Abstract
Background & Aims
Liver fibrosis arises from long-term chronic liver injury, accompanied by an accelerated wound healing response with interstitial accumulation of extracellular matrix (ECM). Activated hepatic stellate cells (HSC) are the main source for ECM production. MicroRNA29a (miR-29a) is a crucial antifibrotic miRNA that is repressed during fibrosis, resulting in up-regulation of collagen synthesis.
Methods
Intracellular and extracellular miRNA levels of primary and immortalized myofibroblastic HSC in response to profibrogenic stimulation by transforming growth factor β (TGFβ) or platelet-derived growth factor-BB (PDGF-BB) or upon inhibition of vesicular transport and autophagy processes were determined by quantitative polymerase chain reaction. Autophagy flux was studied by electron microscopy, flow cytometry, immunoblotting, and immunocytochemistry. Hepatic and serum miR-29a levels were quantified by using both liver tissue and serum samples from a cohort of chronic hepatitis C virus patients and a murine CCl4 induced liver fibrosis model.
Results
In our study, we show that TGFβ and PDGF-BB resulted in decrease of intracellular miR-29a and a pronounced increase of vesicular miR-29a release into the supernatant. Strikingly, miR-29a vesicular release was accompanied by enhanced autophagic activity and up-regulation of the autophagy marker protein LC3. Moreover, autophagy inhibition strongly prevented miR-29a secretion and repressed its targets’ expression such as Col1A1. Consistently, hepatic miR-29a loss and increased LC3 expression in myofibroblastic HSC were associated with increased serum miR-29a levels in CCl4-treated murine liver fibrosis and specimens of hepatitis C virus patients with chronic liver disease.
Conclusions
We provide evidence that activation-associated autophagy in HSC induces release of miR-29a, whereas inhibition of autophagy represses fibrogenic gene expression in part through attenuated miR-29a secretion.
Keywords: Circulating MiRNA, MiR-29, Liver Fibrosis
Abbreviations used in this paper: αSMA, alpha smooth muscle actin; ATG, autophagy-related; DMEM, Dulbecco modified Eagle medium; ECM, extracellular matrix; EV, extracellular vesicles; FCS, fetal calf serum; HCV, hepatitis C virus; HSC, hepatic stellate cells; miRNA, microRNA; mTOR, mammalian target of rapamycin; PCR, polymerase chain reaction; PDGF, platelet-derived growth factor; siRNA, small interfering RNA; TGFβ, transforming growth factor beta; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling
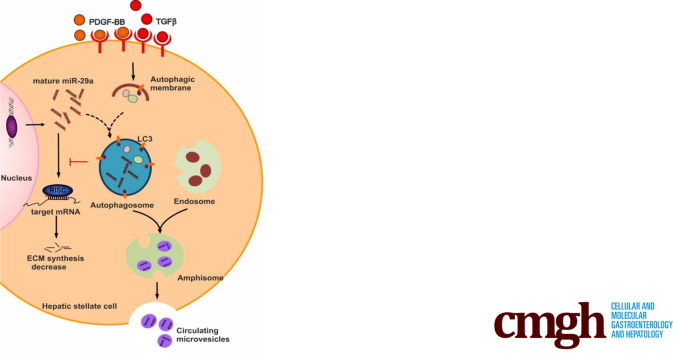
Graphical abstract
Summary.
Cellular miR-29a, inhibiting collagen synthesis, is reduced in activated HSC because of its vesicular release upon autophagy induction by profibrogenic TGFβ or PDGF-BB stimulation. Accordingly, autophagy in HSC of clinical specimens with liver fibrosis was linked to elevated serum miR-29a levels.
