ABSTRACT
Bacterial wilt is accompanied by microbial communities shift and soil acidification. However, the relationship between the changes of bacterial communities and bacterial wilt under the influence of different acidification levels has not been fully elucidated. Here, we analyzed the abundance of Ralstonia solanacearum, rhizosphere bacterial communities and carbon metabolism at differently acidic levels (pH 6.45, pH 5.60, pH 5.35, pH 4.90 and pH 4.45) and soil amendment treatment (CaO). The results indicated that both the abundance of R. solanacearum and the incidence of bacterial wilt showed a significant trend of first increasing and then decreasing with the increase of soil pH. The Firmicutes phylum and potentially beneficial genera Bacillus, Paenibacillus, Flavobacterium and Pseudomonas were significantly enriched at pH 6.45. The metabolic ability in response to the l-arginine and 4-hydroxybenzoic acid was significantly increased at pH 6.45. After using CaO to increase the pH of diseased soil from 5.45 to 6.05, the abundance of R. solanacearum and the incidence of bacterial wilt were significantly reduced, the Firmicutes and potentially beneficial genera Bacillus and Pseudomonas were significantly enriched. Overall, alleviating soil acidification to a slightly acidic level (pH 6.0–6.5) could suppress bacterial wilt by suppressing the growth of R. solanacearum and enriching the rhizosphere potentially beneficial bacteria, and further emphasized the importance of increasing soil pH in biological control of bacterial wilt.
IMPORTANCE The rhizosphere microbiota and soil acidification have been shown to have impacts on bacterial wilt. However, the influence of different acidification levels on the rhizosphere communities and bacterial wilt has not been fully studied. In this study, the potentially beneficial bacteria (Bacillus and Pseudomonas) were significantly enriched in the slightly acidic soil (pH 6.45), leading to the increase of the metabolism of 4-hydroxybenzoic acid and the decrease of pathogenic R. solanacearum, thereby alleviating the occurrence of bacterial wilt. The changes of potentially beneficial bacteria and pathogenic R. solanacearum in strongly acidic soil (pH 5.35) with the highest incidence of bacterial wilt were just the opposite. These findings help clarify the mechanisms by which soil bacteria exert influence on bacterial wilt outbreak under different soil acidification levels.
KEYWORDS: soil acidification, bacterial wilt, bacterial communities, beneficial bacteria
INTRODUCTION
Excess soil acidification is a major problem in worldwide soil deterioration and is becoming increasingly serious in intensive agriculture (1). To better understand the cation‐anion pools in soil, different ranges of soil pH have been employed to determine the variation of soil acidity. Generally, most crops favor soils with pH between 5.5 and 6.5, which belongs to slightly acid (pH 6.0–6.5) and moderately acid (pH 5.5–6.0) (2, 3). However, strongly acidic soil (pH 4.5–5.5) represents 30%–40% of the world’s arable soils, and adversely affects the production of many crops (3, 4). At pH 4.5 or below (extremely acid), the Al3+ predominates in the soil solution and has the greatest impact on plant growth (5). In addition, many researchers revealed that soil acidification is closely related to the occurrence of soilborne disease (6–9).
Bacterial wilt, caused by Ralstonia solanacearum, is a typical soilborne disease that can infect Solanaceae crops (10), such as tobacco (11), tomato (12), and eggplant (13). Previous studies showed that the occurrence of bacterial wilt was related to soil pH. Strongly acidic condition (pH 4.5–5.5) was conductive to R. solanacearum growth in B medium, which aggravated the occurrence of bacterial wilt in the pot experiment (6). Within the range of pH 4.5–6.5, pH had a significantly negative correlation with bacterial wilt infection rate (14). Meanwhile, the occurrence of bacterial wilt can be effectively controlled by anthropogenically increasing soil pH (15–17). Earlier studies indicated that soil amendments calcium oxide (18), rock dust (19) and calcium carbonate (20) were effective for controlling bacterial wilt by increasing the soil pH. However, how soil pH affects the occurrence of bacterial wilt is still unclear.
On the other hand, studies have shown that the occurrence of bacterial wilt is also closely related to the bacterial community composition of rhizosphere soil (11, 14, 21). Gram-positive bacteria Firmicutes and Actinobacteria, have been identified as bacterial wilt disease-suppressing rhizobacteria (22). Meanwhile, diverse beneficial rhizobacterial genera have been identified as disease-suppressing microbes, including the genera of Bacillus (23, 24), Pseudomonas (25), Streptomyces (23), Paenibacillus (26), Flavobacterium (27), and Arthrobacter (28). Moreover, in bacterial wilt-suppression soil, the enrichment of beneficial microbes in soil was closely related to the metabolism of l-arginine and 4-hydroxybenzoic acid (an auto-toxic substance secreted by plant root) (29).
Changes in soil pH can strongly affect the activity and community structure of soil microorganisms (30). And bacterial communities were more strongly influenced by pH than fungal communities (31). Therefore, the occurrence of bacterial wilt is closely related to soil pH and bacterial community composition. However, the bacterial community composition under different soil acidity is still unclear together with the relationship in the suppression of bacterial wilt disease. In this study, we changed the soil acidity of nondiseased soil (experiment I, Fig. S1), then soil amendment CaO (experiment II) was used to improve the pH of acidic bacterial wilt diseased soil, investigated i) the changes in bacterial community composition at different acidity levels, and ii) the relationship between the change in bacterial community composition and the occurrence of bacterial wilt.
RESULTS
Soil chemical properties and incidence of bacterial wilt in experiment I.
The changes in soil chemical properties in response to different soil pH are shown in Table 1. With the decrease of soil pH, the contents of available nitrogen and exchangeable aluminum were significantly increased (P < 0.05), while the exchangeable calcium was significantly decreased (P < 0.05). Moreover, the contents of available phosphorus and exchangeable magnesium decreased first and then increased.
TABLE 1.
The basic chemical properties at different pH levelsa
| Treatment | SOM (g/kg) | AN (mg/kg) |
AP (mg/kg) |
AK (mg/kg) |
AS (mg/kg) |
ExMg (mg/kg) |
ExCa (mg/kg) |
ExAl (mg/kg) |
|---|---|---|---|---|---|---|---|---|
| pH4.45 | 31.40 ± 0.05a | 161.98 ± 1.03d | 38.67 ± 0.23c | 254.16 ± 2.89b | 86.82 ± 1.83c | 64.50 ± 1.50c | 961.50 ± 15.08a | 302.89 ± 1.62e |
| pH4.90 | 36.55 ± 0.08a | 142.04 ± 1.44c | 31.23 ± 0.27b | 255.22 ± 0.29b | 85.19 ± 3.53c | 65.00 ± 2.02c | 952.50 ± 8.80a | 238.15 ± 1.85d |
| pH5.35 | 30.04 ± 0.06a | 139.55 ± 0.01c | 29.46 ± 0.38b | 214.40 ± 1.39a | 63.54 ± 1.73a | 46.75 ± 0.35a | 990.00 ± 3.37b | 76.97 ± 0.79b |
| pH5.60 | 33.99 ± 0.08a | 130.83 ± 2.16a | 24.78 ± 0.16a | 285.38 ± 2.99d | 73.55 ± 5.45b | 50.67 ± 2.45ab | 1036.67 ± 12.25b | 73.35 ± 1.04b |
| pH6.45 | 31.48 ± 4.47a | 132.08 ± 1.18ab | 33.07 ± 2.68bc | 266.14 ± 0.19c | 65.94 ± 1.26a | 58.25 ± 0.19b | 1102.50 ± 2.80c | 40.29 ± 0.64a |
The results are the mean of three measurement replicates ± standard error. Small letters indicate a significant difference among different samples (one-way ANOVA, P < 0.05). SOM, soil organic matter; AN, available nitrogen; AP, available phosphorus; AK, available potassium; AS, available sulfur; ExMg, exchangeable magnesium; ExCa, exchangeable calcium; ExAl, exchangeable aluminum.
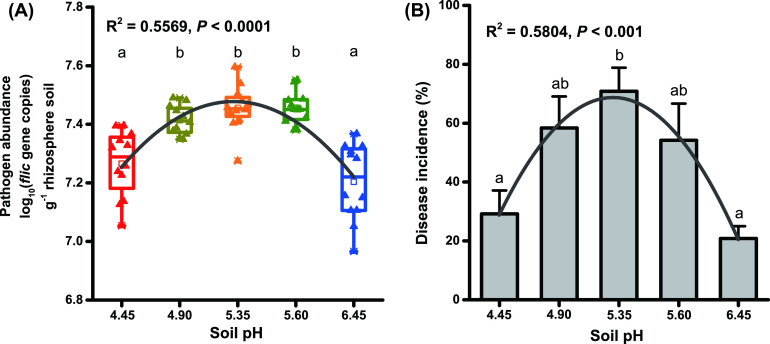
The abundance of R. solanacearum in the rhizosphere soil of pH 4.90 – pH 5.60 was significantly higher than that of pH 4.45 and pH 6.45. And in the acidic range, the abundance of R. solanacearum in the rhizosphere soil showed a trend of increased firstly and then decreased with the increase of soil pH (R2 = 0.5569, P < 0.0001, Fig. 1A). Correspondingly, the incidence of tobacco bacterial wilt also increased first and then decreased with the increase of soil pH (R2 = 0.5804, P < 0.001). Compared with pH 4.45 and pH 6.45, pH 5.35 significantly increased the disease incidence of bacterial wilt by 41.66% and 50.00%, respectively (Fig. 1B, Fig. S2).
FIG 1.
The occurrence of bacterial wilt at different acidification gradients. (A) The abundance of pathogen R. solanacearum in different samples. (B) The disease incidence of different treatments. Different letters indicate significant (P < 0.05) differences according to one-way ANOVA.
Composition of bacterial community at different acidity levels in experiment I.
