Abstract
At the Rowley Shoals in Western Australia, the prominent reef flat becomes exposed on low tide and the stagnant water in the shallow atoll lagoons heats up, creating a natural laboratory for characterizing the mechanisms of coral resilience to climate change. To explore these mechanisms in the reef coral Acropora tenuis, we collected samples from lagoon and reef slope habitats and combined whole-genome sequencing, ITS2 metabarcoding, experimental heat stress, and transcriptomics. Despite high gene flow across the atoll, we identified clear shifts in allele frequencies between habitats at relatively small linked genomic islands. Common garden heat stress assays showed corals from the lagoon to be more resistant to bleaching, and RNA sequencing revealed marked differences in baseline levels of gene expression between habitats. Our results provide new insight into the complex mechanisms of coral resilience to climate change and highlight the potential for spatially varying selection across complex coral reef seascapes to drive pronounced ecological divergence in climate-related traits.
Spatially varying selection drives ecological divergence in climate-related traits amidst high gene flow.
INTRODUCTION
The impacts of anthropogenic climate change and associated extreme weather events are devastating natural ecosystems around the world. Coral reefs are especially threatened, and it is uncertain whether reef-building corals can keep pace with this unprecedented rate of environmental change (1). Fortunately, most coral species have large effective population sizes (2–4) with broad geographic ranges that span strong environmental gradients over which their phenotypes vary. As a result, they tend to harbor an abundance of phenotypic and genetic variation in fitness-related traits that are facing intensified selection with climate change, such as thermal tolerance (5–9). The resulting ecological divergence among populations and associated levels of connectivity will underlie the capacity of species to adapt to rapid environmental change over the coming decades (10–12).
Thermal tolerance in corals is a highly heritable and polygenic trait (9, 13, 14), governed by a complex interplay between host genetics and symbiont community composition (15–17). Studies documenting transcriptome-wide changes in gene expression regularly report hundreds of genes that respond to thermal stress and that are associated with a diverse range of cellular pathways (18–21). Consistent with these patterns, tens to hundreds of loci are often reported in FST-based outlier analyses among corals across large environmental gradients (14, 22, 23). When gene flow is high, as in most broadcast-spawning corals, adaptation favors the tight linkage of small effect loci into regions of reduced genetic distance and recombination (24). In this case, specific genomic regions may play a particularly important role in driving evolutionary change (13, 25). Identifying genomic regions that control complex climate-related traits is central to developing a mechanistic and predictive understanding of climate change resilience in corals. In turn, this information can be used to help design effective spatial management strategies aimed at protecting areas of reef that harbor high levels of stress-tolerant alleles, as well as improve more proactive management interventions such as reef restoration and assisted gene flow initiatives (26–28).
Variation in thermal tolerance occurs across large spatial scales (29, 30), where populations are exposed to contrasting environmental conditions, as well as across fine-spatial scales on the same reef (31–33). A combination of complex bathymetry, hydrodynamics, and water quality can expose some coral communities to more variable or extreme environmental conditions than other areas of reef. Such environments can select for heat resistance and offer important insight into the mechanisms of climate change resilience in corals. Populations from these variable or more extreme habitats are a natural asset for reef management, yet we have a limited understanding of the genetic mechanisms at play.
The lagoon at each of the three reef atolls at Western Australia’s Rowley Shoals is a highly variable habitat that supports extensive coral growth. These atolls have a prominent reef flat that greatly restricts any flushing or exchange of water with the open ocean. As a result, coral populations in the lagoon are regularly exposed to warmer temperature and more stagnant flow dynamics than those on the reef slope (Fig. 1 and figs. S1 to S3). Despite these contrasting environmental conditions, some coral species, such as the widespread and ecologically important Acropora tenuis, are common in both habitats. Here, we combined low-coverage whole-genome sequencing (WGS), Symbiodiniaceae internal transcribed spacer 2 (ITS2) metabarcoding, common garden acute experimental heat stress, and RNA sequencing (RNA-seq) to explore the mechanisms that confer resilience to the periodic environmental extremes of the lagoon habitat at Clerke Reef in the Rowley Shoals. To place levels of divergence among habitats into a broader ecological and evolutionary context, we included samples from the neighboring Scott Reef system, located approximately 400 km to the northeast along the continental shelf (Fig. 1A).
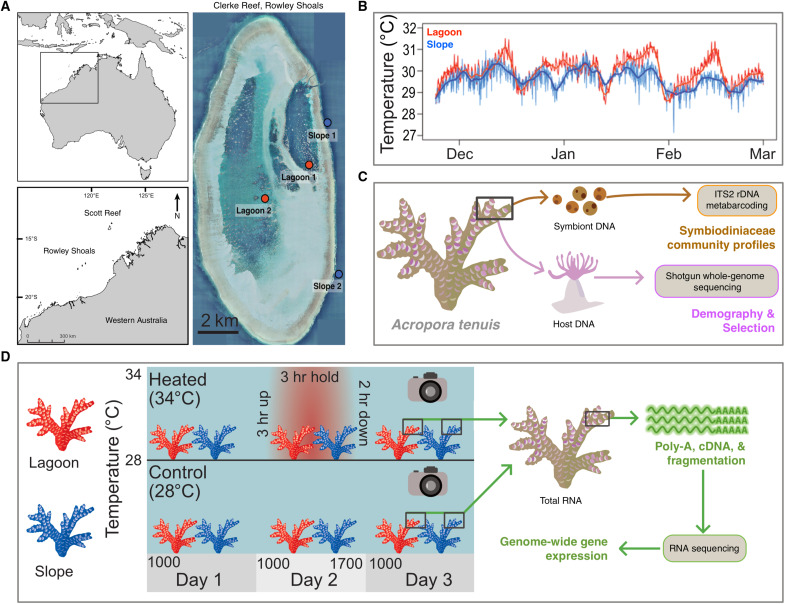
Fig. 1. Sample sites, experimental design, and sequencing methods.
(A) Map of sampling locations in lagoon and slope habitats at Clerke Reef, Rowley Shoals. (B) Time-series temperature plot from the lagoon (L1) and slope (S1) habitats across the 2017/2018 summer. (C) Tissue samples for whole-genome sequencing (WGS) and symbiont ITS2 metabarcoding were collected from A. tenuis at two lagoon (n = 20) and two slope (n = 20) sites (n = 80 total). Samples were also collected from the reef slope habitat at South Scott (n = 10) in the Scott Reef system. (D) Experimental setup for common garden acute heat stress assays. Colony fragments used in experimental heat stress and RNA-seq (n = 10 per habitat) were collected from slope 1 and lagoon 1.
RESULTS AND DISCUSSION
Isolated in space and time
We used a low-coverage WGS approach and mapped 573,937,253 paired-end (250 cycles) shotgun sequence reads from 85 colonies of the spring spawning lineage of A. tenuis (34) to our pseudo-chromosome assembly (13, 35), achieving a mean genome sequencing coverage of 4.2× (± 0.10 SE) per sample (fig. S4 and table S2). Multiple clustering methods based on the nuclear data indicated strong genetic subdivision between Scott Reef and Rowley Shoals (global FST = 0.25; Fig. 2, C and D). We did not, however, observe any clear geographic clustering of samples based on the complete mitochondrial genomes (fig. S6). Considering the strong divergence across the nuclear genome, a lack of clear structure across the mitochondrion likely reflects historical introgression between reef systems and/or the low mutation rates characteristic of cnidarians (36). Estimates of Tajima’s D at the Rowley Shoals showed a strong positive shift relative to Scott Reef, indicative of a recent bottleneck (Fig. 2A). Demographic changes from 50 thousand years (ka) to 5 million years (Ma) ago inferred from the Multiple Sequentially Markovian Coalescent (MSMC) based on a single high-coverage (46×) individual showed a steady decline in Ne at the Rowley Shoals following a broad peak between ~100 ka and 1 Ma ago (Fig. 2B and fig. S7). This peak in effective population size approximately 150,000 years before present is similar to other estimates for this species from the Indo-Pacific (35, 37) and coincides with the Last Interglacial, a period of global warming and rapid sea-level change (38).
