ABSTRACT
Huntington’s disease (HD) is a neurodegenerative disorder caused by a trinucleotide expansion in the HTT gene, which is expressed throughout the brain and body, including the gut epithelium and enteric nervous system. Afflicted individuals suffer from progressive impairments in motor, psychiatric, and cognitive faculties, as well as peripheral deficits, including the alteration of the gut microbiome. However, studies characterizing the gut microbiome in HD have focused entirely on the bacterial component, while the fungal community (mycobiome) has been overlooked. The gut mycobiome has gained recognition for its role in host homeostasis and maintenance of the gut epithelial barrier. We aimed to characterize the gut mycobiome profile in HD using fecal samples collected from the R6/1 transgenic mouse model (and wild-type littermate controls) from 4 to 12 weeks of age, corresponding to presymptomatic through to early disease stages. Shotgun sequencing was performed on fecal DNA samples, followed by metagenomic analyses. The HD gut mycobiome beta diversity was significantly different from that of wild-type littermates at 12 weeks of age, while no genotype differences were observed at the earlier time points. Similarly, greater alpha diversity was observed in the HD mice by 12 weeks of age. Key taxa, including Malassezia restricta, Yarrowia lipolytica, and Aspergillus species, were identified as having a negative association with HD. Furthermore, integration of the bacterial and fungal data sets at 12 weeks of age identified negative correlations between the HD-associated fungal species and Lactobacillus reuteri. These findings provide new insights into gut microbiome alterations in HD and may help identify novel therapeutic targets.
IMPORTANCE Huntington’s disease (HD) is a fatal neurodegenerative disorder affecting both the mind and body. We have recently discovered that gut bacteria are disrupted in HD. The present study provides the first evidence of an altered gut fungal community (mycobiome) in HD. The genomes of many thousands of gut microbes were sequenced and used to assess “metagenomics” in particular the different types of fungal species in the HD versus control gut, in a mouse model. At an early disease stage, before the onset of symptoms, the overall gut mycobiome structure (array of fungi) in HD mice was distinct from that of their wild-type littermates. Alterations of multiple key fungi species were identified as being associated with the onset of disease symptoms, some of which showed strong correlations with the gut bacterial community. This study highlights the potential role of gut fungi in HD and may facilitate the development of novel therapeutic approaches.
KEYWORDS: DNA sequencing, fungal-bacterial interactions, host-cell interactions, metagenomics, mycology
INTRODUCTION
Huntington’s disease (HD) is a heritable neurodegenerative disorder that is characterized by gradual brain atrophy, particularly in the cerebral cortex and striatum, although other parts of the brain and body are affected (1). The afflicted individuals suffer from progressive motor, cognitive, and psychiatric symptoms, leading to death after 15 to 20 years (1). HD is formally diagnosed when motor symptoms become apparent, which typically occurs between 35 and 55 years of age, although other symptoms may manifest 10 to 15 years prior to this (1). The disease is caused by the expansion of CAG repeats in the huntingtin (Htt) gene, which is ubiquitously expressed not only in the brain but also in many tissues throughout the body, including skeletal muscles and gut intestinal epithelial cells (2–4). The expression of the HD mutation profoundly disrupts transcriptional regulation and normal cellular function, which affects overall physiology (5–7). Thus, the afflicted individuals also suffer from peripheral impairments, including skeletal muscle atrophy, significant weight loss, and impaired immune response (8–11). Recent evidence from our group and others has also shown altered gut bacteria (bacteriome) in HD preclinical models and patients, occurring during the early stage of the disease (12–14).
The gut microbiome encompasses a rich and complex ecosystem composed of various microorganisms, including bacteria, fungi, viruses, and archaea. Studies from the last decade have demonstrated the importance of the gut commensal bacteria in the regulation of host food metabolism and energy homeostasis, as well as the host immunological response. Notably, the gut microbiome maintains a bidirectional communication with the brain through the microbiome-gut-brain axis, and it is capable of regulating host brain health and behavior (15). While previous studies have largely focused on the bacterial residents (i.e., the bacteriome) of the gut, the fungal microbiome (i.e., the mycobiome) has been less explored. The mycobiome represents a relatively small fraction of the gut microbiome (0.01% to 0.1%); however, it has recently begun to gain recognition as an integral part of the gut ecosystem in maintaining host health (16–18).
Similar to intestinal bacteria, commensal fungi are an integral component in host immunomodulation and metabolism (19–22). It has recently been demonstrated that the gut fungal community can induce profound changes in the gut bacteriome structure and shape the gut microbiome during early life (23). These fungal taxa could then elicit robust local and systemic immune responses from synergistic interactions with the gut bacterial population and, thus, play a role in the maturation of the host immune system (23, 24). Additionally, certain fungal species are widely used as probiotics, as they can secrete enzymes that inactivate toxins produced by inflammatory intestinal residents and suppress the growth of other potential pathobionts (25–27). For example, Saccharomyces boulardii is prescribed as a probiotic for the management of diarrhea and to prevent intestinal colonization with Clostridium difficile following antibiotic therapy (28–30). Oral administration of Candida kefyr has been shown to alter the gut microbiome composition and attenuate gastrointestinal inflammation (31). Furthermore, fungi are able to synthesize and release neurotransmitters, similarly to many bacteria. Saccharomyces cerevisiae and Penicillium chrysogenum can produce high concentrations of norepinephrine, which is capable of reducing anxiety (32, 33). Candida albicans has been shown to produce histamine, another neuromodulator involved in appetite regulation, sleep-wake rhythm, and cognitive activity (34). These neurotransmitters could also promote or inhibit bacterial and fungal growth, in addition to their neuromodulatory effects on the enteric nervous system, if not the central nervous system (33).
However, intestinal fungi can also elicit deleterious effects on host health and have been associated with a number of disorders, including gastrointestinal diseases like inflammatory bowel disease (IBD), Crohn’s disease (CD), and colorectal cancer (35–38). One study reported increased representation of Candida albicans and overgrowth of other Candida species and decreased proportions of S. cerevisiae in IBD patients (35). In CD, Candida tropicalis was overrepresented in patients and correlated positively with anti-S. cerevisiae antibodies (36). Additionally, altered Basidiomycota/Ascomycota ratios were observed in these diseases (35, 39). Other recent evidence has shown disruption of the gut mycobiome in neurological disorders, including autism spectrum disorder (ASD), multiple sclerosis (MS), mild cognitive impairment, and Alzheimer’s disease (40–45). Moreover, perturbations in the gut fungal-bacterial relationships were observed in these disorders (36, 37, 42, 43, 46).
Remarkably, Alonso et al. reported evidence of yeast-like cells and fungal antigens in postmortem brain slices of HD patients, suggesting that HD patients experience fungal infections more commonly than matched controls (47). Furthermore, the enzyme chitinase 3-like 1 protein, secreted by astrocytes and macrophages, was elevated in the cerebrospinal fluid in HD (48). This enzyme can be activated by chitin, a major component of the fungal cell wall. Given that the gastrointestinal tract is the major source of fungal antigens and infection, we hypothesized that the HD gut mycobiome composition might be altered, which might then contribute to HD pathogenesis and progression.
We have previously shown through shotgun metagenomic sequencing the change in the gut bacteriome profile in the R6/1 transgenic mouse model of HD (herein known as HD mice) at 12 weeks of age, which coincides with the onset of overt motor symptoms (49). Here, we aimed to characterize the gut mycobiome profile in HD mice and determine the unique characteristics that distinguish it from the gut mycobiome profile of healthy controls, as well as to interrogate the fungal-bacterial interactions in these mice.
