ABSTRACT
There is increasing awareness that archaea are interrelated with human diseases (including cancer). Archaea utilize unique metabolic pathways to produce a variety of metabolites that serve as a direct link to host-microbe interactions. However, knowledge on the diversity of human-associated archaea is still extremely limited, and less is known about the pathological effects of their metabolites to the tumor microenvironment and carcinogenesis. In the present study, we performed a large-scale analysis of archaea and their cancer-related metabolites across different body sites using >44,000 contigs with length >1,000 bp. Taxonomy annotation revealed that the occurrence and diversity of archaea are higher in two body sites, the gut and the oral cavity. Unlike other human-associated microbes, the nonmetric multidimensional scaling (NMDS) and permutational multivariate analysis of variance (PERMANOVA) analyses have shown no difference of archaeal compositions between Easterners and Westerners. Likewise, protein annotation suggests that genes encoding cancer-related metabolites (e.g., short-chain fatty acids and polyamines) are more prevalent and diverse in gut and oral samples. Archaea carrying these metabolites are restricted to Euryarchaeota and the TACK superphylum (Thaumarchaeota, Aigarchaeota, Crenarchaeota, and Korarchaeota), especially methanogenic archaea, such as Methanobacteria.
IMPORTANCE More evidence suggests that archaea are associated with human disease, including cancer. Here, we present the first framework of the diversity and distribution of human-associated archaea across human body sites, such as gut and oral cavity, using long contigs. Furthermore, we unveiled the potential archaeal metabolites linking to different lineages that might influence the tumor microenvironment and carcinogenesis. These results could open a new door to the guidance of diagnosing cancer and developing new treatment strategies.
KEYWORDS: archaea, metabolites, carcinogenesis, gut, oral, contig
INTRODUCTION
Accumulating evidence reveals that microbial metabolites produced within the human body serve as an important factor, directly influencing the progression of pathological conditions such as cancer (1–4). As a double-edged sword, some microbial metabolites, such as butyrate, exhibit significant functions in the suppression of inflammation and cancer (5, 6), whereas others, such as secondary bile acids, promote carcinogenesis (7). As a consequence, investigations on metabolites of human microbiome may shed light on the elusive interplay between microbe and human. Despite the majority of microbes in humans being bacteria (8), the emerging evidence of the interactions of minor communities, such as archaea, fungi, parasite, and viruses, with human health and disease suggests an important role of archaea to human individuals (9–14).
Similar to bacteria, archaeal lineage is an important component of life in diverse ecosystems, such as the human body (8, 15). The first human-derived archaeal isolate was obtained from the gastrointestinal tract more than 40 years ago (16–19). Since then, archaea have been isolated from other body sites, including oral mucosa (20), subgingival plaque (20), and human colostrum and milk (21). Up until now, these isolates have been restricted to methanogenic and halophilic archaea, e.g., Methanobrevibacter smithii, Methanomassiliicoccus luminyensis, Methanomethylophilus alvus, and Haloferax massiliensis. Recent analyses with high-throughput sequencing amplicons reveal a more diverse archaeal community present across different human body sites (8, 22). Although there is no direct evidence linking archaea to certain human morbidities (10, 12), archaea share some characteristics with known pathogens, such as ample access to a host and capabilities to colonize and coexist with other species in a host (through components such as membrane-bound adhesin-like proteins) (12, 23). Furthermore, the abundance of halophilic and/or methanogenic archaea has proven to be positively or negatively correlated with human diseases, e.g., periodontal disease (24) and colorectal cancer (25), suggesting a potential effect of archaea on human disease states. The proportion of archaea in the microbial community can increase up to 25% in certain diseases (26). On the contrary, some archaea are beneficial to human health. For instance, the trimethylamine-degrading methanogenic archaea can serve as live biotherapeutic products (also known as “archaebiotics”) to prevent cardiovascular diseases (27). It is therefore important and meaningful to determine the underlying interactions between the archaeal component of the human microbiome and diseases, especially cancer (28).
The relative abundance of archaea is lower than their bacterial counterparts in the human body, and only a minority of archaea can be cultivated or isolated for in-depth analysis, due to their unique metabolic and physiological characteristics (15, 29). The development of high-throughput sequencing techniques in recent years has facilitated the availability and analysis of archaeal sequences. In the present study, we utilized more than 44,000 metagenome-assembled contiguous sequences (contigs) with length of >1,000 bp from publicly available databases to investigate the diversity of archaea, especially the cancer-related metabolites in different body sites. Microbial metabolites are believed to be the essential bridge connecting microbiota, tumor microenvironment, and cancer development (28, 30). Our study provides a framework for better investigating the correlation of archaeal metabolites with cancer.
RESULTS
Composition of archaea in/on human body.
First, we obtained 32,451,655 contigs from 5,632 samples available in public databases (Table S1). These data sets originated from different body sites, including buccal mucosa, dorsum of tongue, external naris, gut, gingiva, hard palate, retroauricular crease, palatine tonsil, cubital fossa, saliva, throat, and vagina. After taxonomic assignment, we obtained 44,760 archaeal contigs (>1,000 bp) from 1,818 samples, with numbers ranging from 183 to 14,994 across different body sites. Specifically, the occurrence of archaeal contigs was higher in body sites like gut samples from Chinese individuals (gut-Chinese; 100%, i.e., present in 100% of the investigated samples), dorsum of tongue (99.1%), gut samples from the Human Biome Project (gut-HMP; 97.2%), and gingiva (94.4%), whereas the average number of archaeal contigs was higher in samples retrieved from right retroauricular crease (105.1), left retroauricular crease (84.3), gut-Chinese (54.9), and gut-HMP (53.4; Fig. 1A and Table S1). Although archaea from the left/right retroauricular creases were almost shared with gut-HMP and external naris at the phylum/class level with a Bray-Curtis dissimilarity of ∼0.05 (Fig. 1B), the archaeal α-diversity of samples from retroauricular creases was much higher than that of those from other body sites (P < 0.001) at the species level (Fig. 1C).
FIG 1.
