ABSTRACT
Symptoms of Clostridioides difficile infection (CDI) are attributed largely to two toxins, TcdA and TcdB. About 17–23% of C. difficile isolates produce binary toxin, which enhances C. difficile pathogenesis. Previously, we engineered the nontoxigenic C. difficile strain CCUG37785 (designated as CCUG37785) to express immunogenic fragments of TcdA and TcdB as an oral mucosal CDI vaccine candidate. In this study, we performed genomic and phenotypic analyses of CCUG37785 and evaluated its potential use for preventing and treating CDI. Whole genome sequencing showed that CCUG37785 is ribotype ST3 and lacks toxin genes. Comparative analyses of PaLoc and CdtLoc loci of CCUG37785 revealed 115-bp and 68-bp conserved fragments in these regions, respectively. Phenotypic comparisons between CCUG37785 and C. difficile R20291 (an epidemic hypervirulent BI/NAPI/027 strain, designated as R20291) found that CCUG37785 exhibited significantly higher adhesion and sporulation, significantly lower spore germination and biofilm formation, and comparable motility to R20291. We also showed that oral inoculation of CCUG37785 spores prior to infection with R20291 spores provided mice almost full protection against developing CDI. However, oral inoculation of CCUG37785 spores after infection with R20291 spores only provided minor protection against CDI. Further analysis showed that mice pretreated with CCUG37785 spores secreted significantly less R20291 spores, while mice treated with CCUG37785 spores after infection with R20291 secreted a comparable amount of R20291 spores to mice infected with R20291 spores only. Our data both highlight the potential use of CCUG37785 for the prevention of primary and recurrent CDI in humans and support its use as an oral mucosal vaccine carrier against CDI.
IMPORTANCE Clostridioides difficile infection (CDI) symptoms range from diarrhea to intestinal inflammation/lesion and death and are mainly caused by two exotoxins, TcdA and TcdB. Active vaccination provides the attractive opportunity to prevent CDI and recurrence. No vaccine against CDI is currently licensed. Tremendous efforts have been devoted to developing vaccines targeting both toxins. However, ideally, vaccines should target both toxins and C. difficile cells/spores that transmit the disease and cause recurrence. Furthermore, C. difficile is an enteric pathogen, and mucosal/oral immunization would be particularly useful to protect the host against CDI considering that the gut is the main site of disease onset and progression. Data in our current study not only highlight the potential use of CCUG37785 to prevent primary and recurrent CDI in humans but also further support its use as an oral mucosal vaccine carrier against CDI.
KEYWORDS: Clostridioides difficile infection (CDI), nontoxigenic Clostridioides difficile (NTCD), spores, therapeutics, microbial colonization, pathogenicity locus, genomics
INTRODUCTION
Clostridioides difficile (C. difficile) is a Gram-positive, toxin-producing and spore-forming anaerobic bacterium (1). C. difficile is part of the normal intestinal microbiota in 1–3% of healthy adults, and 15–20% infants (2), and it is the leading cause of nosocomial antibiotic-associated diarrhea in developed countries. Moreover, a continued rise in the incidence of severe C. difficile infection (CDI) has been observed worldwide. Different studies from North America and Europe suggested that approximately 20–27% of all CDI cases were community-associated (2, 3). C. difficile produces three protein toxins, including toxin A (TcdA), toxin B (TcdB), and binary toxin (CDT), the first two of which are the major virulence factors of C. difficile and are the major drivers of CDI symptoms. CDT exists in about 17–23% C. difficile strains (4) and is associated with the increased severity of CDI (5, 6). According to reports from the Centers for Disease Control and Prevention (CDC) in 2017, there were 223,900 cases in hospitalized patients and 12,800 deaths in the United States.
Genes encoding TcdA (tcdA) and TcdB (tcdB) of toxigenic C. difficile strains are located within the pathogenicity locus (PaLoc) (7). In the nontoxigenic C. difficile (NTCD) strains, the PaLoc region is replaced by a highly conserved 115/75-bp non-coding region (8, 9).
NTCD strains have been explored as a safe and effective preventive for CDI (10–14). A hamster model was used to evaluate the effectiveness of a NTCD strain as a preventive agent for the infection with toxigenic C. difficile (15), and the authors found that the prevention of CDI mortality was correlated closely with the NTCD colonization prior to challenge with toxigenic C. difficile strains. In 2013, the same group found that NTCD colonization was displaced by the toxigenic strain (16) in some hamsters used in experiments, suggesting that toxigenic and NTCD strains may compete for colonization. A possible mechanism by which C. difficile strains establish or lose colonization in the intestine is that strains differ in their ability to adhere to colonic mucosal cells or mucus (14, 17). Both adherence ability and motility are important for C. difficile strains to establish colonization. CDI has very high rates of recurrence/relapse after treatment with antibiotics. NTCD strain administration after CDI treatment may serve as a preventive strategy against recurrent CDI (14, 18).
Previously, we engineered the nontoxigenic strain C. difficile CCUG37785 (designated as CCUG37785) to express mTcd138, which is comprised of the glucosyltransferase and cysteine proteinase domains of TcdB and the receptor binding domain of TcdA, generating the strain NTCD_Tcd138 as an oral mucosal vaccine candidate against CDI. Our data indicate that NTCD_Tcd138 is a promising oral vaccine candidate against CDI (19). It was the first report on the use of nontoxigenic C. difficile strains-based vaccines against CDI. In this study, we aimed to perform comparative genomic and phenotypic analyses of the strain CCUG37785 in comparison with other relevant C. difficile strains and to determine if CCUG37785 can protect mice against infection with a toxigenic C. difficile strain.
RESULTS
Genome analysis of C. difficile CCUG37785.
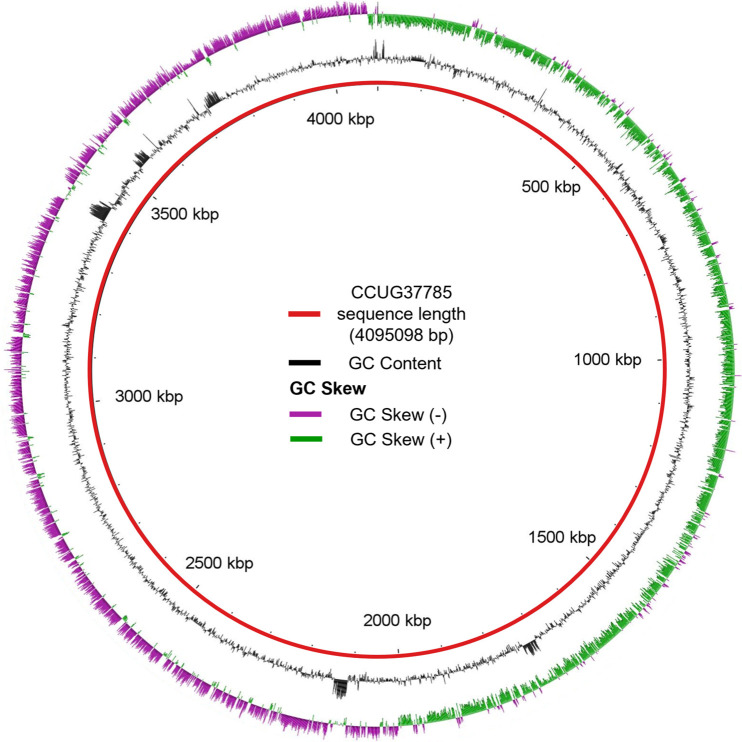
Whole genome sequencing, conducted at a sequencing depth of approximately 300X, yielded high quality reads, which were assembled into a draft genome with an N50 of 323,436 and a maximum contig length of 793,487. Strain CCUG37785 has a genome size of 4,095,098 bp, with a GC content of 28.8% in line with the 28–30% GC content typically observed in C. difficile genomes (Table 1). The GC content and skew graph of CCUG37785 (Fig. 1) are characterized by a high-GC content region (0–2 Mbp) followed by a relatively low-GC content region spanning the remainder of the genome, which is similar to those of CD630 (Fig. 2).
TABLE 1.
