Abstract
Background and objective
The objective of this study was to systematically review and meta-analyze the diagnostic accuracy of current machine learning classifiers for age-related macular degeneration (AMD). Artificial intelligence diagnostic algorithms can automatically detect and diagnose AMD through training data from large sets of fundus or OCT images. The use of AI algorithms is a powerful tool, and it is a method of obtaining a cost-effective, simple, and fast diagnosis of AMD.
Methods
MEDLINE, EMBASE, CINAHL, and ProQuest Dissertations and Theses were searched systematically and thoroughly. Conferences held through Association for Research in Vision and Ophthalmology, American Academy of Ophthalmology, and Canadian Society of Ophthalmology were searched. Studies were screened using Covidence software and data on sensitivity, specificity and area under curve were extracted from the included studies. STATA 15.0 was used to conduct the meta-analysis.
Results
Our search strategy identified 307 records from online databases and 174 records from gray literature. Total of 13 records, 64,798 subjects (and 612,429 images), were used for the quantitative analysis. The pooled estimate for sensitivity was 0.918 [95% CI: 0.678, 0.98] and specificity was 0.888 [95% CI: 0.578, 0.98] for AMD screening using machine learning classifiers. The relative odds of a positive screen test in AMD cases were 89.74 [95% CI: 3.05–2641.59] times more likely than a negative screen test in non-AMD cases. The positive likelihood ratio was 8.22 [95% CI: 1.52–44.48] and the negative likelihood ratio was 0.09 [95% CI: 0.02–0.52].
Conclusion
The included studies show promising results for the diagnostic accuracy of the machine learning classifiers for AMD and its implementation in clinical settings.
Subject terms: Diagnosis, Epidemiology
Introduction
Background
Age-related macular degeneration (AMD) is an eye condition that causes irreversible blindness in adults worldwide. AMD is a neurodegenerative disease that preferentially affects the macula—the central part of the retina [1]. Patients with AMD may experience blurred or distorted vision, and their vision deteriorates over time. A hallmark of clinically identifying AMD is yellowish drusen seen underneath the retinal pigment epithelium [2, 3]. In addition, the presence or absence of choroidal neovascularization is a strong indicator for vascular permeability and fragility. AMD is often further clinically classified into exudative (wet) or non-exudative (dry) AMD after assessing the stage of the disease, and then further into classes [2, 4].
It is projected that there will be a massive increase in the number of AMD cases in aging populations [5]. On a global perspective, AMD currently has a global prevalence of ~170 million cases. The cases of AMD and the Global Burden of Disease in elderly populations will rise dramatically with the world’s aging population [2, 6]. The number of people expected to be above the age of 60 by 2050 is two billion. By 2040, it is expected for the global prevalence of AMD to increase to 288 million cases [5].
Diagnostic techniques and treatment of AMD using artificial intelligence (AI) will become increasingly important as multi-morbidity and patient demands rise. Precise diagnosis and staging of AMD include obtaining imaging techniques of the retina by fundus imaging or optical coherence tomography (OCT) [7–9]. Ophthalmologists rely on these images to confirm diagnosis and provide an individualized treatment plan for the patient. However, the current process for diagnosis is expensive and time consuming [5]. The increase of AMD cases due to the aging population will be a large contributor to potential professional burnout in the field of ophthalmology [10]. Ophthalmologists are continually facing high pressures at work influenced by high demands of patient care and overtime work [11, 12]. Thus, novel methods must be implemented in the workplace to further assist physicians in the diagnosis and treatment of diseases.
AI has been widely used in the medical field to aid healthcare workers and it is one of the most influential technological innovations [13]. The purpose of AI is to simulate a human’s mental process through computers and learn to solve problems similarly to the human brain. Specifically, machine learning (ML) has transformed methods for diagnosing AMD and other eye diseases such as diabetic retinopathy, glaucoma, and cataracts in the past decade [14–16]. Through ML, one can train and teach an algorithm by using vast amounts of data to perform a specific task. Ultimately, these AI diagnostic technologies can automatically detect and diagnose AMD through training and testing data from large sets of fundus or OCT images. The use of AI technology can be a powerful tool, and it is a method of obtaining a cost-effective, simple, and fast diagnosis of AMD [13]. Common ML classifiers include the use of support vector machines (SVM) and convolutional neural networks (CNN) [13–16].
Aims of the study
The recent research and applications of AI in ophthalmology is growing and it is a quickly expanding field. There has been vast literature on ML diagnostic algorithms used for eye diseases. In recent research, there are systematic reviews published on the applications of AI for diagnosing glaucoma and for screening diabetic retinopathy [15, 17]. The objective of this study is to systematically review and meta-analyze the diagnostic accuracy of ML classifiers for all types of AMD among all eyes available in datasets to assess their accuracy and reliability to be implemented in clinical settings.
Methods
The protocol for this systematic review was registered in PROSPERO (CRD42020219313) and this review was conducted according to the PRISMA statement recommendations [18].
