Abstract
MYC regulates multiple gene programs, raising questions about the potential selectivity and downstream transcriptional consequences of MYC inhibitors as cancer therapeutics. Here, we examined the effect of a small-molecule MYC inhibitor, MYCi975, on the MYC/MAX cistromes, epigenome, transcriptome, and tumorigenesis. Integrating these data revealed three major classes of MYCi975-modulated gene targets: type 1 (down-regulated), type 2 (up-regulated), and type 3 (unaltered). While cell cycle and signal transduction pathways were heavily targeted by MYCi, RNA biogenesis and core transcriptional pathway genes were spared. MYCi975 altered chromatin binding of MYC and the MYC network family proteins, and chromatin accessibility and H3K27 acetylation alterations revealed MYCi975 suppression of MYC-regulated lineage factors AR/ARv7, FOXA1, and FOXM1. Consequently, MYCi975 synergistically sensitized resistant prostate cancer cells to enzalutamide and estrogen receptor–positive breast cancer cells to 4-hydroxytamoxifen. Our results demonstrate that MYCi975 selectively inhibits MYC target gene expression and provide a mechanistic rationale for potential combination therapies.
The effect of a small-molecule MYC inhibitor on target genes is selective, preferentially affecting gene programs.
INTRODUCTION
MYC functions as a transcription factor that regulates a diverse set of gene networks including ribosome biogenesis, mRNA translation, microRNA regulation, cell cycle progression, DNA replication and repair, immune response, metabolism, and apoptosis (1–3). Dysregulated MYC expression is strongly implicated in tumorigenesis and is a hallmark of various types of cancer (4–6); however, therapeutic targeting of MYC has been challenging due to the difficulty of generating high-quality small-molecule inhibitors for what is an intrinsically disordered protein. In addition, it remains unclear whether MYC’s key role in regulating basic cellular processes will undermine cancer cell selectivity and thus tolerability of MYC inhibitors. Attempts to target MYC directly have focused on the MYC/MAX heterodimerization domain. A dominant-negative mutant MYC peptide (OmoMYC) was developed that binds directly to MYC and abrogates MYC function (7, 8). Several small-molecule inhibitors (e.g., 10075-G5 and 10058-F4) disrupt MYC/MAX dimerization and have been shown to reduce MYC activity (9, 10). Other approaches have included the use of covalent inhibitors to disrupt MYC function (11). Use of various inhibitors as molecular probes has led to a substantial increase in knowledge regarding MYC function and a continued focus on the potential of MYC inhibitors as cancer therapeutics (10).
Given that MYC generally functions as a transcription factor, it is critical to understand the molecular mechanisms by which MYC inhibitors limit chromatin binding of MYC and the formation of MAX heterodimers and the subsequent effects on target gene expression. Whether MYC inhibitors selectively modulate MYC binding and/or target gene expression is not clear but has important implications for tolerability as a cancer therapeutic. Investigations of MYC function on transcriptional output demonstrate both global and selective gene regulation (12–15). Experimental and mathematical modeling studies have suggested that higher levels of oncogenic MYC promote chromatin binding to activate and repress transcription of specific genes (16). The specificity of MYC chromatin binding and gene expression changes in models of MYC overexpression suggest that variations in promoter affinities and cofactors are underlying determinants that regulate distinct biological processes (17). The genomic distribution of MYC at target gene promoters and enhancers can provide additional clues to the transcriptionally oncogenic properties of MYC. In models of MYC/MAX heterodimer inhibition with OmoMYC peptide, promoter occupancy of MYC target genes was found to be significantly altered (18). Histone H3 lysine 27 acetylation (H3K27ac) is a mark of active promoters and enhancers, and MYC has been shown to recruit histone acetyltransferases and induce genome-wide chromatin acetylation at H3K27 (19, 20). Binding of MYC at enhancers in models of MYC amplification demonstrates enhancer invasion and a variant MYC DNA binding profile (14).
We recently developed and characterized a small-molecule MYC inhibitor, MYCi975, that binds directly to MYC, disrupts MYC/MAX dimerization, and promotes proteasomal-mediated MYC degradation, resulting in decreased tumor growth in vivo (21, 22). MYCi975 was also well tolerated in animal models. In this work, we examined the sensitivity of MYC/MAX binding sites and the global epigenomic landscape to MYCi975, established the extent to which MYCi975 limits MYC genomic function, and provide evidence supporting the rationale for combination cancer therapies.
RESULTS
The MYC cistrome is altered at canonical targets and genome wide in response to MYCi975 to regulate specific gene expression programs
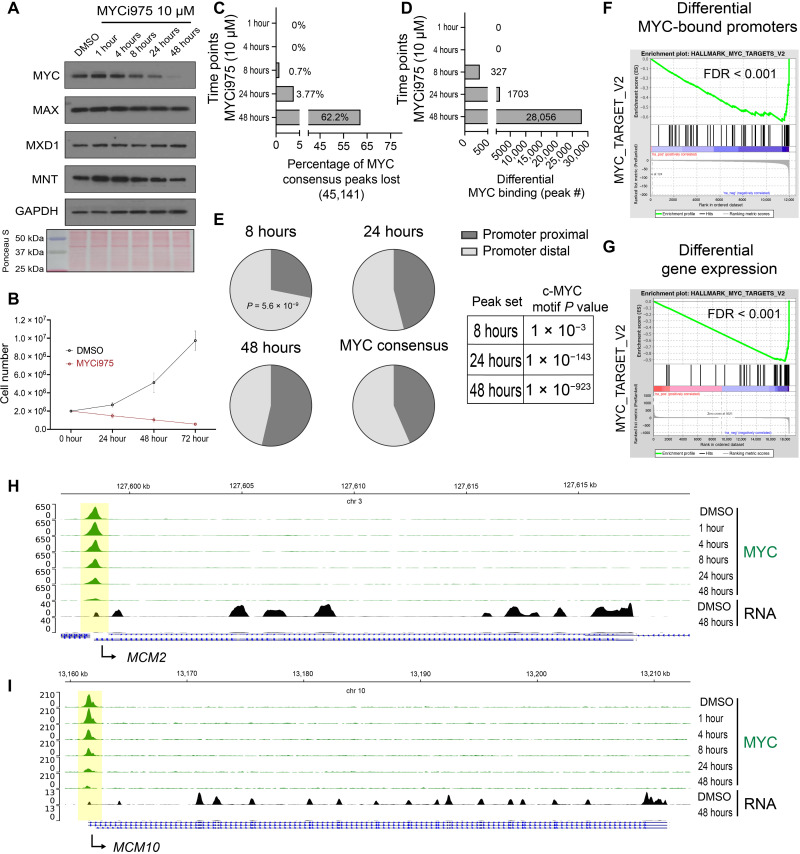
To profile changes in MYC occupancy and target gene expression in response to MYCi975 treatment, we first evaluated the kinetics of MYC protein down-regulation in response to 10 μM MYCi975 for 1, 4, 8, 24, and 48 hours in the high–MYC-expressing 22Rv1 prostate cancer cell line. In a time-dependent manner, MYCi975 induced MYC protein down-regulation, achieving approximately >90% decrease in MYC protein levels by 48 hours (Fig. 1A and fig. S1A). Protein levels of MYC heterodimeric binding partner MAX and related protein MNT were also reduced (~20 to 30% at 48 hours), while MXD1 protein levels remained relatively stable (Fig. 1A and fig. S1A). MYCi975 (10 μM) reduced 22Rv1 cell proliferation by half, representing the median inhibitory concentration (IC50) of 22Rv1 cells, suppressing cell growth and colony formation after 4 days, and reducing cell viability (Fig. 1B and fig. S1, B to D). MYCi975 treatment induced poly(ADP-ribose) polymerase (PARP) cleavage, a hallmark of apoptosis, in 22Rv1 cells (fig. S1E), suggesting apoptosis as a possible mechanism for MYCi975-induced cell death. On the basis of these results, we chose to perform genome-wide analysis on cells treated with MYCi975 for up to 48 hours.
Fig. 1. MYC cistrome is altered at canonical MYC targets and genome wide in response to MYCi975.
(A) 22Rv1 cells were treated with 10 μM MYCi975 for the indicated times followed by immunoblot analyses for the indicated proteins. (B) Total cell number of 22Rv1 cells as determined by trypan blue exclusion assay for both vehicle control and 10 μM MYCi975-treated 22Rv1 cells (n = 4, errors bars represent means ± SEM). (C) Percentage of differential MYC binding peaks lost in response to MYCi975 compared to MYC consensus peaks (n = 4). (D) Total number of differentially bound MYC peaks following differential binding analysis (n = 4) for each time point. (E) Pie chart distribution of 8-, 24-, and 48-hour differential MYC-bound peaks significantly altered (FDR < 0.01) by MYCi975 and MYC consensus peaks from control conditions. The cumulative distribution function of the hypergeometric distribution was used to test for significant differences in differential peak set distribution [promoter proximal (±2 kb from TSS) and promoter distal (>±2 kb from TSS)] using each time point versus MYC consensus peaks. Each peak set was used as input for motif enrichment and the statistical P value reported (right). (F) All MYC-bound annotated promoters were ranked on the basis of differential binding analysis in DMSO- versus 48-hour MYCi975-treated cells (−log10 FDR × log2 fold change) and used as input for GSEA. (G) Differential gene expression analysis was preranked (same as for ChIP-seq data) and used as input for GSEA. Results demonstrated significant enrichment for canonical MYC target genes. (H and I) Gene loci analysis of MCM2 and MCM10 reveals MYC chromatin occupancy loss (green) with associated RNA level loss (black) in response to MYCi975 in 22Rv1 cells.
Next, we performed MYC chromatin immunoprecipitation sequencing (ChIP-seq) in 22Rv1 cells treated with MYCi975 for 1, 4, 8, 24, and 48 hours. Principal components analysis (PCA) demonstrated clustering of the early time points (1-, 4-, and 8-hour groups), whereas continued treatment led to a maximum separation of the 24- and 48-hour groups (fig. S1F). This suggested that the effects of MYCi975 on MYC chromatin occupancy are coincident with MYC protein degradation. Consistent with this possibility, we found that after 1 hour of MYCi975 treatment, both the total peak number and ChIP efficiency remained relatively stable; at all other time points, these values were significantly reduced (fig. S1, G and H). Differential binding analysis comparing each MYCi975 time point to the dimethyl sulfoxide (DMSO)–treated control group revealed time-dependent effects of MYCi975 treatment on MYC chromatin occupancy. Significant changes in MYC ChIP-seq signal were evident as early as 8 hours after MYCi975 treatment [327 differentially bound peaks, false discovery rate (FDR) < 0.01]. Maximal differential MYC binding was found in the 48-hour MYCi975-treated cells (28,056 peaks, FDR < 0.01) where more than 62% of the total MYC cistrome was altered (Fig. 1, C and D). We defined the peaks with statistically significant changes (FDR < 0.01) in MYC occupancy as “MYCi975-sensitive” sites. To investigate both MYCi975-sensitve sites and those with FDR > 0.01 (“MYCi975 insensitive”), we analyzed both MYC and the well-characterized MYC cofactor WDR5 (23). We plotted the normalized tag density of both MYC and WDR5 chromatin binding at MYCi975-sensitive and MYCi975-insensitive peaks and found that both MYC and WDR5 tag density are significantly lower at MYCi975-insensitive sites (fig. S1I). Genome-wide distribution analysis of both peak sets revealed that MYC-bound MYCi975-insensitive sites were predominantly promoter distal (fig. S1J). Unexpectedly, both peak sets contained significant motif enrichment for the canonical c-Myc motif (fig. S1K). MYCi975-insensitive peaks annotated to promoters were used as input for gene ontology analysis demonstrating significant enrichment for nervous system development and cell differentiation, suggesting that downstream MYC-regulated gene pathways may remain unaltered (fig. S1L). Further analysis is focused on the statistically significant MYCi975-sensitive sites. Genome distribution analysis showed that early MYCi975-sensitive sites are significantly enriched for promoter-distal [>2 kb from the nearest transcription start site (TSS)] bound MYC and have low c-Myc motif significance compared to later time points (Fig. 1E). To analyze the global changes in the MYC-bound promoter network in response to MYCi975, we integrated promoter annotation and gene set enrichment analysis (GSEA) (24). MYC consensus peaks were annotated in reference to the nearest TSS to define promoter-proximal (±2 kb from the TSS) and promoter-distal (more than ±2 kb from the TSS) MYC binding sites. MYC-bound promoter-proximal sites were ranked on the basis of fold change and FDR from differential peak analysis. Differential MYC-bound promoters revealed significant loss of canonical MYC target gene programs (“HALLMARK_MYC_TARGETS_V2”; Fig. 1F).