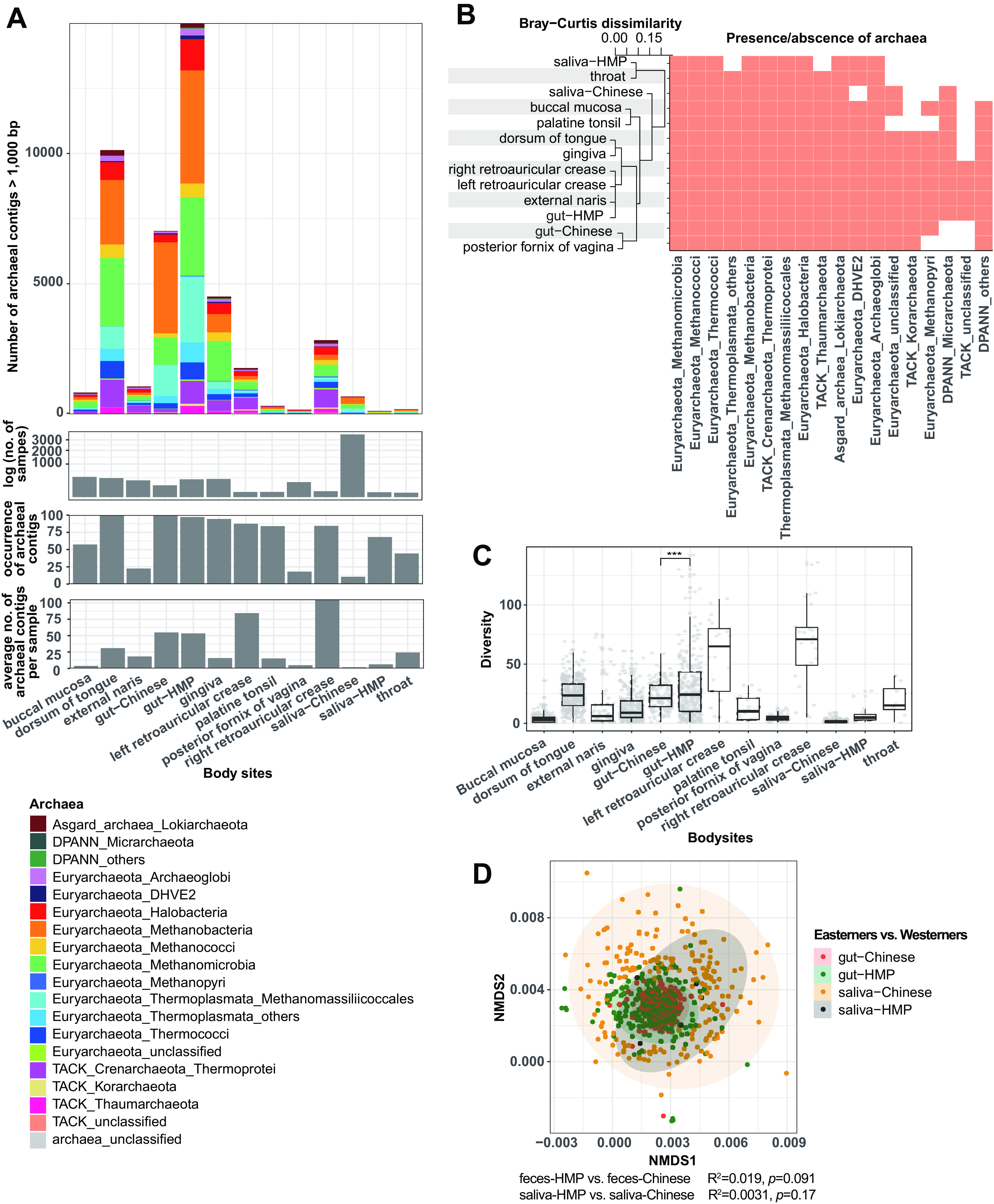
Diversity and distribution of archaea in humans. (A) Number of different archaeal contigs (>1,000 bp) in each body site. The bottom three histograms summarize the total number of samples used for analysis, occurrence of archaeal contigs, and average number of archaeal contigs per sample, respectively. (B) Presence/absence of archaea and their Bray-Curtis dissimilarity across body sites at the species level. (C) Diversity of archaea across different body sites at the species level. ***, P < 0.001. (D) NMDS analysis of the archaea composition at different body sites.
Similarities and differences between Easterners and Westerners.
Important factors influencing the presence and diversity of microorganisms include geography and ethnicity (related to aspects such as host genetics, life history, and diet) (31, 32). Hence, we compared the diversity and similarity of archaeal communities from gut and saliva samples of Easterners and Westerners. We observed that samples from Westerners (gut-HMP) had an archaeal diversity higher than that of samples from Easterners (gut-Chinese; P < 0.001), whereas no difference in archaeal diversity was observed between the saliva-HMP and saliva-Chinese (Fig. 1C). Further, analyses of archaeal composition using nonmetric multidimensional scaling (NMDS) and permutational multivariate analysis of variance (PERMANOVA) showed no difference for fecal (R2 = 0.019, P = 0.091) and saliva (R2 = 0.0031, P = 0.17) samples between Easterners and Westerners at the species level (Fig. 1D).
Archaea and their metabolites related to carcinogenesis.
To determine the potential connections of archaeal metabolites to carcinogenesis, the archaeal contigs were processed to annotate the functional genes with numerous databases and software (see Materials and Methods). We found several types of archaeal metabolites potentially linked to carcinogenesis, including short-chain fatty acids (SCFA; i.e., acetate and lactic acid) (3, 33), indoles (3, 33), polyamines (i.e., cadaverine, putrescine, and spermidine) (3, 33), ammonia (33), secondary bile acids (2°BA) (3, 33), methylglyoxal (34–38), acetaldehyde (3), γ-aminobutyric acid (GABA) (39), and arsenic (40), as well as other metabolites, including H2 and trimethylamine (TMA), that could influence the production of carcinogens H2S (3) and trimethylamine N-oxide (TMAO) (41) (Fig. 2 and Fig. S1 and S2). The known effects of these oncogenic metabolites on the host include influencing microbiota modulation, inflammation, energy supply, and reactive oxygen species (ROS) production, as well as affecting host signaling pathways through modification of histones or essential enzymes in metabolic pathways (Table 1). Although identified in diverse body sites, genes encoding these metabolites, like methylglyoxal and acetaldehyde, were more prevalent and diverse in oral (buccal mucosa, dorsum of tongue, and saliva-Chinese) and gut (gut-Chinese and gut-HMP) samples (Fig. 2), representing 64.7% and 100% of the investigated metabolites, respectively. On the other hand, archaea in other body sites, such as retroauricular creases, throat, and vagina, were more likely to produce polyamines (Fig. 2).
FIG 2.
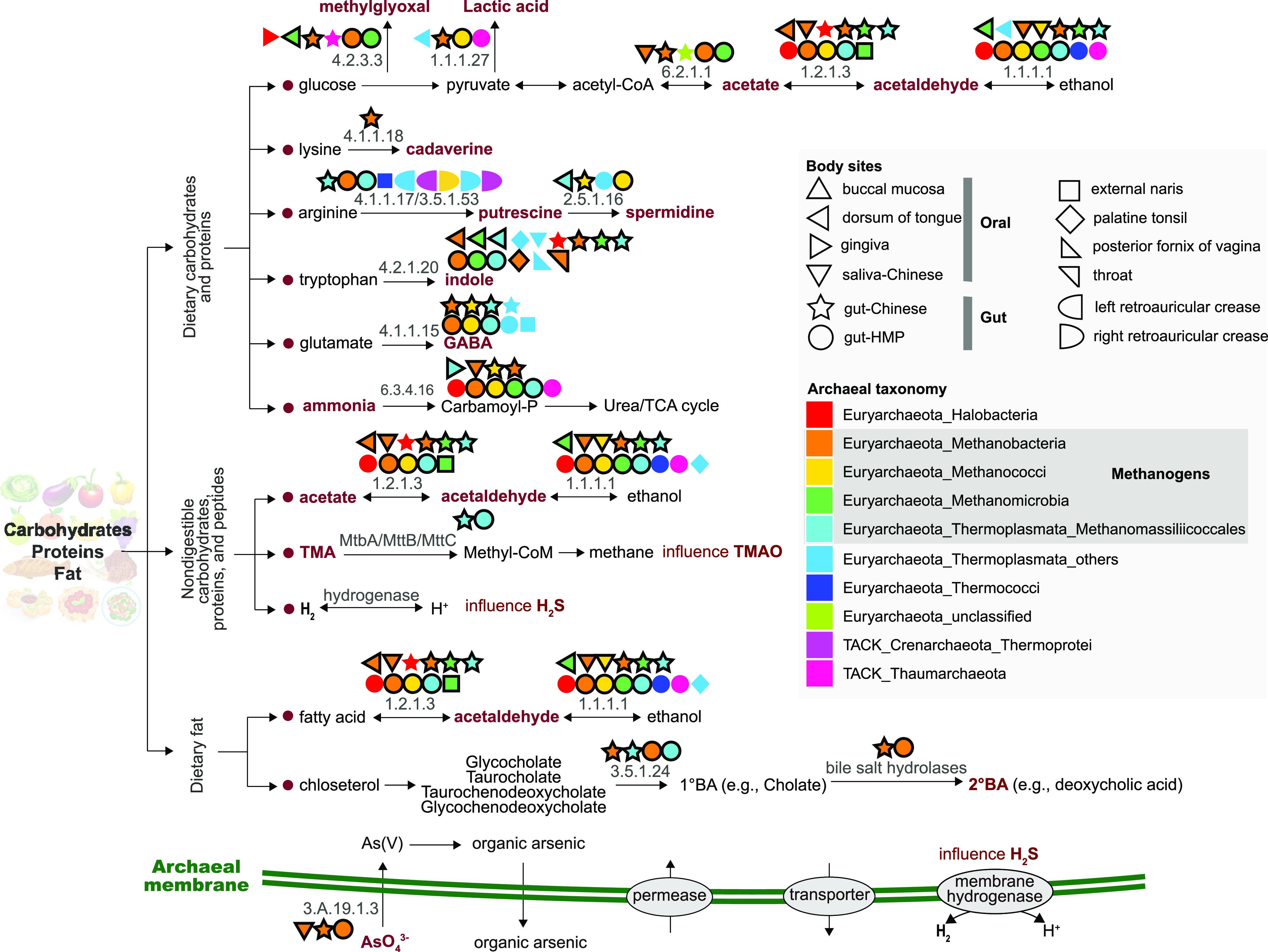
Archaeal metabolites that are directly or indirectly involved in the initiation and/or progression of carcinogenesis. The pertinent enzymes of the metabolites are also shown in the figure. Symbol shape and color represent different body sites and archaeal taxa, respectively. Symbols with black borders denote methanogens. GABA, γ-aminobutyric acid; TMA, trimethylamine; TMAO, trimethylamine N-oxide; 1°BA, primary bile acid; 2°BA, secondary bile acid. Descriptions of enzymes are available in Table S3. Detailed information of the presence of genes and those encoding hydrogenases and TCDB is available in Fig. S1 and S2 and Table S4, respectively.
