Abstract
The first 13.6 kb of the mercury and multidrug resistance transposon Tn1696, which includes the class 1 integron In4, has been sequenced. In4 is 8.33 kb long and contains the 5′-conserved segment (5′-CS) and 2.24 kb of the 3′-conserved segment (3′-CS) flanking four integrated cassettes. The 3′-CS region is followed by one full copy and an adjacent partial copy of the insertion sequence IS6100 flanked, in inverse orientation, by two short segments (123 and 152 bp) from the outer right-hand end of class 1 integrons. This structure is representative of a distinct group of class 1 integrons that differs from In2, found in Tn21, and other related class 1 integrons. In4 does not include transposition genes but is bounded by characteristic 25-bp inverted repeats and flanked by a direct duplication of 5 bp of the target sequence, indicating that it was inserted by a transpositional mechanism. In4 lies between the resII and resI sites of a backbone mercury resistance transposon which is >99.5% identical to Tn5036. Although Tn21 and Tn1696 are both classified as members of the Tn21 subfamily of the Tn3 transposon family, the backbone mercury resistance transposons are only 79 to 96% identical. Tn21 also contains a region of about 0.7 kb not found in Tn1696. The integrons In2 and In4 carrying the antibiotic resistance genes have been inserted at different locations into distinct ancestral mercury resistance transposons. Thus, Tn21 and Tn1696 have independent histories and origins. Other transposons (Tn1403 and Tn1412) that include a class 1 integron also have independent origins. In all except Tn21, the integron is located within the res region of the backbone transposon.
Tn1696 and Tn21 are large transposons that confer resistance to mercuric ions and to more than one antibiotic. Tn1696 was originally found in plasmid R1033 (36), isolated from a Pseudomonas aeruginosa strain (42), and Tn21 was found in plasmid NR1 (R100) (10), isolated from a Shigella flexneri strain (29). Early comparisons revealed similarities (40). One is that the antibiotic resistance determinants are located between the transposition gene (tnpA and tnpR) module and the mercury resistance (mer) module, and despite differences in the restriction maps (10, 18), these two regions form stable heteroduplexes (40). The central regions containing the antibiotic resistance determinants, sul1 (sulfonamide resistance) and aadA1 (streptomycin and spectinomycin resistance) in Tn21 and sul1, aacC1 (gentamicin resistance), aadA2, and cmlA1 (chloramphenicol resistance) in Tn1696, also formed hybrids in which the aacC1 and cmlA1 genes appear as single-stranded loops flanking the aadA genes (Fig. 1A). The central regions are now known to be class 1 integrons, In2 in Tn21 and In4 in Tn1696 (4, 14, 43), with one gene cassette (aadA1) in In2 (19, 43, 45) and four (aacC1, orfE, aadA2, and cmlA1) in In4 (2, 8, 13, 44, 50).
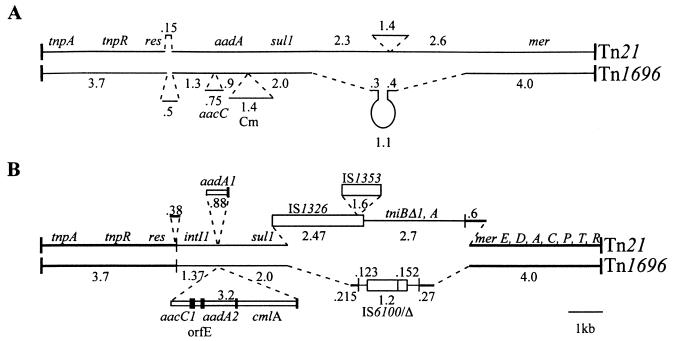
FIG. 1.
Relationship between Tn21 and Tn1696. (A) A schematic representation of the heteroduplex formed by Tn21 and Tn1696. The features shown are from Fig. 4C in reference 40, and gene names and distances are those assigned by the authors. (B) Alignment of Tn21 and Tn1696 sequences. Thick lines represent the backbone transposons and narrow lines the backbones of the integrons. Vertical bars indicate the terminal repeats of the transposons (thick) and integrons (thin). Open boxes show insertion sequences and narrow open boxes with adjacent filled boxes (59-base element) indicate the gene cassettes. Lengths of individual regions are shown in kilobases.
The integrons found in multiply antibiotic-resistant clinical and environmental isolates generally contain one or more integrated cassettes. Most of them have identical integrase genes and belong to the integron class 1. In class 1 integrons, the region located upstream (5′) of the integrated gene cassettes (5′-conserved segment; 5′-CS) encodes the IntI1 integrase that is responsible for cassette insertion, and in the majority, the region 3′ to the cassettes (3′-conserved segment; 3′-CS) includes a sulfonamide resistance determinant, sul1. Integrons of this type are found in many distinct locations (14, 15, 33, 43), including in the plasmids R46 (IncN), R388 (IncW), R751 (IncP), and pVS1 and in the transposons Tn21 and Tn1696, a fact consistent with the notion that class 1 integrons are themselves mobile elements. However, only a single class 1 integron that is an active transposon, namely Tn402, has been found to date. Tn402 (Fig. 2A) contains the complete transposition gene (tni) module consisting of four genes (tniA, B, Q, and R) required for transposition (22, 33) but lacks the 3′-CS and the sul1 gene (33).
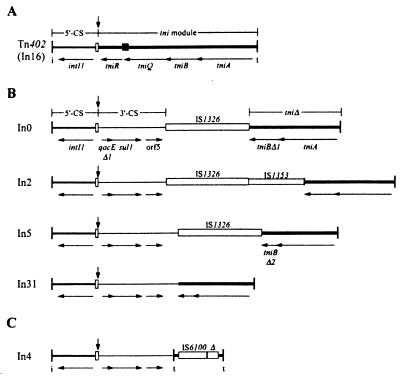
FIG. 2.
