ABSTRACT
Corynebacterium, particularly Corynebacterium kroppenstedtii, has been increasingly recognized as an important pathogen causing mastitis. However, no clear taxonomic, microbiological, or clinical identification for C. kroppenstedtii-related Corynebacterium species is recognized. During the investigation of isolates cultured from female patients with mastitis, 27 lipophilic C. kroppenstedtii-like isolates were obtained from clinical breast specimens from 2017 to 2019 in Guangzhou, China. These isolates were identified by phenotypic characterization, matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS), partial sequencing of the 16S rRNA, rpoB, and fusA genes, and whole-genome sequencing methods. By phylogenetic analyses, two major clusters were identified that were closely related to C. kroppenstedtii DSM 44385T. Comparative genome analyses suggested that these isolates formed two distinct genospecies within the genus Corynebacterium. The digital DNA–DNA hybridization (dDDH) values for the two genospecies were 45.5 to 47.8% between them and 47.4 to 47.7% and 49.9% to C. kroppenstedtii DSM 44385T, respectively. Based on these results, it can be concluded that these isolates need to be recognized as two new species of the genus Corynebacterium, for which we proposed the names Corynebacterium parakroppenstedtii sp. nov. and Corynebacterium pseudokroppenstedtii sp. nov. The type strain for the novel species Corynebacterium parakroppenstedtii is MC-26T (NBRC 115146T; CCTCC AB 2020210T), and that for Corynebacterium pseudokroppenstedtii is MC-17XT (NBRC 115143T; CCTCC AB 2020199T).
IMPORTANCE In this study, we characterized two novel species that were closely related to but hard to distinguish from C. kroppenstedtii by routine identification methods used in clinical laboratories. Since all 27 C. kroppenstedtii-like isolates were obtained from breast specimens of female patients with mastitis, they may be potential pathogens causing mastitis. We hope to perform further epidemiological investigation of these strains and explore their role in mastitis.
KEYWORDS: mastitis, breast pathogens, Corynebacterium, taxonomy, antimicrobial susceptibility
INTRODUCTION
The genus Corynebacterium represents a large group of Gram-positive, non-spore-forming, rod-shaped bacteria within the family Corynebacteriaceae of the order Corynebacteriales (1). It comprises more than 130 species with validly described names and has been detected in various habitats, such as soil, food, animals, humans, and plant surfaces. Except for some particular species that are well-established pathogens of humans and animals, Corynebacterium spp. have often been assigned as opportunistic pathogens. They are common components of the skin microbiome and generally have been dismissed as contaminants when recovered from clinical specimens, although they have been increasingly recognized to be associated with clinical symptoms (2–4).
Several Corynebacterium species, Corynebacterium kroppenstedtii in particular, have been reported to be associated with mastitis, which is a chronic inflammatory disease of unknown etiology in parous women of reproductive age (5). C. kroppenstedtii which lacks the typical mycolic acids of the cell envelope was first documented in 1998 from a human sputum specimen (6). It has occasionally been associated with human infection, mainly breast abscesses and granulomatous mastitis (7–9). Although the virulence of C. kroppenstedtii in mastitis is not fully understood, it is often isolated alone and early in disease progression, suggesting its pathogenic role (9, 10). In recent years, increasing C. kroppenstedtii infection has been reported by newer techniques, such as matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) and 16S rRNA gene sequencing (7, 11–15).
In this study, we describe the characterization of 27 C. kroppenstedtii-like isolates obtained from female patients with mastitis at our hospital from 2017 to 2019 and identify them as two novel species of the genus Corynebacterium.
RESULTS
Clinical information of C. kroppenstedtii-like isolates.
Twenty-seven C. kroppenstedtii-like isolates were obtained from breast specimens of 27 different female patients with mastitis. Most isolates exhibited pure growth like that of C. kroppenstedtii, and some other isolates exhibited mixed growth with coagulase-negative Staphylococcus and other Corynebacterium spp. Further histological analyses were available for 13 patients. Granulomatous mastitis was observed in 12 patients, and suppurative mastitis was observed in 1 patient. The ages of the patients were in the range of 26 to 47 years (mean 34.3 years). Basic information of these 27 cases of C. kroppenstedtii-like infection is presented in Table 1.
TABLE 1.
Data of 27 cases of C. kroppenstedtii-like isolates from Chinese clinical breast specimens
| Strain no. | Isolation date | Age (yr)/sexa | Specimen source | Diagnosisb | Comorbidity | Treatmentc | Prognosis |
|---|---|---|---|---|---|---|---|
| C. kroppenstedtii-like group I | |||||||
| MC-01 | 2017 | 26/F | Pus, tissue | GLM | Hyperprolactinemia | Surgery, TCM, dexamethasone, bromocriptine | Recovery |
| MC-04 | 2017 | 36/F | Pus, tissue | SM | Unknown | Surgery, antibiotic: rifampicin, isoniazid, ethambutol, bromocriptine | Recovery |
| MC-05 | 2017 | 35/F | Pus, tissue | M | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-06 | 2017 | 32/F | Pus, tissue | M | Pulmonary Tuberculosis | TCM, bromocriptine | Transfer to chest hospital |
| MC-08 | 2017 | 42/F | Pus, tissue | M | HBV carrier | Surgery, TCM, dexamethasone, bromocriptine | Recovery |
| MC-09 | 2017 | 42/F | Pus, tissue | GLM | Thalassemia | Surgery, TCM, bromocriptine | Recovery |
| MC-10 | 2017 | 33/F | Pus, tissue | M | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-11 | 2017 | 28/F | Pus, tissue | GLM | Pituitary adenoma | Surgery, TCM, bromocriptine | Recovery |
| MC-12 | 2018 | 35/F | Pus | M | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-13 | 2018 | 34/F | Pus, tissue | GLM | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-15 | 2018 | 31/F | Pus, tissue | GLM | HBV carrier | Surgery, TCM, bromocriptine | Recovery |
| MC-16 | 2018 | 27/F | Pus | M | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-19 | 2018 | 35/F | Secretion | M | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-20 | 2018 | 35/F | Pus | M | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-21 | 2018 | 30/F | Pus, tissue | GLM | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-22 | 2018 | 36/F | Pus, tissue | M | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-23 | 2018 | 33/F | Secretion | M | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-24 | 2019 | 37/F | Pus, tissue | GLM | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-25 | 2019 | 31/F | Pus, tissue | GLM | HBV carrier | Surgery, TCM, bromocriptine | Recovery |
| MC-26 | 2019 | 47/F | Secretion | M | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-27 | 2019 | 47/F | Pus | M | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-28 | 2019 | 38/F | Pus, tissue | GLM | Diabetes mellitus | Surgery, TCM, bromocriptine | Self-discharged |
| MC-29 | 2019 | 31/F | Puncture fluid | M | Unknown | Surgery, TCM, bromocriptine | Recovery |
| C. kroppenstedtii-like group II | |||||||
| MC-02 | 2017 | 42/F | Secretion | GLM | Hyperprolactinemia | Surgery, TCM, bromocriptine | Recovery |
| MC-03 | 2017 | 26/F | Pus, tissue | M | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-07 | 2017 | 31/F | Pus, tissue | GLM | Unknown | Surgery, TCM, bromocriptine | Recovery |
| MC-17X | 2018 | 27/F | Pus | M | Unknown | Surgery, TCM, bromocriptine | Self-discharged |
F, female.
GLM, granulomatous lobular mastitis; SM, suppurative mastitis; M, mastitis.
Surgery included abscess incision drainage and debridement. TCM, traditional Chinese medicine.
Phenotypic testing and MALDI-TOF MS.
All isolates grew on Columbia blood agar plates as grayish, smooth, circular, convex, and nonhemolytic colonies of less than 1 mm in diameter after 72 h of incubation at 35°C in the presence of 5% CO2 atmosphere. On microscopic examination, cells were Gram-positive, rod-shaped, nonmotile, and non-spore-forming with typical coryneform morphology. All isolates were lipophilic, and the growth was enhanced on brain heart infusion broth and Columbia blood agar supplemented with 1% Tween 80. In general, the cellular and colony morphologies of these 27 isolates were similar to those of C. kroppenstedtii DSM 44385T.