TABLE 1.
Summary of major archaeal metabolites and their associations with carcinogenesis
| Archaeal metabolites | Known effects on hosta |
|---|---|
| Acetate | Anti-inflammation, tumor proliferation, intestinal barrier function |
| Lactic acid | ROS production, intestinal barrier function |
| Polyamines (cadaverine, putrescine, and spermidine) | Inflammation, ROS production, genotoxicity, DNA repair/protection |
| Indole | DNA damage, anti-inflammatory |
| Acetaldehyde | Inflammation, DNA damage, aberrant signaling pathways |
| Methylglyoxal | Aberrant signaling pathways |
| GABA | Aberrant signaling pathways |
| Ammonia | ROS production, genotoxicity, tumor proliferation |
| TMA/TMAO | Inflammation, aberrant signaling pathways |
| 2°BA | Microbiota modulation, cellular differentiation, apoptosis, ROS production, genotoxicity |
| H2/H2S | DNA damage, inflammation, ROS production, genotoxicity |
| Inorganic arsenic | DNA damage, ROS production, genotoxicity |
To verify whether the abundance of genes encoding these metabolites is different between healthy individuals and cancer patients, we selected a representative data set for comparison. We uncovered that the abundance of genes encoding enzymes (EC 6.2.3.3, EC 6.2.1.1, EC 1.1.1.1, EC 4.1.1.18, EC 4.2.1.20, EC 4.1.1.15, bile acid hydrolase, and 3.A.19.1.3) for cancer-related metabolites was much higher (P < 0.05) in colorectal cancer patients than in healthy individuals (Fig. S3), suggesting the potential of these archaea-derived metabolites to carcinogenesis.
Archaea potentially producing these metabolites were classified into 10 lineages within the Euryarchaeota and the TACK superphylum (Fig. 2). Although other archaeal lineages, such as Lokiarchaeota (Asgard archaea) and Micrarchaeota (DPANN), were identified in human samples (Fig. 1), they were not discussed in the next-step analyses because these contigs encode mainly housekeeping genes (Table S2). Additionally, the proportion of these species is relatively low in human samples and can seldomly be retrieved through metagenomic amplification. Here, methanogens, including Methanobacteria, Methanococci, Methanomicrobia, and Methanomassiliicoccales, were more frequently associated with the production or consumption of these metabolites, and they may be exclusively responsible for the transformation of several metabolites (such as TMA and 2°BA) in the human body. Specifically, Methanobacteria were the most popular methanogen and carried genes for all of the investigated metabolites. In addition, the presence of metabolites produced by other archaeal lineages (such as Halobacteria, Thermococci, Thermoprotei, and Thaumarchaeota) may also be responsible for the cycling of metabolites related to carcinogenesis (Fig. 2 and Table 1). Further, cooccurrence analysis showed a positive correlation between methanogenic and halophilic archaea and the known cancer-related pathogen Helicobacter pylori (28) (Fig. S4).
In line with these metabolites, we identified the correlated membrane-bound transporters, such as the polyamine exports (PF00324 and PF07690) and 2°BA transporters (PF01758; Fig. S1B and Table S4). Genes encoding these transporters are diversely distributed across different body sites, ensuring that the archaeal metabolites can be transferred into the host cells or to their mutualistic partners, such as bacteria and fungi, and can further influence the host pathophysiological states directly and/or indirectly.
DISCUSSION
Microbial communities interact with human cells through the production and regulation of metabolites to maintain cellular metabolism and the immune and neuronal systems (42, 43). In the past decade, related investigations have highlighted the increasing risk of dysbiosis of the archaeal or bacterial microbiome associated with diseases such as colorectal cancer (25) and breast cancer (2). Archaea (specifically methanogenic archaea) have been indicated to be essential components and represent keystone species in metabolic processes in the human body (22, 44, 45). Although recent studies have revealed their correlations with diseases (11, 12, 25, 29), more research regarding different archaea and their potential pathological roles in carcinogenesis is urgently needed.
The interactions of archaea with cancer can be traced back to as early as the 1980s, when it was unveiled that Africans with higher levels of methanogenic archaea showed a lower risk for large bowel cancer (31). Lately, shotgun metagenomic analyses have indicated that other than the methanogenic archaea, the abundance of halophilic archaea was positively correlated with colorectal cancer (25), underlying their potential impact to humans. Although archaea such as Thaumarchaeota and other unclassified Euryarchaeota were also identified in or on human body (8, 22), to the best of our knowledge, their potentials with cancer have not been reported in depth. For the first time, we have identified genes encoding cancer-related metabolites affiliated with nonmethanogenic or nonhalophilic archaea, such as Thermococci, Thermoprotei, and Thaumarchaeota, at the contig level (Fig. 2). The Thaumarchaeota in the gut may participate in biochemical processes, such as the oxidation of candidate carcinogen ammonia (46, 47) as supplied from the deaminated proteinaceous material, while Thermococci and Thermoprotei may be more responsible for polyamine production (48). Recent reports using 16S rRNA genes have also revealed the presence of these archaea in the gastrointestinal tract, lung, and skin (25). These facts support our observation in this study. However, further molecular experiments are still needed to delineate the participation of these archaea in cancer progression.