Structure of the backbones of class 1 integrons. Each integron is bounded by inverted repeats (vertical bars) designated IRi and IRt (i and t, respectively) and includes the 5′-CS (medium line), which begins with IRi, contains the intI1 gene, and ends at the vertical arrow in the attI1 site (narrow open box). This arrow also marks the position of integrated cassettes. (A) Tn402 (In16) also includes a complete tni module (thick line), with a full set of transposition genes (tniA, B, and Q), a resolvase gene (tniR), and a res site (filled box). Tn402 contains an array of three cassettes, dfrB3-orfD-qacE (not shown). (B) In0, In2, In5, and In31 include the 3′-CS (thin line) and only part of the tni module. The 3′-CS contains qacEΔ1, a truncated version of the qacE cassette, the sul1 sulfonamide resistance determinant, and orf5. Insertion sequences are shown as open boxes. The cassette arrays (not shown) are as follows: In0, no cassettes; In2, aadA1; In 5, aacA(IIa); and In31, blaP3-aacA4-catB6-orfN-qacG. (C) In4 includes two copies of very short regions of the IRt end of the tni module separated by one complete and one partial (Δ) copy of IS6100 (open box). The cassette array is aacC1-orfE-aadA2-cmlA1.
When several independently located class 1 integrons that include the 3′-CS and the sul1 gene were examined, a variety of structures were found (14). All of these integrons include the 1.36-kb 5′-CS (15, 43) and the 2.0-kb 3′-CS region identified by Stokes and Hall (43). Thereafter, the sequences diverge one by one from a conserved sequence which is now known to also be part of the 3′-CS (14), indicating that a variety of different events have shaped the 3′-CS and the region beyond it. Beyond their divergence points, three of the integrons examined, In0 from pVS1, In2 from Tn21, and In5 from pSCH884, are clearly related (4). These three integrons include an insertion sequence, IS1326, and part of the 4-kb segment containing the transposition (tni) genes. In In0, In2, and In5, IS1326 appears to have caused deletions of adjacent sequences, leading to the loss of part of the transposition module (Fig. 2B), and these integrons are thus transposition-defective transposon derivatives (4). A further class 1 integron, In31, is closely related to In5, but the IS1326 element has been lost (23). Similarly, In22, the integron carried by Tn5086, which is a close relative of Tn21, has arisen by the loss of the IS1326 element from the configuration found in In2 or In0 (33). Though members of this group lack some of the tni genes, In0, In2, In5, and In31 are all bounded by inverted repeats of 25 bp (IRi and IRt, at the integrase and tni ends, respectively) that are also found in Tn402 (4, 5, 14, 33), and two (In0 and In2) are flanked by 5-bp direct duplications, suggesting that they moved to their present locations by transposition (5, 14).
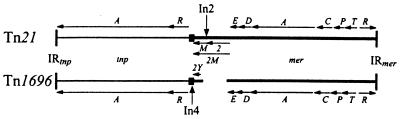
However, not all of the 3′-CS (sul1)-containing class 1 integrons that have been examined in detail have the general structure outlined above. In both In1 (from R46) and In4 (from Tn1696) a region identical to one end of the insertion sequence IS6100 (26) was found beyond the point at which their sequences diverge from the 3′-CS sequence (14). The fact that In2 and In4 sequences diverge within the 3′-CS is consistent with early heteroduplex studies (40) which show substantial differences in the regions flanking the 5′- and 3′-CS (Fig. 1A). Thus, though Tn21 and Tn1696 are clearly related, the integron inserts appear to differ in structure and to be located at slightly different positions in the backbone transposon (Fig. 1A).
Despite the known differences, Tn21 and Tn1696 have generally been regarded as belonging to a single evolutionary pathway in which the insertion of the integron precedes their divergence (3, 12, 27, 41, 50). The sequence of Tn21 has recently been completed (4) and has been compiled (GenBank accession no. AF071413; 25). Here, we have completed the sequence of the first 13.6 kb of Tn1696, which includes all of In4, the tnp module, and part of the mer module of the backbone mercury resistance transposon. Comparison of the two sequences revealed several significant differences between the two transposons, reflecting an independent origin for Tn21 and Tn1696.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli JM109 (Δlac-proAB) supE thi F′ traD36 proAB+ lacIq lacZ ΔM15 was used to propagate plasmid DNA. The plasmids used are listed in Table 1. The pRMH series of plasmids was constructed by randomly cloning fragments from the appropriately digested parental plasmid (pXS2 or R1033) into either pUC18, pUC19 (51), or pACYC184 (6) using standard procedures (37). Plasmids containing the appropriate fragments were identified by screening for antibiotic resistance, by restriction mapping, and by sequencing the fragment ends using universal primer. M13 clones containing the SphI fragment from pRMH484 in both orientations were also constructed. Bacteria were routinely cultured at 37°C in Luria-Bertani medium or Luria-Bertani agar supplemented as appropriate with ampicillin (100 μg ml−1) or chloramphenicol (25 μg ml−1). Antibiotics were obtained from Sigma Chemical Co., St. Louis, Mo.
TABLE 1.
Plasmids
| Plasmid | Description | Relevant phenotype | Reference |
|---|---|---|---|
| R1033 | IncP1; contains Tn1696 | Apr Cmr Gmr Kmr Spr Sur Tcr Hgr | 18 |
| pXS2a | 7.0-kb EcoRI fragment of R1033 in pUC9 | Apr Cmr Sur Hgr | 18 |
| pRMH50 | 9.6-kb SalI fragment of R1033 in pUC18 | Apr Gmr Smr Spr | 14 |
| pRMH483 | 0.8-kb SphI fragment of pXS2 in pUC19 | Apr | This work |
| pRMH484 | 1.7-kb SphI fragment of pXS2 in pUC19 | Apr | This work |
| pRMH493 | 7.3-kb EcoRI fragment of R1033 in pUC19 | Apr Cmr Sur | This work |
| pRMH494 | 0.33-kb PstI fragment of pRMH493 in pUC19 | Apr | This work |
pXS2 was reported to confer Hg but does not contain all of the mer genes.
