ABSTRACT
Two tet(X4)-positive Enterobacter cloacae isolates TECL_1 and TECL_2 were isolated from pigs in China. S1-PFGE and Southern blotting showed that tet(X4) located on plasmids in the size of ∼290 kb and ∼190 kb in TECL_1 and TECL_2, respectively. Conjugation experiment demonstrated that the tet(X4)-harboring plasmid can transfer from the donor strain TECL_1 and TECL_2 to the recipient strain Escherichia coli J53, and the tigecycline resistance of transconjugants was increased by 128-fold and 64-fold compared with E. coli J53, respectively. We obtained the complete plasmid sequence of pTECL_2-190k-tetX4 (190,185 bp) from E. cloacae TECL_2 and found that the plasmid was a hybrid plasmid with replicon types of IncFIA, IncHI1A and IncHI1B. We further analyzed 85 tet(X4)-carrying plasmids in the public database and clarified that pTECL_2-190k-tetX4-like plasmid was widespread in multiple species of Enterobacteriaceae.
IMPORTANCE We identified two tet(X4)-positive E. cloacae isolates, which has not been previously reported. We obtained the complete sequence of pTECL_2-190k-tetX4 and found that it was a hybrid plasmid with multiple replicon types, including IncFIA, IncHI1A and IncHI1B. By comparing all the known tet(X4)-carrying plasmids, we found that pTECL_2-190k-tetX4-like plasmid has been disseminated across various species in China. Our study expanded the identification of tet(X4)-positive species and emphasized that pTECL_2-190k-tetX4-like plasmid has spread widely in various species.
KEYWORDS: tet(X4), tigecycline resistance, Enterobacter cloacae
OBSERVATION
Enterobacter cloacae is one of the members of Enterobacteriaceae, which was reported as an important opportunistic microbial pathogen for a broad range of hospital-acquired infections (1). Tigecycline is a last-resort antibiotic for the treatment of life-threatening infections caused by multidrug-resistant bacteria, such as carbapenem-resistant Enterobacteriaceae (2). In recent years, the tigecycline resistance gene tet(X) has been reported to mediate high-level resistance to all tetracycline antibiotics in isolates from animals, humans and the environment, posing a significant risk to public health (3–5). In 2019, a plasmid-borne high-level tigecycline resistance gene tet(X) was identified (6). Herein, we identified two E. cloacae isolates harboring plasmid-mediated tet(X) gene, which has not been reported, and further analyzed the genetic context of the tet(X4)-carrying plasmid pTECL_2-190k-tetX4.
We collected 590 nonduplicate samples, including 475 pig nasal swabs, 67 pig anal swabs and 48 staff skin swabs from a pig farm and a slaughterhouse in Guangdong Province. Then colonies were selected from BHI plate containing 4 mg/L tigecycline after preinoculation and screened for tet(X4) variants by PCR (7). Finally, we identified two E. cloacae strains TECL_1 and TECL_2 carrying tet(X4). MICs of 14 antimicrobial agents for strains were determined (Table S1). Both strains were resistant to tigecycline, tetracycline, rifamycin, ampicillin, chloramphenicol and ciprofloxacin. In addition, TECL_1 was resistant to fosfomycin; TECL_2 was resistant to ceftazidime, colistin sulfate, cefotaxime, gentamicin, and trimethoprim-sulfamethoxazole. Then the two E. cloacae isolates were subjected to genomic DNA extraction and whole-genome sequencing. The sequencing reads were assembled into contigs using SPAdes version 3.10 (8). Antibiotic resistance genes (ARGs) were predicted using ResFinder v3.2 (9). It showed that TECL_1 harbored nine ARGs including tet(X4), tet(M), aadA2, aadA22, blaTEM-1B, qnrS1, lnu(G), lnu(F) and floR. TECL_2 carried 21 ARGs including tet(X4), tet(A), ant(3'')-Ia, aadA16, aac(6')-Ib-cr, aac(3)-IId, aph(6)-Id, aph(3')-Ia, aph(3'')-Ib, blaCTX-M, blaCMH-3, oqxA, oqxB, qnrS1, lnu(G), fosA, mph(A), sul1, sul2, arr-3 and dfrA27.