All the 27 isolates, along with the type strain C. kroppenstedtii DSM 44385T, were found to be catalase and pyrazinamidase positive but nitrate and urease negative. Exceptions were the case of isolate MC-28, which was pyrazinamidase negative and isolate MC-11 urease positive. They produced acid from glucose but not from sucrose, ribose, or xylose (exception: isolate MC-27 could not produce acid from glucose). Results of API Coryne assay indicated that 25 of the isolates and the type strain showed 87.5 to 99.5% identity to Corynebacterium argentoratense, while 2 isolates were similar to Corynebacterium urealyticum with 60.5% and 8.8% identity, respectively (C. kroppenstedtii is not included in the API database). The characteristics produced by API Coryne strips are shown in Table 2, and the comparative analyses with C. kroppenstedtii DSM 44385T are shown in Table 3.
TABLE 2.
Polyphasic identification results of C. kroppenstedtii-like isolates
| Strain no. | API corynea |
Strain identified by MALDTOF-MSb in database: |
Partial 16S rRNA genec |
Partial rpoB genec |
Partial fusA genec |
Genome GenBank accession no. | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Profile no. obtained | Significant taxon (% ID, Td) | Biotyper database (score value) | Vitek database (confidence value) | Similarity | GenBank accession no. | Similarity | GenBank accession no. | Similarity | GenBank accession no. | ||
| TS | 2040104 | Car (87.5, 0.49) | Ckr CCUG 44504 (2.39) | Ckr 99.9% |
NR_074408.1
|
AY492274.1 | CP001620.1 | CP001620.1 | |||
| C. kroppenstedtii-like group I | |||||||||||
| MC-01 | 2040104 | Car (87.5, 0.49) | Ckr CCUG 49276 (1.643) | Ckr 99.9% | 99.6% | MW819649 | 96.2% | MZ031080 | 97.7% | MZ031107 | JAKJKX000000000 |
| MC-04 | 2040104 | Car (87.5, 0.49) | Gcr DSM 15881T (1.583) | Ckr 99.9% | 99.6% |
MW819652
|
96.2% | MZ031083 | 97.7% | MZ031110 | JAKJKY000000000 |
| MC-05 | 2000104 | Car (99.5, 0.99) | Ckr DSM 44385T (1.579) | Ckr 99.9% |
99.6% |
MW819653
|
96.2% |
MZ031084 | 97.7% | MZ031111 | JAKJKZ000000000 |
| MC-06 | 2000104 | Car (99.5, 0.99) | Ckr CCUG 44504 (2.077) | Ckr 99.9% |
99.6% |
MW819654
|
96.2% |
MZ031085 | 97.7% | MZ031112 | JAKJLA000000000 |
| MC-08 | 2000104 | Car (99.5, 0.99) | Ckr CCUG 49276 (1.384) | Ckr 99.9% |
99.6% |
MW819656
|
96.2% | MZ031087 | 97.7% | MZ031114 | JAKLTI000000000 |
| MC-09 | 2000104 | Car (99.5, 0.99) | Ckr CCUG 44504 (1.401) | Ckr 99.9% |
99.6% |
MW819657 | 96.2% |
MZ031088 | 97.7% | MZ031115 | JAKKNX000000000 |
| MC-10 | 2000104 | Car (99.5, 0.99) | Ckr DSM 44385T (1.687) | Ckr 99.9% |
99.6% |
MW819658
|
96.2% |
MZ031089 | 97.7% | MZ031116 | JAKKNY000000000 |
| MC-11 | 2041104 | Cur (60.5, 0.27) | Ckr CCUG 44504 (1.892) | Ckr 99.9% |
99.9% |
MW819659
|
96.2% |
MZ031090 | 97.6% | MZ031117 | JAFFSY000000000 |
| MC-12 | 2040104 | Car (87.5, 0.49) | Ckr DSM 44385T (1.465) | Ckr 99.9% | 99.6% |
MW819660
|
96.2% | MZ031091 | 97.7% | MZ031118 | JAKJKU000000000 |
| MC-13 | 2040104 | Car (87.5, 0.49) | Ckr CCUG 44504 (1.709) | Ckr 99.9% | 99.6% | MW819661 | 96.2% | MZ031092 | 97.7% | MZ031119 | JAKKFA000000000 |
| MC-15 | 2040104 | Car (87.5, 0.49) | Ckr CCUG 44504 (1.828) | Ckr 99.9% | 99.6% | MW819662 | 96.2% | MZ031093 | 97.7% | MZ031120 | JAKLTJ000000000 |
| MC-16 | 2040104 | Car (87.5, 0.49) | Ckr CCUG 44504 (1.635) | Ckr 99.9% | 99.6% | MW819663 | 96.2% | MZ031094 | 97.7% | MZ031121 | JAKJKV000000000 |
| MC-19 | 2000104 | Car (99.5, 0.99) | Ckr DSM 44385T (1.602) | Ckr 99.9% | 99.6% | MW819665 | 96.2% | MZ031096 | 97.7% | MZ031123 | JAKJKW000000000 |
| MC-20 | 2000104 | Car (99.5, 0.99) | Ckr CCUG 44504 (1.924) | Ckr 99.9% | 99.6% | MW819666 | 96.2% | MZ031097 | 97.7% | MZ031124 | JAKJKP000000000 |
| MC-21 | 2040104 | Car (87.5, 0.49) | Ckr CCUG 44504 (1.927) | Ckr 99.9% | 99.6% | MW819667 | 96.2% | MZ031098 | 97.7% | MZ031125 | JAKJKQ000000000 |
| MC-22 | 2040104 | Car (87.5, 0.49) | Ckr DSM 44385T (1.419) | Ckr 99.9% | 99.6% | MW819668 | 96.2% | MZ031099 | 97.7% | MZ031126 | JAKJKR000000000 |
| MC-23 | 2040104 | Car (87.5, 0.49) | Ckr CCUG 44504 (1.622) | Ckr 99.9% | 99.6% | MW819669 | 96.2% | MZ031100 | 97.7% | MZ031127 | JAKJKS000000000 |
| MC-24 | 2000104 | Car (99.5, 0.99) | Ckr DSM 44385T (1.441) | Ckr 99.9% | 99.7% | MW819670 | 96.2% | MZ031101 | 97.7% | MZ031128 | JAGSNZ000000000 |
| MC-25 | 2000104 | Car (99.5, 0.99) | Ckr CCUG 61180 (1.467) | Ckr 99.9% | 99.6% | MW819671 | 96.2% | MZ031102 | 97.7% | MZ031129 | JAKKOA000000000 |
| MC-26 | 2000104 | Car (99.5, 0.99) | Ckr DSM 44385T (1.423) | Ckr 99.9% | 99.6% | MW819672 | 96.2% | MZ031103 | 97.7% | MZ031130 | JAGSOA000000000 |
| MC-27 | 2040004 | Cur (8.8, 0.44) | Ckr CCUG 44504 (1.966) | Ckr 99.9% | 99.6% | MW819673 | 96.2% | MZ031104 | 97.7% | MZ031131 | JAKJKT000000000 |
| MC-28 | 0000104 | Car (90.3, 0.65) | Ckr DSM 44385T (1.597) | Ckr 99.9% | 99.6% | MW819674 | 98.1% | MZ031105 | 97.4% | MZ031132 | JAGSNY000000000 |
| MC-29 | 2040104 | Car (87.5, 0.49) | Ckr CCUG 49276 (1.603) | Ckr 99.9% | 99.6% | MW819675 | 96.2% | MZ031106 | 97.6% | MZ031133 | JAKJKO000000000 |
| C. kroppenstedtii-like group II | |||||||||||
| MC-02 | 2040104 | Car (87.5, 0.49) | Ckr CCUG 44504 (2.276) | Ckr 99.9% | 99.9% | MW819650 | 96.2% | MZ031081 | 97.3% | MZ031108 | JAKKNZ000000000 |
| MC-03 | 2040104 | Car (87.5, 0.49) | Ckr CCUG 44504 (2.029) | Ckr 99.9% | 99.9% | MW819651 | 96.2% | MZ031082 | 97.3% | MZ031109 | JAKLTK000000000 |
| MC-07 | 2040104 | Car (87.5, 0.49) | Ckr CCUG 44504 (2.316) | Ckr 99.9% | 99.9% | MW819655 | 96.2% | MZ031086 | 97.3% | MZ031113 | JAKJLB000000000 |
| MC-17X | 2040104 | Car (87.5, 0.49) | Ckr CCUG 44504 (2.295) | Ckr 99.9% | 99.9% | MW819664 | 96.2% | MZ031095 | 97.3% | MZ031122 | JAEUWU000000000 |
Car, C. argentoratense; Cur, C. urealyticum.