The gut, oral cavity, skin, and vagina harbor a peculiar set of highly diverse microbial consortia, with diversity highest in gut and oral cavity (49–52). This phenomenon is also applicable to archaea, as we identified an archaeal diversity in oral and gut samples (gut, dorsum of tongue, and gingiva) higher than that in other body sites (Fig. 1). Similarly, genes encoding cancer-related metabolites have been detected in the oral cavity and gut in most cases, but they were seldomly identified in other body sites (Fig. 2). The decrease of beneficial metabolites or the increase of detrimental metabolites to an alarming level has been substantiated to be associated with cancer in the oral cavity and gut (49, 53). Thus, the equilibrium of archaea-related metabolites in the oral cavity and/or gut, especially the archaeon-specific TMA, may be pivotal to human health.
The dysbiosis of gut and oral microbiome is linked to other diseases of different systems, including diabetes (54), pneumonia (55), and cardiovascular disease (56). Meanwhile, recent gut-brain axis studies also observed a significant link between the gut microbiome and some complex diseases, such as Alzheimer’s disease (57) and Parkinson’s disease (58). In the present study, our literature search of these archaeal metabolites potentially links them to different non-oral or non-gut cancers, beneficially or detrimentally (Fig. 3). Carcinogens, including acetaldehyde and methylglyoxal, have been reported to influence human health by regulating protein modifications or perturbing protein/chromatin structure (59, 60). Thus, the produced metabolites in gut and oral cavity might also increase the cancer risk in other organs, such as liver and breast (61). Since archaea are difficult to enrich or isolate, these correlations could be further analyzed using a combination of metagenomics and metatranscriptomics on a larger scale.
FIG 3.
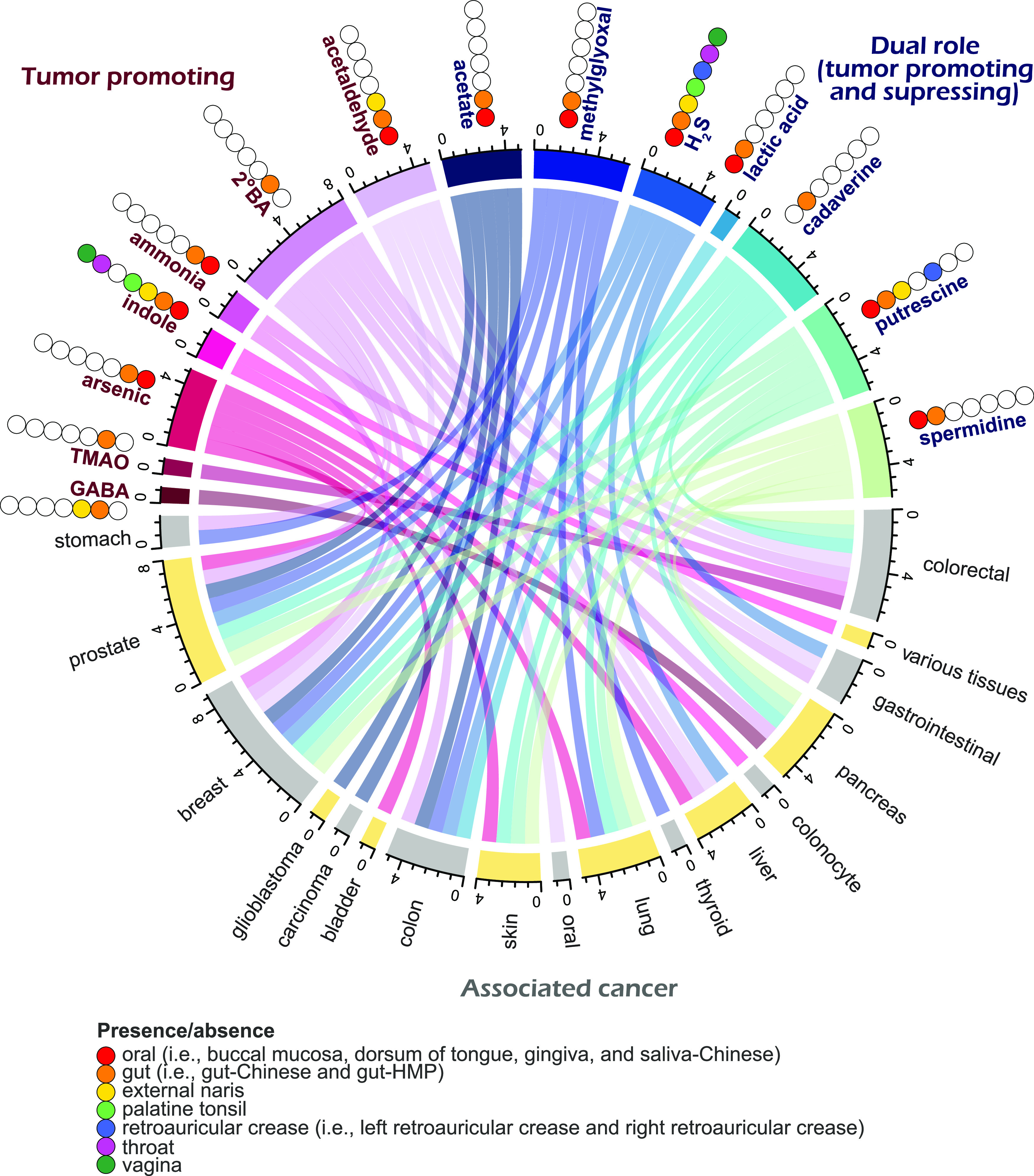
Archaeal metabolites and their potential roles with various cancers. Relations were organized and displayed on the basis of previous reports (7, 40–42, 61, 87–89).
Bacterial biomarkers like Fusobacterium species (62), Porphyromonas asaccharolytica, and Peptostreptococcus stomatis (63) hold promise for early diagnosis of a certain cancer. Similar studies have been performed for archaea, showing that several representatives of methanogenic and halophilic archaea may be used as biomarkers for colorectal tumor development (25). In the present study, screening of contigs has shown that cancer-related archaeal biomarkers are also present in samples from healthy individuals (Fig. S5). A possible reason for this inconsistency is that the taxonomic annotation in the report was assigned using short reads (25), which might not be accurate for taxonomy assignment to the species level. Studies using functional genes have been applied for predictions of diseases, such as dental caries (53) and colorectal cancer (64). Indeed, the application of functional genes as biomarkers is more reasonable, as (i) the human microbiota is fluctuating because factors such as geographic location, diet (31, 32), and age (65) influence the presence, abundance, and diversity of archaea in humans and (ii) archaea may have functional redundancy (i.e., duplicate genes across archaea that maintain biochemical functions over time, independent of variance in taxonomic composition), as reported for microbes in other ecosystems (66). Thus, the identification of archaeal marker genes for cancer, especially those with high-level transcription increases in patients, is needed for early diagnosis of archaea-induced cancer.
The human microbiome is a complex aggregate of the microbes. Usually, microbes like bacteria, archaea, and fungi coexist to interact with their host through an elaborate network (8, 67). In human body, methanogenic archaea have been reported to be positively correlated with bacteria (68), and their mutualism has also been shown in humans and a humanized gnotobiotic mouse model (69). Further, methanogens are capable of syntrophic interactions with bacteria to enhance the production of SCFA (29), which play a role in cancer suppression within the host (33). However, the dysbiosis of microbes could interrupt the mutualism between bacteria and methanogenic archaea and lead to diseases like colorectal cancer (25). As no proof for such archaeal pathogens exists (10, 12), these human-associated archaea are more likely to function in cooperation with other microbes, such as the known cancer-related pathogen Helicobacter pylori (Fig. S4).
