Abstract
An Achromobacter xylosoxydans strain showing broad-spectrum resistance to β-lactams (including carbapenems) and aminoglycosides was isolated at the University Hospital of Verona (Verona, Italy). This strain was found to produce metallo-β-lactamase activity and to harbor a 30-kb nonconjugative plasmid, named pAX22, carrying a blaVIM-1 determinant inserted into a class 1 integron. Characterization of this integron, named In70, revealed an original array of four gene cassettes containing, respectively, the blaVIM-1 gene and three different aminoglycoside resistance determinants, including an aacA4 allele, a new aph-like gene named aphA15, and an aadA1 allele. The aphA15 gene is the first example of an aph-like gene carried on a mobile gene cassette, and its product exhibits close similarity to the APH(3′)-IIa aminoglycoside phosphotransferase encoded by Tn5 (36% amino acid identity) and to an APH(3′)-IIb enzyme from Pseudomonas aeruginosa (38% amino acid identity). Expression of the cloned aphA15 gene in Escherichia coli reduced the susceptibility to kanamycin and neomycin as well as (slightly) to amikacin, netilmicin, and streptomycin. Characterization of the 5′ and 3′ conserved segments of In70 and of their flanking regions showed that In70 belongs to the group of class 1 integrons associated with defective transposon derivatives originating from Tn402-like elements. The structure of the 3′ conserved segment indicates the closest ancestry with members of the In0-In2 lineage. In70, with its array of cassette-borne resistance genes, can mediate broad-spectrum resistance to most β-lactams and aminoglycosides.
VIM-1 and VIM-2 are new metallo-β-lactamases, 90% identical to each other at the sequence level, that have recently been identified in carbapenem-resistant Pseudomonas aeruginosa nosocomial isolates from the Mediterranean area (12, 17, 21, 27). These enzymes exhibit a very broad substrate specificity (including carbapenems and most other β-lactams) and were found to be encoded by determinants carried on mobile gene cassettes inserted into integrons (12, 17). Integrons and gene cassettes are elements that participate in a powerful site-specific recombination system, operating in procaryotic genomes, that plays a major role in spreading of antibiotic resistance genes in the clinical setting (see references 8 and 19 for reviews). For the above reasons, the blaVIM-1 and blaVIM-2 genes could become relevant resistance determinants in the clinical setting.
The blaVIM-1 gene was originally cloned from a P. aeruginosa strain that was isolated at the University Hospital of Verona (Verona, Italy), having caused an outbreak in the Intensive Care Unit of that hospital (14). In that strain, the blaVIM-1 gene was found to be part of a gene cassette carried on a class 1 integron that was only partially characterized (12) and was located on the bacterial chromosome.
Here we report on the finding of a plasmid-borne blaVIM-1 determinant in an Achromobacter xylosoxydans isolate from the same hospital and on the characterization of the blaVIM-1-containing integron carried by that plasmid.
MATERIALS AND METHODS
Bacterial strains and genetic vectors.
A. xylosoxydans subsp. denitrificans AX-22 was isolated in 1998 from a urine specimen of an inpatient at the University Hospital of Verona, Verona, Italy, and identified according to standard procedures (23). Escherichia coli DH5α (GIBCO-BRL, Gaithersburg, Md.) was used as the host for natural and recombinant plasmids. E. coli MKD-135 (argH rpoB18 rpoB19 recA rpsL) (kindly provided by G. Kholodii, Institute for Molecular Genetics, Russian Academy of Sciences, Moscow, Russia) and P. aeruginosa 10145/3 (an rpoB his derivative of strain ATCC 10145T) were used as recipients in conjugation experiments. Bacteria were always grown aerobically at 37°C unless otherwise specified. The plasmids pBC-SK and pBluescript KS (Stratagene, Inc., La Jolla, Calif.) were used as cloning vectors.
In vitro susceptibility testing.
Antibiotics were from commercial sources. MICs were determined by a macrodilution broth method (15), using Mueller-Hinton (MH) broth (Difco Laboratories, Detroit, Mich.) and a bacterial inoculum of 5 × 105 CFU per tube. Results were recorded after incubation for 18 h at 37°C and interpreted according to the guidelines of the National Committee for Clinical Laboratory Standards (16).