Sequenced nontoxigenic C. difficile strains in the literaturea
| Name | Size (bp) | Plasmids | GC% | CDS | tRNA | rRNA | Accession | Type | Reference |
|---|---|---|---|---|---|---|---|---|---|
| CCUG37785 | 4,095,098 | 0 | 28.8 | 3733 | 48 | 5 | JAGKRT010000000 | WGS | This study |
| CD37 | 4010233 | 0 | 28.6 | 3,695 | 16 | 5 | AHJJ00000000.1 | WGS | 21 |
| Z31 | 4298263 | 0 | 29.2 | 4,128 | 58 | 29 | CP013196 | CG | 22 |
| 5.3 | 4009318 | 0 | 28.3 | 3,580 | 82 | 46 | AZSH00000000.1 | WGS | 14 |
| DSM 28666 | 4122585 | 1 | 29.0 | 3802 | 89 | 36 | CP012321.1 | CG | 24 |
| DSM 29637 | 4114585 | 0 | 28.6 | 3833 | 89 | 35 | CP016106.1 | CG | |
| DSM 29688 | 4228964 | 0 | 28.9 | 4022 | 90 | 35 | CP019858.1 | CG | |
| DSM28670 | 4191653 | 0 | 28.8 | 3972 | 91 | 35 | CP012312 | CG | |
| DSM 29629 | 4111235 | 0 | 28.6 | 3799 | 90 | 35 | CP016104 | CG | |
| DSM 28669 | 4137693 | 0 | 28.8 | 3901 | 90 | 35 | CP012323 | CG |
GenBank annotations of nontoxigenic strains were downloaded as either whole-genome shotgun (WGS) sequences or as complete genome (CG) files for use in further analysis.
FIG 1.
The GC content (black graph) and GC skew (green and purple graph) of the CCUG37785 genome. Figure was made with BLAST Ring Image Generator (BRIG).
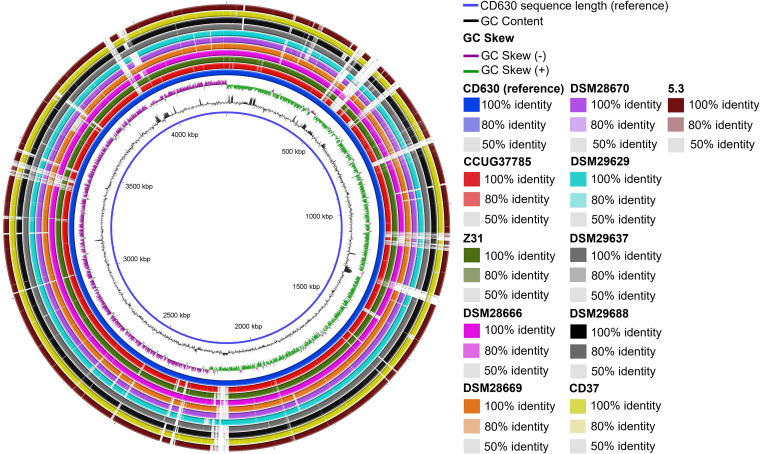
FIG 2.
Sequence identity between nontoxigenic C. difficile strains. The colored rings illustrate shared sequence identity between the indicated nontoxigenic strains and CD630 (reference strain). White space within the rings indicates low (<50%) sequence identity between the given strain and CD630. The inner blue line indicates the length of the CD630 genome. The GC content and GC skew graphs of strain CD630 are also indicated. Figure made with BLAST Ring Image Generator (BRIG).
The sequence type (ST) of this isolate based on public MLST typing was determined to be ST3. The RAST annotation software predicted 48 tRNA genes and 5 rRNA genes in CCUG37785 (Table 1). No plasmids were predicted in CCUG37785 after searching with PlasmidFinder Software (20). Compared with the reference genome of toxigenic strain CD630, the CCUG37785 genome contained none of the toxin genes of the PaLoc (tcdA, tcdB, tcdR, tcdC or tcdE) or CdtLoc (cdtA or cdtB). The result was confirmed by our toxin expression assay by ELISA (Fig. S1 in the supplemental material) and PCR (data not shown). For comparison, sequenced nontoxigenic C. difficile strains were mined from the literature (21–24) (Table 1). Strain CD37, for example, was the first nontoxigenic C. difficile with draft genome analyzed (21), whereas strain Z31 was the first nontoxigenic C. difficile strain with its complete genome sequence reported and studied (22).
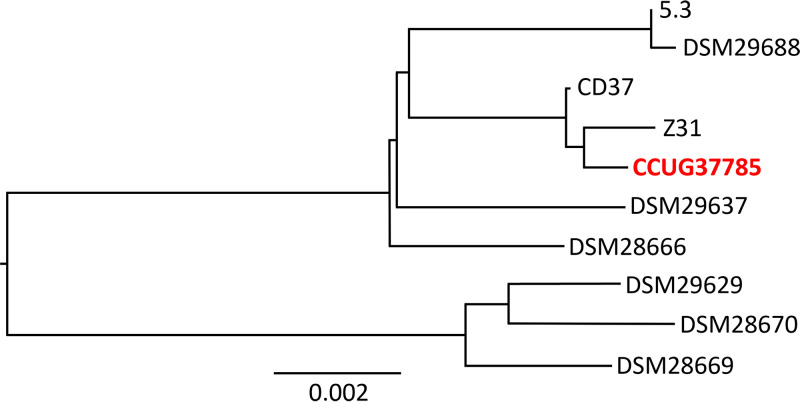
Genomes of all 10 strains in Table 1 were uploaded to the Type Strain Genome Server (TYGS) (25) to generate a minimum evolution phylogenetic tree (Fig. 3). Strain CCUG37785 is most closely related to strains Z31 and CD37. Yet, the scale line in Fig. 3 indicating 0.002 nucleotide substitutions per site suggests that, in fact, all the nontoxigenic strains included in the phylogenetic tree share significant sequence identity with CCUG37785. BLASTn analysis confirms this observation, as using the CCUG37785 genome as a query to search against nontoxigenic strains reveals that other strains share >98% identity and >90% query cover with CCUG37785 (Table S1 in the supplemental material). The selected nontoxigenic strains share significant sequence identity with the toxigenic reference strain CD630 (Fig. 2). Also, while nontoxigenic strains contain some genomic regions with low sequence identity to CD630 (<50%, white regions in each ring), these occur at similar locations in the nontoxigenic strains analyzed (e.g., ∼500 kbp, ∼800 kbp, etc.).
FIG 3.
Phylogenetic tree of nontoxigenic C. difficile strains. A minimum evolution phylogenetic tree was generated using the Type (Strain) Genome Server (TYGS).
Prophages.
Previously reported nontoxigenic C. difficile strains have been found to encode numerous prophage regions, such as those of Z31 (22). In CCUG37785, the PHASTER program predicted 3 incomplete prophage-like regions and one putative complete prophage (Table 2). The intact prophage (region 3) was predicted to have a length of 56.6 Kb with a GC content of 29.2%, which are both in agreement with the length and GC% ranges observed in C. difficile bacteriophages (26). A draft annotation of the intact prophage-like region is provided in Table S2 in the supplemental material.
TABLE 2.
Putative prophage carriage in CCUG37785
| Region | Length | Position | Completeness | Score | # Putative ORFs | %ORFs with an assigned function | GC % |
|---|---|---|---|---|---|---|---|
| 1 | 13.7Kb | 234102-247839 | Incomplete | 30 | 11 | 100 | 30.2 |
| 2 | 27.4Kb | 1313979-1341418 | Incomplete | 60 | 31 | 67.7 | 27.6 |
| 3 | 56.6Kb | 1554571-1611218 | Intact | 150 | 76 | 60.5 | 29.2 |
| 4 | 5.8Kb | 2455294-2461108 | Incomplete | 20 | 19 | 57.9 | 23.8 |
Comparative analysis of PaLoc.