Search strategy
A systematic and thorough database search included MEDLINE/PubMed, EMBASE, CINAHL and ProQuest Dissertations and Theses to find articles on diagnostic AI technologies used in the field of ophthalmology. The search strategy and keywords for each database is detailed in Appendix A and the searches were conducted until September 12, 2020. The search was limited to English and human studies only. We did not place any limits on publication date and study location to maximize our eligible studies. OVID AutoAlerts for MEDLINE and EMBASE databases were used for weekly updates for any new published literature that the search strategy encompassed.
In addition, gray literature searches were conducted in order to obtain a comprehensive search. Conferences held through the American Academy of Ophthalmology, the Association for Research in Vision and Ophthalmology, and the Canadian Society of Ophthalmology were searched in all available years. We searched through the conferences until September 12, 2020. Keywords that were used for the gray literature search consisted of “AI” and “diagnosis”. The search strategy and search results for each conference is displayed in Appendix B. Forward and backward citation tracing were carried out on studies that were included after the full-text screening. Refer to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) flow diagram for more details [18].
Inclusion and exclusion criteria
This review included studies that used AI algorithms to assess the diagnosis of AMD on human eyes. The AI can be in the form of ML, neural networks, or deep learning to assist in the automized diagnosis of AMD. The AI must be an advanced AI—meaning that it involved a training or learning process. We excluded studies that looked at only computers conducting an automated process, without the mention of AI and a learning process. We included studies that used AI algorithm to make the first diagnosis of any AMD type and stage from healthy eyes, or the algorithm was able to differentiate between AMD and non-AMD eyes. We excluded studies that further classified already-diagnosed AMD patients into later stages.
In addition, studies that used any imaging techniques were included, such as fundus imaging and OCT. We included any studies that also showed a performance indicator for the diagnostic ability of the AI technology—this included any reports of sensitivity, specificity, accuracy, or area under curve (AUC) values. The studies must include a reference standard by trained clinicians or ophthalmologists to validate and confirm the AI’s diagnosis. Included publications must be primary studies, and there were no restrictions on study design; ophthalmology news articles, opinion pieces, and case reports were excluded. Only studies in English were included, and there was no restriction placed on study location or publication date.
Screening
Database search results were all imported into Covidence systematic review software (Veritas Health Innovation, Melbourne, Australia). In Covidence, all duplicates were removed, and three levels of systematic screening were conducted by two independent reviewers (R.C. & J.C.). If consensus could not be reached, all disagreements were resolved by discussion and consensus of the two reviewers. The first level of screening was a broad title screening. If the study title did not mention the diagnosis of any eye disease, the study was excluded, and the remaining studies would move on to the next level of screening. The second level of screening was an abstract screening. If the study did not look at investigating an advanced AI with a training process for the diagnosis of any eye disease, the study was excluded. The final level of screening examined the full text to selectively choose relevant studies for the diagnosis of any type and stage of AMD including both dry and wet AMD; remaining included studies moved on to the risk of bias assessment.
Cohen’s kappa (κ) statistic was calculated after each level of screening before conflicts were resolved—the scores were 0.80, 0.62, and 0.82 respectively as shown in Appendix C.
Risk of bias assessment and data extraction
A Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool was used as a risk of bias assessment to assess the level of bias and concerns of applicability of each study after the full text screening [19]. The risk of bias assessment was conducted by two reviewers (R.C. & J.C.). QUADAS-2 considered four domains: patient selection, index tests, reference standard, and flow and timing. All studies were included, and any conflicts were resolved by discussion.
Study data were extracted by two reviewers (R.C. & J.C.). The following data were extracted from each study: author, year of publication, study location, study design, ML classifier, type of imaging, number of study participants, number of images used, number of images labelled normal and non-normal, training and testing process, image database and datasets, AMD diagnosis confirmation, AUC, sensitivity, specificity, and accuracy. If the study used multiple ML classifiers, all data from each classifier were extracted. The model of the imaging camera was recorded if provided in the study. The data extraction table for meta-analysis is outlined in Appendix D.
Statistical analysis
STATA 15.0 (STATA Corp, College Station, Texas, U.S.A.) was used to synthesize and analyze the data for the diagnostic accuracy of ML for AMD. The main outcome of interest was the sensitivity, specificity, and AUC of each ML classifier used in the studies. Hierarchical logistic regression was used to determine the pooled estimates of sensitivity and specificity of diagnostic accuracy.
In addition, summaries of the fitted Hierarchical Summary Receiver Operating Characteristic (HSROC) model, the summary receiver operating characteristics (SROC) curve, and the 95% confidence interval were plotted graphically. The positive/negative likelihood ratios (LR+/LR−) were calculated using bivariate models to generate estimates of the likelihood of a positive or negative test. From this result, the diagnostic odds ratio (DOR) was calculated to determine the relative diagnostic effectiveness. DOR is the ratio of the odds of a positive screen test in an AMD case relative to the odds of a negative screen test in a non-AMD case.
Forest plots showing the within-study estimates and confidence intervals for sensitivity and specificity were plotted separately. For each study, the sensitivity and specificity were aggregated using the fixed or random-effect model based on the absence or presence of heterogeneity to estimate the summary effect.