Fig. 2. Demographic and evolutionary history of A. tenuis from the Rowley Shoals.
(A) Density plot of Tajima’s D in 1-kb windows across the genome for Rowley Shoals (black) and neighboring Scott Reef (blue). For Rowley Shoals, individual black lines (n = 10) represent bootstrap replicates. (B) Changes in population size through time at the Rowley Shoals using partially sequentially Markovian coalescent from deep sequencing (45×) of a single colony. Individual black lines show bootstrap replicates, and bold gray line represents the mean. (C) Scatterplot of the first two principal components (PC) and (D) admixture proportions for samples collected from Scott Reef (blue) and Rowley Shoals (gray) based on 5,493,423 loci (MAF > 0.05).
High gene flow between habitats
We detected low levels of nuclear divergence among lagoon and slope samples at Clerke Reef (weighted FST = 0.007), with no clear spatial clustering of colonies by habitat (Fig. 3B). Principal components analysis (PCA) based on 5,493,423 single-nucleotide polymorphisms (SNPs) [minor allele frequency (MAF) > 0.05] showed all samples to be admixed across habitats, indicating high gene flow. This lack of differentiation between habitats was not surprising considering the spatial proximity of sample sites and the broadcast spawning life history strategy of A. tenuis. It is also consistent with patterns of gene flow between habitats in the congener Acropora digitifera from the Rowley Shoals (39). A lack of any clear genetic structure between habitats at Clerke Reef suggests that local recruitment comes from a mixed larval pool. In such cases, environmental heterogeneity gives rise to patterns of spatially varying selection where different habitats select for genotypes from a common gene pool that match the local conditions (40). In turn, this results in an overall increase in adaptive genetic variation in the broader metapopulation (41). Unlike directional selection, which reduces genetic variation by fixing beneficial alleles, balancing selection maintains genetic variation within a species and plays a crucial role in adaptation in species with high gene flow and that span strong environmental gradients (42).
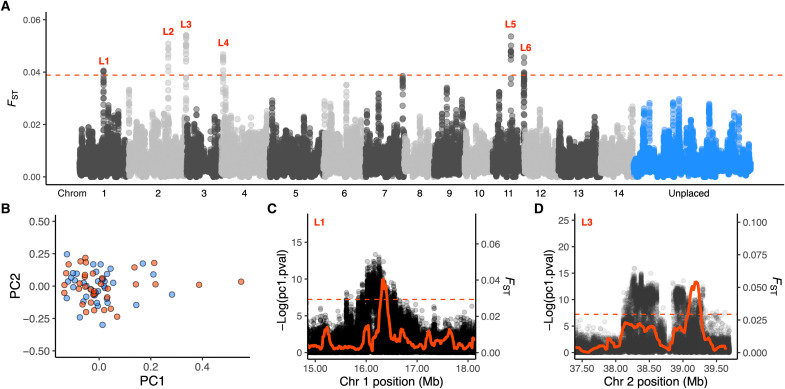
Fig. 3. Genome-wide patterns of differentiation in A. tenuis from lagoon and slope habitats.
(A) Manhattan plot of FST across the genome using 100-kb windows (10-kb step) and based on the unfiltered SFS. Alternating colors indicate different chromosomes. Each point is the average FST for all SNPs in that window. Red dashed line denotes top 0.01% windows. (B) Scatterplot of the first two principal components for colonies from lagoon (red) and slope (blue) habitats. (C and D) Zoomed-in plots of FST (red line, 100-kb windows) and population-independent selection coefficients (PCAngsd) for SNPs (black points) surrounding outlier regions calculated in PCAngsd (44). The program uses posterior expectations of the genotypes to identify SNPs with a distribution that exceeds expectations under neutrality along the first principal component (44). We calculated this selection statistic for each SNP (MAF > 0.05) along each chromosome separately and calculated outlier probabilities using the pchisq function (two-tailed mode) in R.
Signatures of selection amidst high gene flow
When gene flow is high, selection favors the tight linkage of small effect loci into regions of reduced genetic distance and recombination (24, 43). To this end, we searched for signs of adaptation to the warmer lagoon habitat at Clerke Reef by calculating FST between habitats using 100-kb windows with 10-kb steps. Using this sliding window approach, we identified several large regions of the genome that showed high levels of differentiation and were therefore strong candidates for selection (Fig. 3A). The most extreme values genome-wide overlapped six islands of divergence spread across four different chromosomes (chromosomes 1, 2, 4, and 11). Mean FST across these windows was 0.047, compared to a genome-wide average of 0.007 across all windows. To provide further support to these findings, we used a habitat-blind approach and tested whether any of the outlier regions also included SNPs with a distribution that exceeded expectations under neutrality along the first principal component (44), indicating that they were likely candidates for selection. We calculated this selection statistic based on genotype likelihoods for each SNP (MAF > 0.05) along each chromosome separately and then converted this value to an outlier probability using the pchisq function in R (two-tailed mode). This population-blind approach identified two of the six outlier loci identified using the sliding window approach, locus 1 and locus 3, to include a large number of SNPs showing signs of selection, with distributions that exceeded what was expected under neutrality along the first principal component (Fig. 3, C and D, and fig. S8).
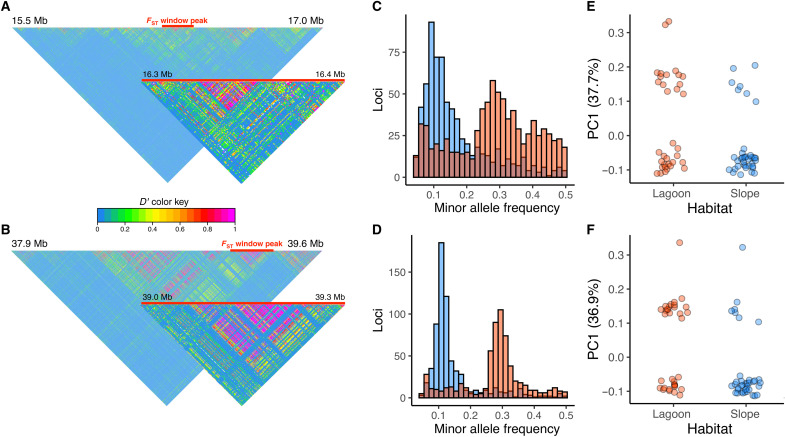
Estimates of linkage disequilibrium (LD) across locus 1 and locus 3 showed FST window peaks to span strong linkage blocks across hundreds of loci (Fig. 4, A and B). Across these regions, we observed a significant increase in MAFs in lagoon colonies relative to those from the reef slope (Welch two-sample t test, P < 0.001; Fig. 4, C and D). Allele frequencies seemed to be concentrated at a particular value in each habitat, a further sign of reduced recombination and supporting the observed patterns of LD. When we carried out a PCA across locus 1 and locus 3, samples clustered strongly into three distinct groups along the first principal component, likely representing the three possible genotypes for each haplotype block (Fig. 4, E and F). Notably, the two dominant genotypes showed different proportions in lagoon and slope samples.