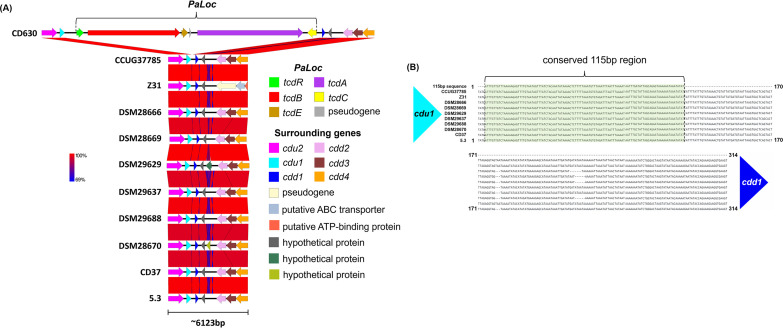
The pathogenicity locus (PaLoc) of C. difficile is made up of a 19.6 kb region encoding five genes including the large clostridial toxins A and B (27), which are the main cause of the negative health effects associated with C. difficile infection (28–30). The other three surrounding genes are tcdR, tcdC, and tcdE. Gene tcdR plays a role in positive transcriptional regulation, tcdC negatively regulates toxin transcription, and tcdE facilitates toxin secretion (27, 31, 32). The PaLoc region is highly conserved, and nontoxigenic strains lack the entire region (33). Instead, nontoxigenic strains contain a conserved 115-bp sequence in place of the PaLoc region that is unique to nontoxigenic strains (8, 9, 27, 31, 34).
Upstream of the PaLoc region (or 115-bp insert in nontoxigenic strains), C. difficile genomes encode the genes cdu1, cdu2, and cdu3, while downstream genes include cdd1, cdd2, cdd3, and cdd4 (31). In light of this knowledge, we also mined the PaLoc and surrounding regions of CCUG37785 in comparison with those of CD630 and the sequenced NTCD strains listed in Table 1. Comparative analysis of PaLoc regions demonstrates how the arrangement of genes adjacent to PaLoc is conserved in toxigenic and nontoxigenic strains (Fig. 4A). We demonstrate that the cdu1 and cdu2 genes upstream of the PaLoc of strain CD630 share high sequence similarity to genes upstream of the PaLoc of CCUG37785 and other nontoxigenic strains. The downstream genes of the CD630 PaLoc also display high similarity with the sampled strains. In between the cdu1 and cdd1 genes, toxigenic strains would normally encode the toxin genes and regulatory genes of PaLoc, but CCUG37785 and other nontoxigenic strains have a conserved 115-bp sequence as reported (31) (Fig. 4B). CCUG37785 genome contains a 115-bp sequence upstream of cdd1 (dark blue) and downstream of cdu1 (light blue) that is identical to the reported sequence of the insert characterized in the reported nontoxigenic strains (Fig. 4B).
FIG 4.
(A) Comparative analysis of PaLoc region. Downstream of cdu2 (pink) and cdu1 (light blue), toxigenic strains like CD630 encode toxin-related genes starting with tcdR (green) and ending with tcdC (yellow). Downstream of the PaLoc, CD630 encodes cdd1 (dark blue), cdd2 (pink), cdd3 (maroon), and cdd4 (orange). Nontoxigenic strains share virtually 100% identity with these aforementioned genes upstream and downstream of the PaLoc region, but nontoxigenic strains substitute the toxin-related genes for a 115 bp insert in between cdu1 and cdd1. (B) Conserved sequences replace the PaLoc in nontoxigenic strains. Between the cdu1 and cdd1 genes, nontoxigenic strains encode a conserved 115 bp sequence in place of the typical PaLoc genes of toxigenic strains. Sequences were mined from their respective genomes and aligned using MAFFT.
Comparative analysis of CdtLoc.
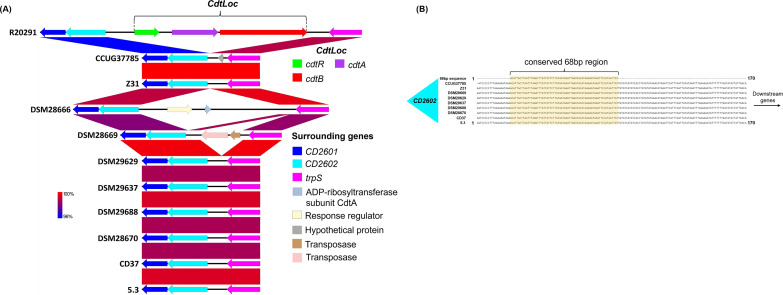
Hypervirulent strains of C. difficile sometimes also encode a binary toxin (CDT), which is associated with greater disease severity (5). The CdtLocus (CdtLoc) is a 6.2-kB region that encodes the CDTb and CDTa toxins along with a regulatory gene cdtR (35, 36). While the PaLoc region can exist in many shortened forms depending on the specific strain (37), CdtLoc is either entirely present or absent in C. difficile genomes (38). C. difficile genomes that lack the CdtLoc regions instead have a conserved 68-bp sequence that is not found in binary toxin-positive strains (36). The CdtLoc or the 68-bp sequence is flanked upstream by CD2601 and CD2602 and downstream by trpS, the response regulator associated with cdtR (5).
Comparative analysis of CdtLoc regions shows that the genes upstream of CdtLoc including CD2601 (dark blue) and CD2602 (light blue) are present in nontoxigenic strains including CCUG37785 (Fig. 5A). trpS, which is encoded downstream of the CdtLoc in R20291, is also present in all the nontoxigenic strains examined. The CdtLoc appears to be more heterogeneous than the PaLoc regions shown in Fig. 4, as DSM28666 and DSM28669 display additional sequences and/or genes between CD2602 and trpS that are dissimilar to other strains. Further analysis of the sequences between CD2602 and trpS is shown in Fig. 5B. The figure illustrates that most of the CDT-negative strains examined, including CCUG37785, contain the conserved 68-bp sequence in place of CdtLoc. DSM28666 is the one exception to this trend, as no significant similarity was found between the 68-bp sequence and the DSM28666 genome.
FIG 5.
(A) Comparative analysis of CdtLoc region. The example binary toxin-positive strain CD196 encodes the genes CD2601 (dark blue) and CD2602 (light blue) upstream of binary toxin-related genes and trpS downstream. Nontoxigenic strains also encode CD2601, CD2602, and trpS, but the binary toxin genes are absent. DSM28666 and DSM28669 do, however, encode genes for a response regulator (cream), hypothetical proteins (gray), and transposases (pink, brown) that are not homologous to sequences at this genomic locus/in this region in other nontoxigenic strains. (B) Conserved sequences replace the CdtLoc in nontoxigenic strains. Between the CD2602 and trpS genes, nontoxigenic strains encode a conserved 68 bp sequence in place of the typical CdtLoc genes of binary toxin-positive strains. Sequences were mined from their respective genomes and aligned using MAFFT.
Strain CCUG37785 shows significantly higher sporulation rates and adherence to human intestinal epithelial cells than R20291.
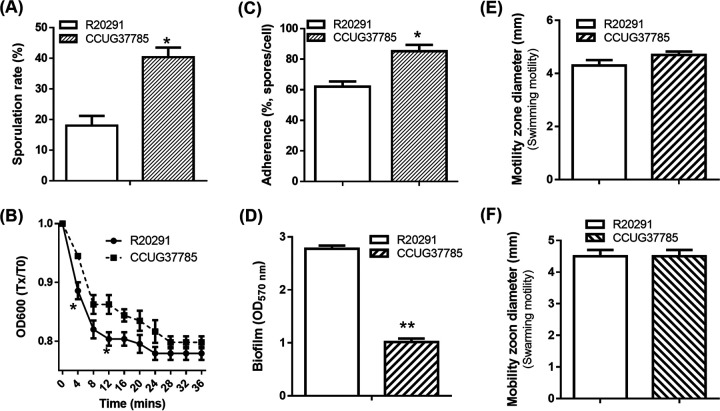
We first compared strain CCUG37785 with R20291 in sporulation and adherence to intestinal epithelial cells. As shown in Fig. 6A, CCUG37785 exhibited a significantly higher frequency of sporulation (40.33 ± 5.38%), than R20291 (18.03 ± 4.73%; *P < 0.05). Purified C. difficile spores were used to evaluate their adherence to HCT-8 cells. CCUG37785 showed a significantly higher adherence rate (85.22 ± 4.15%) than R20291 (62.00 ± 3.47%) (Fig. 6C; *P < 0.05).
FIG 6.
Comparison of C. difficile strains R20291 and CCUG37785 in sulation, germination, adhesion, motility, biofilm formation and motility. (A) Sporulation assay. (B) Germination assay. For C. difficile germination analysis, the purified spores were diluted to an OD600 0f 1.0 in the germination buffer. The value of OD600 was monitored immediately (0 min, t0), and was detected once every 2 min (tx) for 20 min at 37°C. The germination ratio was calculated as OD600 (Tx)/OD600 (T0). Spores in germination buffer without TA was used as the negative control. (C) Adherence of C. difficile vegetative cells to HCT-8 cells in vitro. (D) Biofilm formation analysis. (E and F) Halo diameter of motility zones (swimming analysis on 0.175% agar plate, swarming analysis on 0.3% agar plate). Experiments were independently repeated thrice. One-way analysis of variance (ANOVA) was used for statistical significance. Data are present as “Mean±SD”. ∗, P < 0.05; ∗∗, P < 0.01.
