ABSTRACT
The food we eat not only nourishes our bodies but also provides nutrients to the bacteria living in our guts. Gut bacterial communities are known to be affected by many factors, including diet and bowel cleansing, but the impacts of vegetarian and omnivore diets on fecal bacterial composition are still uncertain. In this study, we analyzed the bacterial compositions of fecal samples from vegetarians and omnivores 5 to 7 days after bowel cleansing, and we correlated specific dietary constituents with the relative abundances of specialized fecal bacteria. A total of 46 participants (23 vegetarians and 23 omnivores) were recruited. All participants underwent standard bowel cleansing before colonoscopy screening. Fecal samples were collected from each participant 5 to 7 days after bowel cleansing, and the fecal microbiota compositions were analyzed with next-generation sequencing. Sixteen participants also provided an image-based dietary record for nutritional assessment. No major differences between dietary groups were observed in terms of fecal bacterial richness, alpha diversity, or beta diversity. A minority of potential pathobionts tended to be elevated in omnivores compared to vegetarians, whereas potential probiotic species tended to be higher in the vegetarians. Detailed dietary assessments further revealed that the plant- and animal-derived proteins may oppositely modulate the relative abundances of pathobionts Bilophila and Lachnoclostridium. However, these results were not statistically significant after multiple-comparison correction. These results suggest that specialized probiotic and pathobiont microbiota constituents are sensitive to the plant- or animal-derived dietary components ingested by vegetarians and omnivores after bowel cleansing.
IMPORTANCE Dietary pattern and food choice are associated with expansion of gut pathobionts and risk for metabolic and colonic disease. However, the effects of dietary interventions on intestinal microbiota remain unclear. After bowel cleansing, potential pathobionts and probiotic bacteria were increased in omnivores and vegetarians, respectively. The pathobionts Bilophila and Lachnoclostridium were oppositely modulated by dietary animal and plant protein. From a clinical perspective, fecal pathobionts that may indicate risk for metabolic and colonic disease can potentially be modulated with dietary interventions.
KEYWORDS: gut microbiome, pathobionts, diet, metabolic disease, colorectal cancer, inflammatory bowel disease
INTRODUCTION
Dietary patterns may exert distinct nutritional effects on both the host and their gut bacteria (1). It is well known that the intestinal and fecal microbial composition responds continually and rapidly to many factors, including diet (1–3), environmental exposures (4), the host genome (5, 6), lifestyle (3, 7), hygiene (8, 9), and use of antibiotics (10, 11). Furthermore, both the intestinal microbiota per se and microbiota-generated metabolites have been shown to mediate interactions between diet and disease (1, 12–14). While vegetarian diets are often perceived by the public as being healthier overall than omnivore diets, the effects of vegetarian diets on intestinal microbiota and disease remain to be fully elucidated (15–17).
Some positive effects of vegetarian diets have been previously reported (1, 18). Early research on vegetarian diets focused on the direct effects of certain dietary components on physiological functions and pathological disease progression, especially with regard to high fiber, potassium and magnesium contents, antioxidant properties of vitamins, and the protective capacities of numerous phytochemicals in plant-based foods (18, 19). Additionally, the roles of certain macronutrients in vegetarian diets, such as plant-based protein, have been associated with longevity and lower risks of mortality from obesity (20), type 2 diabetes (T2D) (21), cardiovascular disease (CVD) (22), and cancer (15, 22). Over the last decade, major advancements have been made in our understanding of intestinal bacteria, and many effects of vegetarian diets on gut microbiota and their metabolites have been reported (1, 16, 17). Furthermore, it has been suggested that there is a positive association between animal protein intake and risks of CVD (23) and colorectal cancer (CRC) (24); these increased risks may be the consequence of high levels of intestinal bacteria-produced hydrogen sulfide (H2S) and trimethylamine-N-oxide (TMAO). However, contradictory results and inconsistent outcomes have been reported in comparisons of vegetarian and omnivore dietary effects on intestinal flora (16, 25). These conflicting results may be attributable to many factors, such as the length of time for dietary intervention, geographical variations, interindividual variability, or the methods applied for feces collection. At present, it therefore remains unclear precisely how intestinal microbiota respond to dietary interventions.
The profile of human gut microbiota is shaped from birth in a process that continues throughout the individual’s life span. The microbial composition in the colon is dynamic and changes rapidly due to many influences (26), including bowel cleansing prior to colonic endoscopic examination, bowel surgery, and fecal microbiota transplantation (27–29). Additionally, different bowel cleansing methods have been shown to influence the fecal microbiota to various degrees (27–30). Previous studies showed that bowel cleansing with polyethylene glycol (PEG) results in an immediate decrease of total microbial load and alters biodiversity; the microbiota are largely restored within 14 days to 1 month, accompanied by slight changes in composition (27, 29). Notably, a small-scale study suggested that only a minority of subjects are susceptible to PEG-induced perturbations (28). Although most studies on vegetarian diets suggest some beneficial effects on the intestinal microbiota (1, 31), the results do not indicate with any certainty which specific bacterial populations are subject to change (16, 25) or how bowel cleansing might affect the intervention. It is reasonable to expect that bowel cleansing prior to collection of fecal samples may reveal more precise associations between dietary pattern and intestinal microbiota composition, as the fecal bacterial changes would not be masked by historical population dynamics. Moreover, a recent report suggested that a constitutive bowel enema during vegetarian dietary intervention may produce a more sustained beneficial effect on the gut microbiota in patients with Parkinson’s disease (32). Thus, bowel cleansing may be useful as a clinical means to accelerate alterations in intestinal microbiota. Nevertheless, whether dietary composition can impact the reestablishment of gut microbiota after bowel cleansing remains unknown.
In the current study, fecal samples were collected from volunteers on vegetarian and omnivore diets 5 to 7 days after bowel cleansing for CRC screening. The compositions of fecal microbiota were determined for each participant. Moreover, detailed assessments of diet were performed on a subset of 16 participants. We then compared the bacterial compositions and relative bacterial abundances between diet groups, and we further evaluated correlations between specialized bacteria and specific dietary constituents.
