Abstract
When released into biological fluids like blood or saliva, brain extracellular vesicles (EVs) might provide a window into otherwise inaccessible tissue, contributing useful biomarkers of neurodegenerative and other central nervous system (CNS) diseases. To enrich for brain EVs in the periphery, however, cell‐specific EV surface markers are needed. The protein that has been used most frequently to obtain EVs of putative neuronal origin is the transmembrane L1 cell adhesion molecule (L1CAM/CD171). In this systematic review, we examine the existing literature on L1CAM and EVs, including investigations of both neurodegenerative disease and cancer through the lens of the minimal information for studies of EVs (MISEV), specifically in the domains of nomenclature usage, EV sources, and EV separation and characterization. Although numerous studies have reported L1CAM‐associated biomarker signatures that correlate with disease, interpretation of these results is complicated since L1CAM expression is not restricted to neurons and is also upregulated during cancer progression. A recent study has suggested that L1CAM epitopes are present in biofluids mostly or entirely as cleaved, soluble protein. Our findings on practices and trends in L1CAM‐mediated EV separation, enrichment, and characterization yield insights that may assist with interpreting results, evaluating rigor, and suggesting avenues for further exploration.
Keywords: ectosomes, exosomes, extracellular vesicles, immunoaffinity, L1CAM, microvesicles, MISEV, neurons, systematic review
1. INTRODUCTION
Extracellular vesicles (EVs) are a heterogeneous group of membrane‐bound particles that are released by all studied cell types (Cocozza, Grisard, Martin‐Jaular, Mathieu, & Théry, 2020; Van Niel, D'Angelo, & Raposo, 2018; Yáñez‐Mó et al., 2015; Yates et al., 2022). Most abundant in the 30–150 nm diameter range, EVs function in waste removal and cell‐cell communication by transporting nucleic acids, lipids, proteins, and other molecules (Harding, Heuser, & Stahl, 1983; Johnstone, 1992; Mateescu et al., 2017; Russell et al., 2019). EVs are commonly classified by biogenesis into plasma membrane‐origin ectosomes (or microvesicles) and endosome‐origin exosomes, although physical separation of these different classes is difficult (Cocucci & Meldolesi, 2015; Lötvall et al., 2014; Meldolesi, 2021; Théry et al., 2018; Witwer & Théry, 2019). EVs have also been reported to cross bodily barriers such as the mammalian blood‐brain barrier (Alvarez‐Erviti et al., 2011; Dickens et al., 2017; Pulliam, Sun, Mustapic, Chawla, & Kapogiannis, 2019; Russell et al., 2019). Due to the relative clinical inaccessibility of the brain, diseases and conditions of the central nervous system (CNS), including neurodegenerative diseases such as Alzheimer's and Parkinson's, present diagnostic, prognostic, and monitoring challenges for which EVs in biofluids are a potential solution (Campbell & Mocchetti, 2021; Coleman & Hill, 2015; Manu, Hohjoh, & Yamamura, 2021; Thompson et al., 2016; Upadhya & Shetty, 2021; Vandendriessche, Bruggeman, Van Cauwenberghe, & Vandenbroucke, 2020) as a kind of ‘liquid biopsy’: if displaying markers of the cell of origin, EVs and their cargo of proteins, nucleic acids, and more can theoretically be traced back to the parent cells and can serve as indicators of cell and tissue health (Pulliam et al., 2019; Shankar, Balaj, Stott, Nahed, & Carter, 2017; Vassileff, Cheng, & Hill, 2020). However, realizing the promise of EVs requires highly specific and abundant surface markers that can be used as molecular handles to separate target EVs from the high background of EVs from other cells in biofluids like blood (Thompson et al., 2016).
To date, the L1 cell adhesion molecule (L1CAM, also known as CD171 or simply L1) has been most frequently used to obtain materials of putative CNS neuronal origin for biomarker discovery and validation studies (Pulliam et al., 2019). L1CAM is a member of the L1 family of adhesion proteins. Usually, expressed as a single‐pass transmembrane protein, L1CAM contains immunoglobulin domains and fibronectin‐like repeat domains in an extracellular, N‐terminal portion, while a cytoplasmic C‐terminal tail participates in signalling within the cell (Samatov, Wicklein, & Tonevitsky, 2016). Splice forms lacking the transmembrane domain have also been described (Angiolini et al., 2019). Various functions in neuronal migration and differentiation have been ascribed to the adhesive and cell transduction properties of the protein (Maness & Schachner, 2007). In investigations of L1CAM‐positive EVs, antibodies to L1CAM are typically mixed with whole or partly processed biofluids, and materials attached to the antibodies are enriched and molecularly profiled. This approach has been used to propose molecular profiles associated with Alzheimer's disease, Parkinson's disease, mild cognitive impairment, HIV‐associated neurocognitive disorders, traumatic brain injury, and more.
However, the utility of L1CAM as a marker of brain neuronal EVs has also been questioned for several reasons (Hill, 2019; Norman et al., 2021). First, L1CAM expression is not restricted to CNS neurons (Hill, 2019). L1CAM was discovered in neurons (Faissner, Kruse, Nieke, & Schachner, 1984), but it may also be expressed by oligodendrocytes in the brain. In the periphery, according to protein and gene expression data, a wide variety of cells, including peripheral neurons and Schwann cells; melanocytes; immune cells such as T cells, B cells, and monocytes; and certain epithelial and endothelial cells; may express L1CAM. The protein is produced in several types of cancer, with higher levels indicating advanced disease and poor prognosis (Altevogt et al., 2020; Colombo & Meldolesi, 2015; Fogel et al., 2003; Gavert, Ben‐Shmuel, Raveh, & Ben‐Ze'ev, 2008). Indeed, the earliest studies of EV L1CAM were in the context of cancer (Gutwein et al., 2003; Gutwein et al., 2005). Second, the membrane association of the L1CAM extracellular portion may be transient since the L1CAM ectodomain is susceptible to proteolysis (Gutwein et al., 2000; Li & Galileo, 2010; Linneberg, Toft, Kjaer‐Sorensen, & Laursen, 2019; Sugawa, Ono, Yasui, Kishi, & Tsumori, 1997; Yang et al., 2009). Recently, Norman et al. reported that L1CAM epitopes are present in blood and cerebrospinal fluid mostly or entirely as cleaved, soluble protein, not as EV transmembrane molecules (Norman et al., 2021). Third, the specificity of at least one L1CAM antibody is unclear. Norman et al. reported cross‐reactivity of the antibody with alpha‐synuclein, a protein commonly assayed after L1CAM immunoprecipitation (IP), potentially explaining the positive biomarker results of some studies (Norman et al., 2021). Fourth, the widely held and oft‐repeated assumption that EVs easily cross the blood‐brain barrier is still supported mostly by indirect evidence (Verweij et al., 2021).
In this review, we assessed the reporting within the EV L1CAM literature through the lens of the minimal information for studies of EVs (MISEV) (Théry et al., 2018; Witwer et al., 2021), specifically in the four domains of nomenclature usage, EV sources, EV separation, and EV characterization. We sought to trace the ‘interpretation landscape’ around published L1CAM EV studies. For each study, we asked several questions. What types of EVs were studied (or presumed to be studied), and how were they defined? What sources of EVs were used? What were the methods of EVs separation and/or concentration? Which anti‐L1CAM antibodies were used? How were EVs characterized? Which EV and non‐EV markers (enriched/depleted), if any, were examined? What components of EVs were profiled? The overall goal was to understand what interpretations of EV L1CAM affinity studies can be admitted or excluded, and what might be done to enhance future studies.
2. RESULTS AND METHODS
2.1. Literature search and selection
A literature search was performed using the three databases PubMed, Web of Science, and Scopus. The following keywords/MeSH terms and operators were used: (L1CAM OR ‘L1 cell adhesion molecule’) AND (exosome OR exosomes OR ‘extracellular vesicle’ OR ‘extracellular vesicles’ OR ectosome OR ectosomes OR microvesicle OR microvesicles). Only English‐language studies were included. All studies published from the start of each database until end of July, 2021 were included. Manuscripts found in two or more databases were included only once, for a total of 59 publications (Figure 1). These were manually curated to select only those with primary research data that reported on putatively L1CAM‐associated EVs. Three review publications (without primary data) were removed. Five studies that did not specifically combine L1CAM and EVs were also removed. A total of 51 articles thus satisfied all criteria (see Figure 1 and list of papers in Table S1) (Anastasi et al., 2021; Athauda et al., 2019; Bhargava et al., 2021; Cha et al., 2019; Chawla et al., 2019; Cressatti et al., 2021; Dagur et al., 2020; Eitan et al., 2017; Fauré et al., 2006; Fu, Jiang, Tofaris, & Davis, 2020; Goetzl et al., 2019; Goetzl et al., 2015; Goetzl, Peltz, Mustapic, Kapogiannis, & Yaffe, 2020; Gomes et al., 2015; Gu et al., 2020; Gutwein et al., 2003; Gutwein et al., 2005; Herrero et al., 2019; Jiang et al., 2020; Jiang et al., 2021; Keller et al., 2009; Kodidela et al., 2020; Kumar et al., 2021; Mansur et al., 2021; Mullins, Mustapic, Goetzl, & Kapogiannis, 2017; Nasca et al., 2021; Nogueras‐Ortiz et al., 2020; Norman et al., 2021; Pace, Dutt, & Galileo, 2019; Patterson, Deep, & Brinkley, 2018; Peltz et al., 2020; Pulliam, Liston, Sun, & Narvid, 2020; Rani et al., 2019; Shi et al., 2016; Shi et al., 2014; Si et al., 2019; Stoeck et al., 2006; Suire et al., 2017; Sun, Dalvi, Abadjian, Tang, & Pulliam, 2017; Sun, Fernandes, & Pulliam, 2019; Trnka, Ivanova, Hiatt, & Matsell, 2012; Walker et al., 2021; Webber et al., 2014; Winston et al., 2019; Winston et al., 2016; Winston, Goetzl, Baker, Vitiello, & Rissman, 2018; Yang et al., 2009; Yang et al., 2011; Yuyama et al., 2019; Zhao et al., 2020; Zou et al., 2020).
FIGURE 1.