Strain CCUG37785 exhibits lower germination rates and biofilm formation, but comparable motility in comparison with R20291.
The germination capability of CCUG37785 spores was evaluated. As shown in Fig. 6B, strain CCUG37785 exhibited significantly lower germination rates than R20291 (*P < 0.05). The capability of C. difficile CCUG37785 to form biofilms was also evaluated by crystal violet (CV) staining after growth in microtiter plates. Strain CCUG37785 exhibited significantly lower levels of biofilm formation than R20291 (Fig. 6D, **P < 0.01). To assess the motility of CCUG37785 vegetative cells, swimming and swarming assays were performed. As shown in Fig. 6E and F, strain CCUG37785 showed similar motility to strain R20291 (swimming assay, P = 0.1265; swarming assay, P < 0.999).
Strain CCUG37785 protects mice against infection with an epidemic C. difficile strain.
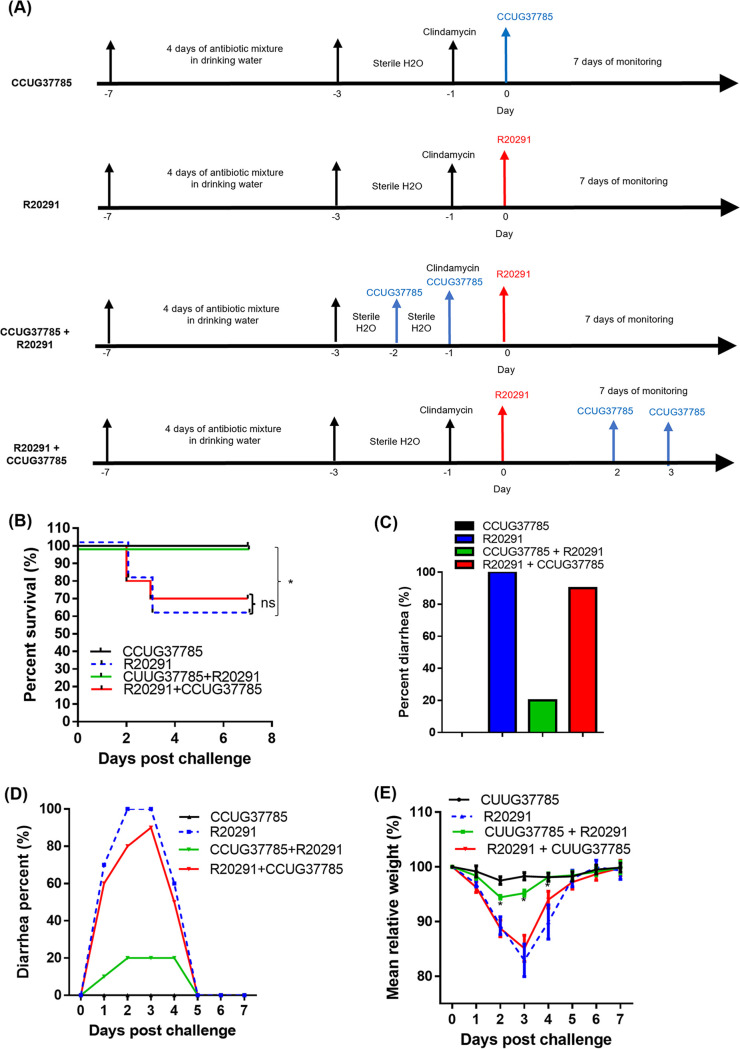
We further evaluated whether or not the strain CCUG37785 can protect mice against infection with the strain R20291. The experimental scheme is illustrated in Fig. 7A. Four groups of mice (n = 10) were used. Group 1 (CCUG37785) was challenged with 106 CCUG37785 spores as a control; Group 2 (R20291) was challenged with 106 R20291 spores as a non-treatment group; Group 3 (CCUG37785/R20291) was given 106 CCUG37785 spores, followed by challenge with 106 R20291 spores; Group 4 (R20291/CCUG37785) was infected with 106 R20291 spores, followed by administering 106 CCUG37785 spores. Four groups of mice were monitored for mortality (Fig. 7B), weight loss (Fig. 7E) and diarrhea (Fig. 7C and D). While all mice in group R20291 developed diarrhea, only 20% mice from the group CCUG37785 + R20291 developed diarrhea (Fig. 7C and D). All mice in group CCUG37785 + R20291 mice survived, which is significantly higher than survival of group R20291 mice (60%) (*P < 0.05) (Fig. 7B). Mice in group R20291 also lost significantly more weight than those in group CCUG37785 + R20291 (P < 0.05, on postinfection days 2 and 3). As expected, mice from the control group CCUG37785 did not develop any signs of disease, neither diarrhea (Fig. 7C and D) nor weight loss (Fig. 7E), and all mice survived (Fig. 7B). In group R20291 + CCUG37785, 90% mice developed diarrhea (Fig. 7C and D) and 70% mice survived (Fig. 7B). Also, there was no significant difference between the weight loss of group R20291 + CCUG37785 and group R20291 (Fig. 7E). These results indicate that mice treated with CCUG37785 spores after infection with R20291 only provided limited protection against challenge with strain R20291.
FIG 7.
C. difficile CCUG37785 protects mice against infection with C. difficile R20291. (A) Experimental scheme of testing if strain CCUG37785 can protect mice against infection with the strain R20291. After 4 days of pretreatment with an antibiotic mixture, mice were given autoclaved water for 2 days, followed by a single dose of intraperitoneal injection with clindamycin (10 mg/kg) 1 day before (day-1) challenge with R20291 spores by gavage (day 0). Group 1 (CCUG37785) was challenged with 106 CCUG37785 spores on day 0; Group 2 (R20291) was challenged with 106 R20291 spores on day 0; Group 3 (CCUG37785/R20291) was given 106 CCUG37785 spores by gavage on days -1 and -2, followed by challenge with 106 R20291 spores on day 0; Group 4 (R20291/CCUG37785) was infected with 106 R20291 spores on day 0, followed by giving 106 CCUG37785 spores by gavage on days 2 and 3. Mice were monitored for disease symptoms for 7 days. (B) Percent survivals, (C) total percentage of diarrhea during the infection period, (D) daily percentage of diarrhea during the infection period, and (E) weight change of four groups of mice were plotted. Data are presented as mean ± SD (n = 10). ∗, P < 0.05, compare between group R20291 and group CUUG37785 + R20291. ns, no significantly.
Pretreatment of mice with CCUG37785 spores before infection with R20291 spores decrease the R20291 spores and toxin levels in feces.
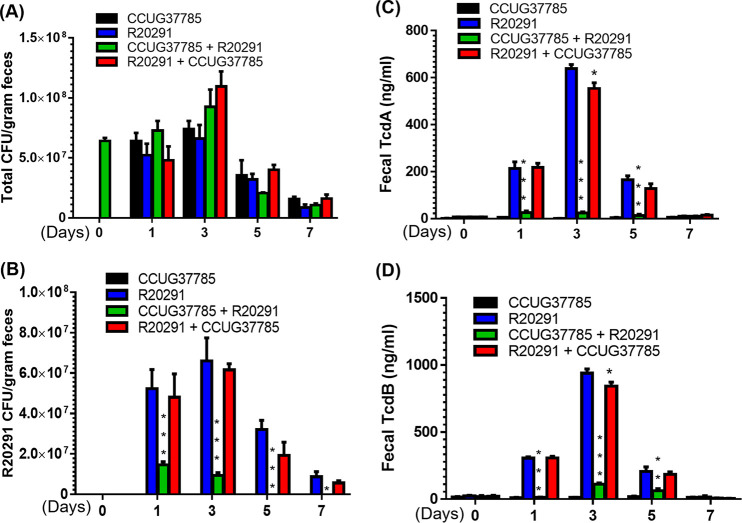
Mice from the group CCUG37785 + R20291 excreted much less toxins in feces as compared with the group R20291 (Fig. 8C and D). No toxins were detected in feces from group CCUG37785 (Fig. 8C and D). However, group R20291 + CCUG37785 excreted less TcdA (Fig. 8C, *P < 0.05) and TcdB (Fig. 8D, *P < 0.05) than group R20291 only in day 3 and provided limited protection.
