ABSTRACT
Since the introduction of the Zika virus (ZIKV) into Brazil in 2015, its transmission dynamics have been intensively studied in many parts of the country, although much is still unknown about its circulation in the midwestern states. Here, using nanopore technology, we obtained 23 novel partial and near-complete ZIKV genomes from the state of Goiás, located in the Midwest of Brazil. Genomic, phylogenetic, and epidemiological approaches were used to retrospectively explore the spatiotemporal evolution of the ZIKV-Asian genotype in this region. As a likely consequence of a gradual accumulation of herd immunity, epidemiological data revealed a decline in the number of reported cases over 2018 to 2021. Phylogenetic reconstructions revealed that multiple independent introductions of the Asian lineage have occurred in Goiás over time and revealed a complex transmission dynamic between epidemic seasons. Together, our results highlight the utility of genomic, epidemiological, and evolutionary methods to understand mosquito-borne epidemics.
IMPORTANCE Despite the considerable morbidity and mortality of arboviral infections in Brazil, such as Zika, chikungunya, dengue fever, and yellow fever, our understanding of these outbreaks is hampered by the limited availability of genomic data to track and control the epidemic. In this study, we provide a retrospective reconstruction of the Zika virus transmission dynamics in the state of Goiás by analyzing genomic data from areas in Midwest Brazil not covered by other previous studies. Our study provides an understanding of how ZIKV initiates transmission in this region and reveals a complex transmission dynamic between epidemic seasons. Together, our results highlight the utility of genomic, epidemiological, and evolutionary methods to understand mosquito-borne epidemics, revealing how this toolkit can be used to help policymakers prioritize areas to be targeted, especially in the context of finite public health resources.
KEYWORDS: Zika virus, Asian lineage, Midwest Brazil, genomic epidemiology
INTRODUCTION
The Zika virus (ZIKV) is a mosquito-borne flavivirus that was first identified in Uganda in 1947 (1). Outbreaks of ZIKV infection have already been recorded in Africa, Asia, the Pacific, and the Americas (2, 3). The first confirmed case of ZIKV infection in the Americas was reported in Northeast Brazil in May 2015 (4), although phylogenetic studies indicate virus introduction much earlier (2013 to 2014) (5). Since then, the virus has spread throughout the Americas, probably due to a combination of several factors, including a completely susceptible population, favorable climatic conditions for the adequability of the Aedes aegypti mosquitoes as main vectors for its transmission, and sustained human mobility (6–8). Between January 2016 and December 2018, the Brazilian Midwestern region, which covers an area of 1.6 million km2 and is inhabited by about 14 million people in 467 municipalities, reported a total number of 54,457 Zika cases (9–14). Most of these cases (55%) were reported in the states of Mato Grosso and Goiás, across several epidemic seasons (9–14). Despite some work done over the large epidemic between 2015 and 2016, there is still a paucity of studies directly investigating the circulation and genetic diversity of the ZIKV in this region. In this study, using our experience with mobile laboratory (15), we used nanopore sequencing to generate ZIKV genomes from infected patients residing in Goiás and provide a retrospective reconstruction of its transmission dynamics in that state.
RESULTS
The 23 sequenced samples obtained in this study were collected from females (65%) and males (35%) (Table S1) with a median age of 30 years (range: 19 to 57). All sequenced samples were collected from different municipalities in the state of Goiás (Table S1, Fig. 1A) and contained sufficient viral genetic material (≥2 ng/μL) for library preparation. Cycle threshold (CT) values were on average 27.96 (range: 25 to 32), and sequences presented a median genome coverage of 82.5% (range: 56.1 to 93.2). Epidemiological data and sequencing statistics are detailed in Table S1.