Our previous study of transcriptomic profiles after exposure to MYCi975 demonstrated disruption of canonical MYC gene programs (21). To determine the time-dependent effects of MYCi975 on gene expression, we treated 22Rv1 cells with MYCi975 for 24 and 48 hours to induce extensive MYC chromatin occupancy loss and then performed RNA sequencing (RNA-seq). To control for RNA abundance levels, we included a spike-in control and isolated the same number of cells for each MYCi975 treatment time point. We filtered gene expression based on the lower limit of detection determined by the spike-in control. We found no significant difference in total RNA yield in 22Rv1 cells treated with MYCi975 for 24 or 48 hours compared to controls (fig. S2, A and B). As a quality control, we used a Euclidean sample distance matrix to assess sample similarity [log2-normalized counts, DESeq2 (25)], which confirmed that the MYCi975-treated samples were similar to each other and distinct from controls (fig. S2C).
We next performed differential gene expression analysis comparing DMSO control versus 24-hour MYCi975 or DMSO control versus 48-hour MYCi975 treatment. The 24- and 48-hour MYCi975-treated cells had 6973 and 9080 differentially expressed (DE) genes, respectively (FDR < 0.01) (table S1). Differential gene expression data were preranked (−log10 FDR × log2 fold change) and used as input for GSEA (24, 26). The top differential gene program HALLMARK_MYC_TARGETS_V2 was suppressed in both 24- and 48-hour MYCi975-treated cells; we found that the same gene set contained promoter-proximal MYCi975-sensitive sites (Fig. 1G and table S1). Together, these findings suggest that MYCi975 treatment results in the loss of MYC chromatin occupancy and associated gene expression at hallmark MYC target genes, including the key cell cycle–associated genes MCM2 (Fig. 1H) and MCM10 (Fig. 1I). From analysis of the 327 MYC lost peaks at 8 hours, 93 are promoter proximal. Gene ontology on the 93 genes reveals the following gene sets that are weakly significant (FDR < 0.01): DNA metabolic process (GO:0006259), heterocycle metabolic process (GO:0046483), and DNA replication (GO:0006260), and MCM10 is identified as one of these early MYCi975 response genes.
To further analyze the MYCi975-regulated transcriptome and identify common dysregulated genes in multiple cancer cell models, we integrated our RNA-seq results comparing DMSO versus 10 μM MYCi975 for 24 hours in 22Rv1 cells with previously reported RNA-seq datasets (PC3: 8 μM for 24 hours; P493-6: 6 μM for 24 hours) (21)). Overlapping DE genes (FDR < 0.01) for each dataset were visualized using a Venn diagram (fig. S2D, top). A total of 1183 genes were dysregulated by MYCi975 in all three datasets. Gene ontology analysis of this common gene list revealed significant enrichment for regulation of nucleic acid metabolism, DNA replication, and cell cycle pathways (fig. S2D, bottom). These data suggest that MYCi975 has a significant effect on a core set of DNA synthesis and cell cycle progression genes.
MYCi975 selectively affects MYC binding to promoters and target gene expression
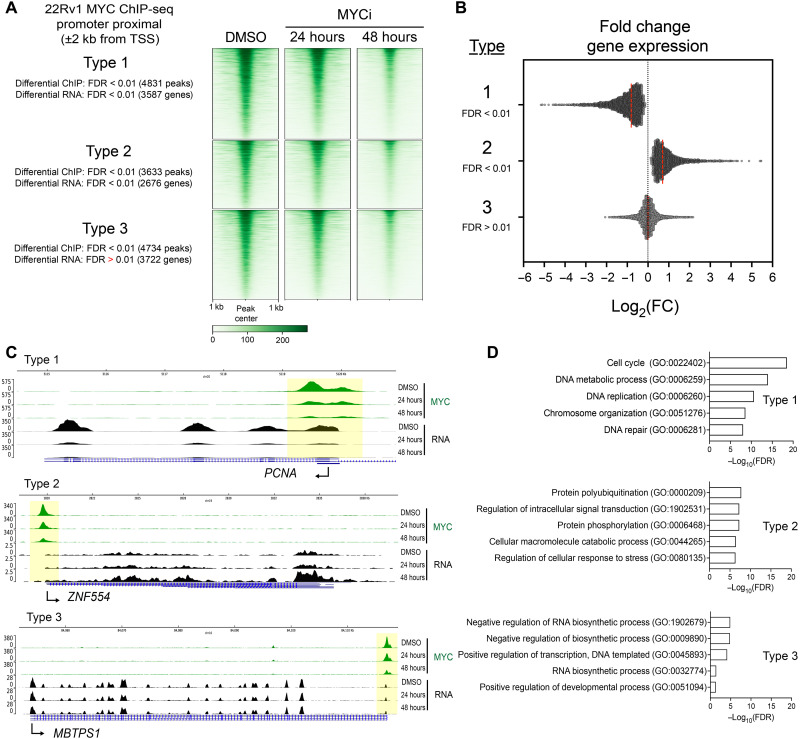
To analyze the global binding patterns of MYC and the downstream transcriptional impact in response to MYCi975, we integrated differential binding analysis at MYC-bound promoters with differential gene expression analysis. We categorized MYC-bound sites into “types” based on whether MYCi975 led to loss of MYC binding and changes/no changes in the RNA levels of target genes (Fig. 2A and table S2). Among the MYC-bound promoters within MYCi975-sensitive sites, we found that associated target gene expression was either decreased (type 1), increased (type 2), or unchanged (type 3) (Fig. 2, A and B). In addition, we investigated the MYCi975-insenstive promoter proximal peaks and integrated differential expression levels. There was a final type (type 4, consisting of MYCi975-insensitive peaks) of target genes that had far fewer tag density when compared to the other three types and for which MYC binding and target gene expression remained unchanged after MYCi975 treatment (fig. S2, E to G). MYC binding and gene expression changes in type 4 were not statistically significant (both differential binding and RNA levels with FDR > 0.01) when compared to the other three types and therefore were not further investigated. Figure 2C shows representative examples of MYCi975 target gene types 1 to 3. Using gene ontology analysis, we found that type 1 genes were significantly enriched for cell cycle and DNA replication gene programs generally important for conferring proliferative advantages to cancer cells (Fig. 2D, top, and table S2). Type 2 genes were strongly enriched for kinase and signal transduction pathways (Fig. 2D). Notably, type 2 genes among others included RORa, a transcription factor known to play a tumor-suppressive role in breast cancer by stabilizing p53 and activating p53 gene transcription (27, 28). Type 3 genes, which demonstrated changes in MYC promoter occupancy with no change in RNA levels, were significantly enriched for basic RNA metabolic processes and core transcriptional pathways important for normal cell function (Fig. 2D, bottom). Overall, these data indicate that the effects of MYCi975 on MYC binding to target gene promoters and subsequent changes in RNA levels are not uniform, with MYC target genes representing cell cycle and DNA replication genes more sensitive to the effects of MYCi975 than MYC target genes involved in basic RNA metabolic and gene transcription processes.
Fig. 2. MYCi975 selectively affects MYC binding to promoters and target gene expression.
(A) Heatmap representation of ChIP-seq signal at annotated promoters of MYCi975-sensitive sites (types 1, 2, and 3). (B) Log2(fold change) of genes for each type as calculated from differential gene expression analysis of DMSO- versus 48-hour MYCi975-treated 22Rv1 cells. These genes have an occurrence of MYC bound at or near the promoter. The red dotted line indicates the mean log2(fold change) for each type. (C) Genome browser tracks of representative genes for each MYCi975 gene type, demonstrating outcomes of MYC ChIP-seq data (green) and RNA-seq data (black). (D) Type 1 to 3 genes were used as input for gene ontology enrichment (http://geneontology.org). The top five results for each gene type are displayed.
The MYCi975-sensitive MYC cistrome differs at promoter-proximal and promoter-distal regions
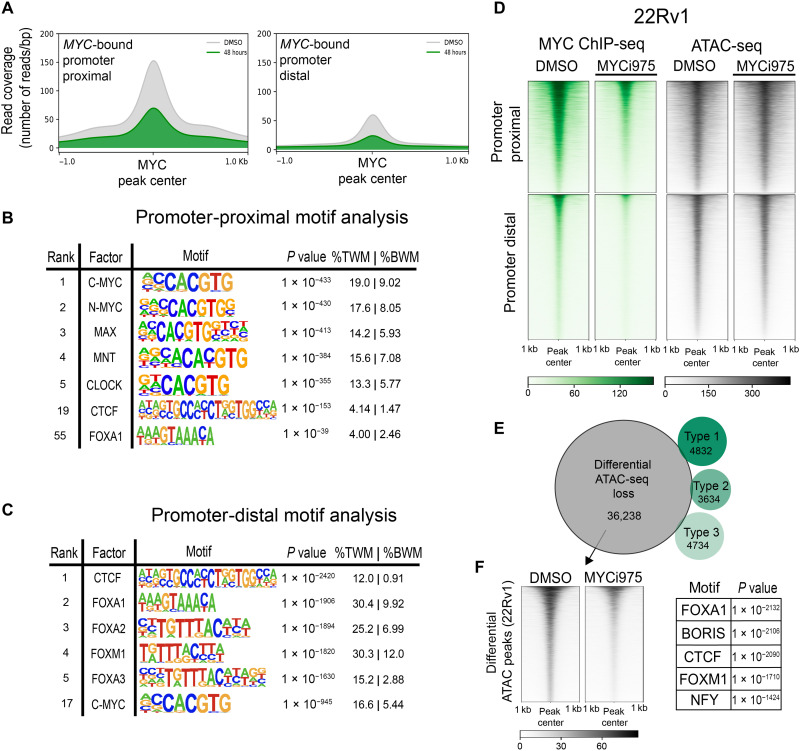
To further analyze MYC chromatin occupancy at genomic regions beyond promoters, we split MYC-bound sites into promoter-proximal and promoter-distal peaks (29). MYC-bound sites in 22Rv1 cells consisted of 20,915 promoter-proximal and 27,108 promoter-distal peaks. To determine the extent to which MYCi975 sensitivity encompasses both genomic regions, we plotted read coverage in both vehicle control and MYCi975-treated cells at MYC-bound promoter-proximal and promoter-distal sites (Fig. 3A). MYC occupancy was reduced at both regions following MYCi975 treatment. Motif analysis of MYC promoter-proximal peaks demonstrated significant enrichment for canonical E-box sequences (Fig. 3B), whereas MYC promoter-distal peaks demonstrated significant enrichment for CTCF and FOX motifs, along with canonical MYC motif enrichment (Fig. 3C), suggesting a possible transcription factor cooperativity at promoter-distal sites in regulating target genes.
Fig. 3. MYC-bound promoter-distal and MYCi975-sensitive chromatin accessibility sites exhibit significant CTCF and FOX factor motif enrichment.
(A) MYC consensus peaks were separated into promoter proximal (±2 kb from TSS) or promoter distal (>±2 kb from TSS). The mean MYC ChIP-seq read coverage is plotted at both promoter-proximal and promoter-distal MYC-bound sites in vehicle control and 48-hour MYCi975-treated 22Rv1 cells. (B) Motif enrichment analysis of MYC-bound promoter-proximal peaks, listing the top five enriched motifs, in addition to CTCF (#19) and FOXA1 (#55) for contrast (%TWM, percentage of targets with motif; %BWM, percentage of background with motif). (C) Motif enrichment analysis of MYC-bound promoter-distal peaks, listing the top five and including c-MYC (#17). (D) Heatmap representation of all MYC-bound sites separated into promoter-proximal and promoter-distal sites, showing MYC ChIP-seq coverage and 22Rv1 ATAC-seq coverage in DMSO- versus 48-hour MYCi975-treated 22Rv1 cells. (E) Venn diagram demonstrating overlap of the differential MYCI975 types 1 to 3 with differential ATAC-seq peaks in DMSO- versus 48-hour MYCi975-treated 22Rv1 cells. (F) Motif enrichment analysis of the differentially lost ATAC-seq peaks (left) in MYCi975-treated 22Rv1 cells. Motif enrichment analysis was run on differential ATAC-seq peaks outside of the MYC cistrome, showing enrichment for both CTCF and FOX factors (right).