RESULTS
The current study investigated the fungal community in the shotgun metagenomic data set constructed from 90 fecal samples of R6/1 HD transgenic mice and their wild-type (WT) littermates sampled at five time points: 4 (young), 6, 8, 10, and 12 (early disease stage) weeks of age (49). On average, 0.1% of the reads aligned to fungal genomes, which primarily consisted of fungi from the Ascomycota (∼61%) and Basidiomycota (∼22%) phyla, with Ascomycota being the most abundant phylum represented. The most abundant genera in the murine gut fungal genera consisted of Aspergillus (∼6%), Batrachochytrium (∼3.27%), Schizosaccharomyces (∼2.15%), and Fusarium (1.7%).
HD mice harbor an altered gut mycobiome composition at week 12.
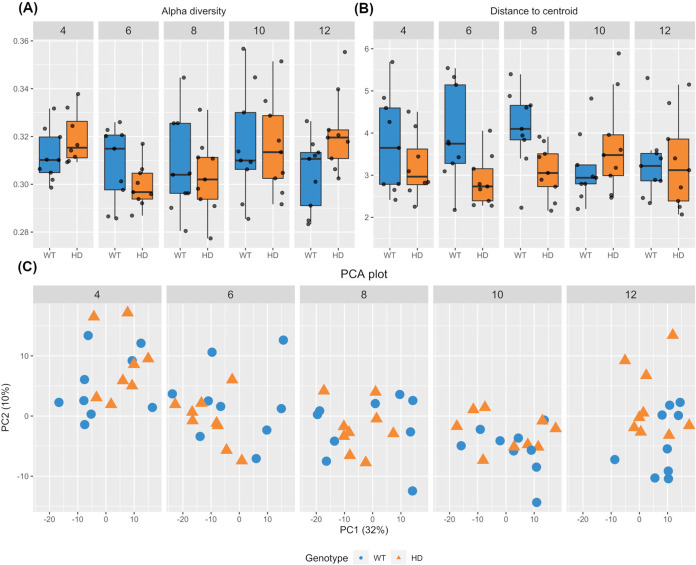
The Ascomycota/Basidiomycota ratio was previously reported as an indicator of healthy gut fungal composition. In our case, no significant differences in these two phyla were observed between the two genotypes. The Shannon alpha diversity index, which measures the evenness and richness of communities within a sample, did not differ between the two genotypes during early life until the young adult age, from week 4 to week 10, but it increased in the HD gut at week 12 (t test, P = 0.034) (Fig. 1A). To determine the variability in the gut fungal communities, the average distance of each sample to the group centroid was calculated and compared. Overall, the fecal mycobiome was less heterogenous in HD mice than in their WT littermates at week 6 (P = 0.018) and week 8 (P = 0.005) but not at any other time points (Fig. 1B). Additionally, mycobiome stability was determined by calculating the Aitchison distance travelled by each sample across time, as previously described (50). There were no significant differences in within-subject fungal stability between WT and HD mice (see the supplemental material). The beta diversity of gut mycobiome was similar between the two genotypes from week 4 to week 10. In contrast, the gut mycobiome composition of HD mice was significantly different from that of their WT counterparts at week 12, as shown by the clustering of samples according to genotype in principal-component analysis (PCA) (permutational multivariate analysis of variance [PERMANOVA], P = 0.025) (Fig. 1C).
FIG 1.
Gut mycobiome alterations in HD mice. (A) Increased alpha diversity of fecal mycobiome in HD mice at week 12, but not any earlier age, compared to their WT littermates (Kruskal-Wallis, P = 0.034). (B) Boxplot showing the distance of each sample to the group centroid. HD mice displayed less heterogeneity in gut mycobiome composition only at week 6 (P = 0.018) and week 8 (P = 0.005). (C) Principal-component analysis (PCA) plot of the gut mycobiome. The HD mouse gut mycobiome composition was significantly different from that of their WT littermates at week 12 but not at any earlier time point (PERMANOVA, P = 0.025).
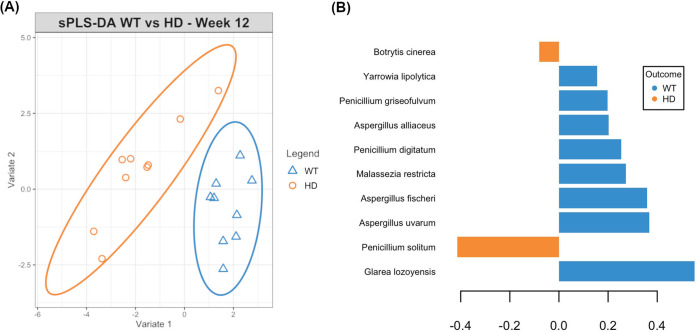
As an overall difference in gut fungal communities was observed only at week 12, the supervised sparse partial least-squares discriminant analysis (sPLS-DA) method was employed to identify the main species driving this difference. A signature consisting of 15 fungal species that were discriminatory between the two genotypes was identified (classification error rate = 0.27) (Fig. 2). In particular, the fungal species identified as having the best predictive value were Glarea lozoyensis, Malassezia restricta, Penicillium digitatum, Yarrowia lipolytica, and Aspergillus spp., including Aspergillus fischeri, Aspergillus uvarum, and Aspergillus alliaceus, all of which were depleted in the HD mouse mycobiome compared to their levels in the WT littermate mycobiome. Furthermore, Penicillium solitum and Meyerozyma guilliermondii were also selected in the signature, both of which were enriched in the HD gut mycobiota.
FIG 2.
Signature gut fungal species discriminatory between WT and HD mice identified by sparse PLS-DA (classification error rate = 0.27). (A) Sample plot (95% confidence ellipses) revealed distinct clustering of WT and HD samples, indicating the predictive power of the selected fungal signature to distinguish between the two genotypes. (B) Contributions of the top 10 fungal species identified to be discriminatory between WT and HD samples. Species with negative contributions were enriched in HD mice (largest mean value in this group), while those with positive contributions were enriched in WT mice.
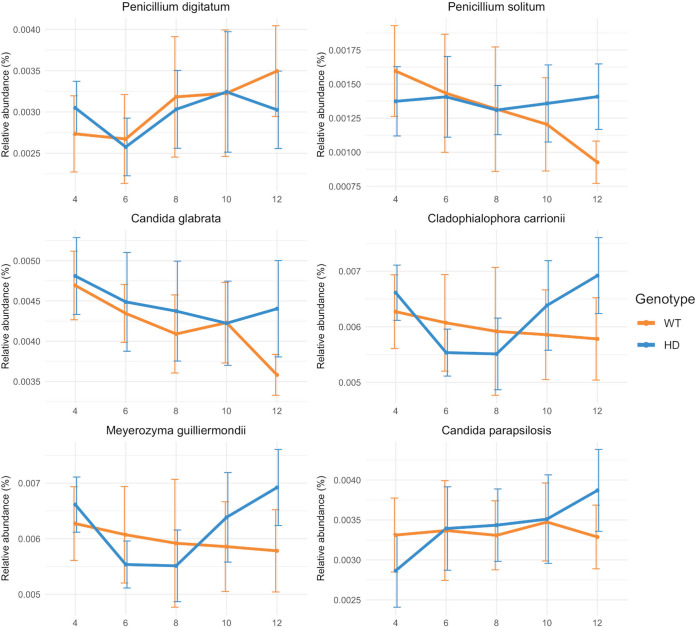
To understand the potential changes in relative abundances underlying the temporal shift in the gut mycobiome, linear mixed-effect model splines (LMMS) modeling and sparse principal-component analysis (sPCA) implemented in the TimeOmics package were applied. Candida glabrata, Candida parapsilosis, and Cladophialophora carrionii displayed coabundance and correlated negatively with P. digitatum and Paracoccidioides lutzii, which was confirmed by the proportionality test (see the supplemental material). Notably, the temporal trends of P. solitum, M. guillerimondii, C. glabrata, and C. parapsilosis were different between the two genotypes (Fig. 3). P. solitum and C. glabrata gradually decreased over time in the WT mice, but their relative abundances remained unchanged in HD mice (Fig. 3). On the other hand, M. guilliermondii and C. parapsilosis remained at stable levels in the WT mice, while they increased gradually in the HD mice (Fig. 3). However, differential-abundance analysis of all LMMS profiles revealed no significant effect of genotype or interaction effect of genotype and time in any of these longitudinal profiles (see the supplemental material).
