ABSTRACT
The emergence of multidrug-resistant bacteria has become a major problem. Cockroaches may play an important role in the spread of those bacteria between the environment and humans. This study was designed to screen extended-spectrum β-lactamase (ESBL)-producing and colistin-resistant strains and to investigate the molecular support of multidrug-resistant Enterobacteriaceae in the external surface and gut homogenates of cockroaches collected from different locations in Tunisia. Between July 2017 and June 2018, 144 Enterobacteriaceae samples were isolated from 115 trapped cockroaches (collective catering, houses, and a hospital). Antibiotic susceptibility testing was performed using the disk diffusion method. Extended-spectrum β-lactamase-encoding genes and the mcr-1 gene were investigated by real-time PCR (RT-PCR) and standard PCR. The genetic relationship among isolates was studied with the help of multilocus sequence type (MLST) analysis. Of the 144 Enterobacteriaceae isolates, 22 strains exhibited a positive ESBL-screening test (73.3%), including 17 Escherichia coli isolates and 5 Klebsiella pneumoniae isolates. Among them, 9 Escherichia coli isolates were resistant to colistin, with an MIC ranging from 8 to16 μg/L, all of which harbored the mcr-1 gene. Eight blaCTX-M-15 genes were detected; two among them were associated with blaTEM-117 and blaTEM-128, and seven blaCTX-M-1 genes were detected that also harbored the mcr-1 gene. Genotyping analysis revealed 7 different sequence types already described in humans and animals. We report the first survey of mcr-1 in ESBL-producing E. coli isolates from cockroaches. Our findings highlight cockroaches as a source of nosocomial infections, and they are a reservoir of colistin-resistant E. coli, which is a carrier of other additional risk genes such as blaESBL, especially in hospitals.
IMPORTANCE Multidrug resistance in Enterobacteriaceae has become a major concern worldwide that is increasingly observed in human, animals, and also cockroaches. In our study, we found that cockroaches may play an important role as a potential vector of multidrug-resistant Enterobacteriaceae in the hospital environment and collective catering. Our study describes the first survey of mcr-1 in ESBL-producing E. coli isolates from hospital cockroaches. Our results further highlight the possibility that mcr-1 may enter humans via cockroach contamination and thereby threaten public health. Our results show that these cockroaches are an important reservoir of colistin-resistant E. coli and carriers of other additional risk genes such as blaESBL, hence the importance of strengthening prevention strategies and of strictly respecting hygiene measures in order to control their distribution and spread in Tunisia.
KEYWORDS: cockroaches, ESBL, Enterobacteriaceae, ST648, Tunisia, colistin resistance, mcr-1
INTRODUCTION
The emergence of multidrug-resistant Enterobacteriaceae has become a major concern worldwide (1). For the past decade, Enterobacteriaceae genes (especially those of Escherichia coli and Klebsiella pneumoniae) have frequently been observed encoding ampicillin-hydrolyzing β-lactamases, leading to the use of third-generation cephalosporins (2). Third-generation cephalosporins are considered clinically important antimicrobials for the treatment of highly dangerous bacterial infections, due to their efficacy in a broad range of clinical treatments. In addition, the side effects of cephalosporins are less significant than those of penicillin antibiotics and other antibacterial agents (3, 4).
Enterobacteriaceae pursue various molecular strategies for development of resistance to β-lactams, and the resistance to various types of β-lactam antibiotics, including the extended-spectrum cephalosporins, is essentially induced by the production of extended-spectrum beta-lactamases (ESBLs) (5). The extended-spectrum β-lactamases are bacterial enzymes that inactivate β-lactam antibiotics. ESBLs can hydrolyze cephalosporins and monobactams but not cephamycins and carbapenems, conferring a high level of multidrug resistance (6).
In recent years, the plasmid-mediated mcr-1 gene has been detected worldwide in human and animal samples (7, 8). In addition, cooccurrence of ESBL genes and mcr-1 has been reported in Enterobacteriaceae (7, 9). A recent report showed that the coharboring of mcr-1 and blaCTX-M-1 found on a conjugative IncHI2 type plasmid, together with other genes, confers multidrug resistance (10, 11). Consequently, antibiotic therapy has become limited for treating bacterial infections in both human and veterinary clinical settings (12).
Insects are known to be sources of transmission and spread of infectious diseases (13). Cockroaches play an important role as potential mechanical vectors in the transmission of pathogenic microorganisms; they are widely distributed in the environment, including houses, food industries, kitchens, and hospitals (14). Blattella germanica, Periplaneta americana, and Blatta orientalis are the most abundant cockroach species, and they are associated with human habitations (15, 16). They can be infected with different species of vertebrate pathogens under natural or in vitro conditions (13).
Cockroaches can carry medically important bacteria on the surface of their exoskeleton (e.g., legs, mouthparts) and in the digestive tract. Therefore, bacterial transmission can occur through regurgitation, defecation, or translocation from the exoskeleton. Furthermore, degrading cockroaches can contaminate the environment with bacteria resistant to antimicrobials, making these insects a threat to human health (17).
Moreover, the transmission and spread of multidrug-resistant Enterobacteriaceae in both hospital and community settings are generally related to poor hygiene, hand contact, and contaminated instruments (18, 19). Nevertheless, cockroaches which colonize these environments have been suggested as potential vectors of pathogenic antimicrobial-resistant bacteria, and they can have a great impact in the epidemiology of nosocomial infections, particularly those caused by strains of ESBL-producing Enterobacteriaceae (15, 20, 21).
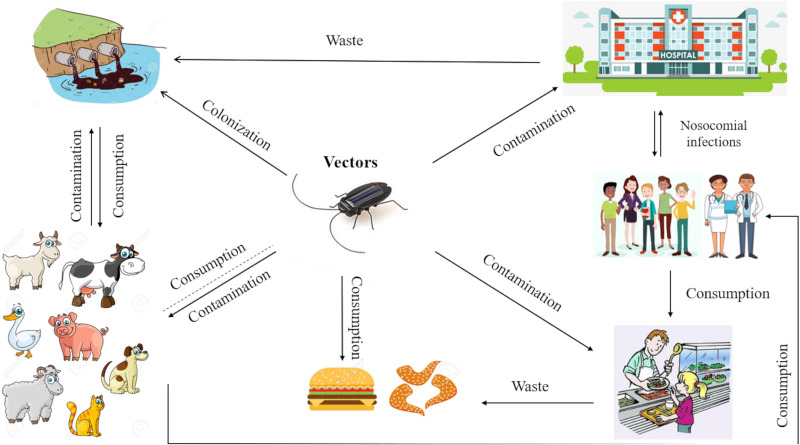
Antimicrobial resistance requires a one-health, one-world approach (10, 22). In particular, resistant bacteria reside within humans, animals, food and the environment, and pathogens can be widely disseminated in agricultural and human waste (Fig. 1). Likewise, there are no barriers to the transmission of resistance genes across bacterial species and compartments (22, 23).
