ABSTRACT
Emergence of pathogens harboring multiple resistance genes incurs great concerns. Cooccurrence of mobile resistance genes conferring resistance to tigecycline, colistin, and carbapenems in Escherichia coli has not been investigated. This study aimed to characterize three E. coli isolates coharboring tet(X4), mcr-1, and blaNDM-5. Isolates coharboring tet(X4), mcr-1, and blaNDM-5 were identified and characterized by PCR, Sanger sequencing, antimicrobial susceptibility testing, conjugation assays, Illumina sequencing, nanopore sequencing, and bioinformatic analysis. Three E. coli isolates carrying tet(X4), mcr-1, and blaNDM-5 were identified from pigeons in China. They were resistant to almost all antimicrobials except enrofloxacin. tet(X4) and blaNDM-5 could be conjugated into E. coli C600, but mcr-1 was nontransferable in three isolates. Three isolates belonged to sequence type 6775 (ST6775), and clonal dissemination of isolates carrying tet(X4), mcr-1, and blaNDM-5 existed in the pigeon farm. Genetic analysis revealed that mcr-1 mediated by the Tn6330 was located on the chromosome, tet(X4) was located on the IncFII plasmid, and blaNDM-5 was located on the IncX3 plasmid. We first characterized the E. coli isolates carrying tet(X4), mcr-1, and blaNDM-5 simultaneously. Relevant measures should be taken to decrease the prevalence of pathogens carrying tet(X4), mcr-1, and blaNDM-5.
IMPORTANCE Tigecycline and colistin are regarded as vital antimicrobials to treat multidrug-resistant (MDR) bacterial infections, such as that caused by carbapenemase-producing Enterobacteriaceae (CPE). Cooccurrence of mobile resistance genes conferring resistance to last-resort antimicrobials in E. coli remains unknown. Here, we characterized E. coli strains coharboring tet(X4), mcr-1, and blaNDM-5 phenotypically and genetically. Resistance genes tet(X4), mcr-1, and blaNDM-5 were located on transposons or plasmids that were mobile genetic elements related to the capture, accumulation, and dissemination of such important resistance genes. The emergence of E. coli isolates carrying tet(X4), mcr-1, and blaNDM-5 highlights the importance of monitoring the coexistence of novel mobile resistance genes in different settings with a One Health approach. Risk of transmission of such MDR pathogens from animals to humans should be evaluated comprehensively.
KEYWORDS: Escherichia coli, ST6775, tet(X4), mcr-1, bla NDM-5 , coexistence
INTRODUCTION
Antimicrobial resistance is considered a global threat to public health. Carbapenems are critical drugs treating Gram-negative bacterial infections. Accordingly, preventing the spread of carbapenemase-producing Enterobacteriaceae (CPE) among different settings continues to present serious challenges (1). New Delhi metallo-β-lactamase (NDM), conferring resistance to almost all β-lactams, is the main carbapenemase, and 40 gene variants, including blaNDM-1 to blaNDM-40 (https://www.ncbi.nlm.nih.gov/pathogens/refgene/#NDM), encode NDM enzymes, among which blaNDM-1 and blaNDM-5 genes are still the most prevalent variants (2). Colistin and tigecycline are two of the last-resort therapies to treat infections caused by carbapenem-resistant bacterial pathogens (3–5). However, the emergence of plasmid-mediated colistin resistance genes mcr-1 to mcr-10 (6, 7) and the plasmid-mediated tigecycline resistance genes tet(X3) to tet(X15) (8, 9) was reported and attracted public attention. The coexistence of tet(X4), mcr-1, and blaNDM-5 will seriously threaten the treatment of carbapenem-resistant bacterial pathogen infections, but no such MDR pathogens are reported currently.
Pigeon is one of the most commonly consumed food animals in Jiangsu, China. Pigeons have close contact to human activities, such as breeders raising pigeons or cleaning pigeon cages. These activities, therefore, may pose potential threats to humans as well as other animals, as pigeons may carry and spread different pathogens, including drug-resistant bacteria. Therefore, the antimicrobial resistance of bacteria isolated from pigeons is worthy of our attention. In this study, we performed an antimicrobial resistance surveillance study of bacteria from pigeons and revealed the molecular characteristics of three ST6775 Escherichia coli isolates coharboring tet(X4), mcr-1, and blaNDM-5 from pigeons in China, indicating that the continuous surveillance of such hazardous MDR bacteria should be strengthened to decrease the spread of such MDR bacteria around the world.
RESULTS
Isolates’ identification and resistance phenotypes.
Among 100 fecal samples, 3 tet(X4)-positive isolates, PT62, PT76, and PT77, were acquired. Strikingly, the tet(X4)-positive isolates coharbored mcr-1 and blaNDM-5. They were identified as E. coli. Antimicrobial susceptibility testing revealed that all three E. coli isolates conferred resistance to tigecycline, colistin, and meropenem (Table 1). In addition, they exhibited the same resistance spectrum and were phenotypically resistant to almost all other antimicrobials tested, including kanamycin, doxycycline, ampicillin, streptomycin, amoxicillin, florfenicol, oxytetracycline, and tetracycline, but no resistance to enrofloxacin (Table 1). Three isolates were MDR bacteria accordingly.
TABLE 1.
