Abstract
Since the advent of COVID-19, the number of deaths has increased exponentially, boosting the requirement for various research studies that may correctly diagnose the illness at an early stage. Using chest X-rays, this study presents deep learning-based algorithms for classifying patients with COVID illness, healthy controls, and pneumonia classes. Data gathering, pre-processing, feature extraction, and classification are the four primary aspects of the approach. The pictures of chest X-rays utilized in this investigation came from various publicly available databases. The pictures were filtered to increase image quality in the pre-processing stage, and the chest X-ray images were de-noised using the empirical wavelet transform (EWT). Following that, four deep learning models were used to extract features. The first two models, Inception-V3 and Resnet-50, are based on transfer learning models. The Resnet-50 is combined with a temporal convolutional neural network (TCN) to create the third model. The fourth model is our suggested RESCOVIDTCNNet model, which integrates EWT, Resnet-50, and TCN. Finally, an artificial neural network (ANN) and a support vector machine were used to classify the data (SVM). Using five-fold cross-validation for 3-class classification, our suggested RESCOVIDTCNNet achieved a 99.5 percent accuracy. Our prototype can be utilized in developing nations where radiologists are in low supply to acquire a diagnosis quickly.
Keywords: COVID-19 diagnosis, X-ray Lung images, Pre-trained CNN methods: Inception-V3 & Resnet-50, TCN, EWT
1. Introduction
A novel kind of viral pneumonia, coronavirus 2 (SARS-CoV-2), has been found in China (Lu et al. 2020). It is caused by the severe acute respiratory syndrome. In February 2020, the World Health Organization (WHO) named the virus COVID-19 and designated it a pandemic in March 2020. COVID-19 has been verified in approximately 239 million people worldwide, with 4.8 million fatalities as of October 15, 2021 (World Health Organization, 2021). Furthermore, many people may be infected with COVID-19 yet have no symptoms. These individuals do not show signs of COVID-19 sickness; instead, they are viral carriers who can disseminate the virus to susceptible persons (Huff and Singh, 2020). Therefore, early diagnosis of the disease, even in the absence of symptoms, can control the spread of the virus and save the patient’s life.
Several indicators can help to diagnose the patient’s health status. The most common test for COVID-19 detection is the Reverse Transcription Polymerase Chain Reaction (RT-PCR) (Wu et al. 2020). However, RT-PCR is relatively low in sensitivity and time‐consuming. Moreover, it is costly and requires specific materials and equipment, which are not easily accessible. Alternatively, various types of radiological imaging exist, such as X-rays and computed tomography (CT), to identify patients affected by Pneumonia. It is reported that COVID-19 patients’ lungs exhibit some visual features, such as markings and spots, that may distinguish COVID-19 positive cases from typical cases using radiological images (Balaha et al. 2021). Unlike RT-PCR and CT, chest X-ray is cheaper, less time-consuming, and readily available for screening. The X-ray has lower ionizing radiations than CT scans, which allows for multiple follow-ups on the effects of COVID-19 on lung tissue. Therefore, chest X-ray images gathered from different databases have been used to detect COVID-19 disease in this paper. Sometimes noises appear in the X-ray images, which affects the diagnosis. To overcome this challenge, time–frequency analysis can be used to remove high-frequency components from the noisy images. Also, the image pre-processing step helps to further improve the model performance.
Many traditional machine learning techniques have been presented in the literature for the early diagnosis of COVID-19 (Heidari et al., 2020, Li et al., 2021, Sharifrazi et al., 2021, Júnior et al., 2021, Khan et al., 2021, Fan et al., 2021, Karthik et al., 2021). The convolutional neural network (CNN) (Le Cun et al. 2015), support vector machine (SVM) (Cortes and Vapnik, 1995), residual exemplar local binary pattern (ResExLBP), iterative ReliefF (Li et al. 2021), and Sobel filter (Sobel and Feldman, 1968) have been used. Moreover, most previous methods used deep learning networks to achieve good classification results. Ozturk et al. (2020) presented a deep learning structure based on a pre-trained model known as DarkCovidNet, and their proposed model was defined as a Darknet-19 classifier. The former models were used to diagnose X-ray lung images infected with COVID-19. Instead of building a model from scratch, this model used the Darknet-19 classifier (Redmon and Farhadi, 2017). Their proposed model was tested with three different classes: COVID cases, normal, and pneumonia. The number of X-ray images was 1000, and they have obtained 98.08% and 87.02% accuracy rates for binary (i.e., normal and COVID-19) and three classes (i.e., normal, COVID-19, and Pneumonia) classifications, respectively. The proposed approach (Toraman et al., 2020, Afshar et al., 2020) used capsule networks to detect COVID-19 disease using X-ray images.
COVID-19 has recently been detected utilizing X-ray pictures using a variety of transfer learning approaches (Apostolopoulos and Mpesiana, 2020, Asif et al., 2020, Brunese et al., 2020, Das et al., 2020, Zhang et al., 2020, Abraham and Nair, 2020, Jain et al., 2020, Ismael and Şengür, 2021, Jin et al., 2021, Quan et al., 2021; Das et al. 2021). The researchers in (Das et al. 2020) used the Xception model to provide an automated deep transfer learning technique for identifying COVID-19 infection from chest X-rays using the Xception model. They used 500 normal, 500 pneumonia, and 125 COVID-19 X-ray images in their study, with a classification accuracy of 97.4%. For multi-classification diagnosis, Apostolopoulos and Mpesiana (2020) combined transfer learning models with a VGG-19 pre-trained model and reported an accuracy of 93.48 percent on test data. Multi-CNN was used to extract characteristics from chest X-ray pictures, and it consisted of several pre-trained CNNs (Abraham and Nair, 2020, Ismael and Şengür, 2021). Das et al. (2021) developed an automated Covid-19 diagnostic technique based on the VGG-16 pre-trained model. On 2,905 chest X-ray pictures, our approach had a 97.67 percent accuracy. Despite all of these efforts, the following are the major research gaps in the automated identification of COVID-19:
-
i.
The accuracy of diagnosis still needs to be improved by reducing misclassification.
-
ii.
Most of the related deep learning models did not give enough attention to the pre-processing stage.
-
iii.
The architecture of the pre-trained models cannot provide high optimal diagnostic results.
Therefore, in this study, we developed a diagnostic method for the automated detection of COVID-19 infection using chest X-rays using the transfer learning method. The main contributions of this paper are-.
-
•
Investigation of various deep learning models for the automated detection of COVID-19, pneumonia, and normal X-ray chest images.
-
•
Development of the RESCOVIDTCNNet model, which combines EWT, Resnet50, and a temporal convolutional neural network (TCN).
-
•
The model was constructed utilizing more than 5000 X-ray chest scans, and the proposed model had the best results.
-
•
The chest X-ray images were pre-processed using the empirical wavelet transform (EWT).
2. Related works
Several studies have recently been modified to diagnose COVID illnesses using chest X-ray pictures. Transfer learning approaches relying on (CNN) can be used for classification, feature extraction, and transfer learning (Zhang et al. 2020; Abraham et al. 2020; Jin et al., 2021, Quan et al., 2021, Das et al., 2021, Ozcan, 2021).Table 1 summarizes the results of studies developed for the automated identification of COVID-19 utilizing X-ray images. Table 1 shows that there is still space to improve the model's classification performance utilizing large, varied datasets. As a result, we've presented a new RESCOVIDTCNNet to categorize three pictures utilizing X-ray images and achieve maximum diagnostic accuracy.Table 2 show the datasets used in our study.
Table 1.
Summary of automated COVID-19 detection systems developed. Unless stated otherwise, all accuracy results are reported according to 3 class classification (Normal, COVID-19, and Pneumonia) (2-Class and multi-class).
