Abstract
Background
Knowing how long SARS-CoV-2-positive individuals can remain infective is crucial for the design of infection prevention and control strategies. Viral culture is the gold standard for detecting an active-replicative virus and evaluating its infectious potential.
Objective
To assess the correlation of SARS-CoV-2 infectivity with the number of days from symptom onset and the Ct value, using culture as a reference method. Also, to describe a detailed protocol for SARS-CoV-2 culture and immunofluorescence confirmation based on our experience with other respiratory viruses.
Study design
100 consecutive respiratory samples positive for SARS-CoV-2 by RT-PCR from different subjects were inoculated into VERO E6 cells.
Results
Viral isolation was successful in 58% of samples. The median number of days from symptom onset for culture-positive samples was 2, and 15 for culture-negative samples. Six positive cultures were obtained in patients ≥14 days after symptom onset, all of whom were immunocompromised or with severe COVID-19. The mean Ct value was 12.64 units higher in culture-negative than in culture-positive samples. The probability of successfully isolating SARS-CoV-2 in samples with a Ct value <22 was 100%, decreasing to 3.1% when >27.
Conclusions
Our findings show a significant positive correlation between the probability of isolating SARS-CoV-2 in culture, fewer days of symptoms and a lower RT-PCR Ct value. SARS-CoV-2 infectivity lasts no more than 14 days from symptom onset in immunocompetent individuals. In contrast, in immunocompromised patients or those with severe COVID-19 infectivity may remain after 14 days. Ct value <22 always indicates infectivity.
Keywords: SARS-CoV-2, COVID-19, Viral culture, Viral isolation, Infectivity, VERO E6 cells
1. Background
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a zoonotic enveloped RNA virus, responsible for coronavirus infectious disease 2019 (COVID-19), which emerged in Wuhan, China, in late 2019 and quickly spread worldwide, causing a global pandemic [1]. Vaccination, early diagnosis, contact tracing and isolation of suspected and confirmed cases are crucial for pandemic control [2,3].
Reverse transcription polymerase chain reaction (RT-PCR) in respiratory samples is the most sensitive and the most frequently used method for COVID-19 diagnosis and in infection control precautions [4, 5]. Long-term shedding of viral RNA (≥14 days from symptom onset) has been reported in COVID-19 patients [6, 7, 8]. SARS-CoV-2 RT-PCR can remain positive for weeks, as it cannot distinguish between infective virus and viral fragments without infectious potential, leading to prolonged periods of isolation or work leave [6,9,10]. Determining how long SARS-CoV-2-positive individuals remain infective is thus crucial for the design of effective infection prevention and control strategies [11].
The cycle threshold (Ct) value obtained in the RT-PCR is inversely related to the viral load and has been employed as a semi-quantitative marker of infectivity and for clinical decision-making [12, 13, 14]. However, the use of Ct values to infer SARS-CoV-2 transmissibility has many limitations, as they can be influenced by a multitude of factors, including sample type, the adequacy of sample collection, transport and storage, or the variety of platforms for RNA extraction and amplification [15, 16].
Viral culture is the gold standard for the detection of an active-replicative virus and the assessment of its infectious potential [17, 18, 19]. As this technique requires laboratory biosafety level 3 facilities (BSL3), experienced staff and a longer turnaround time than RT-PCR, it is not used in routine diagnostic algorithms. Nevertheless, the culture of SARS-CoV-2 plays an important role in providing a more complete understanding of its transmissibility and duration of infectivity. As well as guiding recommendations for infection prevention and control, this information is essential for the development and validation of therapeutic agents and vaccines, and to assess the sensitivity and specificity of molecular detection methods [20]. Monitoring the behaviour of SARS-CoV-2 has become even more urgent with the emergence of new variants, as the impact of each mutation needs to be understood [21].
2. Objective
To gain new insights into the behaviour of SARS-CoV-2 by comparing the results obtained by culture (gold standard) with those of RT-PCR, and to establish the relationship between the Ct value, the number of days from symptom onset, and the infectious potential of the virus. Based on our group's experience in the diagnosis of respiratory viruses by conventional techniques, another objective was to describe a protocol for SARS-CoV-2 culture and confirmation by immunofluorescence.