FIG 1.
Antimicrobial resistance (AMR) interactions and transmission among different habitats.
The aims of this study were first to evaluate the Enterobacteriaceae carrier rate on the external surface and gut homogenates of cockroaches and then to determine their antimicrobial susceptibility patterns, screen for ESBL-producing and colistin-resistant strains, and compare their molecular support in cockroaches collected from a collective catering, a hospital, and houses in Tunisia.
RESULTS
Bacterial isolation.
From July 2017 to June 2018, a total of 115 cockroaches were trapped in collective catering (n = 48), houses (n = 35), and a hospital (n = 32). Eighty-seven isolates from cockroaches were found to be P. americana, 21 were B. germanica, and 7 were B. orientalis.
From the 115 cockroaches trapped, 144 bacterial isolates were obtained. Table 1 illustrates the prevalence of Enterobacteriaceae isolated from different types of cockroaches trapped in different sites. E. coli (34.48%) was the predominant species from external surface and gut homogenates of cockroaches, followed by K. pneumoniae (31.72%) and Enterobacter cloacae (19.31%) (Table 1).
TABLE 1.
Enterobacteriaceae isolates identified from external surface and gut homogenates of cockroaches trapped in different sitesa
| Bacterial isolate | No. and type of cockroach (n = 115) in bacteria: |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
P. americana (n = 87) |
B. germanica (n = 21) |
B. orientalis (n = 7) |
||||||||||||||||||
| EH |
GH |
EH |
GH |
EH |
GH |
|||||||||||||||
| Hospital | CC | House | Hospital | CC | House | Hospital | CC | House | Hospital | CC | House | Hospital | CC | House | Hospital | CC | House | Total | ||
| E. coli | 11 | 8 | 2 | 15 | 3 | 2 | 0 | 2 | 0 | 0 | 2 | 0 | 2 | 1 | 0 | 2 | 0 | 0 | 50 | |
| K. pneumoniae | 2 | 7 | 3 | 4 | 7 | 5 | 0 | 10 | 0 | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 46 | |
| E. cloacae | 6 | 1 | 2 | 1 | 3 | 2 | 0 | 5 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 28 | |
| Serratia marcescens | 0 | 0 | 0 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | |
| Citrobacter freundii | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | |
| Providencia rettgeri | 0 | 1 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | |
| Citrobacter gillenii | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | |
| Citrobacter sedlakii | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | |
| Serratia rubidaea | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | |
| Pantoea calida | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Leclercia adecarboxylata | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Total | 20 | 18 | 9 | 23 | 16 | 13 | 0 | 19 | 0 | 0 | 20 | 0 | 2 | 1 | 0 | 2 | 1 | 0 | 144 | |
EH, external homogenate; GH, gut homogenate; CC, collective catering.
Antimicrobial susceptibility testing.
Antibiotic sensitivity testing was performed for all isolates for 16 antibiotics. A high prevalence of resistance was found to amoxicillin (72.4%), amoxicillin-clavulanic acid (60.7%), and cephalothin (63.4%). Furthermore, 21.4% of strains were resistant to doxycycline and 20.7% to ceftriaxone and nitrofurantoin.
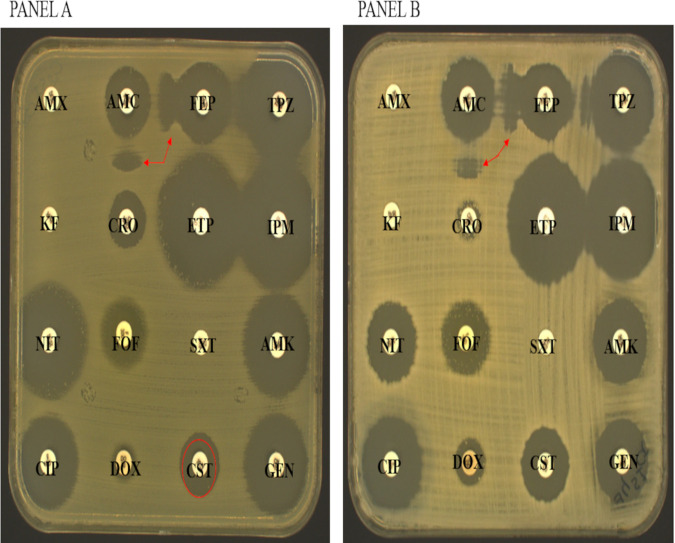
Ceftriaxone-resistant isolates (30/144; 20.8%) were recovered from the different sites analyzed (16/30 in a hospital, 13/30 in collective catering, 1/30 in houses). Among the 30 ceftriaxone-resistant isolates, 22 had a positive ESBL-screening test (73.3%), including 17 E. coli and 5 K. pneumoniae isolates (Fig. 2B).
FIG 2.
Examples of susceptibility testing plates. (A) Antibiogram by diffusion of E. coli (EC2) isolated from hospital expressing blaCTX-M1 and mcr-1. (B) Klebsiella pneumoniae isolated from collective catering expressing blaCTX-M15. AMX: amoxicillin, AMC: amoxicillin-clavulanic acid, TZP: piperacillin-tazobactam, KF: cephalothin, CRO: ceftriaxone, FEP: cefepime, ETP: ertapenem, IMP: imipenem, AMK: amikacin, GEN: gentamicin, CIP: ciprofloxacin, FOF: fosfomycin, NIT: nitrofurantoin, DOX: doxycycline, SXT: trimethoprim-sulfamethoxazole, CST: colistin.
ESBL-positive isolates were also resistant to the non-beta-lactam antibiotics (Table 2), being resistant to trimethoprim-sulfamethoxazole (17/22; 77.2%), doxycycline (17/22; 77.2%), ciprofloxacin (16/22; 72.7%), and colistin (9/22; 40.9%), but all isolates showed susceptibility to carbapenems (ertapenem and imipenem).
TABLE 2.