| Study | Dataset(s) | Classes | Classifier | Accuracy |
|---|---|---|---|---|
| Ozturk et al. 2020 | CO19-Ximage (Cohen, 2020) and Ch-X8image (Wang et al. 2017). |
Normal (5 0 0)COVID-19 (1 2 7) Pneumonia (5 0 0) |
Darknet-19 | 2-Class: 98.08% 3-Class: 87.02% |
| Li et al. 2021 | GitHub and Kaggle | Normal (2 3 4)COVID-19 (87) |
SVM | 100% |
| Asif et al. 2020 | CO19-Ximage (Cohen, 2020) and COVQU (Chowdhury et al., 2020, Rahman et al., 2021) | Normal (1,341)COVID-19 (8 6 4)Pneumonia (1,345) |
Inception-V3. | 98.30% |
| Brunese et al. 2020 | CO19-Ximage (Cohen, 2020), X-Ray Image Dataset (Ozturk et al. 2020), and Ch-X8image (Wang et al. 2017) | Normal (3,520) COVID-19 (2 5 0)VGG16.Pneumonia (2,753) |
VGG16. |
97.00% |
| Das et al. 2020 | X-Ray Image Dataset (Ozturk et al. 2020) | 1,000 chest X-rays images included Normal, COVID-19, and Pneumonia classes. | Xception model | 97.4 |
| Toraman et al. 2020 | CO19-Ximage (Cohen, 2020) and Ch-X8image (Wang et al. 2017) | Normal (1,050)COVID-19 (2 3 1)Pneumonia (1,050) |
capsule neural network | 2-Class: 97.24% 3-Class: 84.22% |
| Zhang et al. 2020 | CO19-Ximage (Cohen, 2020), Ch-X8image (Wang et al. 2017), X-Ray Image Dataset (Ozturk et al. 2020), and Kaggle. |
Normal (5 5 7)COVID-19 (2 3 4)Pneumonia (7 3 0) |
Inception-V3 | 90.00% |
| Abraham et al. 2020 | CO19-Ximage (Cohen, 2020) and Ch-Ximage (Mooney, 2020) | COVID-19 (4 5 3)non-COVID (4 9 7) |
Squeezenet + Darknet-53 + MobilenetV2 + Xception + Shufflenet |
2-Class: 91.16% |
| Jain et al. 2020 | CO19-Ximage (Cohen et al. 2020) and Ch-Ximage (Mooney, 2020) | Normal (3 1 5)COVID-19 (2 5 0)Bacterial Pneumonia (3 0 0)Viral Pneumonia (3 5 0) |
ResNet50 and ResNet-101 | Multi-class: 97.77% |
| Afshar et al. 2020 | CO19-Ximage (Cohen, 2020) and Ch-Ximage (Mooney et al. 2020) | 94,323 chest X-rays images included Normal, COVID-19, Bacterial Pneumonia, and Viral Pneumonia classes. | Capsule Networks | Multi-Class: 95.70% |
| Heidari et al. 2020 | Mendeley Data (Kermany et al. 2018), COVQU (Chowdhury et al., 2020, Rahman et al., 2021), and CO19-Ximage (Cohen, 2020) | Normal (2,880)COVID-19 (4 1 5)Pneumonia (5,179) |
VGG16 | 96.90% |
| Ismael and Şengür, 2021 | CO19-Ximage (Cohen, 2020) and CO19-Ximage (Mooney, 2020). | Normal (2 0 0)COVID-19 (1 8 0) |
ResNet50 + SVM classifier with the Linear kernel function | 2-Class: 94.70% |
| Jin et al. 2021 | COVQU (Chowdhury et al., 2020, Rahman et al., 2021) and CO19-Ximage (Cohen, 2020) | Normal (6 0 0)COVID-19 (5 4 3)Pneumonia (6 0 0) |
AlexNet + ReliefF + SVM | 99.43% |
| Demir, 2021 | Ch-Ximage (Mooney, 2020) and Ch-X8image (Wang et al. 2017) | Normal (2 0 0)COVID-19 (3 6 1)Pneumonia (5 0 0) |
LSTM | 97.11% |
| Sharifrazi et al. 2021 | Omid Hospital in Tehran | Normal (2 5 6)COVID-19 (77) |
CNN + SVM + Sobel filter |
2-Class: 99.02% |
| Quan et al. 2021 | CoronaHack (Praveen, 2020) CO19-Ximage (Cohen, 2020), and COVQU [7, 8] |
Normal (2,917)COVID-19 (7 8 1)Bacterial Pneumonia (2,850)Viral Pneumonia (2,884) |
DenseNet and CapsNet | 90.70% |
| Júnior et al. 2021 | CO19-Ximage (Cohen, 2020) and Ch-Ximage (Mooney 2020) | Normal (2 5 0)COVID-19 (2 5 0) |
CNN + PCA | 2-Class: 97.60–100% |
| Das et al. 2021 | Kaggle | Normal (1,341)COVID-19 (2 1 9)Pneumonia (1,345) |
VGG-16 and ResNet-50 | 97.67% |
| Albahli et al. 2021 | Ch-Ximage datasets (Ahsan et al. 2020) and (Boudrioua et al. 2020) | Normal (8,851)COVID-19 (5 9 0)Pneumonia (6,057) |
DenseNet | 92.00% |
| Ozcan, 2021 | X-Ray Image Dataset (Ozturk et al. 2020) | Normal (5 0 0)COVID-19 (1 2 5)Pneumonia (5 0 0) |
AlexNet + ResNet50 | 2-Class: 99.52% 3-Class: 87.64% |
| Irfan et al. 2021 | GitHub, COVID-19 radiography database, Kaggle, COVID-19 image data collection, and COVID-19 Chest X-ray Dataset | X-ray ImagesNormal (1100)COVID-19 (1900)Pneumonia (2000) CT ImagesNormal (6 0 0)COVID-19 (7 0 0)Pneumonia (1000) |
Hybrid deep neural networks (HDNN) consist of dropout, convolution, max-pooling layer, LSTM blocks, and a fully connected layer | 3-Class: 99% |
| Almalki et al. 2021 | COVID-19 Chest X-ray Dataset, Kaggle repository “Chest X-Ray Images, | A total of 1251 images were taken from the repositoriesNormal (620 samples)Pneumonia (660 samples)Viral-pneumonia (654 samples)Corona (568 samples) |
CoVIR-net Model (Inception + Resnet Models) |
CoVIR-net + Random Forest Multi-class: 97.29% |
COVID-19 X-ray image: CO19-Ximage Chest X-Ray Images: Ch-Ximage ChestX-ray8: Ch-X8image.
Table 2.
provides the details of the three datasets used and the number of selected images from each dataset.
| Datasets | Databases | Number of X-ray Chest Images in each dataset | Amount of X-ray Chest Images Selected for this Study |
|---|---|---|---|
| Ozturk et al. 2020 | COVID-19 X-ray image (Cohen, 2020) | COVID-19: 125 images | COVID-19: 125 images |
| ChestX-ray8 (Wang et al. 2017) | Normal: 500 images Pneumonia: 500 images | Normal: 329 images Pneumonia: 325 images | |
| (Mooney, 2020) | Chest X-Ray Images (Mooney, 2020) | Normal: 1592 images Pneumonia: 4273 images | Normal: 1343 images Pneumonia: 1345 images |
| (Chowdhury et al., 2020, Rahman et al., 2021) | COVQU (Chowdhury et al., 2020, Rahman et al., 2021) | COVID-19: 3616 images Normal: 10,192 images Pneumonia: 1345 images |
COVID-19: 1545 images |
| Total | COVID-19 = 1670 images Normal = 1672 images Pneumonia = 1670 images |
||
Overall, based on the above literature review, the suggested RESCOVIDTCNNet model is unique because it combines EWT with Resnet-50 and TCN. The data-gathering stage begins with collecting chest X-ray images from various sources. Second, EWT is utilized to increase the resolution of the input pictures during pre-processing. Third, feature extraction is proposed using Inception-V3 and Resnet-50, two pre-trained models. Finally, artificial neural networks (ANN) and support vector machines (SVM) are used in the classification stage. All of these strategies were included primarily to increase classification accuracy. The use of EWT and Resnet-50 helps improve the quality of X-ray pictures. The feature extraction stage is aided by a mix of Resnet-50 and Inception-v3 pre-trained models. TCN was then applied to the retrieved features in order to extract even more prominent characteristics from the X-ray pictures.
3. Methodology
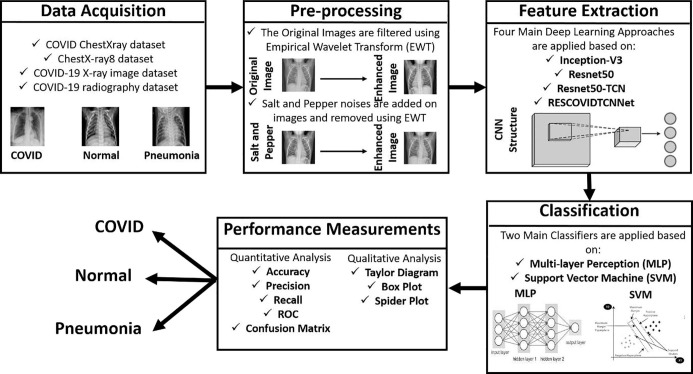
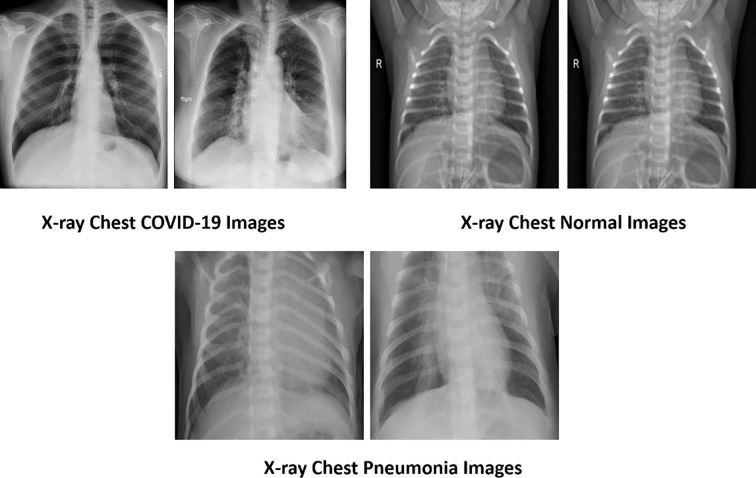
The methodology consists of four main phases essential for constructing the proposed deep learning models, as shown in Fig. 1 . The first phase is the data acquisition phase, and in this step, four main datasets are selected containing X-ray chest images of COVID, normal, and pneumonia. The images are pre-processed using empirical wavelet transform (EWT) in the second filtration phase. Fig. 2 represents the three types of lung X-rays images.
Fig. 1.
Illustration of the proposed methodology for automated COVID-19 detection using chest X-ray images.
Fig. 2.
Typical chest X-ray images of different categories.
Then in the feature extraction step, four main deep learning models are used: Inception-V3, Resnet50, Resnet50-TCN, and RESCOVIDTCNNet. Finally, these extracted features are classified using multilayer perceptron (MLP) and support vector machine (SVM) classifiers.
3.1. COVID19-Xray-Datasets
This section presents various lung X-ray images gathered from publicly available datasets required for COVID-19 diagnosis. We have taken care of two image selection criteria in this work: (i) balance the X-ray images number in each class and (ii) not choose the same image from different datasets twice. The first dataset used in this work was obtained by (Ozturk et al. 2020) and available on the Kaggle portal (Talo, 2020). The dataset in (Ozturk et al. 2020) was obtained from two different databases. The first database was collected (Cohen, 2020), called COVID-19 X-ray image. This database consists of 125 COVID-19 images. The second database was provided by (Wang et al. 2017), called ChestX-ray8, and this database comprises 500 normal and pneumonia X-ray images. This study used 125 COVID-19 images from the COVID-19 X-ray image database, 329 normal, and 325 pneumonia from the ChestX-ray8 database. The second dataset was obtained from (Mooney, 2020) called Chest X-Ray images, and it consists of 1592 normal and 4273 pneumonia images. The number of images selected for this study from this database is 1343 normal and 1345 pneumonia images. The third dataset was obtained from (Rahman et al. 2021) and is available on the Kaggle portal (Rahman et al. 2021).
This dataset is called COVQU. The main aim of this dataset is to obtain the images of the COVID-19 class. This dataset consists of 3616 COVID-19, 10,192 normal, and 1345 viral pneumonia images.
We have selected 1545 COVID-19 images from this third dataset. Finally, we have selected 1672 normal, 1670 COVID-19, and 1670 Pneuomina images from these three datasets for this study.