RESULTS
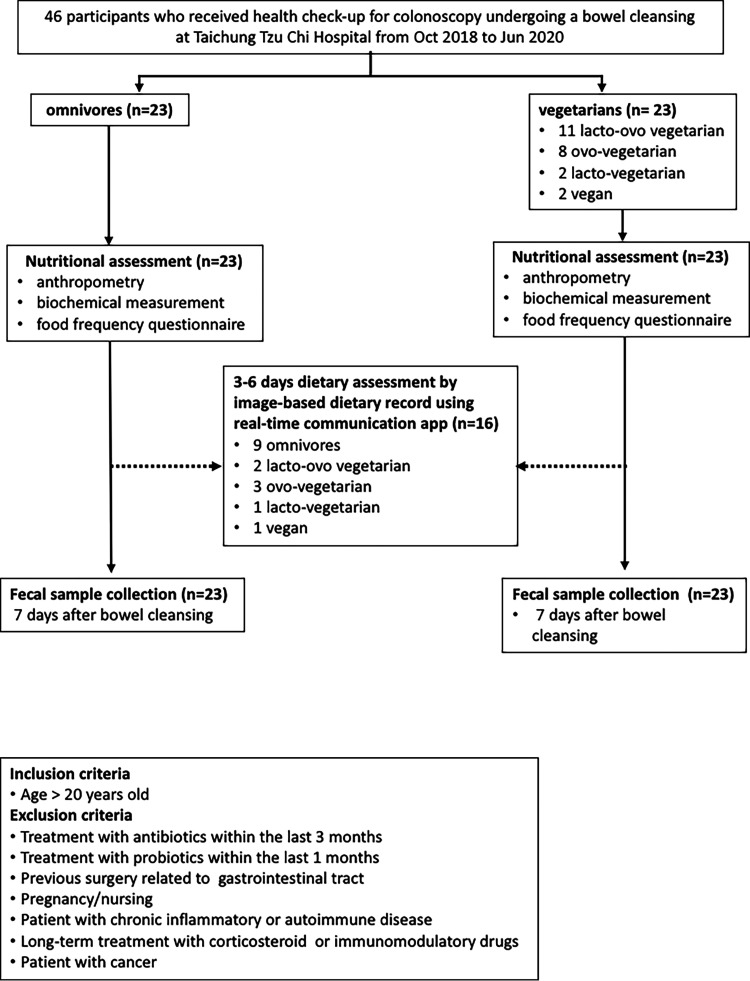
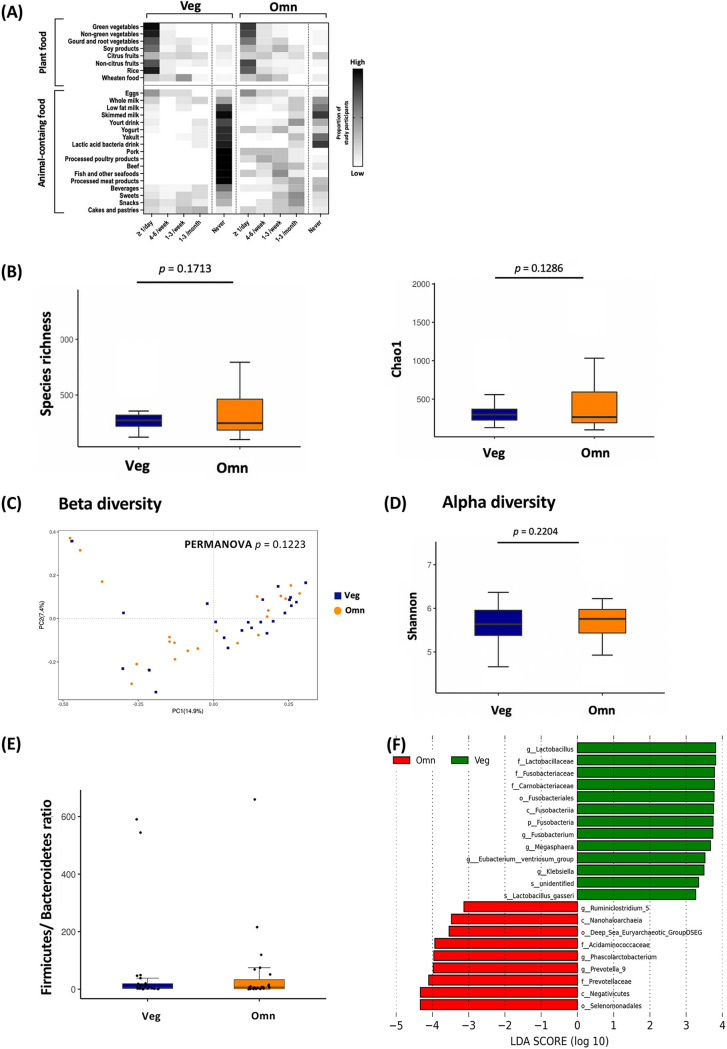
By analyzing the microbiota in fecal samples from participants, we found that the vegetarian diet contributed to only slight differences in fecal bacterial richness and diversity. A total of 23 vegetarian and 23 omnivore participants were enrolled in this study, and 16 participants completed an additional image-based dietary assessment. A flowchart summarizing the participants and study design is shown in Fig. 1. Food frequency questionnaire (FFQ)-based long-term dietary assessments for all participants are summarized in Fig. 2A. Vegetarians consumed mostly plant-based foods, with a small proportion of eggs and dairy products. The omnivores consumed various animal-derived foods with modest amounts of plant-based food. It is worth noting that vegetarians had a higher intake of soy-based foods than omnivores. A comparison of biochemical values revealed significantly higher levels of total cholesterol in omnivores (Table 1).
FIG 1.
Flow diagram of participants included in this study.
FIG 2.
Vegetarians and omnivores displayed no major differences in fecal microbiota composition after bowel cleansing. (A) Heatmap of food intake frequency in vegetarians (Veg) and omnivores (Omn) based on the food frequency questionnaire (FFQ). Each heatmap square indicates the proportion of study participants consuming a specific food with the indicated frequency. Darker shade indicates more people consumed that category of food. On average, vegetarian diets included no meat, low dairy/eggs, and high plant-based food. Rows, food category; columns, intake frequency. (B to E) No significant differences between diet groups were found in fecal bacterial species richness (B), beta diversity by principal coordinate analysis calculated using Bray-Curtis distance (PERMANOVA, P = 0.1223) (C), alpha diversity (D), or Firmicutes/Bacteroidetes ratio (E). (F) Histogram of the linear discriminant analysis (LDA) score demonstrated different taxa in fecal microbiota between Veg (top) and Omn (bottom).
TABLE 1.
