ABSTRACT
Klebsiella pneumoniae is a leading cause of highly drug-resistant infections in hospitals worldwide. Strain-level bacterial identification on the genetic determinants of multidrug resistance and high pathogenicity is critical for the surveillance and treatment of this clinically relevant pathogen. In this study, metagenomic next-generation sequencing was performed for specimens collected from August 2020 to May 2021 in Ruijin Hospital, Ningbo Women and Children’s Hospital, and the Second Affiliated Hospital of Harbin Medical University. Genome biology of K. pneumoniae prevalent in China was characterized based on metagenomic data. Thirty K. pneumoniae strains derived from 14 sequence types were identified by multilocus sequence typing. The hypervirulent ST11 K. pneumoniae strains carrying the KL64 capsular locus were the most prevalent in the hospital population. The phylogenomic analyses revealed that the metagenome-reconstructed strains and public isolate genomes belonging to the same STs were closely related in the phylogenetic tree. Furthermore, the pangenome structure of the detected K. pneumoniae strains was analyzed, particularly focusing on the distribution of antimicrobial resistance genes and virulence genes across the strains. The genes encoding carbapenemases and extended-spectrum beta-lactamases were frequently detected in the strains of ST11 and ST15. The highest numbers of virulence genes were identified in the well-known hypervirulent strains affiliated to ST23 bearing the K1 capsule. In comparison to traditional cultivation and identification, strain-level metagenomics is advantageous to understand the mechanisms underlying resistance and virulence of K. pneumoniae directly from clinical specimens. Our findings should provide novel clues for future research into culture-independent metagenomic surveillance for bacterial pathogens.
IMPORTANCE Routine culture and PCR-based molecular testing in the clinical microbiology laboratory are unable to recognize pathogens at the strain level and to detect strain-specific genetic determinants involved in virulence and resistance. To address this issue, we explored the strain-level profiling of K. pneumoniae prevalent in China based on metagenome-sequenced patient materials. Genome biology of the targeted bacterium can be well characterized through decoding sequence signatures and functional gene profiles at the single-strain resolution. The in-depth metagenomic analysis on strain profiling presented here shall provide a promising perspective for culture-free pathogen surveillance and molecular epidemiology of nosocomial infections.
KEYWORDS: K. pneumoniae, metagenome-reconstructed strains, strain profiling, phylogeny, MLST, capsule typing, antimicrobial resistance genes, virulence genes
INTRODUCTION
Klebsiella pneumoniae is a Gram-negative opportunistic pathogen belonging to the family of Enterobacteriaceae (1). It is one of the most common etiologic agents of nosocomial infections for hospitalized patients all over the world (2, 3). Among the multidrug-resistant (MDR) Enterobacteriaceae isolated in Sri Lanka, K. pneumoniae dominated (80.7%), followed by Citrobacter freundii (7.0%), Escherichia coli (5.3%), Providencia rettgeri (3.5%), Enterobacter cloacae (1.7%), and Klebsiella aerogenes (1.7%) (4). K. pneumoniae that can produce carbapenemases and extended-spectrum beta-lactamases (ESBLs) has been recently judged as a critical threat for public health by the World Health Organization (5, 6). The epidemiology of carbapenemase-producing and ESBL-producing K. pneumoniae has been extensively investigated (7, 8). Previous studies have pointed out that MDR clinical isolates of K. pneumoniae are usually accompanied by high pathogenicity, which can lead to serious systematic infections, including pneumonia, meningitis, urinary tract and bloodstream infections (9, 10). To date, a number of disease-related virulence factors that contribute to K. pneumoniae infections and host immune evasion have been uncovered, e.g., siderophore systems, capsular polysaccharides (CPS), lipopolysaccharides, and fimbriae (10–12).
Whole-genome sequencing (WGS) and high-throughput genomic analyses on hundreds of K. pneumoniae isolates have provided valuable insights into population structure, hypervirulent clones, and resistance mechanisms of this important pathogen (11, 13–15). However, the WGS-based strategy needs conventional culture and not every strains from the targeted bacterial species can be successfully isolated under the clinical setting. It is still difficult for accurate species/strain identification of the K. pneumoniae species complex by bacterial isolation and hospital pathogen assays, like Vitek2 (16). Additionally, PCR-based molecular tests are unable to identify emerging sequence signatures in evolving pathogens (17). Due to the limited abilities of traditional methods for detecting the clinically relevant genotypes of virulence and resistance, metagenomic next-generation sequencing (mNGS) is becoming an auxiliary technique for cultivation-free and unbiased pathogen detection in hospitalized patients with complicated infections (18, 19). The state-of-the-art computational methodologies on assembly-free metagenomics have enabled profiling of putative bacterial strains and their functional potential at single-strain resolution (20–22). For instance, Kleborate developed by Lam et al. has been applied to gut metagenomes for detecting genotype characteristics that are clinically relevant to K. pneumoniae and other members belonging to the species complex (23).
In this study, our aim is to uncover the molecular characterizations of K. pneumoniae strains directly from the metagenome-sequenced specimens. Strain-level population genomics analyses were performed to identify prevalent sequence signatures and phylogenetic relationships of metagenome-recovered K. pneumoniae strains. Furthermore, the pangenome structure and function of these K. pneumoniae strains were investigated, particularly focusing on the distribution of antibiotic resistance genes and virulence genes across the strains.
RESULTS
General features of metagenome-reconstructed strains.
We initially investigated 150 clinical specimens that were subject to mNGS and were positive for K. pneumoniae. Based on species-specific multilocus sequence type (MLST) from metagenomic data, 30 K. pneumoniae strains were detected and designated as metagenome-reconstructed strains (MRSs) herein (Table 1). The sequencing depth of the MRSs was ranged from 5- to 107-fold, with a median depth of 22-fold (Table S1). The reconstructed strains of K. pneumoniae were assigned to 14 different sequence types (STs). Using reference-guided read recruitment and local assembly, 14 capsule types of K. pneumoniae were predicted for the 25 MRSs (Table 1). The most prevalent sequence type was ST11 of nine K. pneumoniae strains, six out of which encoded the CPS loci of KL64. The samples consisting of the ST11 MRSs were distributed in Shanghai (5), Zhejiang (3), and Heilongjiang (1), respectively (Table 1). The second prevalent sequence type was ST15 of five K. pneumoniae strains belonging to KL19 (4) and KL8 (1). The samples of the ST15 MRSs were distributed in Shanghai (4) and Heilongjiang (1). Besides, the taxonomic profiling of species relative abundances showed that K. pneumoniae was the highly abundant bacterial species dominating the communities of the 30 specimens, with a mean abundance of 82%.