Characteristics and antibiotic resistance profile of strains harboring ESBL and mcr-1 genes
| Strain | Isolate source/locationa | Bacteria species | Resistance profileb |
MIC | Gene(s) | MLST | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMX | AMC | TZP | KF | CRO | FEP | ETP | IMP | AMK | GEN | CIP | FOF | NIT | DOX | SXT | CST | ||||||
| EC1 | Hospital/GH | E. coli | R | I | S | R | R | R | S | S | S | S | I | S | S | R | R | R | 16 μg/mL | blaTEM-135; blaCTX-M1; mcr-1 | ST648 |
| EC2 | Hospital/GH | E. coli | R | I | S | R | R | I | S | S | S | S | I | S | S | R | R | R | 8 μg/mL | blaTEM-135; blaCTX-M1; mcr-1 | ST648 |
| EC3 | Hospital/GH | E. coli | R | I | S | R | R | R | S | S | S | S | I | S | S | R | R | R | 16 μg/mL | blaTEM-159; blaCTX-M1; mcr-1 | ST648 |
| EC4 | Hospital/GH | E. coli | R | I | S | R | R | I | S | S | S | S | I | S | S | R | R | R | 8 μg/mL | blaTEM-135; blaCTX-M1; mcr-1 | ST648 |
| EC5 | Hospital/EH | E. coli | R | I | S | R | R | R | S | S | S | S | I | S | S | R | R | R | 8 μg/mL | blaTEM-126; blaCTX-M1; mcr-1 | ST648 |
| EC6 | Hospital/GH | E. coli | R | I | S | R | R | I | S | S | S | S | I | S | S | R | R | R | 16 μg/mL | blaCTX-M1; mcr-1 | ST648 |
| EC7 | Hospital/GH | E. coli | R | I | S | R | R | I | S | S | S | S | I | S | S | R | R | R | 16 μg/mL | blaCTX-M1; mcr-1 | ST648 |
| EC8 | Hospital/EH | E. coli | S | I | S | R | R | I | S | S | S | S | S | S | S | S | S | S | bla CTX-M15 | ST2914 | |
| EC9 | Hospital/EH | E. coli | R | R | S | R | R | R | S | S | S | S | I | S | S | S | S | S | bla CTX-M15 | ST2914 | |
| EC10 | Hospital/GH | E. coli | R | I | S | R | R | I | S | S | S | S | I | S | S | R | R | R | 8 μg/mL | blaTEM-159; mcr-1 | ST648 |
| EC11 | Hospital/GH | E. coli | R | I | S | R | R | R | S | S | S | S | I | S | S | R | R | R | 8 μg/mL | blaTEM-1; mcr-1 | ST648 |
| KP1 | Hospital/GH | K. pneumoniae | R | I | S | R | R | R | S | S | S | R | S | S | S | R | R | S | blaTEM-117; blaCTX-M15 | ST551 | |
| EC12 | Collective catering/GH | E. coli | R | R | S | R | R | R | S | S | S | S | S | S | S | R | R | S | blaTEM-128; blaCTX-M15 | ST8149 | |
| EC13 | Collective catering/EH | E. coli | R | I | S | R | R | R | S | S | S | S | S | S | S | R | R | S | bla CTX-M15 | ST8149 | |
| KP2 | Collective catering/GH | K. pneumoniae | R | S | S | R | R | R | S | S | S | S | I | S | S | R | R | S | bla CTX-M15 | ST2010 | |
| KP3 | Collective catering/GH | K. pneumoniae | R | I | S | R | R | R | S | S | S | S | I | S | S | R | R | S | bla CTX-M15 | ST2010 | |
| EC14 | House/GH | E. coli | R | R | S | R | R | R | S | S | S | S | R | S | S | S | S | S | bla CTX-M15 | ST155 | |
aEH, external homogenate; GH, gut homogenate.
MX, amoxicillin; AMC, amoxicillin-clavulanic acid; TZP, piperacillin-tazobactam; KF, cephalothin; CRO, ceftriaxone; FEP, cefepime; ETP, ertapenem; IMP, imipenem; AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin; FOF, fosfomycin; NIT, nitrofurantoin; DOX, doxycycline; SXT, trimethoprim-sulfamethoxazole; CST, colistin; R, resistant; I, intermediate; S, susceptible; MLST, multilocus sequence typing. Genes: CTX-M, especially blaCTX-15, conferring resistance to all the penicillins and cephalosporins and high-level resistance to other classes of antibiotics, especially fluoroquinolones and cotrimoxazole, but not carbapenems; TEM, conferring resistance to penicillin family antibiotics such as ampicillin and aminopenicillins (especially blaTEM-1/135); MCR-1, conferring resistance to colistin.
Furthermore, nine E. coli isolates were found to be resistant to colistin, with MICs between 8 and 16 μg/L. All isolates were resistant to amoxicillin, amoxicillin-clavulanic acid, cephalothin, ceftriaxone, cefepime, ciprofloxacin, doxycycline, and trimethoprim-sulfamethoxazole (Table 2 and Fig. 2A).
Molecular detection of antibiotic resistance.
Beta-lactamase-encoding genes were detected in 17 strains, corresponding to an ESBL phenotype that is characterized by resistance to penicillins and cephalosporins. Of the 17 ESBL producers, PCR and nucleotide sequencing identified blaCTX-M-15 in 8 E. coli isolates, and 2 of these isolates coharbored blaTEM-117/128. Seven strains harbored the blaCTX-M-1 gene; among them, five strains coharbored blaTEM-135/159/126.
Two strains harbored blaTEM-1/159 genes. In view of the antibiogram results, 5 strains could express other β-lactamases that were not researched in our study (Table 2). None of the isolates harbored blaKPC, blaVIM, blaNDM, and blaOXA-48 genes.
In addition, colistin MIC of strains EC2, EC4, EC5, EC10, and EC11 was 8 μg/mL, and that of EC1, EC3, EC6, and EC7 was 16 μg/mL. Standard PCR results and sequencing analyses showed that all nine colistin-resistant E. coli isolates were positive for the mcr-1 gene, and all the strains were detected in cockroaches from the hospital. These colistin-resistant E. coli isolates coharbored ESBL genes, including 5 strains expressing both blaCTX-M1 and blaTEM, 2 harboring only blaCTX-M1, and 2 possessing blaTEM (Table 2). None of the isolates harbored mcr-2/3/4/5/8 genes.
MLST analysis.