To test heterogeneity, I2 statistics, Z-value, and χ2 statistics were computed. An I2 value of <50% implies low heterogeneity, and in these cases, a fixed-effect model was computed. An I2 statistics of 50% or more represents high heterogeneity, and in these cases a random-effect model was calculated. In addition, a high Z-value, a low p value (<0.01) and a large χ2 value implies significant heterogeneity and therefore, a random-effect model was computed. Forest plots were also generated for each case.
Results
Search results and study characteristics
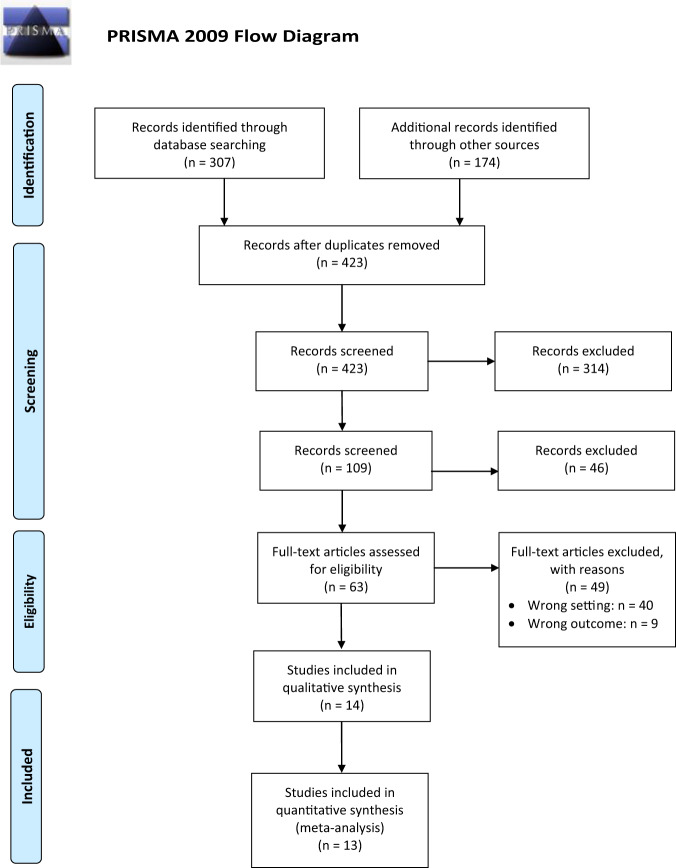
Details on the inclusion process is found in Fig. 1, the PRISMA flow chart [18]. The search strategy on all databases identified 307 articles, and the gray literature search found 174 articles. Fifty-eight of the articles were duplicates, leaving 423 articles for the title screening. After the first level (title screening), 314 studies were excluded, and 109 studies were included. For the second level (abstract) screening, 46 articles were excluded, and 63 studies were included for the final level (full text) screening. Ultimately, 49 studies were excluded, and 14 studies met the inclusion criteria and were included for the risk of bias assessment. All 14 included studies were included for the qualitative synthesis, but only 13 of those studies were included for the meta-analysis [20–33].
Fig. 1. PRISMA flow diagram.
Flow diagram showing the study selection process and reasons for exclusion.
The study characteristics table can be found in Table 1. All included studies were observational studies, which examined ML applications for the diagnosis of AMD. Studies were conducted in North America [20, 21, 25, 28, 29], Europe [22, 31], and Asia [23, 24, 27, 30, 32, 33]. Most of the included studies used OCT images for training and validation [22–33], while only four studies used fundus images [20, 21, 30, 33]. Within the OCT cohort, there was a varied use of ML classifiers; studies utilized CNN [23–25, 27, 31, 32], SVM [22, 28, 29], linear configuration patterns (LCP) [32], AdaBoost [22], random forests [22], and deep learning system (DLS) [30]. In most of the studies that used spectral domain OCT (SD-OCT), retinal images were obtained using the Heidelberg Spectralis from Heidelberg Engineering to feed the AI process. In the fundus imaging cohort, CNNs were used in the four studies [20, 21, 30, 33]. Many of the studies contained both wet and dry AMD types in their study population and four studies specified the AMD stage that was investigated [20, 21, 26, 30].
Table 1.
Study characteristics of included studies.