Ckr, C. kroppenstedtii; Gcr, Glutamicibacter creatinolyticus.
Similarities of partial 16S rRNA, rpoB, and fusA genes are presented with respect to the genome of C. kroppenstedtii DSM 44385T.
TS, the type strain C. kroppenstedtii DSM 44385T; T, typicity index.
TABLE 3.
Phenotypic characteristics of the two groups of C. kroppenstedtii-like isolates and the type strain
| Characteristics | Result for strainsa |
||
|---|---|---|---|
| C. kroppenstedtii-like group I (n = 23) | C. kroppenstedtii-like group II (n = 4) | C. kroppenstedtii DSM 44385T | |
| Lipophilism | 100 | 100 | + |
| Catalase | 100 | 100 | + |
| Nitrate reduction | 0 | 0 | − |
| Hydrolysis of aesculin | 56 | 100 | + |
| Hydrolysis of urea | 4 | 0 | − |
| Enzyme activity | |||
| Pyrazinamidase | 95 | 100 | + |
| Alkaline phosphatase | 0 | 0 | − |
| Acid production from | |||
| Glucose | 95 | 100 | + |
| Sucrose | 0 | 0 | − |
| Ribose | 0 | 0 | − |
| Xylose | 0 | 0 | − |
Numbers represent percentages of positive results. +, positive reaction; −, negative reaction.
By the Vitek MS system (V2.0; bioMérieux, France), all 27 isolates were identified to C. kroppenstedtii with the confidence value of 99.9%. By the Bruker Biotyper system (BDAL Library; Bruker Daltonics, Germany), only 5 isolates (MC-02, MC-03, MC-06, MC-07, MC-17X) had good identification to species level (scores of ≥2.0) for C. kroppenstedtii CCUG 44504 as the “rank 1” identification, and another 6 produced scores of ≥1.7 for C. kroppenstedtii CCUG 44504 as the rank 1 identification. The remainders had values below the accepted score for reliable identification (scores of <1.7). The MALDI-TOF MS results are shown in Table 2.
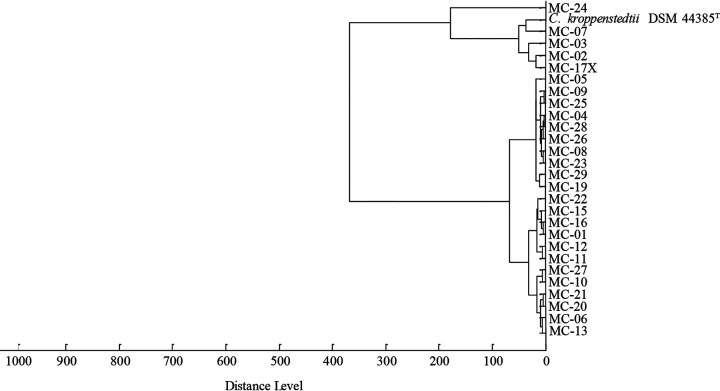
On the basis of the data revealed by MALDI-TOF MS (Bruker Biotyper), 4 isolates (MC-02, MC-03, MC-07, MC-17X) clustered with the type strain C. kroppenstedtii DSM 44385T, 22 isolates (MC-01, MC-04, MC-05, MC-06, MC-08, MC-09, MC-10, MC-11, MC-12, MC-13, MC-15, MC-16, MC-19, MC-20, MC-21, MC-22, MC-23, MC-25, MC-26, MC-27, MC-28, MC-29) constituted a coherent and distinct cluster separate from the type strain, and the remaining one isolate (MC-24) showed a unique position (Fig. 1).
FIG 1.
Dendrogram revealed by MALDI-TOF MS (Bruker Biotyper system) showing the relationship of 27 C. kroppenstedtii-like isolates and the type strain.
Genotypic identification.
To further identify the taxonomic position, the clinical isolates were subjected to partial sequencing of the 16S rRNA gene (about 1,350 bp), rpoB gene (about 420 bp), and fusA gene (about 990 bp) and comparative sequence analysis (Table 2), and clustering analysis of the clinical isolates along with the type strain were done.
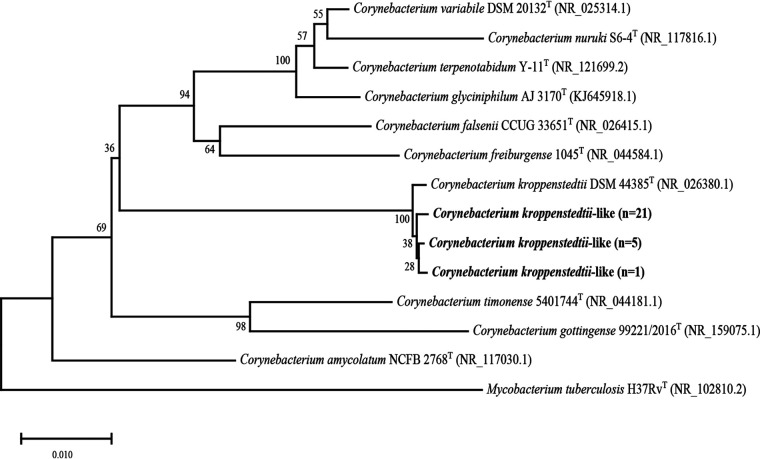
Based on phylogenetic analyses of the partial 16S rRNA gene, all the 27 isolates were closely related to C. kroppenstedtii DSM 44385T (Fig. 2), exhibiting 99.6 to 99.9% similarities. The rpoB and fusA genes of the 27 isolates showed sequences identities of 96.2 to 98.1% and 97.3 to 97.7% with C. kroppenstedtii DSM 44385T, respectively.
FIG 2.
Neighbor-joining tree based on partial 16S rRNA gene showing the phylogenetic relationship of 27 C. kroppenstedtii-like isolates and the most closely related species in the genus Corynebacterium. Bootstrap values based on 1,000 calculations are shown. The scale bar depicts 0.010 substitutions per nucleotide position.