In summary, we performed a large-scale analysis of human-associated archaea and cancer-related metabolites using more than 44,000 contigs across different body sites. Results revealed that although archaea were identified in all body sites, the occurrence and diversity were higher in samples from the oral cavity and gut, with archaeal composition being similar between Easterners and Westerners. Similarly, genes encoding cancer-related metabolites were more diverse and prevalent in oral and gut samples, and these metabolites were affiliated with Euryarchaeota and the TACK superphylum, especially methanogenic archaea. These findings largely improved our understanding of the connections between archaea and human tumor microenvironment, thereby shedding light on early diagnosis and therapeutic design for cancer. However, similar to bacteria, these associations need to be further experimentally verified before instruction or application in therapy.
MATERIALS AND METHODS
Data set acquisition.
Contigs for analysis were retrieved from four different data sets: human saliva samples of Chinese individuals (saliva-Chinese) (70), human fecal samples of Chinese individuals (gut-Chinese) (64), human saliva samples from European individuals (71), and samples from the Human Microbiome Project (HMP) that included body sites like buccal mucosa, dorsum of tongue, throat, and vagina (72). We acquired a total of 5,614 samples (3,591 for saliva-Chinese, 128 for gut-Chinese, and 1,859 for HMP), and these samples were retrieved from healthy Eastern and Western subjects (Table S1). Contigs for the samples were downloaded directly from NCBI (https://www.ncbi.nlm.nih.gov/) based on the published BioProject accession numbers or from the HMP website (https://portal.hmpdacc.org/) with specific filters “HMP” and “Wgs Assembled Seq Set.” It should be noted that the publicly available data sets were from different studies and, thus, the differences in sampling, sequencing, and/or contig binning methods may have potential effect on our results.
Contig taxonomic annotation.
To increase taxonomy assignment accuracy, the downloaded contigs were first filtered using a custom script to remove sequence with length of <1,000 bp. Kaiju (v1.7.3) and Kraken2 (v2.0.8-beta) were both introduced to perform taxonomy annotation with the default parameters against the default databases (e.g., NCBI nonredundant database) (73, 74). The Kaiju software was used because it contained a detailed database for archaea classification, and the Kraken2 software was supplemented for validation. Finally, we had a total of 44,760 archaeal contigs from 1,818 samples (Table S1).
Archaeal protein annotation.
Open reading frames (ORFs) of archaeal contigs were called using the “-p meta” option in Prodigal (v2.6.3) with default parameters (75). For function annotation, all proteins were annotated using the standalone software eggNOG-mapper (v2) (76, 77) and InterProScan (v5.42-78.0) (78). Archaeal clusters of orthologous genes (arCOGs) downloaded in May 2020 were used for archaeal-specific protein annotation using DIAMOND (v0.9.24) (79) with an E value of 1E-10. The annotated putative [NiFe] and [FeFe] hydrogenases sequences were compared against the HydDB database (80) for subgroup classification using DIAMOND with an E value of 1E-10. Metabolite transporters were annotated using the transporter classification database (TCDB) downloaded in May 2020 using DIAMOND with an E value of 1E-10 (81).
Relative abundance of cancer-related genes.
Raw metagenomic reads for colorectal cancer patients (n = 20) and healthy individuals (n = 28) were downloaded from BioProject PRJEB10878 (64). Raw reads were quality-controlled using the sickle (v1.33) with the option “-q 25,” and the human-origin reads were removed using bbmap (v35.85) against the HG19 genome. The abundance of raw reads was processed using the software bwa (v0.7.17), samtools (v1.10), and bbmap (v35.85) with the default setting against the curated archaeal cancer-related genes. Finally, the mapped reads were normalized to per million reads with a custom script. The group comparison was performed with the general linear hypothesis test and Tukey procedures embedded in “multcomp” package.
Cooccurrence of human-associated archaea with pathogens.
A set of 20 publicly available gut metagenomics from colorectal cancer patients were downloaded from the European Nucleotide Archive under the project number PRJEB7774 (Table S5). Raw reads were trimmed using Trimmomatic (v0.39) (82) with the parameter “-phred33 MINLEN:60.” The qualified reads were taxonomically assigned using the Kaiju (v1.7.3) as mentioned above.
To evaluate the cooccurrence patterns of the archaea with pathogens, reads were first tested using the checkerboard score (C-score) through the function “oecosimu” with the parameters “matrix, nestedchecker, method = “swap”, nsimul = 10000,” which also calculated the standardized effect size (SES) to avid biases of raw C-score value. The C-score of 939 and SES score of 1.59 imply that microorganisms in gut are distributed nonrandomly (83). Then, the cooccurrence network analysis was performed at the genus/species level based on the previous method (83, 84). Only correlations with a Spearman’s ρ of >0.6 and Benjamini–Hochberg adjusted P value of <0.01 were kept for analyses. The resulted network was imported into Gephi (version 0.9.2) (85), and the cooccurrence patterns were visualized through calculation with Fruchterman-Reingold algorithm.
Statistical analysis.
Statistical analysis and visualization were performed in the R software (v3.6.3) using the packages “ggplot2,” “vegan,” and “picante,” unless otherwise indicated. Bray-Curtis dissimilarity for archaeal contigs in different body sites was calculated using the package “picante.” Diversity of archaea across different body sites was analyzed at the species level. Multiple comparisons of the presence and absence of archaeal contigs among samples were performed with the general linear hypothesis test and Tukey procedures embedded in “multcomp” package. NMDS analysis of the archaea assembly in different samples at the species level was performed based on Bray-Curtis dissimilarity using the “metaMDS” function in the “vegan” package. The confidential interval for the underlying body site specific ellipse is 0.95. To determine the similarities and differences between samples from Easterners and Westerners, PERMANOVA analysis was performed using the “adonis” function in the “vegan” package with 999 permutations.
Data availability.
The contig data sets that support the findings of this study are available in European Nucleotide Archive under BioProject PRJEB10878 (64), the National Genomics Data Centre with the accession number PRJCA003731 (70), and the HMPDACC Data Portal (https://hmpdacc.org/) (72). The data set for metagenomics is available in European Nucleotide Archive with the accession numbers listed in Table S5 under BioProject PRJEB7774 (86).
ACKNOWLEDGMENTS
This study was supported by OSUCCC startup funds for Q.Z., National Key R&D Program of China (2021YFC2102500) and GuangDong Basic and Applied Basic Research Foundation (2021A1515110334) for M.C., and Shenzhen Bay Laboratory Startup Funds (21230051) for X.T. We also thank Shenzhen Bay Laboratory Supercomputing Center for the platform for data processing.
Footnotes
Supplemental material is available online only.
Contributor Information
Mingwei Cai, Email: caimw@szbl.ac.cn.
Xiaoyu Tang, Email: xtang@szbl.ac.cn.
Qingfei Zheng, Email: Qingfei.Zheng@osumc.edu.
Zhenjiang Zech Xu, Nanchang University.