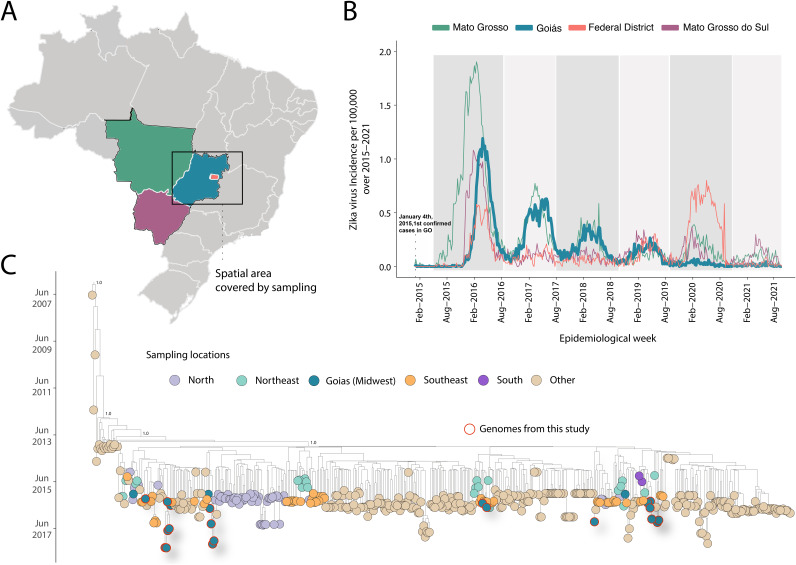
FIG 1.
Genomic epidemiology of ZIKV in Midwest Brazil. (A) Map of Brazil showing the spatial area under investigation. (B) Weekly notified Zika cases normalized per 100,000 individuals in in the Brazilian Midwest region (Federal District and the states of Mato Grosso, Goiás, and Mato Grosso do Sul) between 2015 and 2021. Epidemic curves are colored according to geographical locations. Incidence (cases per 100,000 population) is presented in log10 for visual purposes. (C) Time-scaled maximum clade credibility tree of ZIKV-Asian lineage in Brazil, including the 23 new genomes generated in this study plus n = 479 reference strains sampled worldwide. Tips are colored according to the sample source location. Values around nodes represent posterior probability support of the tree nodes inferred under Bayesian evolutionary analysis using a molecular clock approach.
Figure 1B shows the ZIKV weekly cases normalized per 100,000 individuals notified between 2015 and 2021 in the Brazilian Midwest region (Federal District and the states of Mato Grosso, Goiás, and Mato Grosso do Sul). This weekly reported incidence revealed a major outbreak in the Midwest region during 2015 to 2016, after which ever smaller epidemics took place over the years but the virus persisted through year-round transmission cycles. Overall, the state of Goiás reported the lowest incidence in recent years (2020 to 2021). Interestingly, the Federal District, which experienced the smallest initial outbreaks in 2015 to 2016, later presented a temporary resurgence in 2019 to 2020 (Fig. 1B). Over this period, the cumulative number of cases per 100,000 was 17 for the state of Goiás, 36 for the state of Mato Grosso, and 35 for the state of Mato Grosso do Sul. Although we did not assess the factors dictating the general trend in decreasing incidence over the years, it is likely to have been mediated by the accumulating herd immunity in the region since the virus’s introduction (16). Indeed, some studies have demonstrated this effect in other Brazilian states (16, 17).
To accurately establish evolutionary relationships among the newly generated sequences and other known ZIKV isolates, we subjected a combined data set to phylogenetic inference. A regression of genetic divergence from root to tip against sampling dates confirmed sufficient temporal signal (coefficient correlation = 0.70, r2 = 0.50). Our maximum clade credibility (MCC) tree showed that the newly sequences obtained in this study are scattered throughout the tree and clustered together with viral strains isolated in other Brazilian regions (northeastern and southeastern), suggesting that those regions have likely acted as steppingstone spots for the dissemination of the virus into the state of Goiás (Fig. 1C), which might have been influenced by the increased human mobility and vector suitability. From our time-measured tree, we estimated the time of the most recent common ancestor (TMRCA) to have occurred between mid-February 2014 (95% highest posterior density ranging from 10 February 2014 to 10 October 2014) for the first introduction event and late November 2016 (95% highest posterior density ranging from 30 May 2016 to 1 January 2017) for the last event, suggesting the persistence of the initially introduced virus for the period of 2 years in which reported incidence was highest (2015 to 2016).