MICs (mg/L) of three E. coli isolates cocarrying tet(X4), mcr-1, and blaNDM-5a
| Strain ID | Antimicrobials: |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GEN | KAN | DOX | AMP | ENR | CFF | STR | AMX | RIF | CEF | FFC | MEM | IMP | CST | OXY | TET | TIG | |
| PT62 | >128 | >128 | 128 | >128 | ≤0.125 | >128 | 64 | >128 | 16 | >128 | >128 | 16 | 16 | 8 | >128 | >128 | 128 |
| PT76 | >128 | >128 | 64 | >128 | ≤0.125 | >128 | 32 | >128 | 16 | >128 | >128 | 16 | 16 | 4 | >128 | >128 | 64 |
| PT77 | >128 | >128 | 128 | >128 | ≤0.125 | >128 | 64 | >128 | 32 | >128 | >128 | 16 | 16 | 4 | >128 | >128 | 64 |
| ATCC 25922 | 0.25 | 2 | 0.5 | 4 | ≤0.125 | ≤0.125 | 4 | 4 | 4 | ≤0.125 | 4 | ≤0.125 | ≤0.125 | 0.25 | 4 | 0.5 | ≤0.125 |
GEN, gentamicin; KAN, kanamycin; DOX, doxycycline; AMP, ampicillin; ENR, enrofloxacin; CFF, ceftiofur; STR, streptomycin; AMX, amoxicillin; RIF, rifampicin; CEF, ceftriaxone; FFC, florfenicol; MEM, meropenem; IMP, imipenem; CST, colistin; OXY, oxytetracycline; TET, tetracycline; TIG, tigecycline.
Transferability of tet(X4), mcr-1, and blaNDM-5 genes.
To investigate the transmissibility of tet(X4), mcr-1, and blaNDM-5 genes, we subjected three isolates to conjugation experiments with E. coli C600. Results indicated that resistance genes tet(X4) and blaNDM-5 in three isolates, with their corresponding resistance phenotypes for tigecycline and meropenem, were successfully transferred to E. coli C600, suggesting that tet(X4) and blaNDM-5 genes were located on conjugative plasmids or other mobilizable genetic elements in three isolates. The conjugation frequencies of tet(X4)-bearing genetic structures were high and ranged from (1.07 ± 0.1) × 10−1 to (1.63 ± 0.3) × 10−1 transconjugants per recipient. The transfer of blaNDM-5-harboring genetic structures was at frequencies of (3.95 ± 0.4) × 10−3 to (1.10 ± 0.2) × 10−2 cells per recipient. Cotransfer of tet(X4) and blaNDM-5 was at frequencies of (2.07 ± 0.1) × 10−3 to (3.33 ± 0.3) × 10−3 cells per recipient. However, mcr-1 in three isolates were nontransferable. This indicated that horizontal dissemination of tet(X4) and blaNDM-5 by conjugative plasmids or other mobilizable genetic elements existed in the pigeon farm.
The clonal relationship analysis based on whole-genome sequencing.
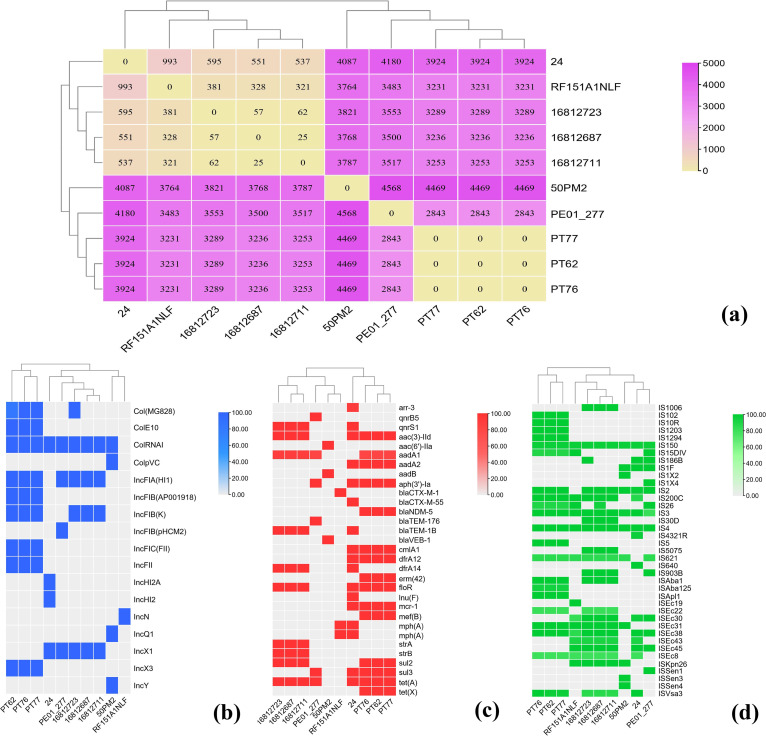
Three isolates were characterized by whole-genome sequencing with the Illumina HiSeq 2500 platform to generate 2 × 150 bp paired-end read data. Multilocus sequence type (MLST) analysis revealed that three isolates positive for tet(X4), mcr-1, and blaNDM-5 belonged to ST6775, which was an infrequent ST type. Analysis of the assembled draft genomes from three isolates coharboring tet(X4), mcr-1, and blaNDM-5 with seven other ST6775 E. coli isolates from the NCBI SRA database provided comprehensive information. We determined the clonal relationship of 10 ST6775 E. coli isolates based on their single-nucleotide polymorphisms (SNPs) of the core genome. The number of differences in SNPs was 0 between PT62, PT76, and PT77 (Fig. 1a). However, the number of SNP differences between the three isolates carrying tet(X4), mcr-1, and blaNDM-5 and the other seven ST6775 E. coli isolates from the NCBI database ranged from 2,843 to 4,469 (Fig. 1a). In addition, all three isolates, PT62, PT76, and PT77, belonged to phylogroup B1 and contained four virulence genes, gad, neuC, terC, and traT. They also contained identical plasmid replicons (Fig. 1b), antimicrobial resistance genes (Fig. 1c), and insertion sequences (Fig. 1d), which suggests that clonal dissemination of isolates carrying tet(X4), mcr-1, and blaNDM-5 existed in the pigeon farm. Apart from tet(X4), mcr-1, and blaNDM-5, 13 resistance genes were identified in each of the three isolates, PT62, PT76, and PT77, including aadA1, aadA2, aph(3′)-Ia, aac(3)-IId, floR, cmlA1, mdf(A), tet(A), sul2, sul3, dfrA12, erm(42), and mef(B) (Fig. 1c). These genes confer resistance to diverse antimicrobials, such as aminoglycosides, phenicol, macrolides, tetracyclines, sulfonamides, and lincosamides. Ten ST6775 E. coli isolates were MDR isolates carrying multiple antimicrobial resistance genes, and each isolate carried at least one β-lactamase gene (Fig. 1c). Isolate 24 from the NCBI database also carried the mcr-1 gene (Fig. 1c). Nine plasmid replicons, Col(MG828), ColE10, ColRNAI, IncFIA(HI1), IncFIB(AP001918), IncFIB(K), IncFIC(FII), IncFII, and IncX3, were found in each of the three isolates PT62, PT76, and PT77 (Fig. 1b). Importantly, we noticed that many other types of plasmids can exist in other ST6775 E. coli from the NCBI database, such as IncHI2 type plasmids where mcr-1 is often located (10) and IncQ1 type plasmids where tet(X4) is often located (11). In addition to PT62, PT76, and PT77, four isolates from the NCBI database also carry ISCR2 or ΔISCR2 (ISVsa3), which provides hot spots for the transposition of tet(X4) (Fig. 1d) (8). Relevant measures should be taken to decrease the prevalence of ST6775 E. coli carrying tet(X4), mcr-1, and blaNDM-5.