According to the MLST analysis, 7 different STs (sequence types) were observed among the 19 sequenced ESBL strains (E. coli [n = 16] and K. pneumoniae [n = 3]). Among the known STs, the most common ST was ST648 (n = 9), followed by ST2914, ST8149, ST2010, and ST155 (2 isolates each), whereas one isolate each of ST23 and ST551 was found (Table 2). MLST analysis showed that the 9 E. coli isolates carrying the mcr-1 gene were assigned to only one sequence type, ST648.
The accession numbers of E. coli and K. pneumoniae ST are available in Table S1.
DISCUSSION
In our study, we found that cockroaches may play an important role as a potential vector of multidrug-resistant bacteria on their external surface and in their gut. In addition to the few reports describing the presence of ESBL-producing Enterobacteriaceae in cockroaches (21, 24–26), this study describes the first detection of the coharboring of mcr-1 and blaESBL genes in nine E. coli strains of lineage ST648 from cockroaches isolated in the hospital.
The present study shows that cockroaches are potential vectors of multidrug-resistant Enterobacteriaceae in the hospital and collective catering environments (21). From the 115 cockroaches trapped, 144 bacterial isolates were obtained. E. coli (34.48%) was the predominant species from external and gut homogenates of cockroaches, followed by K. pneumoniae (31.72%) and E. cloacae (19.31%). Enterobacteriaceae are known as the essential cause of nosocomial and community infections, in particular pneumonia, urinary tract infection, respiratory tract infection, skin infections, septicemia, and gastroenteritis (17, 20). Drug sensitivity results also showed surprisingly high rates of resistance to some of the antibiotics. A high prevalence of resistance was found to amoxicillin (72.4%), amoxicillin-clavulanic acid (60.7%), and cephalothin (63.4%). Also, 21.4% of the strains were resistant to doxycycline, 20.7% to ceftriaxone and nitrofurantoin, and 6.25% to colistin, which is a last-line antibiotic for humans, especially for urinary tract infections (27).
Liu and colleagues (7) described for the first time the plasmid-borne gene mcr-1 encoding resistance to colistin from animals, foodstuffs, and human beings in China. Subsequently, a series of variant mcr genes was identified, including mcr-2 to mcr-10 (28–30). In Tunisia, no study has described the presence of the mcr-1 gene in hospitals. In contrast, the highest mcr-1 prevalence was reported from animals (31–36).
Many studies have shown that ST648 is a developing and generalist lineage that lacks phylogeographic signals and clear host association. ST648 has been reported in human and companion animals in South America, the Middle East, and China. In addition, the combination of multidrug resistance and high-level virulence is the hallmark of ST648 and is similar to the international high-risk clonal lineage of ST131. ST648 became an epidemic vector for circulation/spread of the mcr-1 colistin resistance gene in China (37–39).
In this study, the presence of beta-lactamase-encoding genes was detected in 17 strains, corresponding to an ESBL phenotype which is characterized by resistance to penicillins and cephalosporins. blaCTX-M-15 was the most dominant ESBL type (n = 8). It was found in four isolates from cockroaches collected from collective catering, typed as ST8149 or ST2010 (each n = 2, in E. coli and K. pneumoniae, respectively), those STs having been described in several studies in animal, food, and environmental isolates (40, 41). Three CTX-M15-producing isolates recovered from the hospital were typed as ST2914 (n = 2; E. coli), which has been reported in only one study in isolates of migratory birds in Pakistan (42), or ST551 (n = 1; K. pneumoniae), previously detected in human and environmental isolates (43, 44). Only one CTX-M15-producing isolate recovered from household cockroaches belonged to ST155. ST155 has been found in human clinical isolates, associated with ESBL phenotypes (45).
Consistent with previous studies showing that blaCTX-M-15 is amply present in Tunisia (46), our results suggest that this enzyme is encoded by the gene that mediates resistance to extended-spectrum cephalosporins in our strains. Furthermore, the isolation of ESBL-producing Enterobacteriaceae in household cockroaches is alarming, since the strains could easily spread and trigger a pandemic spread of the clones (24).
It is known that the transfer of resistance genes occurs by different genetic mechanisms, either by horizontal transfer of resistance genes located on different types of mobile DNA elements (transformation, conjugation, or transduction) or by mutations (point mutations, deletions, inversions) (47). ESBL-encoding genes can be transmitted between bacteria by mobile genetic elements, and cockroaches can provide a suitable environment for the exchange of resistance genes between different bacterial species. Therefore, effective prevention and control are needed to reduce nosocomial and foodborne bacterial infections. Cockroach infestation should be of serious concern, given the possible role of cockroaches as reservoirs of antibiotic-resistant bacteria.
Nine E. coli isolates were found to be resistant to colistin, with MICs between 8 and 16 μg/mL. All these strains also harbored the mcr-1 gene and typed as ST648. Seven strains harbored blaCTX-M-1 genes, including five strains that coharbored blaTEM-135/159/126. Two strains harbored blaTEM-1/159. All isolates were resistant to amoxicillin, amoxicillin-clavulanic acid, cephalosporins, doxycycline, and trimethoprim-sulfamethoxazole, and all them were collected from the same hospital. Cockroaches are frequently present in hospital environment; humidity and water damage are important factors of proliferation of insects (48). In general, high relative humidity and low temperature provide the best chances for cockroaches to survive (16). Most strains were isolated from the gut, which could be explained by the great adaptation of these organisms to the intestinal environment (21).
ESBL and mcr genes were frequently detected in several species of Enterobacteriaceae, including K. pneumoniae and E. coli, which are responsible for nosocomial infections in humans and are of particular concern for multidrug resistance (9, 35, 49). Our study describes the first detection of the coharboring of mcr-1 and blaESBL from cockroaches in nine E. coli strains of lineage ST648 isolated from a hospital. This E. coli lineage has emerged as a pandemic clone, and the association between ST648 and ESBL genes has been reported in companion animals (50–52). The presence of the ESBLs and mcr-1 genes in E. coli strains of lineage ST648 in cockroaches is a public health concern, since it could indicate an important and silent diffusion of this clinically significant clone in the human-animal environment interface (51).
The results obtained in several studies, including ours, should better explain a world in which human health, animal health, and the environment are directly or indirectly linked (24, 53, 54).
Cockroaches ingest fluids that can be contaminated with multidrug-resistant bacteria. These bacteria multiply in the digestive tract and are then transferred to the intestine or are regurgitated. As cockroaches share their habitat with animals and humans, transmission of antimicrobial resistance is, therefore, possible (55). Therefore, since cockroaches are a source of multidrug-resistant pathogens and a risk factor for dissemination of these bacteria to humans and/or other animal species in contact with humans, methods to control cockroaches are needed to decrease the spread of nosocomial and foodborne bacteria (16). Surveillance of hospital infections should be complemented by monitoring disinfectants used in the hospital as well as evaluation of infestation of the hospital environment and resistance of cockroaches to biocides, and all water points in intensive care units must be closed (56).
