Abstract
Background
Infections caused by Klebsiella pneumoniae have been difficult to control because of the worldwide emergence of carbapenem-resistant isolates mainly due to carbapenemase production. Information regarding carbapenemase-producing K. pneumoniae is still scarce in Ethiopia. Therefore, the current study aimed to determine the prevalence of carbapenemase-producing K. pneumoniae and to assess the occurrence of blaNDM and blaKPC carbapenemase genes.
Methods
A cross-sectional study was conducted from September 2018 to February 2019 at Tikur Anbessa Specialized Hospital, Addis Ababa, Ethiopia. A total of 132 non-duplicate K. pneumoniae isolates were studied. Phenotypic confirmation of carbapenemase production was done by modified Carbapenem Inactivation Method (mCIM). Multiplex PCR was performed for the detection of carbapenemase-encoding genes blaKPC, and blaNDM.
Results
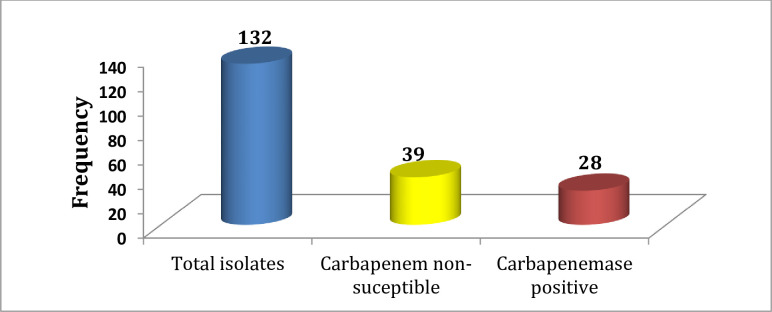
Out of the total 132 K. pneumoniae isolates, 39 (29.6%) were non-susceptible to one or more carbapenems. The prevalence of carbapenemase-producing isolates from the total was 28 (21.2%) with mCIM of which the most dominant gene was blaNDM 26 (92.9%) and one isolate carried blaKPC concomitantly. Carbapenemase-producing K. pneumoniae isolates were 100% non-susceptible to half of the antimicrobials used in the study, including meropenem and ertapenem. Previous use of carbapenems was associated with carbapenemase production (P = 0.004).
Conclusions
The prevalence of carbapenemase-producing K. pneumoniae isolates was worrying in the study area. To our knowledge, the study described the emergence of blaNDM and blaKPC gene carrying K. pneumoniae in Ethiopia for the first time. Further large-scale molecular-based studies, including other carbapenemase genes and sequencing of K. pneumoniae, are warranted to have a clear awareness about the presence of antimicrobial resistance high-risk clones in Ethiopia.
Introduction
Carbapenems have a carbapenem together with the beta-lactam ring which makes them more stable against most β-lactamases [1]. They are the most effective against Gram-positive and Gram-negative bacteria. According to the Clinical and Laboratory Standards Institute (CLSI) guidelines meropenem, imipenem, ertapenem, and doripenem are recommended treatments for infections caused by Enterobacteriaceae [2]. Their effectiveness, stability, and fewer adverse effects compared to other last-line drugs such as polymyxins make them the most reliable last-resort treatments for bacterial infections [1]. The prevalence of ESBL-producing K. pneumoniae such as CTX-M-15-producers continues to impose a serious threat to human health [3], and carbapenems are widely considered as the drugs of choice for the treatment of severe infections caused by Extended-spectrum β-lactamases (ESBL)-producing Enterobacteriaceae [4]. However, in recent years, carbapenem-resistant Enterobacteriaceae particularly K. pneumoniae is rising alarmingly [5, 6].
Resistance to carbapenems is mainly through carbapenemase enzyme production. Productions of other enzymes that have weak carbapenemase activity such as ESBLs and AmpC β-lactamases together with porin alteration, drug efflux pumps, as well as alterations in penicillin-binding proteins are also mentioned as additional resistance mechanisms [1, 5–7]. Based on their molecular structures (Ambler classification system) carbapenemases belong to class A, B, and D of β-lactamases [5–8].
K. pneumoniae has caused hospital outbreaks in different countries [9], requiring early detection of carbapenemases in infected patients and/or carriers to prevent the occurrence of outbreaks. It has been indicated that house flies are potential vectors of antibiotic-resistant K. pneumoniae [10]. Klebsiella pneumoniae carbapenemases (KPCs) are the most common transmissible class A carbapenemase circulating in Enterobacteriaceae predominantly in K. pneumoniae worldwide mainly due to clonal expansion of strains of K. pneumoniae [8]. Unlike KPC the rapid and dramatic dissemination of New Delhi metallo-ß- lactamase (NDM)-producing Enterobacteriaceae is mediated by promiscuous plasmid not associated with dominant clonal strains [5].
Three blaNDM-1-positive Acinetobacter baumannii isolates were reported from Jimma, Ethiopia [11]. However, to the best of our knowledge, no blaNDM- and blaKPC-carrying K. pneumoniae have been described in Ethiopia so far. Therefore, the current study aimed to determine the prevalence of carbapenemase-producing K. pneumoniae and to assess the occurrence of blaNDM and blaKPC carbapenemase genes.