FIG 8.
C. difficile spores and toxin levels in fecal samples of mice in four groups. (A) Total C. difficile spore numbers; (B) C. difficile R20291 spore numbers; (C) Tcd A level; (D) Tcd B level. Comparisons were made between groups (CCUG37785 + R20291) or (R20291 + CCUG37785) and R20291. (Experiments were repeated 3 times, and representative data were shown. Data are present as “Mean±SD”. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001).
Without differentially enumerating CCUG37785 and R20291 spores from mouse feces, we could not determine R20291 spore changes in the gut (Fig. 8A). Therefore, we used PCR to assess the proportion of the R20291 in the C. difficile spores of fecal samples. As shown in Fig. 8B, pretreatment of mice with CCUG37785 before infection (group CCUG37785 + R20291) secreted 70% less amount of C. difficile R20291 spores on day 1, 90% less on day 3, and R20291 spores were not detected on days 5 and 7. Treatment with CCUG37785 spores after infection (group R20291 + CCUG37785), on the other hand, only slightly reduced R20291 spores from day 3 (P = 0.6710). Interestingly, in the group R20291 + CCUG37785, the proportion of the R20291 spores in fecal samples decreased over time; it is about 70% on day 3, 50% on day 5, and 40% on day 7, suggesting that CCUG37785 gradually replaced R20291 in the gut.
DISCUSSION
In this study, we demonstrated that PaLoc and CdtLoc loci in strain CCUG37785 are replaced with a conserved 115-bp fragment and a conserved 68-bp fragment in these two regions, respectively, conforming to most NTCD strains. We phenotypically characterized strain CCUG37785 in comparison with C. difficile R20291 and found that CCUG37785 exhibited significantly higher adhesion and sporulation rates, similar motile ability, but lower spore germination rates and biofilm formation (Fig. 6). The ability to adhere to colonic mucosal cells or mucus is important for C. difficile to establish colonization (14, 17). Therefore, strain CCUG37785 could displace R20291 in the intestine for its significantly higher adhesion and sporulation rates, thereby potentially preventing recurrent CDI.
The data presented here demonstrate that strain CCUG37785 can readily colonize mice but does not cause pathology (Fig. 7). Further, strain CCUG37785 was able to adequately protect mice from subsequent pathogenic C. difficile R20291 infection (Fig. 7), but oral inoculation of CCUG37785 spores after R20291 infection only provided limited protection. These data provide insight into potential mechanisms of protection by strain CCUG37785 possible through colonization resistance. The first colonizing strain of one bacterial species metabolically outcompetes subsequent strains for space and resources (39, 40). In agreement, we found that the CCUG37785 prevention group (CCUG37785 + R20291) secreted much less R20291 spores (Fig. 8B), and much less TcdA and TcdB accordingly (Fig. 8C and D); While the CCUG37785 treatment group (R20291 + CCUG37785) secreted slightly less amount of R20291 spores (Fig. 8B), and slightly less TcdA and TcdB (Fig. 8C and D).
It is known that biofilm formation may confer bacterial cells including C. difficile higher resistance to antibiotics in comparison with planktonic cells. Interestingly, we found that strain CCUG37785 exhibited significantly lower biofilm formation capability than strain R20291, indicating that CCUG37785 could be more susceptible to antibiotic exposure. Indeed, we found that strain R20291 has higher values of MICs on several antibiotics tested (data not shown). However, very limited information is available on clinical effects of C. difficile biofilm formation in vivo.
Taken together, C. difficile CCUG37785 is safe and can be used as a therapeutic strain for the prevention of primary and recurrent CDI in mice; C. difficile CCUG37785 also has advantage of high sporulation rates and high adhesion capability to displace toxigenic strain to establish colonization. Our data is also in support of previous reports (14, 41, 42), which showed that intentional colonization of healthy subjects with NTCD was safe and that NTCD had no major adverse effects when given to healthy subjects with or without antibiotics (41).
In our previous study, we engineered the nontoxigenic strain CCUG37785 to express mTcd138 as a promising oral vaccine candidate against CDI (19). Data in our current study not only highlight the potential use of CCUG37785 to prevent primary and recurrent CDI in humans but also further support its use as an oral mucosal vaccine carrier against CDI. However, toxigenic strain 630Δerm was found to be able to transfer the PaLoc to NTCD strain CD37 in vitro (43), albeit with very low efficiency. Therefore, we should bear this possibility of conversion from NTCD to toxigenic strains in mind although no such conversions have been reported in vivo so far.
MATERIALS AND METHODS
Genome analysis of C. difficile CCUG37785 strain.
Total genomic DNA of the C. difficile CCUG37785 was extracted with a Qiagen genomic DNA extraction kit (Qiagen, USA). After confirming adequate quality of the extracted DNA using agarose gel electrophoresis, and nanodrop- and Qubit- (Thermo Fisher Scientific, USA) based quantification, whole genome sequencing was performed with Illumina technology at Beijing Novogene Bioinformatics Technology Co Ltd. The Illumina platform with massive parallel sequencing (MPS) was used and paired-end (500 bp insert) and mate-pair (5000 bp insert) libraries were constructed.
After assessing the quality of the sequencing reads using FastQC (44), the reads were de novo assembled into contigs using Qiagen CLC Genomics Workbench 11.0.1 (45) using the default settings. Assembled contigs were ordered against the C. difficile reference strain CD630 (GenBank accession no: CP010905.2) using Mauve (version 2.4.0) (46, 47). The draft genome of the CCUG37785 thus obtained was annotated using Rapid Annotation using Subsystems technology (RAST) web server (48). GLIMMER3 (49) was used to call protein-encoding genes, and the tRNAscan-SE tool (50) and “search_for_rnas” tool (developed by Niels Larson, unpublished) were used to call tRNA and rRNA encoding genes, respectively. Web-based Multi Locus Sequence Typing (MLST), using the seven housekeeping genes - adk, atpA, glyA, sodA, dxr, recA and tpi, was conducted with the public MLST tool maintained by the Center for Genomic Epidemiology (51). This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAGKRT000000000. The version described in this paper is version JAGKRT010000000.
Phylogenetic analysis of nontoxigenic C. difficile strains.
Previously isolated and sequenced nontoxigenic C. difficile strains were identified in the literature. For each strain (Table 1), the appropriate genome sequence or accession number was sourced from GenBank. Additional information listed in Table 1 was also gathered from GenBank entries, and the number of plasmids was predicted using PlasmidFinder (20). The sequences and accession numbers were uploaded to the Type Strain Genome Server (TYGS) (25) which compared the genomes using the Genome BLAST Distance Phylogeny approach (GBDP) (52) and generated a minimum evolution tree.
Searching for prophages.
Putative prophages were detected in the bacterial genome using PHASTER (53, 54), which screens for regions with high similarity to a database of phage genes. The software assigned prophage regions a score that determines whether they are considered complete (>90), questionable (70–90), or incomplete (<90).
Annotation of putative prophage-like regions.
Putative prophage FASTAs were downloaded from PHASTER and submitted for automated annotation by the RAST tool kit (RASTtk) annotation engine, an updated version of the Rapid Annotations using Subsystems Technology (RAST) platform (48, 55, 56). The appropriate taxonomy I.D. for C. difficile (1496) was specified, and the “fix frameshift” function was selected. Default settings were used for all other parameters. The functional assignments of each gene were manually curated using the NCBI BLAST-protein tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Genome comparison.
BLAST Ring Image Generator (BRIG) (57) was used to generate comparisons between nontoxigenic strains and the reference strain CD630. The upper, middle, and minimum identity percentage thresholds were set to 100%, 80%, and 50%, respectively.
Analysis of toxin loci.