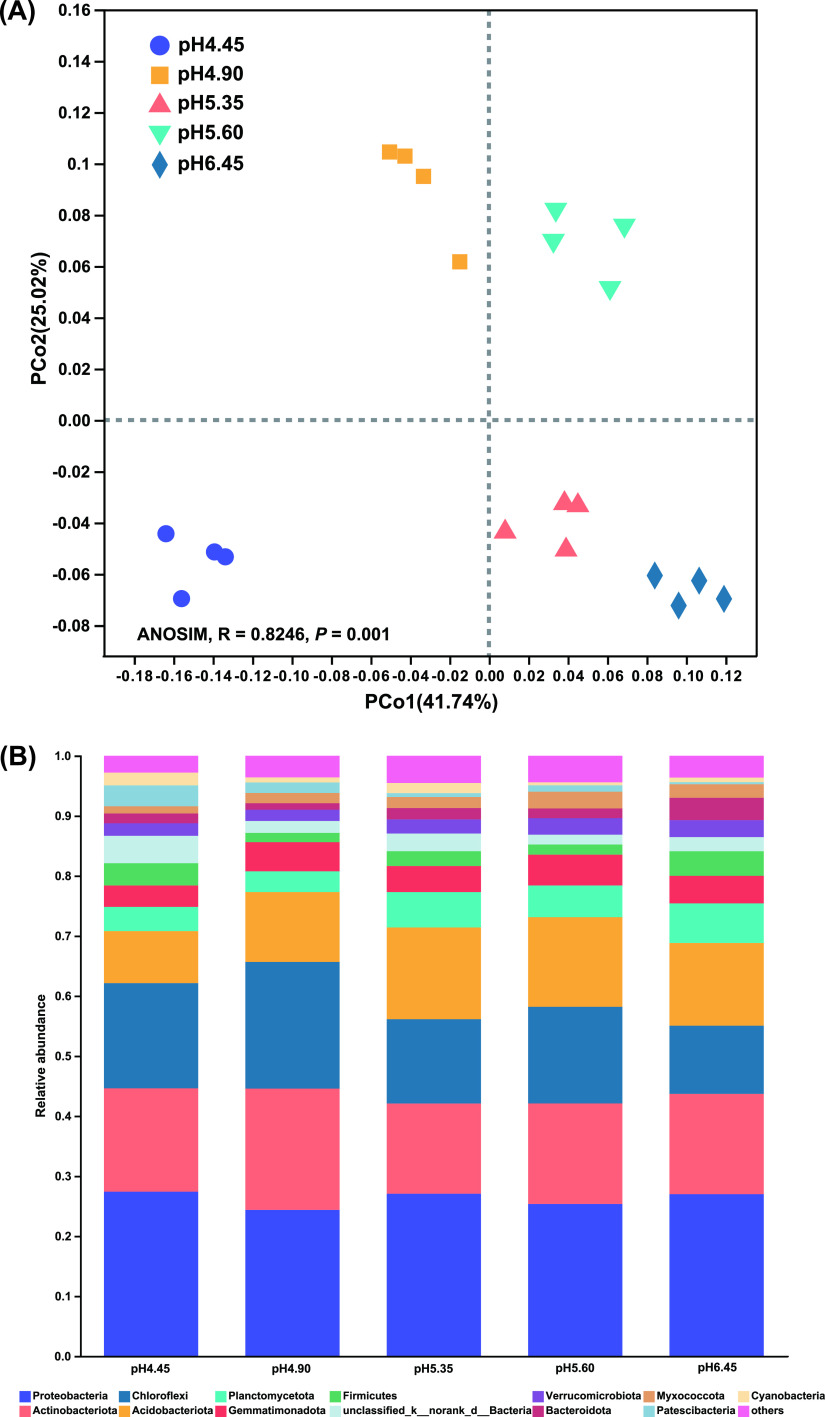
The rarefaction curve showed that the sequencing efforts of bacteria were sufficient for this study as the number of ASVs was saturated (Fig. S3). The bacterial community structure was illustrated using a PCoA plot based on the weighted UniFrac index. The bacterial communities were significantly different among the pH 4.45, pH 4.90, pH 5.35, pH 5.60 and pH 6.45 samples (R = 0.8246, P = 0.001, ANOSIM) (Fig. 2A).
FIG 2.
Comparison of soil microbial community structure among different samples. (A) Principal coordinates analysis (PCoA) by weighted UniFrac of bacterial composition from different soil acidification levels. (B) Relative abundance of bacterial phyla with an abundance greater than 1%.
All bacterial communities were dominated by phyla Proteobacteria, Actinobacteriota, Acidobacteriota, and Chloroflexi with 24.37–27.41%, 15.03–20.20%, 11.35–21.09% and 8.68–15.28% average relative abundance, respectively. The relative abundance of Firmicutes at pH 6.45 was the highest (4.10%), followed by pH 4.45 (3.71%). There was no significant difference in the relative abundance of Firmicutes between pH 6.45 and pH 4.45, however, the relative abundance was significantly higher than pH 4.90 (1.55%), pH 5.35 (2.48%) and pH 5.60 (1.69%) (Fig. 2B).
The linear discriminant analysis (LDA) effect size (LEfSe) method was used to detect bacterial taxa causing significant differences at the different pH levels. At the phylum level, Patescibacteria (LDA = 4.19) and Cyanobacteria (LDA = 3.92) were enriched at pH 4.45. Chloroflexi (LDA = 4.73), Actinobacteriota (LDA = 4.45) and WPS-2 (LDA = 3.41) were enriched at pH 4.90. Acidobacteriota (LDA = 4.56), Armatimonadota (LDA = 3.62), Latescibacterota (LDA = 3.35), Methylomirabilota (LDA = 3.17) and Desulfobacterota (LDA = 3.08) were enriched at pH 5.35. Gemmatimonadota (LDA = 3.97), Myxococcota (LDA = 3.95) and RCP2-54 (LDA = 3.20) were enriched at pH 5.60. Firmicutes (LDA = 4.09) and Bacteroidota (LDA = 4.09) were enriched at pH 6.45 (Fig. 3A).
FIG 3.
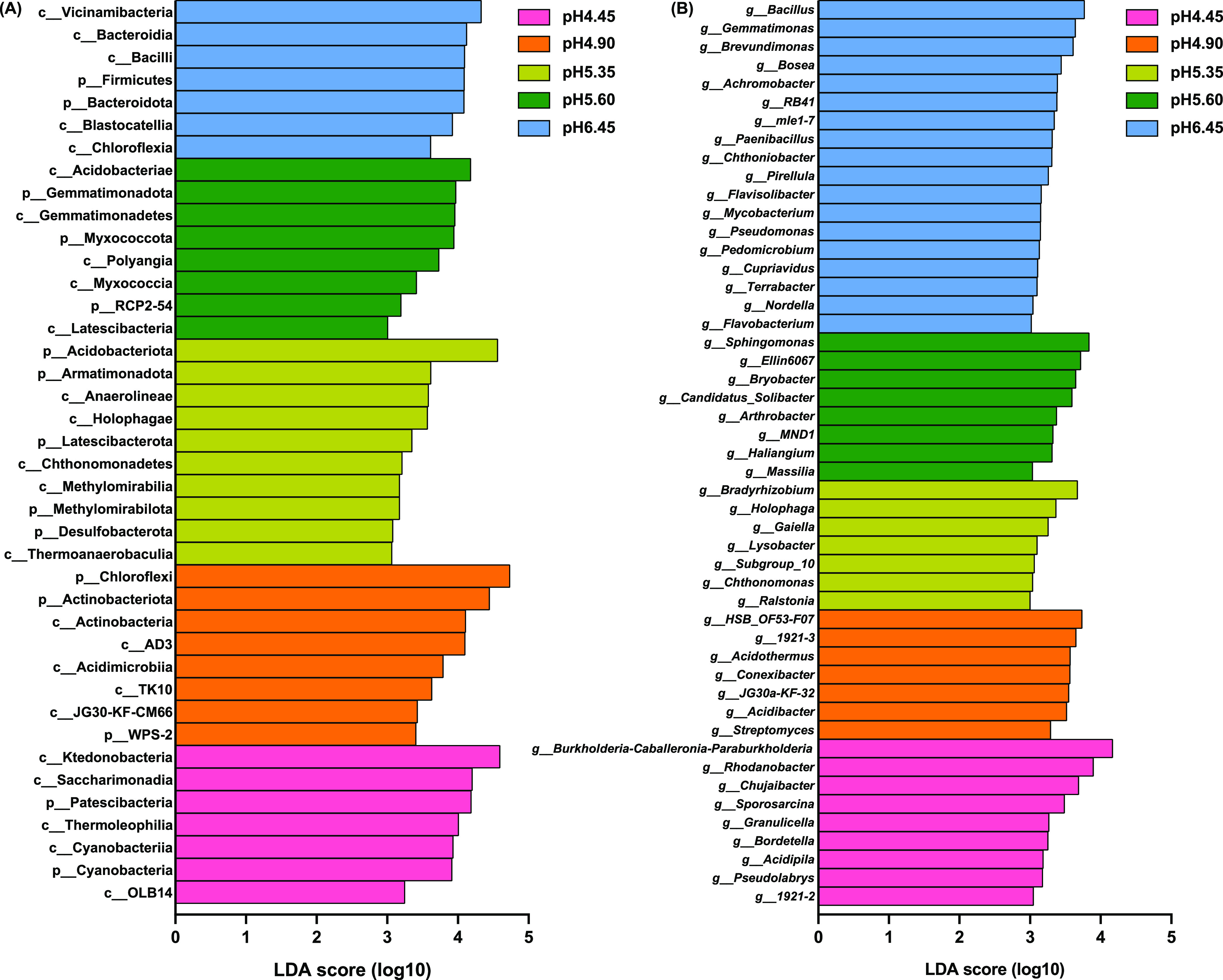
Histogram of the LDA scores computed for differentially abundant bacterial phyla and classes (A) and genera (B, deleted the norank and unclassified taxa) under different acidification levels.
At the genus level (removing norank and unclassified taxa), Ralstonia (LDA = 3.00) was enriched at pH 5.35. Whereas Bacillus (LDA = 3.77), Paenibacillus (LDA = 3.32), Pseudomonas (LDA = 3.15) and Flavobacterium (LDA = 3.02) were significantly enriched at pH 6.45 (Fig. 3B). In the range of acid treatment, with the increase of soil pH, the relative abundance of Flavobacterium increased significantly (R2 = 0.6580, P = 0.0001), and the relative abundance of Paenibacillus (R2 = 0.5374, P = 0.0014) and Bacillus (R2 = 0.6159, P = 0.0003) showed a significant trend of first decreasing and then increasing (Fig. 4). Moreover, spearman correlation analysis showed that there was an extremely significant negative correlation between the relative abundance of Paenibacillus (r = −0.708, P = 0.0004) and Bacillus (r = −0.749, P = 0.0005) with the abundance of R. solanacearum.
FIG 4.