We conducted the first survey of mcr-1 in ESBL-producing E. coli isolates from hospital cockroaches. Our results further highlight the possibility that mcr-1 may enter humans via cockroach contamination and thereby threaten public health. Our results show that these cockroaches are an important reservoir of colistin-resistant E. coli and carriers of other additional risk genes such as blaESBL, hence the importance of strengthening prevention strategies and of strictly respecting hygiene measures in order to control their distribution and spread in Tunisia.
MATERIALS AND METHODS
Collection and identification of cockroaches.
A total of 115 adult cockroaches were collected from July 2017 to June 2018 in Tunisia.
Forty-eight cockroaches were obtained from collective catering, 35 cockroaches were trapped from different parts of houses (kitchens, garden, bathrooms, and toilets), and the remaining 32 were obtained from different parts of a hospital (kitchen and emergency department).
Cockroaches were collected using sterile and separate flasks and transported to our laboratory for bacteriological analysis. After immobilization by freezing at 0°C for 5 min, species identification was done according to the method of Harwood et al. (57).
To wash the external body surface, sterile normal saline (3 mL) was added to each flask and the cockroaches were vigorously washed and transferred to secondary sterile flasks. After washing and decontamination with 70% alcohol, the gut of the cockroaches was dissected and crushed aseptically in a sterile pestle and mortar. The gut was then maintained in a 3 mL sterile physiological saline solution for 5 min to produce a homogenate sample.
Bacterial isolation.
After the external and gut homogenates were prepared and placed in test tubes containing normal saline solution, each sample (1 mL) was preenriched for 24 h at 37°C in buffered peptone water (BPW) and buffered peptone water supplemented with cefotaxime (CTX; 1 mg/L) to isolate cefotaxime-resistant bacteria.
After overnight incubation, the samples were inoculated on MacConkey agar (MCA) and MacConkey agar with 1 mg/L of cefotaxime and then incubated for 24 h at 37°C. Colonies showing Enterobacteriaceae morphology were recovered and identified by matrix-assisted laser desorption ionization–time of flight mass spectrometry.
Antimicrobial susceptibility testing and ESBL identification.
Susceptibility testing was performed using a standard disk diffusion technique according to the recommendations of the Antibiogram Committee of the French Society for Microbiology, 2019. The antimicrobial agents tested included amoxicillin, amoxicillin-clavulanic acid, piperacillin-tazobactam, cephalothin, ceftriaxone, cefepime, ertapenem, imipenem, amikacin, gentamicin, ciprofloxacin, fosfomycin, nitrofurantoin, doxycycline, trimethoprim-sulfamethoxazole, and colistin.
After overnight incubation, the diameter of the zone of inhibition around the discs was measured and interpreted as susceptible, intermediate, and resistant.
To screen for the production of ESBLs, one cefotaxime-resistant isolate obtained per sample was selected and screened for the ESBL phenotype by the double-disk synergy test (DDST).
The detection of colistin-resistant isolates was confirmed for all isolates showing an intermediate and resistant phenotype by disk diffusion by the determination of the MICs of colistin with the UMIC test (Biocentric, Bandol, France). Otherwise, the MICs were determined using Etest strips (bioMérieux, Marcy l’Etoile, France) for the isolates showing carbapenem-non-susceptibility detected with ertapenem and imipenem. The results were interpreted according to CA-SFM/EUCAST (2019) breakpoints.
Characterization of beta-lactamase and colistin resistance genes.
After DNA extraction for all samples by the boiling method, the presence of resistance genes was screened using real-time PCR, with specific primers (ESBL-encoding genes: blaCTX-M, blaSHV, blaTEM; carbapenem resistance-encoding genes: blaKPC, blaNDM, blaVIM, and blaOXA-48) (58).
All colistin-resistant isolates were screened for the presence of mcr-1, mcr-2, mcr-3, mcr-4, mcr-5, and mcr-8 genes (59).
The positive strains detected by real-time PCR were submitted to conventional PCR to confirm the previous results. Subsequently, products were purified and sequenced using the BigDye terminator chemistry on an ABI 3130XL automated sequencer (Thermo Fisher Scientific, Waltham, MA, USA). The obtained sequences were analyzed using ChromasPro and compared to the ARG-ANNOT database and NCBI GenBank database (https://www.ncbi.nlm.nih.gov/).
Phylogenetic analysis.
The determination of a genetic relationship among isolates was done by multilocus sequence typing (MLST) using the seven housekeeping genes. Each gene was amplified and sequenced for each E. coli (adk, fumC, gyr, icd, mdh, purA, and recA) and K. pneumoniae (gapA, infB, mdh, pgi, phoE, rpoB, tonB) strain, and then the sequence types (ST) were assigned in accordance with the online database (https://bigsdb.pasteur.fr/) for E. coli strains; for the strains of K. pneumoniae, we used https://pubmlst.org/.
Data availability.
The accessions numbers of ST found in our study are available in Table S1.
The MLST sequences are available on MLST web database (https://bigsdb.pasteur.fr/) for K. pneumoniae and on https://pubmlst.org/ for E. coli strains.
ACKNOWLEDGMENTS
This work was supported by the French Government under the Investissements d’Avenir (Investments for the Future) program managed by the Agence Nationale de la Recherche (ANR; fr: National Agency for Research) (reference: Méditerranée Infection 10-IAHU-03).
This work was supported by Région Provence-Alpes-Côte d’Azur and European funding FEDER PRIMI.
This work was supported by the research project PEER 7-349 funded by the USAID “Monitoring of antimicrobial resistance of bacteria for a better health of animals in Tunisia.”
We are very grateful to Sihem Chebil for technical assistance. We also thank Mariem Saidani, Assia Kassah, Jouda Kchok, Reem Abdallah, and Rym Lalaoui for their help during the study.
We declare no competing financial interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Jean Marc Rolain, Email: jean-marc.rolain@univ-amu.fr.