3.2. Preprocessing using empirical wavelet transform (EWT)
This paper used EWT (Gilles, 2013) to filter and pre-process the X-ray images. EWT is similar to wavelet transform, in which the transformation based on the EWT results in two main sets of details and the approximation coefficients. Let us consider a signal; the detail coefficients will be defined based on the convolution operation of the signal with EWT to obtain , whereas the approximation coefficients will be known using the convolution of the signal using a scaling function defined as . The details and the approximation coefficients are defined in the following equations, respectively.
| (1) |
| (2) |
The previous equations are considered the details and the approximation of the frequency components obtained from the EWT. Extension of EWT for the 2D image can be defined as 2D Littlewood-Paley EWT, 2D Curvelet EWT, 2D Empirical Ridgelet transform, and 2D tensor EWT. In this study, 2D Littlewood-Paley EWT is applied because it aims to construct of little-Paley wavelet filters in separate scales defined by various concentric rings. It detects the scales first and then detects the angular section with each scale ring. Let us consider to be an image, then the details and the approximation coefficients of 2D Littlewood-Paley EWT are defined using the following equations:
| (3) |
| (4) |
where and are the 2D pseudo-polar Fourier transform and its inverse. Then EWT is applied to each band of the image.
3.3. Transfer learning models based on Resnet50 and Inception-v3
In this study, ResNet-50 and Inception-v3 pre-trained CNN architectures are employed to classify the X-ray chest images into three classes: Normal, COVID-19, and Pneumonia.
ResNet: The residual neural network with 50 layers (ResNet-50) is a convolutional neural network variation of ResNet (He et al. 2016). This architecture was trained using a subset of the ImageNet database (Deng et al 2009), which comprises over one million pictures from 1000 different classes. ResNet-50 is a deep residual learning framework-based network. These residual networks are more customizable, and utilizing deeper models can improve accuracy. ResNet-50′s residual function is made up of three weight layers: 11, 33, and 11 convolutions. The dimensions are reduced thanks to the 11 convolution layers. The 33% layer is used for input/output dimensions that are smaller. The residual connections in the ResNet-50 architecture aid in the maintenance of learnt knowledge and the reduction of model training time.
Inception: The main concept behind the Inception architecture is to determine how to estimate and cover an optimum local sparse structure in a convolutional vision network with conveniently available dense components. Inception-v3 (Szegedy et al. 2016) is widely used for image classification with a pre-trained deep neural network. It consists of two main types of convolutions: factorized and smaller. The former convolutions reduce the computational efficiency by decreasing the number of parameters included in the network. The latter convolutions are used to replace bigger convolutions to achieve faster training. In the Inception-v3 model, the basic convolution block, enhanced Inception modules, and task-specific classifiers are successively concatenated. Basic convolutional operations using 1 × 1 and 3 × 3 kernels are used to learn low-level feature mappings. Next, multi-scale feature representations are concatenated in the Inception module and fed into auxiliary classifiers with various convolution kernels to improve convergence performance. The fully connected layer converts multi-scale feature vectors into a one-dimensional vector, followed by 1 × 1 Inception modules. Finally, the Softmax classifier generates one vector with three classes of probability (i.e., Normal, COVID-19, and Pneumonia).
3.4. The proposed RESCOVIDTCNNet model
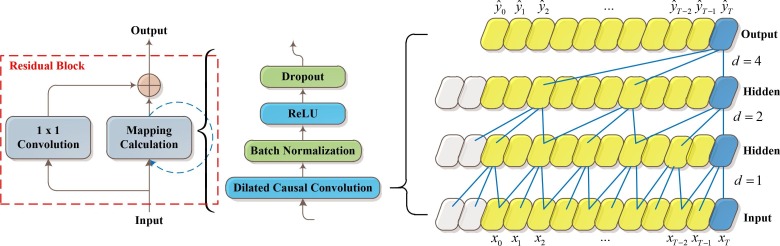
Due to their superior capability to capture temporal relationships in sequential data, recurrent neural networks (RNNs) are preferred for sequence modeling tasks. The RNNs, such as long short-term memory (LSTM) (Hochreiter and Schmidhuber, 1997), convolutional LSTM (ConvLSTM) (Xingjian et al. 2015), WaveNet (Oord et al., 2016), and gated recurrent unit (GRU) (Cho et al. 2014), can capture long-term relationships in sequences and have attained state-of-the-art results in sequence modeling tasks. The TCN model (Bai et al. 2018) modifies the CNN model designed for sequence modeling tasks with causal constraints. The TCN outperformed the RNN and its derivatives LSTM (Breuel, 2015) and GRU (Lynn et al. 2019). Furthermore, the architecture of TCN is simpler and more straightforward than RNN and LSTM networks. In addition, the TCN, like standard CNNs, offers the advantages of convolution kernel sharing and parallel computation, which helps to minimize the calculation time. It may also use a 1D fully-convolutional network (FCN) architecture to transfer a sequence of varying lengths to an output sequence of the same length (Long et al. 2015). Furthermore, the TCN architecture's convolutions are causal convolutions. TCN's typical architecture is seen in Fig. 3 .
Fig. 3.
Schematic diagram of TCN architecture.
The residual network is utilized in the TCN to prevent CNN performance from degrading as the number of layers increases. Each of the m residual blocks in the TCN layer contains two layers of dilated causal convolution. Weight normalization is also done to the convolutional filters in a residual block, with the rectified linear unit (ReLU) (Nair and Hinton, 2015) as the activation function. Because TCN has two dilated causal convolutions and non-linearities, the ReLU activation function must be utilized. After each dilated convolution, a dropout layer is added for regularization. The input and output widths in TCN might be varied. To account for the difference in input–output widths, an 11 convolution is utilized.
The dilated convolution adds certain weights to the convolution kernel while leaving the input data intact, increasing the size of the time series viewed by the network while keeping the amount of calculation relatively constant. Let an input sequence , the dilated convolution operation on the -the element of the 1D sequence (Yan et al. 2020) is defined as.
| (5) |
where indicates the filter with size , the dilation -the coefficient is , and accounts for the direction of the past. As a result, dilation is the same as using a set step size between each pair of adjacent filter taps.
The extended convolution is simplified to a standard convolution with . The output of the top layer may reflect a wider variety of inputs with a bigger expansion, thereby increasing TCN’s receptive field. TCN can extend the receptive field in two ways: by raising the dilation factor or by selecting bigger filter sizes . As an example, Fig. 3 depicts the dilated 1D convolutions with dilation factors and filter size .
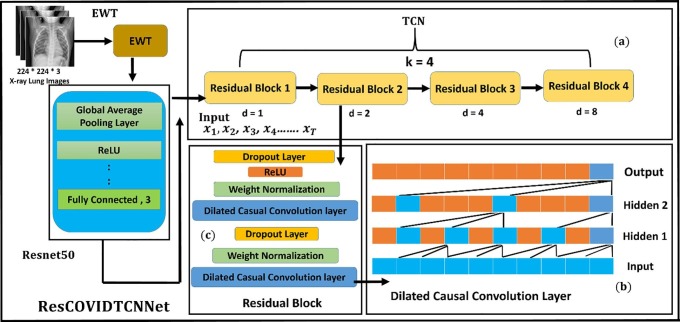
RESCOVIDTCNNet is the combination of Resnet-50 and TCN. This combination is constructed based on selecting the output obtained from the fully connected layer of Resnet50 and feeding it as an input to the TCN. TCN is designed and trained to classify the next input series. Let us consider the results obtained from the fully connected layer of Resnet-50 are defined in the following sequence . The corresponding output that is going to be classified is defined as . The overall structure of the proposed RESCOVIDTCNNet is shown in Fig. 4 , and the pseudocode is provided in algorithm.1.
Fig. 4.
Overview of RESCOVIDTCNNet: (a) TCN structure with its residual block, (b) Structure of dilated causal convolution layer, and (c) Residual block.
Algorithm.1 Pseudocode for the proposed RESCOVIDTCNNet.
|
Input: X-ray Chest Images for different types (Normal, COVID, Pneumonia) Output: Features extracted from RESCOVIDTCNNet EWT (Pre-processing Step) Step1: The X-ray chest images are passed to the EWT for de-nosing and pre-processing. Step2: Two Dimensional Littlewood-Paley EWT is presented To deal with 2D images. Step3: The details and the approximation coefficients based on 2D Dimensional Littlewood-Paley EWT are obtained using the following equations: Step4: The X-ray chest images are reconstructed from the detailed coefficients of EWT. Resnet50 (Feature Extraction) Step5: The Pre-processed EWT X-ray chest images are passed to the transfer learning model known by Resnet50. Step6: The images are passed to 5 main blocks. The first block consists of convolutional and max-pooling layers. Step7: The second block consists of 9 convolutional layers, and the third block consists of 12 convolutional layers. Step8: The output of the third block is based on the fourth block, which consists of 18 convolutional layers, and the output is passed to the fifth block, which consists of convolutional layers. Step9: Then, an average pooling and fully connected layers are applied to the output of the fifth block. Step10: A deep residual learning is presented to create shortcut connections by mapping the layers to residual known by . Step11: The nonlinear layers are mapped to another type of mapping function defined by: : = TCN (Feature Extraction) Step12: The output of the fully connected layer in Resnet50 is passed to the temporal convolutional network (TCN). Step13: The sequence input layer of TCN accepts the output of Resnet50, and it is passed to four main residual blocks. Step14: The output of the residual blocks is passed to the fully connected layer. Step15: The features of the fully connected layer are input to a set of classifiers which are ANN and SVM. |
TCN aims to satisfy two main constraints (Aksan and Hilliges, 2019): (i) the output of the TCN network should be equal to the length of the input, and (ii) it depends on the previous information obtained from the previous steps. TCN residual blocks are based on casual convolution in which each layer at an output time is calculated with the region no later than time step in the past layer. The causal convolutional layers have a problem of respective limited size, and to store a large amount of information, it is required to stack many layers. Therefore, dilated convolution is applied to permit an exponentially large receptive field. The dilated convolutional layers structure is shown in Fig. 4 (b). When the dilated convolutions increase with a dilated factor in an exponent way, this leads to an increase in the size of the receptive field to cover a large number of inputs in the history.