REFERENCES
- 1.Cho I, Blaser MJ. 2012. The human microbiome: at the interface of health and disease. Nat Rev Genet 13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mikó E, Kovács T, Sebő É, Tóth J, Csonka T, Ujlaki G, Sipos A, Szabó J, Méhes G, Bai P. 2019. Microbiome–microbial metabolome–cancer cell interactions in breast cancer–familiar, but unexplored Cells 8:293. doi: 10.3390/cells8040293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Louis P, Hold GL, Flint HJ. 2014. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 4.Johnson CH, Spilker ME, Goetz L, Peterson SN, Siuzdak G. 2016. Metabolite and microbiome interplay in cancer immunotherapy. Cancer Res 76:6146–6152. doi: 10.1158/0008-5472.CAN-16-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He J, Zhang P, Shen L, Niu L, Tan Y, Chen L, Zhao Y, Bai L, Hao X, Li X, Zhang S, Zhu L. 2020. Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int J Mol Sci 21:6356. doi: 10.3390/ijms21176356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu P, Wang Y, Yang G, Zhang Q, Meng L, Xin Y, Jiang X. 2021. The role of short-chain fatty acids in intestinal barrier function, inflammation, oxidative stress, and colonic carcinogenesis. Pharmacol Res 165:105420. doi: 10.1016/j.phrs.2021.105420. [DOI] [PubMed] [Google Scholar]
- 7.Di Ciaula A, Wang DQ-H, Molina-Molina E, Baccetto RL, Calamita G, Palmieri VO, Portincasa P. 2017. Bile acids and cancer: direct and environmental-dependent effects. Ann Hepatol 16:S87–S105. doi: 10.5604/01.3001.0010.5501. [DOI] [PubMed] [Google Scholar]
- 8.Borrel G, Brugère J-F, Gribaldo S, Schmitz RA, Moissl-Eichinger C. 2020. The host-associated archaeome. Nat Rev Microbiol 18:622–636. doi: 10.1038/s41579-020-0407-y. [DOI] [PubMed] [Google Scholar]
- 9.Charlesworth JC, Burns BP. 2015. Untapped resources: biotechnological potential of peptides and secondary metabolites in archaea. Archaea 2015:282035. doi: 10.1155/2015/282035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavicchioli R, Curmi PM, Saunders N, Thomas T. 2003. Pathogenic archaea: do they exist? Bioessays 25:1119–1128. doi: 10.1002/bies.10354. [DOI] [PubMed] [Google Scholar]
- 11.Aminov RI. 2013. Role of archaea in human disease. Front Cell Infect Microbiol 3:42. doi: 10.3389/fcimb.2013.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckburg PB, Lepp PW, Relman DA. 2003. Archaea and their potential role in human disease. Infect Immun 71:591–596. doi: 10.1128/IAI.71.2.591-596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter P. 2013. The secret garden’s gardeners: research increasingly appreciates the crucial role of gut viruses for human health and disease. EMBO Rep 14:683–685. doi: 10.1038/embor.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Limon JJ, Skalski JH, Underhill DM. 2017. Commensal fungi in health and disease. Cell Host Microbe 22:156–165. doi: 10.1016/j.chom.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker BJ, De Anda V, Seitz KW, Dombrowski N, Santoro AE, Lloyd KG. 2020. Diversity, ecology and evolution of Archaea. Nat Microbiol 5:887–900. doi: 10.1038/s41564-020-0715-z. [DOI] [PubMed] [Google Scholar]
- 16.Miller TL, Wolin M, de Macario EC, Macario A. 1982. Isolation of Methanobrevibacter smithii from human feces. Appl Environ Microbiol 43:227–232. doi: 10.1128/aem.43.1.227-232.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Borrel G, Harris HM, Tottey W, Mihajlovski A, Parisot N, Peyretaillade E, Peyret P, Gribaldo S, O'Toole PW, Brugère J-F. 2012. Genome sequence of “Candidatus Methanomethylophilus alvus” Mx1201, a methanogenic archaeon from the human gut belonging to a seventh order of methanogens. J Bacteriol 194:6944–6945. doi: 10.1128/JB.01867-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khelaifia S, Raoult D. 2016. Haloferax massiliensis sp. nov., the first human-associated halophilic archaea. New Microbes New Infect 12:96–98. doi: 10.1016/j.nmni.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seck EH, Senghor B, Merhej V, Bachar D, Cadoret F, Robert C, Azhar EI, Yasir M, Bibi F, Jiman-Fatani AA, Konate DS, Musso D, Doumbo O, Sokhna C, Levasseur A, Lagier JC, Khelaifia S, Million M, Raoult D. 2019. Salt in stools is associated with obesity, gut halophilic microbiota and Akkermansia muciniphila depletion in humans. Int J Obes (Lond) 43:862–871. doi: 10.1038/s41366-018-0201-3. [DOI] [PubMed] [Google Scholar]
- 20.Ferrari A, Brusa T, Rutili A, Canzi E, Biavati B. 1994. Isolation and characterization of Methanobrevibacter oralis sp. nov. Curr Microbiol 29:7–12. doi: 10.1007/BF01570184. [DOI] [Google Scholar]
- 21.Togo AH, Grine G, Khelaifia S, Des Robert C, Brevaut V, Caputo A, Baptiste E, Bonnet M, Levasseur A, Drancourt M, Million M, Raoult D. 2019. Culture of methanogenic archaea from human colostrum and milk. Sci Rep 9:1–10. doi: 10.1038/s41598-019-54759-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koskinen K, Pausan MR, Perras AK, Beck M, Bang C, Mora M, Schilhabel A, Schmitz R, Moissl-Eichinger C. 2017. First insights into the diverse human archaeome: specific detection of archaea in the gastrointestinal tract, lung, and nose and on skin. mBio 8:e00824-17. doi: 10.1128/mBio.00824-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng F, Kittelmann S, Patchett ML, Attwood GT, Janssen PH, Rakonjac J, Gagic D. 2016. An adhesin from hydrogen‐utilizing rumen methanogen Methanobrevibacter ruminantium M 1 binds a broad range of hydrogen‐producing microorganisms. Environ Microbiol 18:3010–3021. doi: 10.1111/1462-2920.13155. [DOI] [PubMed] [Google Scholar]
- 24.Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. 2004. Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci U S A 101:6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coker OO, Wu WKK, Wong SH, Sung JJ, Yu J. 2020. Altered gut archaea composition and interaction with bacteria are associated with colorectal cancer. Gastroenterology 159:1459–1470.e5. doi: 10.1053/j.gastro.2020.06.042. [DOI] [PubMed] [Google Scholar]
- 26.Drancourt M, Nkamga VD, Lakhe NA, Régis J-M, Dufour H, Fournier P-E, Bechah Y, Michael Scheld W, Raoult D. 2017. Evidence of archaeal methanogens in brain abscess. Clin Infect Dis 65:1–5. doi: 10.1093/cid/cix286. [DOI] [PubMed] [Google Scholar]
- 27.Brugère J-F, Borrel G, Gaci N, Tottey W, O’Toole PW, Malpuech-Brugère C. 2014. Archaebiotics. Gut Microbes 5:5–10. doi: 10.4161/gmic.26749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cullin N, Antunes CA, Straussman R, Stein-Thoeringer CK, Elinav E. 2021. Microbiome and cancer. Cancer Cell 39:1317–1341. doi: 10.1016/j.ccell.2021.08.006. [DOI] [PubMed] [Google Scholar]
- 29.Lurie-Weinberger MN, Gophna U. 2015. Archaea in and on the human body: health implications and future directions. PLoS Pathog 11:e1004833. doi: 10.1371/journal.ppat.1004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang ZJ, Wang Y-C, Yang X, Hang HC. 2020. Chemical reporters for exploring microbiology and microbiota mechanisms. Chembiochem 21:19–32. doi: 10.1002/cbic.201900535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segal I, Walker AR, Lord S, Cummings JH. 1988. Breath methane and large bowel cancer risk in contrasting African populations. Gut 29:608–613. doi: 10.1136/gut.29.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morii H, Oda K, Suenaga Y, Nakamura T. 2003. Low methane concentration in the breath of Japanese. J UOEH 25:397–407. (In Japanese). doi: 10.7888/juoeh.25.397. [DOI] [PubMed] [Google Scholar]