3. Material and methods
3.1. Samples
A total of 100 consecutive respiratory samples positive for SARS-CoV-2 by RT-PCR (nasopharyngeal aspirates and swabs, bronchoalveolar lavage), collected in a viral transport medium from different subjects between November 6, 2020 and May 25, 2021 in the Hospital de la Santa Creu i Sant Pau (HSCSP), were selected. Diagnostic RT-PCR in our hospital was performed as soon as the sample was collected using different commercial platforms. All samples were stored at 4 °C until inoculation, which was always performed within 48 h of collection.
3.2. Culture
Sample handling and cell culture procedures were performed in BSL3.
VERO E6 cells (Vircell, Spain) were used for SARS-CoV-2 culture. Before inoculation, each sample was pre-treated with 10% of a mixture of antibiotics (vancomycin-gentamicin) and amphotericin B for 30 min. 300µL of pre-treated sample was inoculated and incubated at 37 °C for up to 10 days.
Cell monolayers were examined daily with an inverted microscope (x40). A positive culture was suspected when a characteristic cytopathic effect (CPE) was observed. Every CPE (clear or doubtful) was confirmed by indirect immunofluorescence (IFI) using a specific monoclonal antibody AntiSARS-CoV-2 (CertTest-BIOTEC, Spain). Culture was considered negative when there was no CPE 10 days after the inoculation or a CPE was not confirmed by IFI.
The complete protocol is provided in the Supplementary Material (S1).
3.3. Statistical analysis
The results are given as number of cases and percentage for categorical data and as median and the other two quartiles for ordinal one. The comparison of Ct values and days of symptoms between positive and negative cultures was done with the nonparametric Mann-Whitney test. The statistical significance level was 5% (a = 0.05), and two-tailed tests were used throughout. All analysis was performed using IBM-SPSS software (version26; SPSS. Inc. Armonk, NY).
3.4. Ethical approval
The study protocol was evaluated and approved by HSCSP Ethics Committee (IIBSP-VIR-2014–41).
4. Results
A total of 100 consecutive RT-PCR positive SARS-CoV-2 respiratory samples from 100 patients were processed by culture; 46 (46%) of them were from paediatric subjects (<18 years) and 54 (54%) from adults (≥18 years). Eleven (11%) samples were from asymptomatic subjects, 59 (59%) corresponded to patients with <14 days from symptom onset and 30 (30%) to patients with ≥14 days from symptom onset. The median number of days (first quartile [Q1]; third quartile [Q3]) from symptom onset was 4 (1; 15). The presence of symptomatology and the number of days from symptom onset refer to the time the samples were collected for RT-PCR.