DNA isolation and restriction mapping.
For large plasmids, plasmid DNA for restriction analysis was isolated using an alkaline lysis method (1). Restriction enzymes were used in accordance with the manufacturers' instructions. Fragments were separated by electrophoresis on 1% (wt/vol) agarose gels and visualized by staining with ethidium bromide. An EcoRI digest of bacteriophage SPP1 (Bresatec, Adelaide, Australia) was used for size markers. Plasmid DNA for sequencing was purified using a Magic Minipreps DNA purification system (Promega Corp., Madison Wis.) or Wizard maxiprep kit (Promega). M13 DNA was prepared by the method of Sanger et al. (38).
DNA sequencing and analysis.
The DNA sequences on both strands of fragments of Tn1696 cloned in M13 or plasmid vectors were determined. Manual DNA sequencing was performed with a Sequenase 2.0 system (46) as recommended by the manufacturer (U.S. Biochemicals, Cleveland, Ohio) using dITP reaction mixtures followed by a 30-min incubation with a 1-mM deoxynucleoside triphosphates mix and terminal deoxynucleotidyl transferase (Boehringer GmbH, Mannheim, Germany). Automated sequencing was performed by SUPAMAC (Sydney University and Royal Prince Alfred Hospital, Sydney, Australia) or by the sequencing facility at the Department of Biological Sciences, Macquarie University Sydney, on an ABI-PRISM 377 sequencer using the Big Dye system. DNA sequences were assembled using Mac Vector 6.5 and AssemblyLIGN (Oxford Molecular Ltd.). GenBank searches were performed using the BLASTN and FastA programs available through WebANGIS (Australian National Genomic Information Service). Programs in the GCG Wisconsin Package, Version 8.1.0, were used via WAG (WebANGIS GCG) to align and analyze DNA sequences.
Nucleotide sequence accession number.
The new sequence has been added to the previous compilation of Tn1696 (In4) under GenBank accession no. U12338.
RESULTS
Structure of In4.
A restriction map of the region of R1033 that includes Tn1696 is shown in Fig. 3. The integron In4 contains four integrated gene cassettes, and the sequence of these cassettes and much of the backbone of In4 has previously been reported (2, 8, 14, 44, 50). The sequence of the remainder of the right-hand (RH) end of In4 was determined (Fig. 4B) and revealed the presence of one complete copy of the insertion sequence IS6100 (26) followed by a partial copy that contains the last 321 bp of IS6100. The partial copy of IS6100 is not present in the clone pXS2 isolated by Hirsch et al. (18) and was presumably lost by homologous recombination after cloning. This explains the loss of a small PstI fragment noted by Hirsch et al. (18). The IS6100-IS6100Δ structure is flanked by short segments from the RH outer end of integrons in inverse orientation, and one of these was identified previously (14). These outer-end segments, 123 bp on the left and 152 bp on the right, both end with the 25-bp sequence that is the RH terminal repeat (IRt) of other class 1 integrons. This configuration is also consistent with the structure seen in electron micrographs (40), namely short inverted repeats (0.1 kb) separated by a region of about 1.1 kb. In4 is thus bounded by the same 25-bp inverted repeats that are found in other class 1 integrons (In2, In0, In5, In31, and Tn402), but it does not contain any of the four tni genes required for the transposition of class 1 integrons and related transposons (22, 33). In4 (Fig. 2C) is thus a transposition-defective transposon derivative.
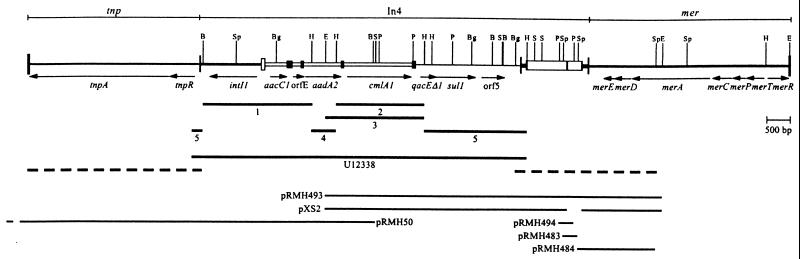
FIG. 3.
Map of Tn1696. The inverted repeats of Tn1696 and In4 are shown as vertical bars. The features of In4 are shown as in Fig. 2C, with each cassette represented as an open box and adjacent filled box (59-base element). Genes are indicated with arrows. Numbered lines show the extent of sequence previously determined by the following: 1, Wohlleben et al. (50); 2, Stokes and Hall (44); 3, Bissonnette et al. (2); 4, Collis and Hall (8); and 5, Hall et al. (14). The line labeled U12338 represents a previous sequence compilation (14), and the regions sequenced during this work are shown by dotted lines. The fragments cloned in various plasmids are also shown. Restriction enzyme sites are as follows: B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; P, PstI; S, SalI; Sp, SphI.