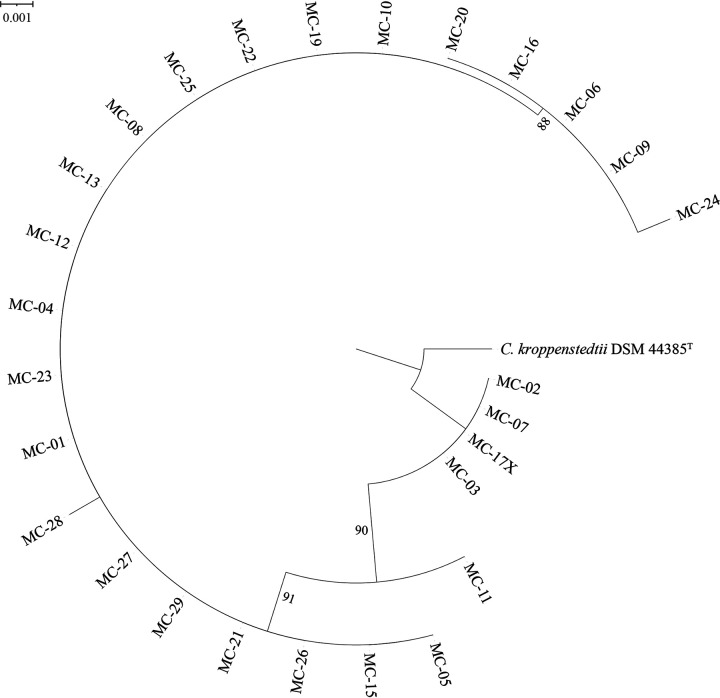
To further confirm the taxonomic identification, the clinical isolates were subjected to whole-genome sequencing. Genomic sequencing results showed that C. kroppenstedtii-like group I and group II had a genome size of 2.50 to 2.96 Mb and 2.47 to 2.80 Mb, respectively. The G+C contents of group I and group II were 54.7 to 57.2% and 55.1 to 57.2%, respectively. The genomic relatedness of these isolates and the type strain were calculated by digital DNA–DNA hybridization (dDDH) and average nucleotide identity (ANI) based on the BLASTN algorithm (ANIb). According to the dDDH analyses, 23 isolates (MC-01, MC-04, MC-05, MC-06, MC-08, MC-09, MC-10, MC-11, MC-12, MC-13, MC-15, MC-16, MC-19, MC-20, MC-21, MC-22, MC-23, MC-24, MC-25, MC-26, MC-27, MC-28, MC-29) were observed to belong to the same novel species-level taxon (C. kroppenstedtii-like group I), exhibiting 75 to 100% sequence identity to each other and 47.4 to 47.7% to the type strain C. kroppenstedtii. Four isolates (MC-02, MC-03, MC-07, MC-17X) were observed to belong to another novel species-level taxon (C. kroppenstedtii-like group II), exhibiting 99.6 to 100% sequence identity to each other, 45.5 to 47.8% identity to group I, and 49.9% identity to the type strain. ANIb analysis showed data similar to that of dDDH analysis. C. kroppenstedtii-like group I shared 96.99 to 99.99% similarities within themselves and 91.83 to 92.28% similarities to the type strain. C. kroppenstedtii-like group II shared 99.92 to 100% similarities between them, 91.83 to 92.28% similarities to group I, and 92.89 to 92.93% similarities to the type strain. Comparative genomic analyses and genomic characteristics of two groups of C. kroppenstedtii-like isolates and the closely related type strains are shown in Table 4. The phylogenomic tree based on concatenation of 18 protein marker genes (Fig. 3) indicated that the clinical isolates constituted two clusters separated from C. kroppenstedtii DSM 44385T, which was consistent with the classification results of dDDH and ANIb.
TABLE 4.
Comparative genomic analysis and genomic characteristics of two groups of C. kroppenstedtii-like isolates and the closest related type strainsa
| Strain | Pairwise comparison resultb |
Genome characteristic |
|||||
|---|---|---|---|---|---|---|---|
| 1 |
2 |
No. of contigs | Size (Mb) | G+C (%) | |||
| dDDH (%) | ANIb (%) | dDDH (%) | ANIb (%) | ||||
| 1 | 97.25 (5.65) | 99.59 (0.68) | 4–356 | 2.50–2.96 | 54.7–57.2 | ||
| 2 | 45.99 (0.38) | 91.97 (0.09) | 99.82 (0.15) | 99.97 (0.03) | 11–108 | 2.47–2.80 | 55.1–57.2 |
| 3 | 47.60 (0.06) | 92.23 (0.27) | 49.90 (0.00) | 92.84 (0.04) | 1 | 2.45 | 57.50 |
| 4 | 21.79 (0.41) | 66.81 (0.02) | 22.05 (0.17) | 66.90 (0.00) | 21 | 2.91 | 49.8 |
| 5 | 21.43 (0.30) | 67.90 (0.11) | 24.78 (0.10) | 68.16 (0.06) | 18 | 2.59 | 66.6 |
Taxa are indicated as 1, Corynebacterium parakroppenstedtii (C. kroppenstedtii-like group I); 2, Corynebacterium pseudokroppenstedtii (C. kroppenstedtii-like group II); 3, Corynebacterium kroppenstedtii DSM 44385T; 4, Corynebacterium freiburgense DSM 45254T; 5, Corynebacterium timonense 5401744T.
Values are mean with standard deviation.
FIG 3.
Protein-concatemer tree based on concatenation of 18 protein markers sequences showing the phylogenetic relationship of 27 C. kroppenstedtii-like isolates and the type strain C. kroppenstedtii DSM 44385T. Branches with bootstrap support of 50% are indicated.
Antimicrobial susceptibility.
Based on the CLSI breakpoints for this genus, most isolates of C. kroppenstedtii-like group I were sensitive to meropenem, cefepime, vancomycin, daptomycin, gentamicin, and linezolid. Nineteen of the 23 isolates (82%) were resistant to erythromycin and clindamycin, 7 isolates (30%) were resistant to trimethoprim-sulfamethoxazole, 5 (21%) were resistant to ciprofloxacin, and 4 (17%) were resistant to ceftriaxone and tetracycline. The majority of isolates of C. kroppenstedtii-like group II were sensitive to meropenem, cefepime, tetracycline, gentamicin, trimethoprim-sulfamethoxazole, linezolid, daptomycin, and vancomycin and resistant to ceftriaxone, ciprofloxacin, erythromycin, and clindamycin. All isolates were intermediate to penicillin. The antimicrobial susceptibility results of the two groups of C. kroppenstedtii-like isolates and C. kroppenstedtii DSM 44385T are shown in Table 5.
TABLE 5.
Antimicrobial susceptibilities of the two groups of C. kroppenstedtii-like isolates and the type strain
| Antimicrobial agent | CLSI breakpointa |
C. kroppenstedtii-like group I MIC (mg/L) (n = 23) |
C. kroppenstedtii-like group II MIC (mg/L) (n = 4) |
C. kroppenstedtii DSM 44385T MIC | |||||
|---|---|---|---|---|---|---|---|---|---|
| MIC50 | MIC90 | Range (% resistant) | MC-02 | MC-03 | MC-07 | MC-17X | |||
| Penicillin | S, ≤0.12; R, ≥4 | 0.5 | 2 | 0.12–2 (0) | 2 | 2 | 0.5 | 1 | 0.12 |
| Ceftriaxone | S, ≤1; R, ≥4 | <1 | 4 | <1–4 (17) | 4 | 4 | <1 | 4 | 2 |
| Cefepime | S, ≤1; R, ≥4 | <1 | <1 | <1 (0) | <1 | <1 | <1 | <1 | 4 |
| Meropenem | S, ≤0.25; R, ≥1 | <0.25 | 0.5 | <0.25–0.5 (0) | <0.25 | <0.25 | <0.25 | <0.25 | <0.25 |
| Vancomycin | S, ≤2 | <0.5 | <0.5 | <0.5 (0) | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 |
| Daptomycin | S, ≤1 | <0.5 | <0.5 | <0.5 (0) | <0.5 | <0.5 | <0.5 | 1 | <0.5 |
| Gentamicin | S, ≤4; R, ≥16 | <4 | <4 | <4 (0) | <4 | <4 | <4 | <4 | <4 |
| Erythromycin | S, ≤0.5; R, ≥2 | >8 | >8 | <0.25 to >8 (82) | >8 | >8 | >8 | >8 | <0.25 |
| Ciprofloxacin | S, ≤1; R, ≥4 | <1 | 4 | <1–4 (21) | 4 | 4 | 4 | >8 | <1 |
| Tetracycline | S, ≤4; R, ≥16 | 8 | >16 | <4 to >16 (17) | <4 | <4 | >16 | <4 | <4 |
| Clindamycin | S, ≤0.5; R, ≥4 | >4 | >4 | <0.25 to >4 (82) | >4 | >4 | >4 | >4 | <0.25 |
| Trimethoprim-sulfamethoxazole | S, ≤2/38; R, ≥4/76 | <0.5/9.5 | >4/76 | <0.5/9.5 to >4/76 (30) | <0.5/9.5 | <0.5/9.5 | >4/76 | <0.5/9.5 | <0.5/9.5 |
| Linezolid | S, ≤2 | <1 | <1 | <1 (0) | <1 | <1 | <1 | <1 | <1 |
| Ampicillin | -b | 0.5 | 2 | <0.25 to 2b | 2 | 2 | 1 | 1 | <0.25 |
| Levofloxacin | -b | <2 | >8 | <2 to >8b | >8 | >8 | >8 | >8 | <2 |
S, susceptible; R, resistant.