The structure of the residual block is shown in Fig. 4 (c). Finally, the RESCOVIDTCNNet model consists of 4 main blocks. Each block consists of two dilated causal convolutional layers, weight normalization layers, and RELU activation functions. The result of each block is an input to the next block. The four blocks are executed, and then the whole model is ended by two main fully connected layers and a classification layer.
4. Experimental results
The outcomes of the suggested approaches achieved in the various subsections are presented in this section. The first section covers the experimental setup as well as the software needed for de-noising, feature extraction, and classification. The outcomes of the EWT filtering procedure are shown in the second subsection. The training parameters necessary for deep learning training models are listed in the third subsection. Finally, the quantitative analysis of performance metrics produced from various deep learning models is described in the last subsection.
4.1. Experiment setup
The presented deep learning models and the classifiers used in this work were implemented using MATLAB software programming language. The classification stage and the performance measurements were obtained using WEKA software. The experiments were performed on a PC with Intel COREi9- 10,232 @ 1.80 GHz 1.99 GHz, 32 GB RAM. The total number of X-ray images (5012) was divided into three parts: training (60%), validation (20%), and test (20%). Five-fold cross-validation (CV) strategy was used to assess the model performance. The classification was performed using two MLP and SVM classifiers to choose the best performing classifier.
4.2. EWT pre-processing results on X-ray images
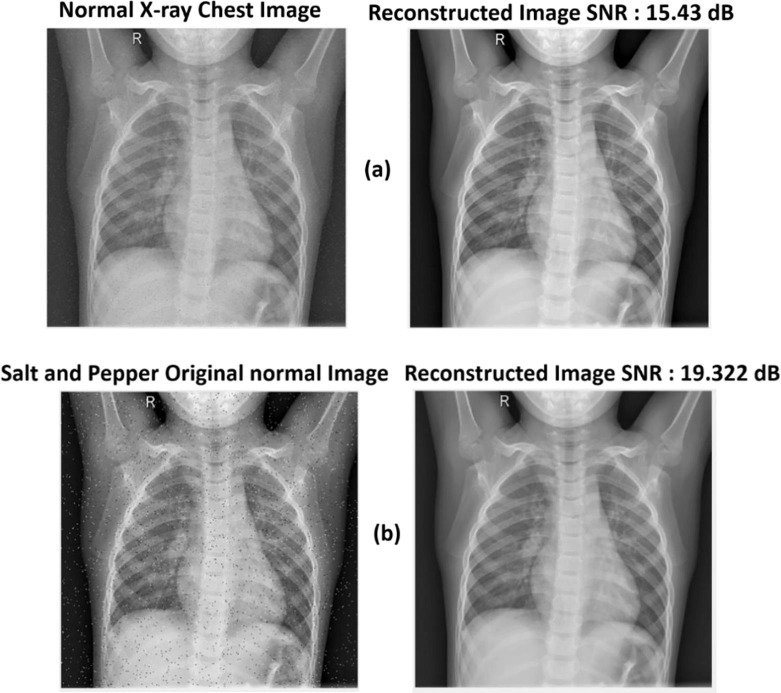
The initial X-ray chest scans revealed that EWT performed well. It increased the original image's performance to get an SNR of 15.43 dB. In addition, salt and pepper noise with a variation of 0.05 is added to the original image to clarify the EWT's performance. The difference is the quantity of salt and pepper noise added to the X-ray lung pictures. The reconstructed picture was of higher quality, free of noise, and had an SNR of 19.322 dB. Fig. 5 depicts EWT findings on the original X-ray picture (a) and the salt and pepper X-ray image (b).
Fig. 5.
Visual representation of images with EWT: (a) original normal X-ray image and its corresponding reconstructed normal X-ray image using EWT, and (b) original normal X-ray after adding salt and pepper noise and its corresponding reconstructed image using EWT.
4.3. Training parameters
Table 3 lists the parameters taken into account during hyper-parameter tuning of deep learning models. The optimizer, momentum, learn rate (drop factor, schedule, and drop period), initially learn rate, L2 regularization, gradient threshold technique and value, maximum epochs, and mini-batch size are all factors to consider. Furthermore, the best parameters for deep learning models are found through experimentation (Rehman et al., 2021, Sultan et al., 2019).
Table 3.
Tuning parameters used for the transfer learning models.
| Training Parameters | InceptionV3 | Resnet-50 | Resnet-50-TCN | |
|---|---|---|---|---|
| Optimizer | (sgdm) | (sgdm) | (sgdm) | Adam (sgdm) |
| Initial Learn rate | 0.05 | 0.001 | 0.001 | 0.1 |
| Learn Rate Schedule | ‘piecewise’ | ‘piecewise’ | ‘piecewise’ | ‘piecewise’ |
| Learn Rate Drop Factor | 0.4 | 0.7 | 0.7 | 0.5 |
| Learn Rate Drop Period | 4 | 10 | 10 | 8 |
| Max Epochs | 200 | 150 | 150 | 4 |
| Mini Batch Size | 16 | 64 | 64 | 8 |
| Verbose | 1 | 1 | 1 | 1 |
| Verbose Frequency (iterations) | 50 | 50 | 50 | 100 |
| Gradient Threshold Method | ‘L2norm’ | ‘L2norm’ | ‘L2norm’ | ‘L2norm’ |
| Gradient Threshold | Inf | Inf | Inf | Inf |
| L2 Regularization | 1 × | 1 × | 1 × |
|
Throughout the training, the stochastic gradient descent with momentum (SGDM) and Adam's optimizers were used. A random collection of data samples is used in the SGDM technique. In our study, the SGDM's momentum value is 0.9, which helps in updating the previous iteration's step to the current iteration. The three fundamental options for calculating the learning rate are schedule, period, and factor. The timetable is classified as 'piecewise.' Every epoch, a given learning rate factor updates the learning rate, whereas the learn rate drop period is the number of epochs used to reduce the learning rate.
The mini-batch size is a parameter that specifies a portion of a training set and may be used to compute the gradient and update the weights of the loss function. Another type of parameter, such as verbose and verbose frequency, is supplied for the progress of the training process and its iterations. The definition of being verbose is 1, but the frequency of being verbose is 50. The gradient threshold and the gradient threshold technique are two more elements to consider while training a model. For gradient values that exceed the threshold, the gradient threshold is a clipping technique. In our experiment, the gradient threshold is Inf, and the threshold approach is L2norm. The major purpose of this parameter is to make the weights as little as possible, but it does not make them zero, resulting in a non-sparse solution. The RESCOVIDTCNNet is trained using the same parameters as Resnet50-TCN, with the difference that the X-ray images are sent to EWT before the deep learning model is fed.
4.4. Performance measurements
To determine the performance of the proposed methods, quantitative and qualitative analysis is applied. They are briefly explained below.
-
•
Quantitative Analysis
In this work, we have used accuracy (ACC), true positive (TP), true positive rate (TPR), false positive (FP), false-positive rate (FPR), kappa (K) (Nair et al. 2010), Matthews correlation coefficient (MCC), F1-score (F1), receiver operating statistic (ROC), precision, recall, and precision-recall curve (PRC) performance parameters to evaluate our proposed model. Each fold's ROC curves were drawn to visually represent the performance (Yan et al. 2020). Table 4 shows the performance obtained for five-fold cross-validation obtained for Inception v3 and Resnet50 deep learning models obtained using MLP and SVM classifiers.
Table 4.
Performance parameters obtained using Inception V3 and Resnet50 models.
| Performance Measurement | Inception V3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MLP | SVM | |||||||||
| 1stFold | 2ndFold | 3rdFold | 4thFold | 5thFold | 1stFold | 2ndFold | 3rdFold | 4thFold | 5thFold | |
| Accuracy | 97.71 | 95.91 | 96.51 | 96.51 | 94.81 | 97.61 | 94.42 | 96.71 | 95.41 | 94.31 |
| True Positive | 980 | 962 | 967 | 967 | 950 | 979 | 947 | 969 | 956 | 945 |
| False Positive | 23 | 41 | 35 | 35 | 52 | 24 | 56 | 33 | 46 | 57 |
| Kappa | 0.966 | 0.939 | 0.948 | 0.948 | 0.922 | 0.964 | 0.916 | 0.951 | 0.931 | 0.915 |
| TP Rate | 0.977 | 0.959 | 0.965 | 0.965 | 0.948 | 0.976 | 0.944 | 0.967 | 0.954 | 0.943 |
| FP Rate | 0.011 | 0.020 | 0.017 | 0.017 | 0.026 | 0.012 | 0.028 | 0.016 | 0.023 | 0.028 |
| Precision | 0.977 | 0.961 | 0.967 | 0.966 | 0.953 | 0.976 | 0.948 | 0.968 | 0.955 | 0.949 |
| Recall | 0.977 | 0.959 | 0.965 | 0.965 | 0.948 | 0.976 | 0.944 | 0.967 | 0.954 | 0.943 |
| F1-measure | 0.977 | 0.959 | 0.965 | 0.965 | 0.948 | 0.976 | 0.944 | 0.967 | 0.954 | 0.943 |
| MCC | 0.966 | 0.940 | 0.949 | 0.948 | 0.925 | 0.964 | 0.918 | 0.951 | 0.931 | 0.918 |
| ROC Area | 0.999 | 0.993 | 0.997 | 0.992 | 0.998 | 0.999 | 0.969 | 0.983 | 0.974 | 0.970 |
| PRC Area | 0.997 | 0.985 | 0.993 | 0.989 | 0.996 | 0.997 | 0.920 | 0.955 | 0.933 | 0.921 |
| Performance Measurement | Resnet50 | |||||||||
| MLP | SVM | |||||||||
| 1stFold | 2ndFold | 3rdFold | 4thFold | 5thFold | 1stFold | 2ndFold | 3rdFold | 4thFold | 5thFold | |
| Accuracy | 98.006 | 96.809 | 97.904 | 97.604 | 98.403 | 97.607 | 96.909 | 97.505 | 96.706 | 98.004 |
| True Positive | 983 | 971 | 981 | 978 | 986 | 979 | 972 | 977 | 969 | 982 |
| False Positive | 20 | 32 | 21 | 24 | 16 | 24 | 31 | 25 | 33 | 20 |
| Kappa | 0.9701 | 0.9521 | 0.9686 | 0.9641 | 0.976 | 0.9641 | 0.9536 | 0.9626 | 0.9506 | 0.9701 |
| TP Rate | 0.980 | 0.968 | 0.979 | 0.976 | 0.984 | 0.976 | 0.969 | 0.975 | 0.967 | 0.980 |
| FP Rate | 0.010 | 0.016 | 0.010 | 0.012 | 0.008 | 0.012 | 0.015 | 0.012 | 0.016 | 0.010 |
| Precision | 0.980 | 0.969 | 0.979 | 0.976 | 0.984 | 0.976 | 0.970 | 0.975 | 0.968 | 0.980 |
| Recall | 0.980 | 0.968 | 0.979 | 0.976 | 0.984 | 0.976 | 0.969 | 0.975 | 0.967 | 0.980 |
| F1-measure | 0.980 | 0.968 | 0.979 | 0.976 | 0.984 | 0.976 | 0.969 | 0.975 | 0.967 | 0.980 |
| MCC | 0.970 | 0.953 | 0.969 | 0.964 | 0.976 | 0.964 | 0.954 | 0.963 | 0.951 | 0.970 |
| ROC Area | 0.998 | 0.996 | 0.996 | 0.998 | 0.999 | 0.987 | 0.979 | 0.987 | 0.981 | 0.991 |
| PRC Area | 0.997 | 0.994 | 0.994 | 0.996 | 0.999 | 0.965 | 0.952 | 0.964 | 0.954 | 0.973 |
The performance of the MLP classifier using the Inception v3 model is greater than the SVM for folds 1, 2, 4, and 5, but the SVM classifier performed better than MLP for fold 3. On the other side, MLP with Resnet50 outperformed the SVM for 1, 3, 4, and 5 folds, whereas SVM outperformed MLP for fold2 only.