DISCUSSION
To retrospectively explore the retrospective spatiotemporal evolution of ZIKV through the Midwestern Brazilian region, we generated 23 partial and near-complete genome sequences from the 2016 to 2018 ZIKV epidemic. Epidemiological data revealed that epidemic waves from the Brazilian Midwest region displayed their largest sizes between 2015 and 2017 (Fig. 1B). This was followed by a reduction in the number of reported cases over 2018 to 2021, likely a consequence of an expected, gradual accumulation of herd immunity, but the persistence of the initially introduced virus lineage through year-round transmission cycles was still indicated.
We found that the ZIKV epidemic in Goiás was ignited by multiple independent introduction events which we infer to have occurred between February 2014 and November 2016, most likely from northeastern and then later from southeastern Brazil, where the virus had already been circulating since late October 2013 (2, 5). Those findings are in line with previous studies that suggested that northeastern Brazil played a significant role in the establishment and dissemination of ZIKV in the Americas (2, 5, 18) and further reveal complex transmission dynamics within Brazilian regions. Since the first ZIKV confirmed case in Goiás was detected on 4 January 2015, our findings further highlight that the virus was cryptically circulated in this region for a period of 11 months, following a pattern that had been observed before during other Zika and dengue epidemics (5, 18).
In summary, our data reveal a complex pattern of ZIKV transmission between epidemic seasons, highlighting that the virus’s interregional spread might have been driven by a combination of several factors, including: (i) a completely susceptible population, (ii) favorable climatic conditions, and (iii) a sustained human mobility, as discussed elsewhere (7, 16). Together, those results highlight the utility of genomic, epidemiological, and evolutionary methods to understand mosquito-borne epidemics.
MATERIALS AND METHODS
Molecular screening.
Serum samples from 23 individuals presenting symptoms compatible with ZIKV infection were submitted to nanopore sequencing during a mobile genomic surveillance activity, which took place in Midwest Brazil in May 2019, under the scope of the ZIBRA-2 project (https://www.zibra2project.org/). Viral RNA was extracted and submitted to a real‐time PCR protocol adapted from reference 19 to confirm the previous diagnosis. Samples were selected for local sequencing based on a PCR cycle threshold (CT) of <32 to maximize genome coverage of clinical samples by nanopore sequencing (20) (Table S1).
cDNA synthesis and multiplex tiling PCR.
Samples were submitted to a cDNA synthesis protocol described previously (20), a multiplex tiling PCR using Q5 high fidelity hot-start DNA polymerase (New England Biolabs), and a ZIKV sequencing primers scheme (20). The thermocycling conditions involved 40 cycles, and reaction conditions were as reported previously (20).
Library preparation and nanopore sequencing.
Amplicons were purified using 1× AMPure XP beads, and cleaned-up PCR products concentrations were measured using Qubit dsDNA HS assay kit. DNA library preparation was carried out using the ligation sequencing kit and the native barcoding kit (NBD104 and NBD114, Oxford Nanopore Technologies, Oxford, UK) (20). Sequencing libraries were loaded into an R9.4 flow cell (Oxford Nanopore Technologies). In each sequencing run, we used negative controls to prevent and check for possible contamination with less than 2% mean coverage.
Generation of consensus sequences.
Raw files were basecalled using Guppy, and barcode demultiplexing was performed using qcat. Consensus sequences were generated by de novo assembling using Genome Detective (https://www.genomedetective.com/) (21).
Phylogenetic and Bayesian analysis.
The 23 new genomic sequences reported in this study were initially submitted to a genotyping analysis using the phylogenetic arbovirus subtyping tool, available at http://genomedetective.com/app/typingtool/zika (22). Genomic data generated in this study were aligned with a worldwide, larger, and updated data set of ZIKV genome sequences (n = 479). Sequences were aligned using MAFFT (23), and preliminary ML-tree was inferred using IQTREE2 (24). Prior to temporal analysis, our data set was also assessed for molecular clock signal in TempEst v1.5.3 (25) following the removal of any potential outliers that may violate the molecular clock assumption. To estimate a time-calibrated phylogeny, we used the Bayesian software package BEASTv.1.10.4 (26), with the Bayesian Skyline tree prior (27) with an uncorrelated relaxed clock and the lognormal distribution (28). Analyses were run in duplicate in BEASTv.1.10.4 (26) for 100 million Markov chain Monte Carlo (MCMC) steps, sampling parameters and trees every 10,000th step. Convergence of MCMC chains was checked using Tracer v.1.7.1 (29). Maximum clade credibility trees were summarized using TreeAnnotator after discarding 10% as burn-in.