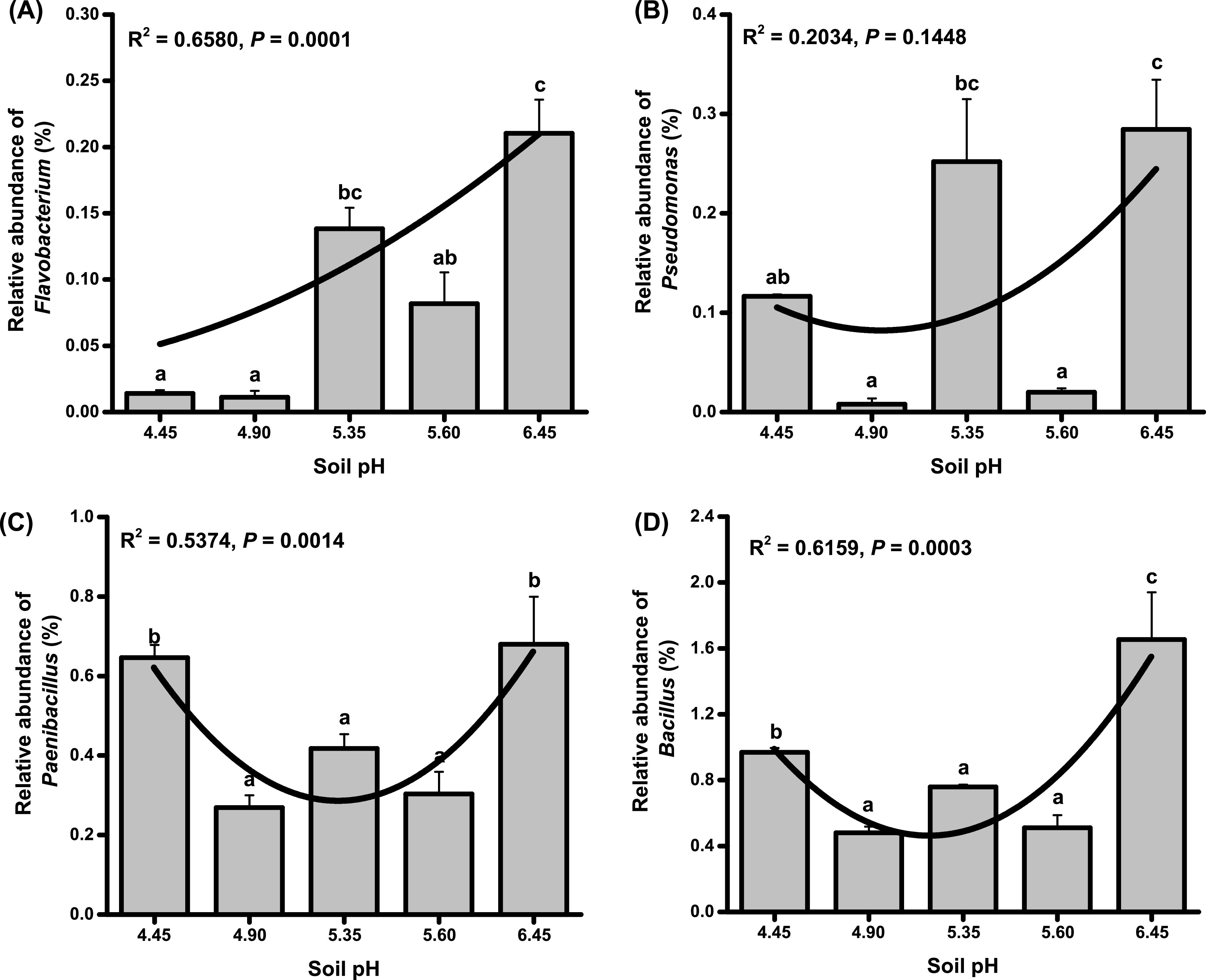
The relative abundance of Flavobacterium (A), Pseudomonas (B), Paenibacillus (C), and Bacillus (D) at different acidification levels. Different letters indicate significant (P < 0.05) differences according to one-way ANOVA.
Relationships between shifts in the bacterial community composition and environmental variables in experiment I.
The effects of environmental variables on the bacterial communities were assessed by redundancy analysis (RDA) (Fig. S4). AN (R2 = 0.9295, P = 0.001), ExAl (R2 = 0.8811, P = 0.001), soil pH (R2 = 0.7899, P = 0.001), AS (R2 = 0.6885, P = 0.001), AP (R2 = 0.6708, P = 0.002), ExCa (R2 = 0.6012, P = 0.001) and ExMg (R2 = 0.3451, P = 0.026) were significantly correlated with bacterial community structures at the different soil pH levels (Fig. S4, Table S1).
Carbon metabolism of the microbial community at different acidity levels in experiment I.
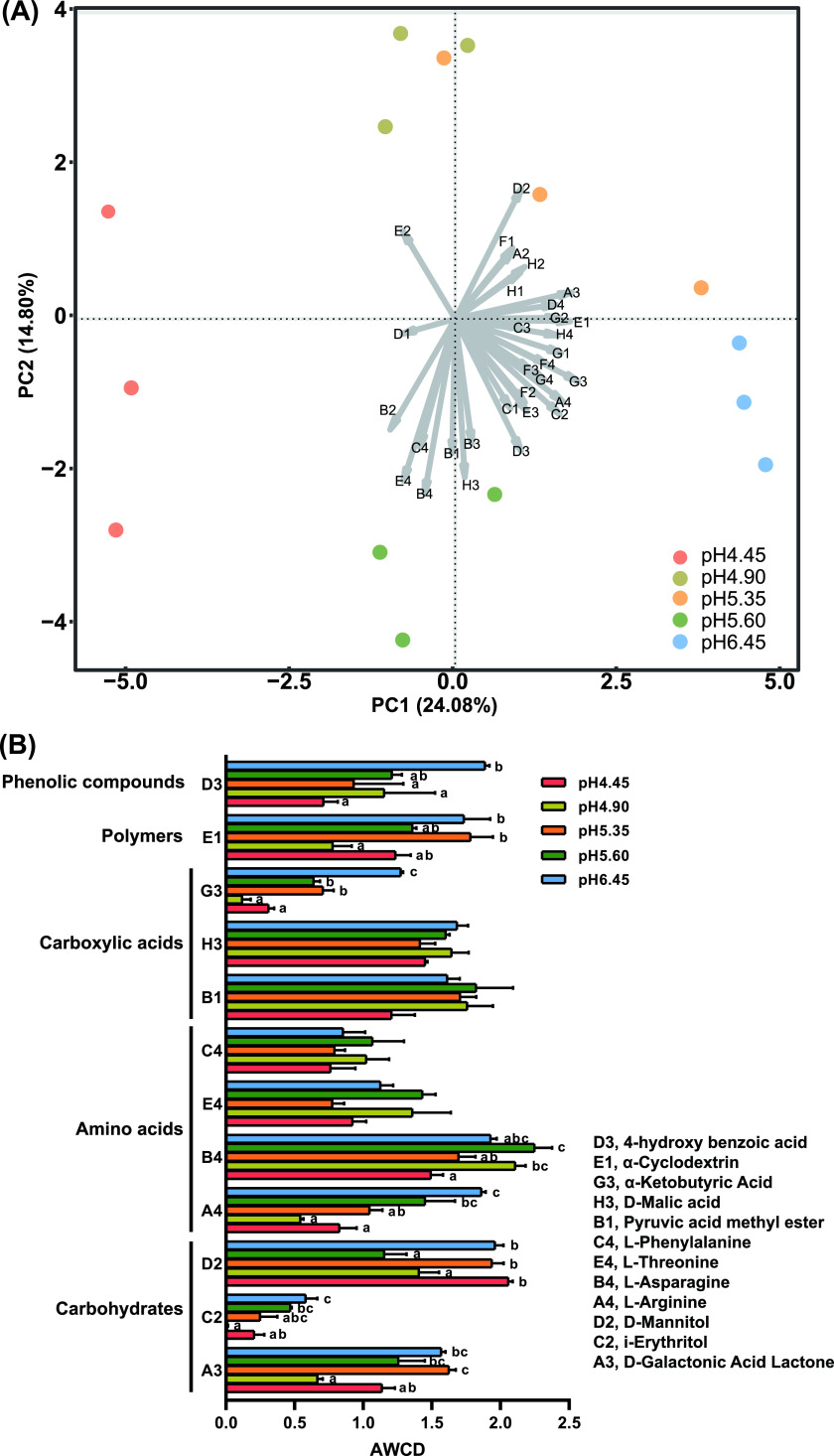
The average well color development (AWCD) values in pH 6.45 were the highest, and those at pH 4.45 were the lowest. As the soil pH increased, the carbon source metabolism capacity of soil microorganisms increased (Fig. S5). In the PCA of the Biolog data at 72 h, the microbial carbon source metabolism was significantly different between pH 4.45 and pH 6.45 (Fig. 5A). Carbon sources that had the arrow length above the average in Fig. 5A were selected to further present their well color development among different treatments in Fig. 5B. With the increase of soil pH, the metabolic capacity of 4-hydroxybenzoic acid, α-ketobutyric acid, l-arginine and i-erythritol were significantly enhanced.
FIG 5.
Microbial carbon source metabolism capacity at different soil pH. A, PCA ordination biplot of the different pH levels according to their carbon source utilization profile. All carbon sources are indicated by arrows. Longer arrows indicate a greater change in carbon source utilization value. B, The average well color development (AWCD) among different treatments (carbon sources which had the arrow length above the average). Different letters indicate significant (P < 0.05) differences according to one-way ANOVA. A2, β-Methyl-d-Glucoside; B2, d-Xylose; B3, d-Galacturonic Acid; C1, Tween 40; C3, 2-Hydroxybenzoic Acid; D1, Tween 80; D4, L-Serine; E2, N-Acetyl-d-Glucosamine; E3, γ-Hydroxybutyric Acid; F1, Glycogen; F2, d-Glucosaminic Acid; F3, Itaconic Acid; F4, Glycyl-L-Glutamic Acid; G1, d-Cellobiose; G2, α-d-Glucose-1-Phosphate; G4, Phenylethylamine; H1, α-d-Lactose; H2, D, L-α-Glycerol Phosphate; H4, Putrescine.
Effect of CaO on the occurrence of bacterial wilt and the rhizosphere bacterial communities in experiment II.
After adding CaO to the acidic soil, the soil pH increased from 5.45 to 6.05. Compared with the control, the disease incidence of bacterial wilt (P = 0.0142, independent-sample t-test) and the abundance of R. solanacearum (P = 0.0002, independent-sample t-test) in CaO treatment were significantly decreased by 45.83% and 1.05-fold, respectively (Fig. 6A and B).
FIG 6.