Materials and methods
Study population
A total of 132 study participants who were visited Tikur Anbessa Specialized Hospital (TASH), Addis Ababa, Ethiopia and became culture-positive for K. pneumoniae over six months (from September 2018 to February 2019) were enrolled conveniently. Preliminary identification of K. pneumoniae was done by inoculating the specimens on MacConkey agar (Oxoid, UK) and 5% sheep blood agar (Oxoid, UK). Further identification was done through Gram stain, and a series of biochemical tests including indole, triple sugar iron agar, citrate utilization, mannitol, malonate, lysine decarboxylase, urea agar, and motility medium. K. pneumoniae is Gram-negative and rod-shaped, lactose fermenter, indole negative, gas and acid producer, hydrogen sulfide negative, citrate positive, mannitol fermenter, malonate positive, lysine decarboxylase positive, urea slow producer, and non-motile [12, 13]. Socio-demographic characteristics and clinical information of the study participants were obtained using a well-designed questionnaire and from their medical records by health care workers.
Antimicrobial susceptibility testing
Using a sterile wire loop, 3–5 single colonies were picked from blood agar and emulsified in 3–4 ml normal saline to prepare a 0.5 McFarland standard using McFarland Densitometer. From the standard, cells were spread onto Muller-Hinton agar (Oxoid, UK) using a sterile swab for the Antimicrobial Susceptibility Testing (AST) [12]. The AST was performed based on the Kirby–Bauer disc diffusion method using the following antimicrobials; Tetracycline (30 μg), Gentamicin (10 μg), Amikacin (30 μg), Ciprofloxacin (5 μg), Chloramphenicol (30 μg), Aztreonam (30 μg), Trimethoprim/sulfamethoxazole (1.25/23.75 μg), Amoxicillin-clavulanate (20/10 μg), Piperacillin/tazobactam (100/10 μg), Cefoxitin (30 μg), Ceftriaxone (30 μg), Imipenem (10 μg), Ertapenem (10 μg) and Meropenem (10 μg) (Oxoid, UK) and (BD, USA). After 16–18 hours of incubation at 35± 2°C, the diameter of the zone of inhibition around antibiotic discs was measured by caliper and interpreted as sensitive, intermediate, or resistant following CLSI (2018) guidelines [2].
Phenotypic confirmatory test for carbapenemase
K. pneumoniae isolates that showed no sensitivity to at least one carbapenems were checked for carbapenemase production using the modified Carbapenem Inactivation Method (mCIM). According to CLSI (2018) guidelines the method has > 99% sensitivity and specificity for detection of carbapenemase among Enterobacteriaceae isolates [2]. Briefly, a suspension was made by taking 1μl loopful of bacteria from an overnight grown culture on a Blood agar plate and then added into 2 ml trypticase soya broth. Subsequently, Meropenem (10 μg) disc was immersed in the suspension and incubated for 4 hours ± 15 minutes at 35°C ± 2°C. After incubation, the disc was removed from the suspension using a 10 μl inoculation loop and placed on a Mueller-Hinton agar plate inoculated with a susceptible E. coli indicator strain (ATCC 29522). Then, the results were read after 18–24 hours of incubation at 35°C ± 2°C. When the bacterial isolate produced carbapenemase, the meropenem disc was inactivated allowing uninhibited growth of the susceptible indicator strain. Discs incubated in suspensions that do not contain carbapenemases yielded a clear inhibition zone. An inhibition zone diameter of 6–15 mm or colonies within a 16–18 mm zone was considered to be a positive result, and a zone of inhibition ≥19 mm was considered to be a negative result [2].
DNA extraction and detection of blaKPC and blaNDM carbapenemase genes
The bacterial DNA was extracted by the boiling lysis method as previously described by El-Badawy et al [14]. Briefly, three to six fresh colonies of the bacteria were suspended in 100μl of DNase-free water in a sterile 1.5ml Eppendorf tube. The bacterial suspension was vortexed for 15 seconds and placed in a boiling Water-bath at 94°C for 10 minutes to lyse the bacterial cells. The lysed bacterial suspension was centrifuged at maximum speed (13,000 ×g) for 5 min. The supernatant, which contains total genomic DNA, was transferred to a new sterile tube using DNase-free tips. The quality and quantity of the extracted DNA were measured using Nanodrop (Thermo Scientific, US) and stored at -20∘C.
Multiplex PCR was performed to detect blaKPC and blaNDM carbapenemase genes using specific primers (Table 1). Briefly, the PCR was performed with approximately 300ng template DNA, 0.2μM of each primer, and 7.5 μl of 2 x QIAGEN Multiplex PCR Master Mix (QIAGEN, Germany) in a final volume of 15μl. Amplification was performed in a thermocycler (Biometra, Germany) with cycling parameters including initial denaturation at 95°C for 15 minutes followed by 35 cycles each of denaturation at 94°C for 30 s, annealing at 58°C for 90 s, extension at 72°C for 90 s, and a final extension at 72°C for 10 minutes. The PCR products were visualized by electrophoresis in 1.5% agarose gel after staining with ethidium bromide. A 100bp ladder molecular weight marker (Promega, US) was used to measure the molecular weight of amplified products. The amplicon was visualized and its size was determined under UV trans-illuminator (Bio-Rad, US).