To validate these findings and enhance rigor, we used an alternative MYC antibody (clone 9E11; see Materials and Methods). More than 90% of 9E11 MYC-bound peaks overlapped with those identified with the clone Y69 antibody used above (19,110 peaks recovered with 9E11 and 45,141 with Y69; fig. S3A). Although the total peak number for the clone 9E11 antibody was lower, motif analysis of both promoter-proximal and promoter-distal sites recapitulated the CTCF and FOX factor enrichment at promoter-distal regions demonstrated with Y69 (fig. S3, B and C). To directly test the extent to which MYC-bound promoter-distal sites are enriched for CTCF, we performed CTCF ChIP-seq following 48-hour MYCi975 treatment, given that MYC occupancy is significantly reduced at tens of thousands of sites at the 48-hour MYCi975 treatment time point. In total, CTCF chromatin occupancy was altered at only 4719 peaks, comprising 6.91% of the CTCF cistrome (4719 of 68,274). To determine whether MYC binding affects CTCF occupancy, we plotted both MYC and CTCF signals centered at MYC-bound MYCi975-sensitive sites (fig. S3, D and E). Differential binding analysis demonstrated that CTCF occupancy at MYC-bound MYCi975-sensitive sites remains relatively stable and unaltered after 48 hours of MYCi975 treatment, suggesting that CTCF binding is not dependent on co-occupancy of MYC (fig. S3E). Of the differential lost CTCF sites, only 85 (0.3%) overlapped with MYCi975-sensitive sites (fig. S3F). To further examine the general occurrence of motif enrichment at promoter-distal MYC peaks in independent datasets, we analyzed motif enrichment for MYC, FOXA1, FOXM1, and CTCF in publicly available MYC ChIP-seq datasets from multiple cancer cell lines representing diverse tissues of origin. Of the 10 cell lines analyzed, 22Rv1 cells from our study and 5 cell lines (MCF7, breast; MCF10A, breast; NB4, leukemia; P493-6, lymphoma; and HeLa, cervical) (30–32) displayed significant motif enrichment for CTCF at MYC-bound promoter-distal peaks, whereas 22Rv1, MCF7, MCF10A, and MycCap cancer cell lines also showed motif enrichment of FOX factors (fig. S3G). Together, these results suggest that binding of MYC, CTCF, and FOX family transcription factors to promoter-distal sites may be necessary for MYC target gene expression in nuclear hormone receptor–positive prostate and breast cancer cells. Supporting our observations, FOX factors have been shown to function as pioneering factors for chromatin accessibility by nuclear receptors such as the androgen receptor (AR) and estrogen receptor (ER) in prostate and breast cancer cells (33, 34). CRISPR-based deletion and HiC-based chromatin looping assays will be necessary to address the possibility of factor cooperativity in future studies.
Chromatin accessibility is a key event in gene regulation. To independently assess the chromatin accessibility changes induced by MYCi975 and determine whether differential MYC binding alters chromatin accessibility, we performed assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) in 48-hour MYCi975-treated 22Rv1 cells. At both promoter-proximal and promoter-distal MYC-bound sites, the majority of ATAC-seq signal remained relatively unchanged, with <9% of the MYCi975-sensitive sites (2381 of 28,056) and <0.3% of MYCi975 type 1 to 3 target genes (37 of 13,200) overlapping with differential ATAC peaks (36,238 differentially lost; Fig. 3, D and E). Given that a large portion of differential ATAC-seq peaks did not overlap with differential MYC binding, we used motif enrichment analysis to investigate the differential ATAC-seq peaks. We found both Forkhead/FOX factor and CTCF motif enrichment (Fig. 3F). To determine whether a similar feature is observed in ER-positive MCF7 breast cancer cells, in which Forkhead factors are critical for ER chromatin occupancy and transcriptional response, we performed ATAC-seq in MCF7 cells and determined whether large-scale chromatin accessibility also changed in response to MYCi975. We found that both CTCF and FOX factors were among the most enriched motifs at differential ATAC sites in MCF7 cells (22,038 differentially lost peaks; fig. S3H). These results suggest that MYCi975 has minimal effects on chromatin accessibility at MYC-only binding sites but alters chromatin accessibility at sites enriched with CTCF and FOX TF binding sites.
MAX cistrome changes in response to MYCi975
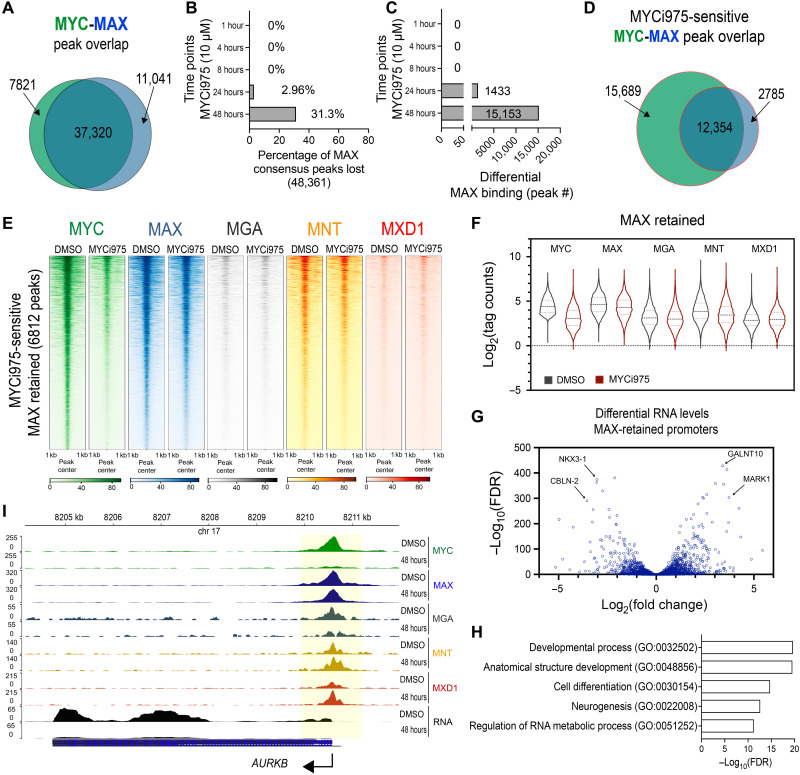
MAX is the key heterodimeric partner of MYC and can also form homodimers and heterodimerize with MNT, MGA, and MXD1. Since ATAC signal did not change significantly at MYC-only binding sites, it was critical to determine the chromatin occupancy of MAX and its heterodimeric partners. In 24- and 48-hour MYCi975-treated cells, we showed that while MYC level decreased significantly, MAX and MNT protein levels decreased slightly, and MXD1 remained relatively stable at varying levels when compared to DMSO-treated control cells (Fig. 1A and fig. S1A). To determine MAX chromatin occupancy in response to MYCi975, we performed MAX ChIP-seq, using the same time points as in the MYC ChIP-seq studies. The total peak number and ChIP efficiency significantly decreased only after 48 hours of MYCi975 treatment (fig. S4, A and B). PCA of differential MAX binding analysis revealed clustering at 24 and 48 hours after MYCi975 treatment (fig. S4C). Considering that MYC heterodimerizes with MAX, we examined whether MYC loss on chromatin is associated with MAX loss. We also determined the extent to which MAX heterodimeric binding partners MNT, MGA, and MXD1 are altered by ChIP-seq (see below). MAX occupancy overlapped with greater than 80% of the MYC consensus cistrome (Fig. 4A), suggesting that MYC-MAX complex is a predominant component of the MYC cistrome. The MAX cistrome was disrupted after 24 and 48 hours of MYCi975 treatment versus DMSO, with 31.3% (15,153 peaks) of the total MAX cistrome altered after 48 hours of MYCi975 treatment (Fig. 4, B and C). These results also indicate that ~70% of the MAX cistrome remained unaltered upon MYCi975 treatment. We next investigated the overlap of MYCi975-sensitive MYC and differential MAX peaks. In MYCi975-treated cells, the total number of differentially bound MYC peaks (28,056) was greater than the number of differentially bound MAX (15,153) peaks, suggesting stable MAX chromatin binding irregardless of significant MYC loss. Peak overlap analysis using a Venn diagram demonstrated that differential MAX occupancy is largely predicted by differential MYC occupancy (Fig. 4D). As expected, at a subset of MYCi975-sensitive MYC sites, MAX occupancy was lost along with MYC occupancy.
Fig. 4. Differential MAX, MGA, and MNT chromatin binding alterations in MYCi975-treated cells.
(A) Peak overlap analysis of MYC and MAX consensus peaks (n = 4). (B) Percentage of differential MAX binding peaks lost in response to MYCi975 compared to MAX consensus peaks (n = 4). (C) Total number of differentially bound MAX peaks following differential binding analysis (n = 4). (D) Peak overlap analysis of differential MYC- and MAX-bound sites (MYCi975 sensitive) in MYCi975-treated 22Rv1 cells. (E) Heatmap representation of MYC, MAX, MGA, MNT, and MXD1 at MYCi975-sensitive sites demonstrating no significant change in MAX occupancy (6812 peaks in total). (F) Log2(normalized tag counts) for MYC, MAX, MGA, MNT, and MXD1 at MAX-retained peaks in 22Rv1 cells. The middle dashed line represents the sample median, and the upper and lower dotted lines represent the upper and lower quartiles, respectively. (G) Differential gene expression analysis of MAX-retained peaks annotated to promoters (±2 kb from the TSS). (H) Gene ontology analysis of promoter-bound MAX-retained peaks. (I) Gene browser tracks of AURKB showing loss of MYC (green), retention of MAX (blue), an increase in MXD1 (red), and loss of AURKB mRNA (black) in 48-hour MYCi975-treated cells. MGA (gray) and MNT (yellow) are also displayed.
To analyze MAX sites co-occupied by MYC and its binding sensitivity to MYCi975, we used the promoter-proximal and promoter-distal MYC peak datasets. MAX occupancy was reduced at both MYC-bound promoter-proximal and promoter-distal peaks (fig. S4D). MYC-independent MAX lost sites (6812 sites) upon MYCi975 treatment may represent MAX homodimer or MAX-MGA/MNT/MXD1 heterodimer binding due to the partial degradation of these proteins in MYCi975-treated cells (Fig. 1A). Given that chromatin accessibility at MYC binding sites remained largely unaffected, it is possible that DNA remains accessible to binding of MAX and its heterodimeric binding partners (as determined by ATAC) upon MYC loss induced by MYCi975 treatment. We identified 6812 MYC/MAX binding sites by overlapping MYCi975-sensitve MYC peaks and unaltered (FDR > 0.01) MAX binding sites (DMSO versus 48-hour MYCi975). Figure 4E shows MAX-retained sites where MAX occupancy (FDR > 0.01) was relatively insensitive to MYCi975 treatment within the MYCi975-sensitive MYC cistrome. At these sites, it is possible that MAX occupancy is stabilized by MAX/MAX homodimers or bound by MAX/MAX family member heterodimers. We plotted MGA, MNT, and MXD1 signal at MAX-retained sites (Fig. 4E). To analyze the occupancy of MYC, MAX, MGA, MNT, and MXD1 at the 6812 MAX-retained peaks, we plotted the log2(normalized tag counts) in DMSO- and 48-hour MYCi975-treated 22Rv1 cells (Fig. 4F). By selecting for MAX-retained sites (differential binding FDR > 0.01), occupancy analysis of both MGA and MNT decreased, whereas that of MXD1 increased (Fig. 4F). These data suggest that MAX retention despite MYC loss is coupled to alterations in canonical MAX heterodimeric partners. In particular, MXD1 increase in tag density suggests a change in the MAX heterodimeric complex formation at MAX-retained sites. To further determine binding overlap between all five transcription factors (MYC, MAX, MNT, MGA, and MXD1), we performed ChIP-seq and used the datasets in an upset plot to analyze replicate peak calls from each transcription factor (35). As expected, the top overlapping result was MYC/MAX; MAX-only sites were extensive, followed by the MYC and MAX heterodimeric binding partners (fig. S4E). We also analyzed globally the occupancy of these factors upon MYCi975 treatment at sites where MYC binding was lost (Fig. 4E). We find that while MAX and MGA remained relatively stable overall, MNT occupancy decreased, whereas MXD1 occupancy increased. Of the MAX-retained binding sites, RNA expression data from promoter-proximal peaks were plotted and demonstrated both down- and up-regulated RNA levels (Fig. 4G). Gene Ontology analysis of promoter-proximal MAX-retained peaks demonstrated enrichment for developmental, cell differentiation, and neurogenesis pathways (Fig. 4H). These data suggest that the loss of MYC accompanied by MAX retention is not uniformly associated with either RNA up- or down-regulation. This is not unexpected, since at these MAX-retained sites, the MYC/MAX complex is no longer predominant, and potentially MAX/MAX homodimers and MAX heterodimers will act antagonistically. In a gene-specific analysis of AURKB, which is highly sensitive to MYCi975 with regard to MYC promoter-proximal occupancy, MAX remained bound after MYCi975 treatment (Fig. 4I). In particular, at the AURKB promoter-proximal site loci, MGA occupancy is lost, whereas MNT remains bound, and MXD1 increases (Fig. 4I). Together, MAX binding and canonical MAX heterodimeric partner binding at MYC binding sites were significantly affected by MYCi975 treatment, although a subset of sites showed relatively stable MAX binding.