Epidemiological data assembly.
Data of weekly notified ZIKV cases in Brazil, available at the Sistema de Informação de Agravos de Notificação (SINAN) (https://portalsinan.saude.gov.br/), were supplied by Brazilian Ministry of Health and were plotted using the R software version 3.5.1.
Data availability.
Newly generated ZIKV sequences have been deposited in GenBank under accession numbers OL423647 to OL423669.
ACKNOWLEDGMENTS
We thank all personnel from Health Surveillance System from the state of Goiás and the Brazilian Ministry of Health that helped with samples, sources, and epidemiological data collection.
Molecular screening and production of ZIKV genomic data: M.G., L.A.P., T.E.R.A., J.X., A.dE.F., S.N.S., P.dS.L., W.dA.M., and S.K.; collected samples and curated metadata: V.F., C.F.C.dA., C.F., C.R.L.P., A.F.M., and V.L.dS.; analyzed the data: M.G., V.F., T.E.R.A., J.L.; helped with study design and data interpretation: M.G., T.E.R.A., L.A.P., V.F., J.L., T.dO., R.V.C., A.F.M., V.L.dS., and L.C.J.A.; wrote the initial manuscript, which was reviewed by all authors: M.G., T.E.R.A., J.L., L.C.J.A.
We have no competing interests to disclose.
This research was reviewed and approved by the Ethical Committee of the Pan American World Health Organization (No. PAHO-2016-08-0029) and the Brazilian Ministry of Health (MoH) as part of the arbovirus genomic surveillance efforts within the terms of Resolution 510/2016 of CONEP (Comissão Nacional de Etica em Pesquisa, Ministerio da Saude; National Ethical Committee for Research, Ministry of Health). Residual anonymized clinical diagnostic samples, with no or minimal risk to patients, were provided for research and surveillance purposes within the terms of Resolution 510/2016 of CONEP. Processing of human samples was approved and the need for participants consent was waived by the Institutional Review Board from the Fundação Oswaldo Cruz/Instituto Oswaldo Cruz (CEP/CAAE: 90249218.6.1001.5248; approval number 2.998.362).
This work was supported by Decit/SCTIE/BrMoH/CNPq (440685/2016-8 to 421598/2018-2), by CAPES (88887.130716/2016-00), and by the European Union’s Horizon 2020 Research and Innovation Program under ZIKAlliance Grant Agreement no. 734548. J.X. is supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001. M.G. is supported by Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ). J.L. was supported by a Research Lectureship by the Department of Zoology, University of Oxford. Funders had no role in study design, data collection and analysis, writing, and/or decision to publish the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Luiz Carlos Junior Alcantara, Email: luiz.alcantara@ioc.fiocruz.br.
Tino Polen, Forschungszentrum Jülich GmbH.