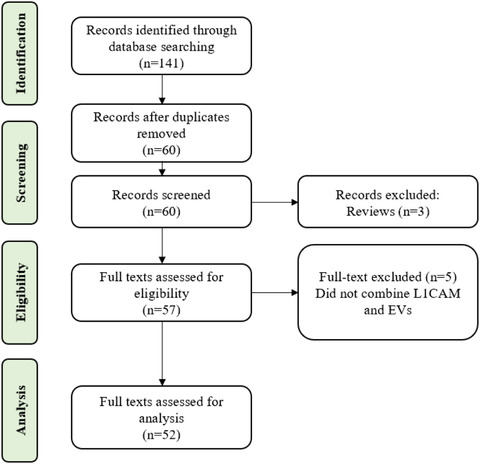
Flow diagram of literature search strategy and inclusion and exclusion criteria
2.2. Types of EVs and nomenclature
The first MISEV category is nomenclature. ISEV recommends the term ‘extracellular vesicle’ unless the biogenetic origin of EVs can be established (Lötvall et al., 2014; Théry et al., 2018): plasma membrane origin for ectosomes (or ‘microvesicles’) and endosomal origin for exosomes. The MISEV2018 consensus guidelines further define EV as the generic term for particles released from the cell, delimited by a phospholipid bilayer, and without a functional nucleus, and suggest that the term ‘EV’ can be used as a scaffold for further descriptors (if clearly defined), such as size, source, surface markers, and conditions of production (Théry et al., 2018). In practice, terms have been used inconsistently and sometimes without clear definition in the literature (Witwer & Théry, 2019). Beyond the consensus, biogenesis‐based definition, the term ‘exosome’ in particular has been used inconsistently to describe any EV, an EV within an arbitrary size range, an EV population positive for one or more tetraspanins or other EV markers, or the products of specific separation/concentration methods, such as ultracentrifugation or PEG precipitation. Since physical and biochemical characteristics of exosomes overlap substantially or entirely with those of other EV classes, most studies that claim to have studied exosomes have instead included a mixture of various EVs and often non‐EV extracellular particles (EPs).
In the L1CAM literature we analysed, we identified the primary term(s) used to describe EVs. In most papers, one term was used exclusively or predominantly, but several articles used terms interchangeably, usually ‘EV’ and ‘exosome.’ 32 articles used the term ‘exosome’ or a modified term like ‘NDE’ for ‘neuron‐derived exosome (Table 1).’ 20 used ‘EV’ or modified terms (Table 1). Other terms, all used before MISEV2014 appeared, were ‘membrane vesicle’, ‘microvesicle’, and ‘exosome‐like vesicle’ (Table 1). In several papers, the term ‘vesicle’ was used loosely in the text to refer to one or more subpopulations, but we did not track this informal usage. The term ‘EV’ was first mentioned in L1CAM studies in 2014 and came into primary usage in a publication in 2015. Since 2018, and with a sharp spike in the number of papers per year, there appears to be a trend toward more use of ‘EV’ and diminished use of ‘exosome’ (Figure 2a).
TABLE 1.
Nomenclature in EV L1CAM studies
| Term | Number | References |
|---|---|---|
| EV | 20 | 28,44,46‐48,51,52,54‐56,65,66,70,71,75,77,87,88,90 |
| Exosome | 32 | 36,39,45,49,50,52,53,55,57‐65,69,72‐74,76,79‐86,89 |
| Other | 4 | 35,76,83,86 |
FIGURE 2.
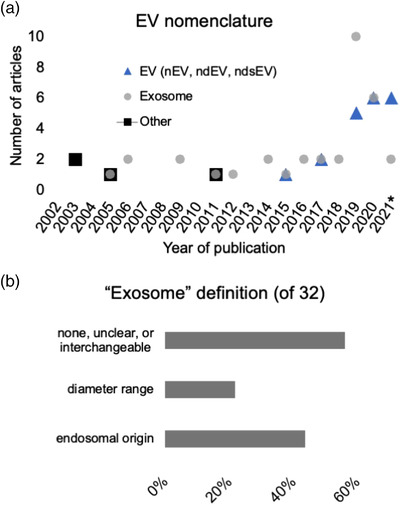
EV nomenclature used in studies of L1CAM and EVs. (a) EV nomenclature by year of publication. *: 2021 articles were analysed only through end of July. (b) Definition of ‘exosome’ for the 32 papers in which the term ‘exosome’ was used primarily. Percentages do not add up to 100% because some papers included multiple definitions
Of 32 papers that reported primarily on ‘exosomes’ or that used the term interchangeably with ‘EVs,’ 14 defined exosomes canonically by endosomal origin, whether or not this origin was investigated or established (Figure 2b). Several articles used diameter/size as a stand‐alone or additional exosome‐defining characteristic, including at least seven that provided specific diameter ranges (Figure 2b)—20–100 nm, 30–150 nm, 40–100 nm, 40–120 nm, 40–140 nm—and others that described exosomes simply as ‘small’. Only rarely did the up‐front exosome definition mention a certain marker, such as CD63, although more papers used markers such as CD63, CD81, TSG101, and Alix to characterize EVs (see below). In contrast, 18 articles did not define ‘exosome’ clearly, gave two or more seemingly contradictory definitions (likely reflecting general confusion in the field at the time of writing), or used terms interchangeably (Figure 2b). At least six defined/used ‘exosome’ as synonymous with ‘EV’. Several papers gave two or more seemingly contradictory definitions of exosomes. One paper used ‘exosomes’ in the title and abstract, then switched to EV throughout methods and results (Athauda et al., 2019), while another used EV throughout, except for a methods section that referred to exosomes (Pulliam et al., 2020).
3. SOURCES OF EVS FOR L1CAM STUDIES
The second MISEV2018 category covers sources of EVs and preprocessing variables (Théry et al., 2018). We did not compare the reporting of preprocessing variables due to a general lack of information and standardized reporting on these variables in most studies. There is thus considerable room for improvement in the reporting of preprocessing variables [a general issue in the EV field, not just for L1CAM studies (Clayton et al., 2019; Erdbrügger et al., 2021; Royo, Théry, Falcón‐Pérez, Nieuwland, & Witwer, 2020; Witwer et al., 2013)] and little information on how these variables might affect the presence or detection of EV‐associated L1CAM in specific sample types. As for sources of EVs, approximately 96% (n = 49) of the 51 articles used human patient or human‐origin cells. Four used samples from animal models: nonhuman primate (Kumar et al., 2021), rat (Dagur et al., 2020), and mouse (Shi et al., 2014; Yuyama et al., 2019). Both articles that used mouse models also used human samples (serum). Blood products (n = 40: serum, plasma, or both) were the most common EV source, consistent with recent field‐wide surveys (Gardiner et al., 2016; Nieuwland, Falcón‐Pérez, Théry, & Witwer, 2020; Royo et al., 2020) and with the ease of collection of blood and perceived potential as a ‘liquid biopsy’ (Table 2) (Clayton et al., 2018). Plasma (n = 31) was used more frequently than serum (n = 10). Of the 40 articles that used blood derivatives, several used other fluids/tissue, such as CSF (Cressatti et al., 2021; Norman et al., 2021), brain tissue (Dagur et al., 2020; Yuyama et al., 2019), urine (Cressatti et al., 2021), ascites fluid (Keller et al., 2009), and saliva (Cressatti et al., 2021) (Table 2). One article each used saliva (Rani et al., 2019), urine (Trnka et al., 2012), and ascites fluid (Gutwein et al., 2005) as the sole EV source (Table 2). Human cell culture‐conditioned medium (CCM) was a source of EVs in 14 articles (Table 2). Use of CCM in L1CAM EV studies has become less prevalent over time: in eight of 12 studies published prior to 2016, but in only six of 37 from 2016 and later.
TABLE 2.
Sources of EVs in L1CAM EV studies
| Source | n (of 51) | References |
|---|---|---|
| Blood products (plasma, serum) | 40 | |
| Serum | 10 | 44,53,55,58,64‐66,74,83,89 |
| Plasma | 31 | 28,44‐52,53,56,57,59,61,63,67‐73,75,76,78,79,85,87‐89 |
| Blood + Other | 10 | |
| Cell culture‐conditioned medium | 14 | 35,39,49,56,57,59,62,85,77,80,82‐84,86 |
| Brain tissue | 2 | 55,58 |
| CSF | 2 | 28,44 |
| Urine | 2 | 44,81 |
| Ascites | 2 | 36,83 |
| Saliva | 2 | 44,60 |
4. SEPARATION AND CONCENTRATION OF L1CAM‐ASSOCIATED EVS
The third MISEV domain is separation and concentration of EVs (Théry et al., 2018). Each separation method discriminates by affinity, size, or density, or some combination thereof, and falls at a different place in the EV specificity/recovery matrix (Cocozza et al., 2020; Théry et al., 2018). Multiple methods may be needed to achieve a desired purity or recovery outcome. Methods that reduce sample volume, recovering most EVs without substantial increases in EV purity, might be considered ‘concentration’ rather than separation methods. Foremost in this category is precipitation or salting‐out, for example, by polyethylene glycol (PEG) or ammonium sulphate. Commercial kits that are advertised as ‘exosome isolation’ reagents are mostly based on this approach. In contrast, affinity capture methods may achieve high specificity but low yield, depending on the antigen that is captured. In L1CAM EV studies, a common workflow first concentrates material by PEG precipitation and then uses antibodies to capture putative L1CAM‐positive material from the precipitated pellet (Table 3, Figure 3).
TABLE 3.
Separation and concentration methods
| Separation/concentration method | n (of 51) | References |
|---|---|---|
| Affinity (immunoprecipitation/capture, FACS) | 35 | 28,44‐51,53‐56,59,61,63‐75,78,79,85,87,88,90 |
| PEG or PEG‐based kit | 30 (26 combining PEG and IP) | 44‐52,56,57,5‐61,63,64,65‐73,75,78,87,88,89 |
| Differential ultracentrifugation | 17 | 28,35,36,39,55,57,58,62,69,76,77,80‐84,86 |
| Density gradient or cushion ultracentrifugation | 9 | 28,35,36,55,58,80,83,84,86 |
| Size exclusion chromatography | 3 | 28,57,85 |
FIGURE 3.
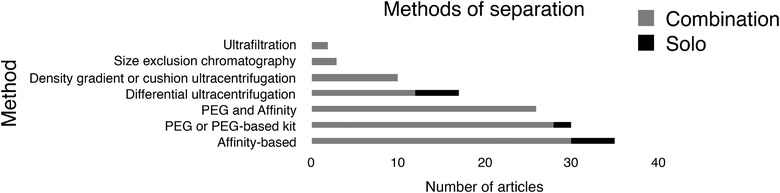
Methods of EV separation
Distribution of concentration/separation methods in n = 51 publications. Several studies used multiple workflows. Some combined more than one method in at least one workflow (n = 39), and these were counted in all respective categories. FACS = fluorescence‐activated cell sorting; PEG = polyethylene glycol. Ultrafiltration of various types was also an occasional component of EV workflows (not shown).