Alterations in FOXA1, AR, and global chromatin H3K27 acetylation in response to MYCi975
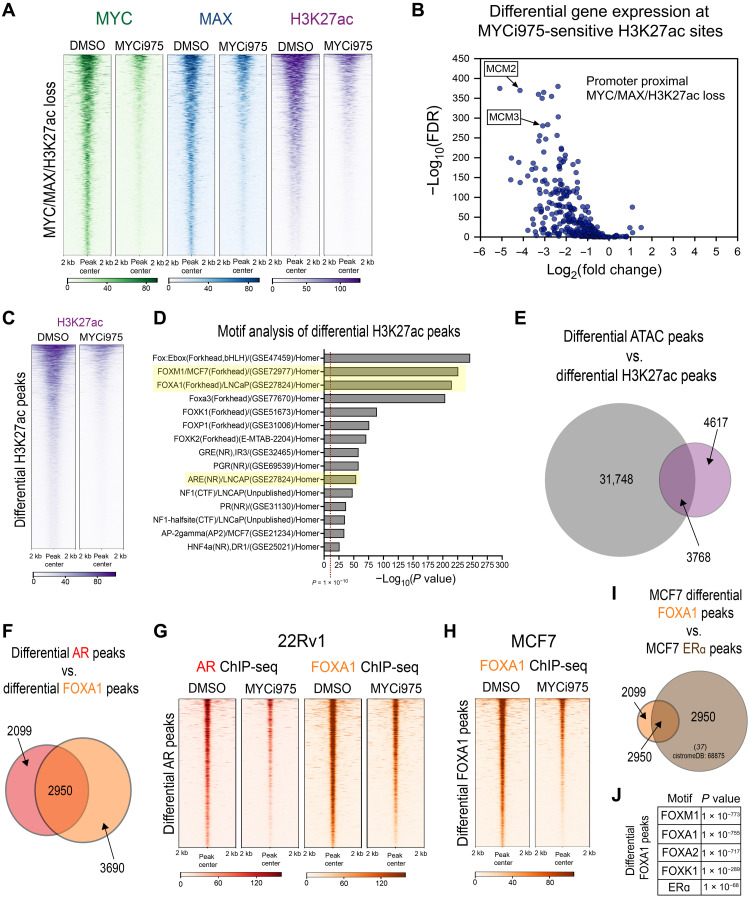
Transcription factors such as MYC regulate gene transcription by recruiting histone acetyltransferases to promote H3K27ac, a mark of active promoters and enhancers (20, 36). To determine the effect of MYCi975 treatment on global H3K27ac, we performed H3K27ac ChIP-seq in MYCi975-treated cells at the same time points used for the MYC and MAX ChIP-seq studies above. PCA analysis demonstrated that maximal H3K27ac signal separation occurred in cells treated with MYCi975 for 24 and 48 hours (fig. S4F). Overall, the total H3K27ac peak number (~37,000 peaks) remained relatively stable; however, IP efficiency significantly increased, suggesting that some H3K27ac peaks increase in occupancy (fig. S4, G and H). We reasoned that type 2 peaks, where RNA levels are up-regulated upon MYCi975 treatment, represent candidate sites for increases in H3K27ac. Heatmap representation of H3K27ac signal at MYCi975 type 2 sites demonstrates an increase in H2K27ac, which is consistent with the IP efficiency increase observed upon MYCi975 treatment (fig. S4I). To integrate changes in MYC/MAX occupancy with changes in H3K27ac, we overlapped MYCi975-sensitive sites in 48-hour–treated cells with the differential H3K27ac signal. At 2218 MYCi975-sensitive sites, we observed significant co-loss of H3K27ac and MYC and MAX occupancy in response to MYCi975 (Fig. 5A). These data suggest that loss of chromatin-bound MYC/MAX induced by MYCi975 leads to specific dysregulation of H3K27ac. Next, we annotated the MYCi975-sensitive differential H3K27ac peaks with MYC/MAX co-loss to promoters, overlapped with DE genes in 48-hour MYCi975-treated cells (304 of 2218). H3K27ac loss at promoters induced by MYCi975 was associated with down-regulation of gene expression, including that of key cell cycle genes (e.g., MCM2 and MCM3; Fig. 5B and see also Fig. 1, H and I).
Fig. 5. MYCi975 alters H3K27ac activity and FOXA1 chromatin occupancy in cancer cells.
(A) Heatmap representation of MYC (green), MAX (blue), and H3K27ac (purple) ChIP-seq signals at overlapping sites, with loss of signal in MYCi975-treated cells (2218 peaks). (B) Differential binding analysis of H3K27ac ChIP-seq was performed and overlapped with MYCi975-sensitive sites. Sites annotated to promoters were cross-referenced with differential gene expression data consisting of 304 genes. The plot demonstrates that loss of MYC/MAX occupancy and H3K27ac signal results in down-regulated gene expression. (C) Heatmap representation of H3K27ac loss of signal in MYCi975-treated cells (8385 peaks). ChIP-seq signal in the MYCi975 column represents 48-hour MYCi975 treatment. (D) Motif enrichment analysis was performed to assess the differential H3K27ac signal, showing enrichment for FOX factors and nuclear receptors [AR, GR, and progesterone receptor (PGR)]. Each P value reported was converted to −log10(P value). (E) Venn diagram representing differential H3K27ac and differential ATAC-seq peak overlap in MYCi975-treated 22Rv1 cells (H2K27ac, n = 2; ATAC-seq, n = 3). (F) Venn diagram representing differential AR and FOXA1 peak overlap in MYCi975-treated 22Rv1 cells. (G) Heatmap representation of AR and FOXA1 ChIP-seq signal at differential AR peaks in MYCi975-treated 22Rv1 cells (n = 3). (H) FOXA1 ChIP-seq signal in MCF7 cells at differential FOXA1 peaks found in MYCi975-treated MCF7 cells (n = 2). (I) MCF7 differential FOXA1 peak overlap with publicly available MCF7 ERa ChIP-seq peaks (http://cistrome.org/db/#/, PMID: 27062924). (J) Motif enrichment analysis results of the differential FOXA1 peaks.
We next investigated motif enrichment of the differential H3K27ac peaks to identify MYC targets modulated by MYCi975 treatment. We analyzed the 8386 promoter-proximal and promoter-distal sites where H3K27ac was lost (Fig. 5C). We reasoned that these sites might represent binding sites of MYC and MYC-regulated transcription factor(s) whose down-regulation after MYCi975 treatment leads to a loss of the H3K27ac active chromatin mark. We performed motif enrichment analysis and found that the most significantly enriched motifs were the FOX family of proteins and nuclear receptors such as AR and glucocorticoid receptor (GR), among others (Fig. 5D). Given that the differential ATAC signal in MYCi975-treated cells also revealed CTCF and FOX factor enrichment, we overlapped both the differential H3K27ac and differential ATAC-seq peaks in MYCi975-treated 22Rv1 cells (Fig. 5E) and found that almost half of differential H3K27ac peaks overlapped with differential ATAC peaks. Notably, CTCF motif enrichment was only within differential ATAC peaks and not within differential H3K27ac peaks (Fig. 5D), whereas FOX factor enrichment was found in both differential ATAC and H3K27ac peaks (Figs. 3F and 5D), suggesting multiple genome-wide chromatin alterations at FOX factor motifs. On the basis of the extensive loss of FOX factor and AR motifs in differential H3K27ac peaks, we performed AR and FOXA1 ChIP-seq in control and 48-hour MYCi975-treated 22Rv1 cells. Peak overlap analysis demonstrated that more than half of the differential AR peaks overlap with differential FOXA1 peaks (Fig. 5F). In total, 5049 AR peaks were lost (~25% of the AR cistrome) while 6640 FOXA1 peaks were lost (~10% of the FOXA1 cistrome) in response to MYCi975 treatment. Plotting the read coverage in heatmap representation of differential AR peaks for both AR and FOXA1 signal demonstrated co-loss of both FOXA1 and AR (Fig. 5G). Both the differential AR and FOXA1 peaks were annotated, and for both factors, peaks were predominantly promoter distal with less than 5% of peaks annotated to promoter-proximal regions. Given that both 22Rv1 and MCF7 cells demonstrated differential chromatin accessibility enriched for FOX factor motifs, we also performed FOXA1 ChIP-seq in 48-hour MYCi975-treated MCF7 cells. Differential binding analysis revealed 2557 differential FOXA1 peaks (~4% of the FOXA1 cistrome; Fig. 5H). To determine whether the differential FOXA1 peaks overlapped with co-occurring ERα peaks, publicly available ERα ChIP-seq data were obtained from cistromeDB [http://cistrome.org/db/#/, cistromeDB: 68875 (37)]. Nearly 60% of the differential FOXA1 peaks overlapped with ERα peaks, and the differential FOXA1 peaks were enriched for both FOXA1 and ERα motifs (Fig. 5, I and J). Together, in both prostate and breast cancer models, we found that FOXA1 binding is altered at hormone receptor co-occupied sites in response to MYCi975 treatment.
MYCi975 enhances the efficacy of antihormone therapy
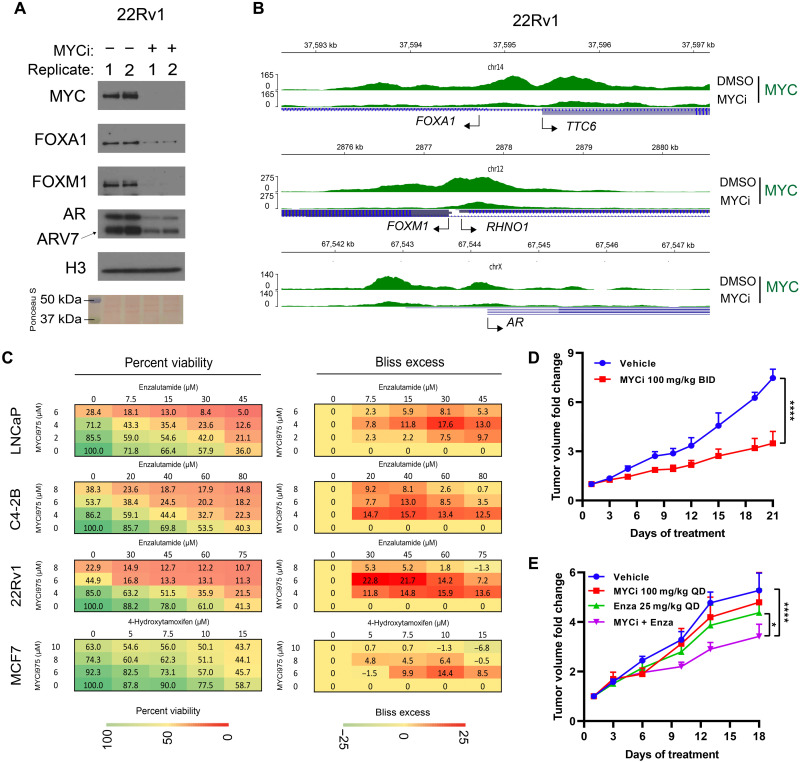
Our RNA-seq results indicated down-regulation of key prostate AR-related lineage transcription factors FOXA1, FOXM1, and AR (table S1), which may contribute to MYCi975 efficacy in the prostate cancer cell model used here. We verified AR, FOXM1, and FOXA1 protein loss in the nuclear fraction of 22Rv1 cells (Fig. 6A). We also found that FOXM1, FOXA1, AR, and the AR splice variant AR variant 7 (ARv7) were all significantly reduced after MYCi975 treatment (Fig. 6A and fig. S4J). Next, we assessed the protein levels of FOXM1, FOXA1, and ERα in MCF7 cells (fig. S4, K and L) and found that FOXM1 was down-regulated, whereas FOXA1 and ERα remained relatively stable. To determine whether the relative loss of these key transcription factors is due to transcriptional regulation by MYC binding, we analyzed MYC occupancies at the promoters of FOXA1, FOXM1, and AR genes in both DMSO- and 48-hour MYCi975-treated 22Rv1 cells. For each target gene, MYC occupied both the proximal and distal sites and was significantly lost in MYCi975-treated cells (Fig. 6B). These results show that MYC occupies regulatory regions of FOXA1, FOXM1, and AR genes, and MYCi975 leads to a decrease in MYC occupancy and a subsequent decrease in their respective mRNA and protein levels.
Fig. 6. MYCi975 enhances the efficacy of antihormone therapy.
(A) Immunoblot showing loss of FOXM1, FOXA1, AR, and AR variant protein levels in sonicated nuclear fractions of MYCi975-treated 22Rv1 cells. Replicates (1 and 2) represent biological replicates of cells treated with 10 μM MYCi975 for 48 hours. (B) Genome browser tracks of MYC (green) ChIP-seq data at the FOXA1, FOXM1, and AR gene loci in MYCi975-treated 22Rv1 cells. TSS of each gene is indicated by an arrow. (C) Representative 4 × 5 dose-response matrices showing percentage viability (left) and Bliss index (right) analysis of predicted versus observed cell viability of prostate cancer cells (LNCaP, C4-2B, and 22RV1) treated with MYCi975 and ENZ or the breast cancer line MCF-7 treated with MYCi975 and 4-OHT (n = 3). Bliss scores > 0 indicate synergy, scores close to zero indicate additivity, and scores < 0 denote antagonism. (D) Fold change tumor volumes of 22Rv1 xenografts in nude mice treated with MYCi975 (100 mg/kg BID p.o.) or vehicle, 5 days a week for 3 weeks. n = 4 to 7 grafts per group (from three to four mice). (E) Fold change tumor volumes of 22Rv1 xenografts in nude mice treated with MYCi975 (100 mg/kg QD p.o.), ENZ (25 mg/kg QD i.p.), combination of MYCi975/ENZ, or vehicle for 18 days. n = 7 to 8 grafts per group (from four to five mice). Error bars represent means ± SEM and analyzed by two-way analysis of variance (ANOVA) in GraphPad Prism for (D) and (E) (*P < 0.05; ****P < 0.0001).