REFERENCES
- 1. Cao-Lormeau V-M, Roche C, Teissier A, Robin E, Berry A-L, Mallet H-P, Sall AA, Musso D. 2014. Zika virus, French Polynesia, South pacific, 2013. Emerg Infect Dis 20:1085–1086. doi: 10.3201/eid2006.140138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faria NR, da Silva Azevedo RdS, Kraemer MUG, Souza R, Cunha MS, Hill SC, Thézé J, Bonsall MB, Bowden TA, Rissanen I, Rocco IM, Nogueira JS, Maeda AY, da Silva Vasami FG, de Lima Macedo FL, Suzuki A, Rodrigues SG, Cruz ACR, Nunes BT, de Almeida Medeiros DB, Rodrigues DSG, Queiroz ALN, da Silva EVP, Henriques DF, da Rosa EST, de Oliveira CS, Martins LC, Vasconcelos HB, Casseb LMN, de Brito Simith D, Messina JP, Abade L, Lourenço J, Alcantara LCJ, de Lima MM, Giovanetti M, Hay SI, de Oliveira RS, da Silva Lemos P, de Oliveira LF, de Lima CPS, da Silva SP, de Vasconcelos JM, Franco L, Cardoso JF, da Silva Gonçalves Vianez-Júnior JL, Mir D, Bello G, Delatorre E, Khan K, et al. 2016. Zika virus in the Americas: early epidemiological and genetic findings. Science 352:345–349. doi: 10.1126/science.aaf5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faria NR, Quick J, Claro IM, Thézé J, de Jesus JG, Giovanetti M, Kraemer MUG, Hill SC, Black A, da Costa AC, Franco LC, Silva SP, Wu C-H, Raghwani J, Cauchemez S, Du Plessis L, Verotti MP, de Oliveira WK, Carmo EH, Coelho GE, Santelli ACFS, Vinhal LC, Henriques CM, Simpson JT, Loose M, Andersen KG, Grubaugh ND, Somasekar S, Chiu CY, Muñoz-Medina JE, Gonzalez-Bonilla CR, Arias CF, Lewis-Ximenez LL, Baylis SA, Chieppe AO, Aguiar SF, Fernandes CA, Lemos PS, Nascimento BLS, Monteiro HAO, Siqueira IC, de Queiroz MG, de Souza TR, Bezerra JF, Lemos MR, Pereira GF, Loudal D, Moura LC, Dhalia R, França RF, et al. 2017. Establishment and cryptic transmission of Zika virus in Brazil and the Americas. Nature 546:406–410. doi: 10.1038/nature22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campos GS, Bandeira AC, Sardi SI. 2015. Zika virus outbreak, Bahia, Brazil. Emerg Infect Dis 21:1885–1886. doi: 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faria NR, da Costa AC, Lourenço J, Loureiro P, Lopes ME, Ribeiro R, Alencar CS, Kraemer MUG, Villabona-Arenas CJ, Wu C-H, Thézé J, Khan K, Brent SE, Romano C, Delwart E, Custer B, Busch MP, Pybus OG, Sabino EC, NHLBI Recipient Epidemiology and Donor Evaluation Study-III (REDS-III). 2017. Genomic and epidemiological characterisation of a dengue virus outbreak among blood donors in Brazil. Sci Rep 7:15216. doi: 10.1038/s41598-017-15152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grubaugh ND, Saraf S, Gangavarapu K, Watts A, Tan AL, Oidtman RJ, Ladner JT, Oliveira G, Matteson NL, Kraemer MUG, Vogels CBF, Hentoff A, Bhatia D, Stanek D, Scott B, Landis V, Stryker I, Cone MR, Kopp EW, Cannons AC, Heberlein-Larson L, White S, Gillis LD, Ricciardi MJ, Kwal J, Lichtenberger PK, Magnani DM, Watkins DI, Palacios G, Hamer DH, Gardner LM, Perkins TA, Baele G, Khan K, Morrison A, Isern S, Michael SF, Andersen KG, GeoSentinel Surveillance Network. 2019. Travel surveillance and genomics uncover a hidden Zika outbreak during the waning epidemic. Cell 178:1057–1071. doi: 10.1016/j.cell.2019.