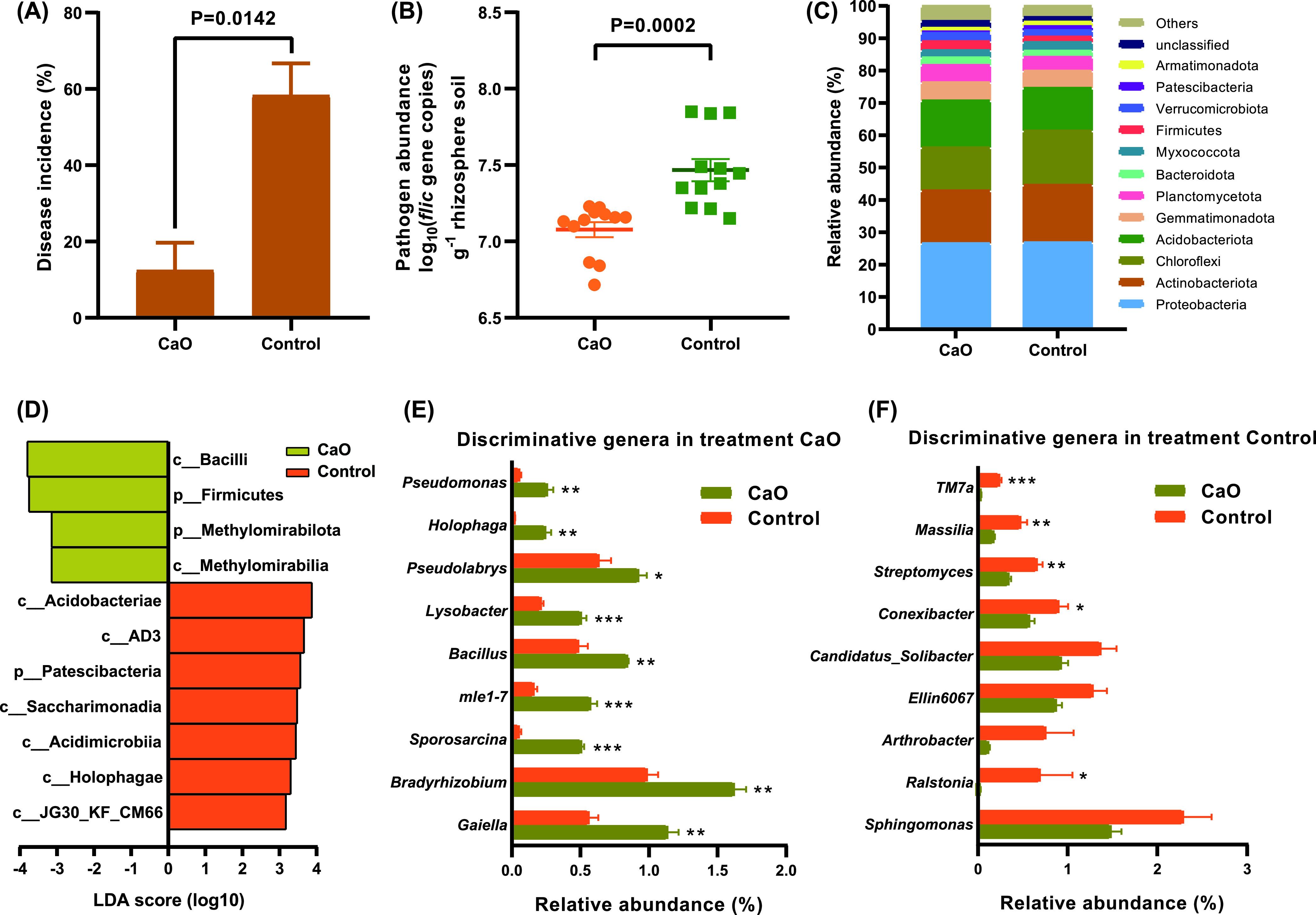
The occurrence of bacterial wilt and comparison of soil community structure between CaO and control in diseased soil. (A) The effect of CaO treatment on the incidence of bacterial wilt. (B) The influence of CaO treatment on the abundance of pathogen R. solanacearum. (C) The relative abundance of bacterial taxa at phylum level. (D) Histogram of the LDA scores computed for differentially abundant bacterial phyla and classes in CaO and control samples. (E–F) The relative abundances of discriminative genera (LDA > 3.0) in CaO and control samples, respectively. The P value and asterisks indicate significantly (* 0.01 < P ≤ 0.05, ** 0.001 < P ≤ 0.01, ***, P ≤ 0.001) difference between CaO and control, as determined by independent-sample t-test.
Principal coordinate analysis, based on the Bray–Curtis dissimilarity index, revealed clear differences between CaO and control samples (Fig. S6). Relative abundance analysis indicated that Proteobacteria (26.58–26.81% average relative abundance), Actinobacteriota (16.31–17.80% average relative abundance), Chloroflexi (13.35–16.80% average relative abundance), and Acidobacteriota (13.19–14.48% average relative abundance) were the main bacterial communities at the phylum level. The relative abundance of Firmicutes in CaO treatment was significantly increased by 1.66-fold compared with the control (Fig. 6C). Moreover, Firmicutes (LDA = 3.75), Methylomirabilota (LDA= 3.15), Bacilli (LDA = 3.80) and Methylomirabilia (LDA = 3.14) were significantly enriched in CaO (Fig. 6D). At the genus level (removing norank and unclassified taxa), Bacillus (LDA = 3.24) and Pseudomonas (LDA = 3.00) were significantly enriched in CaO, and the relative abundances were significantly increased by 1.75-fold and 4.56-fold compared with control, respectively (Fig. 6E). Whereas Ralstonia (LDA = 3.52) was significantly enriched in control, and the relative abundance was increased by 34.50-fold compared with CaO treatment (Fig. 6F).
CaO treatment increased the microbial carbon source metabolism capacity of rhizosphere soil microorganisms (Fig. S7A). The AWCD of L-arginine and 4 – hydroxybenzoic acid in CaO was higher than that of control, however, there was no significant difference between CaO and control (Fig. S7B-C).
DISCUSSION
Soil slightly acidic environment can alleviate the occurrence of bacterial wilt.
Strongly acidic soil (pH 4.5–5.5) is beneficial to the growth of R. solanacearum and aggravates the occurrence of bacterial wilt (6, 32). Some of the control measures of bacterial wilt, such as the addition of biochar (increasing the soil pH from 4.90 to 6.20), rock dust (increasing the soil pH from 5.13 to 6.81) and lime (increasing the soil pH over 6.00), one of the effective factors is improve the soil from strong acidity to slight acidity (pH 6.0–7.0) (2, 6, 15, 19). For some solanaceous crops, the optimum growth pH of tobacco is 6.0 (33), and tomato is pH 6.0–6.8 (34). In this study, the incidence of bacterial wilt with pH 6.45 was the lowest (Fig. 1B). CaO treatment increased the soil pH from 5.45 to 6.05, and compared with control (pH 5.55), the incidence of bacterial wilt was significantly reduced (Fig. 6A). The results showed that when using soil amendments to increase soil pH to control tobacco bacterial wilt, the soil pH is preferably at a slightly acidic level (between 6.0 and 6.5).
Beneficial bacteria increased the soil suppression of bacterial wilt in slightly acidic soil.
Because a steady-state balance of microbial community composition is essential for healthy host-microbe relationships, the enrichment and disruption of the microbial community is an important mechanism for the occurrence of plant diseases (35, 36). Firmicutes and Actinobacteriota abundance in the tomato rhizosphere conferred suppression of bacterial wilt (22). Firmicutes had a positive correlation with plant immunity (37). In this study, compared with pH 6.45, the relative abundance of Firmicutes was significantly decreased at pH 5.35 (Fig. 2B). Moreover, Firmicutes was significantly enriched at pH 6.45 (Fig. 3A), and after CaO (pH 6.05) was used to increase the pH, Firmicutes was also significantly enriched (Fig. 6D).
Bacillus (38–40) and Pseudomonas (41, 42) have been extensively studied for the growth promotion and suppression of bacterial wilt caused by R. solanacearum. Moreover, Bacillus and Pseudomonas were negatively related to the abundance of R. solanacearum (43). In this study, Bacillus had an extremely significant negative correlation with the R. solanacearum, however, there was no significant correlation between the Pseudomonas and R. solanacearum, which may be related to the effect of soil pH. The strains of Paenibacillus have been widely used for the control of bacterial wilt (44, 45). And the Flavobacterium in the rhizosphere of bacterial wilt resistant plants was much higher than that of susceptible plants (27). In this study, the abundance of R. solanacearum was significantly decreased at pH 6.45 (Fig. 1A), the potentially beneficial bacteria, Bacillus, Paenibacillus, Pseudomonas and Flavobacterium were significantly enriched at pH 6.45 (Fig. 3B). Moreover, after increasing soil pH by CaO (pH 6.05), the abundance of R. solanacearum also decreased significantly (Fig. 6B), and the relative abundance of Bacillus and Pseudomonas increased significantly (Fig. 6E). These results suggested that slightly acidic soil could increase the abundance of potentially beneficial bacteria and decrease the pathogen abundance in the rhizosphere, which led to the dominance of beneficial bacteria in the rhizosphere soil, and thus increased the soil suppression of bacterial wilt.
Microorganisms in slightly acidic soil increased their ability to metabolize specific carbon sources.
Soil environments are usually oligotrophic, where microbes compete fiercely for limited nutrients, such as carbon sources (46). Biolog can be used as an indicator of the microbial potential for carbon sources usage and potential changes therein as the results of the changes in environmental conditions (47). l-arginine can not only promote the growth of Bacillus amyloliquefaciens, but also promote the production of antibiotics bacillaene and macrolactin by Bacillus amyloliquefaciens, thereby inhibiting the growth of bacterial wilt (48). The excessive accumulation of 4-hydroxybenzoic acid in the soil inhibits the growth of crops, leading to crop yield reduction, continuous cropping obstacles and destruction of the natural ecological environment (49, 50). 4-hydroxybenzoic acid is one of the major autotoxins secreted by plant roots (51), and (52) indicated that 4-hydroxybenzoic acid was a strong chemoattractant for R. solanacearum. In addition, studies have shown that Thermophilic Bacillus sp. (53) and Pseudomonas sp. (54) can effectively degrade 4-hydroxybenzoic acid in the soil. l-arginine and 4-hydroxybenzoic could not be metabolized by R. solanacearum, whereas they were significantly metabolized in disease-suppressive of bacterial wilt soils, and they may act as indicators for deciphering the bacterial wilt suppression pattern (29). All in all, l-arginine and 4-hydroxybenzoic may promote the growth of the beneficial microbes, instead of the pathogen R. solanacearum. In this study, the microorganisms at pH 6.45 and CaO treatment increased their metabolic ability to the l-arginine and 4-hydroxybenzoic acid (Fig. 5B, Fig. S7B-C). These may be related to the enrichment of the potentially beneficial bacteria, such as Bacillus and Pseudomonas.
