ABSTRACT
Comparative genomic analysis of Vibrio cholerae El Tor associated with endemic cholera in Asia revealed two distinct lineages, one dominant in Bangladesh and the other in India. An in-depth whole-genome study of V. cholerae El Tor strains isolated during endemic cholera in Bangladesh (1991 to 2017) included reference genome sequence data obtained online. Core genome phylogeny established using single nucleotide polymorphisms (SNPs) showed V. cholerae El Tor strains comprised two lineages, BD-1 and BD-2, which, according to Bayesian phylodynamic analysis, originated from paraphyletic group BD-0 around 1981. BD-1 and BD-2 lineages overlapped temporally but were negatively associated as causative agents of cholera during 2004 to 2017. Genome-wide association study (GWAS) revealed 140 SNPs and 31 indels, resulting in gene alleles unique to BD-1 and BD-2. Regression analysis of root to tip distance and year of isolation indicated early BD-0 strains at the base, whereas BD-1 and BD-2 subsequently emerged and progressed by accumulating SNPs. Pangenome analysis provided evidence of gene acquisition by both BD-1 and BD-2, of which six crucial proteins of known function were predominant in BD-2. BD-1 and BD-2 diverged and have distinctively different genomic traits, namely, heterogeneity in VSP-2, VPI-1, mobile elements, toxin encoding elements, and total gene abundance. In addition, the observed phage-inducible chromosomal island-like element (PLE1), and SXT ICE elements (ICETET) in BD-2 presumably provided a fitness advantage for the lineage to outcompete BD-1 as the etiological agent of endemic cholera in Bangladesh, with implications for global cholera epidemiology.
IMPORTANCE Cholera is a global disease with specific reference to the Bay of Bengal Ganges Delta where Vibrio cholerae O1 El Tor, the causative agent of the disease showed two circulating lineages, one dominant in Bangladesh and the other in India. Results of an in-depth genomic study of V. cholerae associated with endemic cholera during the past 27 years (1991 to 2017) indicate emergence and succession of the two lineages, BD-1 and BD-2, arising from a common ancestral paraphyletic group, BD-0, comprising the early strains and short-term evolution of the bacterium in Bangladesh. Among the two V. cholerae lineages, BD-2 supersedes BD-1 and is predominant in the most recent endemic cholera in Bangladesh. The BD-2 lineage contained significantly more SNPs and indels, and showed richness in gene abundance, including antimicrobial resistance genes, gene cassettes, and PLE to fight against bacteriophage infection, acquired over time. These findings have important epidemic implications on a global scale.
KEYWORDS: bacterial evolution, comparative genomics, Vibrio cholerae lineages, antimicrobial resistance, phage-inducible chromosomal island-like elements (PLE)
INTRODUCTION
Cholera is a life-threatening infectious diarrheal disease caused by Vibrio cholerae serogroups O1 and O139 of the Gram-negative gammaproteobacteria (1, 2). The global incidence of cholera is estimated to be 2.9 million cases annually with almost 95,000 deaths (3). In 2017, 34 countries reported a total of 1,227,391 cases and 5,654 deaths (4). Seven cholera pandemics have been recognized since 1817. However, limited information is available regarding the etiological agent for the first five pandemics and no isolates of the causative agent are extant. The sixth pandemic, and possibly those earlier were caused by V. cholerae O1 classical biotype, while the ongoing seventh pandemic is caused by V. cholerae El Tor biotype and began with the displacement of V. cholerae classical biotype in Asia in 1961 (5). V. cholerae El Tor was isolated in Africa in the 1970s and Latin America in 1991 where for more than a century there had been no cholera outbreaks (6). In 1992, a V. cholerae non-O1 strain designated V. cholerae O139 Bengal initiated outbreaks of cholera in coastal areas of India and Bangladesh and subsequently was isolated from patients in several countries of Asia (2). V. cholerae El Tor continues to be the major etiological agent of cholera worldwide.
The severe dehydrating diarrhea characteristic of cholera is associated with several factors, including a toxin and several virulence genes involved in colonization and toxicity and their coordinated expression (1). Cholera toxin (CT) is the virulence factor responsible for secretory diarrhea of cholera and is encoded in the genome of a lysogenic CTX phage. V. cholerae El Tor responsible for the current cholera pandemic harbors the CTX phage classical biotype variant, and the ctxBcla: ctxB genotype 1 (ctxB1) or ctxB7 (7). V. cholerae responsible for the current cholera pandemic has become more virulent by undergoing several shifts in CTX genotype and acquiring virulence-related gene islands (8). Integrative conjugative elements (ICEs) and lysogenic phages are genetic elements that play an important role in the acquisition of virulence, antimicrobial resistance, and heavy metal resistance, which are important components of the pathogenicity of V. cholerae (9, 10). Functions of these elements are important for the pathogen to exert evolutionary advantage and variants can be used as markers of clonal expansion (1). Acquisition of mobile genetic elements (MGEs) through horizontal gene transfer (HGT) and propitious chromosomal mutations are significant landmarks for an evolving bacterium (11).
Whole-genome sequencing of V. cholerae El Tor strains associated with the seventh cholera pandemic revealed three waves, suggesting independent but overlapping paths for the pathogen to spread globally from the Bay of Bengal estuary where cholera has been endemic at least since 1961 but likely for centuries (5). Intercontinental transmission of V. cholerae has been proposed for the 2010 outbreak in Haiti (12). Bangladesh borders on the Bay of Bengal and is considered to be a hot spot of Asiatic cholera, where ca. 100,000 cases and 4,500 deaths are reported each year (13). V. cholerae O1 responsible for endemic cholera in Bangladesh and India has been found to have undergone genetic changes over time, including the acquisition of classical biotype attributes in an El Tor background, thereby becoming more successful as a pathogen (14, 15). A recent whole-genome analysis of Vibrio cholerae El Tor strains isolated between 2009 and 2016 indicated two distinct lineages exist in Bengal (16). The objective of the study reported here was to investigate V. cholerae endemic cholera strains isolated from 1991 to 2017 to understand more completely the emergence and progression of the two lineages in Bangladesh. Virulence and related genomic islands, including toxin and antimicrobial resistance genes differing significantly among the V. cholerae El Tor lineages, were also investigated for potential relevance to the emergence of the lineages.
RESULTS
Phylogenetic analysis.