Fig. 4. Linkage disequilibrium in A. tenuis across FST outlier regions.
(A) Heatmap of linkage disequilibrium (D′) across locus 1 on chromosome 1 and (B) locus 3 on chromosome 2. Warmer colors indicate higher linkage among SNPs. (C) Histogram of MAFs in lagoon (red) and slope (blue) samples from Rowley Shoals across outlier locus 1 and (D) locus 3. (E) Sample loadings onto the first principal component across outlier locus 1 and (F) locus 3 showing different frequencies of the three possible haplotypes in each habitat.
Locus 1 not only directly overlapped genic regions with homologs to two Tolloid-like proteins involved in zinc ion binding and development but also included a number of genes within the broader linkage group that had homology to classic stress response proteins, including E3 ubiquitin protein ligases (Lin41/Cop1), tumor necrosis factor receptors (TFIP8, TRAF2, and TRAF3), and heat shock proteins (HSP70) and chaperones (DnaJ homologs) (table S3), but this gene list was not enriched for any specific molecular function (Padj < 0.05). Locus 3 FST window peak directly overlapped genic regions with homology to phosphatidylinositol-glycan biosynthesis class A proteins. Several other notable genes fell within the broader linkage block upstream of locus 3, including a death-associated protein kinase 3 (DAP kinase 3), a serine/threonine kinase (Kalirin), myosin light chain kinase (MLCK), a heat shock protein (HSP75), and a NACHT domain-containing protein (NLRP5) (table S4). Genes in this broader linkage group upstream of locus 3 were highly enriched for adenosine triphosphate (ATP) binding [Gene Ontology (GO) 0005524, Padj = 0.00026], suggesting a role in enzyme regulation and environmental response pathways.
Areas of high linkage and reduced recombination are often associated with chromosomal rearrangements, such as inversions, which play a key role in driving phenotypic variation and local adaptation (24, 45). The role of structural rearrangements driving adaptation in the marine environment is well documented in teleost fishes (46), but less so for other marine organisms. For example, an approximate 5-Mb chromosomal rearrangement in Atlantic cod (Gadus morhua) enables adaptation of fjord populations to hyposaline conditions despite high gene flow with nearby oceanic populations (47). Whether the patterns observed here are driven by actual inversions remains unclear, and future research that uses long-read sequencing will help elucidate the role of structural rearrangements in the adaptation of reef coral to climate change.
Gene flow in A. tenuis between habitats is high across most of the genome, but barriers to gene flow exist. A number of genomic regions showed strong habitat-specific shifts in allele frequencies across hundreds of linked SNPs. This pattern is consistent with adaptation amidst gene flow, where beneficial genetic variants are consolidated into regions of reduced genetic distance and recombination (43, 48). Our data show that no single locus confers resilience to the warm lagoon, but coordinated changes in allele frequencies at hundreds of loci contribute to survival. However, if those allele frequency shifts confer resilience to certain environment stressors, then that shift must come at a cost; otherwise, the alternate allele would reach fixation in the broader metapopulation, and there would be no variation in this trait. A better understanding of the fitness-related trade-offs is essential future research but points to growth, calcification, and reproduction as primary physiological costs of survival in stressful environments (6, 49, 50).
Symbiont associations are stable across habitats
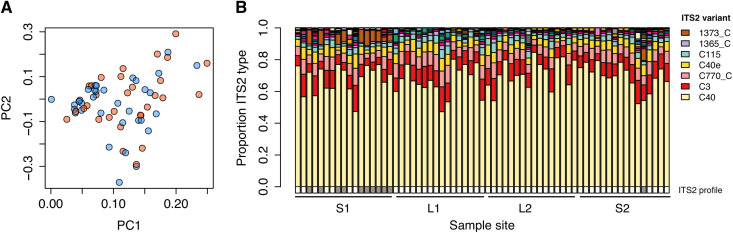
Complex interactions between the coral host and the endosymbiotic algae (family Symbiodiniaceae) are superimposed on the host genetics that drive variation in environmental tolerance (17, 51, 52). Despite the pronounced environmental differences between habitats, metabarcoding of the Symbiodiniaceae ITS2 region of ribosomal DNA (rDNA) for a subset of colonies (n = 65) used for WGS and that were collected across the four sample sites at Clerke Reef revealed no major differences in community composition between habitats (Fig. 5A). All colonies associated exclusively with Cladocopium spp. and were dominated by ITS2 type C40, with various background levels of other Cladocopium ITS2 types (Fig. 5B). An average of 106,213 reads (± 5990 SE) per sample were clustered into 60 distinct ITS2 sequence variants (table S9). Analyses in SymPortal indicated that these ITS2 variants formed two distinct ITS2 type profiles in the data (Fig. 5B), one of which occurred at high frequency in one of the slope sites. This ITS2 type profile was driven by the background ITS2 variant 1373_C, which occurred at moderate frequencies (~7%) in most S1 colonies but was mostly absent from other sites. However, on the basis of the defining intragenomic variant (DIV) profiles, it is possible that more high-resolution loci may reveal ecologically relevant differences in symbiont communities between habitats.
Fig. 5. Symbiont community composition in A. tenuis from lagoon and slope habitats.
(A) Scatterplot of the first two principal components of the symbiont communities (family Symbiodiniaceae) in coral colonies from the lagoon (red) and slope (blue) habitats. (B) Barplot of relative abundance of the different ITS2 types [amplicon sequence variants (ASVs)] at each sample site. Each vertical bar represents an individual colony. Only the most abundant ITS2 variants are displayed in the legend. Boxes below the plot show the distribution of the two symbiont “ITS2 type profiles” identified in the data using SymPortal (77).
Lagoon corals are primed for heat stress
To explore the physiological differences between coral from the two habitats, we combined common garden acute heat stress experiments with RNA-seq. We returned to Clerke Reef a year later and exposed fragments of 15 colonies of A. tenuis (unknown spawning lineage) from the lagoon and slope habitats to acute experimental heat stress using a portable temperature-controlled flow-through seawater system aboard the Australian Institute of Marine Science’s research vessel RV Solander. After 24 hours of acclimation in the tanks, nubbins were exposed to acute heat stress, which consisted of a 3-hour temperature ramp to 34°C starting at 10:00 a.m., followed by a 3-hour hold at 34°C and then a 2-hour ramp back down to ambient temperatures (15, 53, 54). The next morning, 20 hours after the acute heat stress assay began, replicate nubbins from control and heated treatments were photographed to quantify pigment loss using the photographic method (55, 56) and then flash-frozen in liquid nitrogen for gene expression analyses. Samples were first screened using the RNA-seq dataset to ensure that only colonies from the spring cohort were included in any downstream analyses.
Results from the experiment showed that coral colonies from the reef slope were far more pigmented than lagoon corals at the onset of the experiment but suffered greater pigment loss following heat stress (Fig. 6A). These patterns were consistent with our visual scoring method with the CoralWatch Coral Health Chart (fig. S9). Although we did not measure cell density or chlorophyll directly, our photographic method has been shown to correlate strongly with chlorophyll levels (56). Reduced symbiont cell densities and chlorophyll levels have been directly linked to bleaching susceptibility in coral (57, 58), and maintaining lower levels appears to be one mechanism through which lagoon corals minimize photooxidative damage of a shallow high-temperature environment.