| Study | Study location | AI classifier | Imaging technique (Acquisition Machine) | # Study participants | # Images used | AMD type or stage | Training and testing process | Database and datasets | Reference standard |
|---|---|---|---|---|---|---|---|---|---|
| Burlina et al. (2017) | United States | CNN (AlexNet, OverFeat) | Fundus | 4613 | 67,401 | All Types of AMD; Early, Intermediate, and Advanced Stage | 2-class classification; Class 0 – 37,418 images; Class 1 – 29,983 images | National Institutes of Health AREDS | Trained clinical graders |
| Burlina et al. (2019) | United States | CNN | Fundus | 4613 | 133 821 | All Types of AMD; Early, Intermediate, and Advanced Stage | 2-class classification AMD referral challenge derived from the original 4-step AREDS enrolment scale | National Institutes of Health AREDS | 2 Retinal specialists |
| Fraccaro et al. (2015) | Italy | SVM, AdaBoost, Random Forest | Topcon 3D OCT | 487 | 974 | Dry and wet AMD | Linear kernel and the nu-classification, 50 bootstrap tests using out-of-bag predictions | Medical Retina Center of the University Eye Clinic of Genoa | 2 Ophthalmologists |
| Hassan et al. (2018) | Pakistan | CNN | Topcon 3D OCT Heidelberg Spectralis | 499 | 46,913 | Dry and wet AMD | 4992 images used for training, 41,921 images for validation | Duke Dataset - I, Duke Dataset - II, Duke Dataset - III, AFIO Dataset, Amanat Dataset | Ophthalmologists |
| Hwang et al. (2019) | Taiwan | CNN (VGG16 InceptionV3, ResNet50) | Zeiss Cirrus HD OCT, Optovue RTVue-XR Avanti | 747 | 35,900 | Dry, active wet, inactive wet AMD | 28,720 images used for training, 7180 images used for validation | Department of Ophthalmology of Taipei Veterans General Hospital | 2 Retinal specialists |
| Lee et al. (2016) | United States | CNN | SD-OCT Heidelberg Spectralis | 9285 | 101,002 | Dry, wet, and unspecified AMD | 80,839 images used for training, 20,163 images used for validation | Heidelberg Spectralis imaging database | Retinal specialist |
| Li et al. (2019) | China | CNN | SD-OCT Heidelberg Spectralis | 5319 | 109,312 | Early AMD with drusen indicators | 108,312 images used for training, 1000 images for validation | Shiley Eye Institute of the University of California San Diego, the California Retinal Research Foundation, Medical Center Ophthalmology Associates, the Shanghai First People’s Hospital, and the Beijing Tongren Eye Center | 2 Ophthalmologists and a retinal specialist to resolve disagreements |
| Liu et al. (2011) | United States | SVM | Zeiss Cirrus SD-OCT | 136 | 326 | All types of AMD | tenfold cross validation | University of Pittsburgh Medical Center Eye Center, and New England Eye Center | 3 Ophthalmologists |
| Srinivasan et al. (2014) | United States | SVM | SD-OCT Heidelberg Spectralis | 45 | 90 | Dry AMD | Leave-three-out cross-validation | Duke University, Harvard University, and the University of Michigan | Ophthalmologists |
| Tan et al. (2018) | Singapore | CNN | Fundus | – | 1110 | Dry and wet AMD | tenfold cross validation | Ophthalmology Department of Kasturba Medical College (KMC), Manipal, India | Retinal specialists |
| Ting et al. (2017) | Singapore | DLS | Topcon OCT | 38,189 | 108,558 | Intermediate AMD according to AREDS grading system | 72,610 images used for training, 35,948 images used for validation | Singapore National Diabetic Retinopathy Screening Program 2010–2013, Singapore Malay Eye Study, Singapore Indian Eye Study, Singapore Chinese Eye Study, Singapore National Eye Centre AMD | Retinal specialists |
| Treder et al. (2018) | Germany | CNN | SD-OCT Heidelberg Spectralis | 701 | 1112 | Wet AMD | 500 training step procedure with training and validation accuracy | University of Muenster Medical, Germany | – |
| Wang et al. (2016) | China | LCP | 3D SD-OCT | 45 | 3000 | Dry AMD | tenfold cross validation | Duke University, Harvard University, and the University of Michigan | Ophthalmologists |
| Yoo et al. (2019) | South Korea | CNN (VGG-19) | SD-OCT Heidelberg Spectralis, Fundus | 119 | 3000 | Dry and wet AMD | fivefold cross validation | Project Macula Database | 2 Ophthalmologists |
“–” indicates no data reported”.
AdaBoost adaptive boosting, CNN convolutional neural network, DLS deep learning system, OCT optical coherence tomography, SVM support vector machine, LCP linear configuration patterns.
Moreover, Burlina et al. (2017) [20] and Burlina et al. (2019) [21] overlapped in datasets, but their ML training and testing process technique were unique in each study. Srinivasan et al. (2014) [29] and Wang et al. (2016) [33] also used the same dataset, but Srinivasan et al. (2014) did not have any quantitative data to extract that was pertinent to this study. Most studies used either ophthalmologists, retinal specialist, or clinical graders to confirm the patient’s AMD diagnosis with the AI technology. Yoo et al. (2019) [32] was the only included study that investigated both OCT and fundus images for their diagnostic process.
Risk of bias assessment
Most studies had a low risk of bias for the domains of patient selection (64%), index tests (71%), reference standard (64%), and flow and timing (100%). The risk of bias assessment and concerns about applicability are summarized in Appendix E. All studies were included.
Meta-analysis for the diagnostic accuracy of machine learning classifiers
Figure 2 represents the SROC curve in which sensitivity was plotted against the specificity. Each circle in the plot represents a different study and circle size represents the size of the study, that is, the total number of individuals in each study. Figure 2 indicates that majority of included studies are clustered near the top right corner of the plot. Therefore, Fig. 2 shows high specificity and sensitivity of various ML classifiers. The figure shown in Appendix F indicates ML classifiers on an SROC curve. Based on Appendix F, CNN and VGG-19 seems to have high sensitivity, as well as specificity.
Fig. 2. Summary Receiver Operating Characteristic (SROC) plot.