-, Non-species-related CLSI breakpoints.
Detection of resistance genes based on whole-genome sequencing.
Antibiotic resistance genes detected from the genomes of the two groups of C. kroppenstedtii-like isolates were not identical. Aminoglycoside resistance gene APH(3′)-Ia was detected in 14 isolates of group I (60%) and 2 isolates of group II (50%). Aminoglycoside resistance gene APH(3′')-Ib was detected in 18 (78%) and 3 (75%) isolates of C. kroppenstedtii-like group I and group II, respectively. Aminoglycoside resistance gene APH(6)-Id was detected in 18 group I isolates (78%) and 4 group II isolates (100%). Macrolide, lincosamide, and streptogramin resistance gene erm(X) were detected in 17 (74%) and 4 (100%) group I and group II isolates, respectively. Sulfonamide resistance gene sul1 and tetracycline resistance gene tet(W) were detected in 7 (30%) and 17 (74%) C. kroppenstedtii-like group I isolates and 1 (25%) and 2 (50%) C. kroppenstedtii-like group II isolates, respectively. No antibiotic resistance gene was detected in C. kroppenstedtii DSM 44385T. The antibiotic resistance genes detection results of two groups of C. kroppenstedtii-like isolates and C. kroppenstedtii DSM 44385T are shown in Table S1.
TAXONOMY
Description of Corynebacterium parakroppenstedtii sp. nov.
Corynebacterium parakroppenstedtii (pa.ra.krop.pen.stedt’i.i. Gr. pref. para-, besides, alongside, near, like; N. L. gen. masc. n. kroppenstedtii, specific epithet of a Corynebacterium species; N. L. gen. masc. n. parakroppenstedtii, resembling C. kroppenstedtii).
The description of the species is based on 23 strains (C. kroppenstedtii-like group I). Cells are Gram-positive, lipophilic, rod-shaped, nonmotile, non-spore-forming, catalase-positive, and oxidase-negative. Colonies on Columbia blood agar plates are grayish, smooth, circular, convex, and nonhemolytic with less than 1 mm in diameter after 72 h cultivation at 35°C in the presence of 5% CO2 atmosphere. In most strains, acid is produced from glucose but not from sucrose, lactose, xylose, ribose, mannitol, or glycogen. Most strains are positive for pyrazinamidase activity but negative for alkaline phosphatase, β-glucuronidase, β-galactosidase, α-glucosidase, N-acetyl-β-glucosaminidase, and urease activity. Nitrate cannot be reduced. The hydrolysis of gelatin is negative. The hydrolysis of esculin is variable. Most strains are sensitive to meropenem, cefepime, gentamicin, linezolid, daptomycin, and vancomycin but resistant to erythromycin and clindamycin. Intermediate activity to penicillin. Sensitivity to ceftriaxone, ciprofloxacin, tetracycline, and trimethoprim-sulfamethoxazole was strain-dependent.
The type strain is MC-26T (NBRC 115146T; CCTCC AB 2020210T). It has a DNA G+C content of 56.86%. It was isolated from a breast sample of a patient diagnosed with mastitis in Guangdong Provincial Hospital of Traditional Chinese Medicine in 2019.
Description of Corynebacterium pseudokroppenstedtii sp. nov.
Corynebacterium pseudokroppenstedtii (pseu.do.krop.pen.stedt’i.i. Gr. masc./fem. adj. pseudês, false; N. L. gen. masc. n. kroppenstedtii, specific epithet of a Corynebacterium species; N. L. gen. masc. n. pseudokroppenstedtii, a false (Corynebacterium) kroppenstedtii, resembling C. kroppenstedtii).
The description of the species is based on characteristics of 4 strains (C. kroppenstedtii-like group II). Cells are Gram-positive, lipophilic, rod-shaped, nonmotile, non-spore-forming, catalase-positive, and oxidase-negative. Colonies on Columbia blood agar plates are grayish, smooth, circular, convex, and nonhemolytic with less than 1 mm in diameter after 72 h cultivation at 35°C in the presence of 5% CO2 atmosphere. Acid is produced from glucose but not from sucrose, lactose, xylose, ribose, mannitol, or glycogen. Positive for pyrazinamidase activity but negative for alkaline phosphatase, β-glucuronidase, β-galactosidase, α-glucosidase, N-acetyl-β-glucosaminidase, and urease activity. Nitrate reduction and the gelatin hydrolysis tests are negative. The hydrolysis of esculin is positive. Most strains were sensitive to meropenem, cefepime, tetracycline, gentamicin, trimethoprim-sulfamethoxazole, linezolid, daptomycin, and vancomycin but resistant to ceftriaxone, ciprofloxacin, erythromycin, and clindamycin. Intermediate to penicillin.
The type strain is MC-17XT (NBRC 115143T; CCTCC AB 2020199T). It has a DNA G+C content of 57.19%. It was isolated from a breast sample of a patient diagnosed with mastitis in Guangdong Provincial Hospital of Traditional Chinese Medicine in 2018.
DISCUSSION
Available literature and clinical evidence suggest a possible association between Corynebacterium infection and mastitis (7, 8, 10, 13). Among the Corynebacterium species, C. kroppenstedtii was the most common isolate reported in mastitis since its first report published in 2002 (7). Despite increasing data supporting their relationship, the role of C. kroppenstedtii in breast pathologies remains unclear, and further studies are urgently required. There was a report that impaired neutrophil responses to Nod2 agonist were associated with granulomatous mastitis due to corynebacteria (16). Recent emerging data also suggest that hyperprolactinemia may be an important risk factor of mastitis caused by C. kroppenstedtii (12, 14, 17). Prolactin was thought to modulate the inflammatory response and play a role in mastitis pathogenesis (18). It is noteworthy that 2 patients in our study had hyperprolactinemia and 1 patient had pituitary adenoma, which is a common cause of hyperprolactinemia (19). Most patients in our study received treatment of bromocriptine, a drug for hyperprolactinemia, with better curative effects and outcomes. The exact role of hyperprolactinemia in C. kroppenstedtii-related mastitis requires further studies. In our study, all 27 isolates were obtained from breast specimens of female patients with mastitis, supporting the potential pathogenic role of these strains in breast disease and emphasizing that we should pay more attention to the isolation of Corynebacterium species in breast specimens.