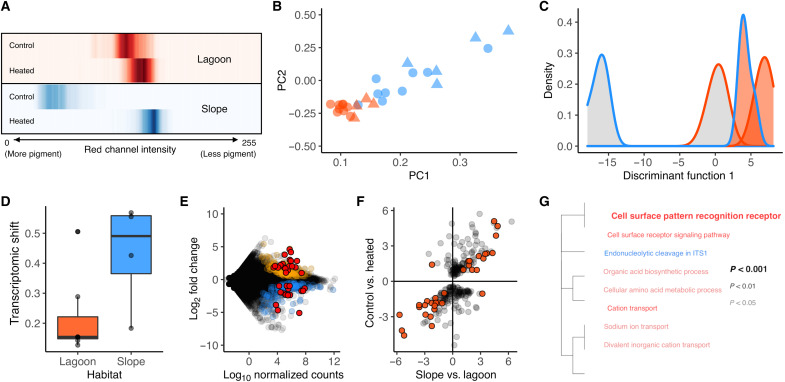
Fig. 6. Physiological differences in A. tenuis from lagoon and slope habitats.
(A) Changes in pigmentation (red channel intensity) of coral nubbins from lagoon (red) and slope (blue) habitats exposed to control (28°C) and heated (34°C) conditions. The darker colors indicate higher density of that pigment value. Values range from 0 (black) to 255 (white), and a positive shift to the right indicates a decline in pigment. (B) Scatterplot of the first two principal components of normalized gene expression in A. tenuis in control (blue) and heated (red) treatments across 21,230 genes with a minimum mean depth per sample of 5. Circles denote samples from lagoon, and triangles denote samples from reef slope. (C) Density distribution of colonies along the first principal component shows control samples from lagoon to cluster closer with the heated treatments than with the other slope controls. (D) Overall transcriptomic shift measured as the distance in multivariate space between heated and control samples for each genotype. (E) Regression of differentially expressed genes between habitats after 2 days of acclimation at ambient temperatures. Each point represents a gene that showed significant differences in baseline expression between habitats. Blue points are down-regulated in the lagoon, and orange points are up-regulated in the lagoon. Red points represent genes that form part of the heat stress response and were also differentially expressed between control and heated treatments across all samples. (F) Linear regression of levels of log fold change in heat-responsive genes between habitats, illustrating that patterns of gene expression under acute heat stress mirror expected changes in expression between habitats. (G) Functional enrichment analyses indicating enriched GO annotations for genes that showed differences in baseline levels of expression between habitats. Blue is up-regulated and red is down-regulated in lagoon samples.
RNA-seq of samples used in the common garden heat stress experiments revealed pronounced differences between corals from the two habitats. Normalized gene expression profiles across 21,230 genes (table S5) showed that lagoon corals held at ambient conditions clustered closely with samples from the heated treatments and experienced a smaller overall transcriptomic shift in response to acute heat stress than corals from the slope (Fig. 6, B to D). In total, we identified 274 genes that differed in baseline levels of expression (Padj < 0.05) between colonies from the two habitats (Fig. 6E and table S6). This gene list was enriched for GO terms related predominantly to transport, metabolism, and signaling (Fig. 6G).
Data from our acute heat stress assay showed that approximately 12% (n = 34) of the genes that differed in baseline expression between habitats were heat-responsive genes that were differentially expressed between heated and control treatments across all samples (Fig. 6E and table S7). This gene set followed intuitive expression profiles based on habitat: Genes that were up-regulated under acute heat stress had higher baseline expression in lagoon corals, and genes that were down-regulated in response to acute heat stress had higher baseline expression in slope corals (Fig. 6F). This gene set included classic stress response genes that repeatedly appear in genomic studies of thermal tolerance in corals (19, 59–61), such as ubiquitin and zinc finger proteins and serine/threonine protein kinases (table S8).
Levels of congruence were relatively low between differentially expressed genes from RNA-seq dataset (habitats or heat stress) and the genomic islands of divergence identified using our low-coverage WGS approach; few heat- or habitat-responsive genes occurred in regions of high genetic differentiation in the WGS datasets. The only exception to this was locus 1 (fig. S11), with an FST window peak that directly overlapped till1, a Tolloid-like proteinase involved in zinc-finger binding and that showed strong changes in regulation under experimental acute heat stress (log2 fold change = 2.62, Padj = 5.8 × 10−14) but that was not differentially expressed between habitats under control or heated conditions (log2 fold change = 0.46, Padj = 0.78). It was also noteworthy that a number of other heat-responsive (n = 7) or habitat-responsive (n = 3) genes occurred up- or downstream of the window peak on locus 1 (fig. S11) in the broader linkage block. However, these genes occurred well outside the FST window peak and were spread among dozens of other genes that did not show habitat- or temperature-driven differences in gene expression.
Corals are capable of large physiological adjustments in response to heat stress, and individuals thriving in environments that are regularly exposed to environmental extremes often show pronounced differences in baseline levels of gene expression than adjacent colonies from more benign environments (15, 62, 63). Higher baseline expression of stress response genes can promote stress tolerance and maintenance of cell homogeneity, while reduced expression can act to limit the negative physiological responses to stress (64). Throughout the daily tidal cycle, corals in the lagoon at Clerke Reef experience warmer water and less current flow than their reef slope counterparts. Our data show that survival in such an environment requires a physiological shift across hundreds of genes. Through this physiological shift, lagoon corals are primed for stressors associated with the daily tidal cycle.
Ecological divergence among populations of coral will be a key driver in their capacity to adapt to a rapidly changing environment. This is particularly true for isolated reef systems, which are largely reliant on local standing genetic variation to adapt to climate-related stressors. In Western Australia, the offshore oceanic atolls of the Rowley Shoals are isolated in space and time, characterized by strong genetic differentiation with neighboring reef systems and a skew in the allele frequency spectrum indicative of a recent bottleneck. Across the Clerke Reef atoll, we identified high gene flow between slope and lagoon habitats but identified a number of genomic islands of divergence that exhibited signs of restricted gene flow and strong LD, suggesting that they may play an important role in adaptation to the marginal lagoon environment. Acute heat stress experiments showed lagoon corals to be more resistant to heat stress than slope corals, and accompanying RNA-seq data showed that this was, in part, achieved through a habitat-specific transcriptomic shift that involved hundreds of genes, a large fraction of which were part of a larger heat stress response. Identifying the genomic regions that confer resilience to different stressors, and understanding the physiological trade-offs associated with those shifts, will allow the development of targeted genetic assays to rapidly screen corals for the complex traits needed to effectively survive climate change.
MATERIALS AND METHODS
Sample collection
A. tenuis forms a species complex in Western Australia with morphologically cryptic colonies that occur in sympatry but spawn in either spring or autumn (34). To avoid confounding our data by including two distinct genetic lineages, we screened 225 colonies collected across four sample sites (two lagoon and two slope) at Clerke Reef with a panel of nine microsatellite loci that can differentiate between the autumn and spring spawning cohort (34). From this analysis, we identified 20 genotypes of the spring cohort from each of the two lagoon and two slope sites at Clerke Reef (n = 80 total; Fig. 1) that we used for downstream sequencing and analyses. The spring cohort was chosen because it was common in both habitats, in contrast to the autumn spawner that was far more dominant on the reef slope. We also included 10 samples from the reef slope of the neighboring Scott Reef system (spring cohort) to place differences among lagoon and slope corals at Clerke Reef into a broader context. All samples were collected along a 200-m transect from colonies separated by at least 1.5 m and preserved in ethanol immediately after collection. DNA was extracted from samples using a QIAGEN DNA Blood and Tissue kit. Allelic variation was measured at nine microsatellite loci (34), and Bayesian admixture analysis in structure (65) was used to assign individuals to either spring or autumn cohort using known reference colony genotypes (table S1).