FIG 3.
Temporal relative abundance of coabundant fungal taxa with different trends between samples from WT and HD mice across time. Mean values and standard deviations are shown. n = 9/group.
Integration of bacterial and fungal data sets hints at the perturbation of interkingdom interaction in HD mice.
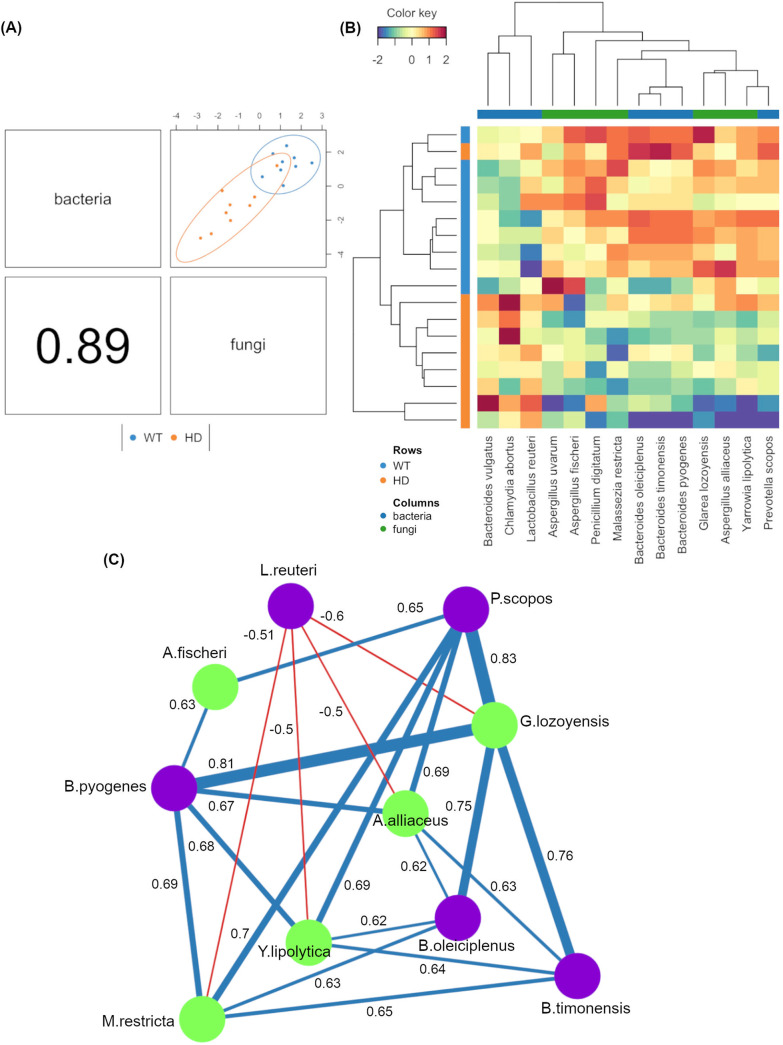
Given that the majority of the changes in gut fungal community structures were observed at the week 12 time point, we focused on this time point to assess potential associations between the gut fungal and bacterial communities. The gut mycobiome data set here was integrated with the previously published gut bacterial data set using DIABLO from the mixOmics R package. With a design matrix of 0.1, which was set to maximize the discrimination between two genotypes, the correlation between the two data sets was still very strong (r = 0.89) (Fig. 4A). Feature selection identified that the fungal species Glarea lozoyensis, M. restricta, A. alliaceus, and Y. lipolytica all possessed strong positive associations (r > 0.6) with the bacterial Bacteroides spp., including Bacteroides pyogenes, Bacteroides oleiciplenus, and Bacteroides timonensis, and with Prevotella scopos (Fig. 4C). Additionally, Lactobacillus reuteri was negatively associated with G. lozoyensis, M. restricta, A. alliaceus, and Y. lipolytica (r < −0.5). A. fischeri was found to positively associate with only Prevotella scopos (r = 0.65) and Bacteroides pyogenes (r = 0.63). Notably, all the selected fungal species were depleted in the HD gut mycobiome.
FIG 4.
Integration of gut bacteriome and mycobiome compositions using DIABLO. (A) The gut bacteriomes and mycobiomes were highly concordant with each other (r = 0.89) and revealed distinct clustering of samples from WT and HD mice. (B) Clustered heatmap showing the abundances of bacterial and fungal species that were found to covary with each other (columns) and were discriminatory between the two genotypes (rows). (C) Correlation network analysis revealed strong positive- and negative-correlation associations between several bacterial and fungal taxa. Green and purple nodes correspond to fungi and bacteria, respectively. Correlations of r > 0.5 (blue lines) or r < −0.5 (red lines) are shown.
DISCUSSION
This study presents the first evidence of the disruption of the gut fungal composition in Huntington’s disease. At 12 weeks of age, which corresponds to an early disease stage and is prior to overt motor symptoms, there was a significant difference in beta diversity between HD mice and their WT littermates that was not observed at any earlier age (51). The HD mice also showed a less heterogenous gut mycobiome configuration than healthy controls at weeks 6 and 8. However, there were more volatile fungal taxa in HD mice. Reduced heterogeneity of the gut microbiome has been observed in a number of diseases, and our present data suggest the presence of particular stressors in HD mice that induce similar ecological changes in gut fungal communities across individuals at these time points (52, 53). An increase in alpha diversity was also observed in HD mice at 12 weeks of age without any alterations at earlier ages. Changes in gut mycobiome alpha diversity can be challenging to interpret, as both elevated and reduced alpha diversity have been associated with adverse health outcomes (43, 45, 46). Therefore, rather than lacking fungal diversity, it is possible that the affected individuals may harbor fewer commensals in favor of proinflammatory pathobionts.
Different longitudinal profiles between the two genotypes were observed for gut commensals Candida parapsilosis, Candida glabrata, Meyerozyma guilliermondii, and Penicillium solitum, with increasing prevalence in the HD gut over time. C. parapsilosis and C. glabrata are both opportunistic fungal pathobionts commonly found in the gut that can have devastating effects, particularly in immunocompromised patients, where they can cause candidiasis and increased mortality (54, 55). C. glabrata and C. parapsilosis overgrowth showed no adverse outcomes in healthy mice but exacerbated gastroinflammation in mice with dextran sulfate sodium (DSS)-induced colitis via stimulation of proinflammatory cytokines, receptor expression, and shifting of major immune cell types (23, 56, 57). Concurrent with our results, enrichment of C. glabrata and C. parapsilosis was reported in the gut mycobiomes of patients with ASD, IBD, and Crohn’s disease (46, 58–60). Individuals with elevated antigens of C. parapsilosis and C. glabrata were associated with higher risk of MS (61). Moreover, it was recently reported that HD patients had fungal structures with C. glabrata antigens in corpora amylacea (CA) in the brain, suggesting that the infection by this fungus occurred at an earlier period long before death, as its secreted proteins were trapped during the formation of CA over the years (47). Increased gut inflammation and permeability, as well as impaired blood-brain barrier, are also comorbidities of HD (62–66). Taken together, our results suggest a bloom in C. glabrata that may aggravate gastrointestinal inflammation in HD and lead to candidemia and Candida infection in the HD brain.