β-lactamase assays.
Carbapenemase activity in crude cell extracts was assayed spectrophotometrically as described previously (12), using 150 μM imipenem as the substrate. One unit was defined as the amount of activity hydrolyzing 1 nmol of substrate per min under the assay conditions. Inhibition of the carbapenemase activity by EDTA was assayed as described previously (12). Protein concentration in solution was assayed by the method of Bradford with a commercial kit (Protein Assay; Bio-Rad, Richmond, Calif.), with bovine serum albumin used as a standard. Analytical isoelectric focusing for detection of β-lactamases was performed as described previously (12).
DNA analysis methodology.
Basic procedures for DNA extraction, analysis, and manipulation were performed as described by Sambrook et al. (22). Hybridization experiments were carried out using nitrocellulose filters (Schleicher & Schuell, Dassel, Germany) and a probe labeled with 32P by the random priming technique (22). The probe was a PCR-generated amplicon comprising the entire blaVIM-1 open reading frame (ORF) (12). P. aeruginosa VR-143/97 (12) and P. aeruginosa ATCC27853 were included as positive and negative hybridization controls, respectively, in colony blot hybridization. Conjugation experiments were performed on MH agar plates. The initial donor/recipient ratio was 0.1. Mating plates were incubated at 30°C for 14 h. E. coli transconjugants were selected on MH agar containing kanamycin (25 μg/ml) plus rifampin (400 μg/ml). P. aeruginosa transconjugants were selected on MH agar containing tobramycin (16 μg/ml) plus rifampin (400 μg/ml). The detection sensitivity of the assay was ≥1 × 10−8 transconjugants/recipient. Electroporation of E. coli was performed with a Gene-Pulser apparatus (Bio-Rad) according to the manufacturer's instructions. DNA sequences were determined on both strands of plasmid templates as described previously (12). Similarity searches against sequence databases were performed using an updated version of the BLAST program at the National Center for Biotechnology Information server (http://www.ncbi.nlm.nih.gov/). Computer analysis of the sequence data and multiple-sequence alignment were performed using the Wisconsin Package (version 8.1; Genetics Computer Group Inc., Madison, Wis.) and the Clustal W program at the server of the Italian European Molecular Biology Laboratory Node of Bari (area.ba.cnr.it).
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to the EMBL, GenBank, and DDBJ sequence databases and assigned the accession numbers AJ278514 and AJ278515.
RESULTS AND DISCUSSION
Identification of A. xylosoxydans clinical isolate producing a plasmid-encoded VIM-like metallo-β-lactamase.
A. xylosoxydans AX22 exhibited broad-spectrum resistance to β-lactams and aminoglycosides (Table 1). The β-lactam resistance pattern (including piperacillin, ceftazidime, and carbapenem resistance) was unusual for this species (5, 25), and the high-level carbapenem resistance suggested the production of an acquired carbapenemase. In fact, carbapenemase activity was detected in a crude extract of AX22 (specific activity, 184 ± 12 U/mg of protein), and this activity was reduced (>80%) after incubation of the crude extract with 2 mM EDTA, suggesting the presence of a metallo-β-lactamase determinant.
TABLE 1.
MICs of various antimicrobial agents for A. xylosoxydans AX22, E. coli DH5α(pAX22), and E. coli DH5α(pMLR-aph70)a
| Antimicrobial agent | MIC (μg/ml) for strain:
|
|||
|---|---|---|---|---|
| AX22 | DH5α(pAX22) | DH5α(pMLR-aph70) | DH5αb | |
| Ampicillin | >256 | >256 | —c | 1 |
| Mezlocillin | >256 | >256 | — | 1 |
| Piperacillin | >256 | 256 | — | 0.5 |
| Cefotaxime | >256 | 128 | — | ≤0.12 |
| Ceftazidime | >256 | 256 | — | 0.12 |
| Cefepime | >256 | 32 | — | ≤0.12 |
| Imipenem | 256 | 4 | — | 0.12 |
| Meropenem | 128 | 2 | — | ≤0.12 |
| Aztreonam | 64 | 0.25 | — | 0.25 |
| Gentamicin | >256 | — | 0.5 | 0.5 |
| Tobramycin | >256 | — | 0.5 | 0.5 |
| Netilmicin | >256 | — | 0.5 | 0.25 |
| Amikacin | 64 | — | 1 | 0.5 |
| Kanamycin | — | — | 16 | 0.5 |
| Streptomycin | — | — | 4 | 2 |
| Neomycin | — | — | 4 | 0.5 |
| Spectinomycin | — | — | 8 | 8 |
The susceptibility of E. coli DH5α is also shown for comparison.