4.1. Precipitation
The second most commonly used separation/concentration method in the literature we surveyed was polymer precipitation (Table 3, Figure 3). Polyethylene glycol‐based precipitation is the presumed basis of numerous commercial kits that are advertised as specific for ‘exosome isolation.’ This approach is, however, not specific for exosomes or EVs, and the precipitate from many EV sources will be predominantly nonvesicular. As such, the method might be best referred to as concentration. PEG or presumed PEG‐based kits were used in 30 of the 51 studies we reviewed, in at least three as a stand‐alone method (Table 2, Figure 3). For 26 of the 35 studies involving L1CAM immunocapture, an initial PEG concentration was followed by immunocapture. This combination was thus the most common workflow overall. Of the 30 PEG/ precipitation protocols, one used a homemade PEG formulation, 27 used kits from System Biosciences, two used a kit from Thermo Fisher, and one protocol each used materials from Nasasbiotech and Cell Guidance Systems. The total 'n' is greater than the number of articles because of a comparison of multiple kits. We note that the Cell Guidance Systems kit combines PEG and SEC. The SEC step may be important for some downstream assays by increasing purity and removing PEG.
4.2. Size‐ and density‐based separation
Differential ultracentrifugation, historically the most widely used EV separation technique (Gardiner et al., 2016; Royo et al., 2020), was employed in at least 17 studies. We did not count a medium‐speed, preclearing step prior to PEG precipitation as differential ultracentrifugation. Ten studies used density gradients ultracentrifugation or cushion ultracentrifugation. Three used size‐exclusion chromatography, and at least two used ultrafiltration (Table 3, Figure 3).
4.3. Affinity‐based separation
L1CAM affinity capture techniques were the most common method of EV separation in the L1CAM literature (n = 35; Table 3, Figure 3). Ectodomains of proteins embedded in the EV lipid bilayer (such as L1CAM) can be recognized by affinity reagents—most commonly, antibodies—and used to select EVs displaying that marker. Antibodies are typically affixed to a surface, such as magnetic beads, nonmagnetic beads, some other matrix material, or a plate, to facilitate separation of bound from unbound material using, for example, a magnet, centrifugation, fluorescence‐activated cell sorting (to sort EV‐binding beads), or plate washing. Captured and purified EVs can then be eluted for subsequent characterization and functional studies or solubilized directly on the capture surface for certain assays.
Of the 35 papers that clearly specified an L1CAM capture strategy, at least 19 used the 5G3 clone that recognizes an N‐terminal portion of the L1 ectodomain (Ig‐like domains), and one used another clone that binds a similar region (Table 4, Figure 4). At least seven papers did not specify the clone, but most or all of these likely used the 5G3 clone based on citation and authorship. Seven used the UJ127 clone that recognizes an ectodomain epitope closer to the plasma membrane (in the fibronectin type III domains; Table 4, Figure 4). One paper appeared to imply that affinity capture was done but gave no related methods (Peltz et al., 2020). Twenty‐five of 35 papers did not specify the catalogue number of the antibody. Although the identity of the clone is most crucial, catalogue number can also be important information, since different formulations are available for some antibodies, and there may be manufacturer‐specific differences. Interestingly, two papers reported immunocapture of L1CAM‐positive EVs using antibodies C2C (2C2) (Zou et al., 2020) and LS‐C470565 (Table 4, Figure 4) (Dagur et al., 2020). These antibodies reportedly recognize epitopes at the C‐terminal end of L1CAM and should thus be intracellular or, for an EV, intraluminal.
TABLE 4.
Anti‐L1CAM antibodies used for reported capture of EVs
| L1CAM antibody clone | n (of 35) | Epitope | Example references |
|---|---|---|---|
| 5G3 | 19 | Ectodomain, immunoglobulin‐like domains 1 to 2; near N‐terminus | 44,45,49,51,59,61,66‐73,78,85,87‐89 |
| EPR23241‐224 | 1 | Similar epitope to 5G3 | 28 |
| UJ127 (UJ127.11) | 7 | Ectodomain, fibronectin type III repeats | 53,64,74,76,79,85,90 |
| C2C (or 2C2) | 1 |
Intracellular, C‐terminus; cross‐reactive with CHL1 |
54 |
| LS‐C470565 | 1 | Intracellular, C‐terminus | 55 |
| Unspecified | 7 | N/A | 46‐48,50,56,63,65 |
FIGURE 4.
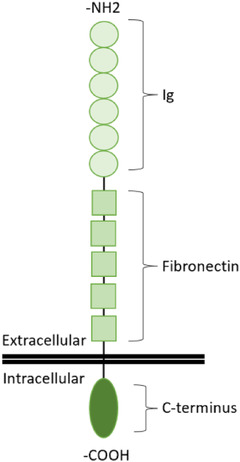
Cartoon of L1CAM's molecular organization relative to the cell membrane. ‐NH2 = amino (N) terminus; ‐COOH = carboxy (C) terminus
5. CHARACTERIZATION OF L1CAM‐ASSOCIATED EVS
Following separation, EV characterization is recommended as the fourth MISEV domain (Théry et al., 2018). Characterization is needed to identify what is present and enriched—and, ideally, what is not present or is depleted—in the EV preparation. Per MISEV2018, characterization should include physical characterization, such as particle size profiling, particle counting, and single‐particle ‘visualization’), and molecular assessment of both positive (enriched) EV markers and negative (or depleted) markers of potentially coseparated non‐EV components. Depending on EV source, the latter may include materials from dead cells, lipoprotein particles, and protein aggregates that overlap in size and/or density with EVs (Théry et al., 2018).
5.1. Physical characterization
For the purposes of this study, we classified physical characterization methods into two broad categories: ‘optical/electrical methods’ (OEM) and ‘nonoptical microscopy methods’ (NOM). The former include nanoparticle tracking analysis, NTA, resistive pulse sensing (RPS), and dynamic light scattering (DLS). These methods can be used to count particles within certain size ranges and, to varying degrees, to determine particle size or features of size distribution. However, they do not on their own discriminate between EVs and non‐EV particles. In contrast, the various forms of electron microscopy (EM) and atomic force microscopy (AFM) can at least theoretically identify EV‐specific morphology or other properties. Using multiple complementary physical characterization methods can be helpful to obtain more information about an EV preparation (Arab et al., 2021; Théry et al., 2018; Vogel et al., 2021) (CITE).
Of the 51 L1CAM articles surveyed, 19 (37%) presented no physical characterization results (Table 5, Figure 5a). In at least three of these, physical methods were mentioned, but no data were shown or described. Twenty‐two studies presented OEM data (Table 5, Figure 5a). Of these, 21 used NTA either as the sole physical characterization method (Meldolesi, 2021) or in combination with other methods. DLS was used by two and RPS by one. Ten studies reported data from more than one physical characterization technique (whether OEM or NOM; Table 5, Figure 5a). Most studies did not provide sufficient information to assess variability across samples, showing, for example, only one NTA profile and/or one electron micrograph.
TABLE 5.
Use of physical characterization techniques
| Technique | n (of 51) | References |
|---|---|---|
| Optical/Electrical Methods (OEM): RPS, NTA, DLS | 22 | 44‐49,52,54,55,57,60,63,66,70,72,75‐77,80,85,87,88 |
| Non‐Optical Microscopy (NOM): TEM, SEM, AFM | 19 | 28,35,46,49,52‐55,57,58,62,64,75,76,79,82,84,85,90 |
| Multiple | 10 | 46,49,52,54,55,57,75‐77,85,90 |
| None (in several cases, techniques were mentioned but results not reported) | 19 | 36,39,50,51,56,59,61,65,67‐69,71,73,74,78,81,83,86,89 |
FIGURE 5.
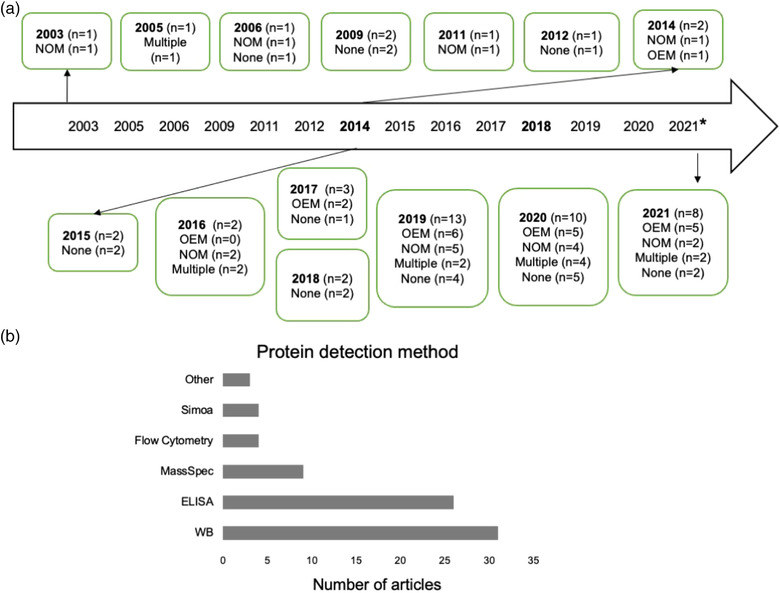
Characterization of EVs. (a). Timeline of usage of physical characterization techniques. OEM = optical/electrical methods: resistive pulse sensing (RPS), nanoparticle tracking analysis (NTA), dynamic light scattering (DLS); NOM = nonoptical microscopy: transmission electron microscopy (TEM), scanning electron microscopy (SEM), atomic force microscopy (AFM). Please note that other methods could be included in these categories, but this list is limited to techniques used in the studies selected for analysis. (b). Methods used for identification of individual EV proteins. WB = Western blot; ELISA = enzyme‐linked immunosorbent assay; MassSpec = mass spectrometry; ‘Other’ techniques included dot blots/antibody arrays
Demarcating the literature by the appearance of MISEV2018, the percentage of papers with no physical characterization was at 50% both pre‐MISEV2014 (five papers, n = 10) and between MISEV2014 and MISEV2018 (five papers, n = 10) but fell to 29% post‐MISEV2018 (nine papers, n = 31; Figure 5a). During these periods, the proportion of articles with multiple physical characterization methods rose from 0% (pre‐MISEV2014) to 20% (from MISEV2014‐MISEV2018, n = 2), and finally to 26% (post‐MISEV2018, n = 8) (Figure 5a).