TABLE 1.
Summary of sequence signatures and gene families of K. pneumoniae strains in the metagenomic samples and study participants
| Strain | Patient ID | Provincea | RA (%)b | ST | K typec | Total genes | Accessory genes | Virulence genes | Resistance genes |
|---|---|---|---|---|---|---|---|---|---|
| Kpn01 | #023 | Heilongjiang | 98.77 | 11 | NA | 5,510 | 2,527 | 324 | 37 |
| Kpn02 | #008 | Shanghai | 94.21 | 11 | KL64 | 5,310 | 2,327 | 318 | 32 |
| Kpn03 | #004 | Shanghai | 99.90 | 11 | NA | 5,272 | 2,289 | 326 | 28 |
| Kpn04 | #108 | Shanghai | 70.05 | 11 | KL64 | 5,536 | 2,553 | 330 | 34 |
| Kpn05 | #114 | Zhejiang | 96.44 | 11 | KL64 | 5,395 | 2,412 | 316 | 36 |
| Kpn06 | #032 | Zhejiang | 99.69 | 11 | KL64 | 5,525 | 2,542 | 329 | 33 |
| Kpn07 | #019 | Shanghai | 97.85 | 11 | NA | 5,629 | 2,646 | 331 | 35 |
| Kpn08 | #088 | Zhejiang | 100.00 | 11 | KL64 | 5,480 | 2,497 | 329 | 31 |
| Kpn09 | #112 | Shanghai | 99.76 | 11 | KL64 | 5,420 | 2,437 | 326 | 33 |
| Kpn10 | #042 | Shanghai | 94.06 | 15 | KL19 | 5,138 | 2,155 | 319 | 32 |
| Kpn11 | #125 | Shanghai | 75.75 | 15 | KL19 | 5,159 | 2,176 | 318 | 34 |
| Kpn12 | #090 | Heilongjiang | 90.21 | 15 | KL8 | 5,107 | 2,124 | 300 | 44 |
| Kpn13 | #022 | Shanghai | 30.72 | 15 | KL19 | 5,149 | 2,166 | 317 | 36 |
| Kpn14 | #079 | Shanghai | 92.81 | 15 | KL19 | 5,149 | 2,166 | 319 | 37 |
| Kpn15 | #021 | Heilongjiang | 76.68 | 23 | KL1 | 5,110 | 2,127 | 351 | 29 |
| Kpn16 | #089 | Heilongjiang | 98.78 | 23 | KL1 | 5,147 | 2,164 | 350 | 27 |
| Kpn17 | #017 | Zhejiang | 81.75 | 29 | KL54 | 5,158 | 2,175 | 337 | 29 |
| Kpn18 | #052 | Shanghai | 89.36 | 29 | NA | 5,240 | 2,257 | 325 | 28 |
| Kpn19 | #059 | Zhejiang | 91.40 | 45 | NA | 4,992 | 2,009 | 314 | 36 |
| Kpn20 | #020 | Heilongjiang | 78.06 | 45 | KL24 | 5,122 | 2,139 | 318 | 40 |
| Kpn21 | #055 | Shanghai | 42.90 | 147 | KL125 | 5,184 | 2,201 | 309 | 46 |
| Kpn22 | #127 | Shanghai | 38.87 | 258 | KL107 | 5,475 | 2,492 | 312 | 40 |
| Kpn23 | #049 | Heilongjiang | 98.85 | 375 | KL2 | 5,048 | 2,065 | 317 | 27 |
| Kpn24 | #041 | Heilongjiang | 95.39 | 412 | KL57 | 5,050 | 2,067 | 308 | 28 |
| Kpn25 | #018 | Zhejiang | 66.33 | 412 | KL57 | 4,984 | 2,001 | 306 | 29 |
| Kpn26 | #006 | Shanghai | 81.21 | 656 | KL149 | 5,150 | 2,167 | 316 | 45 |
| Kpn27 | #061 | Shanghai | 92.68 | 660 | KL16 | 5,001 | 2,018 | 330 | 27 |
| Kpn28 | #074 | Heilongjiang | 57.50 | 902 | KL125 | 5,579 | 2,596 | 303 | 48 |
| Kpn29 | #050 | Shanghai | 89.25 | 1,049 | KL5 | 5,009 | 2,026 | 322 | 27 |
| Kpn30 | #095 | Shanghai | 38.24 | 2,471 | KL53 | 4,818 | 1,835 | 264 | 37 |
The province information of the clinical samples from the three hospitals are shown: Shanghai for Ruijin Hospital, Zhejiang for Ningbo Women and Children’s Hospital, and Heilongjiang for the Second Affiliated Hospital of Harbin Medical University.
The percentage relative abundance denotes the estimated proportion of K. pneumoniae in the bacterial community.
The strains missing the known K types predicted by Kaptive are denoted by NA.
Population-scale phylogeny of K. pneumoniae.