Summary of participants’ demographic and metabolic characteristics
| Characteristic | Value [mean ± SD or no. (%)] for: |
P valuea | q value | |
|---|---|---|---|---|
| Vegetarians (n = 23) | Omnivores (n = 23) | |||
| Age (yr) | 58.83 ± 17.68 | 51.04 ± 4.95 | ||
| <65 | 16 (73.9) | 22 (95.7) | ||
| ≥65 | 6 (26.1) | 1 (4.3) | ||
| Gender | ||||
| Male | 10 (43.5) | 10 (43.5) | ||
| Female | 13 (56.5) | 13 (56.5) | ||
| Body mass index (kg/m2) | 23.18 ± 0.56 | 23.52 ± 2.24 | 0.70 | 0.94 |
| Body fat (%) | 29.53 ± 11.67 | 30.47 ± 12.02 | 0.62 | 1.00 |
| Male | 23.46 ± 1.06 | 26.33 ± 0.78 | 0.17 | 1.00 |
| Female | 34.20 ± 4.95 | 36.66 ± 11.88 | 0.76 | 0.92 |
| Blood pressure (mm Hg) | ||||
| Systolic | 124.09 ± 4.95 | 121.39 ± 14.85 | 0.45 | 0.95 |
| Diastolic | 75.17 ± 19.80 | 73.09 ± 9.90 | 0.47 | 0.85 |
| Plasma biochemical datab | ||||
| Fasting blood glucose (mg/dL) | 100.57 ± 12.73 | 95.00 ± 0.71 | 0.21 | 1.00 |
| HbA1c (%) | 5.79 ± 0.71 | 5.48 ± 0.21 | 0.07 | 1.00 |
| Triglyceride (mg/dL) | 119.57 ± 44.55 | 144.22 ± 69.30 | 0.25 | 1.00 |
| Total cholesterol (mg/dL) | 169.22 ± 36.06 | 202.17 ± 28.99 | 0.08* | 0.23 |
| LDL-C (mg/dL) | 107.13 ± 25.46 | 127.17 ± 17.68 | 0.08 | 1.00 |
| AST (IU/L) | 23.61 ± 10.61 | 21.30 ± 3.54 | 0.31 | 0.91 |
| ALT (IU/L) | 27.09 ± 0.71 | 28.87 ± 2.12 | 0.73 | 0.92 |
| BUN (mg/dL) | 9.04 ± 2.12 | 9.43 ± 4.95 | 0.64 | 1.00 |
| Creatinine | 0.80 ± 0.14 | 0.79 ± 0.35 | 0.86 | 0.98 |
| Uric acid (mg/dL) | 4.98 ± 0.71 | 5.07 ± 0.92 | 0.77 | 0.96 |
The P value is the comparison between vegetarian and omnivore volunteers using Student’s t test. *, P < 0.05.
HbA1c, glycated hemoglobin; LDL-C, low-density lipoprotein-cholesterol; AST, aspartate aminotransferase; ALT, alanine aminotransferase; BUN, blood urea nitrogen.
The fecal bacterial profiles of the vegetarian and omnivore participants did not differ considerably in terms of species richness (Fig. 2B), beta diversity (Fig. 2C), alpha diversity (Fig. 2D), or Firmicutes/Bacteroidetes ratio (Fig. 2E). These results are similar to previous findings (12, 23, 33). In total, we identified 22 fecal bacterial taxa that were mostly distinct between vegetarians and omnivores according to the linear discriminant analysis (LDA) score (LDA score > 3.0) (Fig. 2F; Table S6 in the supplemental material). However, there were no significant results among them after multiple-comparison correction.
Relative abundances of specific fecal bacteria differ between vegetarians and omnivores.
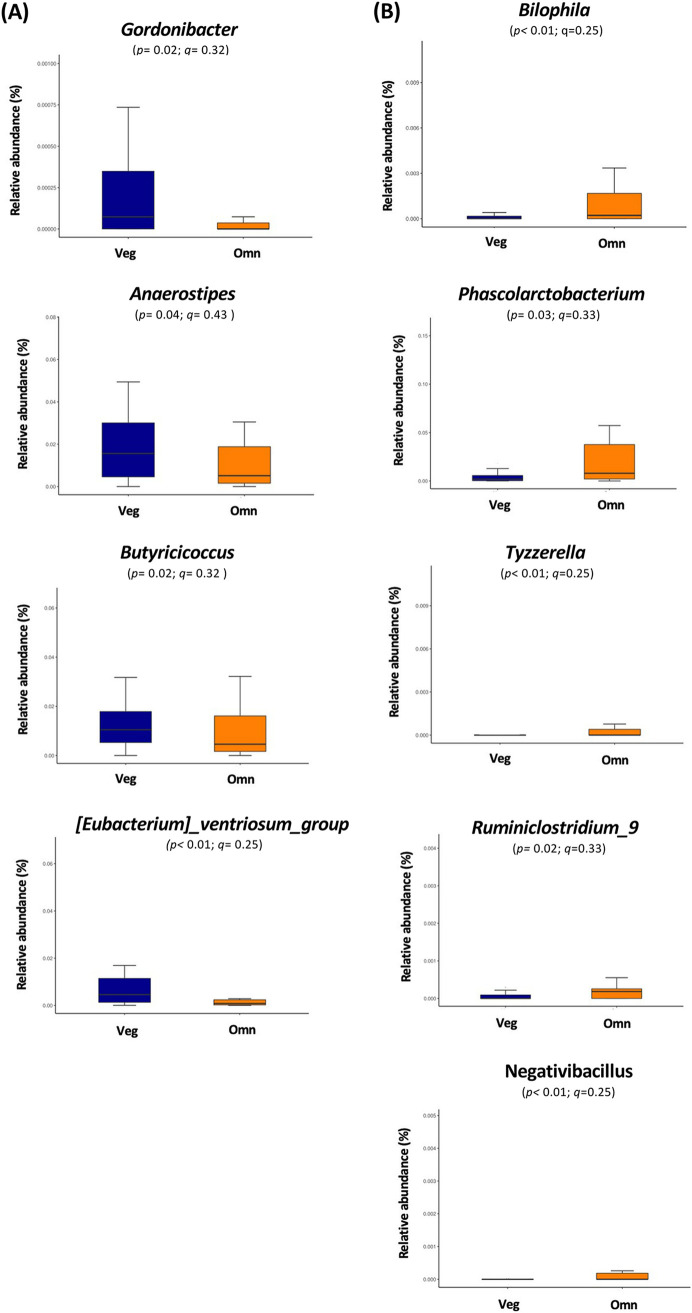
Next, we made a closer examination of the microbiota profiles of all participants by using metagenomeSeq. The differences between vegetarians and omnivores in the relative abundances of specific bacteria at the genus level after bowel cleansing were not statistically significant. The relative abundances of fecal Gordonibacter, Anaerostipes, Butyricicoccus, and (Eubacterium) ventriosum_group tended to be higher in the vegetarians than in omnivores (Fig. 3A). In contrast, the relative abundances of fecal Bilophila, Phascolarctobacterium, Tyzzerella, Ruminiclostridium_9, and Negativibacillus tended to be higher in omnivores than in vegetarians (Fig. 3B). However, there were no significant differences after correction for multiple comparisons.
FIG 3.
MetagenomeSeq revealed differences in certain bacteria between vegetarians and omnivores after bowel cleansing. MetagenomeSeq identified the relative abundances of fecal bacterial genus levels in vegetarians (Veg) and omnivores (Omn) (A), including Gordonibacter, Anaerostipes, Butyricicoccus, and ventriosum_group. The analysis also identified genera that were higher in Omn than Veg (B), including Bilophila, Phascolarctobacterium, Tyzzerella, Ruminiclostridium_9, and Negativibacillus. Adjusted P values are expressed as q values.
Relative abundances of particular fecal bacteria are sensitive to the levels of plant- or animal-derived proteins in the diet.