Soil pH affected rhizosphere bacterial community composition by changing soil chemical properties.
The forces that shape the rhizosphere microbial community cannot be fully understood without discussing the influence of the soil environment (55). Soil variables had an additional, indirect effect on the rhizosphere bacterial communities due to their influence on the composition of the bulk soil bacterial communities (56). In addition, specific soil physicochemical conditions of bulk soil, especially soil pH, may directly select for particular bacterial species in the rhizosphere (57). Soil pH can also affect the rhizosphere bacterial community structure indirectly by influencing nutrient availability (58, 59). And the availability of elements in the soil is closely related to soil pH (2). In this study, soil pH, available nitrogen, available phosphorus, available sulfur, exchangeable calcium, exchangeable magnesium and exchangeable aluminum were significantly correlated with bacterial communities. Moreover, the effects of available nitrogen and exchangeable aluminum ions on soil bacterial communities are stronger than soil pH (Fig. S4). Soil nitrogen availability and soil pH identified as the two most influential soil properties to influence the soil microbial community composition under nitrogen deposition (60). Yang et al. (61) indicated that nitrogen-induced changes in soil pH are an important mechanism driving the ecosystem functions. When the soils with a pH of 5.5 or lower, aluminum ions are dissolved from clay minerals, and there is a significant negative correlation between the aluminum ions and the soil pH (5, 62). These results suggested that soil pH may affect the microbial community composition by changing the availability of soil elements.
An extremely acidic soil environment also reduced the occurrence of bacterial wilt.
Aluminum ions have a toxic effect on the growth of plants (3). The organic acids released from plant roots, such as citric acid, oxalic acid, and malic acid, can chelate Al3+, thereby alleviating aluminum toxicity (63). Aluminum stress changed the rhizosphere bacterial communities (64). When plants are in a stressful environment, root exudates can attract beneficial microbes from the environment, which is called a “cry for help” strategy (65). Aluminum stress can stimulate the increase of Bacillus and Pseudomonas to alleviate ginger aluminum toxicity and bacterial wilt in extremely acidic soil (pH less than 4.5) (7). In this study, in a high-aluminum soil environment with pH 4.45, the growth of tobacco was not significantly affected (Fig. S2). In the acidic range, the abundance of R. solanacearum and the incidence of bacterial wilt showed a significant trend of first increasing and then decreasing with the increase of soil pH (Fig. 1). However, the relative abundances of Paenibacillus and Bacillus showed opposite trends (Fig. 4C and D). This may be the result of the release of root exudates from tobacco to alleviate aluminum toxicity while increasing the relative abundance of potentially beneficial bacteria in the rhizosphere. We are studying the relationship between aluminum stress and bacterial wilt.
In conclusion, slightly acidic soil (pH 6.45) and extremely acidic soil (pH 4.45) suppressed the growth of pathogenic R. solanacearum, thereby alleviating the occurrence of bacterial wilt. Moreover, changes in soil elements availability associated with soil acidic level significantly affected the soil bacterial community structure, leading to the enrichment of the potentially beneficial bacteria and the increase of the metabolism of 4-hydroxybenzoic acid in the slightly acidic soil (pH 6.45), and further the suppression of bacterial wilt (Fig. 7). These findings also explain that biological control of bacterial wilt by adding Bacillus or Pseudomonas, adjusting soil pH to a slightly acidic condition (pH 6.0–6.5) is the prerequisite to achieve a better control effect.
FIG 7.
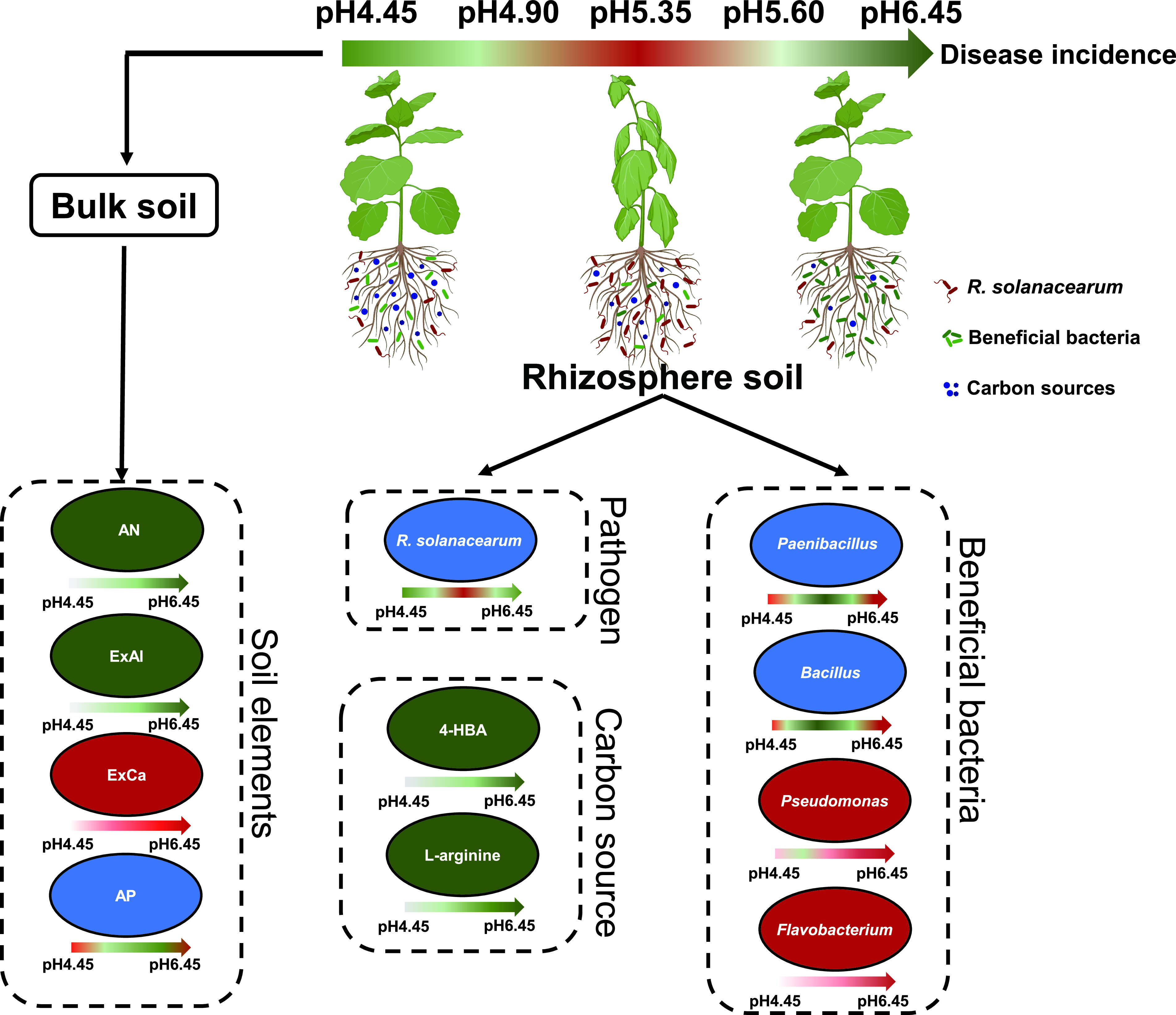
The influence of different acidification levels on various factors. The green color indicates downregulation, the red color indicates upregulation, and the blue color indicates both upregulation and downregulation. The intensity of the color in the arrows is proportional to the extent of the changes. AN, AP, ExCa, and ExAl indicate available soil nitrogen, available phosphorus, soil exchangeable calcium, and exchangeable aluminum, respectively. 4-HBA indicates 4-hydroxybenzoic.
MATERIALS AND METHODS
The effects of differently acidic pH gradients on tobacco rhizosphere microbial community composition were studied in two pot experiments. In the first experiment, nondiseased soil without bacterial wilt was adjusted to different acid gradients. Based on the results of the first experiment, the effect of soil amendment on the rhizosphere bacterial community by increasing the pH of diseased soil with bacterial wilt was further studied in the second experiment.
Soil sampling.
According to the survey, soil without bacterial wilt for continuous cropping 5 years was considered nondiseased soil, whereas the occurrence of bacterial wilt every year for 5 continuous years was considered diseased soil. Nondiseased (107°57.913′ E, 29°10.008′ N, 1315 m) and diseased (107°56.618′ E, 29°08.291′ N, 1219 m) soil samples were collected from Pengshui in Chongqing city, China, in May 2020. The nondiseased (sand-clay-silt, 32.57%–3.56%-63.87%) and diseased (sand-clay-silt, 21.75%–3.67%-74.58%) soil were classified as silt loam (32). Samples were obtained from the 10–20 cm of the soil. The soil samples were filtered through a 2 mm mesh to remove large soil particles and plant root tissue and debris. The soils were temporarily stored at 4°C for subsequent experiments.
Experimental setup.
Experiment I, nondiseased soil was treated with 0.1 mol/L NaOH or H2SO4 to adjust the soil pH to extreme acidity (pH below 4.5), very strong acidity (pH 4.5–5.0), strong acidity (pH 5.0–5.5), moderate acidity (pH 5.5–6.0) and slight acidity (pH 6.0–6.5). During the cultivation period, the pH was checked every 2 days to keep the pH within the set pH range and to keep the soil water holding capacity at 60%. After 30 days of incubation, the pH of the samples was 4.45, 4.90, 5.35, 5.60 and 6.45, and 500 g of soil was collected for chemical property analysis. Nicotiana benthamiana seedlings were then planted in the soil as the first population (1°, n = 32 plants per treatment). The rhizosphere soil of the plants (1°) was taken after 20 days of growth (each treatment had 4 replicates, and each replicate took rhizosphere soil from 8 tobacco seedlings), and the soil samples were stored at −20°C until needed for DNA extraction. Then the second population (2°) of Nicotiana benthamiana was planted in bulk soil with different pH levels (n = 24 plants per treatment). After 7 days of growth, 10 mL of Ralstonia solanacearum wild-type strain CQPS-1 (66) with OD600 = 0.01 was added to each tobacco seedling, and then the disease incidence of bacterial wilt was investigated after 14 days (Fig. S1).