MICs of aminoglycosides for E. coli DH5α(pBluescript KS) were identical to those for DH5α.
—, not assayed.
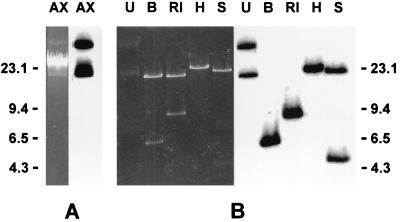
A colony blot hybridization showed that DNA from AX22 was recognized by a blaVIM-1 probe (data not shown). Agarose gel electrophoresis of the total DNA extracted from AX22 revealed the presence of plasmid DNA which was recognized by the blaVIM-1 probe in a Southern blot hybridization (Fig. 1). The blaVIM-containing plasmid, named pAX22, was purified and estimated to be approximately 30 kb in size, based on restriction analysis with various enzymes (Fig. 1). Electroporation of E. coli DH5α with the purified plasmid preparation yielded ampicillin-resistant transformants which contained a plasmid apparently identical to pAX22 (data not shown). DH5α(pAX22) produced carbapenemase activity (specific activity of crude extract, 202 ± 14 U/mg of protein) and, compared to DH5α, exhibited a decreased susceptibility to several β-lactams (Table 1). An isoelectric focusing analysis of a crude extract of DH5α(pAX22) revealed the presence of a single band of β-lactamase activity with an isoelectric pH of 5.1 (data not shown), suggesting that the VIM-like enzyme was the only β-lactamase encoded by pAX22. In fact, the pattern of β-lactam susceptibility shown by DH5α(pAX22) was consistent with that previously observed for the same E. coli host carrying a cloned copy of the blaVIM-1 gene on a multicopy plasmid vector (12).
FIG. 1.
(A) Results of agarose gel electrophoresis of genomic DNA extracted from A. xylosoxydans AX22 are shown at left, and results of Southern blot hybridization with the blaVIM-1 probe are shown at right. AX, AX22. (B) Results of agarose gel electrophoresis of the plasmid DNA preparation from AX22, either undigested or digested with various enzymes are shown at left, and results of Southern blot hybridization with the blaVIM-1 probe are shown at right. U, undigested; B, BamHI; RI, EcoRI; H, HindIII; S, SalI. DNA size standards are reported in kilobase pairs on the sides.
The potential for conjugational transfer of pAX22 was examined in diparental matings using either E. coli MKD-135 or P. aeruginosa 10145/3 as the recipient, but in neither case was conjugational transfer detected. A similar behavior was also observed with other medium-sized plasmids containing integrons carrying cassette-borne metallo-β-lactamase genes (11, 17). It would be interesting to further investigate the biology of these plasmids and their potential role in the dissemination of resistance determinants.
Structure of blaVIM-containing integron carried by plasmid pAX22.
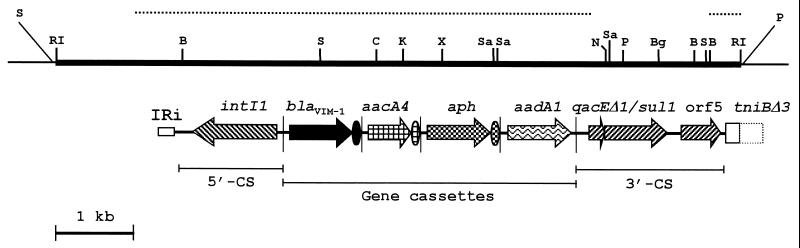
The 8.6-kb EcoRI fragment of pAX22 containing the blaVIM determinant (Fig. 1) was subcloned into plasmid pBC-SK to obtain the recombinant plasmid pLUP-86R (Fig. 2). Restriction mapping and partial sequence analysis of this fragment revealed that the blaVIM determinant was inserted into a class 1 integron which carries an original array of four gene cassettes and was named In70 (Fig. 2).