Furthermore, alterations in the abundance of several key fungal species were identified. Enrichment of Penicillium digitatum and Botrytis cinerea was observed in the HD gut, in line with a previous study reporting the presence of Botrytis cinerea, which may have originated from the intestine, in several brain regions in patients with Alzheimer’s disease (67). On the other hand, Glarea lozoyensis was more prevalent in healthy controls. G. lozoyensis is a natural producer of the antimicrobial compound pneumocandin B0, which is capable of inhibiting the growth of fungal pathobionts like C. albicans (68, 69). Furthermore, Malassezia restricta and Yarrowia lipolytica were underrepresented in the HD gut mycobiome at 12 weeks of age. Although Y. lipolytica is commonly used in the food industry to produce long-chain polyunsaturated fatty acids, it has also been observed in mammalian stool samples and its potential as a probiotic was recently demonstrated (70, 71). Oral supplementation of Y. lipolytica considerably reduced the bacterial load and improved the host immunological response against bacterial infection in fish (72). In turkeys and pigs, dietary administration of this fungus led to enhanced cytokine production and blood leukocyte counts, increased mucosal immunity, and improved overall gut architecture (73–75). As such, supplementation of Y. lipolytica may be beneficial in ameliorating some of the HD symptoms.
Malassezia is a dominant commensal found on the skin but also one of the most prevalent fungal genera in the human gastrointestinal tract, associated with intestinal mucosa (18, 60, 76–79). M. restricta, in particular, is associated with Crohn’s disease and ulcerative colitis (80). Colonization of M. restricta in germ-free mice exacerbated the severity of dextran sodium sulfate-induced colitis (80). The presence of M. restricta stimulated Card9 signaling via the Dectin-2 receptor, which then induced proinflammatory responses toward innate immune cell activation and aggravated intestinal inflammation (80). Recent work demonstrated that fungal signals alone are unable to induce the level of colonic inflammation commonly reported in DSS-treated animals (23), and hence, this activation is potentially dependent on the concomitant presence of bacterial signals.
Our bacteriome-mycobiome integration analysis revealed the negative correlation of Lactobacillus reuteri with a number of fungal species, including M. restricta, Y. lipolytica, G. lozoyensis, and Aspergillus spp., which were identified as differentially abundant between the two genotypes. In line with this, several studies have shown the opposing relationship between L. reuteri and fungal colonization (23, 81–83). Reuterin, a main metabolite secreted by L. reuteri, is a well-known antimicrobial substance that can also suppress fungal growth (83, 84). Additionally, lactic acid and organic acids are also products of L. reuteri that could decrease gut pH and impact fungal growth (85). Apart from that, L. reuteri has neuromodulatory effects in the enteric nervous system since it is capable of producing the neurotransmitter gamma-aminobutyric acid (GABA), which could also directly impact the growth of other residents in the gut (33, 86). Given that a delicate bacteriome-mycobiome balance is required to maintain host health and homeostasis, the altered interkingdom equilibrium in HD could contribute to or modulate gastrointestinal inflammation or the microbiome-gut-brain axis in HD.
The current study investigating the HD gut mycobiome profile is the first to be conducted; however, it possesses several caveats. This was a pilot study with a small sample size, constrained by the cost of metagenomics sequencing, and hence, generalizable extrapolation from it may be limited. Furthermore, the sequencing depth of the investigated shotgun metagenomics data may be insufficient, considering the extremely low abundance of fungi in stool, which may affect the accuracy of the results. The ability of fungi to change morphologically depending on environmental conditions can also often lead to misidentification in fungal databases. In addition, many of the fungal species detected are of environmental origin, and their biological relevance in the mammalian gut is unknown. Nevertheless, the results presented here generated novel hypotheses that could inform future studies aiming to understand the microbiome-gut-brain axis in HD.
In summary, this is the first evidence of the perturbation of gut mycobiome composition in HD. Alterations in the fungal composition and structure of the gut mycobiome were observed at the onset of motor symptoms in HD mice. These changes were characterized by depletion of several key species in the HD gut, including G. lozoyensis, Aspergillus uvarum, Aspergillus fischeri, M. restricta, and P. digitatum. Moreover, the abundance of Lactobacillus reuteri was negatively correlated with those key fungal taxa, suggesting perturbation of interkingdom interactions within the HD gut. The study presented here highlights the need to examine the HD gut mycobiome in more depth, which would advance the understanding of gastrointestinal fungal-bacterial relationships important for host health. This may allow us to identify novel therapeutics for HD and related disorders.
MATERIALS AND METHODS
Taxonomic classification of fungal genomic reads.
Shotgun metagenomic sequences of male R6/1 HD transgenic mice and their wild-type (WT) littermates were previously deposited by our group in NCBI (BioProject accession number PRJNA613182). The data were from 90 stools from 18 mice sampled longitudinally at 5 time points: weeks 4, 6, 8, 10, and 12. Reads that aligned to the mouse genome (mm10) were removed to obtain host-contaminant-free reads with approximately 10,389,634 read pairs of 251 bp per sample. To identify the taxonomic composition, reads were classified using Kaiju ‘Greedy’ mode and matching to the NCBI BLAST nonredundant plus eukaryotic protein database (87). Read counts assigned to the fungal kingdom were then extracted for analysis.
Diversity analysis of fungal DNA reads.
For alpha diversity analysis, data were rarefied to the 4,863,549 sequences per sample before calculating the Shannon index using the phyloseq R package (88). The data were stratified to their age groups before testing for the significance of genotype with the t test, after confirming the equality of variance and normality of the data through Levene’s test and the Shapiro-Wilk test, respectively (89, 90). The P values were then adjusted for multiple comparisons with false discovery rate (FDR) (91). Beta diversity analysis was performed by calculating the Aitchison distance and visualizing it with principal-component analysis (PCA). Similarly, the data were stratified to their respective age groups and then tested for the effect of genotype using Adonis PERMANOVA from the vegan R package (92). The P values were then corrected for multiple testing using FDR. To ascertain the variability in the overall gut mycobiome structure, the distance of each sample from the group centroid was calculated and its significance was determined using the functions betadisper() and permutest(), respectively (92). In all cases, the significance level was set at <0.05.
To identify the fungal signature discriminating between the two genotypes, sparse partial least-squares discriminant analysis (sPLS-DA) from the mixOmics R package was performed on the centered log ratio (CLR)-transformed data (93, 94). The parameters chosen to select a signature of 15 fungal species and “leave-one-out” cross-validation were used to evaluate the classification performance of the method.
Clustering analysis and volatility assessment of fungal temporal profiles.