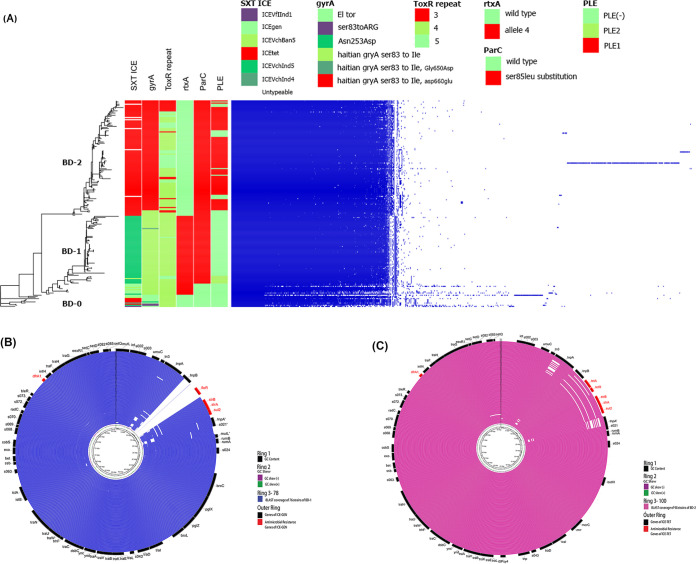
A total of 119 strains were included in the study and their genomes were sequenced using the Illumina platform (MiSeq or HiSeq 2500 sequencer). In addition, 56 strains from our previous study (16) and 17 genomes from the European Nucleotide Archive (17) were used, which are representative of isolates from Bangladesh between 1991 and 2017 (Table S1). Paired-end reads of the 192 genomes were mapped to V. cholerae El Tor N16961 reference strain, a seventh-pandemic V. cholerae O1 El Tor (7PET) strain isolated in Bangladesh in 1975 (18). A total of 1,298 single nucleotide polymorphisms (SNPs) and 413 indels (insertions or deletions) were obtained and, after filtering indels, low call rate, and high-density SNPs, a total of 893 high-quality SNPs were retained for further study. A phylogenetic analysis was conducted to construct a tree based on the 893 high-quality SNPs to evaluate the genetic diversity of the Vibrio cholerae O1 El Tor isolates from Bangladesh. A nested hierarchical structure in the phylogenetic tree was observed, with all but four of the strains isolated between 1999 and 2017 clustering into two major clades, BD-1 (n = 76) and BD-2 (n = 105), shown in green and red, respectively. The remaining strains formed paraphyletic group BD-0 (n = 11) (Fig. 1A, blue). Except for three strains isolated in 2012 that formed a subclade, BD-0 consisted mostly of strains isolated earlier between 1991 and 2000. Dates of isolation of common ancestors of the lineages were inferred using Bayesian Markov Chain Monte Carlo framework Bayesian evolutionary analysis sampling trees (BEAST) (19) (Fig. S1), and maximum clade credibility (MCC) tree was inferred from the posterior distribution of the best fitting model using program TreeAnnotator tool of the BEAST software package. It was estimated from the MCC tree that the most recent common ancestor (MRCA) of lineage BD-1 was isolated in 1987 (95% HPD: 1983 to 1991), and lineage BD-2 in 1997 (95% HPD: 1994 to 2000), where HPD stands for height posterior density. Strains of BD-1 and BD-2 shared genome sequences of strains isolated since 1981 (95% HPD: 1976 to 1986). The number of SNPs in strains of the two clades is relative to reference V. cholerae N16961, which showed strains of BD-0 differed by 107 to 137 SNPs, BD-1 by 123 to 189 SNPs, and BD-2 by 146 to 186 SNPs. An unrooted tree showed SNP diversity among BD-0, BD-1, and BD-2 clades with SNP diversity of BD-2 highest (Fig. 1B). Comparison of isolates in the clades and year of isolation revealed clonal aggregation within the dominant clade and strong temporal signature. Strains of BD-1 and BD-2 were found to be temporally spread but simultaneously isolated during the periods of 2004 to 2011, 2012, 2014 to 2016 (Fig. 1C, Table S2). Strains of BD-1 were mainly isolated during 2004 to 2011 (66.3%, n = 65) while strains of BD-2 were isolated during those years in fewer numbers (33.7%, n = 33) except 2009 when BD-2 strains were dominant (93.33%, n = 14) (Table S1). The following years, from 2012 to 2017, showed BD-2 strains to be dominant (73.5%, n = 72) and BD-1 strains the minority (10.2%, n = 10).
FIG 1.
Phylogenetic analyses of strains showing respective genomic features and year of isolation. (A) Maximum likelihood phylogenetic tree generated from whole-genome SNPs and number of isolated V. cholerae O1 El Tor strains belonging to lineages BD-0, BD-1, and BD-2 rooted from out-group reference strain Vibrio cholerae N16961. Rings show features of the isolates according to the color scheme provided on the left. Tree branches are colored blue, green, and red defining lineages BD-0, BD-1, and BD-2, respectively. (B) Unrooted tree showing independent evolution of BD-1 and BD-2 strains with the number of core genome SNPs of strains in the lineages compared to the N16961 reference strain. (C) Percentage of isolates per year for the three lineages. The size of the circles indicates the percentage of strains belonging to lineages according to the scheme shown.
Genetic variants associated with the clades.
Associations between lineages and the genetic variants were studied using 1298 SNPs and 413 indels, identified by aligning raw reads against V. cholerae N16961 reference genome. Variant annotation using SnpEff (20) showed that among the 1298 SNPs, there were 337 synonymous, 613 nonsynonymous, and 348 variants on intergenic regions (Fig. 2A to C, Table S2). Moreover, of 413 indels, there were 238 frameshift-variants, 107 variants on intergenic regions, and 68 other types of variants (Fig. 2D to F, Table S2). Most of the identified SNPs and indels were in the protein-coding region, many of which function to change the form of a protein. By plotting the distribution of SNP types and indel variants for BD-0 (n = 11), BD-1 (n = 76), and BD-2 (n = 105), it was observed that strains of the clades accumulated SNPs and indels. Strains of BD-2 accumulated more SNPs and indels, increasing genetic distance from BD-0 and BD-1 (Fig. 1B, Fig. 2) and suggesting evolution was occurring compared with reference V. cholerae O1 N16961.
FIG 2.
Box plots of SNPs distribution and indel type in each of three lineage groups. (A) Distribution of 337 synonymous SNP variants. This figure shows that strains of BD-2 lineage accumulated more synonymous SNP variants compared to BD-0 and BD-1 lineages. Notably, synonymous SNP variants do not change the form of protein. (B) Distribution of 613 nonsynonymous SNP variants. These non-synonymous SNP variants include 570 missense variants, 38 stop gained variants, 2 splice-region-variants and stop-retained-variants, 2 stop-lost and splice-region-variants, 1 initiator codon variant. (C) Distribution of 348 upstream/downstream SNP variants. (D) Distribution of 238 frameshift indel variants. (E) Distribution of 107 upstream/downstream indel variants. (F) Distribution of 68 indel variants, including 13 conservative-inframe-insertions, 14 disruptive-inframe-insertions, 11 frameshift-variant and stop-gained, 10 disruptive-inframe-deletions, 10 conservative-inframe-deletions, 1 stop-gained and disruptive-inframe-deletions, 2 feature-elongations, 1 frameshift-variant and stop-lost and splice-region-variant, 1 stop-gained and disruptive-inframe-insertion, 2 frameshift-variant and splice-region-variant, 2 frameshift-variant and start-lost, 1 stop-gained and conservative-inframe-insertion.