5.2. EV cargo analysis methods
EV markers analysed in the L1CAM EV literature were predominantly proteins. Fort nine of 51 articles (96%) used some form of EV protein analysis. To detect specific proteins (Figure 5b), researchers used Western blotting (n = 31, ∼61% of total), regular or multiplexed ELISA (n = 27, 53%), single‐molecule analysis (Simoa, an ultrasensitive ELISA; n = 4, ∼8%), and flow cytometry (n = 4, ∼8%). 24 studies used more than one method (∼47%). Broader protein profiling was done by mass spectrometry in nine studies (∼18%). Other protein detection technologies included immunogold EM, antibody arrays, aptamer affinity, and proximity extension assays. Acetylcholinesterase (AChE) activity detection was used in one study (Nasca et al., 2021), and although AChE is not a generic marker of EVs (Liao et al., 2019), it is possible that neuronal EVs might contain this enzyme; this possibility could be worth further investigation. In contrast with protein, nucleic acids were not widely investigated. Only three papers reported profiling of EV RNA and/or qPCR of individual RNAs (see below).
5.3. Identities of individually targeted molecules
L1CAM and generic EV markers
As stated, proteins were the most frequently targeted molecular class. Naturally for the focus of these studies, L1CAM was among the most commonly probed targets (n = 22 studies, 43%; Figure 6), and usually detected by Western blot. Antibodies commonly used for L1CAM detection are listed in Table S2. For selected examples (nonexhaustive) of other proteins and the techniques used to detect them, see Table S3. As generic surface markers of EVs, the tetraspanins CD9 (n = 13), CD63 (n = 14), and CD81 (n = 17) were commonly used (Figure 6). Internal EV proteins were also probed for in some studies, including Alix (n = 14), TSG101 (n = 11), and syntenin (n = 2) (Figure 6). At least 22 studies specifically measured generic transmembrane and internal proteins, as now recommended by MISEV, including 40% of articles prior to MISEV2014, 20% between MISEV2014 and 2018, and almost 52% of those after 2018.
FIGURE 6.
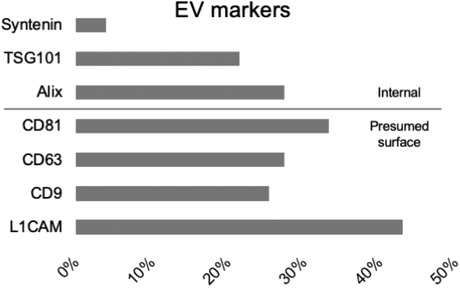
Commonly probed putative EV markers. Bars depict percentage of 51 papers that probed for the indicated marker: internal (above line) or presumed surface markers (below line)
Negative/depleted non‐EV markers
We were able to locate only ten articles in which data for a specifically probed negative/depleted marker of non‐EV material was shown, and all but two of them were published in 2019 or later. Of these ten, four probed for the Golgi marker GM130 (GOLGA2) and five for calnexin, an endoplasmic reticulum marker. These proteins are presumed to be depleted in EVs compared with source cells and may indicate excessive cell death and EV‐like artifacts in cell culture. We were especially impressed with one study in cell culture that measured markers from multiple cellular compartments in EVs and other fractions (Gomes et al., 2015). However, whether these ‘deep cellular’ markers adequately monitor coisolates from biological fluids is perhaps less clear. In this light, one article checked albumin levels (Norman et al., 2021), which may be indicative of protein contamination in biofluid sources including blood, and another study checked Apo‐A1 (Nogueras‐Ortiz et al., 2020), the major protein of high‐density lipoprotein particles (HDL). We refer here only to specifically probed markers. Potentially, information about depleted markers could also be gathered from proteomics data in the cases of studies that reported mass spectrometry results.
In most of the studies analysed here, putative EV cargo molecules were specifically measured or broadly profiled to assess biomarker potential and/or identify possible disease mechanisms, mostly for CNS disorders. We did not design this study to evaluate the strength of the evidence for these measurements and profiling studies and instead focused on the basic MISEV2018 recommendations for EV characterization. However, we can make several observations. ELISAs, including multiplexed ELISAs, as well as single‐molecule analysis (SIMOA, a kind of ultrasensitive ELISA), were commonly used to quantitate known CNS disease markers such as alpha synuclein, amyloid beta, tau (including phosphorylated forms), neurofilament light (NfL), and neurogranin (NRGN), among others, in precipitated and/or selected material. Proximity ligation assays and mass spectrometry were also used. Targeted detection of betaIII‐tubulin was done in one study to interrogate the neuronal origin of selected material (Zou et al., 2020). In another study, NCAM was found in L1CAM‐selected material, although NCAM expression is also not restricted to neurons. These results may be interpreted as evidence that L1CAM reagents select for neuronal EVs. On the other hand, additional controls may be needed to bolster these conclusions. The study that measured betaIII‐tubulin immunoselected using an antibody raised to a luminal L1CAM epitope (Zou et al., 2020), which may complicate interpretation. Another study detected glial fibrillary acidic protein (GFAP) in putative L1CAM+ material (Nogueras‐Ortiz et al., 2020), an unexpected result. Finally, a proteomics study of L1CAM capture identified two proteins that were significantly enriched in brain, but also found significant enrichment of components of common contaminants of EV preparations, including circulating IgG complexes, HDL, very low‐density lipoproteins (VLDL) and chylomicrons (Winston et al., 2016).
RNA analytes
Of the three publications that examined RNA, in one study, several miRNAs were chosen based upon expression in brain tissue. miRs‐212, ‐132, ‐566, ‐182‐5p, and ‐1304‐5p were then quantitated by qPCR, with normalization to miR‐16 (Cha et al., 2019). Interestingly, in this study, miR‐9 and miR‐451a were also assessed, and the authors concluded that miR‐9, a CNS‐enriched miRNA, was also enriched in the L1CAM‐precipitated material (Cha et al., 2019). Another study amplified miR‐155 by qPCR, with normalization to GAPDH mRNA (Anastasi et al., 2021). Finally, one study examined long noncoding RNA by hybridization microarray, followed by validation of one lncRNA by qPCR, with normalization to GAPDH (Zou et al., 2020).
6. CONCLUSIONS
The findings of this review suggest several opportunities for progress in the study of L1CAM and EVs, as seen through the lens of MISEV (Théry et al., 2018). First, EV nomenclature remains somewhat confused, especially in that ‘exosome’ and ‘EV’ have often been used interchangeably despite a lack of investigation of the ratio of EVs with endosomal versus plasma membrane origin. Following consensus nomenclature may enhance clarity: using EV generally and reserving ‘exosome’ and ‘ectosome/microvesicle’ for particles with proven biogenesis.
A second opportunity is to seek better understanding of the influence of sample characteristics and preanalytical variables (such as sample collection, processing, and storage/handling) as well as EV separation steps on the presence and detectability of EV‐associated L1CAM in EV preparations. Here and in later characterization steps, normalization is of particular interest. In most of the studies we reviewed, no normalization was done prior to measurements beyond equalizing starting volumes and/or dilutions. Also, since most of the recent L1CAM EV literature is focused on biomarker development for CNS diseases and injuries, it is interesting to note that L1CAM upregulation in several cancers has also been well established (Altevogt et al., 2020; Colombo & Meldolesi, 2015; Fogel et al., 2003; Gavert et al., 2008). It might therefore be important in future studies to report on and control for cancer burden in patient and control populations. Similarly, instructive will be knowing the extent to which preanalytical variables might affect artifactual detection of non‐membrane‐integral proteins including cleaved L1CAM itself.
Although we could not conduct a full analysis of preanalytical variables here, as they were generally not reported or inconsistently reported, an example of a potentially large influence of a preanalytical variable on L1CAM detection is thrombin‐mediated preclearing of fibrinogen/fibrin from plasma. By aggregation and conglomeration with other factors, fibrinogen is thought to affect plasma EV separation and characterization (Onódi et al., 2018), and this influence could be especially important for concentration methods. At least one precipitation kit manufacturer recommends and provides reagents for a thrombin pretreatment. However, of 26 articles that used PEG or PEG‐based kits to precipitate particles from plasma, only eight specifically indicated thrombin treatment. Enhancing understanding and reporting of such variables would boost reproducibility and comparison between studies.
The MISEV domain of characterization provides additional opportunities for progress (Théry et al., 2018). Overall in the current L1CAM EV literature, the vesicular nature of separated particle populations has not been well supported by physical characterization, with almost 40% of articles reporting no physical characterization and others relying on techniques that do not detect EVs specifically. To be sure, some of these articles use the same methodology as previously published papers, and perhaps the authors felt that it was no longer necessary to perform characterization for an established method.
EV‐enriched and ‐depleted marker characterization has also been limited. In some studies, L1CAM itself was used as the only marker for EV characterization after an L1CAM IP, an approach that cannot rule out artifacts of binding or aggregation. The presence of non‐EV particles in EV preparations was only rarely evaluated. In most cases, only one cellular depleted marker was assessed. Although this may be appropriate for cell culture experiments, complex EV sources such as blood were used in most investigations. Importantly, several studies [e.g., (Anastasi et al., 2021; Peltz et al., 2020)] found evidence that non‐neuronal materials were present in L1CAM‐precipitated material, including broad lipoprotein and free protein contamination (Anastasi et al., 2021). It is thus difficult to assess how non‐EV particles and associated molecules might have contributed to the results of previous studies of L1CAM+ EVs. Investigations with additional controls may be needed to understand these contributions.
However, improvements in characterization and reporting will go only so far with bulk assays. For example, detection of both L1CAM and, say, CD81 in an EV population does not necessarily mean presence on the same particle. Similarly, finding L1CAM and NCAM or NfL in the same population does not necessarily prove colocalization at the particle level. Ultimately, single‐particle, multimarker studies will be needed to establish if L1CAM is truly integral to neuron‐derived EVs, tightly associated, or simply a contaminant of EV preparations.