On the basis of the above results, we reasoned that AR occupancy and gene expression changes induced by MYCi975 may enhance the efficacy of the clinically used AR-targeting compound enzalutamide (ENZ) to suppress cell viability. Using adenosine triphosphate (ATP) luminescence as a surrogate for cell viability (CellTiter-Glo 2.0, Promega, catalog no. G9242), we determined the Bliss index score in 22Rv1, LNCaP, and C4-2B prostate cancer cell lines after treatment with ENZ and MYCi975, alone or in combination (Fig. 6C) (38). These results show that MYCi975 acted synergistically with ENZ to decrease prostate cancer cell viability in all three cell lines. Given that MCF7 cells also exhibit sensitivity to MYCi975 and demonstrate differential FOXA1 occupancy at ER-bound sites (thereby altering ER activity), we also tested for synergy of MYCi975 with clinically used anti-estrogen 4-hydroxytamoxifen (4-OHT). As with the prostate cancer cell lines, MYCi975 and 4-OHT synergized to decrease MCF7 breast cancer cell viability as determined by the Bliss index (Fig. 6C, bottom). Together, both anti-androgen and anti-estrogen treatment enhanced MYCi975 efficacy in prostate and breast cancer cellular models, respectively. These results also suggest that dysregulation of MYC, AR, and FOX gene expression by MYCi975 contributes to decreased cell proliferation and viability and increased cell death (see Fig. 1 and fig. S1).
To extend these observations in vivo, we first assessed MYCi975 efficacy in 22RV1 xenograft–bearing nude mice, treated with MYCi975 (100 mg/kg BID). This treatment led to a significant inhibition of tumor growth without any adverse effect on mouse body weight (Fig. 6D and fig. S4M). We then examined the effect of a lower dose of MYCi975 [100 mg/kg QD (Quaque Die: Once Daily)] alone or in combination with ENZ (25 mg/kg QD) in the same 22RV1 xenograft mouse model. MYCi975 enhanced ENZ efficacy in this model without affecting mouse body weight (Fig. 6E and fig. S4N). These results support our in vitro cell-based studies and provide a plausible mechanism for future development of combinatorial therapeutics for cancer.
DISCUSSION
Successful and specific targeting of MYC has the potential to significantly advance cancer therapeutics. In this study, we leveraged a small-molecule MYC inhibitor, MYCi975, to probe the effects of MYC inhibition on chromatin binding of MYC, MAX, heterodimeric MAX binding partners, the chromatin landscape, and associated gene regulation and cell fate in cancer cells. Through comprehensive profiling of altered chromatin occupancy of MYC, MAX, MNT, MGA, MXD1, AR, and FOXA1; epigenomic H3K27ac marks and chromatin accessibility; and transcriptional response, we characterized selective modulation of chromatin and MYC target genes by MYCi975, yielding important insights into its mechanism of action. In particular, our results indicate a differential effect on mRNA levels of MYC target genes despite uniform chromatin occupancy loss. Type 1 gene targets showed loss of MYC binding and decreased gene expression after MYCi975 treatment. Type 2 targets showed loss of MYC binding and increased gene expression. Type 3 genes showed no change in gene expression after MYCi975, despite loss of MYC binding. Differential effects of MYCi975 on some canonical MYC functions and not others support the concept of the “coalition model” (39), which aims to reconciliate the wide variety of MYC interactors (40) and resultant MYC complexes. In this model, MYC protein complexes, while exhibiting independent functions, cooperate to achieve a collective transcriptional output that reflects the overall MYC function in cells. The fact that some genes, particularly those involved in RNA biogenesis in normal cell function and viability, were not significantly affected by MYCi975 in terms of MYC binding and target gene RNA levels provides support for MYCi975 selectivity and tolerability as a potential cancer therapeutic.
It is notable, by contrast, that MYCi975-sensitive type 1 target genes were enriched for pathways that are hijacked by cancer cells such as cell cycle progression, DNA replication, and DNA repair. The significant loss of MYC binding and reduced gene expression after MYCi975 treatment are consistent with previous work identifying these genes as low-affinity MYC targets (17). The MYCi975 mechanism of action in tumor cells is thus likely to be dependent on the suppression of these pathways. Our findings argue for rational approaches to enhancing MYCi975 function, e.g., by combining them with agents whose efficacy is dependent on suppressing the cell cycle or DNA replication. Alternatively, type 2 genes were highly affected by MYC inhibition with regard to loss of MYC binding but were up-regulated. These genes were enriched for signal transduction and protein ubiquitination/phosphorylation pathways. MYCi975–up-regulated genes, such as pyruvate dehydrogenase kinase 4 (PDK4), has been associated with cancer aggressiveness and positive metabolic regulation of glycolysis (41–43). In addition, the MYCi975–up-regulated gene FTH1 has been identified as a tumor suppressor in both breast (44) and ovarian (45) cancer. Type 3 target genes included members of MYC-regulated pathways that are spared by MYCi975, e.g., RNA metabolism, which could be targeted with other agents in future therapeutic or preclinical studies. Outside of active promoters, particularly in high–MYC-expressing cancer cells, MYC binds low-affinity variant E-box enhancers (14). Our results demonstrated alterations in predominantly promoter-distal H3K27ac-marked sites where MYC/MAX heterodimer binding is significantly lost in MYCi975-treated cells, suggesting a role for MYC/MAX heterodimer binding and H3K27ac at putative enhancers. However, gene expression changes that occur as a consequence of MYCi975-induced loss of putative enhancer-bound MYC/MAX heterodimers and H3K27ac marks have yet to be determined. Alterations in long-range contacts between promoter-distal and promoter-proximal MYC binding, driven by MYC/MAX enhancers, may be induced by MYCi975. Further investigation into the differential enhancer-promoter contacts induced by MYCi975 using promoter capture Hi-C (46, 47) may reveal the role of enhancer-bound MYC/MAX heterodimers in MYCi975-sensitive gene regulation.
Another key finding of this study was the discovery of CTCF and FOX transcription factor enrichment at promoter-distal MYCi975-sensitive sites in a cell type–specific manner. These findings suggest that lineage-specific transcription factors cooperate with MYC in regulating gene programs and may be sensitive to MYCi975. As our study primarily focused on prostate cancer cells, we showed that AR and the AR pioneer factor FOXA1 may collaborate with MYC to promote tumorigenesis (48). MYCi975 treatment reduced the expression of AR, AR splice variant A7 [which is important in driving anti-androgen resistance (49)], and FOXA1. Furthermore, MYCi975 predominantly reduced chromatin occupancy of both AR and FOXA1 at putative enhancers. MYCi975 directly influences the expression of these lineage-specific factors, which, in turn, regulates their target genes, thereby exerting a robust cumulative effect on overall gene expression reprogramming leading to cell proliferation defects and cell death. Consistent with that premise, we found that the loss of AR upon MYCi975 treatment synergistically sensitized multiple prostate cancer cell lines, including castration- and treatment-resistant cells, to the second-generation anti-androgen ENZ. In addition, ENZ antitumor efficacy in 22Rv1 xenografts, which are intrinsically resistant to ENZ due to ARv7 expression, was enhanced by MYCi975 combination treatment. In prostate cancer tumors, c-MYC levels positively correlate with AR isoforms including ARv7, and suppression of c-MYC sensitizes ENZ-resistant cells to ENZ (50). Studies have suggested that FOXA1 plays a role in not only prostate cancer but also breast cancer as a mediator of hormone response (51, 52). Although MYCi975 did not reduce FOXA1 or ERα protein levels in breast cancer cells, FOXA1 occupancy was reduced at co-occurring ERα binding sites, and MYCi975 synergistically sensitized ERα-positive MCF7 cells to anti-estrogen 4-OHT. The mechanism underlying this cell-specific effect of MYCi975 is not currently clear.
MYC and MAX are part of an extended network of transcription factors (53). While the obligate MYC heterodimerization partner MAX is highly sensitive to chromatin loss of MYC caused by MYCi975 treatment, we also observed sites where MYC loss was not associated with MAX loss. This was expected since MAX binding to chromatin, unlike MYC, may occur through multiple dimerization partners and homodimerization (54). Our upset plot of the binding profiles of MYC, MAX, and its heterodimeric partners throughout the genome clearly established MAX homodimer binding as a key event along with MYC-MAX binding. In addition, observed peaks of MYC, MAX, and MAX heterodimeric partners in genomic loci by bulk ChIP-seq may represent cellular heterogeneity of MYC factor binding rather than all factors binding to the same locus. Such an observation underscores the limits of bulk ChIP-seq and suggests that single-cell sequencing approaches may better identify differences in transcription factor occupancy in individual cells. Analysis of MAX heterodimers with MNT and MGA suggests occupancy loss at MYCi975 MAX-retained target genes; however, MXD1 shows a slight increase, and retention of MAX at these sites suggests a role for active repression by MAX homo- and heterodimers (53, 55). Notably, MAX binding to DNA independent of MYC has been associated with differentiation and cell arrest gene programs, competition with canonical MYC/MAX E-box motifs, and interaction with different transcriptional co-regulators (54). Our results are consistent with the notion that MYCi975 establishes a new MAX regulatory cistrome and gene program that promotes cell cycle arrest, cell differentiation, and development. In the context of disease, MAX could function as a tumor suppressor independent of MYC, e.g., in small cell lung cancer (56). Stabilization of the MAX homodimer disrupts MYC function (57). Therefore, use of MAX homodimer–stabilizing compounds along with MYCi975 may establish a dysregulated gene expression network that decreases cell proliferation and promotes cell death.
In summary, this work highlights the impact of a small-molecule inhibitor of MYC on the genome and epigenome of cancer cells, leading to differential regulation of gene programs. As MYC regulates a myriad of gene programs in normal and cancer cells in a pleiotropic manner, our findings highlight the importance of carrying out detailed unbiased molecular analyses to obtain a more complete picture of the mechanism of action of MYC-targeting agents as future cancer therapeutics.
MATERIALS AND METHODS
Cell lines
22Rv1 (CRL-2505), DU145 (HTB-81), MCF7 (HTB-22), MycCap (CRL-3255), and LNCaP (CRL-1740) cells were purchased from the American Type Culture Collection. The C4-2B cell line was a gift from H. Chen of the University of California, Davis. All cell lines in this study were maintained in RPMI (Thermo Fisher Scientific, catalog no. 11875093) supplemented with 10% Gibco fetal bovine serum (FBS; REF:26140-079) in a humidified incubator at 37°C with 5% CO2. All cell lines in this study were maintained at maximum until passage 20 and then discarded.
Antibodies and reagents
Antibodies used in this study were CTCF (Active Motif, catalog no. 61932), c-MYC (Abcam, ab56-9E11), c-MYC (Abcam, ab32072-Y69), WDR5 (Proteintech, catalog no. 15544-1-AP), FOXA1 (Bethyl Laboratories, A305-249A), H3K27ac (Active Motif, catalog no. 39685), AR (Abcam, ab108341), MAX (Proteintech, catalog no. 10426-1-AP), full-length PARP (Cell Signaling Technologies, 46D11, catalog no. 9532T), cleaved PARP (Cell Signaling Technologies, D64E10, catalog no. 5625T), MNT (Bethyl Laboratories, A303-627A), MXD1 (Proteintech, catalog no. 17888-1-AP), MGA (Bethyl Laboratories, A302-865A), FOXM1 (Diagenode, catalog no. C15410232), histone H3 (Abcam, ab10799), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Sigma-Aldrich, G9545). MYCi975 was synthesized as described (21).
Whole-cell extraction and immunoblotting
Cells were washed with ice-cold 1× phosphate-buffered saline (PBS) three times and isolated by cell scraping into ice-cold 1× PBS. Cell pellets were frozen in liquid nitrogen briefly, allowed to thaw for 2 min, and then resuspended in five cell pellet volumes of whole-cell extract lysis buffer (LB) [20 mM tris-HCl (pH 7.6), 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% IGEPAL-CA630, 0.5% sodium deoxycholate, and 0.1% SDS]. The cell resuspension was kept on ice for 20 min with brief vortexing every 5 min. Cell debris was cleared by centrifugation at >20,000g for 25 min at 4°C, and samples were assessed for protein concentration with Bicinchoninic acid (BCA) (Thermo Fisher Scientific, catalog no. 23209). For each sample, 20 μg of protein was separated by SDS–polyacrylamide gel electrophoresis, transferred onto a nitrocellulose blotting membrane (Amersham Protran, catalog no.10600001), and probed with antibodies in PBS, 0.1% Tween 20, and 5% nonfat dry milk [dilutions: (Y69) MYC, 1:10,000; MAX, 1:2000; MNT, 1:2000; MXD1, 1:2000; GAPDH, 1:20,000]. For immunoblotting solubilized nuclear fractions, 15 μg of ChIP input was separated and probed with antibodies exactly as described above [dilutions: (Y69) MYC, 1:10,000; FOXM1, 1:500; FOXA1, 1:1000; AR, 1:2000; histone H3, 1:10,000]. To quantify Western blot exposure, densitometric analysis was conducted with the ImageJ (58) gel analyzer tool and quantified using the area under the curve. Gel loading control protein was set to 100% to quantify the percent changes in protein as compared to loading control.