Fisher exact test (21) was performed for association analysis between genetic variants and the clades BD-1 and BD-2. Association analysis showed that 140 SNPs and 31 indels had a genome-wide significant association (P < 6.40 × 10−9) with BD-1 and BD-2. Among the 140 SNPs were 25 synonymous variants, 53 missense variants, 2 stop gain variants, and 60 variants on intergenic regions (Table S3 and Fig. S2). It was discovered that 21 SNP missense mutations were present in genes with known functions in more than 80% of BD2 strains, resulting in mutant proteins (Table 1). However, there were only seven missense mutations were found in genes with known functions in more than 80% of BD1 strains. Genotype and frequency of 140 significantly associated SNPs, number of SNPs by year of isolation, and root to tip distance showed significant genetic differences between BD-1 and BD-2 (Fig. 3). The number of core genome SNPs by year of isolation was analyzed to detect temporal SNP accumulation patterns of the clades. The number of core genome SNPs did increase over time for both BD-1 and BD-2 (Fig. 3B). Moreover, root-to-tip regression analysis indicated a steady increase in SNP divergence among the strains of the two clades over time (Fig. 3C). Miami plot for frequency of alternative alleles of the 140 significant SNPs showed BD-2 strains had accumulated more clade-specific SNPs, notably in chromosome-2 compared to BD-1 (Fig. 3D).
TABLE 1.
SNPs resulted unique mutant proteins in BD1 and BD2
| SNPa | REF | ALT | FrqBD1 | FrqBD2 | P value | Gene | AA change | Product |
|---|---|---|---|---|---|---|---|---|
| S1_2609994 | G | A | 0 | 105 | 5.61E−53 | nudF_1 | Arg109Cys | ADP-ribose pyrophosphatase |
| S2_266019 | A | G | 0 | 105 | 5.61E−53 | ulaA | Ile354Thr | Ascorbate-specific permease IIC component UlaA |
| S2_1024884 | G | A | 0 | 105 | 5.61E−53 | putA | Ala600Val | Bifunctional protein PutA |
| S2_989172 | C | T | 0 | 105 | 5.61E−53 | yecS | Pro191Ser | YecS |
| S1_798976 | T | C | 0 | 105 | 5.61E−53 | suhB | Glu217Gly | Inositol-1-monophosphatase |
| S1_994229 | G | A | 0 | 105 | 5.61E−53 | stcE_2 | Gly201Asp | Metalloprotease StcE precursor |
| S2_921045 | A | C | 0 | 105 | 5.61E−53 | ctpH_6 | Ile161Ser | Methyl-accepting chemotaxis protein CtpH |
| S1_1622584 | G | A | 0 | 105 | 5.61E−53 | cobB | Pro50Leu | NAD-dependent protein deacetylase |
| S2_773493 | T | A | 0 | 105 | 5.61E−53 | phhA | Gln19Leu | Phenylalanine-4-hydroxylase |
| S1_681574 | G | T | 0 | 105 | 5.61E−53 | glmM | Arg196Leu | Phosphoglucosamine mutase |
| S2_161094 | T | G | 0 | 105 | 5.61E−53 | siaT_5 | Ser241Ala | Sialic acid TRAP transporter permease protein SiaT |
| S1_1452755 | T | C | 0 | 105 | 5.61E−53 | cysG_1 | Val38Ala | Siroheme synthase |
| S1_2731709 | G | A | 0 | 105 | 5.61E−53 | tamA | Thr266Ile | Translocation and assembly module TamA precursor |
| S1_545919 | T | G | 0 | 104 | 4.32E−51 | pctB_1 | Leu249Trp | Methyl-accepting chemotaxis protein PctB |
| S1_2814292 | T | C | 0 | 102 | 4.43E−48 | argG | Thr283Ala | Argininosuccinate synthase |
| S1_1332186 | T | G | 0 | 99 | 1.96E−44 | gyrA | Asp660Glu | DNA gyrase subunit A |
| S1_149686 | G | T | 0 | 99 | 1.96E−44 | murI | Ala137Ser | Glutamate racemase |
| S2_562858 | A | T | 0 | 99 | 1.96E−44 | VCA0627 | Thr6Ser | rRNA methylase |
| S1_628646 | C | T | 0 | 85 | 1.32E−32 | hrpB_1 | Ala782Val | ATP-dependent RNA helicase HrpB |
| S1_673206 | A | G | 0 | 85 | 1.32E−32 | tyrS_2 | Thr393Ala | Tyrosine—tRNA ligase |
| S1_2357516 | G | A | 0 | 79 | 7.24E−29 | angR | Leu227Phe | Anguibactin system regulator |
| S1_2483236 | G | A | 66 | 0 | 4.18E−39 | lysX | Ala150Thr | Alpha-aminoadipate—LysW ligase LysX |
| S1_1682925 | C | T | 67 | 0 | 3.63E−40 | appC | Ala226Thr | Cytochrome bd-II ubiquinol oxidase subunit 1 |
| S1_368119 | T | C | 67 | 0 | 3.63E−40 | mutL | Cys350Arg | DNA mismatch repair protein MutL |
| S1_1359179 | G | A | 67 | 0 | 3.63E−40 | licH | Ala56Thr | putative 6-phospho-beta-glucosidase |
| S1_1060408 | C | T | 71 | 0 | 6.86E−45 | nagA_1 | Asp150Asn | N-acetylglucosamine-6-phosphate deacetylase |
| S1_276112 | G | A | 76 | 0 | 5.61E−53 | mak | Gly116Arg | Fructokinase |
| S1_1782501 | G | A | 76 | 0 | 5.61E−53 | cph2_4 | Leu79Phe | Phytochrome-like protein cph2 |
Here, SNP refers to the SNPs which had alternative alleles uniquely found in more than 80% of BD1 or BD-2 strains, located within proteins of known functions and altered amino acids. SNPs were named according to their chromosomal position. For example, “S1_2609994” is an SNP/indel site, where “S” stands for the site and “2609994” stands for the site's base pair location. Reference allele = REF, alternative allele = ALT, AA change = amino acid change. Freq_BD1 is the frequency of an alternative allele in BD1 and Freq_BD2 is the frequency of an alternative allele in BD2. Note that, the frequencies of alternative alleles of the SNPs are zero for BD-0. P value is from the Fisher exact test.