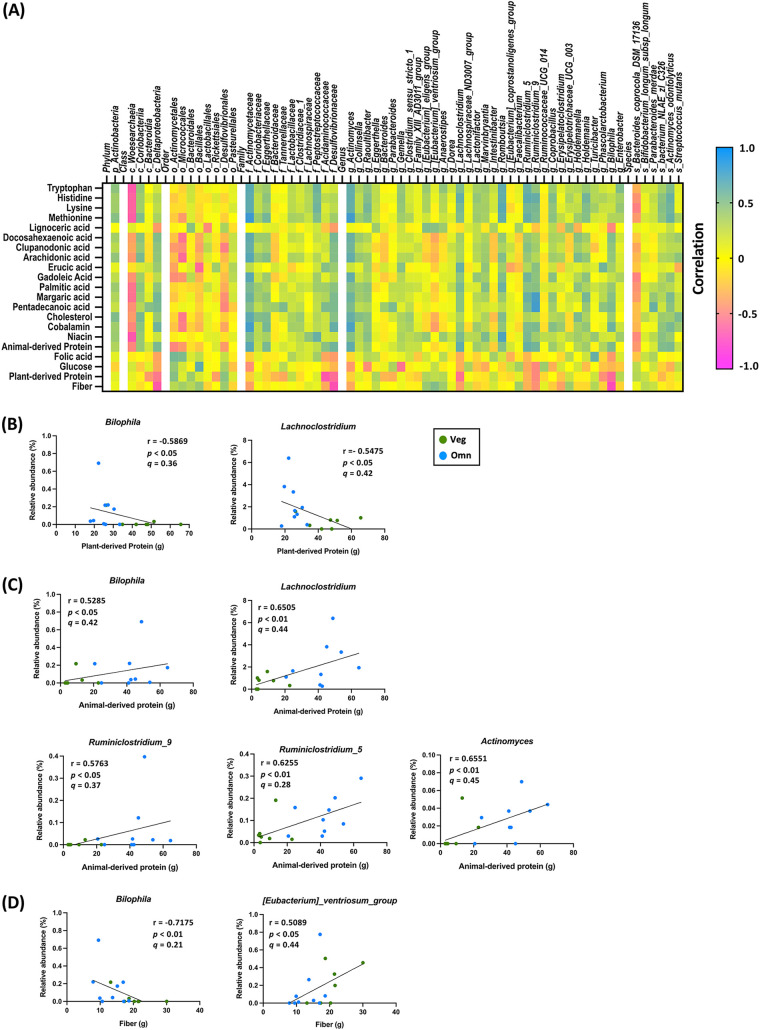
A total of 16 participants (7 vegetarians and 9 omnivores) further completed an average 4-day image-based dietary assessment after bowel cleansing. We analyzed the daily intakes of major nutrients, monosaccharides, fatty acids, amino acids, vitamins, and minerals (Tables S1 to S5). Among these nutrients, intakes of fiber, plant-derived protein, glucose, folic acid, and lignoceric acid were significantly higher in vegetarians than in omnivores. The intakes of animal-derived protein, cholesterol, niacin, cobalamin, several fatty acids, and amino acids were higher for omnivores than for vegetarians. We further analyzed the correlations between nutrients that differed between groups and the relative abundances of fecal bacteria. The results are shown in a heatmap (Fig. 4A); Spearman correlation coefficients, original P values, and adjusted P values (q values) are shown in Table S7. The multiple-comparison correction was performed separately for different taxonomic ranks.
FIG 4.
Certain fecal bacteria were sensitive to distinct dietary nutrients and displayed opposite correlations with plant and animal protein after bowel cleansing. A total of 16 participants, including 7 vegetarians (Veg) and 9 omnivores (Omn), provided image-based dietary records. For nutrients with intakes that were significantly different between the two dietary groups, correlation analyses with fecal bacterial abundances were performed. (A) Heatmap summarizes Spearman correlations for pairwise comparisons of nutrient intake (rows) and bacterial taxa (columns). (B) Correlations between plant-derived protein and relative abundances of Bilophila and Lachnoclostridium. (C) Correlations between animal-derived protein and relative abundances of Bilophila, Lachnoclostridium, Actinomyces, Ruminiclostridium_5, and Ruminiclostridium_9. (D) Correlations between dietary fiber and relative abundances of Bilophila and (Eubacterium) ventriosum_group genera.
Fecal abundances of Bilophila and Lachnoclostridium are correlated with dietary plant and animal proteins.
The detailed dietary assessments revealed no difference in total protein intake. Expectedly, however, the vegetarians had a significantly higher intake of plant protein, while the omnivores had a significantly higher intake of animal protein (Table S1).
To investigate the contributions of distinct protein sources to the bacterial signature, we first correlated dietary plant- and animal-derived protein with bacterial taxa in vegetarians and omnivores. The relative abundances of fecal Bilophila and Lachnoclostridium were inversely correlated with dietary plant protein intakes (r = −0.5869 and −0.5475, respectively; P < 0.05; q not significant) (Fig. 4B) and positively correlated with dietary animal protein intakes (r = 0.5285, P < 0.05 and r = 0.6505, P < 0.01, respectively; q not significant) (Fig. 4C). In addition, the relative abundance of fecal Bilophila was also negatively correlated with dietary fiber (r = −0.7175, q < 0.05) (Fig. 4D). The abundances of other fecal bacteria, including Actinomyces (r = 0.6551, q < 0.05), Ruminiclostridium_5 (r = 0.6255, P < 0.05, q not significant), and Ruminiclostridium_9 (r = 0.6777, q < 0.05), were positively correlated with dietary animal protein (Fig. 4C). (Eubacterium) ventriosum_group was positively correlated with dietary fiber (r = 0.5089, q not significant) (Fig. 4D).
DISCUSSION
In this study, we collected fecal samples from vegetarians and omnivores 5 to 7 days after bowel cleansing. We found that some potential pathobiont species were relatively higher in omnivores, while some potential probiotic species were higher in vegetarians. Moreover, the pathobiont species Bilophila and Lachnoclostridium were positively correlated with dietary animal protein and negatively correlated with dietary plant protein sources. Changes in an individual’s fecal bacterial composition before and after bowel cleansing were not evaluated due to a lack of precleansing samples. Furthermore, the small sample size in this study led to a lack of significant differences according to adjusted P (q) value. Nevertheless, our results implied that a minority of fecal bacteria might be differentially modulated by vegetarian and omnivore diets, since no significant differences in major microbiota composition were found between groups. The health effects of these specific changes remain unknown and warrant further study. Understanding how distinct fecal bacteria differ in individuals with different dietary intakes after bowel cleansing will be important for the clinical application of personalized nutrition in the era of individualized medicine.