Crystal violet staining
Cells were plated into six-well plates at 25,000 cells per well, and the medium was supplemented in triplicate with either DMSO (0.2%) or 10 μM MYCi975 unless noted otherwise. Cells were treated for 4 days with a medium change 48 hours after plating. The medium was aspirated, and ice-cold PBS was carefully added to the side of the wells. The PBS was aspirated, and cell washing was performed once more. After the last wash, 1 ml of crystal violet solution (0.1% crystal violet in 20% ethanol) was added, and the plate was gently rocked for 5 min. Crystal violet was discarded, wells were washed with deionized water three to five times until clear, and then plates were inverted to dry.
Cell viability and IC50 determination
22Rv1, LNCaP, and C4-2B cells were plated into 96-well plates at 2500 cells per well supplemented in triplicate with either DMSO, MYCi975, ENZ (Selleckchem, catalog no. S1250), or both drug treatments (n = 2). 22Rv1 cells were treated with 25 μM ENZ, while LNCaP and C4-2B cells were treated with 10 μM ENZ. Both the LNCaP and C4-2B cell lines express full-length AR, whereas 22Rv1 cells express both full-length and ligand-independent ARv7, which drives a mechanism of anti-androgen resistance (49). Given these differences, cells were treated with different concentrations of ENZ to reflect sensitivity. The plates were incubated at 37°C for 4 days. Medium from wells was removed, 50 μl of CellTiter-Glo 2.0 reagent (Promega, catalog no. G9241) was added, and the plate was protected from light while shaking for 20 min. A 1:1 addition of culture medium was added and allowed to equilibrate for 5 min while shaking, and then the solution was read for luminescence signal. For IC50 curves, 22Rv1 cells were seeded at 2000 cells per well in 96-well plates and analyzed in quadruplicate. Cells were plated and allowed to attach for 24 hours and then treated with the indicated concentration of MYCi975 for 72 hours. IC50 curves were extrapolated from the ATP content signal from 22Rv1 cells using nonlinear regression.
Bliss index analysis
LNCaP, 22RV1, C4-2B, and MCF-7 cell lines were cultured in RPMI 1640 (Gibco) supplemented with 10% heat-inactivated FBS (Gibco) and 1% penicillin-streptomycin (10,000 U/ml; Life Technologies) and grown in a 37°C incubator with 5% CO2. While the prostate cancer lines were plated for drug combination experiments in the same medium, the MCF-7 cell line was plated in phenol red–free RPMI 1640 (Gibco) supplemented with 10% charcoal-stripped FBS 24 hours before drug treatment. ENZ or 4-OHT was combined with MYCi975 in 4 × 5 dose-response matrices in 96-well plates. The prostate cancer lines (LNCaP, 22RV1, and C4-2B) were seeded at 1000 cells per well, while the breast cancer line MCF-7 was seeded at 2000 cells per well. Relative cell viability was determined using the CellTiter-Glo 2.0 Cell Viability Assay (Promega, G9242) after 72 hours of drug treatment as shown, and luminescence signals were acquired using a plate reader (PerkinElmer Victor 3V). For the Bliss matrix synergy experiments, the tested concentrations of the two drugs were chosen such that the effect on cellular viability after 72 hours of treatment is similar for both drugs over the tested concentrations. The goal was to choose a range of concentrations with effects spanning 30 to 100% viability. Concentrations that affect the cell at more than 30% viability were reasoned to be unlikely to show synergy, since most of the effect is due to one drug in this scenario. For the prostate cancer cell lines, the range used for MYCi975 was very similar, which is a result of MYCi975 showing similar IC50 values in these lines. However, these lines behave very differently toward ENZ alone, with LNCaP being ENZ sensitive (IC50 value of ~35 μM), while C42B and 22RV1 are ENZ-resistant lines (IC50 value of ~60 to 70 μM for both). The ENZ concentrations used in this study reflect this behavior, as can be seen in the percent viability matrices. Drug combination data were assessed using the SynergyFinder package using the Bliss independence model (38), which converts percent viability values to fraction affected (FA). The predicted fractional growth inhibition of the drug combination is calculated using the equation FA + FB − (FA × FB), where FA and FB are the fractional growth inhibitions of the drugs A and B at a given dose. Bliss excess is the difference between the expected growth inhibition and the observed inhibition. A Bliss excess score of >0 indicates synergy, close to zero indicates additivity, and <0 denotes antagonism (59).
Chromatin immunoprecipitation sequencing
Cells were plated into 15-cm2 plates at a density of 5 million cells per plate, and MYCi975 was supplemented such that cells would be isolated at the same time. Accordingly, the total number of plates equaled the total number of samples on day 0 of plating (DMSO and 1, 4, 8, 24, and 48 hours). Twenty-four hours after cell plating, MYCi975 was added to one plate for the 48-hour time point. Another 24 hours later (48 hours after cell plating), all plates were given fresh medium. At this time, the plate that was previously treated (48-hour time point) was supplemented with MYCi975 immediately, along with a new plate as the 24-hour time point. On the third day, the 1-, 4-, and 8-hour plates were given MYCi975 such that the end of all time points occurred at the same time. ChIP was carried out as previously described (60) with slight modifications. Once all plates were treated at their respective time points, 1% formaldehyde cross-linking was performed by an addition of 2 ml of 16% paraformaldehyde (Electron Microscopy Sciences, catalog no. 15710) to each plate, which contains 30 ml of culture medium, for 10 min. Glycine was added to a final concentration of 119 mM (1.6 ml of 2.5 M glycine) for 5 min to quench cross-linking. Cells were washed with ice-cold 1× PBS three times and then scraped into PBS and pelleted by centrifugation at 1000g for 10 min at 4°C. The cell pellet was flash-frozen with liquid nitrogen, allowed to thaw on ice, and resuspended in 1 ml of LB1 [50 mM Hepes-KOH (pH 7.6), 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10% glycerol, 0.5% IGEPAL-CA630, and 0.25% Triton X-100]. Then, 2 ml of LB1 was added to the cell resuspension and mixed end over end for 10 min at 4°C and then pelleted by centrifugation at 2000g for 5 min at 4°C. The pellet was resuspended in 1 ml of LB2 [10 mM tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, and 0.5 mM EGTA], and then 2 ml of LB2 was added to cell resuspension and mixed end over end for 10 min at 4°C. The nuclei pellet was resuspended in 0.9 ml of LB3 [10 mM tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% sodium deoxycholate, and 0.5% sarkosyl] and sonicated with a Misonix microtipped sonicator at ~5 W for 12 cycles for 15 s of sonication then 45 s of cooling (tubes are immersed in an ice-water bath). Triton X-100 (10%) was added for a final concentration of 1%. The sonicated nuclear fraction was pelleted for cell debris by centrifugation (>20,000g) for 25 min at 4°C. Protein concentration was determined by BCA (Thermo Fisher Scientific, catalog no. 23209), and 300 μg of chromatin was used for ChIP [c-MYC (Y69), 1 μg; c-MYC (ab56), 2 μg; WDR5, 2 μg; MAX, 2 μg; MNT, 2 μg; MXD1, 2 μg; MGA, 2 μg; H3K27ac, 2 μg; CTCF, 2 μg; FOXA1, 2 μg; AR, 1 μg]. In addition, input DNA was obtained from 10% of ChIP (30 μg) and used as a control.
IP was performed at 4°C with end-over-end mixing for 12 to 16 hours. Protein G Dynabeads (Life Technologies, catalog no. 10004D_3641869636) were used to pull down antibody (20 μl) bound to DNA fragments. IPs were washed four times with ice-cold ChIP-RIPA (radioimmunoprecipitation assay) [50 mM Hepes-KOH (pH 7.6), 500 mM LiCl, 1 mM EDTA, 1.0% IGEPAL-CA630, and 0.7% sodium deoxycholate, filter (0.2 μm)–sterilized before use]. For the fifth wash, 1 ml of TE (Tris-HCl-EDTA)–NaCl [10 mM tris-HCl (pH 8.0), 1 mM EDTA, and 50 mM NaCl, filter (0.2 μm)–sterilized before use] was added to IPs and fully resuspended by end-over-end mixing. Beads were aggregated using a magnet rack, and TE-NaCl was completely aspirated. DNA was eluted with addition 50 μl of elution buffer (0.1 M NaHCO3 and 1% SDS), held at 65°C with vigorous circular mixing for 15 min, and the eluate was collected. Beads were incubated with another 50 μl for a total of two elution steps followed by pooling. Cross-links were reversed by bringing the sample to 65°C in 190 mM NaCl (5 μl of 4 M NaCl into 100 μl of eluate) for 12 hours with vigorous circular mixing. Proteinase K (Invitrogen, catalog no. 25530-015) and ribonuclease (Worthington Biochemical Corp., catalog no. LS002132) digestion was carried out followed by DNA isolation with a Qiagen miniElute polymerase chain reaction (PCR) kit (catalog no. 28004). As input for library preparation, <1 to 5 ng of immunoprecipitated DNA were used. Library preparation was carried out with the KAPA Hyper Prep Kit (catalog no. KK8502). Adapter ligation was carried out for 60 min followed by 0.7 to 0.9× double-sided size selection with Ampure XP beads (catalog no. A63880) and a final 1× cleanup after library amplification (12 cycles in total). Library concentration was determined with Qubit, and library profile was analyzed by an Agilent 2100 Bioanalyzer. Libraries were sequenced in multiplex, and library pool concentrations were calculated with a KAPA Biosystems library quantification kit (catalog no. KK4835). Single-end reads [76 base pairs (bp)] were sequenced using an Illumina NextSeq500.
RNA sequencing
Cells were plated into six-well plates and, 24 hours later, treated with either 10 μM MYCi975 or DMSO (0.2%). Cells were trypsinized and counted in duplicate using Countess II (Thermo Fisher Scientific, catalog no. AMQAF1000) and 0.4% trypan blue (Thermo Fisher Scientific, catalog no. T10282), and 2 × 106 cells from each treatment group were washed in ice-cold PBS and centrifuged at 500g for 5 min at 4°C. RNA was isolated from cell pellets from biological quadruplicates (cells from different passages) with a Qiagen RNeasy kit (catalog no. 74134). For each sample, ~500 ng (equal aliquots of RNA to control for abundance) of RNA was used as input for RNA-seq library preparation. Into each sample, a 1:100 dilution of ERCC92 Spike-In Mix 1 (Thermo Fisher Scientific, catalog no. 4456740) was added and incorporated into the analysis pipeline. To capture a broader scope of RNA species, ribosome depletion library kits were used (KAPA Biosystems, catalog no. KK8560). RNA libraries were multiplexed, and final concentration was calculated with the same method as ChIP-seq. Next-generation sequencing was performed with paired-end reads (2 × 42 bp) on an Illumina NextSeq500.
Assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq)
Cells were plated in 10-cm2 plates and treated with 10 μM MYCi975 or DMSO (0.2%) for 48 hours. ATAC-seq libraries were generated as previously described with slight modifications (61, 62). Treated and control cells were trypsinized, and 1 million cells were washed in ice-cold PBS. Cells were pelleted at 500g for 5 min at 4°C and resuspended in 1 ml of lysis reaction mix [0.1% Tween 20, 0.1% IGEPAL (Sigma-Aldrich, catalog no. I8896), and 0.01% Digitonin (Promega, catalog no. G9441)] in ATAC resuspension buffer [10 mM tris-HCl (pH 8.0), 10 mM NaCl, and 3 mM MgCl2]. The lysis reaction was carried out on ice for 3 min, and then 4 ml of wash buffer (0.1% Tween 20 in ATAC resuspension buffer) was added and mixed end over end. Nuclei were pelleted at 500g for 10 min at 4°C. The pellet was resuspended in 125 μl of ice-cold PBS, and the nuclei were counted and inspected for quality. In total, 12,500 nuclei were aliquoted into the transposase tagmentation mix [2.5 μl of TDE1 enzyme and 25 μl of TD buffer (Illumina, FC-121-1030), 16.5 μl of nuclei in ice-cold PBS, 0.5 μl of 1% Digitonin, 0.5 μl of 10% Tween 20, and 5 μl of molecular biology–grade water]. Tagmentation was carried out at 37°C for 30 min in a Thermomixer (Eppendorf) at 300 rpm. DNA was isolated using the Zymo DNA clean and concentrator (catalog no. D4013) and amplified for 9 cycles with New England Biosystems High Fidelity 2X PCR Master Mix (NEB, catalog no. M0541S) as described (61). PCR-amplified ATAC-seq libraries were purified using the Zymo kit (catalog no. D4013), and Ampure XP beads (catalog no. A63880) were used for 0.6× to 1.8× size selection. Library distribution was analyzed with an Agilent 2100 Bioanalyzer. Paired-end sequencing was performed (2 × 42 bp) using an Illumina NextSeq500.