FIG 1.
Comprehensive information of the assembled draft genomes from 10 ST6775 E. coli isolates. (a) The number of SNP differences among 10 ST6775 E. coli isolates, including three isolates in the study and seven isolates from the NCBI SRA database. (b) The distribution of plasmid replicons in 10 ST6775 E. coli isolates. The blue rectangles indicate the presence of plasmid replicons. (c) The distribution of antimicrobial resistance genes in 10 ST6775 E. coli isolates. The red rectangles indicate the presence of antimicrobial resistance genes. (d) The distribution of insert sequences in 10 ST6775 E. coli isolates. The green rectangles indicate the presence of insert sequences. Legend labels indicates the similarity (%).
The sequence features of QitanTech sequencing.
To learn the genetic contexts of tet(X4), mcr-1, and blaNDM-5, we carried out the representative isolate PT62 with QitanTech nanopore long-read sequencing to obtain complete genome sequences. A total of 109,949 reads with 748.8 Mb were obtained through the QitanTech nanopore long-read sequencing technology. The mean read length and mean read Phred quality of the total sequencing data were 6,810.5 bp and 8.1, respectively. The median read length and median read Phred quality of the total sequencing data were 3,139 bp and 8.2, respectively. The read length, N50, of the total QitanTech sequencing data was 15,338 bp. The longest read length was 114,033 bp. The mean basecall quality scores of the top five longest reads (reads were more than 99 kb) were greater than 8.0. Then, the QitanTech long-read sequencing data and Illumina short-read data of isolate PT62 were used to perform de novo assembly with the hybrid strategy to obtain the completed chromosome and plasmids. Bioinformatic analysis revealed that isolate PT62 harbored a chromosome and seven plasmids comprising pPT62-tetX-108kb, pPT62-106kb, pPT62-NDM-47kb, pPT62-37kb, pPT62-9kb, pPT62-3kb, and pPT62-1kb (Table 2).
TABLE 2.
Basic information of the chromosome and plasmids in E. coli PT62
| Component | Size (bp) | Accession no. | Replicon type(s) | Resistance gene(s) |
|---|---|---|---|---|
| PT62-chromosome | 4,578,540 | CP090448 | mcr-1, mdf(A) | |
| pPT62-tetX-108kb | 108,576 | CP090449 | IncFII | tet(X4), erm(42), sul2, aac(3)-IId, floR |
| pPT62-106kb | 106,761 | CP090450 | IncFIB(AP001918), IncFIC(FII) | aph(3′)-Ia, sul3, mef(B) |
| pPT62-NDM-47kb | 47,849 | CP090451 | IncX3 | bla NDM-5 |
| pPT62-37kb | 37,779 | CP090452 | IncFIA(HI1), IncFIB(K) | aadA2, aadA1, cmlA1, dfrA12, sul3, tet(A) |
| pPT62-9kb | 9,533 | CP090453 | ColE10 | None |
| pPT62-3kb | 2,994 | CP090454 | ColRNAI | None |
| pPT62-1kb | 1,506 | CP090455 | Col(MG828) | None |
Genetic environment analysis of mcr-1.
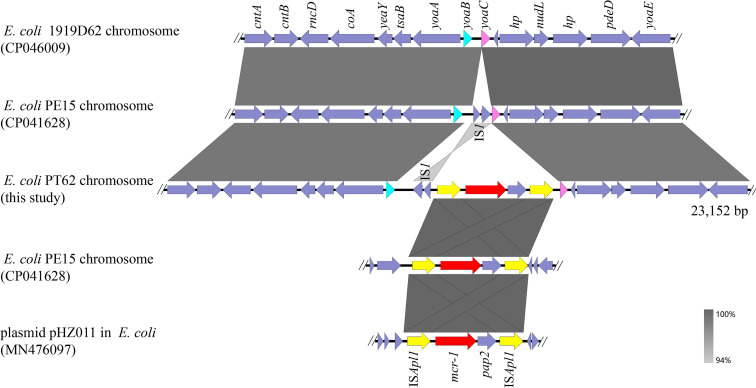
The mcr-1 gene in E. coli PT62 was located on the chromosome and mediated by the Tn6330 (ISApl1-mcr-1-pap2-ISApl1) mobile element. A chromosomal segment (23,152 bp in length) containing Tn6330 was selected to perform online BLASTn analysis, which indicated that the segment exhibited 99% identity at 77% coverage with E. coli 1919D62 chromosome (CP046009) without Tn6330 and 99% identity at 75% coverage against E. coli PE15 chromosome (CP041628) with Tn6330 appearing in another position of PE15 chromosome (Fig. 2). In addition, the Tn6330 could also exist on the plasmids (Fig. 2). It has been reported that Tn6330 could appear on the plasmid and the chromosome in one isolate simultaneously (12). Tn6330 was believed to play an important role in facilitating the transmission of mcr-1 (12, 13), therefore expanding the host range. The E. coli PT62 may acquire mcr-1 by inserting Tn6330 and IS1 between genes yoaB and yoaC (Fig. 2).