Previous studies reported that tet(X4) is mostly located on the plasmid in Enterobacteriaceae (10, 11). To determine the transferability of tet(X4)-harboring plasmids in E. cloacae isolates, we performed the conjugation experiment. It showed that tet(X4) could be successfully transferred from TECL_1 and TECL_2 into the recipient E. coli J53 by filter mating. The transconjugants J53/pTECL_1-290k-tetX4 and J53/pTECL_2-190k-tetX4 were resistant to tigecycline with MIC values of 32 and 16 mg/L, respectively. The tigecycline resistance of two transconjugants was increased by 128-fold and 64-fold compared with E. coli J53 (Table S1). Then PCR and Sanger sequencing demonstrated that the transconjugants carried tet(X4). Furthermore, S1-PFGE and Southern blotting hybridization revealed that tet(X4) located on plasmids in the size of ∼290 kb and ∼190 kb in TECL_1 and TECL_2, respectively (Fig. 1a). The results proved that tet(X4) was located on the plasmid of E. cloacae isolates, and could be transmitted to other species, causing the spread of tigecycline resistance in Enterobacteriaceae.
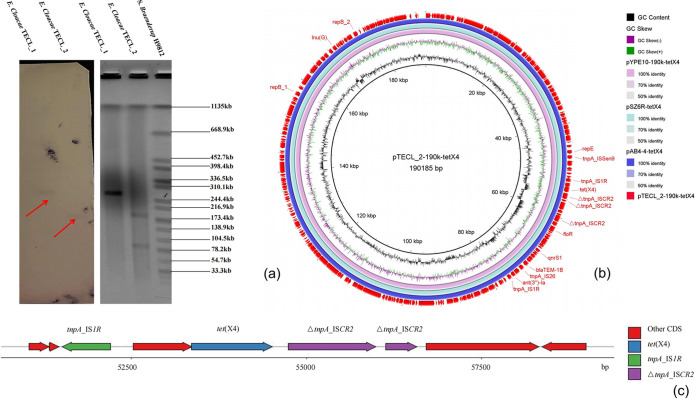
FIG 1.
The plasmid structure of pTECL_2-190k-tetX4. (a) The location of tet(X4) in E. cloacae isolates TECL_1 and TECL_2 by S1-PFGE and Southern blotting. Salmonella Braenderup strain H9812 was used as the marker by XbaI enzyme digestion. (b) The circular genetic map of pTECL_2-190k-tetX4, pYPE10-190k-tetX4 (GenBank accession No. CP041449.1), pSZ6R-tetX4 (GenBank accession No. MW940627.1) and pAB4-4-tetX4 (GenBank accession No. MW940615.1). The arrows on the outer circle represent the genes of replicon, antibiotic resistance, and transposase. The three middle circles show the similarity of three plasmids harboring tet(X4) with pTECL_2-190k-tetX4. The inner arc represents GC skew curve and the next represents GC contents. (c) Major structural features of the tet(X4) gene. The blue arrow represents tet(X4). The green arrow represents the transposase of the IS1R element and the transposase of the ΔISCR2 element is purple. The number marked on the ruler at the bottom of the picture corresponds to the nucleotide position on the plasmid.
