebschner D, Afonine PV, Baker ML, Bunkoczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD. 2019. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol 75:861–877. 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download jvi.00249-22-s0001.pdf, PDF file, 0.08 MB (82.7KB, pdf)
Data Availability Statement
Coordinates and structure factors have been submitted to the Protein Data Bank under accession number 7U0N.