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraemer MUG, Reiner RC, Brady OJ, Messina JP, Gilbert M, Pigott DM, Yi D, Johnson K, Earl L, Marczak LB, Shirude S, Davis Weaver N, Bisanzio D, Perkins TA, Lai S, Lu X, Jones P, Coelho GE, Carvalho RG, Van Bortel W, Marsboom C, Hendrickx G, Schaffner F, Moore CG, Nax HH, Bengtsson L, Wetter E, Tatem AJ, Brownstein JS, Smith DL, Lambrechts L, Cauchemez S, Linard C, Faria NR, Pybus OG, Scott TW, Liu Q, Yu H, Wint GRW, Hay SI, Golding N. 2019. Past and future spread of the arbovirus vectors Aedes aegypti and Aedes albopictus. Nat Microbiol 4:854–863. doi: 10.1038/s41564-019-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thézé J, Li T, Du Plessis L, Bouquet J, Kraemer MUG, Somasekar S, Yu G, de Cesare M, Balmaseda A, Kuan G, Harris E, Wu C-H, Ansari MA, Bowden R, Faria NR, Yagi S, Messenger S, Brooks T, Stone M, Bloch EM, Busch M, Muñoz-Medina JE, González-Bonilla CR, Wolinsky S, López S, Arias CF, Bonsall D, Chiu CY, Pybus OG. 2018. Genomic epidemiology reconstructs the introduction and spread of Zika virus in Central America and Mexico. Cell Host Microbe 23:855–864. doi: 10.1016/j.chom.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brasil Ministério da Saúde. 2017. Monitoramento dos casos de dengue, febre de chikungunya e febre pelo vírus Zika até a Semana Epidemiológica 52, 2016. Brasil Ministério da Saúde, Rio de Janeiro, Brazil. https://bvsms.saude.gov.br/arboviroses/. [Google Scholar]
- 10.Brasil Ministério da Saúde. 2018. Monitoramento dos casos de dengue, febre de chikungunya e febre pelo vírus Zika até a Semana Epidemiológica 52, 2017. Brasil Ministério da Saúde, Rio de Janeiro, Brazil. https://bvsms.saude.gov.br/arboviroses/. [Google Scholar]
- 11.Brasil Ministério da Saúde. 2019. Monitoramento dos casos de dengue, febre de chikungunya e doença aguda pelo vírus Zika até a Semana Epidemiológica 52 de 2018. Brasil Ministério da Saúde, Rio de Janeiro, Brazil. https://bvsms.saude.gov.br/arboviroses/. [Google Scholar]
- 12.Brasil Ministério da Saúde. 2020. Monitoramento dos casos de arboviroses urbanas transmitidas pelo Aedes (dengue, chikungunya e Zika), Semanas Epidemiológicas 01 a 52. Brasil Ministério da Saúde, Rio de Janeiro, Brazil. https://bvsms.saude.gov.br/arboviroses/. [Google Scholar]
- 13.Brasil Ministério da Saúde. 2021. Monitoramento dos casos de arboviroses urbanas causados por vírus transmitidos por Aedes (dengue, chikungunya e zika), semanas epidemiológicas 1 a 53, 2020. Brasil Ministério da Saúde, Rio de Janeiro, Brazil. https://www.gov.br/saude/pt-br/media/pdf/2021/fevereiro/01/boletim_epidemiologico_svs_3.pdf. [Google Scholar]
- 14.Brasil Ministério da Saúde. 2021. Monitoramento dos casos de arboviroses urbanas causados por vírus transmitidos pelo mosquito Aedes (dengue, chikungunya e zika), semanas epidemiológicas 1 a 11, 2021. Brasil Ministério da Saúde, Rio de Janeiro, Brazil. https://www.gov.br/saude/pt-br/media/pdf/2021/marco/26/boletim_epidemiologico_svs_11.pdf. [Google Scholar]
- 15.Adelino TÉR, Giovanetti M, Fonseca V, Xavier J, de Abreu ÁS, do Nascimento VA, Demarchi LHF, Oliveira MAA, da Silva VL, de Mello ALES, Cunha GM, Santos RH, de Oliveira EC, Júnior JAC, de Melo Iani FC, de Filippis AMB, de Abreu AL, de Jesus R, de Albuquerque CFC, Rico JM, do Carmo Said RF, Silva JA, de Moura NFO, Leite P, Frutuoso LCV, Haddad SK, Martínez A, Barreto FK, Vazquez CC, da Cunha RV, Araújo ELL, de Oliveira Tosta SF, de Araújo Fabri A, Chalhoub FLL, da Silva Lemos P, de Bruycker-Nogueira F, de Castro Lichs GG, Zardin MCSU, Segovia FMC, Gonçalves CCM, Fernandez Grillo ZDC, Slavov SN, Pereira LA, Mendonça AF, Pereira FM, da Magalhães JJF, Dos Santos Júnior AdCM, de Lima MM, Nogueira RMR, Góes-Neto Aet al. 2021. Field and classroom initiatives for portable sequence-based monitoring of dengue virus in Brazil. Nat Commun 12:2296. doi: 10.1038/s41467-021-22607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lourenço J, Maia de Lima M, Faria NR, Walker A, Kraemer