Viral isolation was successful in 58% samples. The percentage of positivity in persons <18 years was 60.9%, and ≥18 years, 55.6%. The median number of days (Q1; Q3) from the inoculation of the sample to positive culture was 3 (2; 4). The median (Q1; Q3) RT-PCR Ct value was 23.08 (17.2; 29.45). The overall results are summarised in Table 1 .
Table 1.
Comparative association of SARS-CoV-2 viral culture results with RT-PCR Ct values and days from symptom onset.
| Total | Positive culture | Negative culture | P-value | |
|---|---|---|---|---|
| N | 100 | 58 | 42 | |
| Median RT-PCR Ct value (Q1; Q3) | 23.08 (17.2; 29.45) | 18.15 (15.88; 21.26) | 30.79 (26.79; 34) | <0.001 |
| Asymptomatic (n) | 11 | 6 | 5 | |
|
Days of symptoms (median) 1–2 days (n) 3–7 days (n) 8–13 days (n) 14–30 days (n) > 30 days (n) |
4 39 15 5 22 8 |
2 32 10 4 3 3 |
15 7 5 1 19 5 |
<0.001 |
4.1. Correlation between culture-days of symptoms
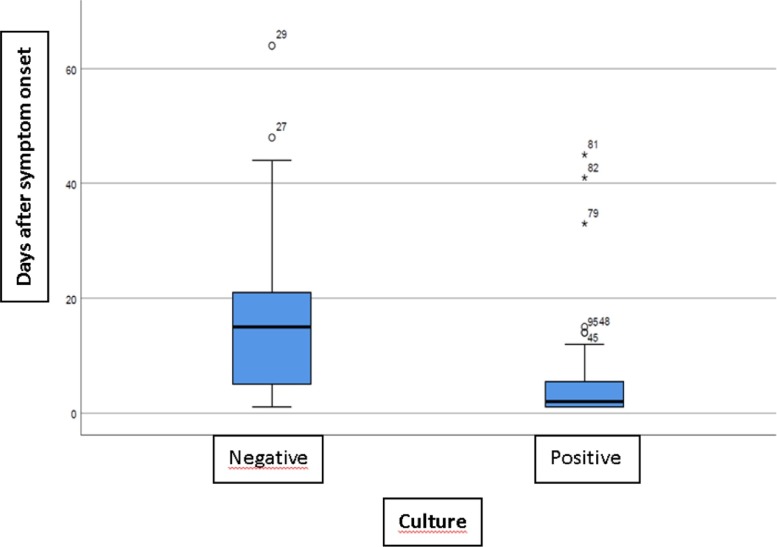
Out of the culture-positive samples, 6 (10.3%) were from asymptomatic subjects, 46 (79.3%) corresponded to patients with <14 days from symptom onset and 6 (10.4%) to patients with ≥14 days from symptom onset. Out of the culture-negative samples, 5 (11.9%) were from asymptomatic subjects, 13 (31%) corresponded to patients with <14 days from symptom onset and 24 (57.1%) to patients with ≥14 days from symptom onset. The median number of days from symptom onset (Q1; Q3) for culture-positive samples was 2 (1; 5.75), and for culture-negative samples, 15 (4.5; 22).
Long-term viral shedding was detected by RT-PCR in 30 samples, but only 6 (20%) of them were culture-positive. Three of these culture-positive samples were from patients who had presented symptoms for more than 30 days (33, 41, 44 days).
4.2. Correlation between culture-RT-PCR Ct value
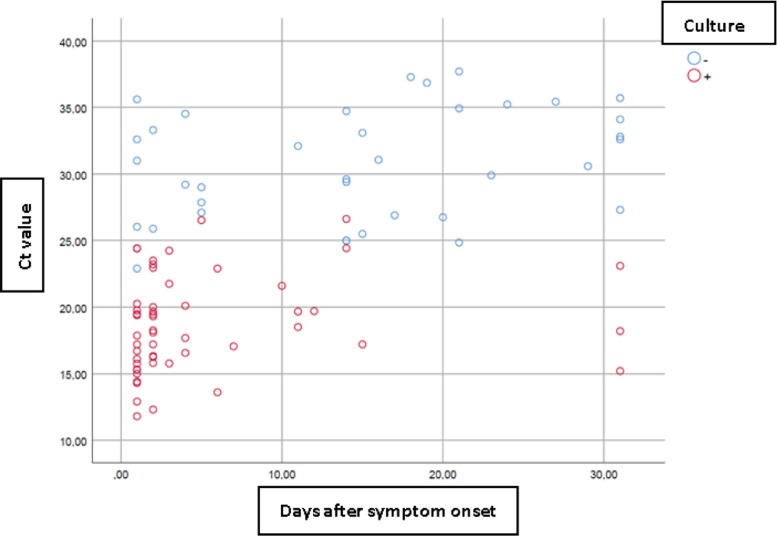
The median RT-PCR Ct value (Q1; Q3) in culture-positive samples was 18.15 (15.88; 21.26), and in culture-negative samples, 30.79 (26.79; 34), being 12.64 units higher in the latter.
The probability of successfully isolating SARS-CoV-2 in samples with a Ct value <22 was 100%. This probability decreased to 3.1% in samples with a Ct value >27, as virus isolation was only achieved in one sample with a Ct value of 31.4. The probability of SARS-CoV-2 isolation according to RT-PCR Ct value is shown in Table 2 .
Table 2.
Probability of isolating SARS-CoV-2 in culture in function of RT-PCR Ct value.
| RT-PCT Ct value (n) | Positive culture (%) | Negative culture (%) |
|---|---|---|
| <22 (45) | 45 (100%) | 0 |
| 22–27 (23) | 12 (52.2%) | 11 (47.8%) |
| >27 (32) | 1 (3.1%) | 31 (96.9%) |
In Table 3 , the Ct values are broken down by the number of days with symptoms and culture. The correlation between culture and days of symptoms is shown in Fig. 1 , and between culture, Ct value and days of symptoms in Fig. 2 .
Table 3.