Courtney J. Robinson, Howard University
REFERENCES
- 1.Doi Y, Paterson DL. 2015. Carbapenemase-producing Enterobacteriaceae. Semin Respir Crit Care Med 36:74–84. doi: 10.1055/s-0035-1544208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dortet L, Poirel L, Nordmann P. 2012. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob Agents Chemother 56:6437–6440. doi: 10.1128/AAC.01395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Randall LP, Clouting C, Horton RA, Coldham NG, Wu G, Clifton-Hadley FA, Davies RH, Teale CJ. 2011. Prevalence of Escherichia coli carrying extended-spectrum beta-lactamases (CTX-M and TEM-52) from broiler chickens and turkeys in Great Britain between 2006 and 2009. J Antimicrob Chemother 66:86–95. doi: 10.1093/jac/dkq396. [DOI] [PubMed] [Google Scholar]
- 4.Das N, Madhavan J, Selvi A, Das D. 2019. An overview of cephalosporin antibiotics as emerging contaminants: a serious environmental concern. 3 Biotech 9:231. doi: 10.1007/s13205-019-1766-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfeifer Y, Cullik A, Witte W. 2010. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol 300:371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Pitout JD, Laupland KB. 2008. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 8.Nakayama T, Kumeda Y, Kawahara R, Yamaguchi T, Yamamoto Y. 2018. Carriage of colistin-resistant, extended-spectrum beta-lactamase-producing Escherichia coli harboring the mcr-1 resistance gene after short-term international travel to Vietnam. Infect Drug Resist 11:391–395. doi: 10.2147/IDR.S153178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuda A, Usui M, Okubo T, Tagaki C, Sukpanyatham N, Tamura Y. 2018. Co-harboring of cephalosporin (bla)/colistin (mcr) resistance genes among Enterobacteriaceae from flies in Thailand. FEMS Microbiol Lett 365:fny178. doi: 10.1093/femsle/fny178. [DOI] [PubMed] [Google Scholar]
- 10.Haenni M, Poirel L, Kieffer N, Chatre P, Saras E, Metayer V, Dumoulin R, Nordmann P, Madec JY. 2016. Co-occurrence of extended spectrum beta lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis 16:281–282. doi: 10.1016/S1473-3099(16)00007-4. [DOI] [PubMed] [Google Scholar]
- 11.Zheng B, Huang C, Xu H, Guo L, Zhang J, Wang X, Jiang X, Yu X, Jin L, Li X, Feng Y, Xiao Y, Li L. 2017. Occurrence and genomic characterization of ESBL-producing, MCR-1-harboring Escherichia coli in farming soil. Front Microbiol 8:2510. doi: 10.3389/fmicb.2017.02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, Huo C, Sun Y, Zhou Y, Xiong Y, Zhao Z, Zhou Q, Sha L, Zhang B, Chen Y. 2019. Emergence and molecular characterization of multidrug-resistant Klebsiella pneumoniae isolates harboring bla CTX-M-15 extended-spectrum beta-lactamases causing ventilator-associated pneumonia in China. Infect Drug Resist 12:33–43. doi: 10.2147/IDR.S189494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zarei O, Shokoohizadeh L, Hossainpour H, Alikhani MY. 2018. Molecular analysis of Pseudomonas aeruginosa isolated from clinical, environmental and cockroach sources by ERIC-PCR. BMC Res Notes 11:668. doi: 10.1186/s13104-018-3765-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anacarso I, Iseppi R, Sabia C, Messi P, Condo C, Bondi M, de Niederhausern S. 2016. Conjugation-mediated transfer of antibiotic-resistance plasmids between Enterobacteriaceae in the digestive tract of Blaberus craniifer (Blattodea: Blaberidae). J Med Entomol 53:591–597. doi: 10.1093/jme/tjw005. [DOI] [PubMed] [Google Scholar]
- 15.Tilahun B, Worku B, Tachbele E, Terefe S, Kloos H, Legesse W. 2012. High load of multi-drug resistant nosocomial neonatal pathogens carried by cockroaches in a neonatal intensive care unit at Tikur Anbessa specialized hospital, Addis Ababa, Ethiopia. Antimicrob Resist Infect Control 1:12. doi: 10.1186/2047-2994-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donkor ES. 2019. Nosocomial pathogens: an in-depth analysis of the vectorial potential of cockroaches. Trop Med Infect Dis 4:14. doi: 10.3390/tropicalmed4010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fakoorziba MR, Eghbal F, Hassanzadeh J, Moemenbellah-Fard MD. 2010. Cockroaches (Periplaneta americana and Blattella germanica) as potential vectors of the pathogenic bacteria found in nosocomial infections. Ann Trop Med Parasitol 104:521–528. doi: 10.1179/136485910X12786389891326. [DOI] [PubMed] [Google Scholar]
- 18.Goroncy-Bermes P, Koburger T, Meyer B. 2010. Impact of the amount of hand rub applied in hygienic hand disinfection on the reduction of microbial counts on hands. J Hosp Infect 74:212–218. doi: 10.1016/j.jhin.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Palmore TN, Henderson DK. 2013. Managing transmission of carbapenem-resistant enterobacteriaceae in healthcare settings: a view from the trenches. Clin Infect Dis 57:1593–1599. doi: 10.1093/cid/cit531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moges F, Eshetie S, Endris M, Huruy K, Muluye D, Feleke T, G/Silassie F, Ayalew G, Nagappan R. 2016. Cockroaches as a source of high bacterial pathogens with multidrug resistant strains in Gondar Town, Ethiopia. Biomed Res Int 2016:1–6. doi: 10.1155/2016/2825056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loucif L, Gacemi-Kirane D, Cherak Z, Chamlal N, Grainat N, Rolain JM. 2016. First report of German cockroaches (Blattella germanica) as reservoirs of CTX-M-15 extended-spectrum-beta-lactamase- and OXA-48 carbapenemase-producing Enterobacteriaceae in Batna University Hospital, Algeria. Antimicrob Agents Chemother 60:6377–6380. doi: 10.1128/AAC.00871-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walsh TR. 2018. A one-health approach to antimicrobial resistance. Nat Microbiol 3:854–855. doi: 10.1038/s41564-018-0208-5. [DOI] [PubMed] [Google Scholar]