With a single protein, L1CAM, at the centre of all studies we examined, affinity reagents for that protein are naturally crucial to rigor and reproducibility, as well as increased vigilance for the possibility that antibodies to L1CAM have nonspecific binding or capture soluble protein and associated material rather than EVs. Based on our observations, there is an opportunity for more transparency on which antibodies and formulations are used. Also, important is knowing which epitope each antibody recognizes and choosing reagents for different purposes based on this binding. For example, after an extracellular domain of L1CAM is used for immunocapture, it might be useful to probe with an antibody to a cytoplasmic C‐terminal domain to show that full‐length L1CAM is present and presumably captured. Similarly, antibodies to an EV‐internal domain could be used as controls. Unexpectedly, internal epitopes were used for capture of putatively L1CAM+ EVs in at least two studies (Dagur et al., 2020; Zou et al., 2020). How these epitopes would permit capture of intact EVs is unclear. Possibly, these antibodies might capture damaged EVs or cellular membrane fragments that form particles with ‘flipped’ membrane topology. In any case, understanding more about the nonspecific binding of anti‐L1CAM antibodies will be valuable.
We would like to conclude with some general remarks about strengths and weaknesses of this study and its findings. It is possible that our literature search may have missed important publications. A systematic review is also only as strong as the analysis and the individuals who contributed to it. With only two authors, our study may be considered weaker than studies with larger numbers of authors. However, we feel that the general conclusions would remain even if several publications or data points were missed. We would furthermore like to note that the need for more detailed method reporting or clearer use of nomenclature is in no way specific to the L1CAM EV literature and is instead a general opportunity for improvement (Van Deun et al., 2017). Our findings are also not a criticism of any particular study or group. Although a general lack of EV characterization may cast doubt on some interpretations, our study does not cast doubt on other aspects of published findings. Especially for the majority of L1CAM EV studies that focus on biomarkers of CNS disease, we must remember that a reproducible biomarker that is captured by an anti‐L1CAM antibody is still a reproducible biomarker. This is true even if L1CAM is not integral to EVs; even if the captured L1CAM did not come from a neuron; and even if the binding is to something other than L1CAM entirely. Even so, new approaches and evidence may be needed to ensure correct interpretation of cell‐specific EV data.
CONFLICT OF INTEREST
KWW is a member of the advisory board of NeuroDex, Inc.
Supporting information
Supplementary information
Supplementary information
Supplementary information
ACKNOWLEDGEMENTS
The authors acknowledge members of the Witwer lab for helpful discussions and are grateful for support of the Department of Molecular and Comparative Pathobiology at The Johns Hopkins University. KWW is supported in part by the National Institute on Drug Abuse (NIDA; R01DA047807), the National Institute of Mental Health (NIMH; R21/R33MH118164), The National Institute of Allergy and Infectious Diseases (NIAID, R01AI144997), the National Cancer Institute (NCI) and Office of the Director (UG3CA241694), the Michael J. Fox Foundation (MJFF; Grant 00900821), and the Richman Family Precision Medicine Centre of Excellence in Alzheimer's Disease at Johns Hopkins Medicine.
Gomes, D. E. , & Witwer, K. W. (2022). L1CAM‐associated extracellular vesicles: A systematic review of nomenclature, sources, separation, and characterization. Journal of Extracellular Biology, 1, e35. 10.1002/jex2.35
REFERENCES
- Altevogt, P. , Ben‐Ze'ev, A. , Gavert, N. , Schumacher, U. , Schäfer, H. , & Sebens, S. (2020). Recent insights into the role of L1CAM in cancer initiation and progression. International Journal of Cancer, 147(12), 3292–6. Available from: https://pubmed.ncbi.nlm.nih.gov/32588424/ [DOI] [PubMed] [Google Scholar]
- Alvarez‐Erviti, L. , Seow, Y. , Yin, H. , Betts, C. , Lakhal, S. , & Wood, M. J. A. (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature Biotechnology, 29(4), 306–9. [DOI] [PubMed] [Google Scholar]
- Anastasi, F. , Masciandaro, S. M. , Carratore, R. D. , Dell'anno, M. T. , Signore, G. , Falleni, A. , Mcdonnell, L. A. , & Bongioanni, P. (2021). Proteomics profiling of neuron‐derived small extracellular vesicles from human plasma: Enabling single‐subject analysis. International Journal of Molecular Sciences, 22(6), 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiolini, F. , Belloni, E. , Giordano, M. , Campioni, M. , Forneris, F. , Paronetto, M. P. , Lupia, M. , Brandas, C. , Pradella, D. , Di Matteo, A. , Giampietro, C. , Jodice, G. , Luise, C. , Bertalot, G. , Freddi, S. , Malinverno, M. , Irimia, M. , Moulton, J. D. , Summerton, J. , … Ghigna, C. (2019). A novel L1CAM isoform with angiogenic activity generated by NOVA2‐mediated alternative splicing. Elife, 8, e44305. Available from: https://pubmed.ncbi.nlm.nih.gov/30829570/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab, T. , Mallick, E. R. , Huang, Y. , Dong, L. , Liao, Z. , Zhao, Z. , Gololobova, O. , Smith, B. , Haughey, N. J. , Pienta, K. J. , Slusher, B. S. , Tarwater, P. M. , Tosar, J. P. , Zivkovic, A. M. , Vreeland, W. N. , Paulaitis, M. E. , & Witwer, K. W. (2021). Characterization of extracellular vesicles and synthetic nanoparticles with four orthogonal single‐particle analysis platforms. Journal of extracellular vesicles, 10(6), e12079. Available from: https://pubmed.ncbi.nlm.nih.gov/33850608/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athauda, D. , Gulyani, S. , Karnati, H. K. , Li, Y. , Tweedie, D. , Mustapic, M. , Chawla, S. , Chowdhury, K. , Skene, S. S. , Greig, N. H. , Kapogiannis, D. , & Foltynie, T. (2019). Utility of neuronal‐derived exosomes to examine molecular mechanisms that affect motor function in patients with parkinson disease: A secondary analysis of the exenatide‐PD trial. JAMA Neurology, 76(4), 420–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhargava, P. , Nogueras‐Ortiz, C. , Kim, S. , Delgado‐Peraza, F. , Calabresi, P. A. , & Kapogiannis, D. (2021). Synaptic and complement markers in extracellular vesicles in multiple sclerosis. Multiple sclerosis Journal, 27(4), 509–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, L. A. , & Mocchetti, I. (2021). Extracellular vesicles and HIV‐associated neurocognitive disorders: Implications in neuropathogenesis and disease diagnosis. Neurotoxicity Research, 39(6), 2098–107. Available from: https://pubmed.ncbi.nlm.nih.gov/34618322/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha, D. J. , Mengel, D. , Mustapic, M. , Liu, W. , Selkoe, D. J. , Kapogiannis, D. , Galasko, D. , Rissman, R. A. , Bennett, D. A. , & Walsh, D. M. (2019). miR‐212 and miR‐132 are downregulated in neurally derived plasma exosomes of Alzheimer's patients. Frontiers in Neuroscience, 13, 1208. Available from: http://www.ncbi.nlm.nih.gov/pubmed/31849573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla, S. , Gulyani, S. , Allen, R. P. , Earley, C. J. , Li, Xu , Van Zijl, P. , & Kapogiannis, D. (2019). Extracellular vesicles reveal abnormalities in neuronal iron metabolism in restless legs syndrome. Sleep, 42(7), zsz079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, A. , Boilard, E. , Buzas, E. I. , Cheng, L. , Falcón‐Perez, J. M. , Gardiner, C. , Gustafson, D. , Gualerzi, A. , Hendrix, An , Hoffman, A. , Jones, J. , Lässer, C. , Lawson, C. , Lenassi, M. , Nazarenko, I. , O'driscoll, L. , Pink, R. , Siljander, P. R.‐.M. , Soekmadji, C. , … Nieuwland, R. (2019). Considerations towards a roadmap for collection, handling and storage of blood extracellular vesicles. Journal of Extracellular Vesicles, 8(1), 1647027. Available from: https://pubmed.ncbi.nlm.nih.gov/31489143/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, A. , Buschmann, D. , Byrd, J. B. , Carter, D. R. F. , Cheng, L. , Compton, C. , Daaboul, G. , Devitt, A. , Falcon‐Perez, J. M. , Gardiner, C. , Gustafson, D. , Harrison, P. , Helmbrecht, C. , Hendrix, An , Hill, A. , Hoffman, A. , Jones, J. C. , Kalluri, R. , Kang, J. Y. , … Nieuwland, R. (2018). Summary of the ISEV workshop on extracellular vesicles as disease biomarkers, held in Birmingham, UK, during December 2017. Journal of Extracellular Vesicles, 7(1), 1473707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocozza, F. , Grisard, E. , Martin‐Jaular, L. , Mathieu, M. , & Théry, C. (2020). SnapShot: Extracellular vesicles. Cell, 182(1), 262–262.e1. Available from: https://pubmed.ncbi.nlm.nih.gov/32649878/ [DOI] [PubMed] [Google Scholar]
- Cocucci, E. , & Meldolesi, J. (2015). Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends in Cell Biology, 25(6), 364–72. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25683921 [DOI] [PubMed] [Google Scholar]
- Coleman, B. M. , & Hill, A. F. (2015). Extracellular vesicles–Their role in the packaging and spread of misfolded proteins associated with neurodegenerative diseases. Seminars in Cell & Developmental Biology, 40, 89–96. Available from: https://pubmed.ncbi.nlm.nih.gov/25704308/ [DOI] [PubMed] [Google Scholar]
- Colombo, F. , & Meldolesi, J. (2015). L1‐CAM and N‐CAM: From adhesion proteins to pharmacological targets. Trends in Pharmacological Sciences, 36(11), 769–81. Available from: https://pubmed.ncbi.nlm.nih.gov/26478212/ [DOI] [PubMed] [Google Scholar]