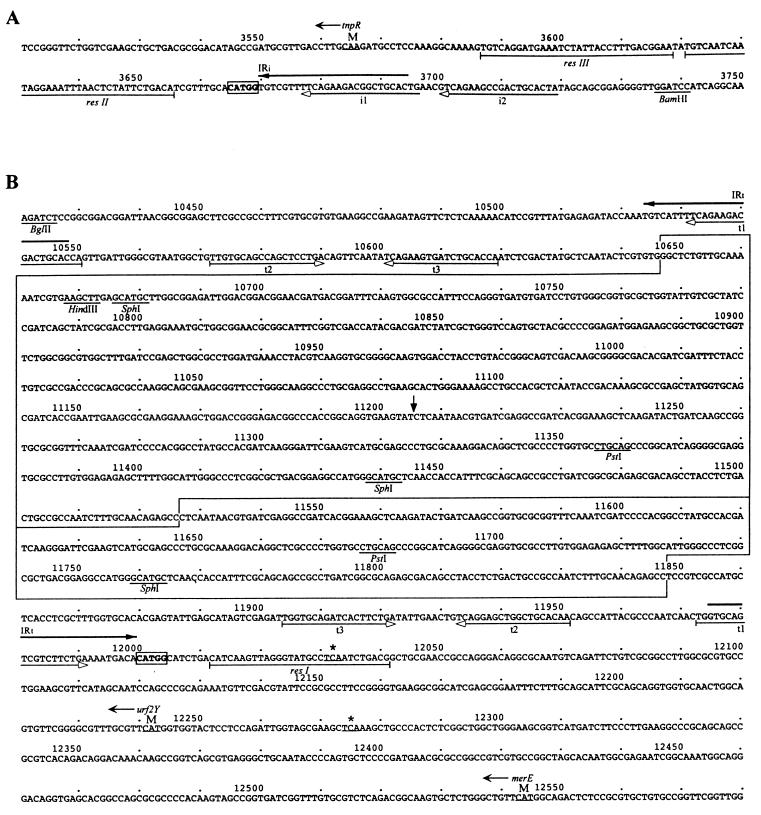
FIG. 4.
Sequence surrounding IRi (A) and IRt (B) of Tn1696. The numbers correspond to those in the extended GenBank entry (accession no. U12338). Thick horizontal arrows indicate the integron ends, IRi and IRt. Five copies of a 19-bp repeat noted by Rådström et al. (33) are indicated by thin arrows below the sequence labeled i1, i2, t1, t2, and t3. The 5-bp direct duplication flanking In4 is shown in bold and is boxed. The three components of the res site are indicated by horizontal bars. The start codons of genes are underlined and indicated by an M, with the name and the direction of the gene indicated above. Positions of stop codons (underlined) are indicated by asterisks. The boxed regions represent IS6100 and IS6100Δ, and the vertical arrow in IS6100 indicates the position corresponding to the first nucleotide of IS6100Δ. Selected restriction enzyme sites are also shown.
Structure of Tn1696 and location of In4.
The sequences of Tn1696 adjacent to the outer ends of In4 (Fig. 4) were compared to other DNA sequences present in GenBank. Both sequences contained regions that are related to sequences found between the tnpR and merE genes in Tn501 and Tn21 and were almost identical to a single continuous region of the mercury resistance transposons Tn5036 (>99.5% identity) isolated from Enterobacter cloacae (accession no. Y09025; 53) and Tn3926 (97% identity) isolated from Yersinia enterocolitica (accession no. X78059; 30). The 3.7-kb region located to the left of In4 extending from IRtnp of Tn1696 includes the tnpA and tnpR genes (Fig. 3; accession no. U12338) and differs from the Tn5036 sequence at only eight positions (Table 2). In R1033, Tn1696 is located between bases 13244 and 13245 in the RP1 (RP4) sequence (accession no. L27758; 32). The 1,588-bp region to the right of In4 sequenced in this study is continuous with the tnp region located to the left of In4 and includes a short open reading frame known as urf2Y, the merD and merE genes, and part of the merA gene. This region also differs from the Tn5036 sequence at very few positions (6 of 1,588). Thus, In4 was inserted into Tn5036 or a close relative to generate Tn1696. In4 is located between the resII and resI regions of the res site, which was identified by comparison to the experimentally defined res sites of Tn21 and Tn1721 (16, 35). In4 is flanked by a direct duplication of 5 bp of the target sequence (boxed in Fig. 4), indicating that it was inserted by a transposition event.
TABLE 2.
Nucleotide and amino acid differences between Tn5036 and the Tn1696 backbone
| Positiona | Nucleotide(s)
|
Location (gene) | Amino acid(s)
|
||
|---|---|---|---|---|---|
| Tn5036 | Tn1696 | Tn5036 | Tn1696 | ||
| 1404 | A | C | tnpA | Ser | Ala |
| 2360 | G | C | tnpA | Ala | Gly |
| 2780 | G | A | tnpA | Ser | Leu |
| 3036 | A | G | tnpR | Thr | Thr |
| 3078 | C | T | tnpR | Gln | Gln |
| 3432 | A | C | tnpR | Leu | Leu |
| 3460 | C | G | tnpR | Cys | Ser |
| 3647 | A | AT | resII | ||
| 3861 | A | T | urf2Y | Val | Glu |
| 4416 | G | A | merD | Phe | Phe |
| 4424 to 4425 | CG | GC | merD | ArgVal | ArgLys |
| 4584 | C | T | |||
| 4682 | C | T | merA | Gln | Gln |
Numbers refer to position in Tn5036 sequence, accession no. Y09025.
In2 and In4 have different locations within distinct but related transposons.
Though the Tn1696 sequence flanking In4 is related to sequences found in Tn21, comparison of the sequences of the genes in the Tn21 backbone and in Tn5036 (25) revealed that the levels of identity of the individual genes range from 79 to 96%. TnX, the ancestral mercury resistance transposon for Tn21, also includes a region of 782 bp located between the urf2Y and merE genes that replaces a 65-bp segment in Tn5036 or Tn1696, and In2 is located within this extra segment (25). The location in the Tn21 sequence corresponding to the position of the In4 insertion is separated by 378 bp from the position of In2 in Tn21 (Fig. 5). In heteroduplexes formed between Tn21 and Tn1696 DNA, Schmidt and Klopfer-Kaul (40) found single-stranded loops adjacent to the IRi of the integrons, and the lengths of these loops were estimated to be 0.15 kb in Tn21 and 0.5 kb in Tn1696 (Fig. 1A). From the sequence data, it is clear that these loops have been incorrectly assigned and that the longer single-stranded loop is part of Tn21, as shown in Fig. 1B. The presence of single-stranded loops in both transposons can be explained if the heteroduplex between the 0.1- to 0.15-kb region adjacent to the integron insertion point in Tn1696 and the corresponding region of Tn21 is not as stable as that of surrounding regions. In the region between the start codon of the tnpR gene and the left end of In4, only 68 of 104 bases (65%) are identical in Tn1696 and Tn21, and this is sufficiently low to explain both the observed loop in Tn1696 and the length of the loop in Tn21.