Ct value according to days after symptom onset and the viral culture result.
| Days of symptoms | Global Ct value | Ct value culture positive | Ct value culture negative |
|---|---|---|---|
| Asymptomatic | 22.74 (16.48; 27.79) | 16.51 (15.75; 21.19) | 24.17 (23.05; 23.61) |
| 1–2 days | 19.3 (15.95; 23.08) | 17.53 (15.64; 19.68) | 31 (25.96; 32.95) |
| 3–7 days | 22.9 (17.37; 27.48) | 18.89 (16.68; 22.61) | 29 (27.86; 29.2) |
| 8–13 days | 19.7 (19.67; 21.6) | 19.69 (19.67; 20.18) | 32.1 (32.1; 32.1) |
| 14–30 days | 29.75(25.78; 34.88) | 24.43 (20.82; 25.53) | 30.58 (26.83; 35.08) |
| >30 days | 29.95 (21.88; 33.13) | 18.2 (16.7; 20.65) | 32.8 (32.6; 34.1) |
Fig. 1.
Viral culture results plotted by days after symptom onset: most of the positive cultures are concentrated within the first 13 days from symptom onset, but 6 outliers are observed beyond 14 days, all of them patients with severe COVID-19 or immunodepressed.
Fig. 2.
Viral culture results plotted by Ct values and days after symptom onset. Most positive cultures (red dots) are concentrated at low Ct values and within a few days of symptom onset. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.).
5. Discussion
VERO E6 cells were selected for the present work as they have been previously found optimal for SARS-CoV-2 multiplication [22, 23]. These cells provide a versatile medium for the recovery of most viruses, including those that are difficult to isolate [24]. They were used to isolate SARS-CoV-2 from the first patients admitted to a Wuhan hospital for COVID-19 pneumonia in December 2019 [1], and for isolation and investigation of SARS-CoV in 2003–2004 [25, 26].
The cytopathic effect of SARS-CoV-2 in VERO E6 cells is easily recognisable and appears relatively quickly. In most cases, small syncytia begin to be observed in the monolayer in the first 48–72 h post-inoculation. As the days pass, the number of syncytia and their size increases, until the entire monolayer is affected, and the cells are destroyed.
One of the main limitations in drawing conclusions from published studies on SARS-CoV-2 culture is the lack of standardisation and the great variability in culture protocols [27], which makes it difficult to compare the results obtained. When implementing SARS-CoV-2 culture, we rely on our own established protocols; the reference technique for the diagnosis of respiratory viruses in our laboratory has been based on culture and immunofluorescence for more than 40 years.
In this study, SARS-CoV-2 was successfully isolated from 58% of the inoculated samples, notably higher than the percentage of culture positivity reported in other studies [28, 29] which ranges mainly from 20 to 40%. An explanation for this higher percentage of positivity is the long experience of our laboratory in the study of respiratory viruses by culture, which has allowed us to optimise this technique and successfully implement it in our diagnostic routine.
When optimizing viral culture, an important point is to store the sample correctly and inoculate it as soon as possible after collection. The longer the period between collection and inoculation, the lower the chances of recovering the virus, due to degradation, especially if optimal storage conditions are not maintained [30]. In our laboratory, all samples were stored at 4 °C and inoculated within 48 h of collection. Another significant aspect is how long the sample should be incubated before the culture can be considered negative. In this work, all samples were incubated for 10 days; although most CPE were observed within 72 h post-inoculation, in some cases positive cultures were obtained after 6 days. Some studies have classified cultures as negative before 6 days [6, 31], which entails a risk of missing positive cultures. A notable difference between our approach and those of other SARS-CoV-2 culture studies is that, instead of confirming CPE with an RT-PCR of the supernatant, the verification was done by immunofluorescence, using a specific monoclonal antibody. RT-PCR of the supernatant can detect RNA from the inoculated sample without the need for virus multiplication, but IFI allows the direct observation of virus-infected cells. The last point to note about this work is that it includes samples taken over a long period of time (more than 6 months), in contrast to 2 months or less in other studies [28, 29].
Besides their differences in viral culture techniques, the studies are also quite diverse in the type of individuals involved (age, symptomatology, days of evolution), which is a further hindrance when attempting to draw conclusions. The majority include only adult and symptomatic patients, with only a few small-scale studies of SARS-CoV-2 in paediatric patients [13, 32]. In the present work, 46% of samples were taken from a paediatric age group and like Singanayagam et al., 2020, a higher percentage of culture positivity was found in paediatric individuals (60.9%) compared to adults (55.6%), although the difference was not significant.