For Resnet50-TCN and our proposed RESCOVIDTCNNet, Table 5 illustrates the performance metrics achieved utilizing five-folds with MLP and SVM classifiers. It can be seen that the performance of the MLP classifier with the Resnet50 model yielded better results than the SVM for folds 1, 2, and 3, while the SVM classifier performed better than MLP for folds 4 and 5.
Table 6.
presents the results of the training and validation accuracies obtained for the five-fold cross-validation of RESCOVIDTCNNet. It can be noted from the table that they are consistent, and validation follows the training highlighting the proper training of the system. Table 6: Training and validation accuracies obtained from Fold 1 to Fold 5 of the RESCOVIDTCNNet.
| Folds | Training Accuracy (%) | Validation Accuracy (%) |
|---|---|---|
| Fold 1 | 99.890% | 99.700% |
| Fold 2 | 99.553% | 99.251% |
| Fold 3 | 99.457% | 99.241% |
| Fold 4 | 99.012% | 98.803% |
| Fold 5 | 99.912% | 99.850% |
Table 5.
Performance parameters obtained using Resnet50-TCN and proposed model.
| Performance Measurements | Resnet50-TCN | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MLP | SVM | |||||||||
| 1stFold | 2ndFold | 3rdFold | 4thFold | 5thFold | 1stFold | 2ndFold | 3rdFold | 4thFold | 5thFold | |
| Accuracy | 99.700 | 99.401 | 99.401 | 98.902 | 98.902 | 99.401 | 98.503 | 98.503 | 99.201 | 99.201 |
| True Positive | 1000 | 996 | 996 | 991 | 991 | 997 | 987 | 987 | 994 | 994 |
| False Positive | 3 | 6 | 6 | 11 | 11 | 6 | 15 | 15 | 8 | 8 |
| Kappa | 0.9955 | 0.991 | 0.991 | 0.9835 | 0.9835 | 0.991 | 0.997 | 0.977 | 0.988 | 0.988 |
| TP Rate | 0.997 | 0.994 | 0.994 | 0.989 | 0.989 | 0.994 | 0.985 | 0.985 | 0.992 | 0.992 |
| FP Rate | 0.001 | 0.003 | 0.003 | 0.005 | 0.005 | 0.003 | 0.007 | 0.007 | 0.004 | 0.004 |
| Precision | 0.997 | 0.994 | 0.994 | 0.989 | 0.989 | 0.994 | 0.985 | 0.985 | 0.992 | 0.992 |
| Recall | 0.997 | 0.994 | 0.994 | 0.989 | 0.989 | 0.994 | 0.985 | 0.985 | 0.992 | 0.992 |
| F1-measure | 0.997 | 0.994 | 0.994 | 0.989 | 0.989 | 0.994 | 0.985 | 0.985 | 0.992 | 0.992 |
| MCC | 0.996 | 0.991 | 0.991 | 0.984 | 0.984 | 0.991 | 0.978 | 0.978 | 0.988 | 0.988 |
| ROC Area | 1.000 | 0.999 | 0.999 | 1.000 | 1.000 | 0.997 | 0.992 | 0.992 | 0.996 | 0.996 |
| PRC Area | 1.000 | 0.999 | 0.999 | 1.000 | 1.000 | 0.992 | 0.977 | 0.977 | 0.989 | 0.989 |
| Performance Measurement | RESCOVIDTCNNet | |||||||||
| MLP | SVM | |||||||||
| 1stFold | 2ndFold | 3rdFold | 4thFold | 5thFold | 1stFold | 2ndFold | 3rdFold | 4thFold | 5thFold | |
| Accuracy | 99.700 | 99.501 | 99.501 | 98.902 | 99.900 | 99.700 | 99.201 | 99.201 | 98.902 | 99.800 |
| True Positive | 1000 | 997 | 997 | 991 | 1001 | 1000 | 994 | 994 | 991 | 1002 |
| False Positive | 3 | 5 | 5 | 11 | 1 | 3 | 8 | 8 | 11 | 0 |
| Kappa | 0.9955 | 0.9925 | 0.9925 | 0.9835 | 0.9983 | 0.995 | 0.988 | 0.988 | 0.9835 | 0.997 |
| TP Rate | 0.997 | 0.995 | 0.995 | 0.989 | 0.999 | 0.997 | 0.992 | 0.992 | 0.989 | 0.998 |
| FP Rate | 0.001 | 0.002 | 0.002 | 0.005 | 0.000 | 0.001 | 0.004 | 0.004 | 0.005 | 0.001 |
| Precision | 0.997 | 0.995 | 0.995 | 0.989 | 0.999 | 0.997 | 0.992 | 0.992 | 0.989 | 0.998 |
| Recall | 0.997 | 0.995 | 0.995 | 0.989 | 0.999 | 0.997 | 0.992 | 0.992 | 0.989 | 0.998 |
| F1-measure | 0.997 | 0.995 | 0.995 | 0.989 | 0.999 | 0.997 | 0.992 | 0.992 | 0.989 | 0.998 |
| MCC | 0.996 | 0.993 | 0.993 | 0.984 | 0.999 | 0.996 | 0.988 | 0.988 | 0.984 | 0.998 |
| ROC Area | 1.000 | 0.999 | 0.999 | 1.000 | 0.999 | 0.999 | 0.996 | 0.996 | 0.993 | 1.000 |
| PRC Area | 1.000 | 0.999 | 0.999 | 0.999 | 0.998 | 0.997 | 0.989 | 0.989 | 0.983 | 1.000 |
On the other hand, it can be noted from Table 7 that MLP performance with Resnet50 is the same as that of SVM for folds 1 and 3, whereas the performance of MLP is higher than SVM for folds 2, 3, and 5. Table 7 shows the performance matrices obtained with an average of the five-fold cross-validation strategy for four deep learning models with MLP and SVM classifiers. It can be realized from the table that our proposed RESCOVIDTCNNet surpassed other deep learning models using an MLP classifier.
Table 7.
Summary of performances obtained with an average of five-fold cross-validation strategy for four deep learning models with MLP and SVM classifiers.
| Classifiers / Models |
Average Five Fold Cross-Validation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | TP | FP | K | TPR | FPR | P | R | F1 | MCC | ROC | PRC | ||
| MLP | Inception V3 | 96.288 | 4826 | 186 | 0.944 | 0.962 | 0.018 | 0.964 | 0.962 | 0.962 | 0.945 | 0.995 | 0.992 |
| Resnet50 | 97.745 | 4899 | 113 | 0.966 | 0.977 | 0.011 | 0.977 | 0.977 | 0.977 | 0.966 | 0.997 | 0.996 | |
| Resnet50 TCN | 99.261 | 4974 | 37 | 0.988 | 0.992 | 0.003 | 0.992 | 0.992 | 0.992 | 0.989 | 0.999 | 0.999 | |
| Proposed | 99.500 | 4987 | 25 | 0.992 | 0.995 | 0.002 | 0.995 | 0.995 | 0.995 | 0.993 | 0.999 | 0.999 | |
| SVM | Inception V3 | 95.690 | 4796 | 216 | 0.935 | 0.956 | 0.021 | 0.959 | 0.956 | 0.956 | 0.936 | 0.979 | 0.945 |
| Resnet50 | 97.346 | 4879 | 133 | 0.960 | 0.973 | 0.013 | 0.973 | 0.973 | 0.973 | 0.960 | 0.985 | 0.961 | |
| Resnet50 TCN | 98.961 | 4959 | 52 | 0.988 | 0.989 | 0.005 | 0.989 | 0.989 | 0.989 | 0.984 | 0.994 | 0.984 | |
| Proposed | 99.360 | 4980 | 32 | 0.990 | 0.993 | 0.003 | 0.993 | 0.993 | 0.993 | 0.990 | 0.996 | 0.991 | |
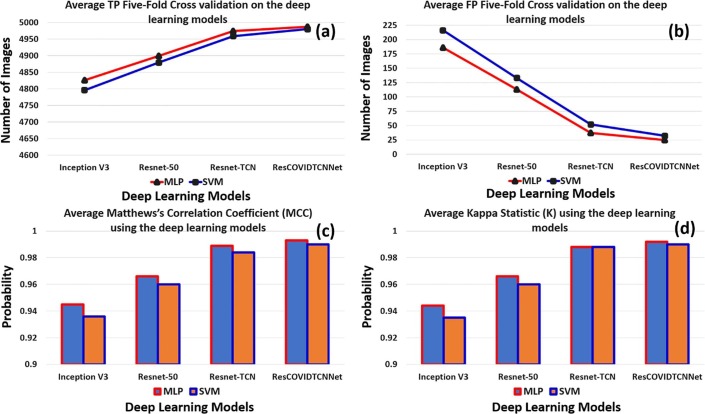
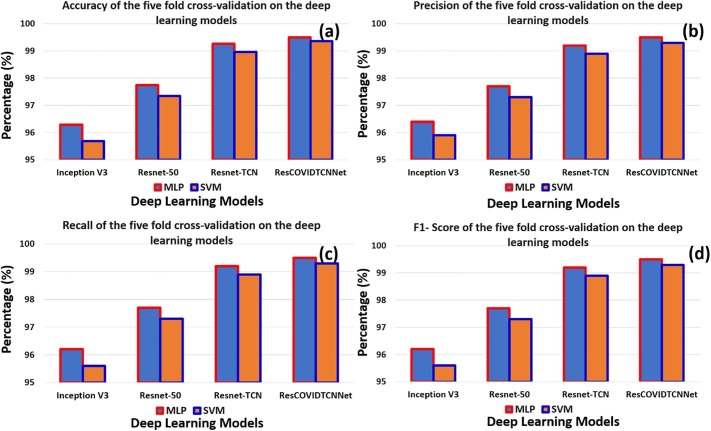
Fig. 6 shows a graphical representation of the performance of four deep learning models using MLP and SVM classifiers. Fig. 6 (a) and (b) indicate the number of correctly and incorrectly classified instances. The red curve represents the MLP accuracy with four deep learning models, while the blue curve represents the SVM accuracy with the deep learning models. Fig. 7 provides better visualization of the average percentages of five-folds using bar graphs. The purple and the orange bars represent the MLP and SVM performance with deep learning models, respectively. Fig. 7 (a) and (b) show the accuracy and precision of the deep learning models using MLP and SVM, respectively. Fig. 7 (c) and (d) describe the Recall and the F1-score of the deep learning models, respectively. It can be seen that the proposed model has the highest performance of the rest of the models.