Isolation is necessary for the successful detection, accurate identification, and antibiotic susceptibility testing of this potential pathogen. The isolation of the lipophilic Corynebacterium can be challenging due to the fastidious growth. In our study, the incubation of breast specimens is always at least 72 h at 35°C with 5% CO2 to best recover the fastidious bacteria, which is also applicable to other clinical laboratories. On account of the lipophilic nature, the addition of a lipid component, such as Tween80, is often used to improve the culture yield. A medium that contains galactose, Tween 80, and fosfomycin has been specifically designed for the isolation of C. kroppenstedtii (20). The accurate identification of Corynebacterium species has become more reliable with the availability of MALDI-TOF MS and molecular techniques in the clinical laboratory (21). MALDI-TOF MS is a powerful tool to identify organisms to both genus and species levels rapidly and accurately and has been widely used in clinical laboratories. API kit assay is another method for rapid identification of fastidious bacteria, but it is not always suitable for species like C. kroppenstedtii. The API Coryne (bioMérieux, France) was designed in the early 1990s, but its databases have been updated only infrequently. Currently, gene sequencing is still the gold standard for microbial identification. In the present study, the use of phenotypic characterization can hardly identify and differentiate the two groups of C. kroppenstedtii-like isolates and the type strain. MALDI-TOF MS using the Vitek MS system and Bruker Biotyper system also failed to distinguish these isolates from C. kroppenstedtii. Based on the dendrogram produced by the Bruker Biotyper system, only C. kroppenstedtii-like group I was separated from the type strain. The partial 16S rRNA gene identity (99.6 to 99.9%) also could not distinguish these isolates from C. kroppenstedtii. These isolates, however, showed 96.2 to 98.1% partial rpoB gene and 97.3 to 97.7% partial fusA gene similarity to C. kroppenstedtii, demonstrating that rpoB and fusA genes sequencing allow more accurate identification for C. kroppenstedtii-like strains, as they are significantly more polymorphic than the 16S rRNA gene. Previous reports have shown that rpoB gene sequencing is a better option to identify and differentiate Corynebacterium species (22, 23). In this study, whole-genome sequencing was found to be the ultimate tool to distinguish and identify two new species belonging to the genus Corynebacterium.
There is no consensus for optimal management of Corynebacterium breast infection with treatment options such as surgical excision, corticosteroids, or antibiotics treatment. However, some reports showed that antibiotics play a marginal role in the natural treatment of this disease (7, 10, 24). Most patients in our study received traditional Chinese medicine, bromocriptine, and surgical debridement treatment. The traditional Chinese medicine decoction used for treatment is Kuijian Xiaoju Tang, which is composed of mainly Radix Bupleuri, Fructus Tribuli, Smilacis Glabrae Rhizoma, Gleditsiae Spina, Angelicae Dahuricae Radix, Radix Trichosanthis, Angelicae Sinensis Radix, Paeoniae Radix Rubra, Radix Rhizoma Glycyrrhizae, and Prunellae Spica. A small number of patients also received dexamethasone and antibiotic treatment like rifampicin, isoniazide, and ethambutol. Most patients had a good outcome after treatment. There is also recent clinical research suggesting that traditional Chinese medicine is effective in treating mastitis, indicating the potential advantage of traditional Chinese medicine (25, 26).
For C. kroppenstedtii breast infection, the data about antimicrobial treatment options are limited, and some studies have reported that C. kroppenstedtii isolates are susceptible to most antibiotics except for fosfomycin using the disk diffusion method or the E test (24, 27–29). Meanwhile, resistance to penicillin (10, 11), imipenem (14), erythromycin (12, 30), trimethoprim-sulfamethoxazole (28, 30), and clindamycin (10, 12, 30) has been reported. In this study, most C. kroppenstedtii-like isolates were susceptible to meropenem, cefepime, vancomycin, daptomycin, gentamicin, and linezolid. Isolates’ resistance to ceftriaxone, ciprofloxacin, erythromycin, tetracycline, clindamycin, and trimethoprim-sulfamethoxazole was found. There is a possibility of a difference in antibiotic susceptibility with different genospecies. Accordingly, correct species identification and antimicrobial susceptibility testing would ideally be performed for all isolates.
The resistance genes detected using the genomes of 27 clinical isolates included APH(3′)-Ia, APH(3′')-Ib, APH(6)-Id, erm(X), sul1, and tet(W), which are known from other corynebacteria (31–33). Comparing the prediction of antibiotic resistance genes with the results of in vitro susceptibility testing, the results can be summarized to the following points. (i) Twenty-three isolates were resistant to both erythromycin and clindamycin, while 21 isolates were found to have the corresponding antibiotic resistance gene erm(X). (ii) Tetracycline and sulfonamide antibiotic resistance genes were detected in 19 isolates and 8 isolates, respectively, but their susceptibility testing of tetracycline and trimethoprim-sulfamethoxazole was variable. (iii) Aminoglycoside resistance genes were detected in 22 isolates, but all of them were sensitive to gentamicin. (iv) None of the isolates were found to have β-lactam and quinolone resistance genes, but the response of all isolates to penicillin was intermediate; some isolates were intermediate or resistant to ceftriaxone, meropenem, and ciprofloxacin. By combining the in vitro susceptibility testing and the prediction of antibiotic resistance genes, we may be able to prioritize treatment with antibiotics other than macrolide, lincosamide, and tetracycline. These results indicate that not only are the antibiotic resistance and antibiotic susceptibility profiles of the same genospecies different, but the prediction of the antibiotic resistance gene profile is also different from the actual antibiotic resistance. This phenomenon may be related to differences in the expression of resistance genes in different strains. Antibiotic resistance genes exist not only in chromosomes but also in plasmids. Some isolates showing antibiotic resistance by in vitro susceptibility testing with no corresponding antibiotic resistance genes being detected may be due to the incompleteness of the draft genome. In addition, the inconsistencies between the antibiotic susceptibility of some isolates and the detected antibiotic resistance genes may be attributed to the low survival pressure of in vitro culture and the variation after serial passages. All these mentioned resistance genes are predicted for reference. The actual existence and expression of resistance genes need to be further verified by molecular methods. For clinical treatment, it may be feasible to classify through large amounts of actual antibiotic susceptibility data.
In conclusion, the C. kroppenstedtii-like clinical isolates associated with mastitis in our study represent two novel genospecies within the genus Corynebacterium, for which the names Corynebacterium parakroppenstedtii sp. nov. (C. kroppenstedtii-like group I) and Corynebacterium pseudokroppenstedtii sp. nov. (C. kroppenstedtii-like group II) are proposed. Our work is an important documentation of the identification of important potential pathogens for mastitis and provides hints that we should pay more attention to the isolation of Corynebacterium species in breast specimens. Our work also provides the antibiotic susceptibility profiles and suitable identification methods for these two novel species. By sharing the descriptions of two novel species as well as our experience in the identification, we hope that further epidemiological investigation of these strains can be performed and that their role in mastitis can be explored.
MATERIALS AND METHODS
Strains.
Twenty-seven C. kroppenstedtii-like isolates were isolated from clinical breast specimens in Guangdong Provincial Hospital of Traditional Chinese Medicine from 2017 to 2019, and the strain C. kroppenstedtii DSM 44385T was included in the present study as a reference type strain. Basic information of these C. kroppenstedtii-like isolates was outlined in Table 1. The purified isolates were routinely maintained by subculturing on Columbia blood agar plates at 35°C in a humidified atmosphere supplemented with 5% CO2 and stored as glycerol suspensions (30%, vol/vol) with 2% blood at −80°C.
Phenotypic testing and MALDI-TOF MS.
These isolates were initially identified by phenotypic characteristics and MALDI-TOF MS. Microscopic characteristics were determined by Gram stain. Lipid requirement was tested by comparing cultures grown on brain heart infusion broth and Columbia blood agar with cultures grown on these media supplemented with 1% Tween 80 after 3 days at 35°C with 5% CO2. Biochemical characterizations were performed using commercial API Coryne (bioMérieux, France) according to the manufacturer’s instructions. API Web was used to interpret the API codes. MALDI-TOF MS was carried out by both Bruker Biotyper system (BDAL Library; Bruker Daltonics, Germany) and Vitek MS system (V2.0; bioMérieux, France) according to the manufacturer’s instructions. The percentage similarities of identical mass peaks obtained by the Bruker Biotyper system (Bruker Daltonics, Germany) were calculated and used to generate dendrogram by the statistical toolbox of Matlab 7.1 (MathWorks Inc., USA) integrated into the MALDI Biotyper 2.0 software.
DNA sequencing and analysis.