Analysis of the longitudinal fungal community structure was performed using the TimeOmics R package (95, 96). Briefly, each feature was fit with the best possible model using linear mixed-effect model splines (LMMS) to cluster the fungal species across time (97). This method accounts for between- and within-individual variability and irregular time sampling while avoiding under- or oversmoothing. The Breusch-Pagan test and mean square error were used to assess the homoskedasticity of residuals to remove profiles that were considered noisy before clustering. Sparse PCA (sPCA) was applied on the smooth profiles to identify and cluster species that contributed to explaining time variability. To determine the significance of differences in each temporal profile between the two genotypes, LMMS for differential expression analysis (LMMSDE) was performed (97). As correlations can be spurious for microbiome data that are compositional, the proportionality distance between species identified as relevant in our method was calculated as a measure of association. Median distances within clusters were calculated and compared to all the median distances between all species.
Supervised integration for interkingdom interaction.
Bacterial counts were extracted from the Kaiju taxonomic assignment output. To identify the associations between the gut bacteriome and mycobiome that were altered in HD, CLR-transformed fungal and bacterial data sets were integrated using the DIABLO framework from the mixOmics package in a supervised analysis (94, 98). A design matrix of 0.1 was used to place higher importance on the discrimination of genotypes rather than maximizing correlation between the two data sets. A signature of 7 bacterial and 7 fungal species was selected for the model. A correlation cutoff of 0.5 was set for the network visualization using Gephi (99).
Supplementary Material
ACKNOWLEDGMENTS
We thank Thibault Renoir for his advice and assistance regarding the HD mouse model.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors. We acknowledge that A.J.H. was supported in part by a National Health and Medical Research Council (NHMRC) Principal Research Fellowship (grant number GNT1117148) and the DHB Foundation (Equity Trustees), and K.-A.L.C. was supported in part by an NHMRC career development fellowship (grant number GNT1159458). The shotgun sequencing was funded by a Computational Biology Research Initiative (CBRI) grant to K.-A.L.C. and A.J.H. from the University of Melbourne.
Footnotes
Supplemental material is available online only.
Contributor Information
Anthony J. Hannan, Email: anthony.hannan@florey.edu.au.
Rebecca S. Shapiro, University of Guelph
REFERENCES
- 1.Bates GP, Dorsey R, Gusella JF, Hayden MR, Kay C, Leavitt BR, Nance M, Ross CA, Scahill RI, Wetzel R, Wild EJ, Tabrizi SJ. 2015. Huntington disease. Nat Rev Dis Primers 1:15005. doi: 10.1038/nrdp.2015.5. [DOI] [PubMed] [Google Scholar]
- 2.The Huntington’s Disease Collaborative Research Group. 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 3.Sathasivam K, Hobbs C, Turmaine M, Mangiarini L, Mahal A, Bertaux F, Wanker EE, Doherty P, Davies SW, Bates GP. 1999. Formation of polyglutamine inclusions in non-CNS tissue. Hum Mol Genet 8:813–822. doi: 10.1093/hmg/8.5.813. [DOI] [PubMed] [Google Scholar]
- 4.Moffitt H, McPhail GD, Woodman B, Hobbs C, Bates GP. 2009. Formation of polyglutamine inclusions in a wide range of non-CNS tissues in the HdhQ150 knock-in mouse model of Huntington’s disease. PLoS One 4:e8025. doi: 10.1371/journal.pone.0008025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luthi-Carter R, Cha J-HJ. 2003. Mechanisms of transcriptional dysregulation in Huntington’s disease. Clin Neurosci Res 3:165–177. doi: 10.1016/S1566-2772(03)00059-8. [DOI] [Google Scholar]
- 6.Chang DTW, Rintoul GL, Pandipati S, Reynolds IJ. 2006. Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiol Dis 22:388–400. doi: 10.1016/j.nbd.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Kim J, Moody JP, Edgerly CK, Bordiuk OL, Cormier K, Smith K, Beal MF, Ferrante RJ. 2010. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum Mol Genet 19:3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zielonka D, Piotrowska I, Marcinkowski JT, Mielcarek M. 2014. Skeletal muscle pathology in Huntington’s disease. Front Physiol 5:380. doi: 10.3389/fphys.2014.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aziz NA, Van Der Burg J, Landwehrmeyer G, Brundin P, Stijnen T, Roos R, EHDI Study Group. 2008. Weight loss in Huntington disease increases with higher CAG repeat number. Neurology 71:1506–1513. doi: 10.1212/01.wnl.0000334276.09729.0e. [DOI] [PubMed] [Google Scholar]
- 10.Lakra P, Aditi K, Agrawal N. 2019. Peripheral expression of mutant huntingtin is a critical determinant of weight loss and metabolic disturbances in Huntington’s disease. Sci Rep 9:1–15. doi: 10.1038/s41598-019-46470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Björkqvist M, Wild EJ, Thiele J, Silvestroni A, Andre R, Lahiri N, Raibon E, Lee RV, Benn CL, Soulet D, Magnusson A, Woodman B, Landles C, Pouladi MA, Hayden MR, Khalili-Shirazi A, Lowdell MW, Brundin P, Bates GP, Leavitt BR, Möller T, Tabrizi SJ. 2008. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington’s disease. J Exp Med 205:1869–1877. doi: 10.1084/jem.20080178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong G, Lê Cao K-A, Judd LM, Li S, Renoir T, Hannan AJ. 2018. Microbiome profiling reveals gut dysbiosis in a transgenic mouse model of Huntington’s disease. Neurobiol Dis 135:104268. doi: 10.1016/j.nbd.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Radulescu CI, Garcia-Miralles M, Sidik H, Bardile CF, Yusof NABM, Lee HU, Ho EXP, Chu CW, Layton E, Low D, De Sessions PF, Pettersson S, Ginhoux F, Pouladi MA. 2019. Manipulation of microbiota reveals altered callosal myelination and white matter plasticity in a model of Huntington disease. Neurobiol Dis 127:65–75. doi: 10.1016/j.nbd.2019.02.011. [DOI] [PubMed] [Google Scholar]
- 14.Wasser CI, Mercieca E-C, Kong G, Hannan AJ, McKeown SJ, Glikmann-Johnston Y, Stout JC. 2020. Gut dysbiosis in Huntington’s disease: associations between gut microbiota, cognitive performance and clinical outcomes. Brain Commun 2:fcaa110. doi: 10.1093/braincomms/fcaa110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cryan JF, O’Mahony SM. 2011. The microbiome‐gut‐brain axis: from bowel to behavior. Neurogastroenterol Motil 23:187–192. doi: 10.1111/j.1365-2982.2010.01664.x. [DOI] [PubMed] [Google Scholar]