Genotype likelihoods
A recently developed reference assembly for A. tenuis is one of the most complete genome assemblies available for corals to date (35). However, it remains fragmented and lacks chromosome-scale resolution. In an attempt to improve this assembly, we used RaGOO (66) to generate pseudo-chromosomes for the A. tenuis assembly (aten_final_0.11.fasta) using the closely related [divergence approximately 15 Ma ago (37)] and recently published chromosome-scale Acropora millepora (13) genome assembly. RaGOO is a reference-guided scaffolding method that orders and orients assembly scaffolds and contigs to a single reference genome (66). We ran RaGOO with default parameters using minimap2 as the aligner (67). A total of 407 Mb of sequence was localized, corresponding to 83.6% of A. tenuis contigs, 94% of which fell within the 14 A. millepora chromosomes. Gene coordinates from the reference A. tenuis assembly were lifted over to the pseudo-chromosomes using the RaGOO utility script lift_over.py included in the software.
Shotgun libraries were prepared and individually barcoded using Nextera Flex Library Preparation Kits (Illumina) and pooled together for sequencing on NovaSeq 6000 (S1, 300 cycles). Of the 90 samples that we processed, five samples from Rowley Shoals (three lagoon and two slope) did not produce libraries of sufficient quality for sequencing and were excluded from this study. Raw FASTQ files were mapped with bwa mem to the A. tenuis pseudo-chromosome-scale assembly (aten.chr.fasta). We used SAMtools (68) to sort and index and Picard tools (http://broadinstitute.github.io/picard) to mark and remove duplicate reads. SAMtools was also used to calculate sequencing depth of each sample at each position across the genome. We used ANGSD (69) to estimate the site frequency spectrum (SFS) for each population based on genotype likelihoods. We masked sites minQ < 20, minMapQ < 30, and sites with less than one-third and greater than two times the mean read depth for each sample site. We also filtered out loci with missing data in more than half of the samples per site. Initial screening of samples using PCA in PCAngsd (44) identified three outlier colonies (two of which appeared to be clones) that were collected from one of the lagoon sites (L2) and that were removed from downstream analyses (fig. S5). These samples likely represent either mis-IDs or cryptic species that were not resolved with the panel of microsatellite markers.
Demographic history of the Rowley Shoals
To provide an overall demographic and evolutionary context of the Rowley Shoals, we calculated genome-wide estimates of Tajima’s D for each reef system using realSFS (69). A positive Tajima’s D indicates an absence of low-frequency alleles, suggestive of a recent bottleneck. These analyses were based on the unfiltered folded SFS (ancestral allele state unknown) using 1-kb nonoverlapping windows. To ensure comparable sample sizes between reefs, we restricted our estimates for Tajima’s D at the Rowley Shoals to individuals from a single sample site (S1) and then carried out replicate (n = 10) bootstrapping to downsample the Rowley Shoals population (S2) to 10 individuals before calculating estimates of nucleotide diversity. These analyses revealed a negligible influence of sample size on our estimates of Tajima’s D.
We also sequenced one individual from the Rowley Shoals reef slope to ~46× coverage and applied the partially sequentially Markovian coalescent method implemented in MSMC (70). SNPable (http://lh3lh3.users.sourceforge.net/snpable.shtml) was first used to mask all regions of the genome where it was not possible to obtain unambiguous read mappings. Variant calling was performed for deep sequenced samples using SAMtools and BCFtools. A distribution of MSMC estimates was obtained by generating 100 bootstrap runs, each of which involved recombining a random subsampling of 500-kb-sized chunks to produce 20 large scaffolds of length 10 Mb. Raw outputs from MSMC were converted to real values using a mutation rate of 1.86 × 10−8 events per base per generation and a generation time of 5 years. This choice of generation time is consistent with the fast growth rate and relatively high turnover of Acropora (35, 37), and the value for μ was estimated on the basis of a divergence time of 15.5 Ma between A. tenuis and A. digitifera (71) using synonymous substitution rates for one-to-one orthologs (70). The values of Ne and time that we infer depend on our assumptions of μ and the generation time, and we note that any uncertainty in these parameters is reflected in the absolute values but will not affect the qualitative patterns of population size changes.
Population structure and signatures of selection
To explore patterns of genetic differentiation between habitats, we excluded variant sites with MAF < 0.05 and snp_value > 1 × 10−6 and used PCAngsd (44) to output a covariance matrix that was used as an input to the R function eigen to carry out PCA. PCAngsd also estimates the optimal number of clusters and uses a matrix factorization algorithm to estimate admixture proportions for each sample. To estimate divergence across the mitochondrial genome, we also mapped reads to the complete A. tenuis mitochondrion (18,659 base pairs) (72) and extracted consensus sequences for each sample with SAMtools. We aligned these consensus sequences using a muscle alignment in Geneious and then built a haplotype network in POPART (73).
We searched for signatures of selection in coral from the warm lagoon habitat at Clerke Reef by calculating FST between habitats using 100-kb windows with 10-kb steps and identified empirical outliers as those occurring in the top 0.01% of values genome wide. This sliding window approach allowed us to hone in on large genomic regions of elevated differentiation, or islands of divergence, rather than focusing on any single variant alone. These analyses were done using the unfiltered SFS for each habitat. We then tested for strong LD across any outlier regions using ngsLD (74), which estimates pairwise linkage among variant sites based on genotype likelihoods. Last, we used a population-blind approach to identify targets of selection using the -selection flag in PCAngsd. These analyses were run with the Scott Reef samples removed from the dataset. The program uses posterior expectations of the genotypes to identify SNPs with a distribution that exceeds expectations under neutrality along the first principal component (44). We calculated this selection statistic for each SNP (MAF > 0.05) along each chromosome separately and calculated outlier probabilities using the pchisq function (two-tailed mode) in R.
Symbiont community profiling with ITS2 metabarcoding
In Western Australia, Acropora have a high level of affinity to the genus Cladocopium (75). To determine whether corals from different habitats at the Clerke Reef associate with different Cladocopium types, we applied metabarcoding of the ITS2 region of the rDNA (76) for a subset of the colonies (n = 65) used for WGS and that were collected across the four sample sites at Clerke Reef. The ITS2 region was amplified using the ITSD (5′-GTGAATTGCAGAACTCCGTG-3′) and ITS2-rev2 (5′-CCTCCGCTTACTTATATGCTT-3′) primer pair. Polymerase chain reaction (PCR) was carried out in 30 μl of reactions containing 15 μl of 2× Phusion Master Mix, 0.5 μl of each primer (10 μM) modified with Nextera Illumina overhang sequences, and 1.5 μl of template DNA and water to volume. Thermal cycling conditions were as follows: denaturation at 98°C for 30 s, followed by 35 cycles of 10 s at 98°C, 30 s at 60°C, and 30 s at 72°C, and a final extension at 72°C for 5 min (performed on a Mastercycler Nexus Gradient, Thermo Fisher Scientific). Amplicons underwent a second round of PCR to attach flow cell adapters and unique barcodes and were then pooled at equal molar concentrations for sequencing on Illumina MiSeq using a V2 600 cycle paired end kit at the Australian Genome Research Facility. Demultiplexed reads were processed through SymPortal (77), which was designed specifically for the analysis of Symbiodiniaceae ITS2 metabarcoding data.