Likewise, there are very few published studies involving individuals who were asymptomatic when testing positive for RT-PCR [13, 33]. We included 11 samples from asymptomatic individuals at the time of sample collection, 6 (54.5%) of which were culture-positive. These individuals were tested because of close contact with positive cases or for hospital pre-admission screening. In agreement with the literature [13, 32, 33], our results show that a high percentage of asymptomatic and paediatric individuals with a positive SARS-CoV-2 RT-PCR are infective, which probably plays an important role in the spread of the virus.
Also in agreement with the literature [28], a significant positive correlation was found between the likelihood of isolating SARS-CoV-2 in culture, fewer days of symptoms and a lower RT-PCR Ct value. A significant difference (13 days) was observed in the median number of symptom days between culture-negative and culture-positive samples (15 days vs. 2 days, respectively). The highest percentage of culture positivity was obtained in samples from people with 1–2 days of symptoms (82.1%). In samples collected within the first 7 days of symptoms, successful virus isolation was achieved in 77.8% of cases, but this percentage decreased to 20% at 14 days or more of symptom onset, with positive cultures obtained in only 6 out of 30 inoculated samples. All 6 were immunocompromised patients who required admission to the Critical Care Unit due to severe complications of COVID-19 (3 patients with haematological malignancies, an untreated HIV patient, a morbidly obese diabetic patient, and a 101-year-old man). Our data are in agreement with most studies [28, 29], which could not detect an infectious virus after 10 days of symptom onset, except in immunosuppressed individuals [34] or those with severe COVID-19 [35]. In these cases, virus isolation was achieved 70 days and 32 days after symptom onset, respectively.
It is well established that the Ct value is inversely related to the probability of obtaining a positive culture, which decreases to 32% for each Ct unit from a Ct of 24 [31]. The mean Ct value was 12.64 units higher in culture-negative than in culture-positive samples, which represents a significant increase. Virus isolation was successful in all samples with a Ct value <22, but not achieved when the Ct value was >27, except in one case with a Ct value of 31.4. This sample was from a 2-year-old girl with acute lymphoid leukaemia who was tested by RT-PCR for pre-admission screening. However, another study [13] found that up to 25% of samples with a Ct>30 corresponded to a potentially infectious virus. Despite these results, the use of the Ct value as an indicator of infectivity is not recommended due to its high variability, mainly conditioned by sample and technical factors [15, 16]. The correlation between the Ct value and culture observed here is specific to our laboratory, and therefore cannot be extrapolated elsewhere.
In summary, culture remains the gold standard for determining the infectious capacity of viruses [17], but its laboriousness, turnaround time and the need for special biosafety facilities make it unsuitable for routine COVID-19 diagnosis. However, specialized laboratories equipped for viral culture and with the relevant expertise are needed to i) increase our knowledge of virus transmissibility and duration of infectivity; ii) monitor changes in the behaviour of emerging variants; and iii) conduct research on treatments, vaccines, or diagnostic techniques. Also required is a single defined protocol for SARS-CoV-2 culture, based on experience of culturing other respiratory viruses, and preferably including confirmation by visual identification via immunofluorescence [17]. Our findings show that the probability of isolating SARS-CoV-2 in culture is significantly and positively correlated with fewer days of symptoms and a lower RT-PCR Ct value. However, due to its high variability, use of the Ct value as a general marker of infectivity is not recommended, and the number of symptom days is a more reliable indicator.
Based on the results, it can be concluded that in immunocompetent individuals with mild-moderate COVID-19, SARS-CoV-2 infectivity does not last more than 14 days from symptom onset. In contrast, in immunocompromised patients or those with severe COVID-19, infectivity may remain beyond 14 days, after which the Ct value can be taken into consideration. A Ct value <22 always indicates infectivity, but when Ct value ≥22 it is inconclusive and culture is recommended.
Funding support
This work was supported by own funding.
Declaration of Competing Interest
The authors declare that they have no financial interests or personal relationships that could have appeared to influence the work presented in this paper.
Acknowledgements
We thank the staff of the Hospital de la Santa Creu i Sant Pau for their contribution to the diagnosis of SARS-CoV-2, as well as to Lucy Brzoska for her contribution to English language editing.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2022.105167.