Experiment II, the initial pH of the diseased soil was 5.45. The diseased soil was equally divided into 2 parts, and then the soil was treated with 1 g/kg of CaO and deionized water (control), respectively. After 30 days of soil treatment, the pH of the soil with CaO and control was 6.05 and 5.55, respectively. Before planting Nicotiana benthamiana seedlings, a small amount of soil was collected for the quantitative detection of R. solanacearum. Then, Nicotiana benthamiana seedlings were planted to investigate the occurrence of bacterial wilt (n = 24). On the 15th day after planting tobacco seedlings, the incidence of bacterial wilt was determined, and then the rhizosphere soil of tobacco seedlings was collected and stored at −20°C for DNA extraction.
Determination of soil chemical properties.
Soil pH was assayed with a pH electrode (InLab Science, Mettler Toledo, Switzerland) in soil water suspensions (1:2.5 weight/volume). The soil organic matter (SOM) content was assayed with acidified potassium dichromate (K2Cr2O7–H2SO4). The alkaline hydrolysis diffusion method was used to determine available soil nitrogen (AN). Available phosphorus (AP) was analyzed by the Olsen method (67). Available potassium (AK) was extracted with NH4OAc and analyzed by flame emission spectrometry. Available sulfur (AS) was analyzed with barium sulfate turbidimetry. Exchangeable calcium (ExCa) and magnesium (ExMg) were extracted with NH4OAc, exchangeable aluminum (ExAl) was extracted with KCl, and measured by the inductively coupled plasma (ICP) method (19).
Soil DNA extraction and quantitative PCR (qPCR).
Total soil genomic DNA was extracted from 500 mg of fresh soil using a FastDNA spin kit (MP Biomedicals, United States) according to the standard protocol. The elution volume for DNA was 100 μL. The DNA was stored at −20°C for subsequent analyses.
We used quantified PCR (qPCR) to quantify the abundance of the pathogen R. solanacearum in the rhizosphere soil. R. solanacearum density was quantified by using specific primers FlicF (5′-GAACGCCAACGGTGCGAACT-3′)/FlicR (5′-GGCGGCCTTCAGGGAGGTC-3′) targeting the fliC gene coding the flagellum subunit (68). The qPCR analyses were carried out with a CFX96 Optical Real-time Detection System (Bio-Rad, United States). The reactions were conducted in a 20 μL mixture containing 10 μL of Pro Taq HS SYBR green (AG11701, Accurate Biotechnology, Hunan, Co., Ltd., China), 1 μL of each primer (10 μmol/L), 1 μL of template, and 7 μL of double-distilled water (ddH2O). The qPCR conditions were performed as described by Hu et al. (69) with some modification: 95°C for 5 min, followed by 30 cycles of 94°C for 30 s, 60°C for 40 s and 72°C for 30 s. Melting curve (60°C to 95°C, increment 0.5°C for 5 s) analysis was performed at the end of the PCR experiment to evaluate the specificity of the amplification. Standard curves were created using 10-fold serial dilutions (103–107) of a plasmid containing a copy of the flic sequence. The coefficient of determination of the standard curve was 0.999, and the efficiency was 89.1%.
Sequencing library construction.
16S rRNA high-throughput sequencing was performed for the rhizosphere soils in experiment I and experiment II. PCR amplifications were conducted with 515 forward primers (5′-GTGCCAGCMGCCGCGG-3′) and 806 reverse primers (5′-GGACTACHVGGGTWTCTAAT-3′), which amplified the V4 region of the 16S rRNA gene (70).
The PCR amplification conditions were as follows: 95°C for 3 min, followed by 27 cycles of 30 s at 95°C, 30 s at 55°C, and 45 s at 72°C, and a final extension was performed at 72°C for 10 min. PCR of 515F_806R was performed with 4 μL of 5 × TransStart FastPfu buffer, 2 μL of 2.5 mM deoxynucleoside triphosphates (dNTPs), 0.8 μL of each primer (5 μM), 0.4 μL of TransStart FastPfu DNA polymerase, 10 ng of extracted DNA, and ddH2O to a final volume of 20 μL. Agarose gel electrophoresis was performed to verify the size of the PCR amplicons. Amplicons were subjected to paired-end sequencing on the NovaSeq 6000 sequencing platform using PE250 chemical at Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China).
A total of 2,274,014 (average read length was 256.15 bp) sequences were obtained from the 20 nondiseased soil samples in experiment I, and 898,729 (average read length was 256.17 bp) sequences were obtained from 8 diseased soil samples in experiment II. The raw reads were deposited into the NCBI short-reads archive database under accession number PRJNA804972 (experiment I) and PRJNA715361 (experiment II).
Bioinformatics analysis.
After demultiplexing, the resulting sequences were merged with FLASH (v1.2.11) (71) and quality filtered with fastp (0.19.6) (72). Then, the high-quality sequences were denoised using the DADA2 (73) plugin in the QIIME2 (74) (version 2020.2) pipeline with recommended parameters, which obtains single nucleotide resolution based on error profiles within samples. DADA2 denoised sequences are usually called amplicon sequence variants (ASVs). Taxonomic assignment of ASVs was performed using the Naive Bayes consensus taxonomy classifier implemented in QIIME 2 and the SILVA 16S rRNA database (v138) for bacteria (threshold 0.7).
To complete the diversity and composition analyses, the sequence of each sample was rarefied to the lowest number of sequences (75). Rarefaction curves of ASVs were drawn to verify whether the sequencing depth was adequate to cover most microbial taxa. The differences of bacterial community structure among different soil pH were determined using analysis of similarities (ANOSIM) and principal-component analysis (PCoA) based on the weighted UniFrac or Bray-Curtis distance in the “vegan” package in the R. In order to identify the correlation between bacterial communities and environment variables, redundancy analysis (RDA) was determined in the vegan package.
Linear discriminant analysis (LDA) effect size (LEfSe) employed the factorial Kruskal–Wallis sum-rank test (α = 0.05) to identify taxa with significant differential abundances between categories (using all-against-all comparisons), followed by LDA to estimate the effect size of each differentially abundant feature (logarithmic LDA score ≥ 3.0). Significant taxa were used to generate taxonomic cladograms, which illustrated the differences between sample classes on the website http://huttenhower.sph.harvard.edu/galaxy.
Microbial carbon source metabolic activity.
Microbial carbon source metabolic activity analysis was performed on the rhizosphere soil of pH 4.45, pH 5.90, pH 5.35, pH 5.60 and pH 6.45 in experiment I and the rhizosphere soil of CaO and control in experiment II. Microbial carbon source metabolism expressed in each Biolog EcoPlate (EcoPlate, Biolog, Hayward, CA, USA) was determined as average well color development (AWCD) (76). The experiment was performed on the day of rhizosphere soil sampling to avoid changes in microbial communities during storage of the soil. The carbon source utilization pattern for each soil sample was determined in accordance with the procedures described by Zhang et al. (77). The AWCD was calculated according to Wang et al. (78). The detailed carbon source usage was measured by the absorbance at 590 nm (79). And principal component analysis (PCA) was used to find the most related carbon sources within different treatments at 72 h of culturing (80). Carbon sources which had the arrow length above the average were selected to further present their well color development among different treatments.
Statistical analyses.
The figures were created using GraphPad Prism 8.0.1. The mean and standard error for each set of data were calculated by independent-sample t-test (P < 0.05) or one-way analysis of variance (ANOVA) with Tukey’s honestly significant difference test (P < 0.05) were performed in SPSS software (version 17.0). Linear models to examine the relationships of pathogen abundance, disease incidence and potentially beneficial genera with soil pH in Origin 9. Spearman's rank correlation coefficient between the potentially beneficial genera with the R. solanacearum abundance was calculated using SPSS v17.0.
ACKNOWLEDGMENTS
This work was supported by the key science and technology projects in Sichuan (SCYC201908 and SCYC202010). We thank the free online platform of the Majorbio Cloud Platform (www.majorbio.com) for analyses of the 16S rRNA microbiome sequencing data.
W.D. organized and supervised the project. S.Z. and W.D. designed the experiment and interpreted the results for the manuscript. S.Z. and L.Z. performed the experiments and harvested the rhizosphere soil samples. L.Z. performed the Biolog ECO experiment. L.D. and W.Z. performed the qRT-PCR analysis. S.Z. and X.L. analyzed the microbial carbon source metabolism data. S.Z. analyzed the microbiome data. S.Z. wrote the manuscript. S.Z., X.L., Y.L., and W.D. edited the manuscript. All authors read and approved the final version of the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Wei Ding, Email: dingw@swu.edu.cn.