- 23.Bordier M, Binot A, Pauchard Q, Nguyen DT, Trung TN, Fortane N, Goutard FL. 2018. Antibiotic resistance in Vietnam: moving towards a One Health surveillance system. BMC Public Health 18:1136. doi: 10.1186/s12889-018-6022-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obeng-Nkrumah N, Labi AK, Blankson H, Awuah-Mensah G, Oduro-Mensah D, Anum J, Teye J, Kwashie SD, Bako E, Ayeh-Kumi PF, Asmah R. 2019. Household cockroaches carry CTX-M-15-, OXA-48- and NDM-1-producing enterobacteria, and share beta-lactam resistance determinants with humans. BMC Microbiol 19:272. doi: 10.1186/s12866-019-1629-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cotton MF, Wasserman E, Pieper CH, Theron DC, van Tubbergh D, Campbell G, Fang FC, Barnes A J. 2000. Invasive disease due to extended spectrum beta-lactamase-producing Klebsiella pneumoniae in a neonatal unit: the possible role of cockroaches. J Hosp Infect 44:13–17. doi: 10.1053/jhin.1999.0650. [DOI] [PubMed] [Google Scholar]
- 26.Doosti A, Pourabbas M, Arshi A, Chehelgerdi M, Kabiri H. 2015. TEM and SHV genes in Klebsiella pneumoniae isolated from cockroaches and their antimicrobial resistance pattern. Osong Public Health Res Perspect 6:3–8. doi: 10.1016/j.phrp.2014.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mediavilla JR, Patrawalla A, Chen L, Chavda KD, Mathema B, Vinnard C, Dever LL, Kreiswirth BN. 2016. Colistin- and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. mBio 7. doi: 10.1128/mBio.01191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuda A, Sato T, Shinagawa M, Takahashi S, Asai T, Yokota SI, Usui M, Tamura Y. 2018. High prevalence of mcr-1, mcr-3 and mcr-5 in Escherichia coli derived from diseased pigs in Japan. Int J Antimicrob Agents 51:163–164. doi: 10.1016/j.ijantimicag.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, Zhang S, Shen J, Shen Z, Wang Y. 2018. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect 7:122. doi: 10.1038/s41426-018-0124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. 2020. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect 9:508–516. doi: 10.1080/22221751.2020.1732231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grami R, Mansour W, Mehri W, Bouallegue O, Boujaafar N, Madec JY, Haenni M. 2016. Impact of food animal trade on the spread of mcr-1-mediated colistin resistance, Tunisia, July 2015. Euro Surveill 21:30144. doi: 10.2807/1560-7917.ES.2016.21.8.30144. [DOI] [PubMed] [Google Scholar]
- 32.Maamar E, Alonso CA, Hamzaoui Z, Dakhli N, Abbassi MS, Ferjani S, Saidani M, Boutiba-Ben Boubaker I, Torres C. 2018. Emergence of plasmid-mediated colistin-resistance in CMY-2-producing Escherichia coli of lineage ST2197 in a Tunisian poultry farm. Int J Food Microbiol 269:60–63. doi: 10.1016/j.ijfoodmicro.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 33.Hassen B, Abbassi MS, Ruiz-Ripa L, Mama OM, Hassen A, Torres C, Hammami S. 2020. High prevalence of mcr-1 encoding colistin resistance and first identification of blaCTX-M-55 in ESBL/CMY-2-producing Escherichia coli isolated from chicken faeces and retail meat in Tunisia. Int J Food Microbiol 318:108478. doi: 10.1016/j.ijfoodmicro.2019.108478. [DOI] [PubMed] [Google Scholar]
- 34.Saidani M, Messadi L, Chaouechi A, Tabib I, Saras E, Soudani A, Daaloul-Jedidi M, Mamlouk A, Ben Chehida F, Chakroun C, Madec JY, Haenni M. 2019. High genetic diversity of Enterobacteriaceae clones and plasmids disseminating resistance to extended-spectrum cephalosporins and colistin in healthy chicken in Tunisia. Microb Drug Resist 25:1507–1513. doi: 10.1089/mdr.2019.0138. [DOI] [PubMed] [Google Scholar]
- 35.Saidani M, Messadi L, Sahmin E, Zouaoui S, Soudani A, Daaloul-Jedidi M, Mamlouk A, Chehida FB, Madec JY, Haenni M. 2019. ESBL- and mcr-1-producing Escherichia coli in veal calves in Tunisia. J Glob Antimicrob Resist 19:104–105. doi: 10.1016/j.jgar.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 36.Hassen B, Saloua B, Abbassi MS, Ruiz-Ripa L, Mama OM, Hassen A, Hammami S, Torres C. 2019. mcr-1 encoding colistin resistance in CTX-M-1/CTX-M-15- producing Escherichia coli isolates of bovine and caprine origins in Tunisia. First report of CTX-M-15-ST394/D E. coli from goats. Comp Immunol Microbiol Infect Dis 67:101366. doi: 10.1016/j.cimid.2019.101366. [DOI] [PubMed] [Google Scholar]
- 37.Zhang H, Seward CH, Wu Z, Ye H, Feng Y. 2016. Genomic insights into the ESBL and MCR-1-producing ST648 Escherichia coli with multi-drug resistance. Sci Bull (Beijing) 61:875–878. doi: 10.1007/s11434-016-1086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang RS, Feng Y, Lv XY, Duan JH, Chen J, Fang LX, Xia J, Liao XP, Sun J, Liu YH. 2016. Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single muscovy duck (Cairina moschata). Antimicrob Agents Chemother 60:6899–6902. doi: 10.1128/AAC.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonnevend A, Ghazawi A, Alqahtani M, Shibl A, Jamal W, Hashmey R, Pal T. 2016. Plasmid-mediated colistin resistance in Escherichia coli from the Arabian Peninsula. Int J Infect Dis 50:85–90. doi: 10.1016/j.ijid.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 40.Chenouf NS, Carvalho I, Messai CR, Ruiz-Ripa L, Mama OM, Titouche Y, Zitouni A, Hakem A, Torres C. 2020. Extended spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae from broiler liver in the Center of Algeria, with detection of CTX-M-55 and B2/ST131-CTX-M-15 in Escherichia coli. Microb Drug Resist 27:268–276. doi: 10.1089/mdr.2020.0024. [DOI] [PubMed] [Google Scholar]
- 41.Dong F, Zhang Y, Yao K, Lu J, Guo L, Lyu S, Yang Y, Wang Y, Zheng H, Song W, Liu G. 2018. Epidemiology of carbapenem-resistant Klebsiella pneumoniae bloodstream infections in a Chinese children’s hospital: predominance of New Delhi metallo-beta-lactamase-1. Microb Drug Resist 24:154–160. doi: 10.1089/mdr.2017.0031. [DOI] [PubMed] [Google Scholar]