Table 1. Primers used for detection of blaKPC and blaNDM carbapenemase genes in K. pneumoniae isolates.
| Gene | Primer | Nucleotide sequence | Annealing Temp °C | Amplicon size (bp) | Reference | |
|---|---|---|---|---|---|---|
| 5`————————-3` | ||||||
| bla KPC | KPC | F | CGTCTAGTTCTGCTGTCTTG | 57.8 | 798 | [15] |
| R | CTTGTCATCCTTGTTAGGCG | 62.2 | ||||
| bla NDM | NDM | F | GGTTTGGCGATCTGGTTTTC | 65.5 | 621 | |
| R | CGGAATGGCTCATCACGATC | 67.8 | ||||
Data quality assurance
The reliability of the study findings was guaranteed by implementing quality control measures throughout the whole process of the laboratory work. Quality control strains of Escherichia coli ATCC® 25922 and Pseudomonas aeruginosa ATCC® 27853 were used for controlling the potency of the drugs. K. pneumoniae ATCC® BAA-1705™ and K. pneumoniae ATCC® BAA-1706 were used as positive and negative controls respectively during mCIM. Laboratory reference blaKPC and blaNDM genes were used as positive controls and Escherichia coli ATCC® 25922 as a negative control during PCR analysis. Each primer pair was checked in monoplex PCR before multiplexing.
Data analysis
Data were checked, cleaned, and double entered into Epidata software version 3.1 (The EpiData Association, Denmark), and then it was exported to Statistical Package for Social Sciences (SPSS version 25.0, IBM Corp., USA) software for analysis. The chi-square test or Fisher’s exact test was used as appropriate. Bivariate logistic regression was carried out and variables with a P-value of less than 0.2 were entered into multivariate logistic regression analysis. A P-value < 0.05 at 95% confidence interval was considered as statistically significant.
Ethical approval
This study was approved by the Ethics Review Committee of Department of Microbiology, Immunology, and Parasitology, School of Medicine, College of Health Sciences, Addis Ababa University (Reference number: DERC/17/18/02-N) and AHRI/ALERT ethical review committee (Protocol number: PO12/18). A permission letter was obtained from TASH. Moreover, before commencing the study, written informed consent/assent was obtained from each study participant. Confidentiality was maintained for all data collected.
Results
Prevalence of carbapenemase-producing K. pneumoniae isolates
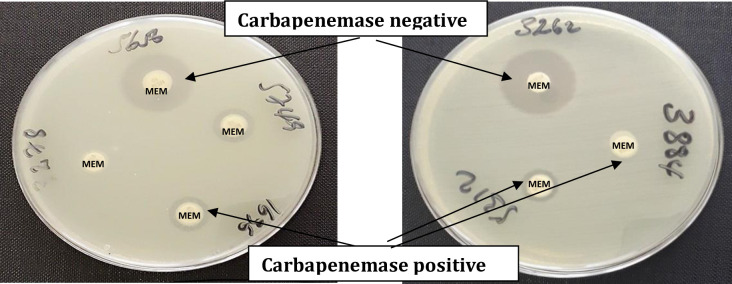
In this study, a total of 132 non-duplicate K. pneumoniae isolates were collected from patients who admitted or attended different departments of TASH. As shown in Fig 1 from the total isolates, 39 (29.6%) showed no sensitivity to one or more carbapenems. Out of these isolates, 28/39 (71.8%) were carbapenemase positive using the modified Carbapenem Inactivation Method (mCIM). The overall prevalence of carbapenemase production from the total isolates was 28/132 (21.2%). Fig 2 indicates the positive and negative results of mCIM.
Fig 1. Frequency of carbapenemase-producing K. pneumoniae isolates.
Fig 2. Carbapenemase positive and negative K. pneumoniae isolates with modified Carbapenem Inactivation Method (mCIM).
MEM: Meropenem.
Distribution of carbapenemase-producing and non-producing isolates among age, sex, and ward type
Among the total K. pneumoniae isolates, 83/132 (62.9%) were recovered from males, of which 18 (21.7%) were carbapenemase positive. Regarding the age of study participants, the majority 74 (56.1%) were below 5 years. The majority of 120 (90.9%) K. pneumoniae isolates were recovered from hospitalized patients, of them, 27 (22.5%) were positive for carbapenemase (Table 2).
Table 2. Distribution of carbapenemase-producing and non-producing isolates among age, sex, and ward type.
| Variables | Total | Carbapenemase | ||
|---|---|---|---|---|
| Positive n (%) | Negative n (%) | |||
| Sex | Male | 83 | 18(21.7) | 65(78.3) |
| Female | 49 | 10(20.4) | 39(79.6) | |
| Age in years | <5 | 74 | 7(9.5) | 67(90.5) |
| 5 to <18 | 20 | 7(35.0) | 13(65.0) | |
| 18 to <45 | 25 | 11(44.0) | 14(56.0) | |
| ≥45 | 13 | 3(23.1) | 10(76.9) | |
| Patient setting | Inpatients | 120 | 27(22.5) | 93(77.5) |
| Outpatient | 12 | 1(8.3) | 11(91.7) | |
| Ward type | ICUs | 46 | 8(17.4) | 38(82.6) |
| Pediatric ward | 53 | 9(17.0) | 44(83.0) | |
| Medical ward | 9 | 6(66.7) | 3(33.3) | |
| Surgical ward | 8 | 4(50.0) | 4(50.0) | |
| Others | 4 | 0(0.0) | 4(100.0) | |
n: number of K. pneumoniae isolates, ICUs: Intensive Care Units.