Data analysis
For single-end ChIP-seq reads, raw fastq files were aligned with bowtie (v1.2.2, settings: -t --best -m 1) using prebuilt genome indexes [National Center for Biotechnology Information (NCBI)] for both hg38 and mm10, downloaded from http://bowtie-bio.sourceforge.net/manual.shtml. For paired-end ATAC-seq, reads were aligned with bowtie2 (v2.2.6, settings: --very-sensitive -X 800). For paired-end RNA-seq, reads were aligned to the hg38 reference genome (NCBI, GRCh38.p12) using STAR (v2.7.5, settings: --alignIntronMin 20 --alignIntronMax 500000) (63). All fastq files were analyzed for quality control using FastQC (version 0.11.9). ERCC92 controls were aligned to the “ERCC92.fa” genome and processed though the RNA-seq pipeline (64). ChIP- and ATAC-seq sequence alignment files were converted to binary format using samtools (version 1.9). ChIP-seq files were used as input for creating HOMER (65) (v4.11.1) tag directories and subsequent peak calling (settings: -tbp 1, style = factor/histone). For ATAC-seq, MACS2 (v2.2.6) (66) was used for peak calling using the DNase-seq enriched cut site method (--nomodel --shift -75 --extsize 150 --nolambda -p 0.01). Using DiffBind [v3.12, settings: minMembers = (# of biological replicates), score=DBA_SCORE_TMM_MINUS_FULL] for differential binding analysis, each time point (1, 4, 8, 24, and 48 hours) was compared to DMSO, and peaks called by HOMER were used to define a consensus peak list (67). Using DESeq2 (25), differences in RNA levels were determined by comparing DMSO versus 24 hours and DMSO versus 48 hours (default settings). Table S2 provides differential binding and differential RNA level results for all MYCi975 target gene types comparing DMSO versus 48-hour MYCi975-treated 22Rv1 cells. To determine the genomic region distribution, we used the HOMER annotatePeaks.pl (hg38) function with the appropriate reference genomes and selected for promoter proximal (±2 kb from TSS) or promoter distal (>±2 kb from TSS). Motif enrichment databases and statistics were calculated using the HOMER findMotifsGenome.pl function.
Visualization for ChIP-seq data including bigwigs, heatmaps, and read coverage histograms was generated with deeptools bamCoverage (settings: --binSize 1 --normalizeUsing RPGC –effectiveGenomeSize mouse/human --extendReads 120 –ignoreDuplicates), plotHeatmap, and plotProfile function (68). Replicates were merged with bigWigMerge (kentUtils, v302), and bedGraphs were converted back to normalized bigwigs with bedGraphToBigWig function using the chrom.sizes file downloaded from UCSC (https://hgdownload.cse.ucsc.edu/goldenpath/hg38/bigZips/). Gene count matrices were generated using featureCounts (69) with the following NCBI reference genome annotation GTF: “hg38.ncbiRefSeq.gtf” (http://hgdownload.soe.ucsc.edu/goldenPath/hg38/bigZips/genes/) (subread v1.6.1). Bigwig files for RNA-seq visualization were created using a similar method as ChIP-seq with merging both the forward and reverse strands using the recommended methods from deeptools (additional commands: samtools sort; bamCoverage –effectiveGenomeSize 2913022398 -e -normalizeUsing CPM v3.3.0) (68). For genome browser tracks, we used pyGenomeTracks (70, 71) with merged bigwig files from biological replicates to display occupancy at certain gene loci. Scale settings were normalized for each experimental ChIP-seq or RNA-seq group (i.e., DMSO and 24 and 48 hours). For any overlapping peaks analyzed (i.e., MYCi975 sensitive versus MYCi975 H3K27ac loss), we used the function mergePeaks (-d 200 when comparing MYC and MAX, -d 200 when comparing ATAC peaks with TF peaks, and -d 1000 when comparing with H3K27ac peaks, HOMER v4.11.1). Tag densities for ChIP-seq signals at each MYCi975 gene target type were calculated with annotatePeaks.pl (settings: hg38, -size 400, -norm 0) using tag directories for each DMSO-treated biological replicate. To compare tag densities of multiple peak sets, each peak tag density (normalized by total tag counts in directory) was log2 transformed. MYCi975 gene target types were selected from MYC-bound annotated promoters. For each peak, differential binding analysis and differential RNA levels were overlapped for the annotated promoters. The differential binding data were sorted on the basis of FDR, and peaks with FDR < 0.01 were used to select MYCi975 gene target types 1 to 3. Type 1 and 2 genes represent all differentially lost MYC-bound promoters with either up- or down-regulated RNA levels, respectively. MYCi975 type 3 genes represent all differentially MYC-bound promoters with differential RNA level data such that FDR > 0.01. Type 4 consists of “MYCi-insensitive” promoter-proximal MYC-bound peaks that represent genes with low tag density, no differential MYC binding, and no differential gene expression (FDR > 0.01). Differential MAX binding was overlapped with MYC binding sites and sorted on the basis of FDR. Of the differential MYC binding sites (MYCi975 sensitive), overlapping MAX analysis determined 6812 peaks where no significant changes in MAX were determined (differential MAX binding FDR > 0.01) and are described as the MAX-retained sites. ATAC-seq alignment files were filtered for mitochondrial reads using sed, and adapters were removed with NGmerge (v0.2_dev) (72). Differential ATAC analysis was carried out using DiffBind with the same score setting as above. MACS2 ATAC-seq peaks were used from each individual replicate for diffBind. Visualization tracks were generated from readpilup files (.bdg, MACS2) from peak calling, and all replicates were merged with the same protocol as above. For GSEA (24) analysis of RNA-seq, each gene was assigned a rank metric (−log10 FDR × log2 fold change), and this preranked list was used as input for GSEA hallmarks v7.4 database. For ChIP-seq data, the rank metric was calculated for promoter-proximal MYC-bound genes and differential MYC binding data using the same hallmarks database as above. For multiple peaks that annotate to a single promoter, the average rank metric of each “multipeak promoter” was used as input in the final preranked list for GSEA.
ENCODE/cistromeDB analysis
Using the ENCODE ChIP-seq matrix (www.encodeproject.org) to locate deposited MYC ChIP-seq datasets, bed files were downloaded. The following is a list of cell lines with MYC ChIP-seq peak datasets that were downloaded from ENCODE using the “transcription factor” and “cell line” tabs selected: NB4 (ENCFF002CZO), K562 (ENCFF002CWI), HeLa (ENCFF950LQM), A549 (ENCFF542GMN), and MCF7 (ENCFF370EQJ). In addition, two MYC ChIP-seq peak datasets were downloaded from the Cistrome Data Browser (http://cistrome.org/db/#/): P493-6 (encode: GSM1036404) and MCF10A (encode: GSM935491). All other cell lines (22Rv1, DU145, and MycCap) were analyzed according to the ChIP-seq protocol above. Peaks were then called with HOMER (v.4.11.1, findPeaks -style factor). All reference genomes were converted to hg38 using CrossMap (v0.3.6) with the “over.chain” files downloaded from source (http://crossmap.sourceforge.net) (73). Once all peaks were converted to the hg38 reference genome, peak files were annotated with HOMER annotatePeaks.pl and then split into either promoter-proximal (HOMER promoter annotated) or promoter-distal (all other HOMER annotations) regions. HOMER motif analysis was run on all peak sets with default settings. Motif enrichment results are displayed as log10(P value) for CTCF, FOXA1, FOXM1, and c-MYC motifs from HOMER.
In vivo experiments
All animal experiments and procedures were performed in compliance with ethical regulations and the approval of the Northwestern University Institutional Animal Care and Use Committee. CD1-Foxn1nu (086) mice were obtained from Charles River Laboratories. All mice were housed in a pathogen-free animal barrier facility. All in vivo experiments were initiated with mice at 6 to 8 weeks of age. The 22Rv1 prostate cancer cells (2 × 106) suspended in 100 μl of BD Matrigel were subcutaneously injected into flanks of mice. Tumor volume (in cubic millimeters) was calculated by the formula (length × width2)/2. When the tumor size reached 150 to 200 mm3, mice were then randomized into groups with similar average tumor size in each group. MYCi975 was prepared and administered as described (21). ENZ was purchased from MedChem Express (HY-70002), prepared in 5% DMSO in corn oil, and given by p.o. (per oral). Tumors were harvested after 18 or 21 days of treatment. Mouse body weight was monitored every 2 to 3 days.
Acknowledgments
We thank A. Jain for synthesis of MYCi975, Y. Omura for high-throughput sequencing of RNA- and ChIP-seq libraries, and J. T. Bass and G. D. Barish (Division of Endocrinology, Feinberg School of Medicine) for providing access to the Illumina NextSeq500 sequencing platform for this study. This research was supported in part through the computational resources and staff contributions provided for the Quest high-performance computing facility at Northwestern University, which is jointly supported by the Office of the Provost, the Office for Research, and Northwestern University Information Technology.
Funding: This work was supported by grants from the National Cancer Institute (R01CA196270, R01CA257258, P50CA180995, F30CA250196, and T32GM008061) and a Prostate Cancer Foundation (PCF) Challenge Award. Part of this work was supported by the Polsky Urologic Cancer Institute and NUSeq Core Facility of Northwestern University.
Author contributions: Conceptualization: D.C., S.A.A., A.G.H., and J.B.P. Methodology: A.G.H., P.N.S., M.I.T., V.S., H.H., J.B.P., and G.E.S. Investigation: A.G.H., J.B.P., M.I.T., and V.S. Visualization: A.G.H., M.I.T., and V.S. Data analysis: A.G.H., M.I.T., and V.S. Supervision: D.C., S.A.A., and J.B.P. Writing—original draft: A.G.H. Writing—review and editing: A.G.H., D.C., S.A.A., J.B.P., V.S., and M.I.T.