- 16.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto J-M, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Doré J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, MetaHIT Consortium, et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dollive S, Chen Y-Y, Grunberg S, Bittinger K, Hoffmann C, Vandivier L, Cuff C, Lewis JD, Wu GD, Bushman FD. 2013. Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment. PLoS One 8:e71806. doi: 10.1371/journal.pone.0071806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nash AK, Auchtung TA, Wong MC, Smith DP, Gesell JR, Ross MC, Stewart CJ, Metcalf GA, Muzny DM, Gibbs RA, Ajami NJ, Petrosino JF. 2017. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome 5:153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiaro TR, Soto R, Zac Stephens W, Kubinak JL, Petersen C, Gogokhia L, Bell R, Delgado JC, Cox J, Voth W, Brown J, Stillman DJ, O’Connell RM, Tebo AE, Round JL. 2017. A member of the gut mycobiota modulates host purine metabolism exacerbating colitis in mice. Sci Transl Med 9:eaaf9044. doi: 10.1126/scitranslmed.aaf9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paterson MJ, Oh S, Underhill DM. 2017. Host–microbe interactions: commensal fungi in the gut. Curr Opin Microbiol 40:131–137. doi: 10.1016/j.mib.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Underhill DM, Iliev ID. 2014. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol 14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang TT, Shao T-Y, Ang WG, Kinder JM, Turner LH, Pham G, Whitt J, Alenghat T, Way SS. 2017. Commensal fungi recapitulate the protective benefits of intestinal bacteria. Cell Host Microbe 22:809–816. doi: 10.1016/j.chom.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Tilburg Bernardes E, Pettersen VK, Gutierrez MW, Laforest-Lapointe I, Jendzjowsky NG, Cavin J-B, Vicentini FA, Keenan CM, Ramay HR, Samara J, MacNaughton WK, Wilson RJA, Kelly MM, McCoy KD, Sharkey KA, Arrieta M-C. 2020. Intestinal fungi are causally implicated in microbiome assembly and immune development in mice. Nat Commun 11:2577. doi: 10.1038/s41467-020-16431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonardi I, Li X, Semon A, Li D, Doron I, Putzel G, Bar A, Prieto D, Rescigno M, McGovern DPB, Pla J, Iliev ID. 2018. CX3CR1+ mononuclear phagocytes control immunity to intestinal fungi. Science 359:232–236. doi: 10.1126/science.aao1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markey L, Shaban L, Green ER, Lemon KP, Mecsas J, Kumamoto CA. 2018. Pre-colonization with the commensal fungus Candida albicans reduces murine susceptibility to Clostridium difficile infection. Gut Microbes 9:497–509. doi: 10.1080/19490976.2018.1465158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buts J-P, Dekeyser N, Stilmant C, Delem E, Smets F, Sokal E. 2006. Saccharomyces boulardii produces in rat small intestine a novel protein phosphatase that inhibits Escherichia coli endotoxin by dephosphorylation. Pediatr Res 60:24–29. doi: 10.1203/01.pdr.0000220322.31940.29. [DOI] [PubMed] [Google Scholar]
- 27.Kumamoto CA, Gresnigt MS, Hube B. 2020. The gut, the bad and the harmless: Candida albicans as a commensal and opportunistic pathogen in the intestine. Curr Opin Microbiol 56:7–15. doi: 10.1016/j.mib.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castagliuolo I, Riegler MF, Valenick L, LaMont JT, Pothoulakis C. 1999. Saccharomyces boulardii protease inhibits the effects of Clostridium difficile toxins A and B in human colonic mucosa. Infect Immun 67:302–307. doi: 10.1128/IAI.67.1.302-307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McFarland LV, Surawicz CM, Greenberg RN, Fekety R, Elmer GW, Moyer KA, Melcher SA, Bowen KE, Cox JL, Noorani Z. 1994. A randomized placebo-controlled trial of Saccharomyces boulardii in combination with standard antibiotics for Clostridium difficile disease. JAMA 271:1913–1918. doi: 10.1001/jama.1994.03510480037031. [DOI] [PubMed] [Google Scholar]
- 30.Billoo AG, Memon M, Khaskheli S, Murtaza G, Iqbal K, Shekhani MS, Siddiqi AQ. 2006. Role of a probiotic (Saccharomyces boulardii) in management and prevention of diarrhoea. World J Gastroenterol 12:4557–4560. doi: 10.3748/wjg.v12.i28.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takata K, Tomita T, Okuno T, Kinoshita M, Koda T, Honorat JA, Takei M, Hagihara K, Sugimoto T, Mochizuki H, Sakoda S, Nakatsuji Y. 2015. Dietary yeasts reduce inflammation in central nerve system via microflora. Ann Clin Transl Neurol 2:56–66. doi: 10.1002/acn3.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsavkelova E, Botvinko I, Kudrin V, Oleskin A. 2000. Detection of neurotransmitter amines in microorganisms with the use of high-performance liquid chromatography. Dokl Biochem 372:115–117. [PubMed] [Google Scholar]
- 33.Liu T, Huang Z. 2019. Evidence-based analysis of neurotransmitter modulation by gut microbiota, p 238–249. In Wang H, Siuly S, Zhou R, Martin-Sanchez F, Zhang Y, Huang Z (ed), Health information science. HIS 2019. Lecture notes in computer science, vol 11837. Springer, Cham, Switzerland. [Google Scholar]
- 34.Voropaeva E. 2002. Resistance to antibiotics and histamine production at the bacteria, isolated from the stomatopharynx of the children with bronchial asthma. Antibiot Khimioter 47:8–13. [PubMed] [Google Scholar]
- 35.Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, Cosnes J, Seksik P, Langella P, Skurnik D, Richard ML, Beaugerie L. 2017. Fungal microbiota dysbiosis in IBD. Gut 66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoarau G, Mukherjee PK, Gower-Rousseau C, Hager C, Chandra J, Retuerto MA, Neut C, Vermeire S, Clemente J, Colombel JF, Fujioka H, Poulain D, Sendid B, Ghannoum MA. 2016. Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial Crohn’s disease. mBio 7:e01250-16. doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das A, O’Herlihy E, Shanahan F, O’Toole PW, Jeffery IB. 2021. The fecal mycobiome in patients with irritable bowel syndrome. Sci Rep 11:124. doi: 10.1038/s41598-020-79478-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coker OO, Nakatsu G, Dai RZ, Wu WKK, Wong SH, Ng SC, Chan FKL, Sung JJY, Yu J. 2019. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 68:654–662. doi: 10.1136/gutjnl-2018-317178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q, Wang C, Tang C, He Q, Li N, Li J. 2014. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J Clin Gastroenterol 48:513–523. doi: 10.1097/MCG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, Jousson O, Leoncini S, Renzi D, Calabrò A, De Filippo C. 2017. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 5:24. doi: 10.1186/s40168-017-0242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iovene MR, Bombace F, Maresca R, Sapone A, Iardino P, Picardi A, Marotta R, Schiraldi C, Siniscalco D, Serra N, de Magistris L, Bravaccio C. 2017. Intestinal dysbiosis and yeast isolation in stool of subjects with autism spectrum disorders. Mycopathologia 182:349–363. doi: 10.1007/s11046-016-0068-6. [DOI] [PubMed] [Google Scholar]
- 42.Shah S, Locca A, Dorsett Y, Cantoni C, Ghezzi L, Lin Q, Bokoliya S, Panier H, Suther C, Gormley M, Liu Y, Evans E, Mikesell R, Obert K, Salter A, Cross AH, Tarr PI, Lovett-Racke A, Piccio L, Zhou Y. 2021. Alterations of the gut mycobiome in patients with MS. EBioMedicine 71:103557. doi: 10.1016/j.ebiom.2021.103557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagpal R, Neth BJ, Wang S, Mishra SP, Craft S, Yadav H. 2020. Gut mycobiome and its interaction with diet, gut bacteria and Alzheimer’s disease markers in subjects with mild cognitive impairment: a pilot study. EBioMedicine 59:102950. doi: 10.1016/j.ebiom.2020.102950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ling Z, Zhu M, Liu X, Shao L, Cheng Y, Yan X, Jiang R, Wu S. 2020. Fecal fungal dysbiosis in Chinese patients with Alzheimer’s disease. Front Cell Dev Biol 8:631460. doi: 10.3389/fcell.2020.631460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou R, Wang Y, Duan M, Guo M, Zhang Q, Zheng H. 2021. Dysbiosis of gut fungal microbiota in children with autism spectrum disorders. J Autism Dev Disord 51:267–275. doi: 10.1007/s10803-020-04543-y. [DOI] [PubMed] [Google Scholar]