Specimen-wise distribution of carbapenemase-producing K. pneumoniae isolates
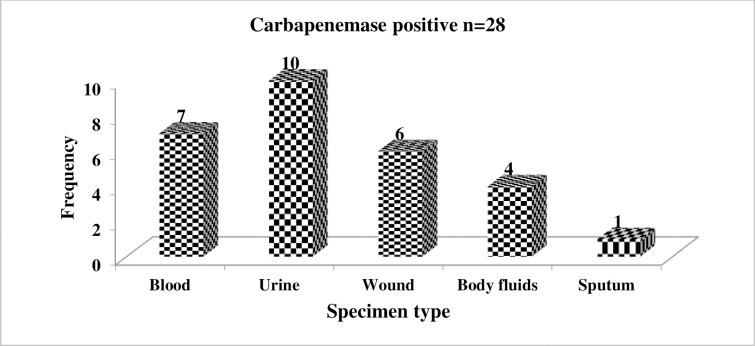
As displayed in Fig 3, urine was the major source of carbapenemase-producing K. pneumoniae isolates with 10/28 (35.7%), while only 1/28 (3.6%) of carbapenemase-producing K. pneumoniae was isolated from sputum.
Fig 3. Specimen-wise distribution of carbapenemase-producing K. pneumoniae isolates.
Antimicrobial susceptibility patterns of carbapenemase-producing and non-producing K. pneumoniae isolates
Resistance of carbapenemase-producing isolates was high to β-lactams as well as other classes of antimicrobials except to amikacin. They were completely non-susceptible to aztreonam, piperacillin-tazobactam, amoxicillin-clavulanate, cefoxitin, ceftriaxone, meropenem, and ertapenem. The susceptibility of carbapenemase producers was 20/28 (7l.4%) to amikacin and from carbapenems least resistance was noted to imipenem 13/28 (46.4%). Carbapenemase positive isolates showed significantly higher resistance to most of the antimicrobials tested including ciprofloxacin (P<0.001), aztreonam (P = 0.016), piperacillin-tazobactam (P<0.001), chloramphenicol (p = 0.018), ceftriaxone (P<0.001), and carbapenems (P<0.001) compared to carbapenemase negative isolates with chi-square test as shown in Table 3. Almost all 130/132 (98.5) K. pneumoniae isolates were multidrug resistance (MDR). The details of susceptibility patterns of each K. pneumoniae isolate are presented in the S1 Table.
Table 3. Antimicrobial susceptibility patterns of carbapenemase-producing and non-producing K. pneumoniae isolates.
| Antimicrobial agents | Carbapenemase positive (n = 28) | Carbapenemase negative (n = 104) | P-value | ||||
|---|---|---|---|---|---|---|---|
| S | I | R | S | I | R | ||
| Tetracycline | 5(17.9) | 4(14.3) | 19(67.9) | 16(15.4) | 11(10.6) | 77(74.0) | 0.791 |
| Gentamicin | 4(14.3) | 0(0.0) | 24(85.7) | 25(24.0) | 8(7.7) | 71(68.3) | 0.133 |
| Amikacin | 20(71.4) | 4(14.3) | 4(14.3) | 103(99.0) | 1(1.0) | 0(0.0) | <0.001* |
| Ciprofloxacin | 1(3.6) | 2(7.1) | 25(89.3) | 57(54.8) | 23(22.1) | 24(23.1) | <0.001* |
| Aztreonam | 0(0.0) | 3(10.7) | 25(89.3) | 13(12.5) | 27(26.0) | 64(61.5) | 0.016* |
| Piperacillin-tazobactam | 0(0.0) | 0(0.0) | 28(100.0) | 55(52.9) | 27(26.0) | 22(21.2) | <0.001* |
| Amoxicillin-clavulanate | 0(0.0) | 1(3.6) | 27(96.4) | 18(17.3) | 31(29.8) | 55(52.9) | <0.001* |
| SXT | 3(10.7) | 1(3.6) | 24(85.7) | 3(2.9) | 1(1.0) | 100(96.2) | 0.121 |
| Chloramphenicol | 7(25.0) | 10(35.7) | 11(39.3) | 52(50.0) | 3(2.9) | 49(47.1) | 0.018* |
| Cefoxitin | 0(0.0) | 0(0.0) | 28(100.0) | 62(59.6) | 12(11.5) | 30(28.8) | <0.001* |
| Ceftriaxone | 0(0.0) | 0(0.0) | 28(100.0) | 4(3.8) | 0(0.0) | 100(96.2) | 0.578 |
| Meropenem | 0(0.0) | 3(10.7) | 25(89.3) | 96(92.3) | 1(1.0) | 7(6.7) | <0.001* |
| Imipenem | 6(21.43) | 9(32.1) | 13(46.4) | 101(97.1) | 0(0.0) | 3(2.9) | <0.001* |
| Ertapenem | 0(0.0) | 2(7.1) | 26(92.9) | 93(89.4) | 3(2.9) | 8(7.7) | <0.001* |
S: Sensitive, I: Intermediate, R: Resistance, SXT: Trimethoprim-sulfamethoxazole
*P-value<0.05.