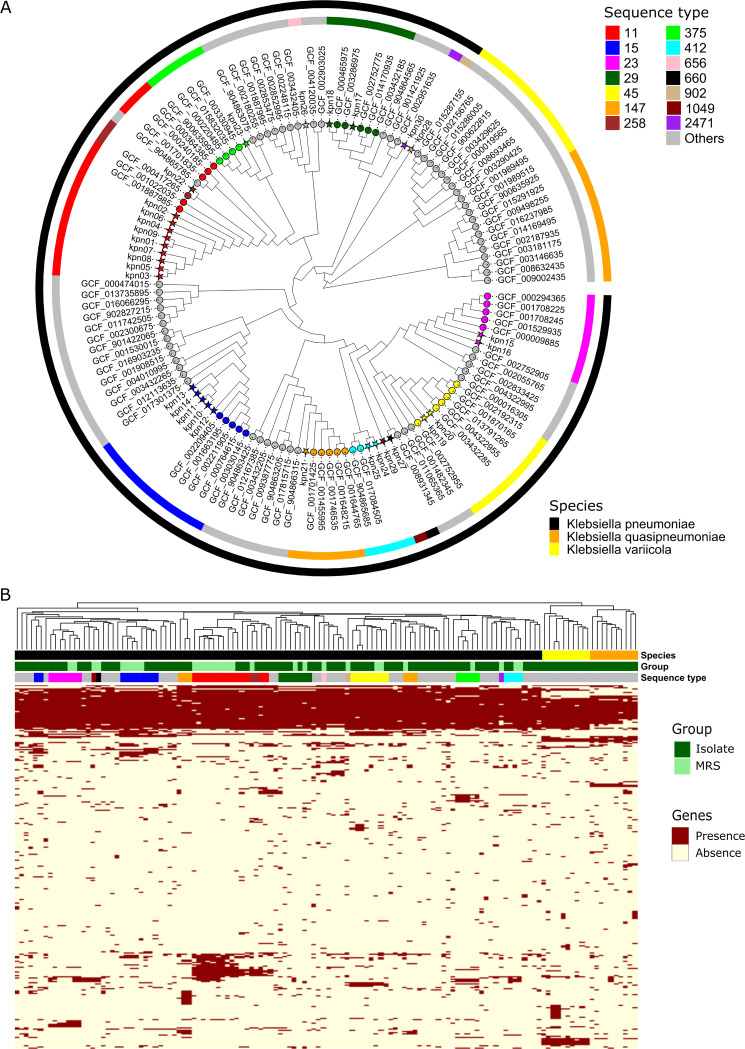
To investigate phylogenetic relationships among bacterial strains, 100 complete isolate genomes of K. pneumoniae species complex were retrieved and compared with 30 metagenome-recovered strains. Using StrainPhlAn, 38 K. pneumoniae-specific marker genes were detected in all the strains. Based on the alignment of single-nucleotide variants (SNVs) in the markers, Fig. 1A displays the phylogenetic tree of the MRSs together with cultivated strains from K. pneumoniae species complex. Obviously, all the metagenome-recovered strains are distributed in the clade of K. pneumoniae and they are distant from the strains in the lineages of K. quasipneumoniae and K. variicola. It was also observed that the metagenome-recovered K. pneumoniae strains and cultivated strains that shared the same STs were more closely related to each other in the phylogenetic tree. For instance, five ST15 MRSs were placed together with the other cultivated strains of ST15 K. pneumoniae in a single clade without the strains from other STs. Two ST412 MRSs and another two cultivated strains of ST412 K. pneumoniae were placed into a single clade. It suggested that MRSs assigned to the identical STs can be grouped into a single phylogenetic lineage, which also comprised the whole-genome sequenced isolates with the related STs, implying that culture-independent and assembly-free metagenomic analyses would be an alternative to genomic surveillance and epidemiology of K. pneumoniae (23).
FIG 1.
Phylogenomic and pangenomic structure of K. pneumoniae. (A) Maximum likelihood phylogeny of K. pneumoniae. The phylogenetic tree was built using the 38 K. pneumoniae-specific marker genes detected in the 30 metagenomic samples and 100 reference genomes from three major members of the K. pneumoniae species complex. (B) Gene family profiles of K. pneumoniae strains from metagenomes as well as isolate genomes. The heatmap displays the presence/absence patterns of the accessory genes across data sets.
Pangenome structure and function of K. pneumoniae.
To better understand the functional potential of bacterial strains in the community, the pangenome analysis was carried out for decoding gene compositions of individual K. pneumoniae strains in the metagenomic samples. Based on the custom K. pneumoniae pangenome consisting of 24,476 gene families, strain-specific gene repertoires were reconstructed for the MRSs detected above. A total of 9,783 gene families were identified in the pangenome of 30 K. pneumoniae MRSs (Table S3). The number of gene families across the strains was ranged from 4,818 to 5,629 (Table 1). Of these, 2,983 were the core gene families present in all the metagenome-recovered strains and cultivated strains. It was apparent that the dendrogram based on genic components can also cluster all the metagenome-recovered strains into the lineage of K. pneumoniae (Fig. 1B). Besides, all the metagenome-recovered strains could be clustered with the cultivated K. pneumoniae strains affiliated to the same STs, implicating these strains may possess similar phenotypic characteristics.
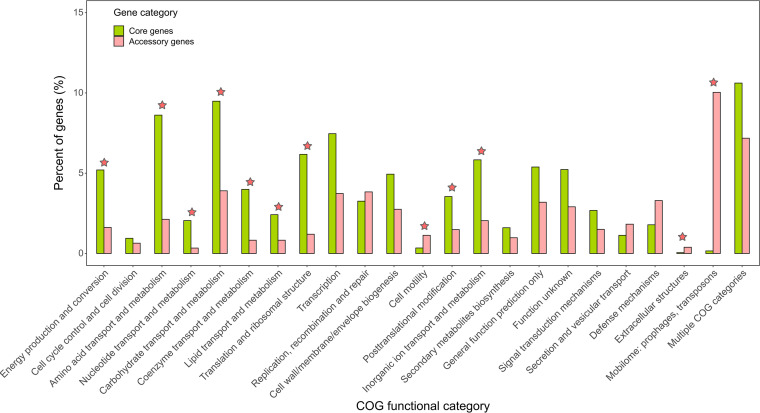
Next, the core and accessory gene families of K. pneumoniae strains present in the metagenomes were classified based on COG functional categories (Table S4). As shown in Fig. 2, nine COG categories were significantly abundant in the set of core genes compared with the set of accessory genes (FDR < 0.001). Among these categories, several were associated with basic cellular activities for bacterial growth and survival, for instance, “carbohydrate transport and metabolism” (OR = 2.43), “amino acid transport and metabolism” (OR = 4.05), “energy production and conversion” (OR = 3.21), and “translation and ribosomal structure” (OR = 5.15). On the contrary, three COG categories were significantly abundant in the set of accessory genes (FDR < 0.001), including “mobilome: prophages, transposons” (OR = 0.02), “cell motility” (OR = 0.30), and “extracellular structures” (OR = 0.14). For the genes encoding hypothetical proteins without homologs in the COG database, significant enrichment was found in the set of accessory genes (OR = 0.17) (Table S4).
FIG 2.
Comparison of COG functional categories between core and accessory gene families in the pangenome of K. pneumoniae MRSs. The asterisk denotes a significant difference in the corresponding category between two genic groups (FDR < 0.001; chi-square test).
Besides, functional annotations of the pangenome gene families were performed for antimicrobial resistance (AMR) genes and virulence-associated genes, respectively. The medium number of AMR genes was 33 with a range from 27 to 48 across the metagenome-recovered strains (Table 1). The most abundant AMR genes were found in the strain Kpn28 belonging to ST902. Additionally, the medium number of virulence genes was 318. The highest numbers of virulence genes were observed in both strains Kpn15 and Kpn16 belonging to ST23 and KL1 K. pneumoniae.
Gene patterns of AMR.