Fig. 6.
(a) and (b) show the number of correctly and incorrectly classified images using five-fold cross-validation with various deep learning models, (c) and (d) describe the MCC and K values obtained with five-fold cross-validation using different deep learning models.
Fig. 7.
(a) and (b) indicate the accuracy and the precision obtained using five-fold cross-using validation for various deep learning models, (c) and (d) shows the recall and F1-score obtained using five-fold cross-validation strategy for various deep learning models.
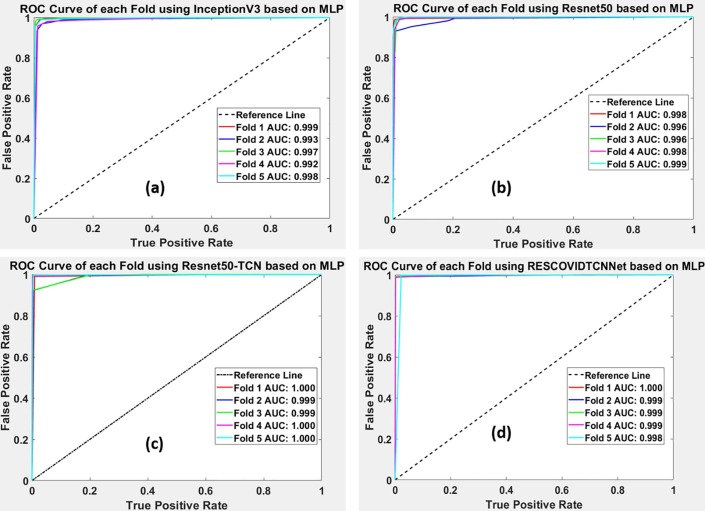
ROC is used to predict the probability of the outcome based on the True positive rate (TPR) and False Positive rate (FPR). Therefore, it is essential to represent the ROC and AUC in each fold to show the results of all five- folds. Fig. 8 represents the ROCs obtained using the deep learning models with the highest performance classifier (MLP). Fig. 8 (a) and (b) represent the results obtained from five-fold cross-validation for the InceptionV3 and Resnet50 models. Fig. 8 (c) and (d) describe the ROCs of Resnet50-TCN and RESCOVIDTCNNet.
Fig. 8.
(a) and (b) represent the ROC obtained with five-fold cross-validation using InceptionV3 and Resnet-50, (c) and (d) describe the ROC for Resnet-50 with TCN and the proposed model using MLP.
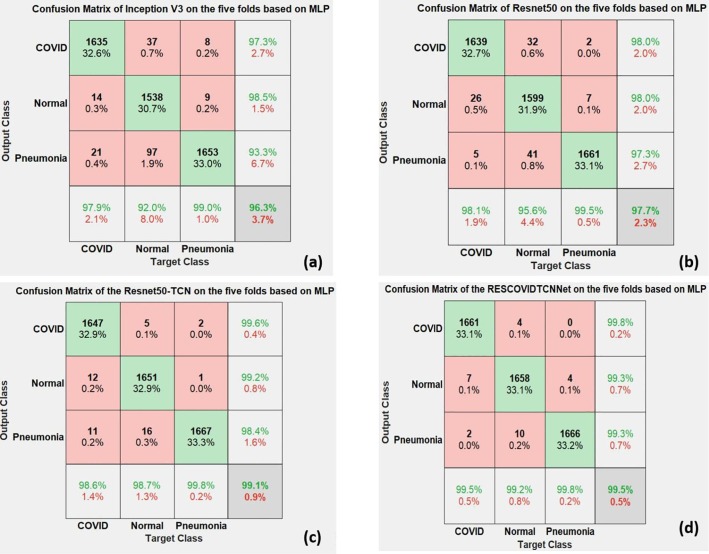
Finally, the confusion matrix of the five folds is represented in Fig. 9 . The confusion matrices obtained for each of the four deep learning models are shown in Figs—9 (a, b, c, and d). The green cells in each matrix represent the number of correctly classified instances during the five-fold cross-validation strategy for each class. In contrast, the red cells represent the misclassified instances. It can be noted from the confusion matrices that Inception V3 showed the lowest accuracy performance based on MLP. At the same time, the proposed RESCOVIDTCNNet achieved the highest accuracy performance with fewer misclassified instances in each class.
-
•
Qualitative Analysis
Fig. 9.
Confusion matrices obtained for four deep learning models used.
This part presents the details of the folds and accuracy distribution in each fold of the deep learning models. The main aim of the qualitative analysis is to provide a good understanding of the results obtained by the deep learning models. Therefore, three different diagrams are illustrated to present the accuracy and detailed results of the deep learning models. The first diagram is the Taylor diagram which indicates several appropriate representations of the systems (Taylor, 2005). Furthermore, it provides a statistical summary of how the models or the system match each other in correlation and the standard deviation. Fig. 10 shows the Taylor diagram designed for the deep learning models. It can be seen from the figure that the highest prediction performance can be realized from the RESCOVIDTCNNet as it has a correlation coefficient of 0.999 and a standard deviation of 0.1.
Fig. 10.
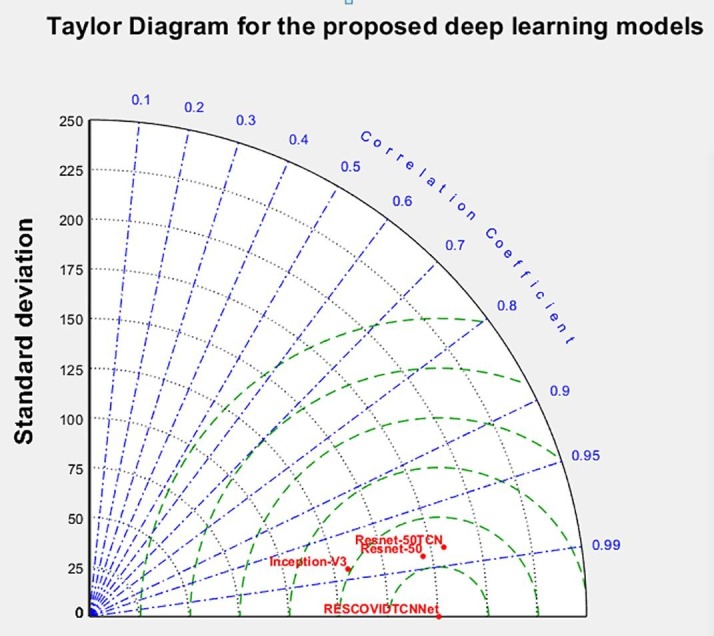
Visual representation of performances obtained with various models using Taylor diagram.
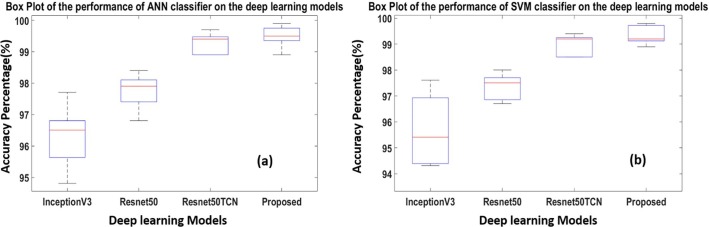
In contrast, Inception-V3 has the lowest performance, with a correlation coefficient of 0.971 and a standard deviation of 19. The second diagram presented is the box plot diagram which better indicates data spread. Our study is used to represent how the accuracies of the folds are spread out using the deep learning models with classifiers.
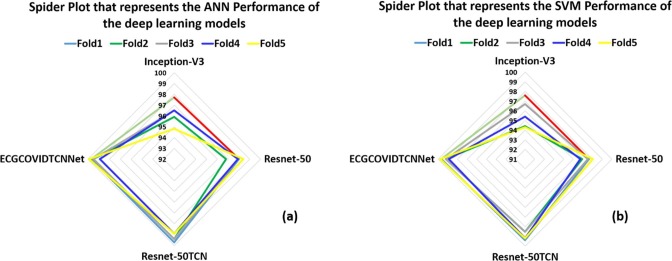
This diagram is drawn for the deep learning model to represent the results of the five-fold cross-validation. In other words, Fig. 11 (a) shows the results of the deep learning models obtained from the five-fold cross-validation using the ANN (MLP) classifier, while Fig. 11 (b) shows the results obtained with the SVM classifier. The third diagram is the spider plot which presents a visual tool that can be used to organize the data logically (Fig. 12 ). It is also defined as a tool that organizes concepts based on space, color, and images. This diagram is drawn in the form of a diamond shape to represent the performance of the deep learning models. Fig. 12 (a) shows the results of the deep learning models based on the folds in five-fold cross-validation using the ANN classifier, whereas (b) illustrates the results of the SVM classifier. It can be manifested clearly that Resnet-50TCN and RESCOVIDTCNNet have approached high accuracies in contrast with the inception-V3 and Resnet-50.