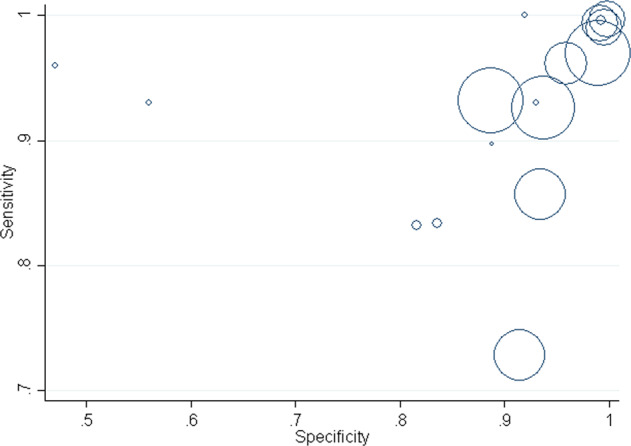
SROC plots the sensitivity against the specificity of the ML classifiers, with each circle representing a study and the size of each circle representing the study size.
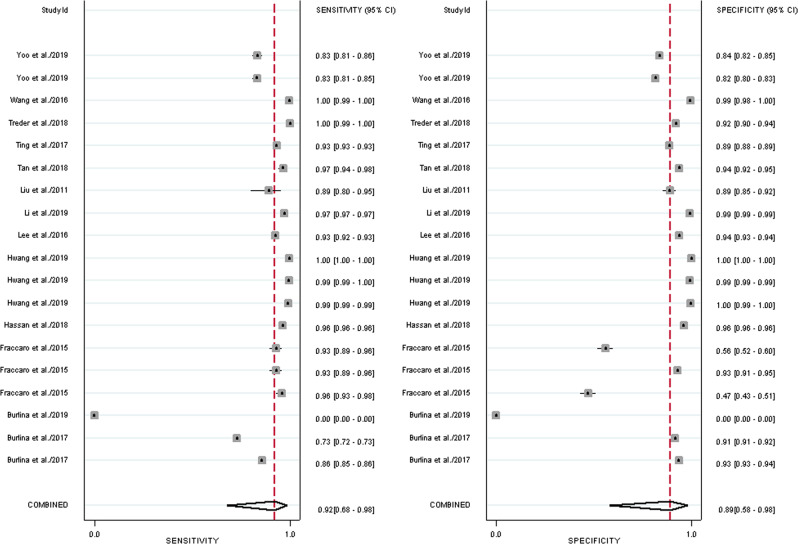
Figure 3 shows the hierarchical summary receiver operating characteristic (HSROC) curve where the circles indicate the study estimates, the HSROC curve or the summary curve, a summary operating point or the summary value for sensitivity and specificity, and the 95% confidence region for the summary operating point. The summary estimate for sensitivity was 0.918 [95% CI: 0.678, 0.98] and specificity was 0.888 [95% CI: 0.578, 0.98] for AMD screening using an ML classifier (Fig. 4). The summary estimates indicate that ML classifier correctly detects 91.8% of AMD cases and correctly classifies 88.8% of those without AMD as AMD-negative. The distribution of the studies in the plot demonstrates the variability of both specificity and sensitivity amongst studies. Five studies fall outside of the 95% confidence interval of the summary estimate.
Fig. 3. Hierarchical Summary Receiver Operating Characteristic (HSROC) plot.
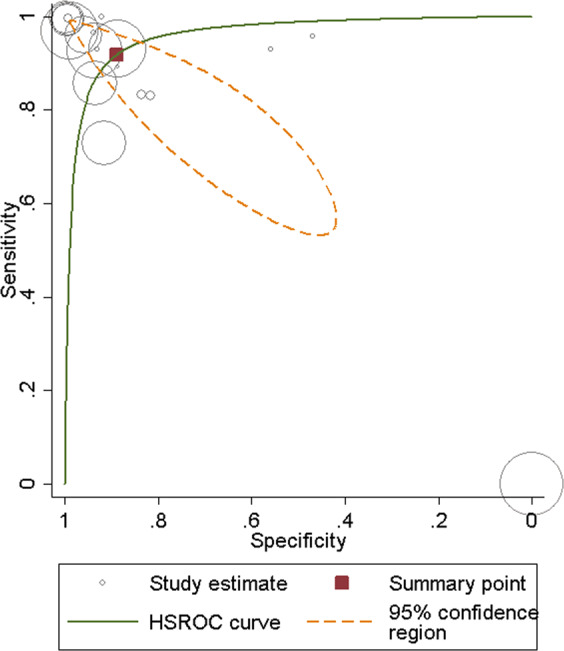
HSROC plot of studies assessing ML classifiers for AMD, including the study estimates, summary point, HSROC curve, and the 95% confidence region.
Fig. 4. Forest plot for the pooled sensitivity and specificity estimates.
Forest plot for the pooled sensitivity and specificity estimates for ML classifiers for AMD using a random effects model.