Common garden heat stress experiment
In 2019, we returned to the reef and collected replicate fragments (n = 2) from 15 colonies of A. tenuis (unknown spawning lineage) from the lagoon (L1) and slope (S1) habitats at Clerke Reef that were exposed to ambient (28°C) and heated (34°C) treatments using a portable temperature-controlled flow-through seawater system aboard our research vessel RV Solander. After 24 hours of acclimation in the tanks, nubbins were exposed to acute heat stress, which consisted of a 3-hour temperature ramp to 34°C starting at 10:00 a.m., followed by a 3-hour hold at 34°C, and then a 2-hour ramp back down to ambient temperatures (5). The next morning, 20 hours after the acute heat stress assay began, replicate nubbins from control and heated treatments were photographed to quantify pigment loss using the photographic method (55), which uses shifts in intensity along the red channel as a proxy for chlorophyll loss. Although an acclimation phase of 1 day in a common garden was designed to remove gene expression changes due to fragmentation or acute environmental responses, it was likely not long enough to erode any long-term plastic effects on gene expression. Samples were then flash-frozen in liquid nitrogen for transcriptome-wide gene expression analyses. Samples from the heat stress experiment were first screened using the RNA-seq dataset (see the next section) to ensure that all analyses were based on a single genetic spawning lineage (spring cohort).
Transcriptome-wide gene expression patterns
Stranded poly-A RNA libraries were prepared using an Agilent’s SureSelect library preparation kit from total RNA extracted from 40 samples (10 heated and 10 control from each habitat with best RNA quality) using a QIAGEN RNeasy Mini kit. The library preparation protocol includes a polyA enrichment followed by fragmentation, reverse transcription, second-strand synthesis with uridine triphosphate, adapted ligation, and 12 cycles of uracil DNA glycosylase (UDG) for indexing. Library quality control (QC) was performed using TapeStation 4100 and repooled on the basis of iSeq data before being sequenced on a NovaSeq SP flow cell in 2 × 50 cycles format to yield approximately 10 million read pairs per sample at Genomics WA. Raw FASTQ files were processed using fastp (78) for adapter trimming and quality filtering (≥Q30), and reads were then mapped using hisat (79) to the A. tenuis genome. As a first quality control step and to avoid including multiple spawning lineages in our analyses of pigmentation and gene expression, we used SAMtools to call variant sites and prcomp in R to carry out PCA. In total, we mapped 462,429,066 paired end reads from 40 complementary DNA (cDNA) to the A. tenuis genome assembly. Initial screening of samples based on a panel 60,941 variant sites across the transcriptome revealed that one individual from the lagoon and five from the reef slope from our heat stress experiment belonged to the autumn A. tenuis spawning lineage (fig. S10). These samples were removed from all of our downstream analyses (including pigmentation analyses with photographic method). Once we isolated a single genetic lineage, differential gene expression analyses were carried out in DESeq2 (80) using a raw counts matrix. Differentially expressed genes were identified using Wald tests after adjusting for multiple comparisons (Padj < 0.05). Functional enrichment based on GO terms was assessed using a two-tailed Mann-Whitney U test in gomwu (81). Last, we measured the magnitude of response to heat stress for each genotype using PCA. For this analysis, we weighted samples by the percent of variation explained by each axis and then calculated the shift (Euclidean distances) in multivariate space between heated and control treatments for each genotype independently.
Acknowledgments
This work was conducted as part of the North West Shoals to Shore Program, which is proudly supported by Santos as part of the company’s commitment to better understand WA’s marine environment. We also acknowledge the support of ARC Linkage Project LP160101508 to explore coral resilience. We gratefully acknowledge the Australian Cancer Research Foundation and the Centre for Advanced Cancer Genomics for making available Illumina Sequencers for the use of Genomics WA. Last, we are extremely grateful to J. Clough and the Jock Clough Marine Foundation for the support.
Funding: J.N.U. was supported by the Woodside Coral Reef Research Fellowship. Genomics WA is supported by Bioplatforms Australia, State Government Western Australia, Australian Cancer Research Foundation, Cancer Research Trust, Harry Perkins Institute of Medical Research, Telethon Kids Institute, and the University of Western Australia.
Author contributions: L.T., J.N.U., and J.P.G. designed the study. L.T., J.N.U., J.P.G., L.D., and C.M.G. carried out experiments and collected data. L.T. conducted laboratory work and analyzed data. L.T. wrote the manuscript with feedback from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper, in the Supplementary Materials, or online via the Open Science Framework (https://osf.io/2ba8p/) and NCBI Sequence Read Archive (PRJNA809112).
Supplementary Materials
This PDF file includes:
Figs. S1 to S12
Tables S10 and S11
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S9
REFERENCES AND NOTES
- 1.Hughes T. P., Barnes M. L., Bellwood D. R., Cinner J. E., Cumming G. S., Jackson J. B. C., Kleypas J., van de Leemput I. A., Lough J. M., Morrison T. H., Palumbi S. R., Van Nes E. H., Scheffer M., Coral reefs in the Anthropocene. Nature 546, 82–90 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Richards Z. T., van Oppen M. J. H., Rarity and genetic diversity in Indo-Pacific Acropora corals. Ecol. Evol. 2, 1867–1888 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drury C., Dale K. E., Panlilio J. M., Miller S. V., Lirman D., Larson E. A., Bartels E., Crawford D. L., Oleksiak M. F., Genomic variation among populations of threatened coral: Acropora cervicornis. BMC Genomics 17, 286 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prada C., Hanna B., Budd A. F., Woodley C. M., Schmutz J., Grimwood J., Iglesias-Prieto R., Pandolfi J. M., Levitan D., Johnson K. G., Knowlton N., Kitano H., DeGiorgio M., Medina M., Empty niches after extinctions increase population sizes of modern corals. Curr. Biol. 26, 3190–3194 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Palkopoulou E., Lipson M., Mallick S., Nielsen S., Rohland N., Baleka S., Karpinski E., Ivancevic A. M., To T.-H., Kortschak R. D., Raison J. M., Qu Z., Chin T.-J., Alt K. W., Claesson S., Dalén L., MacPhee R. D. E., Meller H., Roca A. L., Ryder O. A., Heiman D., Young S., Breen M., Williams C., Aken B. L., Ruffier M., Karlsson E., Johnson J., Di Palma F., Alfoldi J., Adelson D. L., Mailund T., Munch K., Lindblad-Toh K., Hofreiter M., Poinar H., Reich D., A comprehensive genomic history of extinct and living elephants. Proc. Natl. Acad. Sci. U.S.A. 115, E2566–E2574 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenkel C. D., Almanza A. T., Matz M. V., Fine-scale environmental specialization of reef-building corals might be limiting reef recovery in the Florida Keys. Ecology 96, 3197–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Weis V. M., The susceptibility and resilience of corals to thermal stress: Adaptation, acclimatization or both? Mol. Ecol. 19, 1515–1517 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Middlebrook R., Hoegh-Guldberg O., Leggat W., The effect of thermal history on the susceptibility of reef-building corals to thermal stress. J. Exp. Biol. 211, 1050–1056 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Kirk N. L., Howells E. J., Abrego D., Burt J. A., Meyer E., Genomic and transcriptomic signals of thermal tolerance in heat-tolerant corals (Platygyra daedalea) of the Arabian/Persian Gulf. Mol. Ecol. 27, 5180–5194 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Kleypas J. A., Thompson D. M., Castruccio F. S., Curchitser E. N., Pinsky M., Watson J. R., Larval connectivity across temperature gradients and its potential effect on heat tolerance in coral populations. Glob. Chang. Biol. 22, 3539–3549 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Matz M. V., Treml E. A., Aglyamova G. V., Bay L. K., Potential and limits for rapid genetic adaptation to warming in a Great Barrier Reef coral. PLOS Genet. 