Genomic DNAs were extracted using a bacterial genomic DNA extraction kit (AG, China) according to the manufacturer’s instructions. All clinical isolates were subjected to partial sequencing of the 16S rRNA, rpoB, and fusA genes. The protocols and primers of PCR amplification and Sanger sequencing were performed as described previously (22, 34, 35). The sequences were assembled using DNAMAN (version 7) software and compared with those related type strains on the NCBI BLAST website (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic trees were constructed using the neighbor-joining method with 1,000 bootstrap replications in the MEGA (version 7) software (36).
All isolates were subjected to whole-genome sequencing for obtaining a clear species differentiation. Genomic DNAs were extracted using a bacterial genomic DNA extraction kit (AG, China) and sequenced using Illumina NovaSeq PE150. The cleaned data were then assembled using SPAdes version 3.14.0. (37). The assembly was integrated with CISA software (38). The least scaffolds were selected to obtain the draft genome. For the construction of the phylogenomic tree, the marker genes were retrieved from the draft genomes of the 27 clinical isolates and C. kroppenstedtii DSM 44385T using AMPHORA2 (39). The 18 corresponding marker genes were listed as follows: frr, infC, nusA, pyrG, rplA, rplC, rplD, rplE, rplK, rplM, rplT, rpmA, rpsE, rpsJ, rpsK, rpsM, smpB, tsf. The sequences were aligned separately using MUSCLE (40) and concatenated by using a Perl script (https://github.com/nylander/catfasta2phyml). The protein-concatemer tree was established by comparing concatenated amino acids using the RAXML method (41). The tree was visualized through the online Tree of Life program version 6.5 (42). Genomic relatedness of these clinical isolates with the type strain was estimated using digital DNA–DNA hybridization (dDDH) and average nucleotide identity (ANI) based on the BLASTN algorithm (ANIb). dDDH values were calculated using the recommended settings (formula 2) of the Genome-to-Genome Distance Calculator 2.1 (43). ANIb values were calculated using JSpeciesWS (http://jspecies.ribohost.com/jspeciesws/).
Antimicrobial susceptibility testing.
Antibiotic susceptibility testing was carried out with the broth microdilution method by use of Corynebacterium ID&AST kit (TDR, China) according to the manufacturer’s instructions. The CLSI standard for determination and interpretation of antimicrobial MICs for Corynebacterium spp. was applied for the following antibiotics: penicillin, ceftriaxone, cefepime, meropenem, vancomycin, gentamicin, erythromycin, daptomycin, tetracycline, trimethoprim-sulfamethoxazole, ciprofloxacin, clindamycin, and linezolid (44).
Prediction of antibiotic resistance genes from the whole-genome sequencing data.
The Comprehensive Antibiotic Resistance Database (CARD; https://card.mcmaster.ca) (45) was used to predict antibiotic resistance genes in the whole-genome sequences. Resistance Gene Identifier (RGI 5.2.0, CARD 3.1.3) was used with open reading frame (ORF) prediction using Prodigal, homolog detection using DIAMOND, and strict significance based on CARD curated bitscore cutoffs. The “Perfect” and “Strict” default settings for sequence analysis were chosen as selection criteria.
Data availability.
The 16S rRNA, rpoB, and fusA gene sequences of the clinical isolates were deposited in GenBank with accession numbers MW819649 to MW819675 and MZ031080 to MZ031133 (Table 2). The Whole Genome Shotgun projects of the clinical isolates have been deposited at DDBJ/ENA/GenBank under the accession numbers JAFFSY000000000, JAEUWU000000000, JAGSNZ000000000, JAGSOA000000000, JAGSNY000000000, JAKKFA000000000, JAKJKO000000000 to JAKJLB000000000, JAKKNX000000000 to JAKKOA000000000, and JAKLTI000000000 to JAKLTK000000000 (Table 2).
ACKNOWLEDGMENTS
The work was supported by the National Science and Technology Fundamental Resources Investigation Program of China (2021FY100900). We thank Becton, Dickinson, and Company for the work of MALDI-TOF MS analysis. We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Ning Xu, Email: xu_ning21@163.com.
Pinghua Qu, Email: ququtdr@163.com.
Wendy A. Szymczak, Montefiore Medical Center and Albert Einstein College of Medicine
REFERENCES
- 1.Bernard K. 2012. The genus Corynebacterium and other medically relevant coryneform-like bacteria. J Clin Microbiol 50:3152–3158. doi: 10.1128/JCM.00796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soriano F, Fernández-Roblas R. 1988. Infections caused by antibiotic-resistant Corynebacterium group D2. Eur J Clin Microbiol Infect Dis 7:337–341. doi: 10.1007/BF01962333. [DOI] [PubMed] [Google Scholar]
- 3.Coyle MB, Lipsky BA. 1990. Coryneform bacteria in infectious diseases: clinical and laboratory aspects. Clin Microbiol Rev 3:227–246. doi: 10.1128/CMR.3.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard KA, Munro C, Wiebe D, Ongsansoy E. 2002. Characteristics of rare or recently described Corynebacterium species recovered from human clinical material in Canada. J Clin Microbiol 40:4375–4381. doi: 10.1128/JCM.40.11.4375-4381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ocal K, Dag A, Turkmenoglu O, Kara T, Seyit H, Konca K. 2010. Granulomatous mastitis: clinical, pathological features, and management. Breast J 16:176–182. doi: 10.1111/j.1524-4741.2009.00879.x. [DOI] [PubMed] [Google Scholar]
- 6.Collins MD, Falsen E, Akervall E, Sjöden B, Alvarez A. 1998. Corynebacterium kroppenstedtii sp. nov., a novel corynebacterium that does not contain mycolic acids. Int J Syst Bacteriol 48:1449–1454. doi: 10.1099/00207713-48-4-1449. [DOI] [PubMed] [Google Scholar]
- 7.Paviour S, Musaad S, Roberts S, Taylor G, Taylor S, Shore K, Lang S, Holland D. 2002. Corynebacterium species isolated from patients with mastitis. Clin Infect Dis 35:1434–1440. doi: 10.1086/344463. [DOI] [PubMed] [Google Scholar]
- 8.Taylor GB, Paviour SD, Musaad S, Jones WO, Holland DJ. 2003. A clinicopathological review of 34 cases of inflammatory breast disease showing an association between corynebacteria infection and granulomatous mastitis. Pathology 35:109–119. doi: 10.1080/0031302031000082197. [DOI] [PubMed] [Google Scholar]
- 9.Tauch A, Fernández-Natal I, Soriano F. 2016. A microbiological and clinical review on Corynebacterium kroppenstedtii. Int J Infect Dis 48:33–39. doi: 10.1016/j.ijid.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 10.Dobinson HC, Anderson TP, Chambers ST, Doogue MP, Seaward L, Werno AM. 2015. Antimicrobial treatment options for granulomatous mastitis caused by Corynebacterium species. J Clin Microbiol 53:2895–2899. doi: 10.1128/JCM.00760-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goh Z, Tan AL, Madhukhumar P, Yong WS. 2015. Recurrent Corynebacterium kroppenstedtii breast abscess in a young Asian female. Breast J 21:431–432. doi: 10.1111/tbj.12428. [DOI] [PubMed] [Google Scholar]
- 12.Kutsuna S, Mezaki K, Nagamatsu M, Kunimatsu J, Yamamoto K, Fujiya Y, Mawatari M, Takeshita N, Hayakawa K, Kato Y, Kanagawa S, Ohmagari N. 2015. Two cases of granulomatous mastitis caused by Corynebacterium kroppenstedtii infection in nulliparous young women with hyperprolactinemia. Intern Med 54:1815–1818. doi: 10.2169/internalmedicine.54.4254. [DOI] [PubMed] [Google Scholar]