Sequences of nontoxigenic C. difficile were downloaded from GenBank and then the previously annotated pathogenicity locus (PaLoc) and surrounding genes of each strain were color-coded in Artemis. Color-coded annotations were imported into EasyFig software to generate BLAST comparisons. Upstream and downstream genes from PaLoc were identified in the genome annotations based on previous work describing the conserved cdu and cdd genes in these areas (31). The sequences between the cdu1 and cdd1 genes were extracted in Artemis and aligned using MAFFT (58) through the online MPI Bioinformatics Toolkit (59, 60). A similar method was used to analyze the binary toxin locus (Cdt) and surrounding genes. Genes associated with CdtLoc as well as conserved sequenced in nontoxigenic strains were identified using prior studies (5, 36).
Animals.
All studies followed the Guide for the Care and Use of Laboratory Animals of the National Institutes of health and were approved by the Institutes Animal Care and Use Committee (IACUC) at University of South Florida. Wild-type C57BL/6 mice were purchased from Charles River Laboratories.
Preparation of C. difficile spores.
Sporulation of the C. difficile R20291 and CCUG37785 strains were induced in Clospore medium as described previously (61). Briefly, an overnight 20 mL of C. difficile cultured in Columbia Broth was inoculated into 500 mL of Clospore medium and incubated for 1–2 weeks at 37°C in an anaerobic incubator. The spore culture was centrifuged at 10000g for 20 min, and the pellet was washed 5 times with sterile water and suspended in 10 mL of ddH2O. To purify the spores, 1-mL of spore suspension was layered onto the top of 10 mL of 50% (wt/vol) sucrose in water in a 15-mL tube (62). The gradient was centrifuged at 3200 × g for 20 min, and the spore pellet at the bottom was washed 5 times to remove the sucrose and was resuspended in sterile water. Spore preparations were > 99% pure, and spore concentration was determined by serial dilution on TCCA or BHI plates.
Toxin expression assay.
To evaluate the toxin expression in C. difficile CCUG37785 and R20291, 10 mL of C. difficile cultures were collected at 12, 24, 36 and 48 h of post incubation. The cell density was adjusted to the same with fresh BHIS. The collected C. difficile cultures were centrifuged at 4°C, 8000 × g for 15 min, then the supernatants were filtered with 0.22 μm filter and used for ELISA. Anti-TcdA (PCG4.1, Novus Biologicals, USA) and anti-TcdB (AI, Gene Tex, USA) were used as coating antibodies for ELISA, HRP-Chicken anti-TcdA and HRP-Chicken anti-TcdB (Gallus Immunotech, USA) were used as detection antibodies.
Sporulation and germination assay.
C. difficile sporulation and germination analysis were conducted as reported (63). C. difficile strains were cultured to mid-log phase in BHIS medium supplemented with 0.1% taurocholate (Sigma) at 37°C in an anaerobic chamber. A mixture of 70:30 sporulation medium (70% SMC medium and 30% BHIS medium containing 63 g Bacto peptone, 3.5g protease peptone, 11.1 g BHI medium, 1.5 g yeast extract, 1.06 g Tris base, 0.7 g NH4SO4, and 15 g agar per L) was prepared. Cultures were subsequently diluted to an optical density of 0.5 at OD600, then 150 μL of cultures were added and spread over the surface of a 70:30 medium agar plate. Approximately 24 h after the start of stationary phase (T24), C. difficile cells were scraped from the surface of the plate with a sterile inoculating loop and suspend in approximately 5 mL BHIS to an OD600 = approximately 1.0. 500 μl of samples from the sporulation medium were removed from the anaerobic chamber and mixed 1:1 with 95% ethanol for 15 min to kill vegetative cells. The samples were then returned to the anaerobic chamber, 100 μl of the ethanol-treated cultures were mixed with 100 μl of 10% taurocholate, and the mixtures were plated onto BHIS agar to induce C. difficile spore germination. The ethanol resistant CFU/mL was determined after incubation for 24h and was divided by the total CFU/mL of the nonethanol treated cultures.
For C. difficile germination analysis, the purified spores were diluted to an OD600 0f 1.0 in the germination buffer (10 mM Tris (pH 7.5), 150 mM NaCl, 100 mM glycine, 10 mM taurocholic acid [TA]) to detect germination ratio. The value of OD600 was monitored immediately (0 min, t0), and was detected once every 2 min (tx) for 20 min at 37°C. The germination ratio was calculated as OD600 (Tx)/OD600 (T0). Spores in germination buffer without TA was used as the negative control.
Adherence of C. difficile spores to HCT-8 cells.
The adherence of the C. difficile spores to human gut epithelial cells was assessed as described previously (64). Briefly, HCT-8 cells were grown to 95% confluence (5 × 105/well) in a 6-well plate and then moved into the anaerobic chamber, followed by infection with 5 × 106 C. difficile spores at a multiplicity of infection (MOI) of 10:1. The plate was cultured at 37°C for 100 min in an anaerobic chamber. After incubation, the cell-spore mixture was washed three times with 1×PBS via centrifugation at 800 × g for 1 min to remove any unattached spores. The supernatants after centrifugation from each wash step were collected to enumerate any spores that did not adhere to the cells. The spores in the supernatant were enumerated on prereduced BHI agar supplemented with 10% taurocholic acid (wt/vol). The controls included PBS incubated with spores and RPMI incubated with spores, and the adhesion assays were performed in triplicate. The percentage of spore adherence was calculated using the following formula: (initial CFU/mL – eluted CFU/mL)/initial CFU/mL.
Motility.
Motility assays were performed to assess the swimming and swarming behavior of the C. difficile strains (65, 66). C. difficile CCUG37785 and R20291 were cultured to optical OD600 of 0.8. For swimming analysis, 2 μl of different C. difficile cultures were penetrated into soft BHIS agar (0.175%) plates, meanwhile, 2 μl of cultures were dropped onto 0.3% BHIS agar plates for swarming analysis. The swimming assay plates were incubated for 24 h and the swarming plates were incubated for 48 h, respectively. Motility was quantitatively determined by measuring the radius of the zone of motility.
Biofilm assay.
For biofilm analysis, CCUG37785 and R20291 were cultured to OD600 of 0.8, and 1% of C. difficile cultures were inoculated into Reinforced Clostridial Medium (RCM) with 8-well repeats in 96-well plate and incubated in the anaerobic chamber at 37°C for 48 h. The formation of biofilm was analyzed by crystal violet dye. Briefly, C. difficile cultures were removed with pipette carefully. Then 100 μl of 2.5% glutaraldehyde was added into the well to fix the bottom biofilm, and the plate was kept at room temperature for 30 min. Followed, the wells were washed with PBS for 3 times and dyed with 0.25% (wt/vol) crystal violet for 10 min. The crystal violet solution was disposed, and the wells were washed five times with PBS, followed by adding the acetone into wells to dissolve the crystal violet of cells. The dissolved solution was further diluted with ethanol for 2–4 times and then detected at OD570.
Mouse model of C. difficile infection.
A mouse model of C. difficile infection was established as described previously (67, 68). Four groups of C57BL/6 mice (wild-type, n = 10) were given a mixture of five antibiotics including kanamycin (0.4 mg/mL), gentamicin (0.035 mg/mL), colistin (850 U/mL), metronidazole (0.215 mg/mL), and vancomycin (0.045 mg/mL) in the drinking water for 4 days. After 4 days of antibiotic treatment, all mice were given autoclaved water for 2 days, followed by a single dose of clindamycin (10 mg/kg) intraperitoneally (i.p.) 1 day before (day –1) challenge with 106 C. difficile R20291 (for groups 2, 3 and 4) or CCUG37785 (for group 1) spores/mouse by gavage (day 0). Group 1 (CCUG37785) was challenged with 106 C. difficile CCUG37785 spores on day 0; Group 2 (R20291) was challenged with 106 C. difficile R20291 spores on day 0; Group 3 (CCUG37785+R20291) was given 106 C. difficile CCUG37785 spores by gavage on days -1 and -2, followed by challenge with 106 C. difficile R20291 spores on day 0; Group 4 (R20291+CCUG37785) was infected with 106 C. difficile R20291 spores on day 0, followed by given 106 C. difficile CCUG37785 spores by gavage on days 2 and 3. The animals were monitored daily for weight changes, diarrhea and survival, and moribund animals were euthanized. The fecal samples are collected on days 0, 1, 3, 5 and 7 postchallenge.
Evaluation of C. difficile spore numbers and toxin levels in feces.