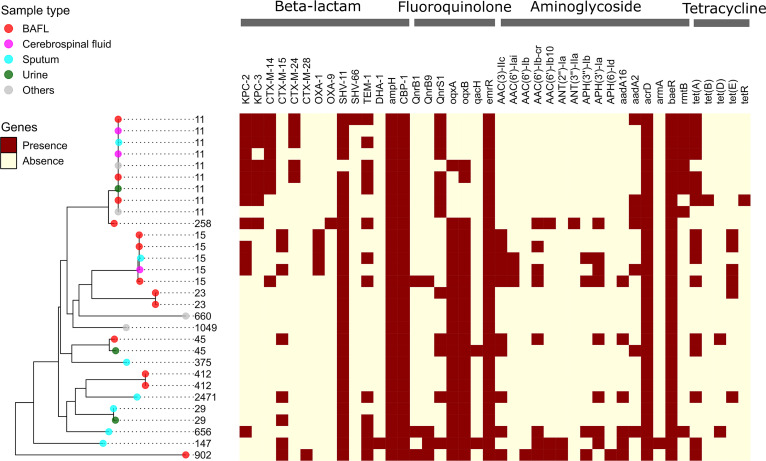
Extensive resistance to common antibiotics has been frequently reported in the infections caused by multidrug-resistant strains of K. pneumoniae (24, 25). In this study, we identified the presence and absence patterns of 77 AMR genes across 30 K. pneumoniae MRSs. These AMR genes were assigned to 36 CARD gene families (Table S5). Approximately three quarters (56 out of 77) of all AMR genes were affiliated to the set of accessory genes. Fig. 3 displays the distribution of the AMR genes associated with four classes of antibiotics (i.e., beta-lactams, fluoroquinolones, aminoglycosides, and tetracyclines) across the strains. Among the genes conferring resistance to carbapenems, both genes blaKPC-2 and blaKPC-3 encoding carbapenemases were detected in most of the strains affiliated to ST11 (7/9) and in one ST258 strain. blaKPC-2 was also found in the strains of ST15 (3/5) and ST656 (1/1). Four gene variants of CTX-M beta-lactamases, which are among the most important ESBLs (26), were detected in 16 strains (53.3%). The gene blaSHV-11 encoding a broad-spectrum beta-lactamase was present in all strains except for a ST11 strain. The mosaic distribution of AMR genes may confer diversified resistance phenotypes to clinical strains of K. pneumoniae.
FIG 3.
Distribution of antimicrobial resistance (AMR) genes across the metagenome-reconstructed strains of K. pneumoniae. The prediction of AMR genes was performed using RGI searching against CARD. The heatmap shows the presence/absence patterns of the genes conferring resistance to β-lactams, fluoroquinolones, aminoglycosides, and tetracyclines. The list of all detected AMR genes is summarized in Table S5.
Gene patterns of virulence factors.
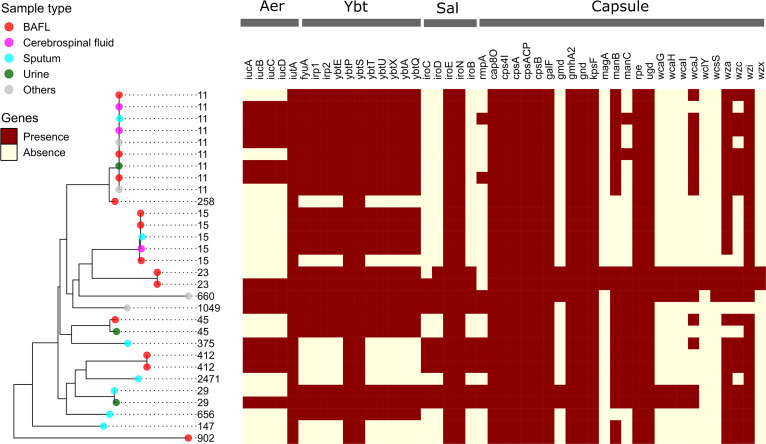
In addition to antimicrobial resistance functions, we also investigated the genetic diversity of virulence determinants of K. pneumoniae strains in the metagenomic samples. Herein, 399 virulence-associated genes encoding a variety of bacterial virulence factors were identified and the details are summarized in Table S6. Fig. 4 shows the distribution of the virulence genes coding for products involved in the biosynthesis of iron-scavenging siderophores (i.e., aerobactin, yersiniabactin, and salmochelin) and capsular polysaccharides. The ybt locus consisting of 11 genes involved in the synthesis of yersiniabactin, which is the best-known K. pneumoniae high-virulence determinant associated with bacteremia and tissue-invasive infections (11, 27), was found in 22 metagenome-recovered strains of ST11 (9/9), ST15 (4/5), ST23 (2/2), ST29 (2/2), ST45 (2/2), ST660 (1/1), ST656 (1/1), and ST1049 (1/1). The genes iucABCD and iutA encoding aerobactin were identified in 13 strains of ST11 (6/9), ST412 (2/2), ST23 (1/2), ST29 (1/2), ST375 (1/1), ST660 (1/1), and ST1049 (1/1). The genes iroBCDE and iroN encoding salmochelin were identified in the strains of ST412 (2/2), ST23 (1/2), ST29 (2/2), ST375 (1/1), ST660 (1/1), and ST1049 (1/1). The genes responsible for the production of enterobactin were identified in nearly all the K. pneumoniae MRSs. Notably, the colibactin synthesis locus clb (including 18 genes), which is adjacent to the ybt locus in the integrative conjugative elements of K. pneumoniae (28), was only present in the two ST23 strains but absent in the metagenome-recovered strains of other STs. Moreover, both ST23 strains encoded an intact KL1 gene cluster for the production of hypercapsule associated with hypervirulent K. pneumoniae (hvKP) strains (10), including cpsA, galF, gmd, gnd, magA, manBC, ugd, wcaGHIJ, wza, wzc, wzi, and wzx (Fig. 4). Besides, the plasmid-borne gene rmpA coding for the regulator of mucoid phenotype A involved in the increase of capsule production was detected in nine metagenome-recovered K. pneumoniae strains of ST11 (2/9), ST412 (2/2), ST29 (2/2), ST375 (1/1), ST660 (1/1), and ST1049 (1/1). Eight out of nine strains carrying the rmpA gene also possessed the Aer locus (Fig. 4), both of which have been characterized as indicators for the presence of K. pneumoniae virulence plasmids in hypervirulent strains (9, 14). The presence/absence patterns of the above virulence-associated genes in certain lineages should keep in line with the previous studies on the K. pneumoniae clinical isolates and more details are discussed below.
FIG 4.