In general, vegetarian diets are purported to be rich in fiber and low in protein and fat. We conducted a thorough dietary assessment for 16 participants and showed that the fiber intake of vegetarians was significantly higher than that of omnivores. However, no significant differences were observed in total protein or fat intake between groups. Compared to vegetarian diets in Western countries, vegetarians in Taiwan have been shown to ingest more plant-derived protein from soy products (34, 35). In the bacterial analysis, we observed no significant changes in species richness, beta diversity, alpha diversity, or Firmicutes/Bacteroidetes ratio. These findings are compatible with previous studies (12, 17, 23, 33) and underscore the importance of performing detailed analysis of individual diet compositions (e.g., image-based records) when studying the relationship between diet and intestinal bacteria. Furthermore, the effects of bowel cleansing should be taken into account when assessing dietary effects on intestinal microbiota. In one recent small cohort study, the gut microbiomes of healthy subjects and patients with Parkinson’s disease were evaluated after a combination of bowel cleansing (oil enema for 8 consecutive days) and dietary intervention (ovo-lacto vegetarian diet plus short-chain fatty acid for 14 days). Remarkably, the bowel cleansing was associated with a positive effect on the gut microbiome and decreased drug dosage in a 1-year follow-up compared to dietary intervention alone (32). While our study would have benefited from larger sample sizes to generate better evidence, the results still provide novel information about how combinations of dietary intervention and bowel cleansing may be applied in the clinic.
Our metagenomeSeq analysis revealed that the fecal loads of Gordonibacter, Anaerostipes, Butyricicoccus, and ventriosum_group in vegetarians tended to be higher than those in omnivores. It has been reported that Butyricicoccus pullicaecorum can metabolize ellagitannin and ellagic acid into urolithins in the intestine, which may have positive effects on health (36–38). Additionally, low relative fecal abundances of butyrate-producing Anaerostipes and ventriosum_group have been reported in patients with CRC (39, 40) compared to their abundances in controls, while reduced richness of Butyricicoccus was found in patients with inflammatory bowel disease (IBD) (41). Butyricicoccus pullicaecorum is a species of the Butyricicoccus genus that has probiotic potential, demonstrated by its amelioration of colitis in a rat model of inflammatory bowel disease (IBD) and strengthening of the epithelial barrier in a Caco-2 cell model, partially via its butyrate-producing ability (41). On the basis of these findings, we suggest that the higher relative abundances of the genera Gordonibacter, Anaerostipes, Butyricicoccus, and ventriosum_group in the feces of vegetarians may positively affect physiological homeostasis. In contrast, the consumption of meat has been associated with the occurrence of CRC, although the detailed mechanism is still under investigation (42). Our results showed that the abundances of the genera Bilophila, Phascolarctobacterium, Tyzzerella, Ruminiclostridium_9, and Negativibacillus in the feces of omnivores tended to be higher than those in vegetarians. Moreover, fecal Actinomyces, Ruminiclostridium_5, and Ruminiclostridium_9 showed a positive correlation with animal protein intakes in omnivores. Previous studies demonstrated that fecal Bilophila (43), Phascolarctobacterium (43, 44), Ruminiclostridium_9 (45), Actinomyces (43), Ruminiclostridium_5 (46), and Ruminiclostridium_9 (45) were relatively enriched in patients with CRC. In addition, the relative abundance of fecal Tyzzerella is associated with lifetime CVD risk (47). Collectively, these data suggest that dietary choice may render an individual prone or resistant to certain diseases by influencing the levels of specific gut bacteria. However, specific criteria for defining healthy versus unhealthy gut flora are still lacking.
In our study, we found that two potential pathobionts, Bilophila and Lachnoclostridium, were sensitive to distinct dietary protein sources. In particular, Bilophila was the only fecal bacterium responsive to dietary fiber, plant-derived protein, and animal-derived protein. In the intestine, Bilophila produces H2S and may be involved in the pathogenesis of CRC (14, 43), as well as IBD (2, 48). Notably, the previous studies of mouse models showed that the expansion of Bilophila wadsworthia synergizes with a high-fat diet to aggravate metabolic dysfunction (49) and IBD (48). Therefore, Bilophila has been suggested as an indicator bacterium for diet-related diseases. Additionally, increased fecal Lachnoclostridium has been linked to the formation of visceral fat (50) and progression of diabetic peripheral neuropathy (51). A recent study in Hong Kong identified a Lachnoclostridium gene as a stool-based noninvasive biomarker for early detection of CRC (13). People from Hong Kong, Taiwan, and other East Asian areas share similar eating habits, so further investigations may assess whether the increase in fecal Lachnoclostridium caused by dietary animal protein is associated with the increased risk of CRC in these locations. Our findings also suggest that fecal Lachnoclostridium might be a useful predictor of metabolic-related disease or CRC occurrence that could be modified with dietary intervention.
In conclusion, our study provides evidence that dietary plant and animal proteins oppositely modulate the fecal abundance of Bilophila and Lachnoclostridium in vegetarians and omnivores after bowel cleansing. Understanding the correlations between specific nutrients and specialized fecal bacteria under certain conditions will pave the way for the development of precision nutrition approaches that involve the use of specialized fecal bacteria as biomarkers to guide interventions.
MATERIALS AND METHODS
Study population and dietary information.
In total, 46 healthy volunteers, including 23 vegetarians (Veg; 11 lacto-ovo vegetarians, 8 ovo-vegetarians, 2 lacto vegetarians, and 2 vegans) and 23 omnivores (Omn), who were scheduled to receive colonoscopy screening at Taichung Tzu Chi Hospital were recruited to the study (Fig. 1). The inclusion and exclusion criteria are indicated in Fig. 1. The Institutional Review Board (IRB) at Taichung Tzu Chi Hospital (REC107-05) approved the human research protocol, and all participants gave written informed consent before the study.
Fecal sample collection after bowel cleansing.
This study was conducted from October 2018 to June 2020 in Taichung Tzu Chi Hospital. The demographic information for participants is summarized in Table 1. In total, 46 subjects (20 males, 26 females) with an average age of 55 years (range 29 to 67 years) were evaluated. Bowklean powder (10 mg picosulfate sodium, 3.5 g magnesium oxide, and 12 g citric acid anhydrous) was used for colon cleaning (52–54). Briefly, Bowklean solution was fresh prepared by dissolving one sachet of the powder in 150 mM water and stirring for 5 min. Before the day of colonoscopy, each subject consumed the first Bowklean powder sachet at 5:00 p.m. (17:00), followed by 1,250 mL of clear liquids within 5 h. On the day of the colonoscopy, a second sachet of Bowklean was consumed 5 h prior to the colonoscopy, followed by 750 mL of clear liquids within 2 h. Fecal samples were collected once from each participant between 5 and 7 days after the colonoscopy.
Dietary assessment. (i) FFQ.
All participants performed a dietary assessment using a semiquantitative food frequency questionnaire (FFQ), adopted from the Nutrition and Health Survey in Taiwan. This survey has been validated in a previous study and can reliably identify major nutrients in the diets of vegetarians and omnivores in Taiwan (55, 56).
(ii) Image-based dietary assessment.
Sixteen participants (7 vegetarians and 9 omnivores) further participated in the image-based dietary assessment. Participants took photographs of their food with a smart phone before each meal and sent the photographs to a dietitian through the Taiwan social communication app LINE for nutritional evaluation. Participants were encouraged to use a Tzu Chi transparent oval bowl (Fig. S1) provided by the project to facilitate accurate nutritional assessment. Participants had an average diet record of 4 days. Follow-up dietary assessment was performed by two independent dietitians, using the Taiwan Food Ingredient Database (Ministry of Health and Welfare, Taiwan, Republic of China) (57).