FIG 3.
SNP analysis of genetic diversity. (A) Phylogenetic treemap of the strains and heat map for genotypes of 140 SNPs are significantly associated with different lineages. The colors delineate four different nucleotides where white represents the missing genotype. Heatmap shows clear differences in the lineages. (B) Number of core genome SNPs referencing the year of isolation. The figure shows the steady accumulation of SNPs of different lineage strains over time. (C) Regression analysis of root-to-tip distance for strains of the lineages. This figure shows the diversity of strains of different lineages. (D) Miami plot of alternative allele frequencies of SNPs for the dominant lineages BD-1 and BD-2. This figure shows the clear difference in SNP accumulation by the two dominant lineages BD-1 and BD-2.
Relative gene abundance.
Pangenome analysis was done using Roary to investigate differences in core and pan genes among the strains of BD-0, BD-1, and BD-2. Roary classified the identified functional genes into four categories: (i) core genes, present in 99 to 100% of the strains; (ii) softcore genes, present in 95 to 99% of the strains; (iii) shell genes, present in 15 to 95% of the strains; and (iv) cloud genes, present in less than 15% of the strains (22). Pangenome analysis revealed significant differences in overall gene composition among the clades (Fig. 4A). According to the definition of core genes in pangenome analysis, the number of core genes largely varied among BD-0, BD-1, and BD-2 (Table S4). Similarly, the number of soft-core genes was also varied. BD-0 is a group of close relatives with a larger genetic distance relative to BD-1 and BD-2. All BD-0 strains and more than 95% of the BD-1 and BD-2 strains had 1102 common genes (Table S5A) most having a known function. About 10% of BD-2 strains had 44 unique genes of which six encoding crucial proteins of known function were found in more than 90% of the BD-2 strains. Those genes are tetracycline repressor protein (tetR), tetracycline resistance protein (tetA), type-I restriction enzyme EcoKI M protein (hsdM), type-I restriction enzyme EcoR124II R protein (hsdR), Mrr restriction system protein (mrr), and 5-methylcytosine-specific restriction enzyme B (mcrB) (Table S5B). In addition, methyl-accepting chemotaxis protein (CtpH) and group_10030 virulence proteins were exclusively found in 60% and 65% of BD-2 strains, respectively. In contrast, about 5 to 15% of the BD-1 strains carried 19 genes that were unique for them (Table S5C). Three genes common to all BD-0 strains were not detected in BD-2 and were present only in 1 to 2 of the BD-1 strains.
FIG 4.
Pangenome analysis showing differences in the abundance of gene clusters among the lineages. (A) Relative gene abundance of lineages identified by Roary. Features of the sequences are shown with bars and details for features listed in Table S1. (B) BLAST coverage of SXT regions of BD-1 isolates compared with ICE-GEN. Rings represent sequentially outwards following Table S1. Outermost ring shows the different genes of ICE-GEN. (C) BLAST coverage of SXT regions of BD-2 isolates compared with ICE-TET. The rings represent strains of BD-2 sequentially outwards following Table S1. The outermost ring shows different genes of ICE-TET.
Next, we conducted Pan-GWAS to identify clade-specific genes by considering gene presence and absence as the explanatory variable and defined lineage groups as the response variable. A total of 92 genes were significantly (P < 4.98 × 10−6) associated with BD-0 and BD-1 (Table S6A). Of these, 62 genes were identified in 54 to 73% of BD-0 but not in BD-1 strains. Of 164 genes associated with BD-0 and BD-2, 46 were found in more than 73% of BD-2, but not in BD-0 strains (Table S6B). In addition, 66 genes were found in more than 45% of BD-0, but not in BD-2 strains. Of 143 genes associated with BD-1 and BD-2 (Table S6C), 29 were found in more than 76% of BD-1, but not in BD-2 strains. Again, 47 genes were found in 22 to 97% of the BD-2, but not in BD-1 strains. These results provide evidence that strains of BD-1 and BD-2 diverged and evolved as two lineages by accumulating genes, after originating from common ancestor BD-0.
Pathogenicity islands and phage inducible chromosomal island-like elements.
V. cholerae strains included in this study were further examined by targeting the pandemic and pathogenicity islands namely, VSP-1, VSP-II, VPI-1, and VPI-2, including the phage inducible chromosomal island-like elements (PLE). Based on the extent of detected regions compared to V. cholerae N16961, five variants of VSP-II (variants 1 to 5 of the wild type) as reported in our recent study (16), and one variant of VPI-1 (variant 1 of the wild type) were observed (Fig. 5). V. cholerae El Tor strains differed in the type of VSP-II and VPI-1 variants. BD-0 had the wild-type of VSP-II, in reference to the El Tor N16961 strain. Most BD-1 strains (except two) had variant-4 VSP-II, with a partial deletion in VC_495 and complete deletion in VC_496 to VC_512, and BD-2 strains carried three VSP-II variants of which ca. 73% had variant-2 VSP-II with partial ORF VC_495 deletion, and complete VC_496 to VC_500 deletion, which appeared consistent with our prior study (16). BD-0 and BD-1 harbored wild type of VPI-1, whereas most of the BD-2 strains (102 of 105 strains) had variant VPI-1 with complete deletion of VC_819 to VC_820 ORFs; and partial deletion in VC_821. All BD-0 strains and 66 of 76 BD-1 strains lacked PLE (Table S1 and S7), while PLE2 was found in 10 BD-1 strains isolated in 2007 possessing the ctxB1 genotype, and one in 2005. Interestingly, most of the BD-2 strains (83 of 103) carried PLE1, but the rest lacked PLE. Thus, BD-2 lineage strains associated with recent Bangladesh endemic cholera are variant-3 VSP-II, variant VPI-1, and the majority possess PLE1.
FIG 5.
Schematic diagram of VSP-II. Schematic alignment view of VSP-II regions for the isolates. Direction of gene transcription is indicated by arrows and gene shadows represent functional annotation. Six types were identified with all BD-0 strains wild-type VSP-II. Two major types, var-2 and var-3, were observed for most BD-2 strains and one major type var-4 for most BD-1 strains.
Variations in SXT/R391 and important genes.