Fig. 11.
Box plot obtained with five- folds cross-validation using deep learning models and classifier (a) ANN & (b) SVM.
Fig. 12.
Spider plot obtained for deep learning model with five-fold cross-validation and classifier: (a) ANN, & (b) SVM.
4.5. Computational complexity
The complexity of four deep learning models depends on several factors: processing or the learning time taken during the training of each of the deep learning models. In addition to this, the number of layers and filters in each layer can result in variances in the complexity between different deep learning models (Gudigar et al., 2021, Bargshady et al., 2022). In our study, the complexity is evaluated based on the learning time, feature extraction from the fully connected layer, and classification of test data in each fold. We have employed five-fold cross-validation, whereby the training time is multiplied by 5. The training time of each Inception-V3, Resnet-50, Resnet-50-TCN, and RESCOVIDTCNNet are 3.5, 1.5, 1.75, and 1.85 h, respectively. The time required to extract the features from the Inception-V3, Resnet-50, Resnet-50-TCN, and RESCOVIDTCNNet are 5.5, 3.2, 3.5, and 3.7 min, respectively. Finally, the time for the classification using the main classifier is the same for all the deep learning models.
5. Discussion
This paper presents various deep learning methodologies used to classify three classes: COVID-19, normal, and pneumonia using X-ray images. The methodology consists of four main stages: data acquisition, pre-processing, feature extraction, and classification. During the data acquisition phase, 5012 X-ray chest images (1670 COVID, 1672 normal, and 1670 pneumonia). Images are filtered using the empirical wavelet transform (EWT) process in the pre-processing stage.
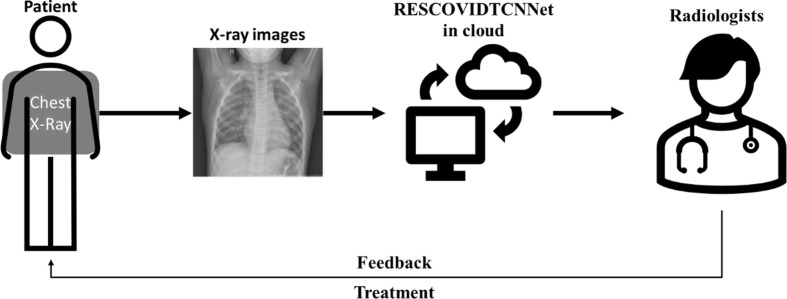
The application of our proposed approach is visualized in Fig. 13 . The proposed RESCOVIDTCNNet with MLP classifier can be placed in the cloud, with an abundance of memory space to store the proposed model and pre-process the chest X-ray images for the classification task. The radiologist, who is the end-user of our proposed model, will be notified of the classification result so that he can confirm it. If a patient is classified as positive for COVID, the radiologist can take immediate action by informing the patient to quarantine him or herself and develop a suitable treatment regime for the patient.
Fig. 13.
Block diagram of cloud-computing for diagnosis of COVID.
It is important to know that the X-ray images do not contain noises. Still, the EWT improves the quality and enhances the proposed model’s classification accuracy. The next stage is the feature extraction, and in this stage, two main pre-trained models InceptionV3 and Resnet50, are used (Barua et al., 2021, Sharifrazi et al., 2021) It was observed that the highest classification accuracy was obtained when fused with the temporal convolutional neural network (TCN). The Resnet50 showed a higher performance than Inception V3. Therefore, it was chosen to fuse with TCN (Resnet50-TCN), also known as RESCOVIDTCNNet. As a result, the classification accuracy has improved from 97% to 99%. Finally, the last classification stage involves two main classifiers, namely ANN (MLP) and SVM, and the highest classification performance was achieved using MLP (Alizadehsani et al., 2021, Shoeibi et al., 2020).
6. Advantages of the proposed model:
-
•
It has enhanced the quality of the X-ray images using the EWT algorithm to reach higher SNR.
-
•
It has improved the diagnosis performance of the novel RESCOVIDTCNNet using transfer learning models.
-
•
Achieved the highest classification performance compared to the other related works developed using Chest X-ray images.
The limitation of our work is that the number of X-ray images used to develop the model is not large enough. Also, our proposed model misclassified 30 images from all folds.
7. Conclusions
Deep learning models based on the CNN architecture are provided in this paper to automatically detect the symptoms of COVID, normal, and pneumonia. Data capture, pre-processing, feature extraction, and classification are the four primary aspects of the suggested technique. The database of healthy, pneumonia, and COVID chest X-ray pictures of both genders and diverse age groups is employed in the first phase (data collecting). The X-ray pictures are filtered using EWT in the second phase (pre-processing), which provided an efficient SNR value to improve the original X-ray images. In the third phase (feature extraction), two transfer learning models (Inception-V3 and Resnet50) and one deep learning model developed based on the ensemble of Resnet50 and TCN are used to compare our proposed RESCOVIDTCNNet model, which is an ensemble of EWT, Resnet50, and TCN. Finally, the fourth phase (classification) uses two classifiers: MLP and SVM. Our results show that RESCOVIDTCNNet exhibited the highest accuracy of 99.5% in classifying the three classes. The main limitation of this work is that the number of images used in each class is not huge. Hence, we plan to validate our generated model using more images taken from diverse races and age groups in the future. Also, we intend to test our proposed system to detect other lung-related diseases like chronic obstructive pulmonary disease (COPD), asthma, and lung cancer. Future work to develop a deep learning model to perform classification tasks instead of a machine learning model like SVM and MLP used in this study will also be considered.
CRediT authorship contribution statement
El-Sayed. A. El-Dahshan: Conceptualization, Methodology, Writing – original draft, Visualization, Writing – review & editing, Supervision. Mahmoud. M. Bassiouni: Methodology, Writing – original draft, Writing – review & editing. Ahmed Hagag: Writing – original draft. Ripon K. Chakrabortty: Writing – review & editing, Visualization. Huiwen. Loh: Writing – review & editing, Visualization. Rajendra U. Acharya: Writing – review & editing, Visualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Abraham B., Nair M.S. Computer-aided detection of COVID-19 from X-ray images using multi-CNN and Bayesnet classifier. Biocybernetics and biomedical engineering. 2020;40(4):1436–1445. doi: 10.1016/j.bbe.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K.N., Mohammadi A. Covid-caps: A capsule network-based framework for identification of covid-19 cases from x-ray images. Pattern Recognition Letters. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahsan, M. M., Gupta, K. D., Islam, M. M., Sen, S., Rahman, M., & Hossain, M. S. (2020). Study of different deep learning approach with explainable ai for screening patients with COVID-19 symptoms: Using ct scan and chest x-ray image dataset. arXiv preprint arXiv:2007.12525.
- Aksan, E., & Hilliges, O. (2019). STCN: Stochastic temporal convolutional networks. arXiv preprint arXiv:1902.06568.
- Albahli S., Ayub N., Shiraz M. Coronavirus disease (COVID-19) detection using X-ray images and enhanced DenseNet. Applied Soft Computing. 2021;110 doi: 10.1016/j.asoc.2021.107645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alizadehsani R., Alizadeh Sani Z., Behjati M., Roshanzamir Z., Hussain S., Abedini N., Islam S.M.S. Risk factors prediction, clinical outcomes, and mortality in COVID-19 patients. Journal of medical virology. 2021;93(4):2307–2320. doi: 10.1002/jmv.26699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almalki, Y. E., Qayyum, A., Irfan, M., Haider, N., Glowacz, A., Alshehri, F. M., & Rahman, S. (2021). A Novel Method for COVID-19 Diagnosis Using Artificial Intelligence in Chest X-ray Images. In Healthcare , 9(5), 522). Multidisciplinary Digital Publishing Institute. https://doi.org/10.3390/healthcare9050522. [DOI] [PMC free article] [PubMed]
- Apostolopoulos I.D., Mpesiana T.A. Covid-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asif S., Wenhui Y., Jin H., Jinhai S. in 2020 IEEE 6th International Conference on Computer and Communications (ICCC) 2020. Classification of COVID-19 from Chest X-ray images using Deep Convolutional Neural Network; pp. 426–433. [DOI] [Google Scholar]
- Bai, S., Kolter, J. Z., & Koltun, V. (2018). An empirical evaluation of generic convolutional and recurrent networks for sequence modeling. arXiv preprint arXiv:1803.01271..
- Balaha H.M., El-Gendy E.M., Saafan M.M. CovH2SD: A COVID-19 detection approach based on Harris Hawks Optimization and stacked deep learning. Expert Systems with Applications. 2021;186 doi: 10.1016/j.eswa.2021.115805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargshady G., Zhou X., Barua P.D., Gururajan R., Li Y., Acharya U.R. Application of CycleGAN and transfer learning techniques for automated detection of COVID-19 using X-ray images. Pattern Recognition Letters. 2022;153:67. doi: 10.1016/j.patrec.2021.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua P.D., Muhammad Gowdh N.F., Rahmat K., Ramli N., Ng W.L., Chan W.Y., Acharya U.R. Automatic COVID-19 detection using exemplar hybrid deep features with X-ray images. International journal of environmental research and public health. 2021;18(15):8052. doi: 10.3390/ijerph18158052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudrioua M.S. COVID-19 detection from chest X-ray images using CNNs models: Further evidence from Deep Transfer Learning. The University of Louisville Journal of Respiratory Infections. 2020;4(1) doi: 10.18297/jri/vol4/iss1/53. [DOI] [Google Scholar]
- Breuel, T. M. (2015). Benchmarking of LSTM networks. arXiv preprint arXiv:1508.02774.
- Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Computer Methods and Programs in Biomedicine. 2020;196 doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, K., Van Merriënboer, B., Gulcehre, C., Bahdanau, D., Bougares, F., Schwenk, H., & Bengio, Y. (2014). Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv preprint arXiv:1406.1078.