Appendix. Supplementary materials
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus Investigating and Research Team A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. Feb 20Epub 2020 Jan 24. PMID: 31978945; PMCID: PMC7092803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kucharski A.J., Klepac P., Conlan A.J.K., Kissler S.M., Tang M.L., Fry H., Gog J.R., Edmunds W.J., CMMID COVID-19 working group Effectiveness of isolation, testing, contact tracing, and physical distancing on reducing transmission of SARS-CoV-2 in different settings: a mathematical modelling study. Lancet Infect. Dis. 2020;20(10):1151–1160. doi: 10.1016/S1473-3099(20)30457-6. OctEpub 2020 Jun 16. PMID: 32559451; PMCID: PMC7511527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alagoz O., Sethi A.K., Patterson B.W., Churpek M., Alhanaee G., Scaria E., Safdar N. The impact of vaccination to control COVID-19 burden in the United States: a simulation modeling approach. PLoS One. 2021;16(7) doi: 10.1371/journal.pone.0254456. Jul 14PMID: 34260633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. JanErratum in: Euro Surveill. 2020 Apr;25(14): Erratum in: Euro Surveill. 2020 Jul;25(30): Erratum in: Euro Surveill. 2021 Feb;26(5): PMID: 31992387; PMCID: PMC6988269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CDC. Discontinuation of Transmission-Based Precautions and Disposition of Patients with COVID-19 in Healthcare Settings (Interim Guidance). https://www.cdc.gov/coronavirus/2019-ncov/hcp/disposition-hospitalized-patients.html.
- 6.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. MayEpub 2020 Apr 1. Erratum in: Nature. 2020 Dec;588(7839):E35. PMID: 32235945. [DOI] [PubMed] [Google Scholar]
- 7.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323(22):2249–2251. doi: 10.1001/jama.2020.8259. Jun 9PMID: 32374370. [DOI] [PubMed] [Google Scholar]
- 8.Ling Y., Xu S.B., Lin Y.X., Tian D., Zhu Z.Q., Dai F.H., Wu F., Song Z.G., Huang W., Chen J., Hu B.J., Wang S., Mao E.Q., Zhu L., Zhang W.H., Lu H.Z. Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med. J. (Engl) 2020;133(9):1039–1043. doi: 10.1097/CM9.0000000000000774. May 5PMID: 32118639; PMCID: PMC7147278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W.D., Chang S.Y., Wang J.T., Tsai M.J., Hung C.C., Hsu C.L., Chang S.C. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J. Infect. 2020;81(2):318–356. doi: 10.1016/j.jinf.2020.03.063. AugEpub 2020 Apr 10. PMID: 32283147; PMCID: PMC7151379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basile K., McPhie K., Carter I., Alderson S., Rahman H., Donovan L., Kumar S., Tran T., Ko D., Sivaruban T., Ngo C., Toi C., O'Sullivan M.V., Sintchenko V., Chen S.C., Maddocks S., Dwyer D.E., Kok J. Cell-based culture of SARS-CoV-2 informs infectivity and safe de-isolation assessments during COVID-19. Clin. Infect. Dis. 2020 Oct 24:ciaa1579. doi: 10.1093/cid/ciaa1579. Epub ahead of print. PMID: 33098412; PMCID: PMC7665383. [DOI] [PMC free article] [PubMed]
- 11.Rhee C., Kanjilal S., Baker M., Klompas M. Duration of Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity: when is it safe to discontinue isolation? Clin. Infect. Dis. 2021;72(8):1467–1474. doi: 10.1093/cid/ciaa1249. Apr 26PMID: 33029620; PMCID: PMC7499497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tom M.R., Mina M.J. To interpret the SARS-CoV-2 test, consider the cycle threshold value. Clin. Infect. Dis. 2020;71(16):2252–2254. doi: 10.1093/cid/ciaa619. Nov 19PMID: 32435816; PMCID: PMC7314112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singanayagam A., Patel M., Charlett A., Lopez Bernal J., Saliba V., Ellis J., Ladhani S., Zambon M., Gopal R. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32) doi: 10.2807/1560-7917.ES.2020.25.32.2001483. AugErratum in: Euro Surveill. 2021 Feb;26(7): PMID: 32794447; PMCID: PMC7427302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao S.N., Manissero D., Steele V.R., Pareja J. A Systematic Review of the Clinical Utility of Cycle Threshold Values in the Context of COVID-19. Infect. Dis. Ther. 2020;9(3):573–586. doi: 10.1007/s40121-020-00324-3. SepEpub 2020 Jul 28. Erratum in: Infect Dis Ther. 2020 Aug 18;: PMID: 32725536; PMCID: PMC7386165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rhoads D., Peaper D.R., She R.C., Nolte F.S., Wojewoda C.M., Anderson N.W., Pritt B.S. College of American pathologists (CAP) microbiology committee perspective: caution must be used in interpreting the cycle threshold (Ct) value. Clin. Infect. Dis. 2021;72(10):e685–e686. doi: 10.1093/cid/ciaa1199. May 18PMID: 32785682. [DOI] [PubMed] [Google Scholar]
- 16.IDSA and AMP statement on the use of SARS-CoV-2 PCR cycle threshold (Ct) values for clinical decision-making. IDSA (infectious Diseases Society of America) and AMP (Association for Molecular Pathology). Updated March 12, 2021.