FIG. 5.
Alignment of the Tn21 and Tn1696 backbones. Vertical bars represent the terminal inverted repeats of the transposons. The tnp regions are shown as thin lines, the mer regions as thick lines, and res sites as filled boxes. Vertical arrows mark the sites of integron insertions. tnp and mer genes are lettered. urf2M (2M) in the Tn21 backbone is split into urf2 (2) and tnpM (M) by the insertion of In2. urf2Y (2Y) of Tn1696 corresponds to the end of urf2M and tnpM.
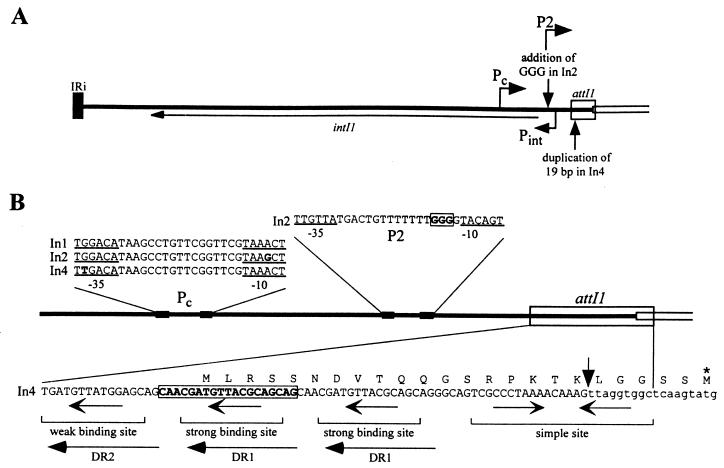
Variation in the 5′-CS of In2 and In4.
The sequence of the 5′-CS has been determined for many different class 1 integrons and little variation has been found. The sequence of In1 found in R46, which was the first complete sequence of the 5′-CS to be reported (accession no. X06046; 15), is used as the baseline for comparisons. Variation is common in the −10 and −35 boxes of the Pc promoter (formerly Pant or P1) that lies within the intI1 gene and directs transcription of the cassette-encoded genes (43). Differences in Pc vary the strength of this promoter (7, 24). The Pc promoter in In1 is of intermediate strength (24), and In2 and In4 contain, respectively, the weak and the strong version of Pc (Fig. 6). However, the 5′-CS of In2 and In4 each includes a further characteristic variation. In In2, a second promoter, P2, has been created by the insertion of three G residues between bp 1238 and 1239 of the 5′-CS (Fig. 6). This insertion increases the spacing between preexisting −35 and −10 boxes from 14 bp, which is outside the range for functional promoters, to 17 bp, which is optimal (17). The P2 promoter has been shown to be functional (7, 39) and is about sixfold stronger than Pc (weak) (7). P2 is thus responsible for the bulk of transcription in In2 and other integrons that contain this configuration. In4 includes a duplication of 19 bp (50) that is located in the attI1 region (Fig. 6). This duplication has two effects; it duplicates the strong binding site for IntI1 (9) and it creates an in-frame ATG start codon that is used to translate the aacC1 gene found in the first cassette of In4 (50). The resultant AAC(3)-Ia protein contains an extension of 23 amino acids (20).
FIG. 6.
Variations in the 5′-CS. (A) Schematic representation of the 5′-CS. The complete 5′-CS (thick line) and part of the adjacent cassette (parallel lines) are shown. The attI1 site is boxed. The extent of the intI1 gene and the positions of promoters Pint and Pc are indicated. Vertical arrows mark the positions of extra residues present in In2 and In4. The P2 promoter is only present in In2. (B) An expanded view of the promoter and attI1 region. The 5′-CS, adjacent cassette, and attI1 site are indicated as in panel A. Filled boxes show the −10 and −35 regions, which are underlined in the relevant sequences. Differences from the standard 5′-CS sequence (In1; accession no. X06046) are shown in bold and bases that are not present in In1 are boxed. The attI1 sequence shown is from In4, with a vertical arrow indicating the boundary between the 5′-CS (uppercase) and the aacC1 cassette (lowercase). The 7-bp core sites found within the IntI1 binding sites are indicated with arrows. DR1 and DR2 are 15-bp direct repeats. The sequence of the 23-amino-acid extension that results from use of the alternative start codon in the duplication is shown above the DNA sequence, and the normal aacC1 start codon within the cassette is shown by an asterisk.
We examined the possibility that the GGG insertion is diagnostic for integrons carried by transposons that are close relatives of Tn21 in that the backbone is the same (TnX) and the integron is in the same position in this backbone. Amongst over 100 sequences of this part of the 5′-CS in the GenBank database, only In2 (Tn21) and 13 additional entries contain the P2 promoter. In all cases P2 is associated with Pc (weak). The additional sequences include In8 in Tn2603 (31), the integron in Tn2426 (54), and In10 in Tn4000 (41), which are known to be closely related to Tn21 on the basis of restriction mapping and heteroduplex analysis (39). In two further cases, In14 (plasmid R) (47) and an unnumbered integron found in the chromosome of a multidrug-resistant S. flexneri strain (34), the sequence adjacent to the IRi of the integron has been reported (14, 34) and is identical to the Tn21 sequence. The location of the remaining eight integrons (accession no. U17586, X64368, AF156486, AF202975, AF231133, AF255921, AJ009819, and AJ237702) is not currently known. Thus, the presence of the GGG insertion may be indicative of an integron derived from In2 by loss or gain of gene cassettes. However, the integrons in other close relatives of Tn21 (Tn2424, Tn2410, and Tn5056) do not include P2, and this may be indicative of cassette exchange via homologous recombination, with one crossover within the 5′-CS and the second within either a cassette, the 3′-CS, or another homologous region. The 19-bp duplication found in In4 is also present in six database entries (accession no. S68049, X64525, X64369, AF202035, AF207065, and AJ009820) and one further published sequence (48). However, little is known about the location of these integrons.