The positive likelihood ratio was 8.22 [95% CI: 1.52–44.48], while the negative likelihood ratio was 0.09 [95% CI: 0.02–0.52] (Appendix G). This shows that the likelihood of a positive diagnosis in an AMD case is greater than the likelihood of negative diagnosis in a non-AMD case. The positive likelihood ratio is >1 and it represents that the positive diagnosis is associated with AMD. Because the negative likelihood ratio is <1, the ML classifier which gave a negative diagnosis that is associated with the absence of AMD. The effectiveness of the diagnostic accuracy of the ML classifiers for AMD given by the DOR is 89.74 [95% CI: 3.05–2641.59] (Appendix G). This demonstrates that the relative odds of a positive screen test in AMD cases are 89.74 times more likely than a negative screen test in a non-AMD case. Thus, the ML classifier is discriminating between the true negative and true positive images correctly and accurately.
Discussion
A thorough systematic review and meta-analysis was conducted of the published literature which resulted in 64,798 subjects and 612,429 images used for training and validation from 13 full text articles. To the best of our knowledge, this is the first review of its kind to summarize the use of ML qualitatively and quantitatively for the diagnosis of AMD.
ML classifiers are advantageous at detecting the true positive and true negative cases for AMD as indicated by its high sensitivity and specificity. With a very high DOR estimate, it suggests that ML classifiers can discriminate between AMD and non-AMD images. The results of the diagnostic accuracy for ML classifiers show promising results for the automated diagnosis of AMD—it demonstrates the possibility that it can be used in clinical practice. AI will assist in tele-ophthalmology practices, especially in rural settings where patients do not have direct access to ophthalmologists [34]. The advantage to using ML classifiers in rural populations is the convenience that it can provide patients in obtaining an early detection of AMD, without the need of a clinician to confirm the diagnosis, and the reduction of patient or clinician travel costs. In urban settings, the use of ML for diagnosis can reduce patient load, wait times, and improve efficiency of ophthalmology clinics [35].
When considering the diagnostic accuracy of any tests, it is important to recognize the trade-offs between specificity and sensitivity. Sensitivity is the ability of the diagnostic test to detect positive cases in those with the disease, whereas specificity is the ability to detect negative cases in those without the disease [36]. Ideally, both sensitivity and specificity should be close to 1.0, though the sensitivity of the test may be of greater importance because it captures the false negatives. Although five studies fell outside of the 95% confidence interval of the summary estimate in the HSROC plot, based on our results, ML classifiers correctly detects 91.8% of AMD cases and correctly classifies 88.8% of those without AMD as AMD-negative. In a future meta-analysis, it would be worthy to investigate the accuracy and performance of AI algorithms to further classify AMD types. Hwang et al. (2019) observed that their AI algorithm sometimes misclassified active wet AMD as inactive wet AMD when the subretinal fluid was shallow, and misclassified dry AMD as inactive wet AMD when the drusenoid pigment epithelial detachment was especially large [29]. Thus, it may be of interest to know which types of AMD are most frequently mistaken for another classification by AI algorithms.
Study limitations for any meta-analysis must be considered before making any inferences. The risk of bias assessment using the QUADAS-2 tool resulted in majority of the studies to have a low risk of bias [19]. However, some of the included studies were retrieved from journals with a high interest in computer science and AI, thus they reported little clinically relevant information such as sensitivity and specificity values. It is in good practice for diagnostic accuracy studies to report not a singular accuracy proportion, but to also report the sensitivity, specificity, true positive, true negative, false positive, and false negative. Burlina et al. (2019) did not report its specificity or sensitivity value, but it was still included in the quantitative analysis because its reported AUC value was used for the SROC plot [21]. These values are important for future related meta-analysis studies and it further informs the readers the accuracy of the classifier.
In addition, meta-analysis of observational studies is influenced by inherent biases [37]. The results of our study could have been influenced by the race and age of the study participants, the ophthalmologist’s number of years of practice in the field, hospital location, imaging technique, camera model, AMD type and stage, and field of expertize within ophthalmology. Furthermore, there may be limitations in the varying use of multiple ML classifiers used in the OCT imaging cohort. Convoluted neural networks were the most reported ML classifier used in our included OCT studies. However, all included studies showed consistent results regardless of the type of classifier used. In addition, the clinical diagnosis and confirmation of AMD may be subject to each ophthalmologist or retinal specialist and study location. All studies should define the diagnosis of AMD in the same manner, but this information was not provided in all the studies.
In conclusion, the diagnostic accuracy of ML classifiers for AMD is very high and it shows very promising diagnostic test performance. The prospects for the use of ML for diagnosis of AMD in the clinical setting is possible, although to what extent our findings can be transferred to a real clinic practice still needs to be established. The advances to have an AI diagnosis system that is completely free of human involvement and supervision seems to still be a thought of the distant future, but these AI algorithms have the potential to ameliorate current medical demands. The use of AI will play a larger role in the regular practice of the diagnosis of eye diseases, and it will advance the important role of telemedicine—specifically tele-ophthalmology. Our study demonstrates just one aspect for the use of ML in the vast field of ophthalmology. The possibilities for the application of AI will be endless as innovative research continues and new algorithms are developed.
Summary
What was known before
The use of ML classifiers for the diagnosis of eye diseases such as diabetic retinopathy is becoming more prevalent in the medical community.
The use of AI algorithms is a powerful tool, and it is a method of obtaining a cost-effective, simple, and fast diagnosis of eye diseases.