- 42.Mohsin M, Raza S, Schaufler K, Roschanski N, Sarwar F, Semmler T, Schierack P, Guenther S. 2017. High prevalence of CTX-M-15-type ESBL-producing E. coli from migratory avian species in Pakistan. Front Microbiol 8:2476. doi: 10.3389/fmicb.2017.02476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higashino M, Murata M, Morinaga Y, Akamatsu N, Matsuda J, Takeda K, Kaku N, Kosai K, Uno N, Hasegawa H, Yanagihara K. 2017. Fluoroquinolone resistance in extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in a Japanese tertiary hospital: silent shifting to CTX-M-15-producing K. pneumoniae. J Med Microbiol 66:1476–1482. doi: 10.1099/jmm.0.000577. [DOI] [PubMed] [Google Scholar]
- 44.Matos AP, Cayo R, Almeida LGP, Streling AP, Nodari CS, Martins W, Narciso AC, Silva RM, Vasconcelos ATR, Gales AC. 2019. Genetic characterization of plasmid-borne bla OXA-58 in distinct Acinetobacter species. mSphere 4. doi: 10.1128/mSphere.00376-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alcala L, Alonso CA, Simon C, Gonzalez-Esteban C, Oros J, Rezusta A, Ortega C, Torres C. 2016. Wild birds, frequent carriers of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli of CTX-M and SHV-12 types. Microb Ecol 72:861–869. doi: 10.1007/s00248-015-0718-0. [DOI] [PubMed] [Google Scholar]
- 46.Maamar E, Hammami S, Alonso CA, Dakhli N, Abbassi MS, Ferjani S, Hamzaoui Z, Saidani M, Torres C, Boutiba-Ben Boubaker I. 2016. High prevalence of extended-spectrum and plasmidic AmpC beta-lactamase-producing Escherichia coli from poultry in Tunisia. Int J Food Microbiol 231:69–75. doi: 10.1016/j.ijfoodmicro.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 47.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31. doi: 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peden D, Reed CE. 2010. Environmental and occupational allergies. J Allergy Clin Immunol 125:S150–S160. doi: 10.1016/j.jaci.2009.10.073. [DOI] [PubMed] [Google Scholar]
- 49.Fournier C, Aires-de-Sousa M, Nordmann P, Poirel L. 2019. Occurrence of CTX-M-15 and MCR-1-producing Enterobacterales in pigs, Portugal; evidences of direct links with antibiotic selective pressure. Int J Antimicrob Agents 55:105802. doi: 10.1016/j.ijantimicag.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 50.Ewers C, Bethe A, Stamm I, Grobbel M, Kopp PA, Guerra B, Stubbe M, Doi Y, Zong Z, Kola A, Schaufler K, Semmler T, Fruth A, Wieler LH, Guenther S. 2014. CTX-M-15-D-ST648 Escherichia coli from companion animals and horses: another pandemic clone combining multiresistance and extraintestinal virulence? J Antimicrob Chemother 69:1224–1230. doi: 10.1093/jac/dkt516. [DOI] [PubMed] [Google Scholar]
- 51.Fernandes MR, Sellera FP, Moura Q, Gaspar VC, Cerdeira L, Lincopan N. 2018. International high-risk clonal lineages of CTX-M-producing Escherichia coli F-ST648 in free-roaming cats, South America. Infect Genet Evol 66:48–51. doi: 10.1016/j.meegid.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 52.Sherchan JB, Hayakawa K, Miyoshi-Akiyama T, Ohmagari N, Kirikae T, Nagamatsu M, Tojo M, Ohara H, Sherchand JB, Tandukar S. 2015. Clinical epidemiology and molecular analysis of extended-spectrum-beta-lactamase-producing Escherichia coli in Nepal: characteristics of sequence types 131 and 648. Antimicrob Agents Chemother 59:3424–3432. doi: 10.1128/AAC.00270-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Q, Wang Y, Hulth A, Xiao Y, Nilsson LE, Li X, Bi Z, Liu Y, Yin H, Luo Y, Nilsson M, Sun C, Zhu Y, Zheng B, Chen B, Sun P, Ding L, Xia X, Ottoson J, Löfmark S, Dyar OJ, Impact Consortium, Börjesson S, Lundborg CS. 2018. Study protocol for One Health data collections, analyses and intervention of the Sino-Swedish integrated multisectoral partnership for antibiotic resistance containment (IMPACT). BMJ Open 8:e017832. doi: 10.1136/bmjopen-2017-017832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trinh P, Zaneveld JR, Safranek S, Rabinowitz PM. 2018. One Health relationships between human, animal, and environmental microbiomes: a mini-review. Front Public Health 6:235. doi: 10.3389/fpubh.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fukuda A, Usui M, Okamura M, Dong-Liang H, Tamura Y. 2019. Role of flies in the maintenance of antimicrobial resistance in farm environments. Microb Drug Resist 25:127–132. doi: 10.1089/mdr.2017.0371. [DOI] [PubMed] [Google Scholar]
- 56.Saitou K, Furuhata K, Kawakami Y, Fukuyama M. 2009. Biofilm formation abilities and disinfectant-resistance of Pseudomonas aeruginosa isolated from cockroaches captured in hospitals. Biocontrol Sci 14:65–68. doi: 10.4265/bio.14.65. [DOI] [PubMed] [Google Scholar]
- 57.Harwood RF, James MT, Herms WB. 1979. Entomology in human and animal health. Macmillan, New York, NY. [Google Scholar]
- 58.Christophy R, Osman M, Mallat H, Achkar M, Ziedeh A, Moukaddem W, Dabboussi F, Hamze M. 2017. Prevalence, antibiotic susceptibility and characterization of antibiotic resistant genes among carbapenem-resistant Gram-negative bacilli and yeast in intestinal flora of cancer patients in North Lebanon. J Infect Public Health 10:716–720. doi: 10.1016/j.jiph.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 59.Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, Hansen IM, Guerra B, Malorny B, Borowiak M, Hammerl JA, Battisti A, Franco A, Alba P, Perrin-Guyomard A, Granier SA, De Frutos Escobar C, Malhotra-Kumar S, Villa L, Carattoli A, Hendriksen RS. 2018. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill 23:17-00672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM00036-21_Supp_1_seq3.pdf, PDF file, 0.1 MB (67.2KB, pdf)
Data Availability Statement
The accessions numbers of ST found in our study are available in Table S1.
The MLST sequences are available on MLST web database (https://bigsdb.pasteur.fr/) for K. pneumoniae and on https://pubmlst.org/ for E. coli strains.