- 46.Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, Wu GD. 2015. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis 21:1948–1956. doi: 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alonso R, Pisa D, Carrasco L. 2019. Brain microbiota in Huntington’s disease patients. Front Microbiol 10:2622. doi: 10.3389/fmicb.2019.02622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vinther-Jensen T, Börnsen L, Budtz-Jørgensen E, Ammitzbøll C, Larsen IU, Hjermind LE, Sellebjerg F, Nielsen JE. 2016. Selected CSF biomarkers indicate no evidence of early neuroinflammation in Huntington disease. Neurol Neuroimmunol Neuroinflamm 3:e287. doi: 10.1212/NXI.0000000000000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong G, Ellul S, Narayana VK, Kanojia K, Ha HTT, Li S, Renoir T, Lê Cao K-A, Hannan AJ. 2020. An integrated metagenomics and metabolomics approach implicates the microbiota-gut-brain axis in the pathogenesis of Huntington’s disease. Neurobiol Dis 148:105199. doi: 10.1016/j.nbd.2020.105199. [DOI] [PubMed] [Google Scholar]
- 50.Bastiaanssen TFS, Gururajan A, van de Wouw M, Moloney GM, Ritz NL, Long-Smith CM, Wiley NC, Murphy AB, Lyte JM, Fouhy F, Stanton C, Claesson MJ, Dinan TG, Cryan JF. 2021. Volatility as a concept to understand the impact of stress on the microbiome. Psychoneuroendocrinology 124:105047. doi: 10.1016/j.psyneuen.2020.105047. [DOI] [PubMed] [Google Scholar]
- 51.Brooks SP, Janghra N, Workman VL, Bayram-Weston Z, Jones L, Dunnett SB. 2012. Longitudinal analysis of the behavioural phenotype in R6/1 (C57BL/6J) Huntington’s disease transgenic mice. Brain Res Bull 88:94–103. doi: 10.1016/j.brainresbull.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 52.Ma ZS. 2020. Testing the Anna Karenina principle in human microbiome-associated diseases. iScience 23:101007. doi: 10.1016/j.isci.2020.101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaneveld JR, McMinds R, Thurber RV. 2017. Stress and stability: applying the Anna Karenina principle to animal microbiomes. Nat Microbiol 2:1–8. doi: 10.1038/nmicrobiol.2017.121. [DOI] [PubMed] [Google Scholar]
- 54.Miranda LN, van der Heijden IM, Costa SF, Sousa API, Sienra RA, Gobara S, Santos CR, Lobo RD, Pessoa VP, Levin AS. 2009. Candida colonisation as a source for candidaemia. J Hosp Infect 72:9–16. doi: 10.1016/j.jhin.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 55.Lionakis MS, Netea MG. 2013. Candida and host determinants of susceptibility to invasive candidiasis. PLoS Pathog 9:e1003079. doi: 10.1371/journal.ppat.1003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jawhara S, Mogensen E, Maggiotto F, Fradin C, Sarazin A, Dubuquoy L, Maes E, Guérardel Y, Janbon G, Poulain D. 2012. Murine model of dextran sulfate sodium-induced colitis reveals Candida glabrata virulence and contribution of β-mannosyltransferases. J Biol Chem 287:11313–11324. doi: 10.1074/jbc.M111.329300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Charlet R, Pruvost Y, Tumba G, Istel F, Poulain D, Kuchler K, Sendid B, Jawhara S. 2018. Remodeling of the Candida glabrata cell wall in the gastrointestinal tract affects the gut microbiota and the immune response. Sci Rep 8:3316. doi: 10.1038/s41598-018-21422-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kantarcioglu AS, Kiraz N, Aydin A. 2016. Microbiota–gut–brain axis: yeast species isolated from stool samples of children with suspected or diagnosed autism spectrum disorders and in vitro susceptibility against nystatin and fluconazole. Mycopathologia 181:1–7. doi: 10.1007/s11046-015-9949-3. [DOI] [PubMed] [Google Scholar]
- 59.Ott SJ, Kühbacher T, Musfeldt M, Rosenstiel P, Hellmig S, Rehman A, Drews O, Weichert W, Timmis KN, Schreiber S. 2008. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol 43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 60.Liguori G, Lamas B, Richard ML, Brandi G, da Costa G, Hoffmann TW, Di Simone MP, Calabrese C, Poggioli G, Langella P, Campieri M, Sokol H. 2016. Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. J Crohns Colitis 10:296–305. doi: 10.1093/ecco-jcc/jjv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benito-León J, Pisa D, Alonso R, Calleja P, Díaz-Sánchez M, Carrasco L. 2010. Association between multiple sclerosis and Candida species: evidence from a case-control study. Eur J Clin Microbiol Infect Dis 29:1139–1145. doi: 10.1007/s10096-010-0979-y. [DOI] [PubMed] [Google Scholar]
- 62.Andrich JE, Wobben M, Klotz P, Goetze O, Saft C. 2009. Upper gastrointestinal findings in Huntington’s disease: patients suffer but do not complain. J Neural Transm (Vienna) 116:1607–1611. doi: 10.1007/s00702-009-0310-1. [DOI] [PubMed] [Google Scholar]
- 63.van der Burg JMM, Winqvist A, Aziz NA, Maat-Schieman MLC, Roos RAC, Bates GP, Brundin P, Björkqvist M, Wierup N. 2011. Gastrointestinal dysfunction contributes to weight loss in Huntington’s disease mice. Neurobiol Dis 44:1–8. doi: 10.1016/j.nbd.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 64.Stan TL, Soylu-Kucharz R, Burleigh S, Prykhodko O, Cao L, Franke N, Sjögren M, Haikal C, Hållenius F, Björkqvist M. 2020. Increased intestinal permeability and gut dysbiosis in the R6/2 mouse model of Huntington’s disease. Sci Rep 10:18270. doi: 10.1038/s41598-020-75229-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Di Pardo A, Amico E, Scalabrì F, Pepe G, Castaldo S, Elifani F, Capocci L, De Sanctis C, Comerci L, Pompeo F, D’Esposito M, Filosa S, Crispi S, Maglione V. 2017. Impairment of blood-brain barrier is an early event in R6/2 mouse model of Huntington disease. Sci Rep 7:41316–41318. doi: 10.1038/srep41316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drouin-Ouellet J, Sawiak SJ, Cisbani G, Lagacé M, Kuan W-L, Saint-Pierre M, Dury RJ, Alata W, St-Amour I, Mason SL, Calon F, Lacroix S, Gowland PA, Francis ST, Barker RA, Cicchetti F. 2015. Cerebrovascular and blood–brain barrier impairments in Huntington’s disease: potential implications for its pathophysiology. Ann Neurol 78:160–177. doi: 10.1002/ana.24406. [DOI] [PubMed] [Google Scholar]
- 67.Alonso R, Pisa D, Marina AI, Morato E, Rabano A, Carrasco L. 2014. Fungal infection in patients with Alzheimer’s disease. J Alzheimers Dis 41:301–311. doi: 10.3233/JAD-132681. [DOI] [PubMed] [Google Scholar]
- 68.Song P, Zhang K, Zhang S, Huang B-Q, Ji X-J, Ren L-J, Gao S, Wen J-P, Huang H. 2018. Enhancement of pneumocandin B0 production in Glarea lozoyensis by low-temperature adaptive laboratory evolution. Front Microbiol 9:2788. doi: 10.3389/fmicb.2018.02788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bouffard FA, Zambias RA, Dropinski JF, Balkovec JM, Hammond ML, Abruzzo GK, Bartizal KF, Marrinan JA, Kurtz MB, McFadden DC. 1994. Synthesis and antifungal activity of novel cationic pneumocandin Bo derivatives. J Med Chem 37:222–225. doi: 10.1021/jm00028a003. [DOI] [PubMed] [Google Scholar]