- 17.Hematian A., Sadeghifard N., Mohebi R., Taherikalani M., Nasrolahi A., Amraei M., Ghafourian S. Traditional and modern cell culture in virus diagnosis. Osong Public Health Res. Perspect. 2016;7(2):77–82. doi: 10.1016/j.phrp.2015.11.011. AprEpub 2016 Jan 8. Erratum in: Osong Public Health Res Perspect. 2020 Aug;11(4):266. PMID: 27169004; PMCID: PMC4850366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hudu S.A., Alshrari A.S., Syahida A., Sekawi Z. Cell culture, technology: enhancing the culture of diagnosing human diseases. J. Clin. Diagn. Res. 2016 Mar;10(3):DE01–DE05. doi: 10.7860/JCDR/2016/15837.7460. Epub 2016 Mar 1. PMID: 27134874; PMCID: PMC4843260. [DOI] [PMC free article] [PubMed]
- 19.Leland D.S., Ginocchio C.C. Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 2007;20(1):49–78. doi: 10.1128/CMR.00002-06. JanPMID: 17223623; PMCID: PMC1797634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dao T.L., Hoang V.T., Colson P., Lagier J.C., Million M., Raoult D., Levasseur A., Gautret P. SARS-CoV-2 infectivity and severity of COVID-19 according to SARS-CoV-2 variants: current evidence. J. Clin. Med. 2021;10(12):2635. doi: 10.3390/jcm10122635. Jun 15PMID: 34203844; PMCID: PMC8232800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young B.E., Ong S.W.X., Ng L.F.P., Anderson D.E., Chia W.N., Chia P.Y., Ang L.W., Mak T.M., Kalimuddin S., Chai L.Y.A., Pada S., Tan S.Y., Sun L., Parthasarathy P., Fong S.W., Chan Y.H., Tan C.W., Lee B., Rötzschke O., Ding Y., Tambyah P., Low J.G.H., Cui L., Barkham T., Lin R.T.P., Leo Y.S., Renia L., Wang L.F., Lye D.C.; Singapore 2019 Novel Coronavirus Outbreak Research team. Viral dynamics and immune correlates of COVID-19 disease severity. Clin. Infect. Dis. 2020 Aug 28:ciaa1280. doi: 10.1093/cid/ciaa1280. Epub ahead of print. PMID: 32856707; PMCID: PMC7499509.