FIG 2.
Genetic environment of mcr-1 in the chromosome. Linear alignment of the selected chromosomal segment (23,152 bp) containing Tn6330 with other homologous sequences available in the NCBI database.
Genetic context analysis of tet(X4).
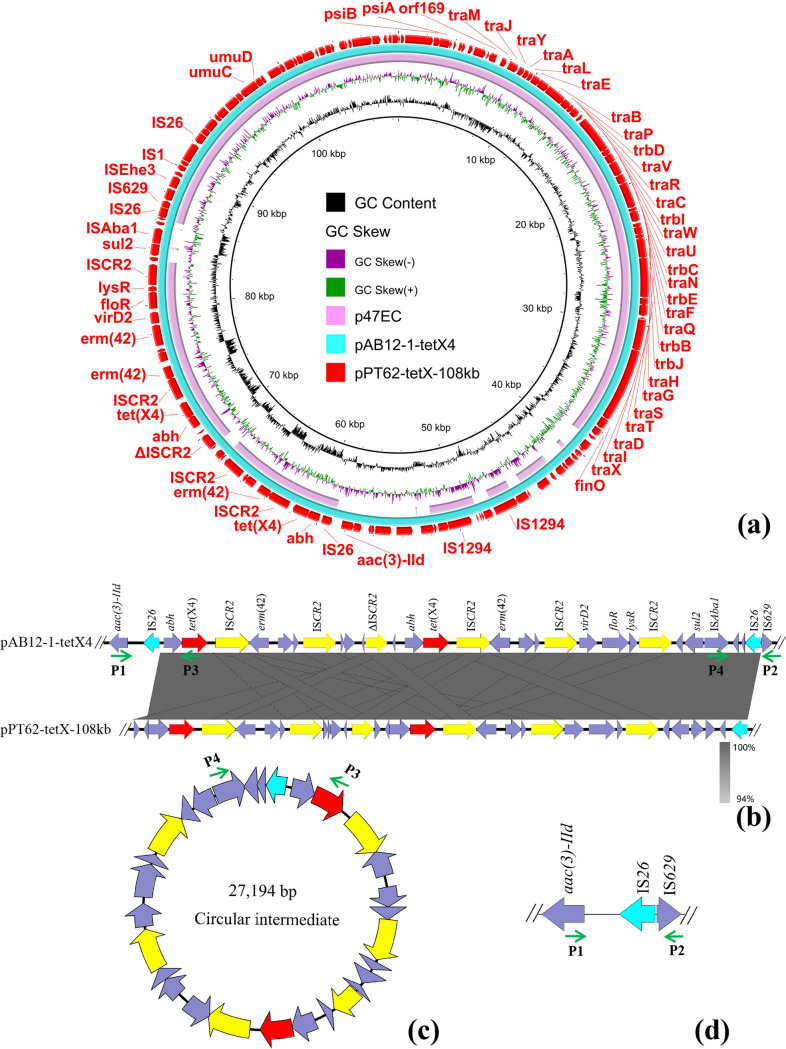
The plasmid pPT62-tetX-108kb was a tet(X4)-bearing IncFII plasmid with an overall 99% nucleotide identity and 100% query coverage to the sequence of plasmid pAB12-1-tetX4 (MZ054177) in E. coli AB12-1. The plasmid pPT62-tetX-108kb also showed 99% identity (85% coverage) to the sequence of plasmid p47EC (MK134376) in E. coli 47EC (Fig. 3a). In addition, two repeats of tet(X4) were found in pPT62-tetX-108kb (Fig. 3a). Various genetic contexts of tet(X4) have been reported previously, and all structures were related to ISCR2 (14). The tet(X4)-harboring genetic context in pPT62-tetX-108kb was abundant with insertion sequences, and its structure was IS26-abh-tet(X4)-ISCR2-erm(42)-orf-hp-ISCR2-hp-ΔISCR2-hp-abh-tet(X4)-ISCR2-erm(42)-orf-hp-ISCR2-virD2-floR-lysR-ISCR2-orf-sul2-ISAba1-orf-hp-IS26 (27,917 bp) (Fig. 3b), which was similar to the genetic structure surrounding tet(X4) found in plasmid pAB12-1-tetX4, but only one copy of IS26 was found on the structure of plasmid pAB12-1-tetX4 (Fig. 3b). He et al. reported that the formation of tet(X4)-carrying circular intermediate is mediated by ISCR2, detected through inverse PCR assays (8). In this study, we designed primers P1, P2, P3, and P4 to verify the formation of tet(X4)-bearing circular intermediate caused by IS26. We proved that the tet(X4)-bearing structure could exist on the plasmid pPT62-tetX-108kb (Fig. 3b) or generate the tet(X4)-bearing circular intermediate mediated by IS26 (Fig. 3c) and leave one copy of IS26 after the circular intermediate fell off (Fig. 3d). Therefore, the structure flanked by IS26 is likely to be responsible for the transfer of tet(X4) among different plasmids or isolates. The plasmid pPT62-tetX-108kb was an IncFII type MDR plasmid carrying genes tet(X4), erm(42), sul2, aac(3)-IId, and floR (Table 2). It has been reported that tet(X4)-harboring IncFII plasmids with high transfer frequency could coexist with mcr-1-bearing IncI2 or IncHI2 plasmids in one isolate (15, 16), implying a potential threat to public health from tet(X4)-bearing IncFII plasmids, and the horizontal spread of tet(X4)-bearing IncFII plasmids should arouse great attention.
FIG 3.
Genetic context of tet(X4) in the IncFII plasmid. (a) Circular comparison between the tet(X)-bearing IncFII plasmid pPT62-tetX-108kb in the study and other homologous plasmids pAB12-1-tetX4 and p47EC available in the NCBI database. The outermost circles indicate the plasmid pPT62-tetX-108kb with genes annotated. (b) Linear alignment of tet(X4)-bearing structure on plasmid pPT62-tetX-108kb with tet(X4)-carrying structure on plasmid pAB12-1-tetX4 available in the NCBI database. Green arrows indicate the directions of the primers used to screen the formation of the circular intermediate, and names of primers are marked next to the arrows. (c) The circular intermediate mediated by IS26. Green arrows indicate the directions of the primers. (d) A copy of IS26 left after the circular intermediate fell off. Green arrows indicate the directions of the primers.