- 70.Gouba N, Drancourt M. 2015. Digestive tract mycobiota: a source of infection. Med Mal Infect 45:9–16. doi: 10.1016/j.medmal.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 71.Zieniuk B, Fabiszewska A. 2019. Yarrowia lipolytica: a beneficious yeast in biotechnology as a rare opportunistic fungal pathogen: a minireview. World J Microbiol Biotechnol 35:1–8. doi: 10.1007/s11274-018-2583-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reyes-Becerril M, Alamillo E, Angulo C. 2021. Probiotic and immunomodulatory activity of marine yeast Yarrowia lipolytica strains and response against Vibrio parahaemolyticus in fish. Probiotics Antimicrob Proteins 13:1292–1305. doi: 10.1007/s12602-021-09769-5. [DOI] [PubMed] [Google Scholar]
- 73.Czech A, Merska M, Ognik K. 2014. Blood immunological and biochemical indicators in turkey hens fed diets with a different content of the yeast Yarrowia lipolytica. Ann Anim Sci 14:935–946. doi: 10.2478/aoas-2014-0057. [DOI] [Google Scholar]
- 74.Czech A, Smolczyk A, Ognik K, Wlazło Ł, Nowakowicz-Dębek B, Kiesz M. 2018. Effect of dietary supplementation with Yarrowia lipolytica or Saccharomyces cerevisiae yeast and probiotic additives on haematological parameters and the gut microbiota in piglets. Res Vet Sci 119:221–227. doi: 10.1016/j.rvsc.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 75.Czech A, Sembratowicz I, Zieba G. 2020. Effect of the use of Yarrowia lipolytica and Saccharomyces cerevisiae yeast with a probiotic in the diet of turkeys on their gut microbiota and immunity. Vet Med (Praha) 65:174–182. doi: 10.17221/145/2019-VETMED. [DOI] [Google Scholar]
- 76.Gouba N, Raoult D, Drancourt M. 2014. Eukaryote culturomics of the gut reveals new species. PLoS One 9:e106994. doi: 10.1371/journal.pone.0106994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Auchtung TA, Fofanova TY, Stewart CJ, Nash AK, Wong MC, Gesell JR, Auchtung JM, Ajami NJ, Petrosino JF. 2018. Investigating colonization of the healthy adult gastrointestinal tract by fungi. mSphere 3:e00092-18. doi: 10.1128/mSphere.00092-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suhr M, Banjara N, Hallen-Adams H. 2016. Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett Appl Microbiol 62:209–215. doi: 10.1111/lam.12539. [DOI] [PubMed] [Google Scholar]
- 79.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Kong HH, Segre JA, NIH Intramural Sequencing Center Comparative Sequencing Program. 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Limon JJ, Tang J, Li D, Wolf AJ, Michelsen KS, Funari V, Gargus M, Nguyen C, Sharma P, Maymi VI, Iliev ID, Skalski JH, Brown J, Landers C, Borneman J, Braun J, Targan SR, McGovern DPB, Underhill DM. 2019. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe 25:377–388.e6. doi: 10.1016/j.chom.2019.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Romeo M, Romeo D, Trovato L, Oliveri S, Palermo F, Cota F, Betta P. 2011. Role of probiotics in the prevention of the enteric colonization by Candida in preterm newborns: incidence of late-onset sepsis and neurological outcome. J Perinatol 31:63–69. doi: 10.1038/jp.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oncel MY, Arayici S, Sari FN, Simsek GK, Yurttutan S, Erdeve O, Saygan S, Uras N, Oguz SS, Dilmen U. 2015. Comparison of Lactobacillus reuteri and nystatin prophylaxis on Candida colonization and infection in very low birth weight infants. J Matern Fetal Neonatal Med 28:1790–1794. doi: 10.3109/14767058.2014.968842. [DOI] [PubMed] [Google Scholar]
- 83.Chew SY, Cheah YK, Seow HF, Sandai D, Than LTL. 2015. Probiotic Lactobacillus rhamnosus GR-1 and Lactobacillus reuteri RC-14 exhibit strong antifungal effects against vulvovaginal candidiasis-causing Candida glabrata isolates. J Appl Microbiol 118:1180–1190. doi: 10.1111/jam.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Axelsson LT, Chung TC, Dobrogosz WJ, Lindgren SE. 1989. Production of a broad spectrum antimicrobial substance by Lactobacillus reuteri. Microb Ecol Health Dis 2:131–136. doi: 10.3109/08910608909140210. [DOI] [Google Scholar]
- 85.Thomas K, Hynes S, Ingledew W. 2001. Effect of lactobacilli on yeast growth, viability and batch and semi‐continuous alcoholic fermentation of corn mash. J Appl Microbiol 90:819–828. doi: 10.1046/j.1365-2672.2001.01311.x. [DOI] [PubMed] [Google Scholar]
- 86.Mu Q, Tavella VJ, Luo XM. 2018. Role of Lactobacillus reuteri in human health and diseases. Front Microbiol 9:757. doi: 10.3389/fmicb.2018.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Menzel P, Ng KL, Krogh A. 2016. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McMurdie PJ, Holmes S. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Levene H. 1961. Robust tests for equality of variances, p 279–292. In Olkin I, Ghurye SG, Hoeffding W, Madow WG, Mann HB (ed), Contributions to probability and statistics essays in honor of Harold Hotelling. Stanford University Press, Palo Alto, CA. [Google Scholar]
- 90.Shapiro SS, Francia R. 1972. An approximate analysis of variance test for normality. J Am Stat Assoc 67:215–216. doi: 10.1080/01621459.1972.10481232. [DOI] [Google Scholar]
- 91.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 92.Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci 14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 93.Le Cao K-A, Costello M-E, Lakis VA, Bartolo F, Chua X-Y, Brazeilles R, Rondeau P. 2016. MixMC: a multivariate statistical framework to gain insight into microbial communities. PLoS One 11:e0160169. doi: 10.1371/journal.pone.0160169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rohart F, Gautier B, Singh A, Lê Cao K-A. 2017. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol 13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bodein A, Chapleur O, Droit A, Lê Cao K-A. 2019. A generic multivariate framework for the integration of microbiome longitudinal studies with other data types. Front Genet 10:963. doi: 10.3389/fgene.2019.00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bodein A, Scott-Boyer M-P, Perin O, Lê Cao K-A, Droit A. 2022. timeOmics: an R package for longitudinal multi-omics data integration. Bioinformatics 38:577–579. doi: 10.1093/bioinformatics/btab664. [DOI] [PubMed] [Google Scholar]
- 97.Straube J, Gorse A-D, Huang BE, Le Cao K-A, PROOF Centre of Excellence Team. 2015. A linear mixed model spline framework for analysing time course ‘omics’ data. PLoS One 10:e0134540. doi: 10.1371/journal.pone.0134540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Singh A, Shannon CP, Gautier B, Rohart F, Vacher M, Tebbutt SJ, Lê Cao K-A. 2019. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 35:3055–3062. doi: 10.1093/bioinformatics/bty1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks. https://gephi.org/publications/gephi-bastian-feb09.pdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM02192-21_Supp_1_seq4.xlsx, PDF file, 0.3 MB (49.7KB, xlsx)