Association of antimicrobial use with carbapenemase production
The possible association of history of antimicrobial consumption (within 3 months) with carbapenemase production was assessed by bivariate and multivariate logistic regression. Based on the local availability, the following antimicrobials were taken by the study participants in each class; 3rd or 4th generation cephalosporins (ceftriaxone, cefotaxime, ceftazidime, and cefepime), carbapenems (meropenem), quinolones (ciprofloxacin), aminoglycosides (gentamicin), and others such as trimethoprim-sulfamethoxazole, erythromycin, augmentin. As revealed in Table 4, from the total study participants, 83/132 (62.9%) have taken 3rd or 4th generation cephalosporins. Out of the total study participants, 20/132 (15.2%) have taken carbapenems of them, 11/20 (55%) were positive for carbapenemase. There was a statistically significant association between carbapenem use and carbapenemase production with multivariate logistic regression (P = 0.004).
Table 4. Association of antimicrobial use and carbapenemase production (N = 132).
| Variable | CP | CN | COR (95%CI) P-value | AOR (95%CI) P-value* | |
|---|---|---|---|---|---|
| Antimicrobial therapy | Yes | 27 | 89 | 4.55 (0.57–36.05) 0.151 | 1.82 (0.19–17.91) 0.607 |
| No | 1 | 15 | 1 | 1 | |
| Carbapenems | Yes | 11 | 9 | 6.83 (2.46–18.96) <0.001 | 6.03 (2.13–17.09) 0.004 |
| No | 17 | 95 | 1 | 1 | |
| 3rd or 4th GCs | Yes | 21 | 62 | 2.03 (0.79–5.21) 0.140 | 1.67 (0.56–5.04) 0.478 |
| No | 7 | 42 | 1 | 1 | |
| Quinolones | Yes | 4 | 10 | 1.57 (0.45–5.43) 0.479 | _ |
| No | 24 | 94 | 1 | ||
| Aminoglycosides | Yes | 12 | 31 | 1.77 (0.75–4.17) 0.194 | 1.70 (0.66–4.38) 0.538 |
| No | 16 | 73 | 1 | 1 | |
CP: Carbapenemase positive, CN: Carbapenemase negative, 3rd or 4th GCs: Third or fourth-generation cephalosporins, COR: Crude odds ratio, AOR: Adjusted odds ratio, CI: Confidence interval
* FDR adjusted P-value.
Detection of blaKPC and blaNDM carbapenemase genes
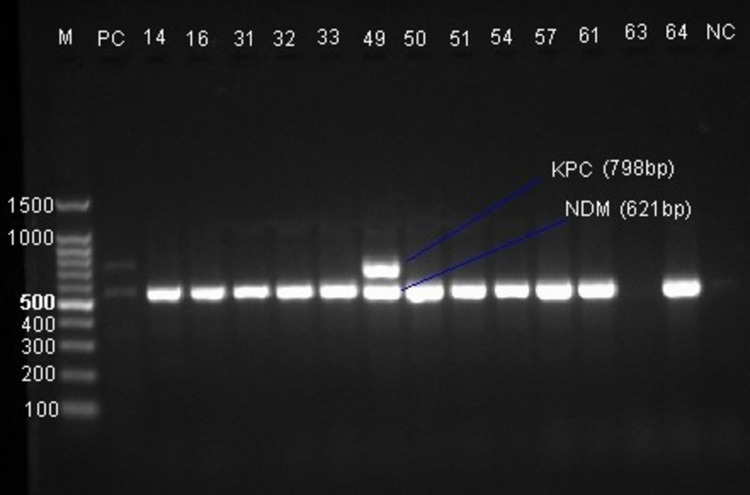
Of the 28 carbapenemase-producing K. pneumoniae isolates with mCIM, 26 (92.9%) were positive for one or both carbapenemase genes in the multiplex PCR. The blaNDM gene was detected in 26 isolates while the blaKPC gene was detected in only one isolate concurrently with blaNDM. More than two-thirds (65.4%) of blaNDM gene-positive isolates were from hospitalized patients in the ICU and pediatric wards (S2 Table). Fig 4 shows the gel image of the blaKPC (798 bp), and blaNDM (621 bp) genes.
Fig 4. Agarose gel electrophoresis of PCR products for carbapenemase genes.
Lane M: 100bp DNA ladder, PC: Positive control, Lanes 14–64: K. pneumoniae isolates, NC: Negative control.
Discussion
There were few studies on carbapenemase-producing bacteria in Ethiopia and almost all of them noted K. pneumoniae as the most common carbapenemase producer compared to other bacterial isolates [16–18]. In the current study, 39 K. pneumoniae isolates were not sensitive to carbapenems, of them, 71.8% were carbapenemase positive phenotypically. Likewise, a study from Sudan showed that 78% of carbapenem-resistant K. pneumoniae isolates were carbapenemase positive [19]. In this study, the overall prevalence of carbapenemase-producing K. pneumoniae was 21.2%, which is comparable with another study conducted in Ethiopia from Bahir Dar (16.5%) [18]. It is lower than a study by Kazemian et al from Iran at which 43.3% of K. pneumoniae from hospitalized patients were carbapenemase producers [20]. Nevertheless, it is higher than a study from Tunisian and Libyan hospitals (11.4%) [21]. Empirical prescription of carbapenems particularly meropenem was very common in the hospital where we did the current study [22] and also in another study by Gebretekle et al a significant amount (38.6%) of meropenem was prescribed when it was not needed [23]. Additionally, in this study, previous carbapenem use has an association with carbapenemase production. This implies that the higher carbapenemase production in the current study could be due to selective pressure created by the indiscriminate use of carbapenems. Furthermore, it might be due to improper infection control practices as the emergence of carbapenemase-producing K. pneumoniae was previously noted in the hospital [17, 24]. A higher number of carbapenemase-producing K. pneumoniae was isolated from urine specimens 10/28 (35.7%), which is similar to a study conducted by Hashemizadeh et al [25].