Distribution of virulence genes across the metagenome-reconstructed strains of K. pneumoniae. The prediction of virulence genes was performed using BLAST searching against VARD. The heatmap shows the presence/absence patterns of the genes associated with the biosynthesis of polysaccharide capsule and three siderophores aerobactin (Aer), yersiniabactin (Ybt), and salmochelin (Sal). The list of all detected virulence genes is summarized in Table S6.
DISCUSSION
As is well known, bacterial infections caused by K. pneumoniae pose a great threat to global public health. Particularly, high pathogenicity and MDR have brought severe challenges to clinically microbiological testing and anti-infection therapy. High-throughput genomic analyses are beneficial for understanding the genetic diversity of clinically relevant genotypes of virulence and drug resistance during spatiotemporal transmission and adaptive evolution of K. pneumoniae. To address these issues, hundreds of studies on bacterial genomics have adopted culture-dependent whole-genome sequencing, which can produce longer reads and deeper genome coverage for high-quality de novo assembly and gene annotation. To explore the application of mNGS to population genomics, we performed integrative analyses to associate K. pneumoniae sequence types or K types with bacterial phylogeny, AMR genes, and virulence genes based on metagenomic sequencing of clinical specimens. Innovative strategies by combining assembly-free and reference-guided local assembly approaches were applied to study strain-level genomics of K. pneumoniae in the clinical setting for the first time.
Thirty K. pneumoniae strains belonging to 14 sequence types were reconstructed based on metagenomic sequencing data in this study. The strain-profiling approaches StrainPhlAn and PanPhlAn were both able to detect and characterize the strains with low sequencing depth even at 5-fold. Our findings further indicated that the distribution of bacterial sequence signatures (i.e., STs and K types) recovered from metagenomes should be well comparable with prior knowledge on the whole-genome-sequenced clinical isolates of K. pneumoniae (15). On the other hand, rpoB has been reported to be a species-specific marker for identification of K. pneumoniae isolates (29). Here, the sequences of the rpoB gene fragments were available for 14 out of 30 specimens and the blast analysis indicated that the amplified rpoB genes belonged to K. pneumoniae (data not shown). Although PCR-based molecular tests and conventional phenotypic methods can support the identification of the targeted species, both exhibit limited utility for providing additional information on clinically relevant lineages (ST) and genotypes. Our study confirms some recent options that metagenomic approaches have enabled culture-free and assembly-free strain profiling analyses for surveillance of the high-risk hvKP clones (20, 22, 30), like ST11, ST15, ST23, ST29, and ST412 detected in the above analysis.
During the past decade, a high prevalence of ST11 carbapenem-resistant K. pneumoniae (CRKP) strains has been reported in the community-acquired and nosocomial infections in China (31–33). Consistently, the most abundant sequence type among all the K. pneumoniae MRSs was found to be ST11, most strains of which encoded the genes blaKPC-2, blaSHV-11, blaCTX-M, qnrS1, and tet(A) mediating resistance to beta-lactams, fluoroquinolones, and tetracycline (Fig. 3). The co-occurrence of these AMR genes has been recognized in an outbreak of 40 CRKP isolates (34). In addition, the analyses of the local assembly and typing of capsular locus demonstrated that all ST11 K. pneumoniae MRSs possessed a single K type KL64, which has been characterized as a newly emerging superbug prevalent in China by a large-scale genome sequence analysis of 364 ST11 isolates (15). Zhou et al. have also pointed out that, among the ST11 population, KL64 replaced KL47 and became the dominant CRKP clone in China since 2016 (35). Furthermore, the other K type KL105 frequently associated with ST11 was absent in the three representative hospitals in China but was prevalent in Poland and Slovakia (23). The evidence again supports the metagenomic approach for surveillance and epidemiology of K. pneumoniae infections.
Besides, the MDR hypervirulent ST23 K. pneumoniae, whose rapid dissemination is driven by diverse plasmids harboring virulence and ARM genes, is another clinically significant lineage that has been paid close attention to by the medical community (13, 36–38). Meanwhile, the highest numbers of virulence genes were observed in the two ST23 strains (Kpn15 and Kpn16) carrying KL1, a well-characterized capsule type highly associated with hvKP strains (39). It was noted that the most abundant genes involved in synthesizing diverse siderophores (i.e., yersiniabactin, aerobactin, salmochelin, colibactin, and enterobactin) were identified in the Kpn16 strain, perhaps playing roles in bacterial hypervirulence and postinfection proliferation for overcoming iron limitations in vivo. Enrichment of these siderophore-related genes in the ST23 lineage has been revealed by a recent study on comparative genomics of K. pneumoniae (11). In particular, the colibactin synthesis locus clb present in the metagenome-recovered strains belonging to ST23 has been detected in 3.5% to 4% of K. pneumoniae, in which colibactin production enables genotoxic effect on host cells by inducing double-strand DNA breaks (28, 36, 40).
In summary, we carried out comprehensive strain-profiling analyses to uncover bacterial sequence types, phylogeny, and pangenomic structure of K. pneumoniae recovered from clinical mNGS data. Genome biology of 30 K. pneumoniae strains was characterized by multilocus sequence typing, phylogenetic reconstruction, and capsule typing. Furthermore, the pangenome structure of metagenome-recovered K. pneumoniae strains was analyzed, particularly the distribution of antimicrobial resistance genes and virulence genes across the strains. Our findings should also provide novel clues for future applications of mNGS to molecular epidemiology and culture-free genomic surveillance of clinically relevant pathogens.
MATERIALS AND METHODS
Clinical specimens.
In this study, we retrospectively investigated 150 clinical samples that were positive for K. pneumoniae according to mNGS testing. The samples were collected from patients in Ruijin Hospital, Ningbo Women and Children’s Hospital, and the Second Affiliated Hospital of Harbin Medical University from August 2020 to May 2021. The information of all samples was listed in Table S1. Ethical approval for the study was obtained from the local Medical Ethics Committee of the Ruijin Hospital (Approval ID KY2021-213).
Metagenomic sequencing and data preprocessing.