To determine the complete sequences of pTECL_2-190k-tetX4, we combined the sequencing data from the genomic DNA and the plasmids and closed predicted gaps within the sequences by PCR and Sanger sequencing using primers listed in Table S2. Finally, we obtained the complete sequence of pTECL_2-190k-tetX4 from E. cloacae strain TECL_2. The sequence was analyzed using the method mentioned in the previous article (12). pTECL_2-190k-tetX4 was a 190,185 bp plasmid with three replicon types IncFIA, IncHI1A and IncHI1B, and encoded 220 predicted ORFs (Fig. 1b). pTECL_2-190k-tetX4 showed a mosaic structure harboring six ARGs, including tet(X4) along with ant(3')-Ia, blaTEM-1B, lnu(G), floR and qnrS1. The tet(X4) gene, was flanked by a complete IS1R element and a truncated ISCR2 element. This IS1R element was located at 1099 bp upstream of tet(X4). The downstream region of tet(X4) was a 223 bp fragment encoding the transposase of ISCR2 element. There is a 136 bp fragment insertion resulting in the truncation of ISCR2 element. (Fig. 1c). BLASTn of pTECL_2-190k-tetX4 against the nr database retrieved similar plasmids from different hosts. The plasmid pTECL_2-190-tetX4 was highly similar with pYPE10-190k-tetX4 isolated from E. coli strain YPE10 (CP041449.1, 99.96% identity at 100% coverage) (13), pSZ6R-tetX4 isolated from Citrobacter braakii strain SZ6R (MW940627.1, 99.92% identity at 100% coverage) (14) and pAB4-4-tetX4 isolated from Klebsiella pneumoniae strain AB4-4 (MW940615.1, 99.92% identity at 100% coverage) (14). The result indicated that the pTECL_2-190k-tetX4-like plasmid might have been widely spread in different species of Enterobacteriaceae.
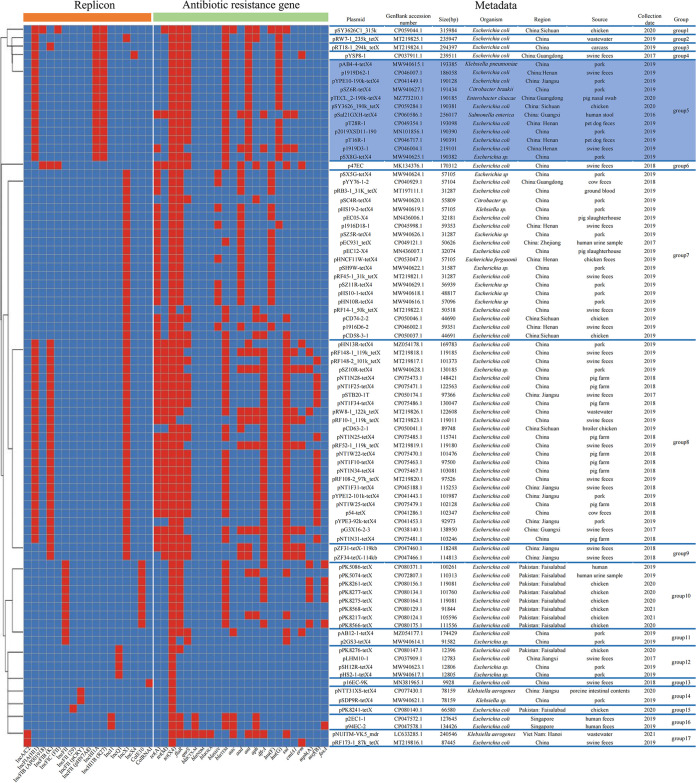
It was reported that the plasmid carrying tet(X4) showed structure diversity (15). We compared all the known tet(X4)-carrying plasmids and tried to clarify the prevalence of plasmids carrying tet(X4), especially the pTECL_2-190k-tetX4-like plasmid. BLASTn of the tet(X4) gene identified 90 publicly available tet(X4)-carrying plasmids, as of 7 August 2021. We excluded three transconjugative plasmids, one plasmid with the sequence of repeated uploads and two plasmids without replicon sequence. The remaining 84 complete plasmid sequences were compared with pTECL_2-190k-tetX4 (Fig. 2). The 85 plasmids carrying tet(X4) were clustered 17 types of plasmid group by the different replicon types. pTECL_2-190k-tetX4-like plasmids (at least 99.91% identity at 92% coverage with pTECL_2-190k-tetX4) are one of the dominant type of plasmids harboring tet(X4) (n = 12). The size of the plasmids is about 200 kb. Besides tet(X4), multiple ARGs were co-existed in these plasmids such as floR, qnrS1, blaTEM-1B, ant(3'')-Ia and lnu(F) (Table S3). Strains carrying pTECL_2-190k-tetX4-like plasmid have been found from swine, pet dog and chicken in China, indicating that this type of plasmid has disseminated widely in animals in China (Fig. 2). E. cloacae TECL_2 was isolated from a pig farm in Guangdong Province, indicating the transmission range of the plasmid in China has been further expanded. In addition, it is noteworthy that the host bacteria of this plasmid are diverse, such as E. coli, E. cloacae, S. enterica, K. pneumoniae and C. braakii, indicating this plasmid has strong host adaptability.