DISCUSSION
The integrons In2 and In4 represent separate evolutionary lineages. The In0-In2-In5 group and the In4 group share the 5′-CS and 3′-CS regions and are bounded by the 25-bp repeats IRi and IRt. However, in the first group IS1326 has caused the loss of parts of the 3′-CS and the tni module, whereas in In4, IS6100 is present and has caused the loss of most of the tni region. The sequences of Tn1696 adjacent to the outer ends of In4 clearly demonstrate both that In4 and In2 were inserted into two transposons that are significantly diverged and that the integron In4 is inserted in Tn1696 at a different location from that of In2 in Tn21. Since the sequences of the conserved segments (5′-CS, 3′-CS, and RH outer-end regions) of In2 and In4 are close to identical, it seems highly unlikely that the transposition regions of Tn21 and Tn1696 diverged after the insertion of the integron. Therefore, the integron insertions are independent events. Thus, Tn21 is not ancestral to all transposons classified as belonging to the Tn501-Tn21 subgroup of the Tn3 transposon family as is frequently claimed (e.g., see references 3, 12, 27, 39, 41, and 50), and In2, the integron found in Tn21, is not ancestral to all other integrons. Tn21 is simply one of many cases where an integron has been acquired by a plasmid or transposon. Nonetheless, Tn21 and its close relatives such as Tn2603 (4) appear to be widely disseminated amongst bacterial species and also globally. They have clearly contributed significantly to the spread of the resistance genes they contain. Evidence for the globalization of mercury resistance transposons, including Tn5036, has also been reported (53). Tn5036 is from an E. cloacae strain isolated from the intestine of a toad found near a mercury mine in Russia and the closely related Tn3926 is from a Y. enterocolitica strain found in milk in France. R1033, which contains Tn1696, was recovered from a clinical Pseudomonas strain isolated in Spain prior to 1975 (42). This emphasizes the connection between environmental, food-borne, and clinical bacteria.
In addition to the many transposons that are known to be close relatives of Tn21, further class II transposons that do not determine resistance to mercuric ions are known to include an integron. One of these is Tn1403, for which the sequences of the left-hand end of the integron (In28) and of a short region adjacent to the IRi have been determined (49). In28 is located within the resI site, and Tn1403 is unable to resolve cointegrates. As In4 also lies within the res site of Tn5036, it is likely that Tn1696 is unable to resolve the cointegrates formed as transpositional intermediates. Tn1412 is another transposon that contains a class 1 integron. In this case, the complete sequence is known (accession no. L36547), and Tn1412 consists of Tn5563 (52), a class II transposon belonging to the Tn3 subgroup (12), and a complex integron structure here designated In32. In32 also lies within the res site (28), which is located between the tnpA and tnpR genes of the Tn5563 backbone. Thus, transposons in the Tn3 family have acquired integrons on at least four independent occasions to create Tn21, Tn1696, Tn1403, and Tn1412, and these events occurred relatively recently in the evolution of these transposons.
The fact that the three integron insertion events that created Tn1696, Tn1403, and Tn1412 occurred within a relatively short segment may reflect the fact that insertion at other sites would inactivate either transposition functions (tnpR and tnpA) or, in the case of Tn1696, the mercury resistance genes (mer). However, an alternate explanation is that the res region is a preferred target for integron insertions. The latter possibility is supported by an examination of the location of all further class 1 integrons where the left-hand boundary (IRi) has been sequenced. In most cases the adjacent gene encodes a member of the resolvase/invertase family (28). Furthermore, the transposon Tn5053, which has a tni module and outer ends closely related to those of class 1 integrons, has been shown to be preferentially inserted into the mrs region of plasmid RP1 (21). The mrs site is the binding site (res site) for a resolvase (ParA) encoded in the par region of RP1 (RP4) (11). Recently, experimental evidence that Tn5053 also preferentially targets res sites in a number of transposons has been reported (28). Insertions occur preferentially at clusters of positions in or near the res site, and the resolvase is essential to this process. Occasionally, Tn5053 insertions occur at some distance from the res site, up to 200 bp. It is possible that the insertion of In2 into TnX to form Tn21 is similar to these rare insertions, though its position, over 350 bp from the res site, is outside the range observed experimentally. As the location of In2 in Tn21 is an exception, in that it lies quite a distance from res, we conclude that Tn1696 is a better examplar of transposons containing integrons. However, the unusual location of In2 permits the conclusion that an integron is unlikely to have been targeted to this position more than once and hence that all of the transposons with an integron in this position are likely to have evolved from a common ancestor.
ACKNOWLEDGMENTS
We thank Diana Brookes for technical assistance.
The project was supported by a grant from the Australian National Health and Medical Research Council.