Genetic environment analysis of blaNDM-5.
The blaNDM-5 gene was located on the IncX3 plasmid pPT62-NDM-47kb. The BLASTn analysis indicated that pPT62-NDM-47kb exhibited 100% identity at 96% coverage with plasmid pEc-MW07_NDM (LC545851) from E. coli Ec-MW07 and plasmid pNDM5_020031 (CP033399) from E. coli WCHEC020031 (Fig. 4). No other antimicrobial resistance genes were identified on the plasmid. The blaNDM-5-bearing genetic structure in pPT62-NDM-47kb was IS26-orf-orf-orf-blaNDM-5-IS5-ISAba125-IS3000-IS1294 (Fig. 4). IncX3 type plasmid was an important vector of blaNDM-5 and has been reported to appear frequently in various sources (17, 18). The similar IncX3 plasmids found in pigeons further highlight the importance of the epidemic IncX3 plasmid in the spread of the blaNDM-5 gene around the ecosystem. The blaNDM-5-bearing IncX3 plasmid was frequently found to coexist with mcr-1-bearing plasmids in single isolates (19, 20). Recently, the blaNDM-5-bearing IncX3 plasmid was reported to coexist with tet(X4)-harboring plasmid in one isolate (21). There was even one strain in which the blaNDM-4, tet(X4) and tmexCD3-toprJ3 genes were in a single IncC-IncX3 hybrid plasmid (22). The emergence of isolates’ resistance to carbapenem and tigecycline makes the clinical treatment of carbapenem-resistant pathogens challenging.
FIG 4.
Genetic environment of blaNDM-5 in the IncX3 plasmid. Circular comparison of three blaNDM-5-bearing plasmids. The blaNDM-5-harboring IncX3 plasmid pPT62-NDM-47kb in this study was compared with two plasmids pEc-MW07_NDM and pNDM5_020031 in the NCBI database. The outermost circles indicate the plasmid pPT62-NDM-47kb with genes annotated.
DISCUSSION
The emergence of E. coli isolates conferring resistance to carbapenem and tigecycline from pigeons in this study is alarming because carbapenems and tigecycline are not licensed for use in animals worldwide; however, colistin may have been used in feedstuff as the commercial feed additive before May 2017. Colistin and tigecycline are two of the last-resort therapies to treat infections caused by carbapenem-resistant bacterial pathogens. The presence of ST6775 E. coli isolates carrying tigecycline resistance gene tet(X4), colistin resistance gene mcr-1, and carbapenem resistance gene blaNDM-5 from pigeons posed a great public health concern, as therapeutic choices in such cases are very limited. In fact, ST6775 is an infrequent sequence type. It is worth thinking about whether the accumulation of tet(X4), mcr-1, and blaNDM-5 in an ST6775 E. coli isolate is related to certain molecular mechanisms or just a coincidence, which warrants further investigations. It is reported that ST410 E. coli isolates could carry blaNDM-5-bearing IncX3 plasmids (23) or tet(X4)-bearing IncFII plasmids (15). Also, chromosomal location of mcr-1 was found in ST410 E. coli isolate (24). The ST48 E. coli is frequently found to carry the blaNDM-5-bearing IncX3 plasmid (17) or the mcr-1-harboring IncX4 plasmid (25). The coexistence of tet(X4)-harboring IncFII plasmid and mcr-1-bearing IncHI2 plasmid in an ST48 or ST6751 E. coli isolate has even been reported (16). The blaNDM-5-bearing IncX3 plasmids were widely detected in ST6751 E. coli isolates (26). Isolates with similar situations are too numerous to mention one by one. It is possible that these ST-type isolates are potential targets for the convergence of three genes tet(X4), mcr-1, and blaNDM-5. Preventing the prevalence of isolates with these three genes coexisting is one of our important tasks to curb the development of antimicrobial resistance.
There have been many studies on plasmid fusion mediated by insert sequences, such as IS26 (14) and ISCR2 (27). In the study, tet(X4)-bearing structure flanking two copies of IS26 could generate the circular intermediate mediated by IS26, suggesting that the structure flanked by IS26 is likely to be responsible for the transfer of tet(X4) among different plasmids or isolates. The tet(X4)-harboring genetic context was abundant with ISCR2 and ΔISCR2; the formation of tet(X4)-carrying minicircles mediated by ISCR2 is also possible (8). Therefore, tet(X4)-bearing structure may be able to form recombinant plasmids with other plasmids carrying ISCR2 or IS26, which frequently appear on many MDR plasmids, such as blaNDM-5-bearing IncX3 plasmids or mcr-1-harboring IncHI2 plasmids. The cocarriage of tet(X6) and mcr-1 by a single IncHI2 plasmid (28), and the cocarriage of blaNDM-4, tet(X4), and tmexCD3-toprJ3 genes by a single IncC-IncX3 hybrid plasmid, have been found previously (22). Previously, we also proved that a single hybrid plasmid bearing mcr-1 and tet(X4) may appear under the mediation of insertion sequences (29). With the emergence of isolates carrying tet(X4), mcr-1, and blaNDM-5, the factors affecting plasmid recombination need to be further studied to prevent the emergence and evolution of the plasmids cocarrying tet(X4), mcr-1, and blaNDM-5.