Microbiota analysis.
Methods for fecal sample collection and genomic DNA extraction, 16S rRNA gene amplicon sequencing, and MiSeq-based high-throughput sequencing are described in detail in the supplemental material.
Statistical analysis. (i) Fecal samples.
The abundances of species at various taxonomic levels were compared among groups using differential-abundance analysis with a zero-inflated Gaussian (ZIG) log-normal model. The analysis was implemented with the “fitFeatureModel” function of the Bioconductor metagenomeSeq package (58). Welch’s t test was performed with STAMP software (version 2.1.3) (59). Bacterial richness, alpha diversity, and Firmicutes/Bacteroidetes ratio between groups were examined by Wilcoxon rank sum tests. Permutational multivariate analysis of variance (PERMANOVA) was used to analyze the significance of differences in beta diversity (PCoA). Statistically significant biomarkers were identified from linear discriminant analysis effect size (LEfSe) analysis (60). The LEfSe method involves conducting a nonparametric Kruskal-Wallis test and Wilcoxon rank sum test to determine the bacterial taxa with significantly different relative abundances between groups. Those taxa with an LDA score (log10) of >3.0 were represented in a bar plot, but does not mean that they are statistically significantly different. The data analyses were conducted by Biotools Co., Ltd. (New Taipei City, Taiwan).
(ii) Dietary parameters.
When comparing the nutrient intakes of the two groups (vegetarian and omnivore), the nonparametric Wilcoxon rank sum test was used. Spearman’s correlation was used to examine associations between specific nutrients and fecal bacteria. Statistical tests were performed using GraphPad Prism (version 8).
For all statistical analyses, the significance level α was set as 0.05, and adjusted P values (q values) were calculated for multiple comparisons using the Benjamini-Hochberg false discovery rate (FDR) correction (61).
Data availability.
The sequencing data have been submitted to the NCBI SRA database under accession number PRJNA786088.
ACKNOWLEDGMENTS
This work was supported by the Taichung Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation, under grant number TTCRD107-07, by the ENABLE project, National Chung Hsing University, under grant number 109ST001E, and by the Taiwan Ministry of Science and Technology under grant number 109-2320-B-005-007.
We thank Ching-Yao Wu and Tzu-Yu Chen, undergraduate students in the Department of Food Science and Biotechnology, National Chung Hsing University, for all their efforts in graphic preparation.
Footnotes
Supplemental material is available online only.
Contributor Information
Ching-Ying Huang, Email: cyh@nchu.edu.tw.
Wei-Hua Chen, Huazhong University of Science and Technology.
REFERENCES
- 1.Zmora N, Suez J, Elinav E. 2019. You are what you eat: diet, health and the gut microbiota. Nat Rev Gastroenterol Hepatol 16:35–56. doi: 10.1038/s41575-018-0061-2. [DOI] [PubMed] [Google Scholar]
- 2.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhong H, Penders J, Shi Z, Ren H, Cai K, Fang C, Ding Q, Thijs C, Blaak EE, Stehouwer CDA, Xu X, Yang H, Wang J, Wang J, Jonkers D, Masclee AAM, Brix S, Li J, Arts ICW, Kristiansen K. 2019. Impact of early events and lifestyle on the gut microbiota and metabolic phenotypes in young school-age children. Microbiome 7:2. doi: 10.1186/s40168-018-0608-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claus SP, Guillou H, Ellero-Simatos S. 2016. The gut microbiota: a major player in the toxicity of environmental pollutants? NPJ Biofilms Microbiomes 2:16003. doi: 10.1038/npjbiofilms.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen J, Sears CL. 2019. Impact of the gut microbiome on the genome and epigenome of colon epithelial cells: contributions to colorectal cancer development. Genome Med 11:11. doi: 10.1186/s13073-019-0621-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van Dullemen HM, Ter Steege RWF, Huttenhower C, Dijkstra G, Xavier RJ, Festen EAM, Wijmenga C, Zhernakova A, Weersma RK. 2018. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut 67:108–119. doi: 10.1136/gutjnl-2016-312135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta VK, Paul S, Dutta C. 2017. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol 8:1162. doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azad MB, Konya T, Maughan H, Guttman DS, Field CJ, Sears MR, Becker AB, Scott JA, Kozyrskyj AL. 2013. Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin Immunol 9:15. doi: 10.1186/1710-1492-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay BB, Amato KR, Azad M, Blaser MJ, Bosch TCG, Chu H, Dominguez-Bello MG, Ehrlich SD, Elinav E, Geva-Zatorsky N, Gros P, Guillemin K, Keck F, Korem T, McFall-Ngai MJ, Melby MK, Nichter M, Pettersson S, Poinar H, Rees T, Tropini C, Zhao L, Giles-Vernick T. 2021. The hygiene hypothesis, the COVID pandemic, and consequences for the human microbiome. Proc Natl Acad Sci USA 118:e2010217118. doi: 10.1073/pnas.2010217118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmermann P, Curtis N. 2019. The effect of antibiotics on the composition of the intestinal microbiota—a systematic review. J Infect 79:471–489. doi: 10.1016/j.jinf.2019.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Ianiro G, Tilg H, Gasbarrini A. 2016. Antibiotics as deep modulators of gut microbiota: between good and evil. Gut 65:1906–1915. doi: 10.1136/gutjnl-2016-312297. [DOI] [PubMed] [Google Scholar]
- 12.Wu WK, Chen CC, Liu PY, Panyod S, Liao BY, Chen PC, Kao HL, Kuo HC, Kuo CH, Chiu THT, Chen RA, Chuang HL, Huang YT, Zou HB, Hsu CC, Chang TY, Lin CL, Ho CT, Yu HT, Sheen LY, Wu MS. 2019. Identification of TMAO-producer phenotype and host-diet-gut dysbiosis by carnitine challenge test in human and germ-free mice. Gut 68:1439–1449. doi: 10.1136/gutjnl-2018-317155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang JQ, Li T, Nakatsu G, Chen YX, Yau TO, Chu E, Wong S, Szeto CH, Ng SC, Chan FKL, Fang JY, Sung JJY, Yu J. 2020. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut 69:1248–1257. doi: 10.1136/gutjnl-2019-318532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peck SC, Denger K, Burrichter A, Irwin SM, Balskus EP, Schleheck D. 2019. A glycyl radical enzyme enables hydrogen sulfide production by the human intestinal bacterium Bilophila wadsworthia. Proc Natl Acad Sci USA 116:3171–3176. doi: 10.1073/pnas.1815661116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naghshi S, Sadeghi O, Willett WC, Esmaillzadeh A. 2020. Dietary intake of total, animal, and plant proteins and risk of all cause, cardiovascular, and cancer mortality: systematic review and dose-response meta-analysis of prospective cohort studies. BMJ 370:m2412. doi: 10.1136/bmj.m2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trefflich I, Jabakhanji A, Menzel J, Blaut M, Michalsen A, Lampen A, Abraham K, Weikert C. 2020. Is a vegan or a vegetarian diet associated with the microbiota composition in the gut? Results of a new cross-sectional study and systematic review. Crit Rev Food Sci Nutr 60:2990–3004. doi: 10.1080/10408398.2019.1676697. [DOI] [PubMed] [Google Scholar]