14, e1007220 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bay R. A., Rose N. H., Logan C. A., Palumbi S. R., Genomic models predict successful coral adaptation if future ocean warming rates are reduced. Sci. Adv. 3, e1791413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller Z. L., Mocellin V. J. L., Morris L. A., Cantin N., Shepherd J., Sarre L., Peng J., Liao Y., Pickrell J., Andolfatto P., Matz M., Bay L. K., Przeworski M., Population genetics of the coral Acropora millepora: Toward genomic prediction of bleaching. Science 369, eaba4674 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Bay R. A., Palumbi S. R., Multilocus adaptation associated with heat resistance in reef-building corals. Curr. Biol. 24, 2952–2956 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Thomas L., Rose N. H., Bay R. A., López E. H., Morikawa M. K., Ruiz-Jones L., Palumbi S. R., Mechanisms of thermal tolerance in reef-building corals across a fine-grained environmental mosaic: Lessons from Ofu, American Samoa. Front. Mar. Sci. 4, 434 (2018). [Google Scholar]
- 16.Ziegler M., Seneca F. O., Yum L. K., Palumbi S. R., Voolstra C. R., Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat. Commun. 8, 14213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berkelmans R., van Oppen M. J. H., The role of zooxanthellae in the thermal tolerance of corals: A “nugget of hope” for coral reefs in an era of climate change. Proc. R. Soc. B Biol. Sci. 273, 2305–2312 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kenkel C. D., Meyer E., Matz M. V., Gene expression under chronic heat stress in populations of the mustard hill coral (Porites astreoides) from different thermal environments. Mol. Ecol. 22, 4322–4334 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Rose N. H., Seneca F. O., Palumbi S. R., Gene networks in the wild: Identifying transcriptional modules that mediate coral resistance to experimental heat stress. Genome Biol. Evol. 8, 243–252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas L., López E. H., Morikawa M. K., Palumbi S. R., Transcriptomic resilience, symbiont shuffling, and vulnerability to recurrent bleaching in reef-building corals. Mol. Ecol. 28, 3371–3382 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Dixon G., Abbott E., Matz M., Meta-analysis of the coral environmental stress response: Acropora corals show opposing responses depending on stress intensity. Mol. Ecol. 29, 2855–2870 (2020). [DOI] [PubMed] [Google Scholar]
- 22.van Oppen M. J. H., Bongaerts P., Frade P., Peplow L. M., Boyd S. E., Nim H. T., Bay L. K., Adaptation to reef habitats through selection on the coral animal and its associated microbiome. Mol. Ecol. 27, 2956–2971 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Thomas L., Kennington W. J., Evans R. D., Kendrick G. A., Stat M., Restricted gene flow and local adaptation highlight the vulnerability of high-latitude reefs to rapid environmental change. Glob. Chang. Biol. 23, 2197–2205 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Thompson M. J., Jiggins C. D., Supergenes and their role in evolution. Heredity 113, 1–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dixon G., Davies S., Aglyamova G., Meyer E., Bay L. K., Matz M. V., Genomic determinants of coral heat tolerance across latitudes. Science 348, 1460–1462 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Anthony K., Bay L. K., Costanza R., Firn J., Gunn J., Harrison P., Heyward A., Lundgren P., Mead D., Moore T., Mumby P. J., van Oppen M. J. H., Robertson J., Runge M. C., Suggett D. J., Schaffelke B., Wachenfeld D., Walshe T., New interventions are needed to save coral reefs. Nat. Ecol. Evol. 1, 1420–1422 (2017). [DOI] [PubMed] [Google Scholar]
- 27.van Oppen M. J. H., Oliver J. K., Putnam H. M., Gates R. D., Building coral reef resilience through assisted evolution. Proc. Natl. Acad. Sci. U.S.A. 112, 2307–2313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quigley K. M., Bay L. K., van Oppen M. J. H., The active spread of adaptive variation for reef resilience. Ecol. Evol. 9, 11122–11135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howells E. J., Beltran V. H., Larsen N. W., Bay L. K., Willis B. L., van Oppen M. J. H., Coral thermal tolerance shaped by local adaptation of photosymbionts. Nat. Clim. Chang. 2, 116–120 (2011). [Google Scholar]
- 30.Jurriaans S., Hoogenboom M. O., Thermal performance of scleractinian corals along a latitudinal gradient on the Great Barrier Reef. Philos. Trans. R. Soc. B Biol. Sci. 374, 20180546 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schoepf V., Stat M., Falter J. L., McCulloch M. T., Limits to the thermal tolerance of corals adapted to a highly fluctuating, naturally extreme temperature environment. Sci. Rep. 5, 17639 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Camp E. F., Schoepf V., Mumby P. J., Suggett D. J., The future of coral reefs subject to rapid climate change: Lessons from natural extreme environments. Front. Mar. Sci. 5, 4 (2018). [Google Scholar]
- 33.Barshis D. J., Stillman J. H., Gates R. D., Toonen R. J., Smith L. W., Birkeland C., Protein expression and genetic structure of the coral Porites lobata in an environmentally extreme Samoan back reef: Does host genotype limit phenotypic plasticity? Mol. Ecol. 19, 1705–1720 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Gilmour J. P., Underwood J. N., Howells E. J., Gates E., Heyward A. J., Biannual spawning and temporal reproductive isolation in Acropora corals. PLOS ONE 11, e0150916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooke I., Ying H., Forêt S., Bongaerts P., Strugnell J. M., Simakov O., Zhang J., Field M. A., Rodriguez-Lanetty M., Bell S. C., Bourne D. G., van Oppen M. J. H., Ragan M. A., Miller D. J., Genomic signatures in the coral holobiont reveal host adaptations driven by Holocene climate change and reef specific symbionts. Sci. Adv. 6, eabc6318 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shearer T. L., Van Oppen M. J. H., Romano S. L., Wörheide G., Slow mitochondrial DNA sequence evolution in the Anthozoa (Cnidaria). Mol. Ecol. 11, 2475–2487 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Mao Y., Economo E. P., Satoh N., The roles of introgression and climate change in the rise to dominance of Acropora corals. Curr. Biol. 28, 3373–3382.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 38.B. R. Wilson, The Biogeography of the Australian North West Shelf (Elsevier, ed. 2013, 2013). [Google Scholar]
- 39.Thomas L., Underwood J. N., Adam A. A. S., Richards Z. T., Dugal L., Miller K. J., Gilmour J. P., Contrasting patterns of genetic connectivity in brooding and spawning corals across a remote atoll system in northwest Australia. Coral Reefs 39, 55–60 (2020). [Google Scholar]
- 40.Levene H., Genetic equilibrium when more than one ecological niche is available. Am. Nat. 87, 331–333 (1953). [Google Scholar]