REFERENCES
- 1.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bissonnette L, Champetier S, Buisson J-P, Roy P H. Characterization of the nonenzymatic chloramphenicol resistance (cmlA) gene of the In4 integron of Tn1696: similarity of the product to transmembrane transport proteins. J Bacteriol. 1991;173:4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bissonnette L, Roy P H. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of gram-negative bacteria. J Bacteriol. 1992;174:1248–1257. doi: 10.1128/jb.174.4.1248-1257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown H J, Stokes H W, Hall R M. The integrons In0, In2, and In5 are defective transposon derivatives. J Bacteriol. 1996;178:4429–4437. doi: 10.1128/jb.178.15.4429-4437.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown N L, Misra T K, Winnie J N, Schmidt A, Seiff M, Silver S. The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol Gen Genet. 1986;202:143–151. doi: 10.1007/BF00330531. [DOI] [PubMed] [Google Scholar]
- 6.Chang A Y C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collis C M, Hall R M. Expression of antibiotic resistance genes in the integrated cassettes of integrons. Antimicrob Agents Chemother. 1995;39:155–162. doi: 10.1128/aac.39.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collis C M, Hall R M. Site-specific deletion and rearrangement of integron insert genes catalyzed by the integron DNA integrase. J Bacteriol. 1992;174:1574–1585. doi: 10.1128/jb.174.5.1574-1585.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collis C M, Kim M-J, Stokes H W, Hall R M. Binding of the purified integron DNA integrase IntI1 to integron- and cassette-associated recombination sites. Mol Microbiol. 1998;29:477–490. doi: 10.1046/j.1365-2958.1998.00936.x. [DOI] [PubMed] [Google Scholar]
- 10.de la Cruz F, Grinsted J. Genetic and molecular characterization of Tn21, a multiple resistance transposon from R100.1. J Bacteriol. 1982;151:222–228. doi: 10.1128/jb.151.1.222-228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eberl L, Kristensen C S, Givskov M, Grohmann E, Gerlitz M, Schwab H. Analysis of the multimer resolution system encoded by the parCBA operon of broad-host-range plasmid RP4. Mol Microbiol. 1994;12:131–141. doi: 10.1111/j.1365-2958.1994.tb01002.x. [DOI] [PubMed] [Google Scholar]
- 12.Grinsted J, de la Cruz F, Schmitt R. The Tn21 subgroup of bacterial transposable elements. Plasmid. 1990;24:163–189. doi: 10.1016/0147-619x(90)90001-s. [DOI] [PubMed] [Google Scholar]
- 13.Hall R M, Brookes D E, Stokes H W. Site-specific insertion of genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 14.Hall R M, Brown H J, Brookes D E, Stokes H W. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J Bacteriol. 1994;176:6286–6294. doi: 10.1128/jb.176.20.6286-6294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall R M, Vockler C. The region of the IncN plasmid R46 coding for resistance to β-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res. 1987;15:7491–7501. doi: 10.1093/nar/15.18.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hall S C, Halford S E. Specificity of DNA recognition in the nucleoprotein complex for site-specific recombination by Tn21 resolvase. Nucleic Acids Res. 1993;21:5712–5719. doi: 10.1093/nar/21.24.5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harley C B, Reynolds R P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirsch P R, Wang C L, Woodward M J. Construction of a Tn5 derivative determining resistance to gentamicin and spectinomycin using a fragment cloned from R1033. Gene. 1986;48:203–209. doi: 10.1016/0378-1119(86)90078-8. [DOI] [PubMed] [Google Scholar]
- 19.Hollingshead S, Vapnek D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenylyltransferase. Plasmid. 1985;13:17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- 20.Hsiang M W, White T J, Davies J E. NH2-terminal sequence of the aminoglycoside acetyltransferase (3)-I mediated by plasmid RIP 135. FEBS Lett. 1978;92:97–99. doi: 10.1016/0014-5793(78)80730-3. [DOI] [PubMed] [Google Scholar]
- 21.Kholodii G Y, Gorlenko Z M, Lomovskaya O L, Mindlin S Z, Yurieva O V, Nikiforov V G. Molecular characterization of an aberrant mercury resistance transposable element from an environmental Acinetobacter strain. Plasmid. 1993;30:303–308. doi: 10.1006/plas.1993.1064. [DOI] [PubMed] [Google Scholar]
- 22.Kholodii G Y, Mindlin S Z, Bass I A, Yurieva O V, Minakhina S V, Nikiforov V G. Four genes, two ends, and a res region are involved in transposition of Tn5053: a paradigm for a novel family of transposons carrying either a mer operon or an integron. Mol Microbiol. 1995;17:1189–1200. doi: 10.1111/j.1365-2958.1995.mmi_17061189.x. [DOI] [PubMed] [Google Scholar]
- 23.Laraki N, Galleni M, Thamm I, Riccio M L, Amicosante G, Frère J-M, Rossolini G M. Structure of In31, a blaIMP-containing Pseudomonas aeruginosa integron phyletically related to In5, which carries an unusual array of gene cassettes. Antimicrob Agents Chemother. 1999;43:890–901. doi: 10.1128/aac.43.4.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lévesque C, Brassard S, Lapointe J, Roy P H. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene. 1994;142:49–54. doi: 10.1016/0378-1119(94)90353-0. [DOI] [PubMed] [Google Scholar]
- 25.Liebert C A, Hall R M, Summers A O. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63:507–522. doi: 10.1128/mmbr.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin C, Timm J, Rauzier J, Gomez-Lus R, Davies J, Gicquel B. Transposition of an antibiotic resistance element in mycobacteria. Nature. 1990;345:739–743. doi: 10.1038/345739a0. [DOI] [PubMed] [Google Scholar]
- 27.Mercier J, Lachapelle J, Couture F, Lafond M, Vézina G, Boissinot M, Levesque R C. Structural and functional characterization of tnpI, a recombinase locus in Tn21 and related β-lactamase transposons. J Bacteriol. 1990;172:3745–3757. doi: 10.1128/jb.172.7.3745-3757.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minakhina S, Kholodii G, Mindlin S, Yurieva O, Nikiforov V. Tn5053 family transposons are res site hunters sensing plasmidal res sites occupied by cognate resolvases. Mol Microbiol. 1999;33:1059–1068. doi: 10.1046/j.1365-2958.1999.01548.x. [DOI] [PubMed] [Google Scholar]