- 17.Wong MW, Yi CH, Liu TT, Lei WY, Hung JS, Lin CL, Lin SZ, Chen CL. 2018. Impact of vegan diets on gut microbiota: an update on the clinical implications. Ci Ji Yi Xue Za Zhi 30:200–203. doi: 10.4103/tcmj.tcmj_21_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craig WJ. 2009. Health effects of vegan diets. Am J Clin Nutr 89:1627S–1633S. doi: 10.3945/ajcn.2009.26736N. [DOI] [PubMed] [Google Scholar]
- 19.Lampe JW. 2003. Spicing up a vegetarian diet: chemopreventive effects of phytochemicals. Am J Clin Nutr 78:579S–583S. doi: 10.1093/ajcn/78.3.579S. [DOI] [PubMed] [Google Scholar]
- 20.Kahleova H, Fleeman R, Hlozkova A, Holubkov R, Barnard ND. 2018. A plant-based diet in overweight individuals in a 16-week randomized clinical trial: metabolic benefits of plant protein. Nutr Diabetes 8:58. doi: 10.1038/s41387-018-0067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMacken M, Shah S. 2017. A plant-based diet for the prevention and treatment of type 2 diabetes. J Geriatr Cardiol 14:342–354. doi: 10.11909/j.issn.1671-5411.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarty MF. 1999. Vegan proteins may reduce risk of cancer, obesity, and cardiovascular disease by promoting increased glucagon activity. Med Hypotheses 53:459–485. doi: 10.1054/mehy.1999.0784. [DOI] [PubMed] [Google Scholar]
- 23.do Rosario VA, Fernandes R, Trindade EB. 2016. Vegetarian diets and gut microbiota: important shifts in markers of metabolism and cardiovascular disease. Nutr Rev 74:444–454. doi: 10.1093/nutrit/nuw012. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Yu J. 2018. The association of diet, gut microbiota and colorectal cancer: what we eat may imply what we get. Protein Cell 9:474–487. doi: 10.1007/s13238-018-0543-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, Ackermann G, Aksenov AA, Behsaz B, Brennan C, Chen Y, DeRight Goldasich L, Dorrestein PC, Dunn RR, Fahimipour AK, Gaffney J, Gilbert JA, Gogul G, Green JL, Hugenholtz P, Humphrey G, Huttenhower C, Jackson MA, Janssen S, Jeste DV, Jiang L, Kelley ST, Knights D, Kosciolek T, Ladau J, Leach J, Marotz C, Meleshko D, Melnik AV, Metcalf JL, Mohimani H, Montassier E, Navas-Molina J, Nguyen TT, Peddada S, Pevzner P, Pollard KS, Rahnavard G, Robbins-Pianka A, Sangwan N, Shorenstein J, Smarr L, Song SJ, Spector T, Swafford AD, Thackray VG, The American Gut Consortium, et al. 2018. American Gut: an open platform for citizen science microbiome research. mSystems 3:e00031-18. doi: 10.1128/mSystems.00031-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quigley EMM. 2017. Gut microbiome as a clinical tool in gastrointestinal disease management: are we there yet? Nat Rev Gastroenterol Hepatol 14:315–320. doi: 10.1038/nrgastro.2017.29. [DOI] [PubMed] [Google Scholar]
- 27.Jalanka J, Salonen A, Salojarvi J, Ritari J, Immonen O, Marciani L, Gowland P, Hoad C, Garsed K, Lam C, Palva A, Spiller RC, de Vos WM. 2015. Effects of bowel cleansing on the intestinal microbiota. Gut 64:1562–1568. doi: 10.1136/gutjnl-2014-307240. [DOI] [PubMed] [Google Scholar]
- 28.O’Brien CL, Allison GE, Grimpen F, Pavli P. 2013. Impact of colonoscopy bowel preparation on intestinal microbiota. PLoS One 8:e62815. doi: 10.1371/journal.pone.0062815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drago L, Toscano M, De Grandi R, Casini V, Pace F. 2016. Persisting changes of intestinal microbiota after bowel lavage and colonoscopy. Eur J Gastroenterol Hepatol 28:532–537. doi: 10.1097/MEG.0000000000000581. [DOI] [PubMed] [Google Scholar]
- 30.Green JE, Davis JA, Berk M, Hair C, Loughman A, Castle D, Athan E, Nierenberg AA, Cryan JF, Jacka F, Marx W. 2020. Efficacy and safety of fecal microbiota transplantation for the treatment of diseases other than Clostridium difficile infection: a systematic review and meta-analysis. Gut Microbes 12:1854640. doi: 10.1080/19490976.2020.1854640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomova A, Bukovsky I, Rembert E, Yonas W, Alwarith J, Barnard ND, Kahleova H. 2019. The effects of vegetarian and vegan diets on gut microbiota. Front Nutr 6:47. doi: 10.3389/fnut.2019.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hegelmaier T, Lebbing M, Duscha A, Tomaske L, Tönges L, Holm JB, Bjørn Nielsen H, Gatermann SG, Przuntek H, Haghikia A. 2020. Interventional influence of the intestinal microbiome through dietary intervention and bowel cleansing might improve motor symptoms in Parkinson’s disease. Cells 9:376. doi: 10.3390/cells9020376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu GD, Compher C, Chen EZ, Smith SA, Shah RD, Bittinger K, Chehoud C, Albenberg LG, Nessel L, Gilroy E, Star J, Weljie AM, Flint HJ, Metz DC, Bennett MJ, Li H, Bushman FD, Lewis JD. 2016. Comparative metabolomics in vegans and omnivores reveal constraints on diet-dependent gut microbiota metabolite production. Gut 65:63–72. doi: 10.1136/gutjnl-2014-308209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiu TH, Huang HY, Chiu YF, Pan WH, Kao HY, Chiu JP, Lin MN, Lin CL. 2014. Taiwanese vegetarians and omnivores: dietary composition, prevalence of diabetes and IFG. PLoS One 9:e88547. doi: 10.1371/journal.pone.0088547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orlich MJ, Chiu THT, Dhillon PK, Key TJ, Fraser GE, Shridhar K, Agrawal S, Kinra S. 2019. Vegetarian epidemiology: review and discussion of findings from geographically diverse cohorts. Adv Nutr 10:S284–S295. doi: 10.1093/advances/nmy109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Selma MV, Beltran D, Garcia-Villalba R, Espin JC, Tomas-Barberan FA. 2014. Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food Funct 5:1779–1784. doi: 10.1039/c4fo00092g. [DOI] [PubMed] [Google Scholar]