In our study, almost all (98.5%) K. pneumoniae isolates were non-susceptible to at least three antimicrobials belonging to different categories and, hence, defined as MDR according to Magiorakos et al [26]. Concerning carbapenemase-producing isolates, relatively higher sensitivity (71.4%) was noted to amikacin, which is comparable with a study in Taiwan (78.8%) [27] suggesting the possible use of this drug against carbapenemase producers. Nonetheless, complete non-susceptibility of carbapenemase-producing K. pneumoniae isolates was observed to aztreonam, amoxicillin-clavulanate, piperacillin-tazobactam, cefoxitin, ceftriaxone, cefotaxime, ceftazidime, cefepime, meropenem, and ertapenem, which is in line with a report from Taiwan [27]. It is indicated that carbapenemase-producing Gram-negative bacteria, in particular, are resistant to all or virtually all beta-lactams, fluoroquinolones, and/or aminoglycosides concomitantly [1]. This is mainly due to the simultaneous presence of several resistance genes in these isolates. K. pneumoniae strains with a high prevalence of resistance against many antimicrobials including imipenem, amoxicillin/clavulanic acid, ceftazidime, piperacillin/tazobactam, tobramycin, ciprofloxacin, co-trimoxazole, and aztreonam, harboring genes encoding multi-drug efflux pump (AcrAB-TolC) and porins (OmpK35 and OmpK36) has been also reported [11, 28].
Karaiskos and Giamarellou (2014) described the worldwide emergence of carbapenemases mediated carbapenem resistance in Klebsiella pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa with a mortality exceeding 50%, is attributed mainly to the lack of effective antimicrobial regimens [29]. Generally, although they have limitations concerning accessibility, resistance, clinical efficacy, and adverse effects; colistin, tigecycline, fosfomycin, temocillin, and newer underdevelopment antimicrobials including carbapenemase inhibitors alone or in combination exhibit promising and/or effective antibacterial activity in vitro and some in vivo against infections caused by carbapenemase-producing bacteria [6, 29].
Two isolates that were positive by mCIM, harbored neither blaKPC nor blaNDM gene. This could be due to other carbapenemase genes, including blaVIM and blaIMP which belong to class B β-lactamases based on the Ambler classification system and blaOXA-48, a class D β-lactamase that are reported in K. pneumoniae increasingly [5, 8]. In this study, blaNDM was detected in 26 out of 28 carbapenemase positive isolates which is the dominant carbapenemase-encoding gene compared to blaKPC noticed only in one isolate. Similarly, in a study from Sudan, (70.7%) of K. pneumoniae isolates were positive for blaNDM with no blaKPC gene [30], and in a study from Egypt, blaNDM-1 was the most predominant carbapenemase gene in K. pneumoniae (74.4%) compared to blaKPC (48.8%) [31]. The first reported blaNDM-1 in Kenya was in K. pneumoniae and corresponds to the first report of NDM-1 producers in Africa [32]. There was also a previous report of blaNDM-1 positive Acinetobacter baumannii in Jimma, Ethiopia [11]. It has been described that K. pneumoniae is the most common species among Enterobacteriaceae that harbors blaNDM and the rapid spread of the gene from its initial emergence in India to all continents is significantly associated with global travel [33]. Since Ethiopians make travel to India and other countries for medical purposes and other reasons there is a possibility to acquire carbapenemase genes. However, it is difficult to conclude without taking a detailed history of patients. Although there are high-risk KPC-carrying K. pneumoniae clones such as ST258 and ST11 as well as most common NDM positive K. pneumoniae lineages such as ST11 and ST14 [33], this study has limitations in that sequencing wasn’t done due to resource constraint.
Conclusion and recommendations
In this study, the prevalence of carbapenemase-producing K. pneumoniae isolates was a matter of great concern. Carbapenemase-producing isolates were highly resistant to many of the antimicrobials used in this study. Only amikacin was relatively active against carbapenemase-producing isolates. Moreover, previous use of carbapenems was associated with carbapenemase production suggesting the need to implement effective antimicrobial stewardship practices in the hospital. To our knowledge, the study described the emergence of blaNDM and blaKPC gene carrying K. pneumoniae in Ethiopia for the first time. Further large-scale molecular-based studies, including other carbapenemase genes and sequencing of K. pneumoniae are warranted, to have a clear awareness about the presence of antimicrobial resistance high-risk clones in Ethiopia.
Supporting information
S: Sensitive, I: Intermediate, R: Resistant, TET: Tetracycline, GM: Gentamicin, AN: Amikacin, CIP: Ciprofloxacin, ATM: Aztreonam, PTZ: Piperacillin-tazobactam, AMC: Amoxicillin-clavulanate, SXT: Trimethoprim-sulfamethoxazole, CHL: Chloramphenicol, CXT: Cefoxitin, CRO: Ceftriaxone, MEM: Meropenem, IMP: Imipenem, ETM: Ertapenem.
(PDF)
mCIM: modified Carbapenem Inactivation Method, ESBL: Extended-spectrum β-lactamase, Interp: Interpretation, ZI in mm: Zone of inhibition in millimeter, M: Male, F: Female, CSF: Cerebrospinal fluid, ICU: Intensive Care Unit.