- Chowdhury M.E., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., Islam M.T. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- Cohen J. P. (2020, 20 July 2021). COVID-19 Image Data Collection. Available: https://github.com/ieee8023/COVID-chestxray-dataset.
- Cortes C., Vapnik V. Support-vector networks. Machine learning. 1995;20:273–297. [Google Scholar]
- Das N.N., Kumar N., Kaur M., Kumar V., Singh D. Automated deep transfer learning-based approach for detection of COVID-19 infection in chest X-rays. Irbm. 2020 doi: 10.1016/j.irbm.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A.K., Kalam S., Kumar C., Sinha D. TLCoV-An automated Covid-19 screening model using Transfer Learning from chest X-ray images. Chaos, Solitons & Fractals. 2021;144 doi: 10.1016/j.chaos.2021.110713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demir F. DeepCoroNet: A deep LSTM approach for automated detection of COVID-19 cases from chest Xray images. Applied Soft Computing. 2021;103 doi: 10.1016/j.asoc.2021.107160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Dong W., Socher R., Li L.J., Li K., Fei-Fei L. June). Imagenet: A large-scale hierarchical image database. Ieee; 2009. pp. 248–255. [DOI] [Google Scholar]
- Fan Y., Liu J., Yao R., Yuan X. COVID-19 Detection from X-ray Images using Multi-Kernel-Size Spatial-Channel Attention Network. Pattern Recognition. 2021;199 doi: 10.1016/j.patcog.2021.108055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilles J. Empirical wavelet transform. IEEE transactions on signal processing. 2013;61(16):3999–4010. doi: 10.1109/TSP.2013.2265222. [DOI] [Google Scholar]
- Gudigar A., Raghavendra U., Nayak S., Ooi C.P., Chan W.Y., Gangavarapu M.R., Acharya U.R. Role of Artificial Intelligence in COVID-19 Detection. Sensors. 2021;21(23):8045. doi: 10.3390/s21238045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K., Zhang X., Ren S., Sun J. In Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [DOI] [Google Scholar]
- Heidari M., Mirniaharikandehei S., Khuzani A. Z., Danala G., Qiu Y., & Zheng B., (2020). Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. International journal of medical informatics, 144, p. 104284, 2020. doi: https://doi.org/10.1016/j.ijmedinf.2020.104284. [DOI] [PMC free article] [PubMed]
- Hochreiter S., Schmidhuber J. Long short-term memory. Neural computation. 1997;9(8):1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- Huff H.V., Singh A. Asymptomatic transmission during the coronavirus disease 2019 pandemic and implications for public health strategies. Clinical Infectious Diseases. 2020;71:2752–2756. doi: 10.1093/cid/ciaa654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfan M., Iftikhar M.A., Yasin S., Draz U., Ali T., Hussain S., Althobiani F. Role of Hybrid Deep Neural Networks (HDNNs), Computed Tomography, and Chest X-rays for the Detection of COVID-19. International Journal of Environmental Research and Public Health. 2021;18(6):3056. doi: 10.3390/ijerph18063056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismael A.M., Şengür A. Deep learning approaches for COVID-19 detection based on chest X-ray images. Expert Systems with Applications. 2021;164 doi: 10.1016/j.eswa.2020.114054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain G., Mittal D., Thakur D., Mittal M.K. A deep learning approach to detect Covid-19 coronavirus with X-ray images. Biocybernetics and biomedical engineering. 2020;40(4):1391–1405. doi: 10.1016/j.bbe.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin W., Dong S., Dong C., Ye X. Hybrid ensemble model for differential diagnosis between COVID-19 and common viral pneumonia by chest X-ray radiograph. Computers in biology and medicine. 2021;131 doi: 10.1016/j.compbiomed.2021.104252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Júnior D.A.D., da Cruz L.B., Diniz J.O.B., da Silva G.L.F., Junior G.B., Silva A.C., Gattass M. Automatic method for classifying COVID-19 patients based on chest X-ray images, using deep features and PSO-optimized XGBoost. Expert Systems with Applications. 2021;183 doi: 10.1016/j.eswa.2021.115452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthik R., Menaka R., Hariharan M. Learning distinctive filters for COVID-19 detection from chest X-ray using shuffled residual CNN. Applied Soft Computing. 2021;99 doi: 10.1016/j.asoc.2020.106744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermany D.S., Goldbaum M., Cai W., Valentim C.C., Liang H., Baxter S.L.…Zhang K. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- Khan M., Mehran M.T., Haq Z.U., Ullah Z., Naqvi S.R. Applications of artificial intelligence in COVID-19 pandemic: A comprehensive review. Expert Systems with Applications. 2021;185 doi: 10.1016/j.eswa.2021.115695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cun Y., Bengio Y., Hinton G. Deep learning. nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- Li Z., Zhao S., Chen Y., Luo F., Kang Z., Cai S., Li Y. A deep-learning-based framework for severity assessment of COVID-19 with CT images. Expert Systems with Applications. 2021;185 doi: 10.1016/j.eswa.2021.115616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Shelhamer E., Darrell T. In Proceedings of the IEEE conference on computer vision and pattern recognition. 2015. Fully convolutional networks for semantic segmentation; pp. 3431–3440. [DOI] [Google Scholar]
- Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. Journal of medical virology. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn H.M., Pan S.B., Kim P. A deep bidirectional GRU network model for biometric electrocardiogram classification based on recurrent neural networks. IEEE Access. 2019;7:145395–145405. doi: 10.1109/ACCESS.2019.2939947. [DOI] [Google Scholar]
- Mooney. P. (2020, 20 July 2021). Chest X-Ray Images (Pneumonia). Available: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia.
- Nair V., Hinton G.E. Icml. 2010. Rectified linear units improve restricted boltzmann machines. [Google Scholar]
- Nair, A., Srinivasan, P., Blackwell, S., Alcicek, C., Fearon, R., De Maria, A., ... & Silver, D. (2015). Massively parallel methods for deep reinforcement learning. arXiv preprint arXiv:1507.04296.
- Oord, A. V. D., Dieleman, S., Zen, H., Simonyan, K., Vinyals, O., Graves, A., ... & Kavukcuoglu, K. (2016). Wavenet: A generative model for raw audio. arXiv preprint arXiv:1609.03499.
- Ozcan T. A new composite approach for COVID-19 detection in X-ray images using deep features. Applied Soft Computing. 2021;111 doi: 10.1016/j.asoc.2021.107669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Computers in biology and medicine. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveen. (2020, 22 July 2020). Corona hack: chest X-ray-dataset Available: https://www.kaggle.com/praveengovi/coronahack-chest-xraydataset.
- Quan H., Xu X., Zheng T., Li Z., Zhao M., Cui X. DenseCapsNet: Detection of COVID-19 from X-ray images using a capsule neural network. Computers in biology and medicine. 2021;133 doi: 10.1016/j.compbiomed.2021.104399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman T., Khandakar A., Qiblawey Y., Tahir A., Kiranyaz S., Kashem S.B.A.…Chowdhury M.E. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Computers in biology and medicine. 2021;132 doi: 10.1016/j.compbiomed.2021.104319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmon J. & Farhadi A., YOLO9000: better, faster, stronger, in Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 7263-7271. doi:10.1109/CVPR.2017.690.
- Rehman N.U., Zia M.S., Meraj T., Rauf H.T., Damaševičius R., El-Sherbeeny A.M., El-Meligy M.A. A Self-Activated CNN Approach for Multi-Class Chest-Related COVID-19 Detection. Applied Sciences. 2021;11(19):9023. doi: 10.3390/app11199023. [DOI] [Google Scholar]
- Sharifrazi D., Alizadehsani R., Roshanzamir M., Joloudari J.H., Shoeibi A., Jafari M., Acharya U.R. Fusion of convolution neural network, support vector machine and Sobel filter for accurate detection of COVID-19 patients using X-ray images. Biomedical Signal Processing and Control. 2021;68 doi: 10.1016/j.bspc.2021.102622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoeibi, A., Khodatars, M., Alizadehsani, R., Ghassemi, N., Jafari, M., Moridian, P., ... & Shi, P. (2020). Automated detection and forecasting of covid-19 using deep learning techniques: A review. arXiv preprint arXiv:2007.10785.
- Sobel I. & Feldman G., (1968) A 3x3 isotropic gradient operator for image processing, a talk at the Stanford Artificial Project in, 271-272.
- Sultan H.H., Salem N.M., Al-Atabany W. Multi-classification of brain tumor images using deep neural network. IEEE Access. 2019;7:69215–69225. doi: 10.1109/ACCESS.2019.2919122. [DOI] [Google Scholar]
- Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. In Proceedings of the IEEE conference on computer vision and pattern recognition. 2016. Rethinking the inception architecture for computer vision; pp. 2818–2826. [DOI] [Google Scholar]
- Talo. M. (2020, 20 July 2021). COVID-19 Dataset. Available: https://github.com/muhammedtalo/COVID-19.
- Taylor K.E. Taylor diagram primer. Work. Pap. 2005:1–4. [Google Scholar]
- Toraman S., Alakus T.B., Turkoglu I. Convolutional capsnet: A novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks, Chaos. Solitons & Fractals. 2020;140 doi: 10.1016/j.chaos.2020.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- X. Wang Y. Peng L. Lu Z. Lu M. Bagheri R.M. Summers Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases In Proceedings of the IEEE conference on computer vision and pattern recognition 2017 2097 2106.
- World Health Organization 17 October 2021) WHO situation report. 2021 Available: https://covid19.who.int/ Last accessed (19-11-2021).
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xingjian, S. H. I., Chen, Z., Wang, H., Yeung, D. Y., Wong, W. K., & Woo, W. C. (2015). Convolutional LSTM network: A machine learning approach for precipitation nowcasting. In Advances in neural information processing systems (pp. 802-810).
- Yan J., Mu L., Wang L., Ranjan R., Zomaya A.Y. Temporal convolutional networks for the advance prediction of ENSO. Scientific reports. 2020;10(1):1–15. doi: 10.1038/s41598-020-65070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Xie, Y., Li, Y., Shen, C., & Xia, Y. (2020). Covid-19 screening on chest x-ray images using deep learning based anomaly detection, arXiv preprint arXiv:2003, 27, 12338.