Kristen M. DeAngelis, University of Massachusetts Amherst
REFERENCES
- 1.Dai Z, Zhang X, Tang C, Muhammad N, Wu J, Brookes PC, Xu J. 2017. Potential role of biochars in decreasing soil acidification: A critical review. Sci Total Environ 581-582:601–611. doi: 10.1016/j.scitotenv.2016.12.169. [DOI] [PubMed] [Google Scholar]
- 2.Jones JB. 2012. Plant nutrition and soil fertility manual Second Edition. Soil pH: its determination and interpretation. CRC Press; pp 57–64. doi: 10.1201/b11577-12. [DOI] [Google Scholar]
- 3.Aggarwal A, Ezaki B, Munjal A, Tripathi BN. 2015. Physiology and biochemistry of aluminum toxicity and tolerance in crops. stress responses in plants pp 35–57. https://link.springer.com/chapter/10.1007/978-3-319-13368-3_2. [Google Scholar]
- 4.Zhou G, Delhaize E, Zhou M, Ryan PR. 2011. Biotechnological solutions for enhancing the aluminum resistance of crop plants. In Shanker A, Venkateswarlu B (ed), Abiotic stress in plants—mechanisms and adaptations. InTech, Rijeka: pp 119–142. [Google Scholar]
- 5.Bojorquez-Quintal E, Escalante-Magana C, Echevarria-Machado I, Martinez-Estevez M. 2017. Aluminum, a friend or foe of higher plants in acid soils. Front Plant Sci 8:1767. doi: 10.3389/fpls.2017.01767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, Liu Y, Wang J, Yang L, Zhang S, Xu C, Ding W. 2017. Soil acidification aggravates the occurrence of bacterial wilt in South China. Front Microbiol 8:703. doi: 10.3389/fmicb.2017.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang S, Jiang Q, Liu X, Liu L, Ding W. 2020. Plant growth promoting Rhizobacteria alleviate aluminum toxicity and ginger bacterial wilt in acidic continuous cropping soil. Front Microbiol 11. doi: 10.3389/fmicb.2020.569512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang X, You MP, Barbetti MJ. 2012. Reduced severity and impact of Fusarium wilt on strawberry by manipulation of soil pH, soil organic amendments and crop rotation. Eur J Plant Pathol 134:619–629. doi: 10.1007/s10658-012-0042-1. [DOI] [Google Scholar]
- 9.Rahman KA, Othman R. 2020. Influence of pH levels on disease development in oil palm seedling roots infected with Ganoderma boninensis. Rhizosphere 13:100181. doi: 10.1016/j.rhisph.2019.100181. [DOI] [Google Scholar]
- 10.Genin S, Denny TP. 2012. Pathogenomics of the Ralstonia solanacearum Species Complex. Annu Rev Phytopathol 50:67–89. doi: 10.1146/annurev-phyto-081211-173000. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Zhang S, Jiang Q, Bai Y, Shen G, Li S, Ding W. 2016. Using community analysis to explore bacterial indicators for disease suppression of tobacco bacterial wilt. Sci Rep 6:36773. doi: 10.1038/srep36773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng X, Liu B, Zhu Y, Wang J, Zhang H, Wang Z. 2019. Bacterial community diversity associated with the severity of bacterial wilt disease in tomato fields in southeast China. Can J Microbiol 65:538–549. doi: 10.1139/cjm-2018-0637. [DOI] [PubMed] [Google Scholar]
- 13.Nakahara H, Mori T, Sadakari N, Matsusaki H, Matsuzoe N. 2016. Selection of effective non-pathogenic Ralstonia solanacearum as biocontrol agents against bacterial wilt in eggplant. J Plant Dis Prot 123:119–124. doi: 10.1007/s41348-016-0019-y. [DOI] [Google Scholar]
- 14.Yang H, Li J, Xiao Y, Gu Y, Liu H, Liang Y, Liu X, Hu J, Meng D, Yin H. 2017. An integrated insight into the relationship between soil microbial community and tobacco bacterial wilt disease. Front Microbiol 8:2179. doi: 10.3389/fmicb.2017.02179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C, Lin Y, Tian X, Xu Q, Chen Z, Lin W. 2017. Tobacco bacterial wilt suppression with biochar soil addition associates to improved soil physiochemical properties and increased rhizosphere bacteria abundance. Appl Soil Ecol 112:90–96. doi: 10.1016/j.apsoil.2016.12.005. [DOI] [Google Scholar]
- 16.Gao Y, Lu Y, Lin W, Tian J, Cai K. 2019. Biochar suppresses bacterial wilt of tomato by improving soil chemical properties and shifting soil microbial community. Microorganisms 7:676. doi: 10.3390/microorganisms7120676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen G, Zhang S, Liu X, Jiang Q, Ding W. 2018. Soil acidification amendments change the rhizosphere bacterial community of tobacco in a bacterial wilt affected field. Appl Microbiol Biotechnol 102:9781–9791. doi: 10.1007/s00253-018-9347-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michel VV, Mew TW. 1998. Effect of a soil amendment on the survival of Ralstonia solanacearum in different soils. Phytopathology 88:300–305. doi: 10.1094/PHYTO.1998.88.4.300. [DOI] [PubMed] [Google Scholar]
- 19.Li J-G, Dong Y-H. 2013. Effect of a rock dust amendment on disease severity of tomato bacterial wilt. Anton Leeuw Int J G 103:11–22. doi: 10.1007/s10482-012-9781-4. [DOI] [PubMed] [Google Scholar]
- 20.He K, Yang S-Y, Li H, Wang H, Li Z-L. 2014. Effects of calcium carbonate on the survival of Ralstonia solanacearum in soil and control of tobacco bacterial wilt. Eur J Plant Pathol 140:665–675. doi: 10.1007/s10658-014-0496-4. [DOI] [Google Scholar]
- 21.Niu J, Chao J, Xiao Y, Chen W, Zhang C, Liu X, Rang Z, Yin H, Dai L. 2017. Insight into the effects of different cropping systems on soil bacterial community and tobacco bacterial wilt rate. J Basic Microbiol 57:3–11. doi: 10.1002/jobm.201600222. [DOI] [PubMed] [Google Scholar]
- 22.Lee SM, Kong HG, Song GC, Ryu CM. 2021. Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. ISME J 15:330–347. doi: 10.1038/s41396-020-00785-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cha J-Y, Han S, Hong H-J, Cho H, Kim D, Kwon Y, Kwon S-K, Crüsemann M, Bok Lee Y, Kim JF, Giaever G, Nislow C, Moore BS, Thomashow LS, Weller DM, Kwak Y-S. 2016. Microbial and biochemical basis of a Fusarium wilt-suppressive soil. ISME J 10:119–129. doi: 10.1038/ismej.2015.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu D, Li K, Hu J, Wang W, Liu X, Gao Z. 2019. Biocontrol and action mechanism of Bacillus amyloliquefaciens and Bacillus subtilis in soybean phytophthora blight. Int J Mol Sci 20:2908. doi: 10.3390/ijms20122908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raaijmakers JM, Weller DM. 1998. Natural plant protection by 2,4-Diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. MPMI 11:144–152. doi: 10.1094/MPMI.1998.11.2.144. [DOI] [Google Scholar]
- 26.Haggag WM, Timmusk S. 2008. Colonization of peanut roots by biofilm‐forming Paenibacillus polymyxa initiates biocontrol against crown rot disease. J Appl Microbiol 104:961–969. doi: 10.1111/j.1365-2672.2007.03611.x. [DOI] [PubMed] [Google Scholar]
- 27.Kwak MJ, Kong HG, Choi K, Kwon SK, Song JY, Lee J, Lee PA, Choi SY, Seo M, Lee HJ, Jung EJ, Park H, Roy N, Kim H, Lee MM, Rubin EM, Lee SW, Kim JF. 2018. Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat Biotechnol 36:1100–1109. doi: 10.1038/nbt.4232. [DOI] [PubMed] [Google Scholar]
- 28.Xiao Y, Liu X, Meng D, Tao J, Gu Y, Yin H, Li J. 2018. The role of soil bacterial community during winter fallow period in the incidence of tobacco bacterial wilt disease. Appl Microbiol Biotechnol 102:2399–2412. doi: 10.1007/s00253-018-8757-3. [DOI] [PubMed] [Google Scholar]
- 29.Liu X, Jiang Q, Hu X, Zhang S, Liu Y, Huang W, Ding W. 2019. Soil microbial carbon metabolism reveals a disease suppression pattern in continuous ginger mono-cropping fields. Appl Soil Ecol 144:165–169. doi: 10.1016/j.apsoil.2019.07.020. [DOI] [Google Scholar]
- 30.Zhalnina K, Dias R, de Quadros PD, Davis-Richardson A, Camargo FA, Clark IM, Mcgrath SP, Hirsch PR, Triplett EW. 2015. Soil pH determines microbial diversity and composition in the park grass experiment. Microb Ecol 69:395–406. doi: 10.1007/s00248-014-0530-2. [DOI] [PubMed] [Google Scholar]
- 31.Rousk J, Baath E, Brookes PC, Lauber CL, Lozupone C, Caporaso JG, Knight R, Fierer N. 2010. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4:1340–1351. doi: 10.1038/ismej.2010.58. [DOI] [PubMed] [Google Scholar]
- 32.Jones 2012. Plant nutrition and soil fertility manual, second edition physical and physiochemical characteristics of soil. CRC Press; 577:43–44. doi: 10.1201/b11577-10. [DOI] [Google Scholar]
- 33.Chen J, Deng HH, Li YH, Yang LL, Deng YS, Deng XH, Zhou ZC, Peng SG, Liu YJ, Tian MH, Chen ZF. 2020. Strong acidic conditions impaired photosynthesis, root system and seedling growth of flue-cured tobacco. Int J Agric Biol 24:645–650. [Google Scholar]
- 34.Kimura S, Sinha N. 2008. How to grow tomatoes. CSH Protoc. doi: 10.1101/pdb.prot5081. [DOI] [PubMed] [Google Scholar]
- 35.Hacquard S, Garrido-Oter R, González A, Spaepen S, Ackermann G, Lebeis S, McHardy AC, Dangl JL, Knight R, Ley R, Schulze-Lefert P. 2015. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 17:603–616. doi: 10.1016/j.chom.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Fernández-González AJ, Martínez-Hidalgo P, Cobo-Díaz JF, Villadas PJ, Martínez-Molina E, Toro N, Tringe SG, Fernández-López M. 2017. The rhizosphere microbiome of burned holm-oak: potential role of the genus Arthrobacter in the recovery of burned soils. Sci Rep 7:6008. doi: 10.1038/s41598-017-06112-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M, Malfatti S, Glavina del Rio T, Jones CD, Tringe SG, Dangl JL. 2015. PLANT MICROBIOME. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349:860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 38.Tan S, Dong Y, Liao H, Huang J, Song S, Xu Y, Shen Q. 2013. Antagonistic bacterium Bacillus amyloliquefaciens induces resistance and controls the bacterial wilt of tomato. Pest Manag Sci 69:1245–1252. doi: 10.1002/ps.3491. [DOI] [PubMed] [Google Scholar]
- 39.Hyakumachi M, Nishimura M, Arakawa T, Asano S, Yoshida S, Tsushima S, Takahashi H. 2013. Bacillus thuringiensis suppresses bacterial wilt disease caused by Ralstonia solanacearum with systemic induction of defense-related gene expression in tomato. Microbes Environ 28:128–134. doi: 10.1264/jsme2.me12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Y, Yan F, Chai Y, Liu H, Kolter R, Losick R, Guo JH. 2013. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ Microbiol 15:848–864. doi: 10.1111/j.1462-2920.2012.02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vanitha SC, Niranjana SR, Mortensen CN, Umesha S. 2009. Bacterial wilt of tomato in Karnataka and its management by Pseudomonas fluorescens. Biocontrol 54:685–695. doi: 10.1007/s10526-009-9217-x. [DOI] [Google Scholar]