FIG. 2.
Physical map of the 8.6-kb EcoRI insert of plasmid pLUP-86R. The thick line represents the DNA insert; the thin lines represent vector sequences. Abbreviations: S, SalI; RI, EcoRI; B, BamHI; Bg, BglII; C, ClaI; K, KpnI; N, NcoI; P, PstI; Sa, SacII; X, XhoI. The horizontal dotted lines above the map indicate the sequenced regions. The structure of In70 and the locations of the various integron components are shown below the map. ORFs are indicated by arrows (truncated when not complete), the 59-be's of the gene cassettes are indicated by ovals, and the cassette boundaries are indicated by vertical bars. The 3′-CS has been sequenced only at its junctions, while the structure of the internal region has been deduced by comparison of the restriction map with that of the 3′-CS of other integrons (9).
The 5′ conserved segment (5′-CS) of In70 contains an intI1 allele typical of class 1 integrons (19) and is identical to that of In0 (2) except for a single G→T transversion in the −35 hexamer of the P1 promoter (formerly Pant). Therefore, the configuration of the P1 promoter of In70 is hybrid, with a TTGACA −35 hexamer typical of the strong P1 promoter and a TAAGCT −10 hexamer typical of the weak P1 promoter (13), and is identical to that of the promoter previously found in the integron from plasmid pBWH301 (4) but different from the configurations of the promoters found in most other integrons (reference 13 and references therein, as well as results of a BLAST search on updated sequence databases). Compared to the blaVIM-1-containing integron cloned from P. aeruginosa VR-143/97 (12), the 5′-CS of In70 exhibits the same G→T transversion in the −35 hexamer that differentiates the P1 promoter of In70 from that of In0 (see above), as well as a G→C transversion in the region between the −35 and −10 hexamers of the same promoter. A similar finding may indicate cassette exchange rather than integron transfer in the dissemination process of the blaVIM-1 cassette.
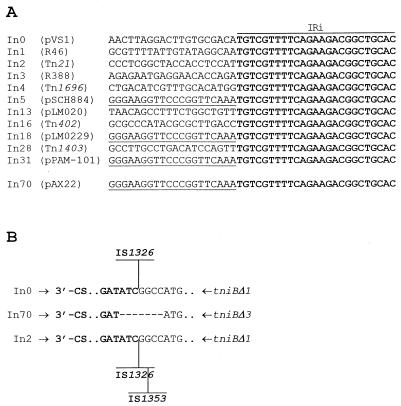
At the left-hand end, the 5′-CS of In70 is bounded by a 25-bp IRi sequence identical to that associated with other integrons of the same family (9, 18). The nucleotide sequence flanking IRi was found to be identical to that flanking IRi of In5, of In18, and of In31 but different from that flanking the IRi of other integrons (Fig. 3A).
FIG. 3.
(A) Comparison of the IRi (boldfaced) and flanking region of In70 with those of other integrons of the same family. Identical flanking sequences are underlined. The sequences of In1, In2, In3, In4, and In5 (9); of In13, In16, and In18 (18); and of In31 (11) have all been reported previously. (B) Nucleotide sequence at the junction between the 3′-CS (boldfaced) of In70 and the truncated tni module, and comparison with the corresponding regions of In0 and In2 (3). The sites of insertion of IS1326 in In0 and in In2 and that of IS1353 in In2 are indicated.
The 3′ conserved segment (3′-CS) of In70 closely resembles that of In0 and In2 (3) but directly merges with a truncated tni module typical of Tn402-like elements, with no insertion sequences present at the 3′-CS-tniBΔ junction (Fig. 2). Compared to In0 and In2 (3), the 3′-CS-tniBΔ junction of In70 exhibits a small deletion, apparently involving both sides (Fig. 3B), which could have been generated following an imprecise excision of IS1326. For this reason, the truncated tniB allele present in In70 was named tniBΔ3.