- 37.Romo-Vaquero M, García-Villalba R, González-Sarrías A, Beltrán D, Tomás-Barberán FA, Espín JC, Selma MV. 2015. Interindividual variability in the human metabolism of ellagic acid: contribution of Gordonibacter to urolithin production. J Funct Foods 17:785–791. doi: 10.1016/j.jff.2015.06.040. [DOI] [Google Scholar]
- 38.Landete JM, Gaya P, Rodriguez E, Langa S, Peiroten A, Medina M, Arques JL. 2017. Probiotic bacteria for healthier aging: immunomodulation and metabolism of phytoestrogens. Biomed Res Int 2017:5939818. doi: 10.1155/2017/5939818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori G, Rampelli S, Orena BS, Rengucci C, De Maio G, Barbieri G, Passardi A, Casadei Gardini A, Frassineti GL, Gaiarsa S, Albertini AM, Ranzani GN, Calistri D, Pasca MR. 2018. Shifts of faecal microbiota during sporadic colorectal carcinogenesis. Sci Rep 8:10329. doi: 10.1038/s41598-018-28671-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ai D, Pan H, Li X, Gao Y, Liu G, Xia LC. 2019. Identifying gut microbiota associated with colorectal cancer using a zero-inflated lognormal model. Front Microbiol 10:826. doi: 10.3389/fmicb.2019.00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eeckhaut V, Machiels K, Perrier C, Romero C, Maes S, Flahou B, Steppe M, Haesebrouck F, Sas B, Ducatelle R, Vermeire S, Van Immerseel F. 2013. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut 62:1745–1752. doi: 10.1136/gutjnl-2012-303611. [DOI] [PubMed] [Google Scholar]
- 42.Cross AJ, Ferrucci LM, Risch A, Graubard BI, Ward MH, Park Y, Hollenbeck AR, Schatzkin A, Sinha R. 2010. A large prospective study of meat consumption and colorectal cancer risk: an investigation of potential mechanisms underlying this association. Cancer Res 70:2406–2414. doi: 10.1158/0008-5472.CAN-09-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, Hosoda F, Rokutan H, Matsumoto M, Takamaru H, Yamada M, Matsuda T, Iwasaki M, Yamaji T, Yachida T, Soga T, Kurokawa K, Toyoda A, Ogura Y, Hayashi T, Hatakeyama M, Nakagama H, Saito Y, Fukuda S, Shibata T, Yamada T. 2019. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med 25:968–976. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 44.Sheng Q, Du H, Cheng X, Cheng X, Tang Y, Pan L, Wang Q, Lin J. 2019. Characteristics of fecal gut microbiota in patients with colorectal cancer at different stages and different sites. Oncol Lett 18:4834–4844. doi: 10.3892/ol.2019.10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y, Jiao N, Zhu R, Zhang Y, Wu D, Wang AJ, Fang S, Tao L, Li Y, Cheng S, He X, Lan P, Tian C, Liu NN, Zhu L. 2021. Identification of microbial markers across populations in early detection of colorectal cancer. Nat Commun 12:3063. doi: 10.1038/s41467-021-23265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim DJ, Yang J, Seo H, Lee WH, Ho Lee D, Kym S, Park YS, Kim JG, Jang IJ, Kim YK, Cho JY. 2020. Colorectal cancer diagnostic model utilizing metagenomic and metabolomic data of stool microbial extracellular vesicles. Sci Rep 10:2860. doi: 10.1038/s41598-020-59529-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly TN, Bazzano LA, Ajami NJ, He H, Zhao J, Petrosino JF, Correa A, He J. 2016. Gut microbiome associates with lifetime cardiovascular disease risk profile among Bogalusa Heart Study participants. Circ Res 119:956–964. doi: 10.1161/CIRCRESAHA.116.309219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. 2012. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Natividad JM, Lamas B, Pham HP, Michel ML, Rainteau D, Bridonneau C, da Costa G, van Hylckama Vlieg J, Sovran B, Chamignon C, Planchais J, Richard ML, Langella P, Veiga P, Sokol H. 2018. Bilophila wadsworthia aggravates high fat diet induced metabolic dysfunctions in mice. Nat Commun 9:2802. doi: 10.1038/s41467-018-05249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nogal A, Louca P, Zhang X, Wells PM, Steves CJ, Spector TD, Falchi M, Valdes AM, Menni C. 2021. Circulating levels of the short-chain fatty acid acetate mediate the effect of the gut microbiome on visceral fat. Front Microbiol 12:711359. doi: 10.3389/fmicb.2021.711359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y, Ye X, Ding D, Lu Y. 2020. Characteristics of the intestinal flora in patients with peripheral neuropathy associated with type 2 diabetes. J Int Med Res 48:300060520936806. doi: 10.1177/0300060520936806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hung SY, Chen HC, Chen WT. 2020. A randomized trial comparing the bowel cleansing efficacy of sodium picosulfate/magnesium citrate and polyethylene glycol/bisacodyl (The Bowklean Study). Sci Rep 10:5604. doi: 10.1038/s41598-020-62120-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rex DK, Katz PO, Bertiger G, Vanner S, Hookey LC, Alderfer V, Joseph RE. 2013. Split-dose administration of a dual-action, low-volume bowel cleanser for colonoscopy: the SEE CLEAR I study. Gastrointest Endosc 78:132–141. doi: 10.1016/j.gie.2013.02.024. [DOI] [PubMed] [Google Scholar]
- 54.Liu FX, Wang L, Yan WJ, Zou LC, Cao YA, Lin XC. 2021. Cleansing efficacy and safety of bowel preparation protocol using sodium picosulfate/magnesium citrate considering subjective experiences: an observational study. World J Clin Cases 9:3586–3596. doi: 10.12998/wjcc.v9.i15.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiu TH, Huang HY, Chen KJ, Wu YR, Chiu JP, Li YH, Chiu BC, Lin CL, Lin MN. 2014. Relative validity and reproducibility of a quantitative FFQ for assessing nutrient intakes of vegetarians in Taiwan. Public Health Nutr 17:1459–1466. doi: 10.1017/S1368980013001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin CL, Wang JH, Chang CC, Chiu THT, Lin MN. 2019. Vegetarian diets and medical expenditure in Taiwan—a matched cohort study. Nutrients 11:2688. doi: 10.3390/nu11112688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ministry of Health and Welfare. 2021. Taiwan food ingredient database. Ministry of Health and Welfare, Taiwan, Republic of China. http://www.dietitians.org.tw/download.
- 58.Paulson JN, Stine OC, Bravo HC, Pop M. 2013. Differential abundance analysis for microbial marker-gene surveys. Nat Methods 10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. 2014. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM02047-21_Supp_1_seq7.pdf, PDF file, 0.8 MB (790.5KB, pdf)
Data Availability Statement
The sequencing data have been submitted to the NCBI SRA database under accession number PRJNA786088.