Competing interests: H.H., G.E.S., and S.A.A. are coinventors on patent applications covering the methods and assays to identify and characterize MYC inhibitors and derivatives. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. In addition, all genome-wide raw and processed datasets were submitted to GEO under the accession number GSE178869 (access below). To review GEO accession GSE178869: (i) Go to https://urldefense.com/v3/__https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE178869__;!!Dq0X2DkFhyF93HkjWTBQKhk!EMHgP_Z9YRL3EeJ0Btc_hqCnOtHuhF8zjMNqBf6xNnxJ2EdXx1d_DkkbG8xAkDZrHqDNrJfOcg$ and (ii) enter token “odejyomyvfcttkp” into the box.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.McMahon S. B., MYC and the control of apoptosis. Cold Spring Harb. Perspect. Med. 4, a014407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eilers M., Eisenman R. N., Myc’s broad reach. Genes Dev. 22, 2755–2766 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Riggelen J., Yetil A., Felsher D. W., MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer 10, 301–309 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Gabay M., Li Y., Felsher D. W., MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 4, a014241 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang C. V., MYC on the path to cancer. Cell 149, 22–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Soucek L., Jucker R., Panacchia L., Ricordy R., Tatò F., Nasi S., Omomyc, a potential Myc dominant negative, enhances Myc-induced apoptosis. Cancer Res. 62, 3507–3510 (2002). [PubMed] [Google Scholar]
- 8.Savino M., Annibali D., Carucci N., Favuzzi E., Cole M. D., Gerard I., Soucek L., Nasi S., The action mechanism of the myc inhibitor termed omomyc may give clues on how to target Myc for cancer therapy. PLOS ONE 6, e22284 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carabet L. A., Rennie P. S., Cherkasov A., Therapeutic inhibition of myc in cancer. Structural bases and computer-aided drug discovery approaches. Int. J. Mol. Sci. 20, 120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKeown M. R., Bradner J. E., Therapeutic strategies to inhibit MYC. Cold Spring Harb. Perspect. Med. 4, a014266 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boike L., Cioffi A. G., Majewski F. C., Co J., Henning N. J., Jones M. D., Liu G., McKenna J. M., Tallarico J. A., Schirle M., Nomura D. K., Discovery of a functional covalent ligand targeting an intrinsically disordered cysteine within MYC. Cell Chem. Biol. 28, 4–13.e17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sabò A., Kress T. R., Pelizzola M., De Pretis S., Gorski M. M., Tesi A., Morelli M. J., Bora P., Doni M., Verrecchia A., Tonelli C., Fagà G., Bianchi V., Ronchi A., Low D., Müller H., Guccione E., Campaner S., Amati B., Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511, 488–492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muhar M., Ebert A., Neumann T., Umkehrer C., Jude J., Wieshofer C., Rescheneder P., Lipp J. J., Herzog V. A., Reichholf B., Cisneros D. A., Hoffmann T., Schlapansky M. F., Bhat P., Von Haeseler A., Köcher T., Obenauf A. C., Popow J., Ameres S. L., Zuber J., SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science 805, 800–805 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin C. Y., Lovén J., Rahl P. B., Paranal R. M., Burge C. B., Bradner J. E., Lee T. I., Young R. A., Transcriptional amplification in tumor cells with elevated c-Myc. Cell 151, 56–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kress T. R., Sabò A., Amati B., MYC: Connecting selective transcriptional control to global RNA production. Nat. Rev. Cancer 15, 593–607 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Walz S., Lorenzin F., Morton J., Wiese K. E., Von Eyss B., Herold S., Rycak L., Dumay-Odelot H., Karim S., Bartkuhn M., Roels F., Wüstefeld T., Fischer M., Teichmann M., Zender L., Wei C. L., Sansom O., Wolf E., Eilers M., Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511, 483–487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lorenzin F., Benary U., Baluapuri A., Walz S., Jung L. A., von Eyss B., Kisker C., Wolf J., Eilers M., Wolf E., Different promoter affinities account for specificity in MYC-dependent gene regulation. eLife 5, e15161 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung L. A., Gebhardt A., Koelmel W., Ade C. P., Walz S., Kuper J., Von Eyss B., Letschert S., Redel C., D’Artista L., Biankin A., Zender L., Sauer M., Wolf E., Evan G., Kisker C., Eilers M., OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors. Oncogene 36, 1911–1924 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Frank S. R., Parisi T., Taubert S., Fernandez P., Fuchs M., Chan H. M., Livingston D. M., Amati B., MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4, 575–580 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinato F., Cesaroni M., Amati B., Guccione E., Analysis of Myc-induced histone modifications on target chromatin. PLOS ONE 3, e3650 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han H., Jain A. D., Truica M. I., Izquierdo-Ferrer J., Anker J. F., Lysy B., Sagar V., Luan Y., Chalmers Z. R., Unno K., Mok H., Vatapalli R., Yoo Y. A., Rodriguez Y., Kandela I., Parker J. B., Chakravarti D., Mishra R. K., Schiltz G. E., Abdulkadir S. A., Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell 36, 483–497.e15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Truica M. I., Burns M. C., Han H., Abdulkadir S. A., Turning up the heat on MYC: Progress in small-molecule inhibitors. Cancer Res. 81, 248–253 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas L. R., Wang Q., Fesik S. W., Tansey W. P., Thomas L. R., Wang Q., Grieb B. C., Phan J., Foshage A. M., Sun Q., Olejniczak E. T., Clark T., Dey S., Lorey S., Alicie B., Howard G. C., Cawthon B., Ess K. C., Eischen C. M., Zhao Z., Fesik S. W., Tansey W. P., Interaction with WDR5 promotes target gene recognition and tumorigenesis by MYC. Mol. Cell 58, 440–452 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C., PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Du J., Xu R., RORα, a potential tumor suppressor and therapeutic target of breast cancer. Int. J. Mol. Sci. 13, 15755–15766 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H., Lee J. M., Lee G., Bhin J., Oh S. K., Kim K., Pyo K. E., Lee J. S., Yim H. Y., Il Kim K., Hwang D., Chung J., Baek S. H., DNA damage-induced RORα is crucial for p53 stabilization and increased apoptosis. Mol. Cell 44, 797–810 (2011). [DOI] [PubMed] [Google Scholar]
- 29.McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M., Bejerano G., GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis C. A., Hitz B. C., Sloan C. A., Chan E. T., Davidson J. M., Gabdank I., Hilton J. A., Jain K., Baymuradov U. K., Narayanan A. K., Onate K. C., Graham K., Miyasato S. R., Dreszer T. R., Strattan J. S., Jolanki O., Tanaka F. Y., Cherry J. M., The Encyclopedia of DNA elements (ENCODE): Data portal update. Nucleic Acids Res. 46, D794–D801 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng R., Wan C., Mei S., Qin Q., Wu Q., Sun H., Chen C. H., Brown M., Zhang X., Meyer C. A., Liu X. S., Cistrome Data Browser: Expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 47, D729–D735 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mei S., Qin Q., Wu Q., Sun H., Zheng R., Zang C., Zhu M., Wu J., Shi X., Taing L., Liu T., Brown M., Meyer C. A., Liu X. S., Cistrome Data Browser: A data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 45, D658–D662 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaret K. S., Carroll J. S., Pioneer transcription factors: Establishing competence for gene expression. Genes Dev. 25, 2227–2241 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayran A., Drouin J., Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem. 293, 13795–13804 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conway J. R., Lex A., Gehlenborg N., UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vervoorts J., Lüscher-Firzlaff J. M., Rottmann S., Lilischkis R., Walsemann G., Dohmann K., Austen M., Lüscher B., Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 4, 484–490 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swinstead E. E., Miranda T. B., Paakinaho V., Baek S., Goldstein I., Hawkins M., Karpova T. S., Ball D., Mazza D., Lavis L. D., Grimm J. B., Morisaki T., Grøntved L., Presman D. M., Hager G. L., Steroid receptors reprogram FoxA1 occupancy through dynamic chromatin transitions. Cell 165, 593–605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng S., Wang W., Aldahdooh J., Malyutina A., Shadbahr T., Tanoli Z., Pessia A., Tang J., SynergyFinder Plus: Toward better interpretation and annotation of drug combination screening datasets. Genom. Proteom. Bioinform. S1672-0229(22)00008-0 (2022), doi:10.1016/j.gpb.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lourenco C., Resetca D., Redel C., Lin P., MacDonald A. S., Ciaccio R., Kenney T. M. G., Wei Y., Andrews D. W., Sunnerhagen M., Arrowsmith C. H., Raught B., Penn L. Z., MYC protein interactors in gene transcription and cancer. Nat. Rev. Cancer 21, 579–591 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Kalkat M., Resetca D., Lourenco C., Chan P. K., Wei Y., Shiah Y. J., Vitkin N., Tong Y., Sunnerhagen M., Done S. J., Boutros P. C., Raught B., Penn L. Z., MYC protein interactome profiling reveals functionally distinct regions that cooperate to drive tumorigenesis. Mol. Cell 72, 836–848.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Leclerc D., Pham D. N. T., Lévesque N., Truongcao M., Foulkes W. D., Sapienza C., Rozen R., Oncogenic role of PDK4 in human colon cancer cells. Br. J. Cancer 116, 930–936 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atas E., Oberhuber M., Kenner L., The implications of PDK1–4 on tumor energy metabolism, aggressiveness and therapy resistance. Front. Oncol. 10, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woolbright B. L., Choudhary D., Mikhalyuk A., Trammel C., Shanmugam S., Abbott E., Pilbeam C. C., Taylor J. A., The role of pyruvate dehydrogenase kinase-4 (PDK4) in bladder cancer and chemoresistance. Mol. Cancer Ther. 17, 2004–2012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aversa I., Zolea F., Ieranò C., Bulotta S., Trotta A. M., Faniello M. C., De Marco C., Malanga D., Biamonte F., Viglietto G., Cuda G., Scala S., Costanzo F., Epithelial-to-mesenchymal transition in FHC-silenced cells: The role of CXCR4/CXCL12 axis. J. Exp. Clin. Cancer Res. 36, 1–15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lobello N., Biamonte F., Pisanu M. E., Faniello M. C., Jakopin Ž., Chiarella E., Giovannone E. D., Mancini R., Ciliberto G., Cuda G., Costanzo F., Ferritin heavy chain is a negative regulator of ovarian cancer stem cell expansion and epithelial to mesenchymal transition. Oncotarget 7, 62019–62033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siersbæk R., Madsen J. G. S., Javierre B. M., Nielsen R., Bagge E. K., Cairns J., Wingett S. W., Traynor S., Spivakov M., Fraser P., Mandrup S., Dynamic rewiring of promoter-anchored chromatin loops during adipocyte differentiation. Mol. Cell 66, 420–435.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Rubin A. J., Barajas B. C., Furlan-Magaril M., Lopez-Pajares V., Mumbach M. R., Howard I., Kim D. S., Boxer L. D., Cairns J., Spivakov M., Wingett S. W., Shi M., Zhao Z., Greenleaf W. J., Kundaje A., Snyder M., Chang H. Y., Fraser P., Khavari P. A., Lineage-specific dynamic and pre-established enhancer-promoter contacts cooperate in terminal differentiation. Nat. Genet. 49, 1522–1528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ni M., Chen Y., Fei T., Li D., Lim E., Liu X. S., Brown M., Amplitude modulation of androgen signaling by c-MYC. Genes Dev. 27, 734–748 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan S. C., Li Y., Dehm S. M., Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J. Biol. Chem. 287, 19736–19749 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bai S., Cao S., Jin L., Kobelski M., Schouest B., Wang X., Ungerleider N., Baddoo M., Zhang W., Corey E., Vessella R. L., Dong X., Zhang K., Yu X., Flemington E. K., Dong Y., A positive role of c-Myc in regulating androgen receptor and its splice variants in prostate cancer. Oncogene 38, 4977–4989 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson J. L. L., Carroll J. S., FoxA1 is a key mediator of hormonal response in breast and prostate cancer. Front. Endocrinol. (Lausanne). 3, 1–6 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hurtado A., Holmes K. A., Ross-Innes C. S., Schmidt D., Carroll J. S., FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat. Genet. 43, 27–33 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carroll P. A., Freie B. W., Mathsyaraja H., Eisenman R. N., The MYC transcription factor network: Balancing metabolism, proliferation and oncogenesis. Front. Med. 12, 412–425 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conacci-Sorrell M., McFerrin L., Eisenman R. N., An overview of MYC and its interactome. Cold Spring Harb. Perspect. Med. 4, a014357 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mukherjee B., Morgenbesser S. D., DePinho R. A., Myc family oncoproteins function through a common pathway to transform normal cells in culture: Cross-interference by Max and trans-acting dominant mutants. Genes Dev. 6, 1480–1492 (1992). [DOI] [PubMed] [Google Scholar]
- 56.Augert A., Mathsyaraja H., Ibrahim A. H., Freie B., Geuenich M. J., Cheng P.-F., Alibeckoff S. P., Wu N., Hiatt J. B., Basom R., Gazdar A., Sullivan L. B., Eisenman R. N., MacPherson D., MAX functions as a tumor suppressor and rewires metabolism in small cell lung cancer. Cancer Cell 38, 97–114.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Struntz N. B., Chen A., Deutzmann A., Lin C. Y., Felsher D. W., Koehler A. N., Struntz N. B., Chen A., Deutzmann A., Wilson R. M., Stefan E., Evans H. L., Stabilization of the Max homodimer with a small molecule attenuates myc-driven transcription article stabilization of the max homodimer with a small molecule attenuates myc-driven transcription. Cell Chem. Biol. 26, 711–723.e14 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kunder R., Velyunskiy M., Dunne S. F., Cho B.-K., Kanojia D., Begg L., Orriols A. M., Fleming-Trujillo E., Vadlamani P., Vialichka A., Bolin R., Perrino J. N., Roth D., Clutter M. R., Zielinski-Mozny N. A., Goo Y. A., Cristofanilli M., Mendillo M. L., Vassilopoulos A., Horiuchi D., Synergistic PIM kinase and proteasome inhibition as a therapeutic strategy for MYC-overexpressing triple-negative breast cancer. Cell Chem. Biol. 29, 358–372 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Parker J. B., Palchaudhuri S., Yin H., Wei J., Chakravarti D., A transcriptional regulatory role of the THAP11-HCF-1 complex in colon cancer cell function. Mol. Cell. Biol. 32, 1654–1670 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corces M. R., Trevino A. E., Hamilton E. G., Greenside P. G., Sinnott-Armstrong N. A., Vesuna S., Satpathy A. T., Rubin A. J., Montine K. S., Wu B., Kathiria A., Cho S. W., Mumbach M. R., Carter A. C., Kasowski M., Orloff L. A., Risca V. I., Kundaje A., Khavari P. A., Montine T. J., Greenleaf W. J., Chang H. Y., An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Risso D., Ngai J., Speed T. P., Dudoit S., Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 32, 896–902 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K., Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W., Shirley X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.R. Stark, G. Brown, DiffBind: Differential binding analysis of ChIP-Seq peak data. 1–29 (2016). https://bioc.ism.ac.jp/packages/3.3/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf.
- 68.Ramírez F., Ryan D. P., Grüning B., Bhardwaj V., Kilpert F., Richter A. S., Heyne S., Dündar F., Manke T., DeepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liao Y., Smyth G. K., Shi W., FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Lopez-Delisle L., Rabbani L., Wolff J., Bhardwaj V., Backofen R., Grüning B., Ramírez F., Manke T., pyGenomeTracks: Reproducible plots for multivariate genomic datasets. Bioinformatics 37, 422–423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramírez F., Bhardwaj V., Arrigoni L., Lam K. C., Grüning B. A., Villaveces J., Habermann B., Akhtar A., Manke T., High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 9, 189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gaspar J. M., NGmerge: Merging paired-end reads via novel empirically-derived models of sequencing errors. BMC Bioinformatics. 19, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao H., Sun Z., Wang J., Huang H., Kocher J. P., Wang L., CrossMap: A versatile tool for coordinate conversion between genome assemblies. Bioinformatics 30, 1006–1007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Tables S1 and S2