MU, Villabona-Arenas CJ, Lambert B, Marques de Cerqueira E, Pybus OG, Alcantara LC, Recker M. 2017. Epidemiological and ecological determinants of Zika virus transmission in an urban setting. Elife 9:6–29. doi: 10.7554/eLife.29820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Netto EM, Moreira-Soto A, Pedroso C, Höser C, Funk S, Kucharski AJ, Rockstroh A, Kümmerer BM, Sampaio GS, Luz E, Vaz SN, Dias JP, Bastos FA, Cabral R, Kistemann T, Ulbert S, de Lamballerie X, Jaenisch T, Brady OJ, Drosten C, Sarno M, Brites C, Drexler JF. 2017. High Zika virus seroprevalence in Salvador, Northeastern Brazil limits the potential for further outbreaks. mBio 8:e013390-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giovanetti M, Faria NR, Lourenço J, Goes de Jesus J, Xavier J, Claro IM, Kraemer MUG, Fonseca V, Dellicour S, Thézé J, da Silva Salles F, Gräf T, Silveira PP, do Nascimento VA, Costa de Souza V, de Melo Iani FC, Castilho-Martins EA, Cruz LN, Wallau G, Fabri A, Levy F, Quick J, de Azevedo V, Aguiar RS, de Oliveira T, Bôtto de Menezes C, da Costa Castilho M, Terra TM, Souza da Silva M, Bispo de Filippis AM, Luiz de Abreu A, Oliveira WK, Croda J, Campelo de Albuquerque CF, Nunes MRT, Sabino EC, Loman N, Naveca FG, Pybus OG, Alcantara LC. 2020. Genomic and epidemiological surveillance of Zika virus in the Amazon region. Cell Rep 30:2275–2283. [DOI] [PubMed] [Google Scholar]
- 19.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quick J, Grubaugh ND, Pullan ST, Claro IM, Smith AD, Gangavarapu K, Oliveira G, Robles-Sikisaka R, Rogers TF, Beutler NA, Burton DR, Lewis-Ximenez LL, de Jesus JG, Giovanetti M, Hill SC, Black A, Bedford T, Carroll MW, Nunes M, Alcantara LC, Sabino EC, Baylis SA, Faria NR, Loose M, Simpson JT, Pybus OG, Andersen KG, Loman NJ. 2017. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc 12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vilsker M, Moosa Y, Nooij S, Fonseca V, Ghysens Y, Dumon K, Pauwels R, Alcantara LC, Vanden Eynden E, Vandamme A-M, Deforche K, de Oliveira T. 2019. Genome Detective: an automated system for virus identification from high-throughput sequencing data. Bioinformatics 35:871–873. doi: 10.1093/bioinformatics/bty695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fonseca V, Libin PJK, Theys K, Faria NR, Nunes MRT, Restovic MI, Freire M, Giovanetti M, Cuypers L, Nowé A, Abecasis A, Deforche K, Santiago GA, de Siqueira IC, San EJ, Machado KCB, Azevedo V, Bispo-de Filippis AM, da Cunha RV, Pybus OG, Vandamme A-M, Alcantara LCJ, de Oliveira T. 2019. A computational method for the identification of Dengue, Zika and Chikungunya virus species and genotypes. PLoS Negl Trop Dis 13:e0007231. doi: 10.1371/journal.pntd.0007231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh K, Kuma K, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res 33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2:25–98. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baele G, Li WLS, Drummond AJ, Suchard MA, Lemey P. 2013. Accurate model selection of relaxed molecular clocks in bayesian phylogenetics. Mol Biol Evol 30:239–243. doi: 10.1093/molbev/mss243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gill MS, Lemey P, Faria NR, Rambaut A, Shapiro B, Suchard MA. 2013. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Mol Biol Evol 30:713–724. doi: 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00155-22_Supp_1_seq3.xlsx, XLSX file, 0.01 MB (11.3KB, xlsx)
Data Availability Statement
Newly generated ZIKV sequences have been deposited in GenBank under accession numbers OL423647 to OL423669.