Altogether, these findings indicate that In70 is another member of the group of class 1 integrons associated with defective transposon derivatives originating from Tn402-like elements. Among them, In70 apparently shares the closest ancestry with members of the In0-In2 lineage (2, 3). However, the finding of an IRi-flanking region which is different from that of either In0 or In2 but identical to those of integrons of the In5-In31 lineage (11) raises the question of the mobility of the defective transposon carrying the integron.
Gene cassettes in In70.
In70 contains four gene cassettes, each including a resistance gene. The first cassette carries a blaVIM-1 determinant (Fig. 2) and is identical to the blaVIM-1 cassette from P. aeruginosa VR-143/97, the VIM-1 index strain previously isolated in the same hospital (12). This finding suggests a common origin for the two determinants, although the original source remains unknown. It also indicates that blaVIM-1, similar to blaVIM-2 (17), can even be found on plasmids.
The second cassette of In70 contains an aacA4 allele (Fig. 2) encoding an AAC(6′)-II aminoglycoside acetyltransferase identical to that encoded by the aacA4 allele from plasmid pIP1855 of Pseudomonas fluorescens BM2687 (10).
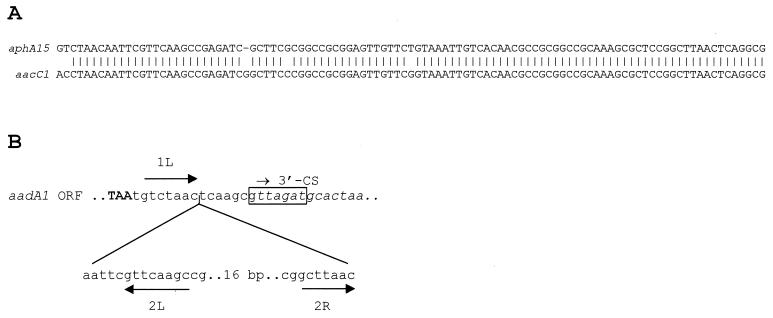
The third cassette of In70 is original and contains a 795-bp ORF encoding a protein which exhibits the closest sequence similarities with an APH(3′)-IIb aminoglycoside phosphotransferase from P. aeruginosa (7) (38% identity) and with the APH(3′)-IIa enzyme encoded by Tn5 (1) (36% identity). The similarity to the above proteins is evident over the entire sequence and includes the three highly conserved motifs and the invariant or highly conserved residues typical of the APH enzymes (24) (Fig. 4). The gene was named aphA15. To analyze whether the aphA15 product was functional in conferring aminoglycoside resistance, a 1-kb KpnI-SacII fragment of In70 including the gene (Fig. 2) was subcloned into the plasmid vector pBluescript KS to obtain recombinant plasmid pMLR-aph70, in which the aphA15 determinant was located downstream of the Plac promoter flanking the vector polylinker. E. coli DH5α(pMLR-aph70) showed a significant reduction of kanamycin and neomycin susceptibility, while the MICs of other aminoglycosides, including gentamicin, tobramycin, netilmicin, amikacin, streptomycin, and spectinomycin, were apparently unaffected or only slightly increased (Table 1), revealing a pattern consistent with that of APH(3′) enzymes (24). The 59-base element (59-be [also named attC]) of the aphA15 cassette of In70 was found to be nearly identical to that of the aacC1 cassette inserted in In4 of Tn1696 (Fig. 5A). A similar situation of gene cassettes containing different resistance determinants but closely related 59-be's has also been reported in other cases (20) and supports the view that gene cassettes can be assembled from pools of different resistance genes and 59-be's and/or that 59-be's can be shuffled between different cassettes. It should be noted that association with gene cassettes has been typically reported for aac and aad but not for aph aminoglycoside resistance determinants (7, 19, 24). Identification of this new cassette demonstrated that also aph-like determinants can be cassette-borne and can exploit the potential of the integron-mediated recombination system for dissemination.
FIG. 4.