Conclusions. In conclusion, to the best of our knowledge, this is the first report of the coexistence of tet(X4), mcr-1, and blaNDM-5 in ST6775 E. coli isolates of animal origin in China. Alarmingly, tet(X4), mcr-1, and blaNDM-5 were located on transposons or plasmids that were mobile genetic elements related to the capture, accumulation, and dissemination of resistance genes. This may lead to increasing prevalence of isolates coharboring tet(X4), mcr-1, and blaNDM-5 in the future. Such MDR isolates pose a significant challenge to public health; thus, comprehensive surveillance of such MDR bacteria among different sources as a One Health approach should be utilized. Furthermore, relevant measures should be taken to decrease the prevalence of E. coli carrying tet(X4), mcr-1, and blaNDM-5.
MATERIALS AND METHODS
Bacterial isolates and identification.
In June 2021, a total of 100 fresh fecal samples were randomly collected from a pigeon farm located in Jiangsu Province, China. Individual fresh fecal samples were collected from 100 different pigeon breeding cages using sterile swabs. Subsequently, each sample was placed in brain heart infusion broth containing tigecycline (2 mg/L) and incubated for 4 h at 37°C with 200 rpm constant shaking. Then, the cultures were plated on MacConkey agar plates supplemented with tigecycline (2 mg/L) and incubated for 14 h at 37°C. One or more colonies with different morphological characteristics from each MacConkey agar plate were purified and subsequently screened for the presence of tet(X) by PCR with specific primers and Sanger sequencing as reported previously (8). mcr and blaNDM were further identified in tet(X)-positive isolates using primers described earlier (17, 30). 16S rRNA gene sequencing was performed to confirm bacterial species.
Antimicrobial susceptibility testing.
The MICs of E. coli isolates coharboring tet(X), mcr, and blaNDM against the antimicrobials (gentamicin, kanamycin, doxycycline, ampicillin, enrofloxacin, ceftiofur, streptomycin, amoxicillin, rifampicin, ceftriaxone, florfenicol, meropenem, imipenem, colistin, oxytetracycline, tetracycline, and tigecycline) were determined using the broth microdilution method in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines (31). Results were interpreted according to the CLSI standards and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints (http://www.eucast.org/clinical_breakpoints/). E. coli ATCC 25922 served as the quality control strain.
Conjugation experiments.
To investigate the transferability of tet(X), mcr, and blaNDM, we performed conjugation assays as described previously (15). Briefly, each isolate carrying tet(X), mcr, and blaNDM was used as the donor and E. coli C600 (resistant to rifampin) was used as the recipient. Cultures of donors and recipient that reached the 0.5 McFarland culture density were mixed at a ratio of 1:1, respectively. Then, 0.1 mL of the mixed cultures was applied onto LB agar plates. After cultures were kept at 37°C for 12 h, we subsequently collected the bacterial cultures on plates and diluted them in sterile saline. We screened transconjugants carrying tet(X), mcr, or (and) blaNDM on LB agar plates containing corresponding antimicrobials. Antimicrobials were used at the following concentrations: tigecycline, 2 mg/L; colistin, 2 mg/L; meropenem, 2 mg/L; rifampicin, 300 mg/L. The presence of tet(X), mcr, and blaNDM in transconjugants was confirmed by PCR and corresponding resistance phenotyping. Frequencies of conjugation transfer were calculated by the number of transconjugants per recipient.
Genome DNA sequencing.
The genomic DNA of three isolates were extracted using the FastPure bacteria DNA isolation minikit (Vazyme, China) in accordance with the manufacturer’s recommendations. The concentration and quantity of genome DNA were assessed using the Colibri LB 915 spectrophotometer (Titertek-Berthold, Germany) and gel electrophoresis. The short-read DNA sequencing was performed via Illumina HiSeq 2500 platform, and one representative isolate was further sent out for QitanTech nanopore single-molecule long-read sequencing, which is an emerging nanopore sequencing technology launched by Qitan Technology (QitanTech) in China (32). Short-read and long-read data were acquired with fastq format for further analysis. The quality of sequence data generated by QitanTech nanopore long-read sequencing technology was evaluated using Nanoplot of NanoPack v1.25.0 (33).
Bioinformatic analysis.
The short-read Illumina raw sequences of three isolates were assembled using SPAdes v3.13.1 (34). Illumina short-read and QitanTech nanopore long-read data were used to perform de novo assembly with Unicycler v0.4.8 (35, 36). The Rapid Annotation using Subsystems Technology annotation website server (https://rast.nmpdr.org/rast.cgi) was then used to annotate the assembled genomes (37). The multilocus sequence typing (MLST) was determined using online tool MLST v2.0 (https://cge.cbs.dtu.dk/services/MLST/). Seven available ST6775 E. coli genomes according to the EnteroBase (https://enterobase.warwick.ac.uk/) were acquired from the NCBI SRA database. The plasmid replicons, antimicrobial resistance genes, and virulence genes were analyzed using PlasmidFinder v2.1, ResFinder v4.1, and VirulenceFinder v2.0 (https://cge.cbs.dtu.dk/services/). Insertion sequences were identified using ISfinder v2.0 (https://www-is.biotoul.fr). The assembly genomes were also annotated using Prokka v1.12 (38). A pan-genome analysis was conducted on the samples categorized as ST6775 E. coli isolates using the Roary v3.13.0 (39). SNPs distances were compared within ST6775 E. coli isolates using snp-dists v0.7.0 (https://github.com/tseemann/snp-dists). TBtools v1.098661 was used to visualize the SNPs distances as well as the distributions of antimicrobial resistance genes, plasmid replicons, and insertion sequences (40). Circular comparisons between plasmids were performed using the BRIG v0.95 tool (41). To visualize the genetic comparison features, we used Easyfig v2.2.3 to generate linear comparison figures (42).
Verification of the circular intermediate formation.
Four primers, P1 (5′-TCTCCTCGTAGGGTGATCGG-3′), P2 (5′-AGCCTTCCCAGCAATCGTC-3′), P3 (5′-TAGTCAGTCCAACGGGTCCA-3′), and P4 (5′-TTCAAATGGCGATTCAGCGT-3′), were designed to determine whether homologous recombination between two copies of IS26 in the same direction could result in the formation of tet(X4)-bearing circular intermediate. Because the complete tet(X4)-containing structure is too long to be directly amplified by PCR, primer pairs P1 and P3 (PCR product was 2,753 bp), and P2 and P4 (2,021 bp) were employed to prove that the structure exists on the tet(X4)-bearing plasmid. Primer pair P3 and P4 (2,679 bp) was used to confirm the formation of the circular intermediate. Primer pair P1 and P2 (2,095 bp) was employed to prove that only one copy of IS26 was left after the circular intermediate fell off. The PCR products were further purified and then confirmed by Sanger sequencing using the same primers.