- 22.Ogando N.S., Dalebout T.J., Zevenhoven-Dobbe J.C., Limpens R.W.A.L., van der Meer Y., Caly L., Druce J., de Vries J.J.C., Kikkert M., Bárcena M., Sidorov I., Snijder E.J. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J. Gen. Virol. 2020;101(9):925–940. doi: 10.1099/jgv.0.001453. SepPMID: 32568027; PMCID: PMC7654748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., Li Y., Uehara A., Wang H., Goldsmith C., Bullock H.A., Wang L., Whitaker B., Lynch B., Gautam R., Schindewolf C., Lokugamage K.G., Scharton D., Plante J.A., Mirchandani D., Widen S.G., Narayanan K., Makino S., Ksiazek T.G., Plante K.S., Weaver S.C., Lindstrom S., Tong S., Menachery V.D., Thornburg N.J. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 2020;26(6):1266–1273. doi: 10.3201/eid2606.200516. JunEpub 2020 Jun 17. PMID: 32160149; PMCID: PMC7258473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hudu S.A., Alshrari A.S., Syahida A., Sekawi Z. Cell culture, technology: enhancing the culture of diagnosing human diseases. J. Clin. Diagn. Res. 2016;10(3):DE01–DE05. doi: 10.7860/JCDR/2016/15837.7460. MarEpub 2016 Mar 1. PMID: 27134874; PMCID: PMC4843260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. May 15Epub 2003 Apr 10. PMID: 12690092. [DOI] [PubMed] [Google Scholar]
- 26.Qinfen Z., Jinming C., Xiaojun H., Huanying Z., Jicheng H., Ling F., Kunpeng L., Jingqiang Z. The life cycle of SARS coronavirus in Vero E6 cells. J. Med. Virol. 2004;73(3):332–337. doi: 10.1002/jmv.20095. JulPMID: 15170625; PMCID: PMC7166737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cimolai N. Not all viral culture approaches are equal. Clin. Infect. Dis. 2021;73(7):e1787–e1788. doi: 10.1093/cid/ciaa1632. Oct 5PMID: 33104181. [DOI] [PubMed] [Google Scholar]
- 28.Jefferson T., Spencer E.A., Brassey J., Heneghan C. Viral cultures for COVID-19 infectious potential assessment - a systematic review. Clin. Infect. Dis. 2020 Dec 3:ciaa1764. doi: 10.1093/cid/ciaa1764. Epub ahead of print. PMID: 33270107; PMCID: PMC7799320. [DOI] [PMC free article] [PubMed]
- 29.Walsh K.A., Spillane S., Comber L., Cardwell K., Harrington P., Connell J., Teljeur C., Broderick N., de Gascun C.F., Smith S.M., Ryan M., O'Neill M. The duration of infectiousness of individuals infected with SARS-CoV-2. J. Infect. 2020;81(6):847–856. doi: 10.1016/j.jinf.2020.10.009. DecEpub 2020 Oct 10. PMID: 33049331; PMCID: PMC7547320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt N.J. In: Diagnostic Procedures For viral, rickettsial, and Chlamydial Infections. 5th ed. Lennette EH, Schmidt, editors. American Public Health Association; Washington, DC: 1979. Cell culture techniques for diagnostic virology. [Google Scholar]
- 31.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., Boodman C., et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin. Infect. Dis. 2020 May 22:ciaa638. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed]
- 32.L'Huillier A.G., Torriani G., Pigny F., Kaiser L., Eckerle I. Culture-competent SARS-CoV-2 in nasopharynx of symptomatic neonates, children, and adolescents. Emerg. Infect. Dis. 2020;26(10):2494–2497. doi: 10.3201/eid2610.202403. OctEpub 2020 Jun 30. PMID: 32603290; PMCID: PMC7510703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., Taylor J., Spicer K., Bardossy A.C., Oakley L.P., Tanwar S., Dyal J.W., Harney J., Chisty Z., Bell J.M., Methner M., Paul P., Carlson C.M., McLaughlin H.P., Thornburg N., Tong S., Tamin A., Tao Y., Uehara A., Harcourt J., Clark S., Brostrom-Smith C., Page L.C., Kay M., Lewis J., Montgomery P., Stone N.D., Clark T.A., Honein M.A., Duchin J.S., Jernigan J.A. Public health–seattle and king county and CDC COVID-19 investigation team. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N. Engl. J. Med. 2020;382(22):2081–2090. doi: 10.1056/NEJMoa2008457. May 28Epub 2020 Apr 24. PMID: 32329971; PMCID: PMC7200056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., Bowden T.A., de Wit E., Riedo F.X., Munster V.J. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell. 2020;183(7):1901–1912. doi: 10.1016/j.cell.2020.10.049. Dec 23e9Epub 2020 Nov 4. PMID: 33248470; PMCID: PMC7640888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Folgueira M.D., Luczkowiak J., Lasala F., Pérez-Rivilla A., Delgado R. Prolonged SARS-CoV-2 cell culture replication in respiratory samples from patients with severe COVID-19. Clin. Microbiol. Infect. 2021;27(6):886–891. doi: 10.1016/j.cmi.2021.02.014. JunEpub 2021 Feb 22. PMID: 33631334; PMCID: PMC7898982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.