- 42.Yendyo S, Ramesh GC, Pandey BR, Coutinho TA, Gauchan DP, Coutinho TA. 2017. Evaluation of Trichoderma spp., Pseudomonas fluorescens and Bacillus subtilis for biological control of Ralstonia wilt of tomato. F1000Res 6:2028. doi: 10.12688/f1000research.12448.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wen T, Zhao M, Liu T, Huang Q, Yuan J, Shen Q. 2020. High abundance of Ralstonia solanacearum changed tomato rhizosphere microbiome and metabolome. BMC Plant Biol 20:166. doi: 10.1186/s12870-020-02365-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li B, Yu RR, Tang QM, Su T, Chen XL, Zhu B, Wang YL, Xie GL, Sun GC. 2011. Biofilm formation ability of Paenibacillus polymyxa and Paenibacillus macerans and their inhibitory effect against tomato bacterial wilt. Afr J Microbiol Res 5:4260–4266. doi: 10.5897/ajmr10.549. [DOI] [Google Scholar]
- 45.Yi J, Zhang D, Cheng Y, Tan J, Luo Y. 2019. The impact of Paenibacillus polymyxa HY96-2 luxS on biofilm formation and control of tomato bacterial wilt. Appl Microbiol Biotechnol 103:9643–9657. doi: 10.1007/s00253-019-10162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemanceau P, Maron PA, Mazurier S, Mougel C, Pivato B, Plassart P, Ranjard L, Revellin C, Tardy V, Wipf D. 2015. Understanding and managing soil biodiversity: a major challenge in agroecology. Agron Sustain Dev 35:67–81. doi: 10.1007/s13593-014-0247-0. [DOI] [Google Scholar]
- 47.Drost SM, Rutgers M, Wouterse M, de Boer W, Bodelier PLE. 2020. Decomposition of mixtures of cover crop residues increases microbial functional diversity. Geoderma 361:114060. doi: 10.1016/j.geoderma.2019.114060. [DOI] [Google Scholar]
- 48.Yang C, Dong Y, Friman VP, Jousset A, Wei Z, Xu Y, Shen Q, Hart M. 2019. Carbon resource richness shapes bacterial competitive interactions by alleviating growth‐antibiosis trade‐off. Funct Ecol 33:868–875. doi: 10.1111/1365-2435.13292. [DOI] [Google Scholar]
- 49.Bais HP, Vepachedu R, Gilroy S, Callaway RM, Vivanco JM. 2003. Allelopathy and exotic plant invasion: from molecules and genes to species interactions. Science 301:1377–1380. doi: 10.1126/science.1083245. [DOI] [PubMed] [Google Scholar]
- 50.Chen P, Wang Y-z, Liu Q-z, Zhang Y-t, Li X-y, Li H-q, Li W-h. 2020. Phase changes of continuous cropping obstacles in strawberry (Fragaria × ananassa Duch.) production. Appl Soil Ecol 155:103626. doi: 10.1016/j.apsoil.2020.103626. [DOI] [Google Scholar]
- 51.Liu Q, Li K, Guo X, Ma L, Guo Y, Liu Z. 2019. Developmental characteristics of grapevine seedlings root border cells and their response to ρ-hydroxybenzoic acid. Plant Soil 443:199–218. doi: 10.1007/s11104-019-04220-9. [DOI] [Google Scholar]
- 52.Hasegawa T, Kato Y, Okabe A, Itoi C, Ooshiro A, Kawaide H, Natsume M. 2019. Effect of Secondary Metabolites of Tomato (Solanum lycopersicum) on Chemotaxis of Ralstonia solanacearum, Pathogen of Bacterial Wilt Disease. J Agric Food Chem 67:1807–1813. doi: 10.1021/acs.jafc.8b06245. [DOI] [PubMed] [Google Scholar]
- 53.Peng X, Misawa N, Harayama S. 2003. Isolation and characterization of thermophilic bacilli degrading cinnamic, 4-coumaric, and ferulic acids. Appl Environ Microbiol 69:1417–1427. doi: 10.1128/AEM.69.3.1417-1427.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jaigeeth D, Suvekbala V, Gautham V, Phale PS. 2007. Metabolism of 2-, 3- and 4-hydroxybenzoates by soil isolates Alcaligenes sp. strain PPH and Pseudomonas sp. strain PPD. FEMS Microbiol Lett 268:59–66. doi: 10.1111/j.1574-6968.2006.00561.x. [DOI] [PubMed] [Google Scholar]
- 55.Chaparro JM, Sheflin AM, Manter DK, Vivanco JM. 2012. Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fertil Soils 48:489–499. doi: 10.1007/s00374-012-0691-4. [DOI] [Google Scholar]
- 56.Vieira S, Sikorski J, Dietz S, Herz K, Schrumpf M, Bruelheide H, Scheel D, Friedrich MW, Overmann J. 2020. Drivers of the composition of active rhizosphere bacterial communities in temperate grasslands. ISME J 14:463–475. doi: 10.1038/s41396-019-0543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Na X, Xu T, Li M, Zhou Z, Ma S, Wang J, He J, Jiao B, Ma F. 2018. Variations of bacterial community diversity within the rhizosphere of three phylogenetically related perennial shrub plant species across environmental gradients. Front Microbiol 9:709. doi: 10.3389/fmicb.2018.00709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marschner P, Crowley D, Yang CH. 2004. Development of specific rhizosphere bacterial communities in relation to plant species, nutrition and soil type. Plant Soil 261:199–208. doi: 10.1023/B:PLSO.0000035569.80747.c5. [DOI] [Google Scholar]
- 59.Vives-Peris V, de Ollas C, Gomez-Cadenas A, Perez-Clemente RM. 2020. Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep 39:3–17. doi: 10.1007/s00299-019-02447-5. [DOI] [PubMed] [Google Scholar]
- 60.Wang J, Shi X, Zheng C, Suter H, Huang Z. 2021. Different responses of soil bacterial and fungal communities to nitrogen deposition in a subtropical forest. Sci Total Environ 755:142449. doi: 10.1016/j.scitotenv.2020.142449. [DOI] [PubMed] [Google Scholar]
- 61.Yang F, Zhang Z, Barberán A, Yang Y, Hu S, Guo H. 2021. Nitrogen-induced acidification plays a vital role driving ecosystem functions: insights from a 6-year nitrogen enrichment experiment in a Tibetan alpine meadow. Soil Biol Biochem 153:108107. doi: 10.1016/j.soilbio.2020.108107. [DOI] [Google Scholar]
- 62.Kochian LV, Hoekenga OA, Pineros MA. 2004. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55:459–493. doi: 10.1146/annurev.arplant.55.031903.141655. [DOI] [PubMed] [Google Scholar]
- 63.Kochian LV, Pineros MA, Liu J, Magalhaes JV. 2015. Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu Rev Plant Biol 66:571–598. doi: 10.1146/annurev-arplant-043014-114822. [DOI] [PubMed] [Google Scholar]
- 64.Shi Q, Jin J, Liu Y, Zhang Y, Cai Z, Ma Q, Cheng Y, Wen R, Nian H, Lian T. 2020. High aluminum drives different rhizobacterial communities between aluminum-tolerant and aluminum-sensitive wild soybean. Front Microbiol 11:1996. doi: 10.3389/fmicb.2020.01996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu H, Brettell LE, Qiu Z, Singh BK. 2020. Microbiome-mediated stress resistance in plants. Trends Plant Sci 25:733–743. doi: 10.1016/j.tplants.2020.03.014. [DOI] [PubMed] [Google Scholar]
- 66.Liu Y, Tang Y, Qin X, Yang L, Jiang G, Li S, Ding W. 2017. Genome sequencing of Ralstonia solanacearum CQPS-1, a phylotype I strain collected from a highland area with continuous cropping of tobacco. Front Microbiol 8:974. doi: 10.3389/fmicb.2017.00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Olsen SR. 1954. Estimation of available phosphorus in soils by extraction with sodium bicarbonate. Miscellaneous Paper Institute for Agricultural Research Samaru. [Google Scholar]
- 68.Schonfeld J, Heuer H, Van Elsas JD, Smalla K. 2003. Specific and sensitive detection of Ralstonia solanacearum in soil on the basis of PCR amplification of fliC fragments. Appl Environ Microbiol 69:7248–7256. doi: 10.1128/AEM.69.12.7248-7256.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu J, Wei Z, Friman VP, Gu SH, Wang XF, Eisenhauer N, Yang TJ, Ma J, Shen QR, Xu YC, Jousset A. 2016. Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. mBio 7:e01790-16. doi: 10.1128/mBio.01790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liao X, Chen C, Zhang J, Dai Y, Zhang X, Xie S. 2015. Operational performance, biomass and microbial community structure: impacts of backwashing on drinking water biofilter. Environ Sci Pollut Res Int 22:546–554. doi: 10.1007/s11356-014-3393-7. [DOI] [PubMed] [Google Scholar]
- 71.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1101/274100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JE, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Durall DM, Duvallet C, Edwardson CF, Ernst M, Estaki M, Fouquier J, Gauglitz JM, Gibbons SM, Gibson DL, Gonzalez A, Gorlick K, Guo J, Hillmann B, Holmes S, Holste H, Huttenhower C, Huttley GA, Janssen S, Jarmusch AK, Jiang L, Kaehler BD, Kang KB, Keefe CR, Keim P, Kelley ST, Knights D, et al. 2019. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lavergne C, Bovio-Winkler P, Etchebehere C, Garcia-Gen S. 2020. Towards centralized biogas plants: Co-digestion of sewage sludge and pig manure maintains process performance and active microbiome diversity. Bioresour Technol 297:122442. doi: 10.1016/j.biortech.2019.122442. [DOI] [PubMed] [Google Scholar]
- 76.Garland JL. 1996. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization - ScienceDirect. Soil Biol Biochem 28:213–221. doi: 10.1016/0038-0717(95)00112-3. [DOI] [Google Scholar]
- 77.Zhang S, Liu X, Jiang Q, Shen G, Ding W. 2017. Legacy effects of continuous chloropicrin-fumigation for 3-years on soil microbial community composition and metabolic activity. AMB Express 7:178. doi: 10.1186/s13568-017-0475-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang X, Zhang W, Gu J, Gao H, Qin Q. 2016. Effects of different bulking agents on the maturity, enzymatic activity, and microbial community functional diversity of kitchen waste compost. Environ Technol 20:1–28. doi: 10.1080/09593330.2016.1155650. [DOI] [PubMed] [Google Scholar]
- 79.Garland J, Mills AL. 1991. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol 57:2351–2359. doi: 10.1002/bit.260380413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.White C, Tardif JC, Adkins A, Staniforth R. 2005. Functional diversity of microbial communities in the mixed boreal plain forest of central Canada. Soil Biol Biochem 37:1359–1372. doi: 10.1016/j.soilbio.2004.12.007. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM02333-21_Supp_1_seq12.pdf, PDF file, 0.7 MB (693.2KB, pdf)