Amino acid sequence comparison among the putative APH(3′) enzyme encoded by the third cassette of In70 [APH(3′)-In70] and its closest relatives, the APH(3′)-IIa enzyme encoded by Tn5 [APH(3′)-IIa] (1) and the APH(3′)-IIb enzyme from P. aeruginosa SCH84043005 [APH(3′)-IIb] (7). Identical residues are each indicated by an asterisk; conservative amino acid substitutions are each indicated by a dot. The three highly conserved motifs that are known to be involved in catalytic activity of the APH enzymes are overlined, and the invariant or highly conserved residues typical of these enzymes (24) are shaded.
FIG. 5.
(A) Comparison of the 59-be of the aphA15 cassette of In70 (aphA15) with that of the aacC1 cassette of In4 from Tn1696 (19). Identical residues are indicated by vertical bars. (B) Nucleotide sequence at the junction between the aadA1 cassette and the the 3′-CS internal boundary of In70, showing the structure of the deleted 59-be of the aadA1 cassette. The termination codon of the aadA1 coding sequence is capitalized; the sequence of the 3′-CS is italicized, and the conserved recombination core site located at the 3′-CS internal boundary is boxed. The 1L internal core site of the 59-be (26) is overlined by an arrow. The deleted region, in comparison with the 59-be of the aadA1 cassette of In2 (19), is indicated below the sequence and the 2L and 2R internal core sites (26) are underlined by arrows.
The fourth cassette of In70 contains an aadA1 allele encoding an AAD(3") aminoglycoside adenylyl transferase (24) and a partially deleted 59-be (Fig. 5B). Compared to the 59-be of the aadA1 cassette of In2, the deletion involves a 40-bp region precisely located between the 1L (retained) and 2R (deleted) core sites of the 59-be (26) (Fig. 5B), suggesting that it was likely generated by an unusual integrase-mediated recombination event between 1L and 2R.
Concluding remarks.
In70 is a new class 1 integron that can mediate broad spectrum β-lactam and aminoglycoside resistance owing to the simultaneous carriage of a gene cassette encoding the VIM-1 metallo-β-lactamase, which can degrade virtually all β-lactams except monobactams (6), and of three gene cassettes encoding three different modifying enzymes whose combined substrate spectrum includes most aminoglycosides. This fact, together with the plasmid-borne nature, could render In70 a powerful vehicle for spreading multidrug resistance among gram-negative nosocomial pathogens. We are currently analyzing the structures of the blaVIM-containing integrons present on the chromosome of P. aeruginosa VR-143/97 (12) and of other VIM-1-producing P. aeruginosa isolates (21) to compare their structures with that of In70.
ACKNOWLEDGMENTS
This work was supported by the European research network on metallo-β-lactamases within the TMR program (contract no. FMRX-CT98-0232) and by grant no. 9906404271 from MURST (PRIN99).
REFERENCES
- 1.Beck E, Ludwig G, Auerswald E A, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 2.Bissonnette L, Roy P H. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J Bacteriol. 1992;174:1248–1257. doi: 10.1128/jb.174.4.1248-1257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown H J, Stokes H W, Hall R M. The integrons In0, In2, and In5 are defective transposon derivatives. J Bacteriol. 1996;178:4429–4437. doi: 10.1128/jb.178.15.4429-4437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bunny K L, Hall R M, Stokes H W. New mobile gene cassettes containing an aminoglycoside resistance gene, aacA7, and a chloramphenicol resistance gene, catB3, in an integron in pBWH301. Antimicrob Agents Chemother. 1995;39:686–693. doi: 10.1128/AAC.39.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fass R J, Barnishan J, Solomon M C, Ayers L W. In vitro activities of quinolones, β-lactams, tobramycin, and trimethoprim-sulfamethoxazole against nonfermentative gram-negative bacilli. Antimicrob Agents Chemother. 1996;40:1412–1418. doi: 10.1128/aac.40.6.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franceschini N, Caravelli B, Docquier J-D, Galleni M, Frére J-M, Amicosante G, Rossolini G M. Purification and biochemical characterization of the VIM-1 metallo-β-lactamase. Antimicrob Agents Chemother. 