The experiments of mNGS were carried out at Genoxor Inc., China. Microbial DNA was extracted and enriched by streamlined host DNA depletion using HostZERO Microbial DNA Kit (Zymo, United States). Extracted DNA was sheared to 300 bp fragments with Covaris M220 (Covaris, MA, United States) following the manufacturer’s protocol. Metagenomic libraries were then constructed using the NEBNext Ultra DNA Library Prep Kit for Illumina. Multiplexed libraries were sequenced in a 75-bp single-end mode using a NextSeq 550 system (Illumina Inc., USA). Raw sequencing data were demultiplexed into Fastq-formatted reads using bcl2fastq v2.20 (Illumina). Trimming adaptor sequences and filtering low-quality bases/reads were then performed using Trimmomatic v0.36 with the options LEADING:15 TRAILING:15 SLIDINGWINDOW:5:20 MINLEN:36 AVGQUAL:20 (41). Human-derived DNA contamination was subtracted through aligning reads to the human reference genome GRCh37 using Bowtie v2.2.6 with the options –threads 32 –end-to-end (42). Species identification of K. pneumoniae was performed by Kraken v2.0.9 (43). Sequencing depth and genome coverage were estimated by mapping reads to the complete genome of K. pneumoniae HS11286 using BBmap v38.18 (44). To estimate the relative abundance of K. pneumoniae reads in the bacterial community, the taxonomic profile of species abundance was calculated using Bracken v2.2 (45).
Analyses of MLST and capsule serotype.
For strain identification and typing of K. pneumoniae from the metagenomic sample, the MLST was detected using the pipeline metaMLST v1.2.2 (46). Briefly, the metaMLST database curated from pubMLST (47) was used to generate a bowtie2 database of K. pneumoniae allelic sequences from seven housekeeping genes: gapA, infB, mdh, pgi, phoE, rpoB, and tonB. The consensus sequences of K. pneumoniae MLST loci present in the metagenomic sample were reconstructed by read alignment against the bowtie2 database and then by a majority rule consensus approach implemented by the mpileup utility in the SAMtools package v0.1.19 (48). The resulting allele sequences were used to assign known and novel ST numbers to individual samples according to the organism-specific MLST protocol. The strains assigned with known STs were defined as the MRSs in the individual samples.
The capsule serotype (K type) was determined by an integrative pipeline based on read recruitment, local assembly, and capsule typing. Briefly, metagenomic reads per sample were first recruited to the Klebsiella capsule locus reference database in the Kaptive package v0.7.3 (49) using BBmap v38.18. Only mapped reads were extracted to assemble the capsule locus using Megahit v1.2.9 (50). The K type was predicted for the resulting genomic assembles using Kaptive (49).
Strain-level profiling analyses.
We employed StrainPhlAn (21), a strain-level profiling tool integrated with the pipeline MetaPhlAn v3.0, for tracking targeted strains in the clinical samples. Briefly, Bowtie2 was used to align metagenomic reads to the MetaPhlAn marker database comprising of ∼1.1 million unique clade-specific marker genes from ∼17,000 species (51). The resulting SAM files were used to reconstruct the sequences of marker genes from all species strains in each sample. Additionally, 46 K. pneumoniae-specific marker genes were extracted from the MetaPhlAn database for detection of the corresponding genes in the reference genomes of isolates using blast v2.10.0 (52). The markers present in the MRSs and isolate genomes were selected and their sequences were concatenated. A multiple sequence alignment was then produced for reconstruction of the maximum-likelihood phylogenetic tree using the package PhyloPhlAn v3.0.60 (53).
On the other hand, PanPhlAn v1.3 (20), which is also a strain-level metagenomic profiling tool, was used to investigate the genic compositions of the MRSs. Firstly, a custom pangenome was created with 100 complete reference genomes derived from three members within the K. pneumoniae species complex, including 80 genomes from K. pneumoniae (KpI), 10 genomes from K. variicola (KpII), and 10 genomes from K. quasipneumoniae (KpIII). The genome sequences of cultured isolates were downloaded from the NCBI Assembly database. The STs of individual isolates were predicted by using MLST v2.16.2 with the seven housekeeping alleles: gapA, infB, mdh, pgi, phoE, rpoB, and tonB (Table S2). The isolate genomes were preferentially selected based on the STs related to the MRSs. Roary v3.13.0 was used for the generation of the PanPhlAn pangenome database of K. pneumoniae. Next, the gene presence/absence patterns of individual strains were scanned by PanPhlAn with the options –min_coverage 2 –left_max 1.25 –right_min 0.75.
Functional annotation of gene family.
Protein functional classification for the pangenome gene families was conducted based on sequence similarity searching against the Clusters of Orthologous Groups (COG) database (54) with blastp v2.10.0. Annotation of AMR genes was performed using the Comprehensive Antibiotic Resistance Database (CARD) and the related program RGI v5.1.1 (55). The query sequences were annotated by the two RGI paradigms perfect and strict. Annotation of virulence-associated genes was performed using the Virulence Factor Database (VFDB) (56) and blastp v2.10.0. The query sequences were annotated by the top hit with the maximum E-value of 1e-20.
Statistical analyses and data visualization.
Comparison of the gene count data was estimated using odds ratio (OR) and chi-square test implemented by the R package Epitools v 0.5–10.1 (57). The phylogenetic tree integrated with the other metadata (sequence types and taxonomic origins of strains) was visualized using the R package ggtree v3.0.2 (58). Hierarchical clustering on a binary matrix of pangenome gene families across the strains was performed and visualized using the R package ComplexHeatmap v2.8.0 (59). The statistical analyses and data visualization were carried out in R v4.1.0 (60).
Data availability.
The microbial reads produced in this study have been deposited in the NCBI Sequence Read Archive (SRA) database under the BioProject accession PRJNA758247.
ACKNOWLEDGMENTS
J.L., Z.X., H.L., Y.F., and D.C. conceived the work and wrote the manuscript; J.L., H.L., F.C., K.H., and X.H. contributed to sample collection; J.L., Z.X., H.L., F.C., K.H., and Y.F. performed metagenomic sequencing and data analysis; K.H., Y.F., and D.C. revised the manuscript.
This work was supported by the National Nature Science Foundation of China (81971869 and 81873944), and the Technical Standard Project and Technology Innovation Action Plan of Shanghai (20DZ2200500).
The funding source had no role in study design, data collection, data analysis, data interpretation, or writing of the report. We declare that we have no conflicts of interest relevant to this article.
Footnotes
Supplemental material is available online only.
Contributor Information
Yuan Fang, Email: yuan.fang@genoxor.com.
Dechang Chen, Email: cdc12064@rjh.cm.cn.
Wei-Hua Chen, Huazhong University of Science and Technology.