FIG 2.
The comparison of the publicly available plasmids carrying tet(X4). The average-linkage clustering method was used to cluster 85 plasmids carrying tet(X4) according to the replicon type. The groups were separated by blue horizontal lines in the figure. The prominent part of the blue block is the group which pTECL_2-190k-tetX4 belongs. The distribution of all replicon types and antibiotic resistance genes were presented by heatmap. In terms of whether the corresponding plasmid replicon and antibiotic resistance genes is present in the plasmid, red represents presence and blue represents absence. The metadata of plasmids is shown on the right of figure, including the name, GenBank accession number, size, and source of plasmids.
In conclusion, the identification of two tet(X4)-positive E. cloacae isolates indicates that the host range of tet(X4) has been further expanded. In addition, the widespread of pTECL_2-190k-tetX4-like plasmid in Enterobacteriaceae must be concerned.
Data availability.
This complete sequence for pTECL_2-190k-tetX4 has been deposited in GenBank under the accession number MZ773210.1.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant number 82061128001, 81830103, 81902123), Guangdong Natural Science Foundation (grant number 2017A030306012), Project of high-level health teams of Zhuhai at 2018 (The Innovation Team for Antimicrobial Resistance and Clinical Infection), 111 Project (grant number B12003), Science, Technology & Innovation Commission of Shenzhen Municipality (JCYJ20190807151601699), and China Postdoctoral Science Foundation (grant number 2019M653192).
The authors report no conflicts of interest in this work.
Footnotes
Supplemental material is available online only.
Contributor Information
Kang Liao, Email: liaokang@mail.sysu.edu.cn.
Yongqiang Yang, Email: yangyongqiang2000@126.com.
Guo-Bao Tian, Email: tiangb@mail.sysu.edu.cn.
Tino Polen, Forschungszentrum Jülich GmbH.
REFERENCES
- 1.Davin-Regli A, Pages JM. 2015. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol 6:392. doi: 10.3389/fmicb.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tasina E, Haidich AB, Kokkali S, Arvanitidou M. 2011. Efficacy and safety of tigecycline for the treatment of infectious diseases: a meta-analysis. Lancet Infect Dis 11:834–844. doi: 10.1016/S1473-3099(11)70177-3. [DOI] [PubMed] [Google Scholar]
- 3.Fang L-X, Chen C, Cui C-Y, Li X-P, Zhang Y, Liao X-P, Sun J, Liu Y-H. 2020. Emerging high-level tigecycline resistance: novel tetracycline destructases spread via the mobile tet(X). Bioessays 42:e2000014. doi: 10.1002/bies.202000014. [DOI] [PubMed] [Google Scholar]
- 4.Chen C, Cui C-Y, Yu J-J, He Q, Wu X-T, He Y-Z, Cui Z-H, Li C, Jia Q-L, Shen X-G, Sun R-Y, Wang X-R, Wang M-G, Tang T, Zhang Y, Liao X-P, Kreiswirth BN, Zhou S-D, Huang B, Du H, Sun J, Chen L, Liu Y-H. 2020. Genetic diversity and characteristics of high-level tigecycline resistance Tet(X) in Acinetobacter species. Genome Med 12:111. doi: 10.1186/s13073-020-00807-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun J, Chen C, Cui C-Y, Zhang Y, Liu X, Cui Z-H, Ma X-Y, Feng Y, Fang L-X, Lian X-L, Zhang R-M, Tang Y-Z, Zhang K-X, Liu H-M, Zhuang Z-H, Zhou S-D, Lv J-N, Du H, Huang B, Yu F-Y, Mathema B, Kreiswirth BN, Liao X-P, Chen L, Liu Y-H. 2019. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat Microbiol 4:1457–1464. doi: 10.1038/s41564-019-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, Ke Y, Ji Q, Wei R, Liu Z, Shen Y, Wang G, Sun L, Lei L, Lv Z, Li Y, Pang M, Wang L, Sun Q, Fu Y, Song H, Hao Y, Shen Z, Wang S, Chen G, Wu C, Shen J, Wang Y. 2019. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol 4:1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- 7.Ding Y, Saw W-Y, Tan LWL, Moong DKN, Nagarajan N, Teo YY, Seedorf H. 2020. Emergence of tigecycline- and eravacycline-resistant Tet(X4)-producing Enterobacteriaceae in the gut microbiota of healthy Singaporeans. J Antimicrob Chemother 75:3480–3484. doi: 10.1093/jac/dkaa372. [DOI] [PubMed] [Google Scholar]
- 8.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun C, Cui M, Zhang S, Liu D, Fu B, Li Z, Bai R, Wang Y, Wang H, Song L, Zhang C, Zhao Q, Shen J, Xu S, Wu C, Wang Y. 2020. Genomic epidemiology of animal-derived tigecycline-resistant Escherichia coli across China reveals recent endemic plasmid-encoded tet(X4) gene. Commun Biol 3:412. doi: 10.1038/s42003-020-01148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun C, Cui M, Zhang S, Wang H, Song L, Zhang C, Zhao Q, Liu D, Wang Y, Shen J, Xu S, Wu C. 2019. Plasmid-mediated tigecycline-resistant gene tet(X4) in Escherichia coli from food-producing animals, China, 2008–2018. Emerg Microbes Infect 8:1524–1527. doi: 10.1080/22221751.2019.1678367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He R, Yang Y, Wu Y, Zhong L-L, Yang Y, Chen G, Qin M, Liang X, Ahmed MAE-GE-S, Lin M, Yan B, Xia Y, Dai M, Chen H, Tian G-B. 2021. Characterization of a plasmid-encoded resistance-nodulation-division efflux pump in Klebsiella pneumoniae and Klebsiella quasipneumoniae from patients in China. Antimicrob Agents Chemother 65. doi: 10.1128/AAC.02075-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li R, Lu X, Liu Z, Liu Y, Xiao X, Wang Z. 2020. Rapid detection and characterization of tet(X4)-positive Escherichia coli strains with nanopore sequencing. J Antimicrob Chemother 75:1068–1070. doi: 10.1093/jac/dkz528. [DOI] [PubMed] [Google Scholar]
- 14.Li R, Li Y, Peng K, Yin Y, Liu Y, He T, Bai L, Wang Z. 2021. Comprehensive genomic investigation of tigecycline resistance gene tet(X4)-bearing strains expanding among different settings. Microbiol Spectr 9:e0163321. doi: 10.1128/spectrum.01633-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, Lu X, Peng K, Liu Z, Li Y, Liu Y, Xiao X, Wang Z. 2020. Deciphering the Structural Diversity and Classification of the Mobile Tigecycline Resistance Gene tet(X)-Bearing Plasmidome among. Bacteria mSystems 5:e00134-20. doi: 10.1128/mSystems.00134-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM02064-21_Supp_1_seq7.pdf, PDF file, 0.3 MB (333.3KB, pdf)
Data Availability Statement
This complete sequence for pTECL_2-190k-tetX4 has been deposited in GenBank under the accession number MZ773210.1.