Nucleotide sequence accession numbers.
The nucleotide sequences of the chromosome and plasmids of E. coli PT62 have been deposited in the NCBI database with GenBank accession numbers CP090448 to CP090455 (Table 2). The draft genomes of E. coli PT76 and PT77 are also available in the NCBI database (BioProject accession number PRJNA780102).
ACKNOWLEDGMENTS
This work was supported by the Jiangsu Agricultural Science and Technology Innovation Fund [CX(21)2010 and CX(21)3138], the China Postdoctoral Science Foundation (no. 2020M671632), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). We declare no conflicts of interest.
Contributor Information
Zhiqiang Wang, Email: zqwang@yzu.edu.cn.
Ruichao Li, Email: rchl88@yzu.edu.cn.
Tino Polen, Forschungszentrum Jülich GmbH.
REFERENCES
- 1.Potter RF, D'Souza AW, Dantas G. 2016. The rapid spread of carbapenem-resistant Enterobacteriaceae. Drug Resist Updat 29:30–46. doi: 10.1016/j.drup.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu W, Feng Y, Tang G, Qiao F, Mcnally A, Zong Z. 2019. NDM metallo-beta-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev 32:e00115-18. doi: 10.1128/CMR.00115-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paul M, Daikos GL, Durante-Mangoni E, Yahav D, Carmeli Y, Benattar YD, Skiada A, Andini R, Eliakim-Raz N, Nutman A, Zusman O, Antoniadou A, Pafundi PC, Adler A, Dickstein Y, Pavleas I, Zampino R, Daitch V, Bitterman R, Zayyad H, Koppel F, Levi I, Babich T, Friberg LE, Mouton JW, Theuretzbacher U, Leibovici L. 2018. Colistin alone versus colistin plus meropenem for treatment of severe infections caused by carbapenem-resistant Gram-negative bacteria: an open-label, randomised controlled trial. Lancet Infect Dis 18:391–400. doi: 10.1016/S1473-3099(18)30099-9. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez-Bano J, Gutierrez-Gutierrez B, Machuca I, Pascual A. 2018. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin Microbiol Rev 31. doi: 10.1128/CMR.00079-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seifert H, Blondeau J, Dowzicky MJ. 2018. In vitro activity of tigecycline and comparators (2014–2016) among key WHO 'priority pathogens' and longitudinal assessment (2004–2016) of antimicrobial resistance: a report from the T.E.S.T. study. Int J Antimicrob Agents 52:474–484. doi: 10.1016/j.ijantimicag.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. 2020. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect 9:508–516. doi: 10.1080/22221751.2020.1732231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He T, Wang R, Liu D, Walsh TR, Zhang R, Lv Y, Ke Y, Ji Q, Wei R, Liu Z, Shen Y, Wang G, Sun L, Lei L, Lv Z, Li Y, Pang M, Wang L, Sun Q, Fu Y, Song H, Hao Y, Shen Z, Wang S, Chen G, Wu C, Shen J, Wang Y. 2019. Emergence of plasmid-mediated high-level tigecycline resistance genes in animals and humans. Nat Microbiol 4:1450–1456. doi: 10.1038/s41564-019-0445-2. [DOI] [PubMed] [Google Scholar]
- 9.Li R, Peng K, Xiao X, Wang Y, Wang Z. 2021. Characterization of novel ISAba1-bounded tet(X15)-bearing composite transposon Tn6866 in Acinetobacter variabilis. J Antimicrob Chemother 76:2481–2483. doi: 10.1093/jac/dkab182. [DOI] [PubMed] [Google Scholar]
- 10.Lu X, Xiao X, Liu Y, Huang S, Li R, Wang Z. 2020. Widespread prevalence of plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli from Père David's deer in China. mSphere 5. doi: 10.1128/mSphere.01221-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun J, Chen C, Cui C-Y, Zhang Y, Liu X, Cui Z-H, Ma X-Y, Feng Y, Fang L-X, Lian X-L, Zhang R-M, Tang Y-Z, Zhang K-X, Liu H-M, Zhuang Z-H, Zhou S-D, Lv J-N, Du H, Huang B, Yu F-Y, Mathema B, Kreiswirth BN, Liao X-P, Chen L, Liu Y-H. 2019. Plasmid-encoded tet(X) genes that confer high-level tigecycline resistance in Escherichia coli. Nat Microbiol 4:1457–1464. doi: 10.1038/s41564-019-0496-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li R, Chen K, Chan EW, Chen S. 2019. Characterization of the stability and dynamics of Tn6330 in an Escherichia coli strain by nanopore long reads. J Antimicrob Chemother 74:1807–1811. doi: 10.1093/jac/dkz117. [DOI] [PubMed] [Google Scholar]
- 13.He Y-Z, Li X-P, Miao Y-Y, Lin J, Sun R-Y, Wang X-P, Guo Y-Y, Liao X-P, Liu Y-H, Feng Y, Sun J. 2019. The ISApl1 2 dimer circular intermediate participates in mcr-1 transposition. Front Microbiol 10:15. doi: 10.3389/fmicb.2019.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li R, Lu X, Peng K, Liu Z, Li Y, Liu Y, Xiao X, Wang Z. 2020. Deciphering the structural diversity and classification of the mobile tigecycline resistance gene tet(X)-bearing plasmidome among Bacteria mSystems 5. doi: 10.1128/mSystems.00134-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li R, Mohsin M, Lu X, Abdullah S, Munir A, Wang Z. 2021. Emergence of plasmid-mediated resistance genes tet(X) and mcr-1 in Escherichia coli clinical isolates from Pakistan. mSphere 6:e0069521. doi: 10.1128/mSphere.00695-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li R, Lu X, Munir A, Abdullah S, Liu Y, Xiao X, Wang Z, Mohsin M. 2022. Widespread prevalence and molecular epidemiology of tet(X4) and mcr-1 harboring Escherichia coli isolated from chickens in Pakistan. Sci Total Environ 806:150689. doi: 10.1016/j.scitotenv.2021.150689. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z, Xiao X, Li Y, Liu Y, Li R, Wang Z. 2019. Emergence of IncX3 plasmid-harboring bla NDM- 5 dominated by Escherichia coli ST48 in a goose farm in Jiangsu, China Front Microbiol 10:2002. doi: 10.3389/fmicb.2019.02002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tian D, Wang B, Zhang H, Pan F, Wang C, Shi Y, Sun Y. 2020. Dissemination of the blaNDM-5 gene via IncX3-type plasmid among Enterobacteriaceae in Children mSphere 5:e00699-19. doi: 10.1128/mSphere.00699-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han H, Liu W, Cui X, Cheng X, Jiang X. 2020. Co-existence of mcr-1 and bla NDM-5 in an Escherichia coli strain isolated from the pharmaceutical industry, WWTP. Infect Drug Resist 13:851–854. doi: 10.2147/IDR.S245047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang R-S, Feng Y, Lv X-Y, Duan J-H, Chen J, Fang L-X, Xia J, Liao X-P, Sun J, Liu Y-H. 2016. Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single Muscovy duck (Cairina moschata). Antimicrob Agents Chemother 60:6899–6902. doi: 10.1128/AAC.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun H, Zhai W, Fu Y, Li R, Du P, Bai L. 2021. Co-occurrence of plasmid-mediated resistance genes tet(X4) and blaNDM-5 in a multidrug-resistant Escherichia coli isolate recovered from chicken in China. J Glob Antimicrob Resist 24:415–417. doi: 10.1016/j.jgar.2021.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Hirabayashi A, Dao TD, Takemura T, Hasebe F, Trang LT, Thanh NH, Tran HH, Shibayama K, Kasuga I, Suzuki M. 2021. A transferable IncC-IncX3 hybrid plasmid cocarrying blaNDM-4, tet(X), and tmexCD3-toprJ3 confers resistance to carbapenem and tigecycline. mSphere 6. doi: 10.1128/mSphere.00592-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramadan H, Gupta SK, Sharma P, Ahmed M, Hiott LM, Barrett JB, Woodley TA, Frye JG, Jackson CR. 2020. Circulation of emerging NDM-5-producing Escherichia coli among humans and dogs in Egypt. Zoonoses Public Health 67:324–329. doi: 10.1111/zph.12676. [DOI] [PubMed] [Google Scholar]
- 24.Falgenhauer L, Waezsada S-E, Gwozdzinski K, Ghosh H, Doijad S, Bunk B, Spröer C, Imirzalioglu C, Seifert H, Irrgang A, Fischer J, Guerra B, Käsbohrer A, Overmann J, Goesmann A, Chakraborty T. 2016. Chromosomal locations of mcr-1 and blaCTX-M-15 in fluoroquinolone-resistant Escherichia coli ST410. Emerg Infect Dis 22:1689–1691. doi: 10.3201/eid2209.160692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Liu H, Wang Q, Du X, Yu Y, Jiang Y. 2020. Coexistence of blaKPC-2-IncN and mcr-1-IncX4 plasmids in a ST48 Escherichia coli strain in China. J Glob Antimicrob Resist 23:149–153. doi: 10.1016/j.jgar.2020.08.023. [DOI] [PubMed] [Google Scholar]
- 26.Zhai R, Fu B, Shi X, Sun C, Liu Z, Wang S, Shen Z, Walsh TR, Cai C, Wang Y, Wu C. 2020. Contaminated in-house environment contributes to the persistence and transmission of NDM-producing bacteria in a Chinese poultry farm. Environ Int 139:105715. doi: 10.1016/j.envint.2020.105715. [DOI] [PubMed] [Google Scholar]
- 27.Li R, Lu X, Peng K, Liu Y, Xiao X, Wang Z. 2020. Reorganization of mcr-1-bearing large MDR plasmids resolved by nanopore sequencing. J Antimicrob Chemother 75:1645–1647. doi: 10.1093/jac/dkaa046. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y, Liu L, Zhang H, Feng Y. 2021. Co-production of Tet(X) and MCR-1, two resistance enzymes by a single plasmid. Environ Microbiol. [DOI] [PubMed] [Google Scholar]
- 29.Lu X, Xiao X, Liu Y, Li R, Wang Z. 2021. Emerging opportunity and destiny of mcr-1- and tet(X4)-coharboring plasmids in Escherichia coli. Microbiol Spectr 9:e0152021. doi: 10.1128/Spectrum.01520-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, Hansen IM, Guerra B, Malorny B, Borowiak M, Hammerl JA, Battisti A, Franco A, Alba P, Perrin-Guyomard A, Granier SA, Escobar CDF, Malhotra-Kumar S, Villa L, Carattoli A, Hendriksen RS. 2018. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill 23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed. CLSI supplement M100. CLSI, Wayne, PA. [Google Scholar]
- 32.Wang Z, Qin L, Liu J, Jiang L, Zou X, Chen X, Song F, Dai H, Hou Y. 2022. Forensic nanopore sequencing of microhaplotype markers using QitanTech's QNome. Forensic Sci Int Genet 57:102657. doi: 10.1016/j.fsigen.2021.102657. [DOI] [PubMed] [Google Scholar]
- 33.De Coster W, D'Hert S, Schultz DT, Cruts M, Van Broeckhoven C. 2018. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, Xie M, Dong N, Lin D, Yang X, Wong MHY, Chan EW-C, Chen S. 2018. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience 7:1–9. doi: 10.1093/gigascience/gix132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 39.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. 2020. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202. doi: 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 41.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]