REFERENCES
- 1.Adeolu M, Alnajar S, Naushad S, Gupta RS. 2016. Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol 66:5575–5599. doi: 10.1099/ijsem.0.001485. [DOI] [PubMed] [Google Scholar]
- 2.Pendleton JN, Gorman SP, Gilmore BF. 2013. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther 11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 3.Xiao S-Z, Wang S, Wu W-M, Zhao S-Y, Gu F-F, Ni Y-X, Guo X-K, Qu J-M, Han L-Z. 2017. The resistance phenotype and molecular epidemiology of Klebsiella pneumoniae in bloodstream infections in Shanghai, China, 2012-2015. Front Microbiol 8:250–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumudunie WGM, Wijesooriya LI, Namalie KD, Sunil-Chandra NP, Wijayasinghe YS. 2020. Epidemiology of multidrug-resistant Enterobacteriaceae in Sri Lanka: first evidence of blaKPC harboring Klebsiella pneumoniae. J Infect Public Health 13:1330–1335. doi: 10.1016/j.jiph.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Wyres KL, Lam MMC, Holt KE. 2020. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol 18:344–359. doi: 10.1038/s41579-019-0315-1. [DOI] [PubMed] [Google Scholar]
- 6.WHO. (ed). 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 7.Aires-de-Sousa M, Ortiz de la Rosa JM, Gonçalves ML, Pereira AL, Nordmann P, Poirel L. 2019. Epidemiology of Carbapenemase-producing Klebsiella pneumoniae in a hospital, Portugal. Emerg Infect Dis 25:1632–1638. doi: 10.3201/eid2509.190656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fils PEL, Cholley P, Gbaguidi-Haore H, Hocquet D, Sauget M, Bertrand X. 2021. ESBL-producing Klebsiella pneumoniae in a university hospital: molecular features, diffusion of epidemic clones and evaluation of cross-transmission. PLoS One 16:e0247875. doi: 10.1371/journal.pone.0247875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shon AS, Bajwa RP, Russo TA. 2013. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence 4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paczosa MK, Mecsas J. 2016. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev 80:629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NTK, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci USA 112:E3574–E3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Follador R, Heinz E, Wyres KL, Ellington MJ, Kowarik M, Holt KE, Thomson NR. 2016. The diversity of Klebsiella pneumoniae surface polysaccharides. Microb Genom 2:e000073. doi: 10.1099/mgen.0.000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam MMC, Wyres KL, Duchêne S, Wick RR, Judd LM, Gan YH, Hoh CH, Archuleta S, Molton JS, Kalimuddin S, Koh TH, Passet V, Brisse S, Holt KE. 2018. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun 9:2703. doi: 10.1038/s41467-018-05114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heinz E, Ejaz H, Bartholdson Scott J, Wang N, Gujaran S, Pickard D, Wilksch J, Cao H, Haq IU, Dougan G, Strugnell RA. 2019. Resistance mechanisms and population structure of highly drug resistant Klebsiella in Pakistan during the introduction of the carbapenemase NDM-1. Sci Rep 9:2392. doi: 10.1038/s41598-019-38943-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao J, Liu C, Liu Y, Zhang Y, Xiong Z, Fan Y, Zou X, Lu B, Cao B. 2020. Genomic characteristics of clinically important ST11 Klebsiella pneumoniae strains worldwide. J Glob Antimicrob Resist 22:519–526. doi: 10.1016/j.jgar.2020.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Lu B, Lin C, Liu H, Zhang X, Tian Y, Huang Y, Yan H, Qu M, Jia L, Wang Q. 2020. Molecular characteristics of Klebsiella pneumoniae isolates from outpatients in sentinel hospitals, Beijing, China, 2010-2019. Front Cell Infect Microbiol 10:85. doi: 10.3389/fcimb.2020.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deurenberg RH, Bathoorn E, Chlebowicz MA, Couto N, Ferdous M, García-Cobos S, Kooistra-Smid AM, Raangs EC, Rosema S, Veloo AC, Zhou K, Friedrich AW, Rossen JW. 2017. Application of next generation sequencing in clinical microbiology and infection prevention. J Biotechnol 243:16–24. doi: 10.1016/j.jbiotec.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 18.Wilson MR, Sample HA, Zorn KC, Arevalo S, Yu G, Neuhaus J, Federman S, Stryke D, Briggs B, Langelier C, Berger A, Douglas V, Josephson SA, Chow FC, Fulton BD, DeRisi JL, Gelfand JM, Naccache SN, Bender J, Dien Bard J, Murkey J, Carlson M, Vespa PM, Vijayan T, Allyn PR, Campeau S, Humphries RM, Klausner JD, Ganzon CD, Memar F, Ocampo NA, Zimmermann LL, Cohen SH, Polage CR, DeBiasi RL, Haller B, Dallas R, Maron G, Hayden R, Messacar K, Dominguez SR, Miller S, Chiu CY. 2019. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N Engl J Med 380:2327–2340. doi: 10.1056/NEJMoa1803396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Han Y, Feng J. 2019. Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm Med 19:252. doi: 10.1186/s12890-019-1022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholz M, Ward DV, Pasolli E, Tolio T, Zolfo M, Asnicar F, Truong DT, Tett A, Morrow AL, Segata N. 2016. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods 13:435–438. doi: 10.1038/nmeth.3802. [DOI] [PubMed] [Google Scholar]
- 21.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. 2017. Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res 27:626–638. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zolfo M, Asnicar F, Manghi P, Pasolli E, Tett A, Segata N. 2018. Profiling microbial strains in urban environments using metagenomic sequencing data. Biol Direct 13:9. doi: 10.1186/s13062-018-0211-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. 2021. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun 12:4188. doi: 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin RM, Bachman MA. 2018. Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol 8:4. doi: 10.3389/fcimb.2018.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreira RL, da Silva BCM, Rezende GS, Nakamura-Silva R, Pitondo-Silva A, Campanini EB, Brito MCA, da Silva EML, Freire CCdM, Cunha AFd, Pranchevicius M-CS. 2019. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-Lactamase encoding genes in a Brazilian intensive care unit. Front Microbiol 9. doi: 10.3389/fmicb.2018.03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Younes A, Hamouda A, Dave J, Amyes SGB. 2011. Prevalence of transferable blaCTX-M-15 from hospital- and community-acquired Klebsiella pneumoniae isolates in Scotland. J Antimicrob Chemother 66:313–318. doi: 10.1093/jac/dkq453. [DOI] [PubMed] [Google Scholar]