- Cressatti, M. , Galindez, J. M. , Juwara, L. , Orlovetskie, N. , Velly, A. M. , Eintracht, S. , Liberman, A. , Gornitsky, M. , & Schipper, H. M. (2021). Characterization and heme oxygenase‐1 content of extracellular vesicles in human biofluids. Journal of Neurochemistry, 157(6), 2195–209. [DOI] [PubMed] [Google Scholar]
- Dagur, R. S. , Liao, Ke , Sil, S. , Niu, F. , Sun, Z. , Lyubchenko, Y. L. , Peeples, E. S. , Hu, G. , & Buch, S. (2020). Neuronal‐derived extracellular vesicles are enriched in the brain and serum of HIV‐1 transgenic rats. Journal of Extracellular Vesicles, 9(1), 1703249. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32002168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens, A. M. , Tovar‐Y‐Romo, L. B. , Yoo, S.‐W. , Trout, A. L. , Bae, M. , Kanmogne, M. , Megra, B. , Williams, D. W. , Witwer, K. W. , Gacias, M. , Tabatadze, N. , Cole, R. N. , Casaccia, P. , Berman, J. W. , Anthony, D. C. , & Haughey, N. J. (2017). Astrocyte‐shed extracellular vesicles regulate the peripheral leukocyte response to inflammatory brain lesions. Science Signaling, 10(473), eaai7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitan, E. , Tosti, V. , Suire, C. N. , Cava, E. , Berkowitz, S. , Bertozzi, B. , Raefsky, S. M. , Veronese, N. , Spangler, R. , Spelta, F. , Mustapic, M. , Kapogiannis, D. , Mattson, M. P. , & Fontana, L. (2017). In a randomized trial in prostate cancer patients, dietary protein restriction modifies markers of leptin and insulin signaling in plasma extracellular vesicles. Aging Cell, 16(6), 1430–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdbrügger, U. , Blijdorp, C. J. , Bijnsdorp, I. V. , Borràs, F. E. , Burger, D. , Bussolati, B. , Byrd, J. B. , Clayton, A. , Dear, J. W. , Falcón‐Pérez, J. M. , Grange, C. , Hill, A. F. , Holthöfer, H. , Hoorn, E. J. , Jenster, G. , Jimenez, C. R. , Junker, K. , Klein, J. , Knepper, M. A. , … Martens‐Uzunova, E. S. (2021). Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles. Journal of extracellular vesicles, 10(7), e12093. Available from: https://pubmed.ncbi.nlm.nih.gov/34035881/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faissner, A. , Kruse, J. , Nieke, J. , & Schachner, M. (1984). Expression of neural cell adhesion molecule L1 during development, in neurological mutants and in the peripheral nervous system. Brain Research, 317(1), 69–82. Available from: https://pubmed.ncbi.nlm.nih.gov/6467033/ [DOI] [PubMed] [Google Scholar]
- Fauré, J. , Lachenal, G. , Court, M. , Hirrlinger, J. , Chatellard‐Causse, C. , Blot, B. , Grange, J. , Schoehn, G. , Goldberg, Y. , Boyer, V. , Kirchhoff, F. , Raposo, G. , Garin, J. , & Sadoul, R. (2006). Exosomes are released by cultured cortical neurones. Molecular and Cellular Neuroscience, 31(4), 642–8. Available from: http://www.ncbi.nlm.nih.gov/pubmed/16446100 [DOI] [PubMed] [Google Scholar]
- Fogel, M. , Gutwein, P. , Mechtersheimer, S. , Riedle, S. , Stoeck, A. , Smirnov, A. , Edler, L. , Ben‐Arie, A. , Huszar, M. , & Altevogt, P. (2003). L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet, 362(9387), 869–75. Available from: http://www.ncbi.nlm.nih.gov/pubmed/13678974 [DOI] [PubMed] [Google Scholar]
- Fu, Y. , Jiang, C. , Tofaris, G. K. , & Davis, J. J. (2020). Facile impedimetric analysis of neuronal exosome markers in Parkinson's disease diagnostics. Analytical Chemistry, 92(20), 13647–51. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32945162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, C. , Vizio, D. D.i , Sahoo, S. , Théry, C. , Witwer, K. W. , Wauben, M. , & Hill, A. F. (2016). Techniques used for the isolation and characterization of extracellular vesicles: Results of a worldwide survey. Journal of Extracellular Vesicles, 5, 32945. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27802845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavert, N. , Ben‐Shmuel, A. , Raveh, S. , & Ben‐Ze'ev, A. (2008). L1‐CAM in cancerous tissues. Expert Opinion on Biological Therapy, 8(11), 1749–57. Available from: https://pubmed.ncbi.nlm.nih.gov/18847309/ [DOI] [PubMed] [Google Scholar]
- Goetzl, E. J. , Boxer, A. , Schwartz, J. B. , Abner, E. L. , Petersen, R. C. , Miller, B. L. , & Kapogiannis, D. (2015). Altered lysosomal proteins in neural‐derived plasma exosomes in preclinical Alzheimer disease. Neurology, 85(1), 40–7. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26062630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzl, E. J. , Elahi, F. M. , Mustapic, M. , Kapogiannis, D. , Pryhoda, M. , Gilmore, A. , Gorgens, K. A. , Davidson, B. , Granholm, A. ‐. C. , & Ledreux, A. (2019). Altered levels of plasma neuron‐derived exosomes and their cargo proteins characterize acute and chronic mild traumatic brain injury. Faseb Journal, 33(4), 5082–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetzl, E. J. , Peltz, C. B. , Mustapic, M. , Kapogiannis, D. , & Yaffe, K. (2020). Neuron‐derived plasma exosome proteins after remote traumatic brain injury. Journal of Neurotrauma, 37(2), 382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes, J. , Gomes‐Alves, P. , Carvalho, S. , Peixoto, C. , Alves, P. , Altevogt, P. , & Costa, J. (2015). Extracellular vesicles from ovarian carcinoma cells display specific glycosignatures. Biomolecules, 5(3), 1741–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, D. , Liu, F. , Meng, M. , Zhang, L. , Gordon, M. L. , Wang, Y. , Cai, Li , & Zhang, N. (2020). Elevated matrix metalloproteinase‐9 levels in neuronal extracellular vesicles in Alzheimer's disease. Annals of Clinical and Translational Neurology, 7(9), 1681–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutwein, P. , Mechtersheimer, S. , Riedle, S. , Stoeck, A. , Gast, D. , Joumaa, S. , Zentgraf, H. , Fogel, M. , & Altevogt, P. (2003). ADAM10‐mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. Faseb Journal, 17(2), 292–4. [DOI] [PubMed] [Google Scholar]
- Gutwein, P. , Oleszewski, M. , Mechtersheimer, S. , Agmon‐Levin, N. , Krauss, K. , & Altevogt, P. (2000). Role of Src kinases in the ADAM‐mediated release of L1 adhesion molecule from human tumor cells. Journal of Biological Chemistry, 275(20), 15490–7. [DOI] [PubMed] [Google Scholar]
- Gutwein, P. , Stoeck, A. , Riedle, S. , Gast, D. , Runz, S. , Condon, T. P. , Marmé, A. , Phong, M.‐C. , Linderkamp, O. , Skorokhod, A. , & Altevogt, P. (2005). Cleavage of L1 in exosomes and apoptotic membrane vesicles released from ovarian carcinoma cells. Clinical Cancer Research, 11(7), 2492–501. [DOI] [PubMed] [Google Scholar]
- Harding, C. , Heuser, J. , & Stahl, P. (1983). Receptor‐mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. Journal of Cell Biology, 97(2), 329–39. Available from: http://www.ncbi.nlm.nih.gov/pubmed/6309857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero, C. , De La Fuente, A. , Casas‐Arozamena, C. , Sebastian, V. , Prieto, M. , Arruebo, M. , Abalo, A. , Colás, E. , Moreno‐Bueno, G. , Gil‐Moreno, A. , Vilar, A. , Cueva, J. , Abal, M. , & Muinelo‐Romay, L. (2019). Extracellular vesicles‐based biomarkers represent a promising liquid biopsy in endometrial cancer. Cancers (Basel), 11(12), Available from: http://www.ncbi.nlm.nih.gov/pubmed/31842290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, A. F. (2019). Extracellular vesicles and neurodegenerative diseases. Journal of Neuroscience, 39(47), 9269–73. Available from: https://pubmed.ncbi.nlm.nih.gov/31748282/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C. , Hopfner, F. , Berg, D. , Hu, M. T. , Pilotto, A. , Borroni, B. , Davis, J. J. , & Tofaris, G. K. (2021). Validation of α‐synuclein in L1CAM‐immunocaptured exosomes as a biomarker for the stratification of Parkinsonian syndromes. Movement Disorders, 36(11), 2663–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C. , Hopfner, F. , Katsikoudi, A. , Hein, R. , Catli, C. , Evetts, S. , Huang, Y. , Wang, H. , Ryder, J. W. , Kuhlenbaeumer, G. , Deuschl, G. , Padovani, A. , Berg, D. , Borroni, B. , Hu, M. T. , Davis, J. J. , & Tofaris, G. K. (2020). Serum neuronal exosomes predict and differentiate Parkinson's disease from atypical parkinsonism. Journal of Neurology, Neurosurgery, and Psychiatry, 91(7), 720–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone, R. M. (1992). The Jeanne Manery‐Fisher Memorial Lecture 1991. Maturation of reticulocytes: Formation of exosomes as a mechanism for shedding membrane proteins. Biochemistry and Cell Biology = Biochimie Et Biologie Cellulaire, 70(3‐4), 179–90. vailable from: http://www.ncbi.nlm.nih.gov/pubmed/1515120 [DOI] [PubMed] [Google Scholar]
- Keller, S. , König, A.‐K. , Marmé, F. , Runz, S. , Wolterink, S. , Koensgen, D. , Mustea, A. , Sehouli, J. , & Altevogt, P. (2009). Systemic presence and tumor‐growth promoting effect of ovarian carcinoma released exosomes. Cancer Letters, 278(1), 73–81. [DOI] [PubMed] [Google Scholar]