(PDF)
Lane M: 100bp DNA ladder; PC: Positive control, Lanes 67–114: K. pneumoniae isolates, NC: Negative control.
(TIF)
Lane M: 100bp DNA ladder, PC1, PC2 & PC3: Positive control, Lanes 67–120: K. pneumoniae isolates, NC: Negative control.
(TIF)
Acknowledgments
We thank Tikur Anbessa Specialized Hospital for permitting us to conduct this study. Our deepest gratitude goes to Mr. Berhanu Yitayew for his overall technical assistance and for donating control resistance genes. Our countless acknowledgments go to the study participants for their willingness to participate.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Armauer Hansen Research Institute. It had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 2016;3(1):15–21. doi: 10.1177/2049936115621709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CLSI. Performance standards for antimicrobial susceptibility testing. 28th ed. CLSI supplement M100-S28.Wayne, PA. Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 3.Ranjbar R, Memariani H, Sorouri R, Memariani M. Distribution of virulence genes and genotyping of CTX-M-15-producing Klebsiella pneumoniae isolated from patients with community-acquired urinary tract infection (CA-UTI). Microb Pathog. 2016;100:244–249. doi: 10.1016/j.micpath.2016.10.002 [DOI] [PubMed] [Google Scholar]
- 4.Harada S, Ishii Y, Yamaguchi K. Extended-spectrum β-lactamases: implications for the clinical laboratory and therapy. Korean J Lab Med. 2008;28(6):401–12. doi: 10.3343/kjlm.2008.28.6.401 [DOI] [PubMed] [Google Scholar]
- 5.Logan LK, Weinstein RA. The Epidemiology of Carbapenem-Resistant Enterobacteriaceae: The Impact and Evolution of a Global Menace. J Infect Dis. 2017;215(suppl_1):S28–S36. doi: 10.1093/infdis/jiw282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–84. doi: 10.1128/AAC.01019-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aslam B, Rasool M, Muzammil S, Siddique AB, Nawaz Z, Shafique M, et al. Carbapenem resistance: Mechanisms and drivers of global menace. Pathog Bact. 2020. [Google Scholar]
- 8.Walsh TR. Emerging carbapenemases: a global perspective. Int J Antimicrob Agents. 2010;36:S8–S14. doi: 10.1016/S0924-8579(10)70004-2 [DOI] [PubMed] [Google Scholar]
- 9.Nordmann P, Naas T, Poirel L. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791. doi: 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ranjbar R, Izadi M, Hafshejani TT, Khamesipour F. Molecular detection and antimicrobial resistance of Klebsiella pneumoniae from house flies (Musca domestica) in kitchens, farms, hospitals and slaughterhouses. J Infect Public Health. 2016;9(4):499–505. doi: 10.1016/j.jiph.2015.12.012 [DOI] [PubMed] [Google Scholar]
- 11.Pritsch M, Zeynudin A, Messerer M, Baumer S, Liegl G, Schubert S, et al. First report on bla NDM-1-producing Acinetobacter baumannii in three clinical isolates from Ethiopia. BMC Infect Dis. 2017;17(1):1–7. doi: 10.1186/s12879-016-2122-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheesbrough M. Cheesbrough M. District laboratory practice in tropical countries: Part 2. 2nd ed. New York: Cambridge University Press; 2006. [Google Scholar]
- 13.Ranjbar R, Kelishadrokhi AF, Chehelgerdi M. Molecular characterization, serotypes and phenotypic and genotypic evaluation of antibiotic resistance of the Klebsiella pneumoniae strains isolated from different types of hospital-acquired infections. Infect Drug Resis. 2019;12:603. doi: 10.2147/IDR.S199639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Badawy MF, Tawakol WM, El-Far SW, Maghrabi IA, Al-Ghamdi SA, Mansy MS, et al. Molecular identification of aminoglycoside-modifying enzymes and plasmid-mediated quinolone resistance genes among Klebsiella pneumoniae clinical isolates recovered from Egyptian patients. Int J Microbiol. 2017;2017:8050432. doi: 10.1155/2017/8050432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel L, Walshb RT, Cuvilliera V, Nordmanna P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70:119–23. doi: 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 16.Eshetie S, Unakal C, Gelaw A, Ayelign B, Endris M, Moges F. Multidrug resistant and carbapenemase producing Enterobacteriaceae among patients with urinary tract infection at referral Hospital, Northwest Ethiopia. Antimicrob Resist Infect Control. 2015;4(1):1–8. doi: 10.1186/s13756-015-0054-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legese MH, Weldearegay GM, Asrat D. Extended-spectrum beta-lactamase-and carbapenemase-producing Enterobacteriaceae among Ethiopian children. Infect Drug Resis. 2017;10:27–34. doi: 10.2147/IDR.S127177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moges F, Eshetie S, Abebe W, Mekonnen F, Dagnew M, Endale A, et al. High prevalence of extended-spectrum beta-lactamase-producing Gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara region. PLoS One. 2019;14(4):e0215177. doi: 10.1371/journal.pone.0215177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dirar M, Bilal N, Ibrahim ME, Hamid M. Resistance Patterns and Phenotypic Detection of β-lactamase Enzymes among Enterobacteriaceae Isolates from Referral Hospitals in Khartoum State, Sudan. Cureus. 