- 27.Lin T-L, Lee C-Z, Hsieh P-F, Tsai S-F, Wang J-T. 2008. Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J Bacteriol 190:515–526. doi: 10.1128/JB.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam MMC, Wick RR, Wyres KL, Gorrie CL, Judd LM, Jenney AWJ, Brisse S, Holt KE. 2018. Genetic diversity, mobilisation and spread of the yersiniabactin-encoding mobile element ICEKp in Klebsiella pneumoniae populations. Microb Genom 4. doi: 10.1099/mgen.0.000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He Y, Guo X, Xiang S, Li J, Li X, Xiang H, He J, Chen D, Chen J. 2016. Comparative analyses of phenotypic methods and 16S rRNA, khe, rpoB genes sequencing for identification of clinical isolates of Klebsiella pneumoniae. Antonie Van Leeuwenhoek 109:1029–1040. doi: 10.1007/s10482-016-0702-9. [DOI] [PubMed] [Google Scholar]
- 30.Van Rossum T, Ferretti P, Maistrenko OM, Bork P. 2020. Diversity within species: interpreting strains in microbiomes. Nat Rev Microbiol 18:491–506. doi: 10.1038/s41579-020-0368-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. 2011. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother 66:307–312. doi: 10.1093/jac/dkq431. [DOI] [PubMed] [Google Scholar]
- 32.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 33.Liao W, Liu Y, Zhang W. 2020. Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: a review over the last 10 years. J Glob Antimicrob Resist 23:174–180. doi: 10.1016/j.jgar.2020.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Zhang R, Wang XD, Cai JC, Zhou HW, Lv HX, Hu QF, Chen G-X. 2011. Outbreak of Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae with high qnr prevalence in a Chinese hospital. J Med Microbiol 60:977–982. doi: 10.1099/jmm.0.015826-0. [DOI] [PubMed] [Google Scholar]
- 35.Zhou K, Xiao T, David S, Wang Q, Zhou Y, Guo L, Aanensen D, Holt KE, Thomson NR, Grundmann H, Shen P, Xiao Y. 2020. Novel subclone of Carbapenem-resistant Klebsiella pneumoniae sequence type 11 with enhanced virulence and transmissibility, China. Emerg Infect Dis 26:289–297. doi: 10.3201/eid2602.190594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shankar C, Jacob JJ, Vasudevan K, Biswas R, Manesh A, Sethuvel DPM, Varughese S, Biswas I, Veeraraghavan B. 2020. Emergence of multidrug resistant hypervirulent ST23 Klebsiella pneumoniae: multidrug resistant plasmid acquisition drives evolution. Front Cell Infect Microbiol 10:575289. doi: 10.3389/fcimb.2020.575289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen D, Ma G, Li C, Jia X, Qin C, Yang T, Wang L, Jiang X, Ding N, Zhang X, Yue L, Yin Z, Zeng L, Zhao Y, Zhou D, Chen F. 2019. Emergence of a multidrug-resistant hypervirulent Klebsiella pneumoniae sequence type 23 strain with a Rare bla (CTX-M-24)-harboring virulence plasmid. Antimicrob Agents Chemother 63. doi: 10.1128/AAC.02273-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan R, Lu Y, Zhu Y, Lan P, Jiang S, Lu J, Shen P, Yu Y, Zhou J, Jiang Y. 2021. A sequence type 23 hypervirulent Klebsiella pneumoniae strain presenting carbapenem resistance by acquiring an IncP1 blaKPC-2 Plasmid. Front Cell Infect Microbiol 11. doi: 10.3389/fcimb.2021.641830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeh KM, Kurup A, Siu LK, Koh YL, Fung CP, Lin JC, Chen TL, Chang FY, Koh TH. 2007. Capsular serotype K1 or K2, rather than magA and rmpA, is a major virulence determinant for Klebsiella pneumoniae liver abscess in Singapore and Taiwan. J Clin Microbiol 45:466–471. doi: 10.1128/JCM.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nougayrède J-P, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. 2006. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 41.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bushnell B. 2014. BBMap: a fast, accurate, splice-aware aligner. Lawrence Berkeley National Lab. (LBNL), Berkeley, CA, USA. [Google Scholar]
- 45.Lu J, Breitwieser FP, Thielen P, Salzberg SL. 2017. Bracken: estimating species abundance in metagenomics data. PeerJ Computer Science 3:e104. doi: 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- 46.Zolfo M, Tett A, Jousson O, Donati C, Segata N. 2017. MetaMLST: multi-locus strain-level bacterial typing from metagenomic samples. Nucleic Acids Res 45:e7. doi: 10.1093/nar/gkw837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124–124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE. 2016. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li D, Liu CM, Luo R, Sadakane K, Lam TW. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 51.Beghini F, McIver LJ, Blanco-Míguez A, Dubois L, Asnicar F, Maharjan S, Mailyan A, Manghi P, Scholz M, Thomas AM, Valles-Colomer M, Weingart G, Zhang Y, Zolfo M, Huttenhower C, Franzosa EA, Segata N. 2021. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 10:e65088. doi: 10.7554/eLife.65088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segata N, Börnigen D, Morgan XC, Huttenhower C. 2013. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun 4:2304–2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41–41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FSL, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45:D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu B, Zheng D, Jin Q, Chen L, Yang J. 2019. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aragon TJ. 2020. epitools: epidemiology tools.
- 58.Yu G, Lam TT-Y, Zhu H, Guan Y. 2018. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol Biol Evol 35:3041–3043. doi: 10.1093/molbev/msy194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 60.R Core Team. 2021. R: a language and environment for statistical computing, on R Foundation for Statistical Computing. https://www.R-project.org/. Accessed November 1, 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM02190-21_Supp_1_seq3.xlsx, XLSX file, 0.03 MB (30KB, xlsx)
Supplemental material. Download SPECTRUM02190-21_Supp_2_seq6.xlsx, XLSX file, 0.01 MB (14.3KB, xlsx)
Supplemental material. Download SPECTRUM02190-21_Supp_3_seq8.xlsx, XLSX file, 1.0 MB (1MB, xlsx)
Supplemental material. Download SPECTRUM02190-21_Supp_4_seq9.xlsx, XLSX file, 0.01 MB (12.9KB, xlsx)
Supplemental material. Download SPECTRUM02190-21_Supp_5_seq10.xlsx, XLSX file, 0.02 MB (20.2KB, xlsx)
Supplemental material. Download SPECTRUM02190-21_Supp_6_seq11.xlsx, XLSX file, 0.06 MB (63.7KB, xlsx)
Supplemental material. Download SPECTRUM02190-21_Supp_7_seq14.pdf, PDF file, 0.2 MB (180.3KB, pdf)
Data Availability Statement
The microbial reads produced in this study have been deposited in the NCBI Sequence Read Archive (SRA) database under the BioProject accession PRJNA758247.