- Kodidela, S. , Gerth, K. , Sinha, N. , Kumar, A. , Kumar, P. , & Kumar, S. (2020). Circulatory astrocyte and neuronal evs as potential biomarkers of neurological dysfunction in HIV‐infected subjects and alcohol/tobacco users. Diagnostics (Basel, Switzerland), 10(6), 349. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32481515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, A. , Kim, S. , Su, Y. , Sharma, M. , Kumar, P. , Singh, S. , Lee, J. , Furdui, C. M. , Singh, R. , Hsu, F.‐C. , Kim, J. , Whitlow, C. T. , Nader, M. A. , & Deep, G. (2021). Brain cell‐derived exosomes in plasma serve as neurodegeneration biomarkers in male cynomolgus monkeys self‐administrating oxycodone. EBioMedicine, 63, 103192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , & Galileo, D. S. (2010). Soluble L1CAM promotes breast cancer cell adhesion and migration in vitro, but not invasion. Cancer Cell International, 10:34. Available from: http://www.ncbi.nlm.nih.gov/pubmed/20840789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao, Z. , Jaular, L. M. , Soueidi, E. , Jouve, M. , Muth, D. C. , Schøyen, T. H. , Seale, T. , Haughey, N. J. , Ostrowski, M. , Théry, C. , & Witwer, K. W. (2019). Acetylcholinesterase is not a generic marker of extracellular vesicles. Journal of Extracellular Vesicles, 8(1), 1628592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linneberg, C. , Toft, C. L. F. , Kjaer‐Sorensen, K. , & Laursen, L. S. (2019). L1cam‐mediated developmental processes of the nervous system are differentially regulated by proteolytic processing. Science Reports, 9(1), 3716. Available from: https://pubmed.ncbi.nlm.nih.gov/30842511/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lötvall, J. , Hill, A. F. , Hochberg, F. , Buzás, E. I. , Di Vizio, D. , Gardiner, C. , Gho, Y. S. , Kurochkin, I. V. , Mathivanan, S. , Quesenberry, P. , Sahoo, S. , Tahara, H. , Wauben, M. H. , Witwer, K. W. , & Théry, C. (2014). Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. Journal of Extracellular Vesicles, 3(1), 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maness, P. F. , & Schachner, M. (2007). Neural recognition molecules of the immunoglobulin superfamily: Signaling transducers of axon guidance and neuronal migration. Nature Neuroscience, 10(1), 19–26. Available from: https://pubmed.ncbi.nlm.nih.gov/17189949/ [DOI] [PubMed] [Google Scholar]
- Mansur, R. B. , Delgado‐Peraza, F. , Subramaniapillai, M. , Lee, Y. , Iacobucci, M. , Nasri, F. , Rodrigues, N. , Rosenblat, J. D. , Brietzke, E. , Cosgrove, V. E. , Kramer, N. E. , Suppes, T. , Raison, C. L. , Fagiolini, A. , Rasgon, N. , Chawla, S. , Nogueras‐Ortiz, C. , Kapogiannis, D. , & Mcintyre, R. S. (2021). Exploring brain insulin resistance in adults with bipolar depression using extracellular vesicles of neuronal origin. Journal of Psychiatric Research, 133:82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manu, M. S. , Hohjoh, H. , & Yamamura, T. (2021). Extracellular Vesicles as Pro‐ and Anti‐inflammatory Mediators, Biomarkers and Potential Therapeutic Agents in Multiple Sclerosis. Aging and Disease, 12(6), 1451–61. Available from: https://pubmed.ncbi.nlm.nih.gov/34527421/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu, B. , Kowal, E. J. K. , Van Balkom, B. W. M. , Bartel, S. , Bhattacharyya, S. N. , Buzás, E. I. , Buck, A. H. , De Candia, P. , Chow, F. W. N. , Das, S. , Driedonks, T. A. P. , Fernández‐Messina, L. , Haderk, F. , Hill, A. F. , Jones, J. C. , Van Keuren‐Jensen, K. R. , Lai, C. P. , Lässer, C. , Di Liegro, I. , … Nolte‐‘T Hoen, E. N. M. (2017). Obstacles and opportunities in the functional analysis of extracellular vesicle RNA – An ISEV position paper. Journal of Extracellular Vesicles, 6(1), 1286095. Available from: https://www.tandfonline.com/doi/full/10.1080/20013078.2017.1286095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldolesi, J. (2021). Extracellular vesicles (exosomes and ectosomes) play key roles in the pathology of brain diseases. Molecular biomedicine, 2(1), Available from: https://pubmed.ncbi.nlm.nih.gov/35006460/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins, R. J. , Mustapic, M. , Goetzl, E. J. , & Kapogiannis, D. (2017). Exosomal biomarkers of brain insulin resistance associated with regional atrophy in Alzheimer's disease. Human brain mapping, 38(4), 1933–40. Available from: http://doi.wiley.com/10.1002/hbm.23494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasca, C. , Dobbin, J. , Bigio, B. , Watson, K. , De Angelis, P. , Kautz, M. , Cochran, A. , Mathé, A. A. , Kocsis, J. H. , Lee, F. S. , Murrough, J. W. , Mcewen, B. S. , & Rasgon, N. (2021). Insulin receptor substrate in brain‐enriched exosomes in subjects with major depression: On the path of creation of biosignatures of central insulin resistance. Molecular Psychiatry, 26(9), 5140–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwland, R. , Falcón‐Pérez, J. M. , Théry, C. , & Witwer, K. W. (2020). Rigor and standardization of extracellular vesicle research: Paving the road towards robustness. Journal of Extracellular Vesicles, 10(2), e12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueras‐Ortiz, C. J. , Mahairaki, V. , Delgado‐Peraza, F. , Das, D. , Avgerinos, K. , Eren, E. , Hentschel, M. , Goetzl, E. J. , Mattson, M. P. , & Kapogiannis, D. (2020). Astrocyte‐ and neuron‐derived extracellular vesicles from Alzheimer's disease patients effect complement‐mediated neurotoxicity. Cells, 9(7), 1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman, M. , Ter‐Ovanesyan, D. , Trieu, W. , Lazarovits, R. , Kowal, E. J. K. , Lee, Ju H. , Chen‐Plotkin, A. S. , Regev, A. , Church, G. M. , & Walt, D. R. (2021). L1CAM is not associated with extracellular vesicles in human cerebrospinal fluid or plasma. Nature Methods, 18(6), 631–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onódi, Z. , Pelyhe, C. , Terézia Nagy, C. , Brenner, G. B. , Almási, L. , Kittel, Á. , Manček‐Keber, M. , Ferdinandy, P. , Buzás, E. I. , & Giricz, Z. (2018). Isolation of high‐purity extracellular vesicles by the combination of iodixanol density gradient ultracentrifugation and bind‐elute chromatography from blood plasma. Frontiers in Physiology, 9(OCT), 1479. Available from: https://pubmed.ncbi.nlm.nih.gov/30405435/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace, K. R. , Dutt, R. , & Galileo, D. S. (2019). Exosomal L1CAM stimulates glioblastoma cell motility, proliferation, and invasiveness. International Journal of Molecular Sciences, 20(16), 3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, S. A. , Deep, G. , & Brinkley, T. E. (2018). Detection of the receptor for advanced glycation endproducts in neuronally‐derived exosomes in plasma. Biochemical and Biophysical Research Communications, 500(4), 892–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltz, C. B. , Kenney, K. , Gill, J. , Diaz‐Arrastia, R. , Gardner, R. C. , & Yaffe, K. (2020). Blood biomarkers of traumatic brain injury and cognitive impairment in older veterans. Neurology, 95(9), e1126–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam, L. , Liston, M. , Sun, B. , & Narvid, J. (2020). Using neuronal extracellular vesicles and machine learning to predict cognitive deficits in HIV. Journal of Neurovirology, 26(6), 880–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulliam, L. , Sun, B. , Mustapic, M. , Chawla, S. , & Kapogiannis, D. (2019). Plasma neuronal exosomes serve as biomarkers of cognitive impairment in HIV infection and Alzheimer's disease. Journal of Neurovirology, 25(5), 702–709. Available from: http://link.springer.com/10.1007/s13365‐018‐0695‐4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani, K. , Mukherjee, R. , Singh, E. , Kumar, S. , Sharma, V. , Vishwakarma, P. , Bharti, P. S. , Nikolajeff, F. , Dinda, A. K. , Goyal, V. , & Kumar, S. (2019). Neuronal exosomes in saliva of Parkinson's disease patients: A pilot study. Parkinson's Related Disorders, 67:21–3. [DOI] [PubMed] [Google Scholar]
- Royo, F. , Théry, C. , Falcón‐Pérez, J. M. , Nieuwland, R. , & Witwer, K. W. (2020). Methods for separation and characterization of extracellular vesicles: Results of a worldwide survey performed by the ISEV rigor and standardization subcommittee. Cells, 9(9), 1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, A. E. , Sneider, A. , Witwer, K. W. , Bergese, P. , Bhattacharyya, S. N. , Cocks, A. , Cocucci, E. , Erdbrügger, U. , Falcon‐Perez, J. M. , Freeman, D. W. , Gallagher, T. M. , Hu, S. , Huang, Y. , Jay, S. M. , Kano, S. ‐. I. , Lavieu, G. , Leszczynska, A. , Llorente, A. M. , Lu, Q. , … Vader, P. (2019). Biological membranes in EV biogenesis, stability, uptake, and cargo transfer: An ISEV position paper arising from the ISEV membranes and EVs workshop. Journal of Extracellular Vesicles, 8(1), 1684862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatov, T. R. , Wicklein, D. , & Tonevitsky, A. G. (2016). L1CAM: Cell adhesion and more. Progress in Histochemistry and Cytochemistry, 51(2), 25–32. Available from: https://pubmed.ncbi.nlm.nih.gov/27267927/ [DOI] [PubMed] [Google Scholar]
- Shankar, G. M. , Balaj, L. , Stott, S. L. , Nahed, B. , & Carter, B. S. (2017). Liquid biopsy for brain tumors. Expert Review of Molecular Diagnostics, 17(10), 943–7. Available from: https://pubmed.ncbi.nlm.nih.gov/28875730/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, M. , Kovac, A. , Korff, A. , Cook, T. J. , Ginghina, C. , Bullock, K. M. , Yang, Li , Stewart, T. , Zheng, D. , Aro, P. , Atik, A. , Kerr, K. F. , Zabetian, C. P. , Peskind, E. R. , Hu, S. ‐. C. , Quinn, J. F. , Galasko, D. R. , Montine, T. J. , Banks, W. A. , & Zhang, J. (2016). CNS tau efflux via exosomes is likely increased in Parkinson's disease but not in Alzheimer's disease. Alzheimer's Dement, 12(11), 1125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, M. , Liu, C. , Cook, T. J. , Bullock, K. M. , Zhao, Y. , Ginghina, C. , Li, Y. , Aro, P. , Dator, R. , He, C. , Hipp, M. J. , Zabetian, C. P. , Peskind, E. R. , Hu, S.‐C. , Quinn, J. F. , Galasko, D. R. , Banks, W. A. , & Zhang, J. (2014). Plasma exosomal α‐synuclein is likely CNS‐derived and increased in Parkinson's disease. Acta Neuropathologica, 128(5), 639–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si, X. , Tian, J. , Chen, Y. , Yan, Y. , Pu, J. , & Zhang, B. (2019). Central nervous system‐derived exosomal alpha‐synuclein in serum may be a biomarker in Parkinson's disease. Neuroscience, 413:308–16. [DOI] [PubMed] [Google Scholar]
- Stoeck, A. , Keller, S. , Riedle, S. , Sanderson, M. P. , Runz, S. , Le Naour, F. , Gutwein, P. , Ludwig, A. , Rubinstein, E. , & Altevogt, P. (2006). A role for exosomes in the constitutive and stimulus‐induced ectodomain cleavage of L1 and CD44. Biochemical Journal, 393(3), 609–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawa, M. , Ono, K. , Yasui, Y. , Kishi, T. , & Tsumori, T. (1997). Enhancement of neurite outgrowth by the soluble form of human L1 (neural cell adhesion molecule). Neuroreport, 8(14), 3157–62. [DOI] [PubMed] [Google Scholar]