Although differences in SXT/R391, ctxB, gyrA, rtxA, and parC across two lineages (BD-1, an analog of lineage-2; BD-2, an analog of lineage-1) were investigated in our recent study (16), these important genetic elements were rechecked to draw overall conclusions for all strains included in this investigation. Moreover, variation in ToxR binding repeats was checked across strains of different lineages. Integrative and conjugative elements (ICEs) were targeted from whole-genome sequences by aligning raw reads or contigs with five publicly available sequences of the ICE element (accession no. GQ463140.1, GQ463141.1, GQ463142.1, MK165649.1, and MK165650.1). Nucleotide blast was used to match extracted sequences with ICE element sequences and typed based on the highest bit score. Four strains of BD-0 blast search yielded high bit scores when aligned with ICEGEN (MK165650.1), ICEVchInd5 (GQ463142.1), or ICEVchBan5 (GQ463140.1). Bit scores were highest for the other BD-0 strains when aligned with ICETET (accession no. MK165649.1), which has genomic characteristics similar to ICEVchVhn2255 (accession no. KT151660). For all BD-1 strain bit scores were high when aligned with ICEGEN, ICEVchInd5, or ICEVchBan5, and for BD-2 strains bit scores were highest when aligned with ICETET, which is consistent with our previous results. All BD-1 and BD-2 strains contained mutant gyrA with an amino acid alteration Ser83Ile, whereas 99 (94.28%) of the 105 BD-2 strains exhibited Asp660Glu, which was not present in BD-1 or BD-0, also supporting our previous findings.
V. cholerae O1 El Tor strains in this study were CTX positive, and each carried a single copy of CTXФ with a particular ctxB genotype. Three variants, ctxB1 (classical genotype), ctxB3 (typical El Tor genotype), and ctxB7 (Haitian variant), of the cholera toxin gene, were detected and found associated with the clades (Fig. 1A). Similar to previous findings, all BD-2 strains had ctxB1 genotype, majority of BD-1 strains had ctxB7 genotype, and all but two BD-1 strains possessed rtxA that differed from El Tor reference N16961 by a single SNP at position 13602 of 1563748 bp (NCBI accession no. NC002505.1), corresponding to rtxA allele 4 (23). However, in this study, it was observed that early BD-1 strains had the ctxB1 genotype, and over time gained the ctxB7 genotype.
A prior study showed that Kolkata strains had four heptad repeats (TTTTGAT), whereas Haitian strains had five heptad repeats (24). All BD-0 strains had four heptad repeats (Table S1), while most BD-1 strains (93.4%; n = 71) had four repeats, and only 5.3% (n = 4) strains had five repeats. As a result, most BD-1 strains with ctxB7 genotypes differed from Haitian strains in ToxR binding repeats. BD-2 strains had more diversity in ToxR binding repeats with 59.0% (n = 62) carrying heptad repeats, 24.8% (n = 26) five repeats, and 16.2% (n = 17) three repeats.
DISCUSSION
Vibrio cholerae biotype El Tor, the causative agent of the 7th cholera pandemic has increased transmissibility and virulence with the acquisition of classical biotype traits (14, 15). The 7th pandemic strains of cholera circulating in Asia comprises two El Tor clades, one dominant in Bangladesh and the other in India (16). Genomic analyses that included additional strains and publicly available genome sequences of wave-2 and wave-3 strains (6, 12) provide a detailed view of longitudinally and temporally representative V. cholerae clades associated with endemic cholera in Bangladesh over 27 years (1991 to 2017). The results provide new insights potentially interpretable as origin and progression, based on differences in SNPs, indels, and gene acquisition, including antibiotic resistance cassettes in BD-1 and BD-2, the latter having gained ascendency and dominance as the agent of Bangladesh endemic cholera.
Results of whole-genome sequencing (16), combined with additional genome sequence data for V. cholerae El Tor isolates of Bangladesh endemic cholera, allowed identification of two lineages, designated BD-1 and BD-2. The two clades appear to have originated from a common ancestor of paraphyletic group BD-0, as early as 1981 (95% HPD: 1976 to 1986). According to Mutreja et al. (12), seven strains of BD-0 isolated between 1991 and 2000 represent wave-2 strains, and only one strain isolated in 1994, wave-3 with a most recent common ancestor (MRCA) for BD-1 and BD-2. The BD-1 and BD-2 clades may belong to wave-3. Although BD-0 consisted predominantly of wave-2 strains, three sequenced strains isolated in 2012 shared a wave-2-like genetic background (6), suggesting wave-2 strains may have already been present. Almost all wave-3 strains from a previous study (12) were grouped with strains belonging to BD-1. Consistent with the results of a previous study (16), significant differences were noted between BD-1 and BD-2, which varied in temporal predominance as the causal agent of Bangladesh endemic cholera. Most (n = 62; 82%) BD-1 strains had been isolated between 2007 and 2012, with predominance during that time. Between 2005 and 2017, 105 strains belonging to BD-2 were reported, with 97 obtained between 2009 and 2017, implying BD-2 association with recent Bangladesh endemic cholera until 2017. Phylodynamic analysis using BEAST (19) revealed strains of BD-1 had been isolated in Bangladesh roughly 10 years before BD-2 strains (Fig. S1), and previously identified as Asian lineage-2 and Asian lineage-1, respectively (16).
BD-1 and BD-2 strains appear to have advanced by accumulating different SNPs and indels. Fisher exact test (21) identified 140 SNPs and 31 indel differences between BD-1 and BD-2, resulting in gene alleles unique to them (Fig. 3). Most of the SNPs and indels were components of protein-coding genes, suggesting a possible crucial role in their adaption in Bangladesh. Regression analysis of the number of SNPs and year of isolation suggested that both clades consistently accumulated SNPs over time, implying evolution in response to environmental selective pressure.
Pangenome analysis using Roary (22) provided evidence of gene acquisition by strains of the clades. A recent study of V. cholerae O1 strains isolated in Pakistan found evidence of gene acquisition, where the number of core and accessory genes varied among different lineages (25). According to the results of the analysis reported here, the number of core and accessory genes varied significantly among strains of BD-0, BD-1, and BD-2 in Bangladesh (Fig. 4A). The Pan-GWAS approach helped to identify genes unique for each clade which could be considered contributing to virulence and/or niche adaptation (26).
Phage inducible chromosomal island-like elements (PLE) protect V. cholerae populations from ICP1 infection by acting as an abortive infection system (27). In this study, the observed predominance in BD-2 of PLE1, not found in BD-0 and BD-1, could have provided a selective advantage for the lineage over BD-1, establishing dominance as an etiological agent of endemic cholera in Bangladesh in recent years.