- 41.Gagnaire P.-A., Normandeau E., Côté C., Hansen M. M., Bernatchez L., The genetic consequences of spatially varying selection in the panmictic american eel (Anguilla rostrata). Genetics 190, 725–736 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernatchez L., On the maintenance of genetic variation and adaptation to environmental change: Considerations from population genomics in fishes. J. Fish Biol. 89, 2519–2556 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Samuk K., Owens G. L., Delmore K. E., Miller S. E., Rennison D. J., Schluter D., Gene flow and selection interact to promote adaptive divergence in regions of low recombination. Mol. Ecol. 26, 4378–4390 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Meisner J., Albrechtsen A., Inferring population structure and admixture proportions in low-depth NGS data. Genetics 210, 719–731 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mérot C., Berdan E., Cayuela H., Djambazian H., Ferchaud A.-L., Laporte M., Normandeau E., Ragoussis J., Wellenreuther M., Bernatchez L., Locally adaptive inversions modulate genetic variation at different geographic scales in a seaweed fly. Mol. Biol. Evol. 38, 3953–3971 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wellenreuther M., Bernatchez L., Eco-evolutionary genomics of chromosomal inversions. Trends Ecol. Evol. 33, 427–440 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Barth J. M. I., Berg P. R., Jonsson P. R., Bonanomi S., Corell H., Hemmer-Hansen J., Jakobsen K. S., Johannesson K., Jorde P. E., Knutsen H., Moksnes P. O., Star B., Stenseth N. C., Svedäng H., Jentoft S., André C., Genome architecture enables local adaptation of Atlantic cod despite high connectivity. Mol. Ecol. 26, 4452–4466 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Tigano A., Friesen V. L., Genomics of local adaptation with gene flow. Mol. Ecol. 25, 2144–2164 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Bay R. A., Palumbi S. R., Transcriptome predictors of coral survival and growth in a highly variable environment. Ecol. Evol. 7, 4794–4803 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howells E. J., Berkelmans R., van Oppen M. J. H., Willis B. L., Bay L. K., Historical thermal regimes define limits to coral acclimatization. Ecology 94, 1078–1088 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Stat M., Gates R. D., Clade D Symbiodinium in scleractinian corals: A “nugget” of hope, a selfish opportunist, an ominous sign, or all of the above? J. Mar. Biol. 2011, 1–9 (2011). [Google Scholar]
- 52.Oliver T., Palumbi S. R., Many corals host thermally resistant symbionts in high-temperature habitat. Coral Reefs 30, 241–250 (2011). [Google Scholar]
- 53.Seneca F. O., Palumbi S. R., The role of transcriptome resilience in resistance of corals to bleaching. Mol. Ecol. 24, 1467–1484 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Palumbi S. R., Barshis D. J., Traylor-Knowles N., Bay R. A., Mechanisms of reef coral resistance to future climate change. Science 344, 895–898 (2014). [DOI] [PubMed] [Google Scholar]
- 55.Winters G., Holzman R., Blekhman A., Beer S., Loya Y., Photographic assessment of coral chlorophyll contents: Implications for ecophysiological studies and coral monitoring. J. Exp. Mar. Biol. Ecol. 380, 25–35 (2009). [Google Scholar]
- 56.Hirayama K., Takayama K., Haruta S., Ishibashi H., Takeuchi I., Effect of low concentrations of Irgarol 1051 on RGB (R, red; G, green; B, blue) colour values of the hard-coral Acropora tenuis. Mar. Pollut. Bull. 124, 678–686 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Cunning R., Baker A. C., Excess algal symbionts increase the susceptibility of reef corals to bleaching. Nat. Clim. Chang. 3, 259–262 (2013). [Google Scholar]
- 58.Cornwell B., Armstrong K., Walker N., Lippert M., Nestor V., Golbuu Y., Palumbi S. R., Widespread variation in heat tolerance and symbiont load are associated with growth tradeoffs in the coral Acropora hyacinthus in Palau. eLife 10, e64790 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thomas L., Palumbi S. R., The genomics of recovery from coral bleaching. Proc. Biol. Sci. 284, 20171790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Savary R., Barshis D. J., Voolstra C. R., Cárdenas A., Evensen N. R., Banc-Prandi G., Fine M., Meibom A., Fast and pervasive transcriptomic resilience and acclimation of extremely heat-tolerant coral holobionts from the northern Red Sea. Proc. Natl. Acad. Sci. U.S.A. 118, e2023298118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voolstra C. R., Buitrago-López C., Perna G., Cárdenas A., Hume B. C. C., Rädecker N., Barshis D. J., Standardized short-term acute heat stress assays resolve historical differences in coral thermotolerance across microhabitat reef sites. Glob. Chang. Biol. 26, 4328–4343 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Barshis D. J., Ladner J. T., Oliver T., Seneca F. O., Traylor-Knowles N., Palumbi S. R., Genomic basis for coral resilience to climate change. Proc. Natl. Acad. Sci. U.S.A. 110, 1387–1392 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kenkel C. D., Matz M. V., Gene expression plasticity as a mechanism of coral adaptation to a variable environment. Nat. Ecol. Evol. 1, 0014 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Rivera H. E., Aichelman H. E., Fifer J. E., Kriefall N. G., Wuitchik D. M., Wuitchik S. J. S., Davies S. W., A framework for understanding gene expression plasticity and its influence on stress tolerance. Mol. Ecol. 30, 1381–1397 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Pritchard J. K., Stephens M., Donnelly P., Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alonge M., Soyk S., Ramakrishnan S., Wang X., Goodwin S., Sedlazeck F. J., Lippman Z. B., Schatz M. C., RaGOO: Fast and accurate reference-guided scaffolding of draft genomes. Genome Biol. 20, 224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H., Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schiffels S., Durbin R., Inferring human population size and separation history from multiple genome sequences. Nat. Genet. 46, 919–925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller D. J., Ball E. E., Forêt S., Satoh N., Coral genomics and transcriptomics—Ushering in a new era in coral biology. J. Exp. Mar. Biol. Ecol. 408, 114–119 (2011). [Google Scholar]
- 72.Van Oppen M. J. H., Catmull J., McDonald B. J., Hislop N. R., Hagerman P. J., Miller D. J., The mitochondrial genome of Acropora tenuis (Cnidaria; Scleractinia) contains a large group I intron and a candidate control region. J. Mol. Evol. 55, 1–13 (2002). [DOI] [PubMed] [Google Scholar]
- 73.Leigh J. W., Bryant D., POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116 (2015). [Google Scholar]
- 74.Fox E. A., Wright A. E., Fumagalli M., Vieira F. G., NgsLD: Evaluating linkage disequilibrium using genotype likelihoods. Bioinformatics 35, 3855–3856 (2019). [DOI] [PubMed] [Google Scholar]
- 75.Thomas L., Kendrick G., Kennington W. J., Richards Z. T., Stat M., Exploring Symbiodinium diversity and host specificity in Acropora corals from geographical extremes of Western Australia with 454 amplicon pyrosequencing. Mol. Ecol. 23, 3113–3126 (2014). [DOI] [PubMed] [Google Scholar]
- 76.Pochon X., Pawlowski J., Zaninetti L., Rowan R., High genetic diversity and relative specificity among Symbiodinium-like endosymbiotic dinoflagellates in soritid foraminiferans. Mar. Biol. 139, 1069–1078 (2001). [Google Scholar]
- 77.Hume B. C. C., Smith E. G., Ziegler M., Warrington H. J. M., Burt J. A., LaJeunesse T. C., Wiedenmann J., Voolstra C. R., SymPortal: A novel analytical framework and platform for coral algal symbiont next-generation sequencing ITS2 profiling. Mol. Ecol. Resour. 19, 1063–1080 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen S., Zhou Y., Chen Y., Gu J., Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim D., Langmead B., Salzberg S. L., HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wright R. M., Aglyamova G. V., Meyer E., Matz M. V., Gene expression associated with white syndromes in a reef building coral, Acropora hyacinthus. BMC Genomics 16, 371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S12
Tables S10 and S11
Tables S1 to S9