- Suire, C. N. , Eitan, E. , Shaffer, N. C. , Tian, Qu , Studenski, S. , Mattson, M. P. , & Kapogiannis, D. (2017). Walking speed decline in older adults is associated with elevated pro‐BDNF in plasma extracellular vesicles. Experimental Gerontology, 98:209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, B. , Dalvi, P. , Abadjian, L. , Tang, N. , & Pulliam, L. (2017). Blood neuron‐derived exosomes as biomarkers of cognitive impairment in HIV. Aids, 31(14), F9–17. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28692534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, B. , Fernandes, N. , & Pulliam, L. (2019). Profile of neuronal exosomes in HIV cognitive impairment exposes sex differences. Aids, 33(11), 1683–92. [DOI] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J.‐M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , … Zuba‐Surma, E. K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7(1), 1535750. Available from: https://www.tandfonline.com/doi/full/10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, A. G. , Gray, E. , Heman‐Ackah, S. M. , Mäger, I. , Talbot, K. , El Andaloussi, S. , Wood, M. J. , & Turner, M. R. (2016). Extracellular vesicles in neurodegenerative disease – pathogenesis to biomarkers. Nature Reviews Neurology, 12(6), 346–57. Available from: https://pubmed.ncbi.nlm.nih.gov/27174238/ [DOI] [PubMed] [Google Scholar]
- Trnka, P. , Ivanova, L. , Hiatt, M .J. , & Matsell, D. G. (2012) .Urinary biomarkers in obstructive nephropathy. Clinical Journal of American Society of Nephrology, 7(10), 1567–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya, R. , & Shetty, A. K. (2021).Extracellular vesicles for the diagnosis and treatment of Parkinson's disease. Aging and disease, 12(6), 1438–50. Available from: https://pubmed.ncbi.nlm.nih.gov/34527420/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deun, J. , Mestdagh, P. , Agostinis, P. , Akay, Ö. , Anand, S. , & Anckaert, J. et al. (2017).EV‐TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nature Methods, 14(3), 228–32. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28245209 [DOI] [PubMed] [Google Scholar]
- Van Niel, G. , D'Angelo, G. , & Raposo, G. (2018).Shedding light on the cell biology of extracellular vesicles. Vol. 19, Nature Reviews Molecular C ell Biology. Nature Publishing Group; p. 213–28. [DOI] [PubMed]
- Vandendriessche, C. , Bruggeman, A. , Van Cauwenberghe, C. , & Vandenbroucke, R. E. (2020). Extracellular vesicles in Alzheimer's and Parkinson's Disease: Small entities with large consequences. Cells, 9(11), 2485. Available from: https://pubmed.ncbi.nlm.nih.gov/33203181/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassileff, N. , Cheng, L. , & Hill, A. F. (2020). Extracellular vesicles – propagators of neuropathology and sources of potential biomarkers and therapeutics for neurodegenerative diseases. Journal of Cell Science, 133(23), jcs243139. Available from: https://pubmed.ncbi.nlm.nih.gov/33310868/ [DOI] [PubMed] [Google Scholar]
- Verweij, F. J. , Balaj, L. , Boulanger, C. M. , Carter, D. R. F. , Compeer, E. B. , D'angelo, G. , El Andaloussi, S. , Goetz, J. G. , Gross, J. C. , Hyenne, V. , Krämer‐Albers, E.‐M. , Lai, C. P. , Loyer, X. , Marki, A. , Momma, S. , Nolte‐‘T Hoen, E. N. M. , Pegtel, D. M. , Peinado, H. , Raposo, G. , … Van Niel, G. (2021). The power of imaging to understand extracellular vesicle biology in vivo. Nature Methods, 18(9), 1013–26. Available from: https://pubmed.ncbi.nlm.nih.gov/34446922/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel, R. , Savage, J. , Muzard, J. , Della Camera, G. , Vella, G. , Law, A. , Marchioni, M. , Mehn, D. , Geiss, O. , Peacock, B. , Aubert, D. , Calzolai, L. , Caputo, F. , & Prina‐Mello, A. (2021). Measuring particle concentration of multimodal synthetic reference materials and extracellular vesicles with orthogonal techniques: Who is up to the challenge? Journal of extracellular vesicles, 10(3), Available from: https://pubmed.ncbi.nlm.nih.gov/33473263/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, K. A. , Chawla, S. , Nogueras‐Ortiz, C. , Coresh, J. , Sharrett, A. R. , Wong, D. F. , Jack, C. R. , Spychalla, A. J. , Gottesman, R. F. , & Kapogiannis, D. (2021). Neuronal insulin signaling and brain structure in nondemented older adults: The atherosclerosis risk in communities study. Neurobiology of Aging, 97:65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber, J. , Stone, T. C. , Katilius, E. , Smith, B. C. , Gordon, B. , & Mason, M. D. , Tabi, Z. , Brewis, I. A. , Clayton, A. (2014). Proteomics analysis of cancer exosomes using a novel modified aptamer‐based array (somascantm) platform. Molecular & Cellular Proteomics, 13(4), 1050–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, C. N. , Goetzl, E. J. , Baker, L. D. , Vitiello, M. V. , & Rissman, R. A. (2018). Growth hormone‐releasing hormone modulation of neuronal exosome biomarkers in mild cognitive impairment. Journal of Alzheimer's Disease, 66(3), 971–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, C. N. , Romero, H. K. , Ellisman, M. , Nauss, S. , Julovich, D. A. , Conger, T. , Hall, J. R. , Campana, W. , O'bryant, S. E. , Nievergelt, C. M. , Baker, D. G. , Risbrough, V. B. , & Rissman, R. A. (2019). Assessing neuronal and astrocyte derived exosomes from individuals with mild traumatic brain injury for markers of neurodegeneration and cytotoxic activity. Frontiers in Neuroscience, 13, 1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston, C. N. , Goetzl, E. J. , Akers, J. C. , Carter, B. S. , Rockenstein, E. M. , & Galasko, D. , Masliah, E. , Rissman, R. A. (2016). Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile. Alzheimer's Dement (Amsterdam, Netherlands), 3, 63–72. Available from: http://www.ncbi.nlm.nih.gov/pubmed/27408937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. , Buzás, E. I. , Bemis, L. T. , Bora, A. , Lässer, C. , Lötvall, J. , Nolte‐‘T Hoen, E. N. , Piper, M. G. , Sivaraman, S. , Skog, J. , Théry, C. , Wauben, M. H. , & Hochberg, F. (2013). Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. Journal of Extracellular Vesicles, 2:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. , & Théry, C. (2019). On primacy, precision, and popularity influencing a choice of nomenclature. Journal of Extracellular Vesicles, 8(1), 1648167. Available from: https://www.tandfonline.com/doi/full/10.1080/20013078.2019.1648167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witwer, K. W. , Goberdhan, D. C.i , O'driscoll, L. , Théry, C. , Welsh, J. A. , Blenkiron, C. , Buzás, E. I. , Di Vizio, D. , Erdbrügger, U. , Falcón‐Pérez, J. M. , Fu, Q. ‐. L. , Hill, A. F. , Lenassi, M. , Lötvall, J. , Nieuwland, R. , Ochiya, T. , Rome, S. , Sahoo, S. , & Zheng, L. (2021). Updating MISEV: Evolving the minimal requirements for studies of extracellular vesicles. Journal of Extracellular Vesicles, 10(14), e12182. Available from: https://pubmed.ncbi.nlm.nih.gov/34953156/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez‐Mó, M. , Siljander, P. R.‐M. , Andreu, Z. , Bedina Zavec, A. , Borràs, F. E. , Buzas, E. I. , Buzas, K. , Casal, E. , Cappello, F. , Carvalho, J. , Colás, E. , Cordeiro‐Da Silva, A. , Fais, S. , Falcon‐Perez, J. M. , Ghobrial, I. M. , Giebel, B. , Gimona, M. , Graner, M. , Gursel, I. , … De Wever, O. (2015). Biological properties of extracellular vesicles and their physiological functions. Journal of extracellular vesicles, 4, 27066. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4433489&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Adla, S. , Temburni, M. K. , Patel, V. P. , Lagow, E. L. , Brady, O. A. , Tian, J. , Boulos, M. I. , & Galileo, D. S. (2009). Stimulation of glioma cell motility by expression, proteolysis, and release of the L1 neural cell recognition molecule. Cancer Cell International, 9, 27. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19874583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, M. , Li, Y. , Chilukuri, K. , Brady, O. A. , Boulos, M. I. , Kappes, J. C. , & Galileo, D. S. (2011). L1 stimulation of human glioma cell motility correlates with FAK activation. Journal of Neuro‐Oncology, 105(1), 27–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates, A. G. , Pink, R. C. , Erdbrügger, U. , Siljander, P. R. , Dellar, E. R. , & Pantazi, P. , Akbar, N. , Cooke, W. R. , Vatish, M. , Dias‐Neto, E. , Anthony, D. C. , Couch, Y. (2022). In sickness and in health: The functional role of extracellular vesicles in physiology and pathology in vivo: Part I: Health and Normal Physiology: Part I: Health and Normal Physiology. Journal of extracellular vesicles, 11(1), e12151. Available from: https://pubmed.ncbi.nlm.nih.gov/35041249/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuyama, K. , Takahashi, K. , Usuki, S. , Mikami, D. , Sun, H. , & Hanamatsu, H. et al. (2019). Plant sphingolipids promote extracellular vesicle release and alleviate amyloid‐β pathologies in a mouse model of Alzheimer's disease. Science Reports, 9(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, A. , Li, Y. , Yan, Yi , Qiu, Y. , Li, B. , Xu, W. , Wang, Y. , Liu, J. , & Deng, Y. (2020). Increased prediction value of biomarker combinations for the conversion of mild cognitive impairment to Alzheimer's dementia. Translational Neurodegeneration, 9(1), 30. Available from: http://www.ncbi.nlm.nih.gov/pubmed/32741361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J. , Guo, Y. , Wei, L. , Yu, F. , Yu, Bo , & Xu, A. (2020). Long noncoding RNA POU3F3 and α‐synuclein in plasma L1CAM exosomes combined with β‐glucocerebrosidase activity: Potential predictors of Parkinson's disease. Neurotherapeutics, 17(3), 1104–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Supplementary information
Supplementary information