- 33.Cox-York K, Stoecker E, Hamm AK, Weir TL. 2019. Microbial metabolites in cancer promotion or prevention, p 317–346. Microbiome and cancer. Springer, Cham, Switzerland. [Google Scholar]
- 34.Zheng Q, Osunsade A, David Y. 2020. Protein arginine deiminase 4 antagonizes methylglyoxal-induced histone glycation. Nat Commun 11:8. doi: 10.1038/s41467-020-17066-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng Q, Omans ND, Leicher R, Osunsade A, Agustinus AS, Finkin-Groner E, D’Ambrosio H, Liu B, Chandarlapaty S, Liu S, David Y. 2019. Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat Commun 10:12. doi: 10.1038/s41467-019-09192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Q, Maksimovic I, Upad A, David Y. 2020. Non-enzymatic covalent modifications: a new link between metabolism and epigenetics. Protein Cell 11:401–416. doi: 10.1007/s13238-020-00722-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng Q, Prescott NA, Maksimovic I, David Y. 2019. (De)Toxifying the epigenetic code. Chem Res Toxicol 32:796–807. doi: 10.1021/acs.chemrestox.9b00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nokin M-J, Durieux F, Peixoto P, Chiavarina B, Peulen O, Blomme A, Turtoi A, Costanza B, Smargiasso N, Baiwir D, Scheijen JL, Schalkwijk CG, Leenders J, De Tullio P, Bianchi E, Thiry M, Uchida K, Spiegel DA, Cochrane JR, Hutton CA, De Pauw E, Delvenne P, Belpomme D, Castronovo V, Bellahcène A. 2016. Methylglyoxal, a glycolysis side-product, induces Hsp90 glycation and YAP-mediated tumor growth and metastasis. Elife 5:e19375. doi: 10.7554/eLife.19375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takehara A, Hosokawa M, Eguchi H, Ohigashi H, Ishikawa O, Nakamura Y, Nakagawa H. 2007. γ-Aminobutyric acid (GABA) stimulates pancreatic cancer growth through overexpressing GABAA receptor π subunit. Cancer Res 67:9704–9712. doi: 10.1158/0008-5472.CAN-07-2099. [DOI] [PubMed] [Google Scholar]
- 40.Martinez VD, Vucic EA, Becker-Santos DD, Gil L, Lam WL. 2011. Arsenic exposure and the induction of human cancers. J Toxicol 2011:431287. doi: 10.1155/2011/431287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guertin KA, Li XS, Graubard BI, Albanes D, Weinstein SJ, Goedert JJ, Wang Z, Hazen SL, Sinha R. 2017. Serum trimethylamine N-oxide, carnitine, choline, and betaine in relation to colorectal cancer risk in the alpha tocopherol, beta carotene cancer prevention study. Cancer Epidemiol Biomarkers Prev 26:945–952. doi: 10.1158/1055-9965.EPI-16-0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossi T, Vergara D, Fanini F, Maffia M, Bravaccini S, Pirini F. 2020. Microbiota-derived metabolites in tumor progression and metastasis. Int J Mol Sci 21:5786. doi: 10.3390/ijms21165786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yousuf B, Mishra A. 2019. Exploring human bacterial diversity toward prevention of infectious disease and health promotion, p 519–533, Microbial Diversity in the Genomic Era. Elsevier Academic Press, Cambridge, MA. [Google Scholar]
- 44.Nkamga VD, Henrissat B, Drancourt M. 2017. Archaea: essential inhabitants of the human digestive microbiota. Hum Microbiome J 3:1–8. doi: 10.1016/j.humic.2016.11.005. [DOI] [Google Scholar]
- 45.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 46.Alonso-Saez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL, Lovejoy C, Tremblay J-E, Potvin M, Heinrich F, Estrada M, Riemann L, Bork P, Pedros-Alio C, Bertilsson S. 2012. Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci U S A 109:17989–17994. doi: 10.1073/pnas.1201914109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clinton SK, Bostwick DG, Olson LM, Mangian HJ, Visek WJ. 1988. Effects of ammonium acetate and sodium cholate on N-methyl-N′-nitro-N-nitrosoguanidine-induced colon carcinogenesis of rats. Cancer Res 48:3035–3039. [PubMed] [Google Scholar]
- 48.Hamana K, Tanaka T, Hosoya R, Niitsu M, Itoh T. 2003. Cellular polyamines of the acidophilic, thermophilic and thermoacidophilic archaebacteria, Acidilobus, Ferroplasma, Pyrobaculum, Pyrococcus, Staphylothermus, Thermococcus, Thermodiscus and Vulcanisaeta. J Gen Appl Microbiol 49:287–293. doi: 10.2323/jgam.49.287. [DOI] [PubMed] [Google Scholar]
- 49.Shreiner AB, Kao JY, Young VB. 2015. The gut microbiome in health and in disease. Curr Opin Gastroenterol 31:69–75. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moon J-H, Lee J-H. 2016. Probing the diversity of healthy oral microbiome with bioinformatics approaches. BMB Rep 49:662–670. doi: 10.5483/bmbrep.2016.49.12.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White BA, Creedon DJ, Nelson KE, Wilson BA. 2011. The vaginal microbiome in health and disease. Trends Endocrinol Metab 22:389–393. doi: 10.1016/j.tem.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grice EA, Segre JA. 2011. The skin microbiome. Nat Rev Microbiol 9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belda-Ferre P, Alcaraz LD, Cabrera-Rubio R, Romero H, Simon-Soro A, Pignatelli M, Mira A. 2012. The oral metagenome in health and disease. ISME J 6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Genco RJ, Grossi SG, Ho A, Nishimura F, Murayama Y. 2005. A proposed model linking inflammation to obesity, diabetes, and periodontal infections. J Periodontol 76:2075–2084. doi: 10.1902/jop.2005.76.11-S.2075. [DOI] [PubMed] [Google Scholar]
- 55.Awano S, Ansai T, Takata Y, Soh I, Akifusa S, Hamasaki T, Yoshida A, Sonoki K, Fujisawa K, Takehara T. 2008. Oral health and mortality risk from pneumonia in the elderly. J Dent Res 87:334–339. doi: 10.1177/154405910808700418. [DOI] [PubMed] [Google Scholar]
- 56.Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, Nielsen J, Bäckhed F. 2013. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 57.Vogt NM, Kerby RL, Dill-McFarland KA, Harding SJ, Merluzzi AP, Johnson SC, Carlsson CM, Asthana S, Zetterberg H, Blennow K, Bendlin BB, Rey FE. 2017. Gut microbiome alterations in Alzheimer’s disease. Sci Rep 7:1–11. doi: 10.1038/s41598-017-13601-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheperjans F, Aho V, Pereira PAB, Koskinen K, Paulin L, Pekkonen E, Haapaniemi E, Kaakkola S, Eerola-Rautio J, Pohja M, Kinnunen E, Murros K, Auvinen P. 2015. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov Disord 30:350–358. doi: 10.1002/mds.26069. [DOI] [PubMed] [Google Scholar]