What this study adds
To our knowledge, this is the first meta-analysis to analyze the diagnostic accuracy of ML classifiers for AMD among all eyes available in datasets to assess their accuracy and reliability to be implemented in clinical settings.
The pooled estimate for sensitivity was 0.918 [95% CI: 0.678, 0.98] and specificity was 0.888 [95% CI: 0.578, 0.98] for AMD screening using ML classifiers.
The included studies show promising results for the diagnostic accuracy of the ML classifiers for AMD and its implementation in clinical settings.
Supplementary information
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Funding
The authors received no financial support for the research, authorship, and/or publication of this article.
Author contributions
RC was responsible for the conceptualization of the research topic, designing and writing the protocol, conducting the database and gray literature search, screening the studies, conducting the risk of bias assessment, curating the data, extraction of data, interpreting the results, writing, and editing the paper. JC was responsible for the screening of the studies, conducting the risk of bias assessment, curating the data, reviewing, and editing the paper. TS was involved in the conceptualization of the research topic, contributed to the general methodology of the report, provided feedback on the report, reviewed and edited the paper. MM was involved in the conceptualization of the research topic, contributed to the general methodology of the report, provided feedback on the report, reviewed and edited the paper. MM-M was responsible for the conceptualization of the research topic, designing the review protocol, analyzing and interpreting the data using statistical software, validation of results, writing and editing the paper. She was also the senior supervisor for the project and was responsible for the project administration.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41433-021-01540-y.
References
- 1.Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358:2606–17. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 2.Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. Lancet. 2012;379:1728–38. doi: 10.1016/S0140-6736(12)60282-7. [DOI] [PubMed] [Google Scholar]
- 3.Pennington KL, DeAngelis MM. Epidemiology of age-related macular degeneration (AMD): associations with cardiovascular disease phenotypes and lipid factors. Eye Vis (Lond, Engl) 2016;3:34. doi: 10.1186/s40662-016-0063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Group A-REDSR. A simplified severity scale for age-related macular degeneration: AREDS report no. 18. Arch Ophthalmol. 2005;123:1570. doi: 10.1001/archopht.123.11.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou C-F, Frances Cotch M, Vitale S, Zhang X, Klein R, Friedman DS, et al. Age-related eye diseases and visual impairment among U.S. adults. Am J Prev Med. 2013;45:29–35. doi: 10.1016/j.amepre.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rein DB, Wittenborn JS, Zhang X, Honeycutt AA, Lesesne SB, Saaddine J. Forecasting age-related macular degeneration through the year 2050: the potential impact of new treatments. Arch Ophthalmol. 2009;127:533–40. doi: 10.1001/archophthalmol.2009.58. [DOI] [PubMed] [Google Scholar]
- 7.Yannuzzi LA, Ober MD, Slakter JS, Spaide RF, Fisher YL, Flower RW, et al. Ophthalmic fundus imaging: today and beyond. Am J Ophthalmol. 2004;137:511–24. doi: 10.1016/j.ajo.2003.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Lamoureux EL, Mitchell P, Rees G, Cheung G, Yeo I, Lee SY, et al. Impact of early and late age-related macular degeneration on vision-specific functioning. Br J Ophthalmol. 2011;95:666–70. doi: 10.1136/bjo.2010.185207. [DOI] [PubMed] [Google Scholar]
- 9.Kanagasingam Y, Bhuiyan A, Abràmoff MD, Smith RT, Goldschmidt L, Wong TY. Progress on retinal image analysis for age related macular degeneration. Prog Retin Eye Res. 2014;38:20–42. doi: 10.1016/j.preteyeres.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Shanafelt TD, Boone S, Tan L, Dyrbye LN, Sotile W, Satele D, et al. Burnout and satisfaction with work-life balance among US physicians relative to the general US population. Arch Intern Med. 2012;172:1377–85. doi: 10.1001/archinternmed.2012.3199. [DOI] [PubMed] [Google Scholar]
- 11.Cheung R, Yu B, Iordanous Y, Malvankar-Mehta MS. The prevalence of occupational burnout among ophthalmologists: a systematic review and meta-analysis. Psychol Rep. 2020; e-pub ahead of print; 10.1177/0033294120954135. [DOI] [PubMed]
- 12.Dewa CS, Loong D, Bonato S, Thanh NX, Jacobs P. How does burnout affect physician productivity? A systematic literature review. BMC Health Serv Res. 2014;14:325. doi: 10.1186/1472-6963-14-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu W, Tong Y, Yu Y, Xing Y, Chen C, Shen Y. Applications of artificial intelligence in ophthalmology: general overview. J Ophthalmol. 2018;11:1–15. [DOI] [PMC free article] [PubMed]
- 14.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–10. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 15.Murtagh P, Greene G, O’Brien C. Current applications of machine learning in the screening and diagnosis of glaucoma: a systematic review and meta-analysis. Int J Ophthalmol. 2020;13:149–62. doi: 10.18240/ijo.2020.01.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X, Lin S, Wong TY. Automatic feature learning to grade nuclear cataracts based on deep learning. IEEE Trans Biomed Eng. 2015;62:2693–701. doi: 10.1109/TBME.2015.2444389. [DOI] [PubMed] [Google Scholar]