2000;44:3003–3007. doi: 10.1128/aac.44.11.3003-3007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hächler H, Santanam P, Kayser F H. Sequence and characterization of a novel chromosomal aminoglycoside phosphotransferase gene, aph(3′)-IIb, in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:1254–1256. doi: 10.1128/aac.40.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 9.Hall R M, Brown H J, Brookes D E, Stokes H W. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J Bacteriol. 1994;176:6286–6294. doi: 10.1128/jb.176.20.6286-6294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lambert T, Ploy M-C, Courvalin P. A spontaneous mutation in the aac(6′)-Ib gene results in altered substrate specificity of aminoglycoside 6′-N-acetyltransferase of a Pseudomonas fluorescens strain. FEMS Microbiol Lett. 1994;115:297–304. doi: 10.1111/j.1574-6968.1994.tb06654.x. [DOI] [PubMed] [Google Scholar]
- 11.Laraki N, Galleni M, Thamm I, Riccio M L, Amicosante G, Frère J-M, Rossolini G M. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob Agents Chemother. 1999;43:890–901. doi: 10.1128/aac.43.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauretti L, Riccio M L, Mazzariol A, Cornaglia G, Amicosante G, Fontana R, Rossolini G M. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob Agents Chemother. 1999;43:1584–1590. doi: 10.1128/aac.43.7.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lévesque C, Brassard S, Lapointe J, Roy P H. Diversity and relative strength of tandem promoters for the antibiotic resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 14.Mazzariol A, Cornaglia G, Piccoli P, Lauretti L, Riccio M L, Rossolini G M, Fontana R. Carbapenem-hydrolyzing β-lactamases in Pseudomonas aeruginosa. Eur J Clin Microbiol Infect Dis. 1999;18:455–456. doi: 10.1007/s100960050320. [DOI] [PubMed] [Google Scholar]
- 15.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard. NCCLS document M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Supplement M100–S9. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 17.Poirel L, Naas T, Nicholas D, Collet L, Bellais S, Cavallo J-D, Nordmann P. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000;44:891–897. doi: 10.1128/aac.44.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rådström P, Sköld O, Swedberg G, Flensburg J, Roy P H, Sundström L. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J Bacteriol. 1994;176:3257–3268. doi: 10.1128/jb.176.11.3257-3268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 20.Recchia G D, Hall R M. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 1997;5:389–394. doi: 10.1016/S0966-842X(97)01123-2. [DOI] [PubMed] [Google Scholar]
- 21.Rossolini G M, Riccio M L, Cornaglia G, Pagani L, Lagatolla C, Selan L, Fontana R. Carbapenem-resistant Pseudomonas aeruginosa with acquired blaVIM metallo-β-lactamase determinants, Italy. Emerg Infect Dis. 2000;6:312–313. doi: 10.3201/eid0603.000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Schreckenberger P C, von Graevenitz A. Acinetobacter, Achromobacter, Alcaligenes, Moraxella, Methylobacterium, and other nonfermentative gram-negative rods. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 539–560. [Google Scholar]
- 24.Shaw K J, Rather P N, Hare R S, Miller G H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol Rev. 1993;57:138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spangler S K, Visalli M A, Jacobs M R, Appelbaum P C. Susceptibilities of non-Pseudomonas aeruginosa gram-negative nonfermentative rods to ciprofloxacin, ofloxacin, levofloxacin, d-ofloxacin, sparfloxacin, ceftazidime, piperacillin, piperacillin-tazobactam, trimethoprim-sulphamethoxazole, and imipenem. Antimicrob Agents Chemother. 1996;40:772–775. doi: 10.1128/aac.40.3.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stokes H W, O'Gorman D B, Recchia G D, Parsekhian M, Hall R M. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol Microbiol. 1997;26:731–745. doi: 10.1046/j.1365-2958.1997.6091980.x. [DOI] [PubMed] [Google Scholar]
- 27.Tsakris A, Pournaras S, Woodford N, Palepou M-F I, Babini G S, Douboyas J, Livermore D M. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J Clin Microbiol. 2000;38:1290–1292. doi: 10.1128/jcm.38.3.1290-1292.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]