- 59.Chen D, Fang L, Li H, Jin C. 2018. The effects of acetaldehyde exposure on histone modifications and chromatin structure in human lung bronchial epithelial cells. Environ Mol Mutagen 59:375–385. doi: 10.1002/em.22187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gaffney DO, Jennings EQ, Anderson CC, Marentette JO, Shi T, Schou Oxvig A-M, Streeter MD, Johannsen M, Spiegel DA, Chapman E, Roede JR, Galligan JJ. 2020. Non-enzymatic lysine lactoylation of glycolytic enzymes. Cell Chem Biol 27:206–213.e6. doi: 10.1016/j.chembiol.2019.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seitz HK, Stickel F. 2010. Acetaldehyde as an underestimated risk factor for cancer development: role of genetics in ethanol metabolism. Genes Nutr 5:121–128. doi: 10.1007/s12263-009-0154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeller G, Tap J, Voigt AY, Sunagawa S, Kultima JR, Costea PI, Amiot A, Böhm J, Brunetti F, Habermann N, Hercog R, Koch M, Luciani A, Mende DR, Schneider MA, Schrotz-King P, Tournigand C, Tran Van Nhieu J, Yamada T, Zimmermann J, Benes V, Kloor M, Ulrich CM, von Knebel Doeberitz M, Sobhani I, Bork P. 2014. Potential of fecal microbiota for early‐stage detection of colorectal cancer. Mol Syst Biol 10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, Tang L, Zhao H, Stenvang J, Li Y, Wang X, Xu X, Chen N, Wu WKK, Al-Aama J, Nielsen HJ, Kiilerich P, Jensen BAH, Yau TO, Lan Z, Jia H, Li J, Xiao L, Lam TYT, Ng SC, Cheng AS-L, Wong VW-S, Chan FKL, Xu X, Yang H, Madsen L, Datz C, Tilg H, Wang J, Brünner N, Kristiansen K, Arumugam M, Sung JJ-Y, Wang J. 2017. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 66:70–78. doi: 10.1136/gutjnl-2015-309800. [DOI] [PubMed] [Google Scholar]
- 65.Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao J-z, Abe F, Osawa R. 2016. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol 16:1–12. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cai M, Wilkins D, Chen J, Ng S-K, Lu H, Jia Y, Lee PK. 2016. Metagenomic reconstruction of key anaerobic digestion pathways in municipal sludge and industrial wastewater biogas-producing systems. Front Microbiol 7:778. doi: 10.3389/fmicb.2016.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hoffmann C, Dollive S, Grunberg S, Chen J, Li H, Wu GD, Lewis JD, Bushman FD. 2013. Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hansen EE, Lozupone CA, Rey FE, Wu M, Guruge JL, Narra A, Goodfellow J, Zaneveld JR, McDonald DT, Goodrich JA, Heath AC, Knight R, Gordon JI. 2011. Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc Natl Acad Sci U S A 108:4599–4606. doi: 10.1073/pnas.1000071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Samuel BS, Gordon JI. 2006. A humanized gnotobiotic mouse model of host–archaeal–bacterial mutualism. Proc Natl Acad Sci U S A 103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu J, Tian L, Chen P, Han M, Song L, Tong X, Sun X, Yang F, Lin Z, Liu X, Liu C, Wang X, Lin Y, Cai K, Hou Y, Xu X, Yang H, Wang J, Kristiansen K, Xiao L, Zhang T, Jia H, Jie Z. 2021. Over 50,000 metagenomically assembled draft genomes for the human oral microbiome reveal new taxa. Genomics Proteomics Bioinformatics S1672-0229:00176-5. doi: 10.1016/j.gpb.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Belstrøm D, Constancias F, Liu Y, Yang L, Drautz-Moses DI, Schuster SC, Kohli GS, Jakobsen TH, Holmstrup P, Givskov M. 2017. Metagenomic and metatranscriptomic analysis of saliva reveals disease-associated microbiota in patients with periodontitis and dental caries. NPJ Biofilms Microbiomes 3:23. doi: 10.1038/s41522-017-0031-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Creasy HH, Felix V, Aluvathingal J, Crabtree J, Ifeonu O, Matsumura J, McCracken C, Nickel L, Orvis J, Schor M, Giglio M, Mahurkar A, White O. 2021. HMPDACC: a human microbiome project multi-omic data resource. Nucleic Acids Res 49:D734–D742. doi: 10.1093/nar/gkaa996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Menzel P, Ng KL, Krogh A. 2016. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7:11257. doi: 10.1038/ncomms11257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salazar G, Paoli L, Alberti A, Huerta-Cepas J, Ruscheweyh H-J, Cuenca M, Field CM, Coelho LP, Cruaud C, Engelen S, Gregory AC, Labadie K, Marec C, Pelletier E, Royo-Llonch M, Roux S, Sánchez P, Uehara H, Zayed AA, Zeller G, Carmichael M, Dimier C, Ferland J, Kandels S, Picheral M, Pisarev S, Poulain J, Acinas SG, Babin M, Bork P, Bowler C, de Vargas C, Guidi L, Hingamp P, Iudicone D, Karp-Boss L, Karsenti E, Ogata H, Pesant S, Speich S, Sullivan MB, Wincker P, Sunagawa S, Tara Oceans Coordinators. 2019. Gene expression changes and community turnover differentially shape the global ocean metatranscriptome. Cell 179:1068–1083.e21. doi: 10.1016/j.cell.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Necci M, Piovesan D, Dosztányi Z, Tosatto SCE. 2017. MobiDB-lite: fast and highly specific consensus prediction of intrinsic disorder in proteins. Bioinformatics 33:1402–1404. doi: 10.1093/bioinformatics/btx015. [DOI] [PubMed] [Google Scholar]
- 79.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 80.Søndergaard D, Pedersen CNS, Greening C. 2016. HydDB: a web tool for hydrogenase classification and analysis. Sci Rep 6:34212. doi: 10.1038/srep34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saier MH, Jr, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G. 2016. The transporter classification database (TCDB): recent advances. Nucleic Acids Res 44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ju F, Xia Y, Guo F, Wang Z, Zhang T. 2014. Taxonomic relatedness shapes bacterial assembly in activated sludge of globally distributed wastewater treatment plants. Environ Microbiol 16:2421–2432. doi: 10.1111/1462-2920.12355. [DOI] [PubMed] [Google Scholar]
- 84.Barberán A, Bates ST, Casamayor EO, Fierer N. 2012. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks. In Proceedings of the Third International AAAI Conference on Web and Social Media, Association for the Advancement of Artificial Intelligence, San Jose, California. [Google Scholar]
- 86.Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, Zhang D, Xia H, Xu X, Jie Z, Su L, Li X, Li X, Li J, Xiao L, Huber-Schönauer U, Niederseer D, Xu X, Al-Aama JY, Yang H, Wang J, Kristiansen K, Arumugam M, Tilg H, Datz C, Wang J. 2015. Gut microbiome development along the colorectal adenoma–carcinoma sequence. Nat Commun 6:13. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 87.Li J, Meng Y, Wu X, Sun Y. 2020. Polyamines and related signaling pathways in cancer. Cancer Cell Int 20:1–16. doi: 10.1186/s12935-020-01545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Leone A, Nigro C, Nicolò A, Prevenzano I, Formisano P, Beguinot F, Miele C. 2021. The dual-role of methylglyoxal in tumor progression–novel therapeutic approaches. Front Oncol 11:645686. doi: 10.3389/fonc.2021.645686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu D, Si W, Wang M, Lv S, Ji A, Li Y. 2015. Hydrogen sulfide in cancer: friend or foe? Nitric Oxide 50:38–45. doi: 10.1016/j.niox.2015.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download Spectrum02367-21_supplemental_file1.xlsx, XLSX file, 15.2 MB (15.6MB, xlsx)
Supplemental material. Download Spectrum02367-21_supplemental_file2.pdf, PDF file, 1.1 MB (1.1MB, pdf)
Data Availability Statement
The contig data sets that support the findings of this study are available in European Nucleotide Archive under BioProject PRJEB10878 (64), the National Genomics Data Centre with the accession number PRJCA003731 (70), and the HMPDACC Data Portal (https://hmpdacc.org/) (72). The data set for metagenomics is available in European Nucleotide Archive with the accession numbers listed in Table S5 under BioProject PRJEB7774 (86).