- 13.Johnstone KJ, Robson J, Cherian SG, Wan Sai Cheong J, Kerr K, Bligh JF. 2017. Cystic neutrophilic granulomatous mastitis associated with Corynebacterium including Corynebacterium kroppenstedtii. Pathology 49:405–412. doi: 10.1016/j.pathol.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 14.Saraiya N, Corpuz M. 2019. Corynebacterium kroppenstedtii: a challenging culprit in breast abscesses and granulomatous mastitis. Curr Opin Obstet Gynecol 31:325–332. doi: 10.1097/GCO.0000000000000541. [DOI] [PubMed] [Google Scholar]
- 15.Tan C, Lu FI, Aftanas P, Tsang K, Mubareka S, Chan A, Kozak R. 2021. Whole genome sequence of Corynebacterium kroppenstedtii isolated from a case of recurrent granulomatous mastitis. IDCases 23:e01034. doi: 10.1016/j.idcr.2020.e01034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bercot B, Kannengiesser C, Oudin C, Grandchamp B, Sanson-Le Pors MJ, Mouly S, Elbim C. 2009. First description of NOD2 variant associated with defective neutrophil responses in a woman with granulomatous mastitis related to corynebacteria. J Clin Microbiol 47:3034–3037. doi: 10.1128/JCM.00561-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong SCY, Poon RWS, Chen JHK, Tse H, Lo JYC, Ng TK, Au JCK, Tse CWS, Cheung IYY, Yuk MT, Luk WK, Yuen KY. 2017. Corynebacterium kroppenstedtii is an emerging cause of mastitis especially in patients with psychiatric illness on antipsychotic medication. Open Forum Infect Dis 4:ofx096. doi: 10.1093/ofid/ofx096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutet P, Sulon J, Closset R, Detilleux J, Beckers JF, Bureau F, Lekeux P. 2007. Prolactin-induced activation of nuclear factor κB in bovine mammary epithelial cells: role in chronic mastitis. J Dairy Sci 90:155–164. doi: 10.3168/jds.S0022-0302(07)72617-6. [DOI] [PubMed] [Google Scholar]
- 19.Capozzi A, Scambia G, Pontecorvi A, Lello S. 2015. Hyperprolactinemia: pathophysiology and therapeutic approach. Gynecol Endocrinol 31:506–510. doi: 10.3109/09513590.2015.1017810. [DOI] [PubMed] [Google Scholar]
- 20.Wong SCY, Poon RWS, Foo CH, Ngan AHY, Tse H, Lam VCM, Leung THY, Wong CP, Cheng VCC, Chen JHK, Yuen KY. 2018. Novel selective medium for the isolation of Corynebacterium kroppenstedtii from heavily colonised clinical specimens. J Clin Pathol 71:781–786. doi: 10.1136/jclinpath-2017-204834. [DOI] [PubMed] [Google Scholar]
- 21.Alibi S, Ferjani A, Gaillot O, Marzouk M, Courcol R, Boukadida J. 2015. Identification of clinically relevant Corynebacterium strains by Api Coryne, MALDI-TOF-mass spectrometry and molecular approaches. Pathol Biol (Paris) 63:153–157. doi: 10.1016/j.patbio.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Khamis A, Raoult D, La Scola B. 2004. rpoB gene sequencing for identification of Corynebacterium species. J Clin Microbiol 42:3925–3931. doi: 10.1128/JCM.42.9.3925-3931.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khamis A, Raoult D, La Scola B. 2005. Comparison between rpoB and 16S rRNA gene sequencing for molecular identification of 168 clinical isolates of Corynebacterium. J Clin Microbiol 43:1934–1936. doi: 10.1128/JCM.43.4.1934-1936.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson MG, Leal S, Plongla R, Leone PA, Gilligan PH. 2016. The brief case: recurrent granulomatous mastitis due to Corynebacterium kroppenstedtii. J Clin Microbiol 54:1938–1941. doi: 10.1128/JCM.03131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xue JX, Ye B, Liu S, Cao SH, Bian WH, Yao C. 2020. Treatment efficacy of Chuang Ling Ye, a traditional Chinese herbal medicine compound, on idiopathic granulomatous mastitis: a randomized controlled trial. Evid Based Complement Alternat Med 2020:6964801. doi: 10.1155/2020/6964801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Li J, Hu XJ. 2020. Postoperative Yanghe decoction regimen improves outcomes for idiopathic granulomatous mastitis: a retrospective cohort study. Medicine 99:e23136. doi: 10.1097/MD.0000000000023136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riegel P, Liégeois P, Chenard MP, Mathelin C, Monteil H. 2004. Isolations of Corynebacterium kroppenstedtii from a breast abscess. Int J Med Microbiol 294:413–416. doi: 10.1016/j.ijmm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 28.Le Flèche-Matéos A, Berthet N, Lomprez F, Arnoux Y, Le Guern AS, Leclercq I, Burguière AM, Manuguerra JC. 2012. Recurrent breast abscesses due to Corynebacterium kroppenstedtii, a human pathogen uncommon in Caucasian women. Case Rep Infect Dis 2012:120968. doi: 10.1155/2012/120968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hagemann JB, Essig A, Herrmann M, Liebold A, Quader MA. 2015. Early prosthetic valve endocarditis caused by Corynebacterium kroppenstedtii. Int J Med Microbiol 305:957–959. doi: 10.1016/j.ijmm.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Poojary I, Kurian A, V A J, Devapriya J D, M A T. 2017. Corynebacterium species causing breast abscesses among patients attending a tertiary care hospital in Chennai, South India. Infect Dis (Lond) 49:528–531. doi: 10.1080/23744235.2017.1296184. [DOI] [PubMed] [Google Scholar]
- 31.Rosato AE, Lee BS, Nash KA. 2001. Inducible macrolide resistance in Corynebacterium jeikeium. Antimicrob Agents Chemother 45:1982–1989. doi: 10.1128/AAC.45.7.1982-1989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schröder J, Maus I, Meyer K, Wördemann S, Blom J, Jaenicke S, Schneider J, Trost E, Tauch A. 2012. Complete genome sequence, lifestyle, and multi-drug resistance of the human pathogen Corynebacterium resistens DSM 45100 isolated from blood samples of a leukemia patient. BMC Genomics 13:141. doi: 10.1186/1471-2164-13-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tauch A, Sandbote J. 2014. The family Corynebacteriaceae. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes. Springer, Berlin, Heidelberg. [Google Scholar]
- 34.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Busse HJ, Kleinhagauer T, Glaeser SP, Spergser J, Kämpfer P, Rückert C. 2019. Classification of three corynebacterial strains isolated from the Northern Bald Ibis (Geronticus eremita): proposal of Corynebacterium choanae sp. nov., Corynebacterium pseudopelargi sp. nov., and Corynebacterium gerontici sp. nov. Int J Syst Evol Microbiol 69:2928–2935. doi: 10.1099/ijsem.0.003580. [DOI] [PubMed] [Google Scholar]
- 36.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin SH, Liao YC. 2013. CISA: contig integrator for sequence assembly of bacterial genomes. PLoS One 8:e60843. doi: 10.1371/journal.pone.0060843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu M, Scott AJ. 2012. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28:1033–1034. doi: 10.1093/bioinformatics/bts079. [DOI] [PubMed] [Google Scholar]
- 40.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clinical and Laboratory Standards Institute. 2015. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria, 3rd ed. CLSI guideline M45. Clinical and Laboratory Standards Institute, Wayne, PA. [DOI] [PubMed] [Google Scholar]
- 45.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen AV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM01372-21_Supp_1_seq4.pdf, PDF file, 0.1 MB (134KB, pdf)
Data Availability Statement
The 16S rRNA, rpoB, and fusA gene sequences of the clinical isolates were deposited in GenBank with accession numbers MW819649 to MW819675 and MZ031080 to MZ031133 (Table 2). The Whole Genome Shotgun projects of the clinical isolates have been deposited at DDBJ/ENA/GenBank under the accession numbers JAFFSY000000000, JAEUWU000000000, JAGSNZ000000000, JAGSOA000000000, JAGSNY000000000, JAKKFA000000000, JAKJKO000000000 to JAKJLB000000000, JAKKNX000000000 to JAKKOA000000000, and JAKLTI000000000 to JAKLTK000000000 (Table 2).