Two BD-0 strains carried CTX phage with ctxB3, while other strains carried CTX phage with typical ctxB1. Strains at the base of BD-1 had CTX with ctxB1 isolated before 2007 and comprised multiple clusters. Moreover, the CTX phage of all BD-2 strains contained classical ctxB1. A mutation in rtxA creating a premature stop codon disabled toxin function in emerging V. cholerae El Tor strains bearing ctxB1 (24). As in the classical strains, altered El Tor pandemic strains eliminated rtxA after acquiring classical ctxB. In this study, BD-0 and BD-2 strains contained the wild-type rtxA allele 1 (Fig. 3A) described by Dolores and Satchell (23). None contained deletions in the rstB gene when reads were compared to V. cholerae N16961 reference genome, indicating rstB of Bangladeshi V. cholerae O1 El Tor isolates does not resemble that of the Haitian outbreak isolates that have been analyzed.
ToxR is a global transcriptional regulator of virulence gene expression, and this repeated sequence is required for ToxR binding and activation of the ctxAB promoter. The ToxR-binding site is located immediately upstream of ctxAB and the affinity of ToxR binding is influenced by the repeat sequences (28). The presence of an increased number of ToxR binding repeats located between zot and ctxA has been hypothesized to correlate with a severe form of cholera (28). In this study, variation was detected in the number of ToxR binding repeats (TTTTGAT) among sequences of the V. cholerae El Tor isolates. All BD-0 strains had four heptad repeats observed in 93.4% of BD-1 and 59% of BD-2 strains. For BD-2 strains, however, greater variation was observed in ToxR binding repeats as ca. 24.8% (n = 26) of BD-2 strains contained five heptad repeats, whereas 16.2% (n = 17) had three heptad repeats, suggesting the robustness of the clade.
Targets of quinolones are type II topoisomerases of DNA gyrase, a heterotetramer composed of two A and two B subunits, encoded by gyrA and gyrB genes, respectively (29). It was observed that all BD-1 and BD-2 strains had a common mutation Ser83 to Ile in gyrA, while 94.29% (99/105) BD-2 had an additional mutation Asp660 to Glu. Furthermore, 87% (66/76) of BD-1 strains exhibited a mutation Ser85 to Leu parC, whereas all BD-2 strains (105/105) had this mutation. In Haitian V. cholerae strains, gyrA and parC genes had two point mutations: Ser83 to Ile in gyrA and Ser85 to Leu in parC. Both are linked to quinolone resistance in V. cholerae strains associated with recent cholera outbreaks in India, Nigeria, and Cameroon (30).
SXT/R391 family ICEs are transferable elements associated with antimicrobial resistance in V. cholerae (31). The SXT-ICE regions of the isolates included in this study were compared with five sequences of the elements to the type SXT/R391 family ICEs belonging to strains associated with cholera (V. cholerae O1 and O139) (9, 32). Four BD-0 strains exhibited ICE elements similar to ICEGEN, ICEVchInd5, or ICEVchBan5, whereas the rest had ICE elements similar to ICETET. Interestingly, ICE elements of BD-1 strains included ICEGEN, ICEVchInd5, or ICEVchBan5-like ICE elements, whereas BD-2 strains differed completely from the others, with only ICETET-like ICE elements.
The results of the study reported here included BD-1 and BD-2 isolated during the Bangladesh endemic cholera of 2004 onwards and that, while existing together, with each subsequent year they exhibited different dominance. BD-2 diverged while retaining the ability to produce multifunctional-autoprocessing repeats-in-toxin (MARTX) and acquiring SXT element ICETET containing tetracycline resistance genes. This observation hints at a selective advantage of BD-2 strains over BD-1 strains for robustness. It is evident from the results of the analyses that BD-1 and BD-2 differ significantly, owing to gene composition and SNPs, and may have evolved independently due to selection pressures. The use of antibiotics, including tetracycline, can exert selection pressure in evolution (16, 33), while strains stopping to produce MARTX along with other variations in the genome might provide a selective advantage. According to suggestions from studies of the dynamics of V. cholerae, immunocompetence of the host against V. cholerae strains may contribute to the dynamics of V. cholerae, hence producing an effect from interaction with humans in the selection and cannot be ruled out (34).
Cholera globally is influenced by thriving populations of V. cholerae occurring naturally in the Ganges Delta of the Bay of Bengal (GDBB) (1, 2, 5, 14). Overall results presented here suggest means of emergence and progression of the two clades in evolution from a progenitor V. cholerae El Tor initiating the seventh pandemic in Asia (5) and reflecting short-term evolution of V. cholerae El Tor associated with Bangladesh endemic cholera in the GDBB (14, 31). BD-2 is concluded to have emerged relatively recently and evolved by acquiring SNPs over time. Also, BD-2 strains showed diversity in indels, possessing SXT/R391 family ICE-elements, PLE1, tetR, and several other important genetic elements, and predominantly associated with recent Bangladesh endemic cholera. As is apparent from our results, BD-1 appears to be an analog of a previously reported lineage 2 from Asia, the major causative agent of cholera in India, Yemen, and Haiti (16). In contrast, BD-2 strains of the present study appear to be an analog of Asian lineage 1, which successfully outcompeted BD-1 (Asian lineage 2) and established predominance as an etiological agent of cholera in a historical hot spot of the disease, Bangladesh. It can be concluded that this reflects the robustness of BD-2 as an epidemic clone emerging locally with the potential to transmit globally and underscoring the need to track the two successful V. cholerae El Tor clades.
MATERIALS AND METHODS
Bacterial isolates.
A total of 119 V. cholerae O1 strains from the icddr,b collection of strains isolated in Bangladesh between 2004 and 2017 (Table S1) were sequenced. Paired-end Illumina short reads for the isolated strains were generated (150 bp, 150 bp) using MiSeq or Hiseq 2500 sequencer as described in our recent study (16). Publicly available paired-end raw reads of 17 strains isolated in Bangladesh between 1991 and 2007 (see the study flow chart in Fig. S3) and 56 strains from our recent study (16) were included in the analysis.
Genome assembly, CTX-prophage typing, and gene annotation.
An ultrafast FASTQ preprocessor implemented in FASTP (35), was used to inspect raw paired-end reads and filter bad ligation or adapter parts. De novo genome assembly implemented in VelvetOptimizer (36) was used to build contigs by optimizing the parameter N50, a metric for assessing the continuity of an assembly. PHASTER, a rapid prophage sequence identification and annotation web server (37), was used to extract CTX-prophage, which was subsequently typed according to mutations in rstA, rstB, and ctxB (7). The bacterial genome annotation tool, Prokka (38), was used for whole-genome gene annotation. ResFinder (39) was used to find the antimicrobial-resistant gene profiles for all the strains.
SNP identification and phylogenetic analysis.