Fecal samples were collected on postinfection days 0, 1, 3, 5 and 7, and stored in –20°C freezer. Prior to use, 50 mg of feces were dissolved with 500 μl PBS (0.1 g/mL) and treated with 500 μl of absolute ethanol (Sigma-Aldrich) for 60 min at room temperature. Then, samples were thoroughly suspended, serially diluted, and plated onto selective medium supplemented with taurocholate (0.1% wt/vol), Cefoxitin (16 μg/mL), d-cycloserine (250 μg/mL) (TCCA plates). The plates were incubated anaerobically at 37°C for 48 h before the colonies were counted, and the results were expressed in CFU/gram. To determine the toxin levels in feces, 0.1 g/mL of fecal solutions containing protease inhibitor cocktail were diluted 2-fold with PBS and used for determination of TcdA and TcdB levels by ELISA.
To determine the proportion of R20291 spores in the C. difficile spores of fecal samples, colony PCR was performed to amplify the tcdB gene in R20291 to distinguish R20291 from NTCD. Quick C. difficile genomic DNA extraction was conducted as reported earlier (69). Briefly, a C. difficile colony was suspended in a microcentrifuge tube containing 1× PCR buffer with fresh lysozyme solution (500 μg/mL final concentration), incubate 15 min at room temperature. Then, proteinase K (200 μg/mL final concentration) was added and incubated for 1 h at 58°C, followed by heating for 15 min at 90°C. Crude genomic extract was used for PCR using primers: TcdB-F (GTATTACCTAATGCTCCAA) and TcdB-R (CACCTTCATAGTTATCTCTT).
Statistical analysis.
Data were analyzed by Kaplan-Meier survival analysis with a log rank test of significance, by analysis of variance (ANOVA), and by one-way or two-way ANOVA followed with Bonferroni post-tests using the Prism statistical software program. Results are expressed as means ± standard errors of means. Differences were considered statistically significant if P < 0.05.
Data availability.
The data that support the findings of this study are available from the corresponding author, X.S., upon reasonable request. The Whole Genome Shotgun project for the nontoxigenic C. difficile strain CCUG37785 has been deposited at DDBJ/ENA/GenBank under the accession JAGKRT000000000. The version described in this paper is version JAGKRT010000000.
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health grants (R01-AI132711, R01-AI149852). We thank Abhraham L. Sonnenshein at Tufts University for providing strain C. difficile CCUG37785.
We declare that we have no conflict of interest.
The manuscript was written through contributions of all authors. X.S. designed the project and participated in data analysis. S.W., X.S., and J.H. wrote and revised the manuscript. S.W. performed experiments and data analysis. J.H. and I.W. performed sequence analysis. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Xingmin Sun, Email: sun5@usf.edu.
Francisco Uzal, University of California, Davis.
REFERENCES
- 1.Moreno MA, Furtner F, Rivara FP. 2013. Clostridium difficile: a cause of diarrhea in children. JAMA Pediatr 167:592. doi: 10.1001/jamapediatrics.2013.2551. [DOI] [PubMed] [Google Scholar]
- 2.Goudarzi M, Seyedjavadi SS, Goudarzi H, Mehdizadeh Aghdam E, Nazeri S. 2014. Clostridium difficile Infection: Epidemiology, pathogenesis, risk factors, and therapeutic options. Scientifica (Cairo) 2014:916826. doi: 10.1155/2014/916826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuijper EJ, Coignard B, Tull P, Esgcd E. 2006. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect 12:2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 4.Eckert C, Emirian A, Le Monnier A, Cathala L, De Montclos H, Goret J, Berger P, Petit A, De Chevigny A, Jean-Pierre H, Nebbad B, Camiade S, Meckenstock R, Lalande V, Marchandin H, Barbut F. 2015. Prevalence and pathogenicity of binary toxin-positive Clostridium difficile strains that do not produce toxins A and B. New Microbes New Infect 3:12–17. doi: 10.1016/j.nmni.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerding DN, Johnson S, Rupnik M, Aktories K. 2014. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes 5:15–27. doi: 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–U97. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 7.Rupnik M, Avesani V, Janc M, von Eichel-Streiber C, Delmee M. 1998. A novel toxinotyping scheme and correlation of toxinotypes with serogroups of Clostridium difficile isolates. J Clin Microbiol 36:2240–2247. doi: 10.1128/JCM.36.8.2240-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monot M, Eckert C, Lemire A, Hamiot A, Dubois T, Tessier C, Dumoulard B, Hamel B, Petit A, Lalande V, Ma L, Bouchier C, Barbut F, Dupuy B. 2015. Clostridium difficile: New insights into the evolution of the pathogenicity locus. Sci Rep 5:15023. doi: 10.1038/srep15023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingle KE, Elliott B, Robinson E, Griffiths D, Eyre DW, Stoesser N, Vaughan A, Golubchik T, Fawley WN, Wilcox MH, Peto TE, Walker AS, Riley TV, Crook DW, Didelot X. 2014. Evolutionary history of the Clostridium difficile pathogenicity locus. Genome Biol Evol 6:36–52. doi: 10.1093/gbe/evt204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borriello SP, Barclay FE. 1985. Protection of hamsters against Clostridium difficile ileocaecitis by prior colonisation with non-pathogenic strains. J Med Microbiol 19:339–350. doi: 10.1099/00222615-19-3-339. [DOI] [PubMed] [Google Scholar]
- 11.Borriello SP, Welch AR, Barclay FE, Davies HA. 1988. Mucosal association by Clostridium difficile in the hamster gastrointestinal tract. J Med Microbiol 25:191–196. doi: 10.1099/00222615-25-3-191. [DOI] [PubMed] [Google Scholar]
- 12.Wilson KH, Sheagren JN. 1983. Antagonism of toxigenic Clostridium difficile by nontoxigenic C. difficile. J Infect Dis 147:733–736. doi: 10.1093/infdis/147.4.733. [DOI] [PubMed] [Google Scholar]
- 13.Seal D, Borriello SP, Barclay F, Welch A, Piper M, Bonnycastle M. 1987. Treatment of relapsing Clostridium difficile diarrhoea by administration of a nontoxigenic strain. Eur J Clin Microbiol 6:51–53. doi: 10.1007/BF02097191. [DOI] [PubMed] [Google Scholar]
- 14.Gerding DN, Sambol SP, Johnson S. 2018. Non-toxigenic Clostridioides (formerly Clostridium) difficile for prevention of C. difficile infection: From bench to bedside back to bench and back to bedside. Front Microbiol 9. doi: 10.3389/fmicb.2018.01700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambol SP, Merrigan MM, Tang JK, Johnson S, Gerding DN. 2002. Colonization for the prevention of Clostridium difficile disease in hamsters. J Infect Dis 186:1781–1789. doi: 10.1086/345676. [DOI] [PubMed] [Google Scholar]
- 16.Nagaro KJ, Phillips ST, Cheknis AK, Sambol SP, Zukowski WE, Johnson S, Gerding DN. 2013. Nontoxigenic Clostridium difficile protects hamsters against challenge with historic and epidemic strains of toxigenic BI/NAP1/027 C. difficile. Antimicrob Agents Chemother 57:5266–5270. doi: 10.1128/AAC.00580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrigan MM, Venugopal A, Roxas JL, Anwar F, Mallozzi MJ, Roxas BAP, Gerding DN, Viswanathan VK, Vedantam G. 2013. Surface-layer protein A (SlpA) is a major contributor to host-cell adherence of Clostridium difficile. PLoS One 8:e78404. doi: 10.1371/journal.pone.0078404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swanson RN, Hardy DJ, Shipkowitz NL, Hanson CW, Ramer NC, Fernandes PB, Clement JJ. 1991. Invitro and invivo evaluation of Tiacumicin-B and Tiacumicin-C against Clostridium-difficile. Antimicrob Agents Chemother 35:1108–1111. doi: 10.1128/AAC.35.6.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Wang S, Bouillaut L, Li C, Duan Z, Zhang K, Ju X, Tzipori S, Sonenshein AL, Sun X. 2018. Oral immunization with nontoxigenic Clostridium difficile strains expressing chimeric fragments of TcdA and TcdB elicits protective immunity against C. difficile infection in both mice and hamsters. Infect Immun 86. doi: 10.1128/IAI.00489-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carattoli A, Zankari E, Garcìa-Fernandez A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. PlasmidFinder and pMLST: in silico detection and typing of plasmids. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brouwer MS, Allan E, Mullany P, Roberts AP. 2012. Draft genome sequence of the nontoxigenic Clostridium difficile strain CD37. Am Soc Microbiol 8:2125–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pereira FL, Júnior CAO, Silva RO, Dorella FA, Carvalho AF, Almeida GM, Leal CA, Lobato FC, Figueiredo H. 2016. Complete genome sequence of Peptoclostridium difficile strain Z31. Gut Pathog 8:11–17. doi: 10.1186/s13099-016-0095-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darling AE, Worden P, Chapman TA, Chowdhury PR, Charles IG, Djordjevic S. 2014. The genome of Clostridium difficile 5.3. Gut Pathog 6:4–4. doi: 10.1186/1757-4749-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riedel T, Wetzel D, Hofmann JD, Plorin SPEO, Dannheim H, Berges M, Zimmermann O, Bunk B, Schober I, Spröer C, Liesegang H, Jahn D, Overmann J, Groß U, Neumann-Schaal M. 2017. High metabolic versatility of different toxigenic and non-toxigenic Clostridioides difficile isolates. Int J Med Microbiol 307:311–320. doi: 10.1016/j.ijmm.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 25.Meier-Kolthoff JP, Göker M. 2019. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:1–10. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heuler J, Fortier L-C, Sun X. 2021. Clostridioides difficile phage biology and application. FEMS Microbiol Rev 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond GA, Johnson J. 1995. The toxigenic element of Clostridium difficile strain VPI 10463. Microbial Pathogenesis 19:203–213. doi: 10.1016/S0882-4010(95)90263-5. [DOI] [PubMed] [Google Scholar]
- 28.Voth DE, Ballard JD. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 18:247–263. doi: 10.1128/CMR.18.2.247-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyras D, O'Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN, Rood JI. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179. doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawson PA, Citron DM, Tyrrell KL, Finegold SM. 2016. Reclassification of clostridium difficile as clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe 40:95–99. doi: 10.1016/j.anaerobe.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Braun V, Hundsberger T, Leukel P, Sauerborn M, von Eichel-Streiber C. 1996. Definition of the single integration site of the pathogenicity locus in Clostridium difficile. Gene 181:29–38. doi: 10.1016/S0378-1119(96)00398-8. [DOI] [PubMed] [Google Scholar]
- 32.Hundsberger T, Braun V, Weidmann M, Leukel P, Sauerborn M, Von Eichel‐Streiber C. 1997. Transcription analysis of the genes tcdA‐E of the pathogenicity locus of Clostridium difficile. Eur J Biochem 244:735–742. doi: 10.1111/j.1432-1033.1997.t01-1-00735.x. [DOI] [PubMed] [Google Scholar]
- 33.Cohen SH, Tang YJ, Silva J. Jr, 2000. Analysis of the pathogenicity locus in Clostridium difficile strains. J Infect Dis 181:659–663. doi: 10.1086/315248. [DOI] [PubMed] [Google Scholar]
- 34.Maslanka JR, Gu CH, Zarin I, Denny JE, Broadaway S, Fett B, Mattei LM, Walk ST, Abt MC. 2020. Detection and elimination of a novel non-toxigenic Clostridioides difficile strain from the microbiota of a mouse colony. Gut Microbes 12:1–15. doi: 10.1080/19490976.2020.1851999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perelle S, Gibert M, Bourlioux P, Corthier G, Popoff MR. 1997. Production of a complete binary toxin (actin-specific ADP-ribosyltransferase) by Clostridium difficile CD196. Infect Immun 65:1402–1407. doi: 10.1128/iai.65.4.1402-1407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter GP, Lyras D, Allen DL, Mackin KE, Howarth PM, O'connor JR, Rood J. 2007. Binary toxin production in Clostridium difficile is regulated by CdtR, a LytTR family response regulator. J Bacteriol 189:7290–7301. doi: 10.1128/JB.00731-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rupnik M. 2008. Heterogeneity of large clostridial toxins: importance of Clostridium difficile toxinotypes. FEMS Microbiol Rev 32:541–555. doi: 10.1111/j.1574-6976.2008.00110.x. [DOI] [PubMed] [Google Scholar]
- 38.Stare BG, Delmee M, Rupnik M. 2007. Variant forms of the binary toxin CDT locus and tcdC gene in Clostridium difficile strains. J Med Microbiol 56:329–335. doi: 10.1099/jmm.0.46931-0. [DOI] [PubMed] [Google Scholar]
- 39.Litvak Y, Baumler AJ. 2019. The founder hypothesis: A basis for microbiota resistance, diversity in taxa carriage, and colonization resistance against pathogens. PLoS Pathog 15:e1007563. doi: 10.1371/journal.ppat.1007563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. 2013. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villano SA, Seiberling M, Tatarowicz W, Monnot-Chase E, Gerding DN. 2012. Evaluation of an Oral Suspension of VP20621, spores of nontoxigenic Clostridium difficile strain M3, in healthy subjects. Antimicrob Agents Chemother 56:5224–5229. doi: 10.1128/AAC.00913-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Natarajan M, Walk ST, Young VB, Aronoff DM. 2013. A clinical and epidemiological review of non-toxigenic Clostridium difficile. Anaerobe 22:1–5. doi: 10.1016/j.anaerobe.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brouwer MS, Roberts AP, Hussain H, Williams RJ, Allan E, Mullany P. 2013. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun 4:2601. doi: 10.1038/ncomms3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom. [Google Scholar]
- 45.QIAGEN. 2018. CLC Genomics Workbench 11.0.1. https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-clc-genomics-workbench/.
- 46.Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. 2009. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu S, Wu H. 2013. More powerful significant testing for time course gene expression data using functional principal component analysis approaches. BMC Bioinformatics 14:6–13. doi: 10.1186/1471-2105-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: A fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365–8366. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alikhan N-F, Petty NK, Zakour NLB, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402–410. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. 2018. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 60.Gabler F, Nam SZ, Till S, Mirdita M, Steinegger M, Söding J, Lupas AN, Alva V. 2020. Protein sequence analysis using the MPI Bioinformatics Toolkit. Curr Protoc Bioinformatics 72:e108. doi: 10.1002/cpbi.108. [DOI] [PubMed] [Google Scholar]
- 61.Perez J, Springthorpe VS, Sattar SA. 2011. Clospore: a liquid medium for producing high titers of semi-purified spores of Clostridium difficile. J AOAC Int 94:618–626. doi: 10.1093/jaoac/94.2.618. [DOI] [PubMed] [Google Scholar]
- 62.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu D, Bullock J, He Y, Sun X. 2019. Cwp22, a novel peptidoglycan cross-linking enzyme, plays pleiotropic roles in Clostridioides difficile. Environ Microbiol 21:3076–3090. doi: 10.1111/1462-2920.14706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Joshi LT, Phillips DS, Williams CF, Alyousef A, Baillie L. 2012. Contribution of spores to the ability of Clostridium difficile to adhere to surfaces. Appl Environ Microbiol 78:7671–7679. doi: 10.1128/AEM.01862-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rashid MH, Kornberg A. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA 97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baban ST, Kuehne SA, Barketi-Klai A, Cartman ST, Kelly ML, Hardie KR, Kansau I, Collignon A, Minton NP. 2013. The role of flagella in Clostridium difficile pathogenesis: comparison between a non-epidemic and an epidemic strain. PLoS One 8:e73026. doi: 10.1371/journal.pone.0073026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, Kelly CP. 2008. A mouse model of Clostridium difficile-associated disease. Gastroenterology 135:1984–1992. doi: 10.1053/j.gastro.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 68.Sun XM, Wang HY, Zhang YR, Chen K, Davis B, Feng HP. 2011. Mouse relapse model of Clostridium difficile infection. Infect Immun 79:2856–2864. doi: 10.1128/IAI.01336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bouillaut L, McBride SM, Sorg JA. 2011. Genetic manipulation of Clostridium difficile. Curr Protoc Microbiol Chapter 9:Unit 9A 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM01788-21_Supp_1_seq12.pdf, PDF file, 0.2 MB (200.2KB, pdf)
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, X.S., upon reasonable request. The Whole Genome Shotgun project for the nontoxigenic C. difficile strain CCUG37785 has been deposited at DDBJ/ENA/GenBank under the accession JAGKRT000000000. The version described in this paper is version JAGKRT010000000.