2020;12(3). doi: 10.7759/cureus.7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazemian H, Heidari H, Ghanavati R, Ghafourian S, Yazdani F, Sadeghifard N, et al. Phenotypic and Genotypic Characterization of ESBL-, AmpC-, and Carbapenemase-Producing Klebsiella pneumoniae and Escherichia coli Isolates. Med Princ Pract. 2019;28(6):547–51. doi: 10.1159/000500311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathlouthi N, Al-Bayssari C, El Salabi A, Bakour S, Gwierif SB, Zorgani AA, et al. Carbapenemases and extended-spectrum β-lactamases producing Enterobacteriaceae isolated from Tunisian and Libyan hospitals. J Infect Dev Ctries. 2016;10(07):718–27. doi: 10.3855/jidc.7426 [DOI] [PubMed] [Google Scholar]
- 22.Fenta T, Engidawork E, Amogne W, Berha AB. Evaluation of current practice of antimicrobial use and clinical outcome of patients with pneumonia at a tertiary care hospital in Ethiopia: A prospective observational study. PLoS One. 2020;15(1):e0227736. doi: 10.1371/journal.pone.0227736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gebretekle GB, Haile Mariam D, Abebe Taye W, Mulu Fentie A, Amogne Degu W, Alemayehu T, et al. Half of Prescribed Antibiotics Are Not Needed: A Pharmacist-Led Antimicrobial Stewardship Intervention and Clinical Outcomes in a Referral Hospital in Ethiopia. Front public health. 2020;8:109. doi: 10.3389/fpubh.2020.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desta K, Woldeamanuel Y, Azazh A, Mohammod H, Desalegn D, Shimelis D, et al. High gastrointestinal colonization rate with extended-Spectrum β-lactamase-producing Enterobacteriaceae in hospitalized patients: emergence of Carbapenemase-Producing K. pneumoniae in Ethiopia. PLoS One. 2016;11(8):e0161685. doi: 10.1371/journal.pone.0161685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashemizadeh Z, Hosseinzadeh Z, Azimzadeh N, Motamedifar M. Dissemination Pattern of Multidrug Resistant Carbapenemase Producing Klebsiella pneumoniae Isolates Using Pulsed-Field Gel Electrophoresis in Southwestern Iran. Infect Drug Resist. 2020;13:921. doi: 10.2147/IDR.S227955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magiorakos AP, Srinivasan A, Carey R, Carmeli Y, Falagas M, Giske C, et al. Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012; 18: 268–281. doi: 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- 27.Chiu SK, Ma L, Chan MC, Lin YT, Fung CP, Wu TL, et al. Carbapenem nonsusceptible Klebsiella pneumoniae in Taiwan: dissemination and increasing resistance of carbapenemase producers during 2012–2015. Sci Rep. 2018;8(1):8468. doi: 10.1038/s41598-018-26691-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shakib P, Ghafourian S, Zolfaghary MR, Hushmandfar R, Ranjbar R, Sadeghifard N. Prevalence of OmpK35 and OmpK36 porin expression in beta-lactamase and non-betalactamase-producing Klebsiella pneumoniae. Biologics. 2012;6:1. doi: 10.2147/BTT.S27582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karaiskos I, Giamarellou H. Multidrug-resistant and extensively drug-resistant Gram-negative pathogens: current and emerging therapeutic approaches. Expert Opin Pharmacother. 2014;15(10):1351–70. doi: 10.1517/14656566.2014.914172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elbadawi HS, Elhag KM, Mahgoub E, Altayb HN, Ntoumi F, Elton L, et al. Detection and characterization of carbapenem resistant Gram‐negative bacilli isolates recovered from hospitalized patients at Soba University Hospital, Sudan. BMC Microbiol. 2021;21(1):1–9. doi: 10.1186/s12866-020-02060-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammad HA, Hadiya S, El-Feky MA, Aly SA. Co-occurrence of Plasmid-mediated Quinolone Resistance and Carbapenemases in Klebsiella pneumoniae Isolates in Assiut, Egypt. Egypt J Med Microbiol. 2017;38(5792):1–7. [Google Scholar]
- 32.Poirel L, Revathi G, Bernabeu S, Nordmann P. Detection of NDM-1-producing Klebsiella pneumoniae in Kenya. Antimicrob Agents Chemother. 2011;55(2):934–6. doi: 10.1128/AAC.01247-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev. 2019;32(2). doi: 10.1128/CMR.00115-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S: Sensitive, I: Intermediate, R: Resistant, TET: Tetracycline, GM: Gentamicin, AN: Amikacin, CIP: Ciprofloxacin, ATM: Aztreonam, PTZ: Piperacillin-tazobactam, AMC: Amoxicillin-clavulanate, SXT: Trimethoprim-sulfamethoxazole, CHL: Chloramphenicol, CXT: Cefoxitin, CRO: Ceftriaxone, MEM: Meropenem, IMP: Imipenem, ETM: Ertapenem.
(PDF)
mCIM: modified Carbapenem Inactivation Method, ESBL: Extended-spectrum β-lactamase, Interp: Interpretation, ZI in mm: Zone of inhibition in millimeter, M: Male, F: Female, CSF: Cerebrospinal fluid, ICU: Intensive Care Unit.
(PDF)
Lane M: 100bp DNA ladder; PC: Positive control, Lanes 67–114: K. pneumoniae isolates, NC: Negative control.
(TIF)
Lane M: 100bp DNA ladder, PC1, PC2 & PC3: Positive control, Lanes 67–120: K. pneumoniae isolates, NC: Negative control.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.