- 29.Nakaya R, Nakamura A, Murata Y. Resistance transfer agents in Shigella. Biochem Biophys Res Commun. 1960;3:654–659. doi: 10.1016/0006-291x(60)90081-4. [DOI] [PubMed] [Google Scholar]
- 30.Osbourn S E V, Turner A K, Grinsted J. Nucleotide sequence within Tn3926 confirms this as a Tn21-like transposable element and provides evidence for the origin of the mer operon carried by plasmid pKLH2. Plasmid. 1995;33:65–69. doi: 10.1006/plas.1995.1008. [DOI] [PubMed] [Google Scholar]
- 31.Ouellette M, Bissonnette L, Roy P H. Precise insertion of antibiotic resistance determinants into Tn21-like transposons: nucleotide sequence of the OXA-1 β-lactamase gene. Proc Natl Acad Sci USA. 1987;84:7378–7382. doi: 10.1073/pnas.84.21.7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pansegrau W, Lanka E, Barth P T, Figurski D H, Guiney D G, Haas D, Helinski D R, Schwab H, Stanisich V, Thomas C M. Complete nucleotide sequence of Birmingham IncPα plasmids. Compilation and comparative analysis. J Mol Biol. 1994;239:623–663. doi: 10.1006/jmbi.1994.1404. [DOI] [PubMed] [Google Scholar]
- 33.Rådström P, Sköld O, Swedberg G, Flensburg J, Roy P H, Sundström L. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J Bacteriol. 1994;176:3257–3268. doi: 10.1128/jb.176.11.3257-3268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajakumar K, Bulach D, Davies J, Ambrose L, Sasakawa C, Adler B. Identification of a chromosomal Shigella flexneri multi-antibiotic resistance locus which shares sequence and organizational similarity with the resistance region of the plasmid NR1. Plasmid. 1997;37:159–168. doi: 10.1006/plas.1997.1280. [DOI] [PubMed] [Google Scholar]
- 35.Rogowsky P, Halford S E, Schmitt R. Definition of three resolvase binding sites at the res loci of Tn21 and Tn1721. EMBO J. 1985;4:2135–2141. doi: 10.1002/j.1460-2075.1985.tb03904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubens C E, McNeill W F, Farrar W E., Jr Transposable plasmid deoxyribonucleic acid sequence in Pseudomonas aeruginosa which mediates resistance to gentamicin and four other antimicrobial agents. J Bacteriol. 1979;139:877–882. doi: 10.1128/jb.139.3.877-882.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt F. The role of insertions, deletions, and substitutions in the evolution of R6 related plasmids encoding aminoglycoside transferase ANT-(2") Mol Gen Genet. 1984;194:248–259. doi: 10.1007/BF00383524. [DOI] [PubMed] [Google Scholar]
- 40.Schmidt F, Klopfer-Kaul I. Evolutionary relationship between Tn21-like elements and pBP201, a plasmid from Klebsiella pneumoniae mediating resistance to gentamicin and eight other drugs. Mol Gen Genet. 1984;197:109–119. doi: 10.1007/BF00327930. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt F R J, Nücken E J, Henschke R B. Structure and function of hot spots providing signals for site-directed specific recombination and gene expression in Tn21 transposons. Mol Microbiol. 1989;3:1545–1555. doi: 10.1111/j.1365-2958.1989.tb00140.x. [DOI] [PubMed] [Google Scholar]
- 42.Smith D I, Gomez Lus R, Rubio Calvo M C, Datta N, Jacob A E, Hedges R W. Third type of plasmid conferring gentamicin resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1975;8:227–230. doi: 10.1128/aac.8.3.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stokes H W, Hall R M. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 44.Stokes H W, Hall R M. Sequence analysis of the inducible chloramphenicol resistance determinant in the Tn1696 integron suggests regulation by translational attenuation. Plasmid. 1991;26:10–19. doi: 10.1016/0147-619x(91)90032-r. [DOI] [PubMed] [Google Scholar]
- 45.Sundström L, Rådström P, Swedberg G, Sköld O. Site-specific recombination promotes linkage between trimethoprim- and sulfonamide resistance genes. Sequence characterization of dhfrV and sull and a recombination active locus of Tn21. Mol Gen Genet. 1988;213:191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- 46.Tabor S, Richardson C C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tenover F C, Filpula D, Phillips K L, Plorde J L. Cloning and sequencing of a gene encoding an aminoglycoside 6′-N-acetyltransferase from an R factor of Citrobacter diversus. J Bacteriol. 1988;170:471–473. doi: 10.1128/jb.170.1.471-473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenover F C, Phillips K L, Gilbert T, Lockhart P, O'Hara P J, Plorde J J. Development of a DNA probe from the deoxyribonucleotide sequence of a 3-N-aminoglycoside acetyltransferase [AAC(3)-I] resistance gene. Antimicrob Agents Chemother. 1989;33:551–559. doi: 10.1128/aac.33.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vézina G, Levesque R C. Molecular characterization of the class II multiresistance transposable element Tn1403 from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35:313–321. doi: 10.1128/aac.35.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wohlleben W, Arnold W, Bissonnette L, Pelletier A, Tanguay A, Roy P H, Gamboa G C, Barry G F, Aubert E, Davies J, Kagan S A. On the evolution of Tn21-like multiresistance transposons: sequence analysis of the gene (aacCI) for gentamicin acetyltransferase-3-I(AAC(3)-I), another member of the Tn21-based expression cassette. Mol Gen Genet. 1989;217:202–208. doi: 10.1007/BF02464882. [DOI] [PubMed] [Google Scholar]
- 51.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 52.Yeo C C, Tham J M, Kwong S M, Yiin S, Poh C L. Tn5563, a transposon encoding putative mercuric ion transport proteins located on plasmid pRA2 of Pseudomonas alcaligenes. FEMS Microbiol Lett. 1998;165:253–260. doi: 10.1111/j.1574-6968.1998.tb13154.x. [DOI] [PubMed] [Google Scholar]
- 53.Yurieva O, Kholodii G, Minakhin L, Gorlenko Z, Kalyaeva E, Mindlin S, Nikiforov V. Intercontinental spread of promiscuous mercury-resistance transposons in environmental bacteria. Mol Microbiol. 1997;24:321–329. doi: 10.1046/j.1365-2958.1997.3261688.x. [DOI] [PubMed] [Google Scholar]
- 54.Zühlsdorf M T, Wiedemann B. Functional and physiological characterization of the Tn21 cassette for resistance genes in Tn2426. J Gen Microbiol. 1993;139:995–1002. doi: 10.1099/00221287-139-5-995. [DOI] [PubMed] [Google Scholar]