- 17.Ullah W, Pathan SK, Panchal A, Anandan S, Saleem K, Sattar Y, et al. Cost-effectiveness and diagnostic accuracy of telemedicine in macular disease and diabetic retinopathy: a systematic review and meta-analysis. Medicine (Baltimore). 2020;99:e20306. [DOI] [PMC free article] [PubMed]
- 18.Moher D, Liberati A, Tetzlaff J, Altman DG, Group TP. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLOS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whiting PF, Rutjes AWS, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36.20. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 20.Burlina PM, Joshi N, Pekala M, Pacheco KD, Freund DE, Bressler NM. Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol. 2017;135:1170–6. doi: 10.1001/jamaophthalmol.2017.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burlina PM, Joshi N, Pacheco KD, Liu TYA, Bressler NM. Assessment of deep generative models for high-resolution synthetic retinal image generation of age-related macular degeneration. JAMA Ophthalmol. 2019;137:258–64. doi: 10.1001/jamaophthalmol.2018.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ting DSW, Cheung CY-L, Lim G, Tan GSW, Quang ND, Gan A, et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211–23. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Treder M, Lauermann JL, Eter N. Automated detection of exudative age-related macular degeneration in spectral domain optical coherence tomography using deep learning. Graefe’s Arch Clin Exp Ophthalmol. 2018;256:259–65. doi: 10.1007/s00417-017-3850-3. [DOI] [PubMed] [Google Scholar]
- 24.Tan JH, Bhandary SV, Sivaprasad S, Hagiwara Y, Bagchi A, Raghavendra U, et al. Age-related macular degeneration detection using deep convolutional neural network. Futur Gener Comput Syst. 2018;87:127–35. doi: 10.1016/j.future.2018.05.001. [DOI] [Google Scholar]
- 25.Yoo TK, Choi JY, Seo JG, Ramasubramanian B, Selvaperumal S, Kim DW. The possibility of the combination of OCT and fundus images for improving the diagnostic accuracy of deep learning for age-related macular degeneration: a preliminary experiment. Med Biol Eng Comput. 2019;57:677–87. doi: 10.1007/s11517-018-1915-z. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, Zhang Y, Yao Z, Zhao R, Zhou F. Machine learning based detection of age-related macular degeneration (AMD) and diabetic macular edema (DME) from optical coherence tomography (OCT) images. Biomed Opt Express. 2016;7:4928–40. doi: 10.1364/BOE.7.004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraccaro P, Nicolo M, Bonetto M, Giacomini M, Weller P, Traverso CE, et al. Combining macula clinical signs and patient characteristics for age-related macular degeneration diagnosis: a machine learning approach. BMC Ophthalmol. 2015;15:10. doi: 10.1186/1471-2415-15-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hassan T, Akram MU, Askhtar M, Khan SA, Yasin U. Multilayered deep structure tensor delaunay triangulation and morphing based automated diagnosis and 3D presentation of human macula. J Med Syst. 2018;42:223. doi: 10.1007/s10916-018-1078-3. [DOI] [PubMed] [Google Scholar]
- 29.Hwang D-K, Hsu C-C, Chang K-J, Chao D, Sun C-H, Jheng Y-C, et al. Artificial intelligence-based decision-making for age-related macular degeneration. Theranostics. 2019;9:232–45. doi: 10.7150/thno.28447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee CS, Baughman DM, Lee AY. Deep learning is effective for classifying normal versus age-related macular degeneration OCT images. Ophthalmol Reti. 2017;1:322–7. doi: 10.1016/j.oret.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li F, Chen H, Liu Z, Zhang X, Wu Z. Fully automated detection of retinal disorders by image-based deep learning. Graefes Arch Clin Exp Ophthalmol. 2019;257:495–505. doi: 10.1007/s00417-018-04224-8. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y-Y, Ishikawa H, Chen M, Wollstein G, Duker JS, Fujimoto JG, et al. Computerized macular pathology diagnosis in spectral domain optical coherence tomography scans based on multiscale texture and shape features. Investig Ophthalmol Vis Sci. 2011;52:8316–22. doi: 10.1167/iovs.10-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srinivasan PP, Kim LA, Mettu PS, Cousins SW, Comer GM, Izatt JA, et al. Fully automated detection of diabetic macular edema and dry age-related macular degeneration from optical coherence tomography images. Biomed Opt Express. 2014;5:3568–77. doi: 10.1364/BOE.5.003568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamminen H, Voipio V, Ruohonen K, Uusitalo H. Telemedicine in ophthalmology. Acta Ophthalmol Scand. 2003;81:105–9. doi: 10.1034/j.1600-0420.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- 35.Saleem SM, Pasquale LR, Sidoti PA, Tsai JC. Virtual ophthalmology: telemedicine in a COVID-19 era. Am J Ophthalmol. 2020;216:237–42. doi: 10.1016/j.ajo.2020.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altman DG, Bland JM. Diagnostic tests. 1: sensitivity and specificity. BMJ. 1994;308:1552. doi: 10.1136/bmj.308.6943.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.