Bowtie2 (40) was used to align high-quality reads with the reference genome sequence of V. cholerae N16961 El Tor (NCBI accession no. NC_002505.1 and NC_002506.1) for variant calling. Samtools (41) and Bcftools (42) were used to call genome variants. A maximum-likelihood phylogeny was inferred on an alignment of concatenated SNPs evenly distributed across a nonrepetitive, nonrecombinant core genome using IQ-TREE v1.6.1 (43). Trees were visualized in FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/) or Interactive Tree of Life online tool (44).
Bayesian phylogenetic inference.
The Bayesian Evolutionary Analysis Sampling Trees (BEAST) v.2.4.4 software package (19) was used for temporal analysis to estimate the divergence date of V. cholerae O1 isolates in Bangladesh. The date of isolation of each strain was used as tip data. A random clock model was implemented using Markov Chain Monte Carlo (MCMC) chains run for 100 million generations with 10% burn-in and sampled every 1000 generations. A GTR nucleotide substitution model was used. Tree data were summarized using TreeAnotator, a tool of BEAST software package, to generate the maximum clade credibility tree.
Pangenome analysis.
A pangenome was constructed using Roary (22) from annotated assemblies of the sample set with a percentage protein identity of 95%. The protein sequences were first extracted and iteratively preclustered with cd-hit (version 4.6) down to 98% identity. An all against all blast (version 2.2.31) was performed on the remaining nonclustered sequences and a single representative sequence from each cd-hit cluster was selected. The data were used by MCL (45) (version 11-294) to cluster the sequences. The preclusters and MCL clusters were merged, and paralogs split by inspecting the conserved gene neighborhood around each sequence (5 genes on either side). Each sequence for each cluster was independently aligned using PRANK (46) (version 0.140603) and combined to form a multi-FASTA alignment of the core genes. Sequences of SXT elements were compared with ICEGEN and ICETET using BRIG 0.95 with 70% BLAST identity (47).
Data availability.
Nucleotide sequence data generated in this study are available in the DDBJ/EMBL/GenBank databases under BioProject ID PRJDB12727.
ACKNOWLEDGMENTS
This work was supported in part by icddr,b, National Institutes of Infectious Diseases (NIID), Tokyo, and the Research Program on Emerging and Reemerging Infectious Diseases (JP21fk0108139) from the Japan Agency for Medical Research and Development (AMED).
We acknowledge icddr,b hospital, and laboratory staff for their support. icddr,b gratefully acknowledges the following donors for providing unrestricted support: Governments of the People’s Republic of Bangladesh, Global Affairs Canada (GAC), Swedish International Development Cooperation Agency (Sida), and the Foreign Commonwealth & Development Office (FCDO), UK. All the authors read and approved the final manuscript.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Munirul Alam, Email: munirul@icddrb.org.
Cheryl P. Andam, University at Albany, State University of New York
REFERENCES
- 1.Reidl J, Klose KE. 2002. Vibrio cholerae and cholera: out of the water and into the host. FEMS Microbiol Rev 26:125–139. doi: 10.1111/j.1574-6976.2002.tb00605.x. [DOI] [PubMed] [Google Scholar]
- 2.Albert MJ, Siddique A, Islam M, Faruque A, Ansaruzzaman M, Faruque S, Sack RB. 1993. Large outbreak of clinical cholera due to Vibrio cholerae non-01 in Bangladesh. Lancet 341:704. doi: 10.1016/0140-6736(93)90481-U. [DOI] [PubMed] [Google Scholar]
- 3.Ramamurthy T, Das B, Chakraborty S, Mukhopadhyay AK, Sack DA. 2020. Diagnostic techniques for rapid detection of Vibrio cholerae O1/O139. Vaccine 38:A73–A82. doi: 10.1016/j.vaccine.2019.07.099. [DOI] [PubMed] [Google Scholar]
- 4.Clemens JD, Nair GB, Ahmed T, Qadri F, Holmgren J. 2017. Cholera. Lancet 390:1539–1549. doi: 10.1016/S0140-6736(17)30559-7. [DOI] [PubMed] [Google Scholar]
- 5.Hu D, Liu B, Feng L, Ding P, Guo X, Wang M, Cao B, Reeves PR, Wang L. 2016. Origins of the current seventh cholera pandemic. Proc Natl Acad Sci USA 113:E7730–E7739. doi: 10.1073/pnas.1608732113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domman D, Quilici ML, Dorman MJ, Njamkepo E, Mutreja A, Mather AE, Delgado G, Morales-Espinosa R, Grimont PAD, Lizarraga-Partida ML, Bouchier C, Aanensen DM, Kuri-Morales P, Tarr CL, Dougan G, Parkhill J, Campos J, Cravioto A, Weill FX, Thomson NR. 2017. Integrated view of Vibrio cholerae in the Americas. Science 358:789–793. doi: 10.1126/science.aao2136. [DOI] [PubMed] [Google Scholar]
- 7.Kim EJ, Lee D, Moon SH, Lee CH, Kim SJ, Lee JH, Kim JO, Song M, Das B, Clemens JD, Pape JW, Nair GB, Kim DW. 2014. Molecular insights into the evolutionary pathway of Vibrio cholerae O1 atypical El Tor variants. PLoS Pathog 10:e1004384. doi: 10.1371/journal.ppat.1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rashid MU, Rashed SM, Islam T, Johura FT, Watanabe H, Ohnishi M, Alam M. 2016. CtxB1 outcompetes CtxB7 in Vibrio cholerae O1, Bangladesh. J Med Microbiol 65:101–103. doi: 10.1099/jmm.0.000190. [DOI] [PubMed] [Google Scholar]
- 9.Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Dery C, Burrus V, Waldor MK. 2009. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet 5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faruque SM, Mekalanos JJ. 2003. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol 11:505–510. doi: 10.1016/j.tim.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Murphy RA, Boyd EF. 2008. Three pathogenicity islands of Vibrio cholerae can excise from the chromosome and form circular intermediates. J Bacteriol 190:636–647. doi: 10.1128/JB.00562-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mutreja A, Kim DW, Thomson NR, Connor TR, Lee JH, Kariuki S, Croucher NJ, Choi SY, Harris SR, Lebens M, Niyogi SK, Kim EJ, Ramamurthy T, Chun J, Wood JL, Clemens JD, Czerkinsky C, Nair GB, Holmgren J, Parkhill J, Dougan G. 2011. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature 477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Islam MT, Clemens JD, Qadri F. 2018. Cholera Control and Prevention in Bangladesh: an Evaluation of the Situation and Solutions. J Infect Dis 218:S171–S172. doi: 10.1093/infdis/jiy470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nair GB, Qadri F, Holmgren J, Svennerholm AM, Safa A, Bhuiyan NA, Ahmad QS, Faruque SM, Faruque AS, Takeda Y, Sack DA. 2006. Cholera due to altered El Tor strains of Vibrio cholerae O1 in Bangladesh. J Clin Microbiol 44:4211–4213. doi: 10.1128/JCM.01304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taneja N, Mishra A, Sangar G, Singh G, Sharma M. 2009. Outbreaks caused by new variants of Vibrio cholerae O1 El Tor, India. Emerg Infect Dis 15:352–354. doi: 10.3201/eid1502.080943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morita D, Morita M, Alam M, Mukhopadhyay AK, Johura F-t, Sultana M, Monira S, Ahmed N, Chowdhury G, Dutta S, Ramamurthy T, Samanta P, Takahashi E, Okamoto K, Izumiya H, Ohnishi M. 2020. Whole-genome analysis of clinical Vibrio cholerae O1 in Kolkata, India, and Dhaka, Bangladesh, reveals two lineages of circulating strains, indicating variation in genomic attributes. mBio 11:e01227-20. doi: 10.1128/mBio.01227-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leinonen R, Akhtar R, Birney E, Bower L, Cerdeno-Tarraga A, Cheng Y, Cleland I, Faruque N, Goodgame N, Gibson R, Hoad G, Jang M, Pakseresht N, Plaister S, Radhakrishnan R, Reddy K, Sobhany S, Ten Hoopen P, Vaughan R, Zalunin V, Cochrane G. 2011. The European nucleotide archive. Nucleic Acids Res 39:D28–31. doi: 10.1093/nar/gkq967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baddam R, Sarker N, Ahmed D, Mazumder R, Abdullah A, Morshed R, Hussain A, Begum S, Shahrin L, Khan AI, Islam MS, Ahmed T, Alam M, Clemens JD, Ahmed N. 2020. Genome Dynamics of Vibrio cholerae isolates linked to seasonal outbreaks of cholera in Dhaka, Bangladesh. mBio 11:e03339-19. doi: 10.1128/mBio.03339-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cingolani P, Platts A, Wang Le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. 2012. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raymond M, Rousset F. 1995. An exact test for population differentiation. Evolution 49:1280–1283. doi: 10.2307/2410454. [DOI] [PubMed] [Google Scholar]
- 22.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolores J, Satchell KJ. 2013. Analysis of Vibrio cholerae genome sequences reveals unique rtxA variants in environmental strains and an rtxA-null mutation in recent altered El Tor isolates. mBio 4:e00624. doi: 10.1128/mBio.00624-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh P, Naha A, Pazhani GP, Ramamurthy T, Mukhopadhyay AK. 2014. Genetic traits of Vibrio cholerae O1 Haitian isolates that are absent in contemporary strains from Kolkata, India. PLoS One 9:e112973. doi: 10.1371/journal.pone.0112973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeb S, Gulfam SM, Bokhari H. 2020. Comparative core/pan genome analysis of Vibrio cholerae isolates from Pakistan. Infect Genet Evol 82:104316. doi: 10.1016/j.meegid.2020.104316. [DOI] [PubMed] [Google Scholar]
- 26.Gori A, Harrison OB, Mlia E, Nishihara Y, Chan JM, Msefula J, Mallewa M, Dube Q, Swarthout TD, Nobbs AH, Maiden MCJ, French N, Heyderman RS. 2020. Pan-GWAS of Streptococcus agalactiae highlights lineage-specific genes associated with virulence and niche adaptation. mBio 11:e00728-20. doi: 10.1128/mBio.00728-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hays SG, Seed KD. 2020. Dominant Vibrio cholerae phage exhibits lysis inhibition sensitive to disruption by a defensive phage satellite. Elife 9:e53200. doi: 10.7554/eLife.53200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfau JD, Taylor RK. 1996. Genetic footprint on the ToxR-binding site in the promoter for cholera toxin. Mol Microbiol 20:213–222. doi: 10.1111/j.1365-2958.1996.tb02502.x. [DOI] [PubMed] [Google Scholar]
- 29.Hooper DC. 1998. Clinical applications of quinolones. Biochim Biophys Acta 1400:45–61. doi: 10.1016/s0167-4781(98)00127-4. [DOI] [PubMed] [Google Scholar]
- 30.Hasan NA, Choi SY, Eppinger M, Clark PW, Chen A, Alam M, Haley BJ, Taviani E, Hine E, Su Q, Tallon LJ, Prosper JB, Furth K, Hoq MM, Li H, Fraser-Liggett CM, Cravioto A, Huq A, Ravel J, Cebula TA, Colwell RR. 2012. Genomic diversity of 2010 Haitian cholera outbreak strains. Proc Natl Acad Sci USA 109:E2010–7. doi: 10.1073/pnas.1207359109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weill FX, Domman D, Njamkepo E, Almesbahi AA, Naji M, Nasher SS, Rakesh A, Assiri AM, Sharma NC, Kariuki S, Pourshafie MR, Rauzier J, Abubakar A, Carter JY, Wamala JF, Seguin C, Bouchier C, Malliavin T, Bakhshi B, Abulmaali HHN, Kumar D, Njoroge SM, Malik MR, Kiiru J, Luquero FJ, Azman AS, Ramamurthy T, Thomson NR, Quilici ML. 2019. Genomic insights into the 2016-2017 cholera epidemic in Yemen. Nature 565:230–233. doi: 10.1038/s41586-018-0818-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarkar A, Morita D, Ghosh A, Chowdhury G, Mukhopadhyay AK, Okamoto K, Ramamurthy T. 2019. Altered integrative and conjugative elements (ICEs) in recent Vibrio cholerae O1 isolated from cholera cases, Kolkata, India. Front Microbiol 10:2072. doi: 10.3389/fmicb.2019.02072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tello A, Austin B, Telfer TC. 2012. Selective pressure of antibiotic pollution on bacteria of importance to public health. Environ Health Perspect 120:1100–1106. doi: 10.1289/ehp.1104650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakraborty S, Mukhopadhyay AK, Bhadra RK, Ghosh AN, Mitra R, Shimada T, Yamasaki S, Faruque SM, Takeda Y, Colwell RR, Nair GB. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl Environ Microbiol 66:4022–4028. doi: 10.1128/AEM.66.9.4022-4028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 39.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S . 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Enright AJ, Van Dongen S, Ouzounis CA. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Loytynoja A. 2014. Phylogeny-aware alignment with PRANK. Methods Mol Biol 1079:155–170. doi: 10.1007/978-1-62703-646-7_10. [DOI] [PubMed] [Google Scholar]
- 47.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00391-22_Supp_1_seq1.pdf, PDF file, 2.2 MB (2.2MB, pdf)
Data Availability Statement
Nucleotide sequence data generated in this study are available in the DDBJ/EMBL/GenBank databases under BioProject ID PRJDB12727.