Liver fibrosis represents a major cause of morbidity and mortality worldwide. It is the pathophysiological consequence of different types of chronic inflammatory liver diseases, including liver injury upon viral hepatitis, alcoholic or drug abuse, in response to metabolic syndrome and fat accumulation, or upon autoimmune or genetic disorders.1 Transforming growth factor β (TGFβ) is the central mediator that is responsible for the imbalance between extracellular matrix (ECM) synthesis and degradation in human and experimental liver fibrosis.2,3 TGFβ signaling includes various pathways such as the non-canonical TAK1/JNK and the RAS/RAF/MAPK pathways as well as the canonical SMAD pathway, leading to induction of ECM synthesis.4,5 In response to liver injury, increased TGFβ levels stimulate hepatic stellate cells (HSC) to transdifferentiate into myofibroblasts,6,7 which are then the main source of ECM production in the fibrotic liver. Among the ECM proteins of fibrotic liver, fibrillar collagens such as collagen I and III are highly increased.2,3,8
TGFβ-induced myofibroblastic HSC activation is not only associated with ECM synthesis but also with an enhanced expression of profibrogenic growth factors such as TGFβ itself, connective tissue growth factor, and platelet-derived growth factor (PDGF).2,3 PDGF is a mitogenic growth factor whose polypeptide chains A and B are arranged in AA, AB, or in BB combinations. Autocrine PDGF-BB stimulation is most potent to induce HSC proliferation.9,10 Hence, the autocrine stimulation of HSC by the altered growth factor profile, in particular by TGFβ and PDGF-BB, triggers HSC activation and maintenance of liver fibrogenesis.2,3,8 Furthermore, exposure of HSC to both mediators potently decreases miR-29.11,12 MicroRNAs (miRNAs) are small non-coding RNAs, which participate in regulation of gene expression by their interaction with transcripts and subsequent inhibition of mRNA translation. Because the interacting region of miRNAs, the so-called seed region, is only 7–8 base pairs long, one mRNA is often regulated by several miRNAs, and one miRNA can interact with more than one mRNA transcript.13,14 The members of the miR-29 family are known to target transcripts that encode various ECM members (eg, elastin, fibrillin, and various collagen subunits). Therefore, miR-29 acts as an antifibrotic miRNA, which is markedly diminished during acute and chronic liver disease.12,15, 16, 17, 18, 19, 20
In the present study, we show that TGFβ- and PDGF-BB–mediated miR-29 decrease in HSC is associated with autophagy-related vesicular release. Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved cellular mechanism, which recycles the cytoplasm components, generates energy under stress conditions, removes superfluous and damaged organelles, adapts to changing nutrient conditions, and maintains cellular homeostasis. Autophagy involves the coordination of numerous autophagy-related (ATG) proteins and the formation of double membrane autophagosomes.
Recent studies have linked liver damage and fibrogenesis to autophagy, affecting hepatic cell types in a different manner.21,22 Whereas in macrophages and endothelial cells inhibition of autophagy accelerates fibrogenesis, in hepatocytes autophagy is important for cellular homeostasis and inhibition leads to development of liver tumors.23,24 In HSC, autophagy promotes the activation and transdifferentiation processes into myofibroblasts, associated with increased ECM synthesis and the enhanced catabolism of fatty acids, providing then the necessary energy for cell activation.25 Hence, the blockade of autophagy during liver fibrogenesis by HSC-specific ATG7 deletion maintains resistance to its activation and liver fibrosis.26 TGFβ stimulates autophagy and is a central driver of autophagy in fibroblasts and in HSC.27,28 Interestingly, not only TGFβ but also PDGF-BB stimulates autophagy in HSC,25 and inhibition of autophagy represses collagen synthesis.29
In this study, we demonstrate that autophagy, induced by TGFβ and PDGF-BB, directly regulates cellular miR-29a levels in myofibroblastic HSC. Notably, autophagy lowers cellular miR-29a levels by inducing its release into secreted extracellular vesicles (EV), thereby attenuating intracellular antifibrotic functions of miR-29a, including inhibition of collagen I synthesis. Moreover, autophagy-induced miR-29a increases circulating miR-29 levels in serum samples of patients with hepatitis C virus (HCV)-associated fibrosis. Increased extracellular miR-29a levels are significantly correlated with fibrosis progression of patients with chronic HCV liver disease.
Results
TGFβ and PDGF-BB Stimulation Results in Vesicular Release From HSC
Because isolated primary HSC in culture spontaneously transdifferentiate into myofibroblasts, we first studied the miR-29a levels in primary mouse HSC after taking them into culture. Whereas in freshly isolated HSC miR-29a was highly abundant, miR-29a levels were decreased during culture and much lower in HSC, which were maintained in culture for 7 days and expressed the myofibrobast-specific marker α-smooth muscle actin (αSMA) (Figure 1A). Previous studies have shown that TGFβ and PDGF-BB are main stimulators of myofibroblastic transition. In addition, they lead to miR-29 decrease and to enhanced ECM deposition.15,30,31 After profibrogenic stimulation with TGFβ or PDGF-BB treatment, cellular miR-29a was reduced in immortalized myofibroblastic HSC by more than 70% as shown in Figure 1B and C (left panel). Surprisingly, analysis of the supernatants of TGFβ and PDGF-BB stimulated HSC revealed that the supernatants from stimulated HSC contained significantly more extracellular miR-29a than the corresponding controls (Figure 1B and C, middle panel). Cellular release of miRNA can be caused by cell apoptosis/necrosis followed by generation of apoptotic bodies, but also by actively regulated vesicular release.32 To rule out the possibility that miR-29 release is due to cell death, we controlled the extent of apoptosis and necrosis. Deoxyuride-5′-triphosphate biotin nick end labeling (TUNEL) assays (Figure 1D) and annexin V cytometry (Figure 1E) clearly showed that there was no increase of apoptosis or necrosis, indicating that miR-29a release upon fibrogenic stimulation is a cell-regulated mechanism.
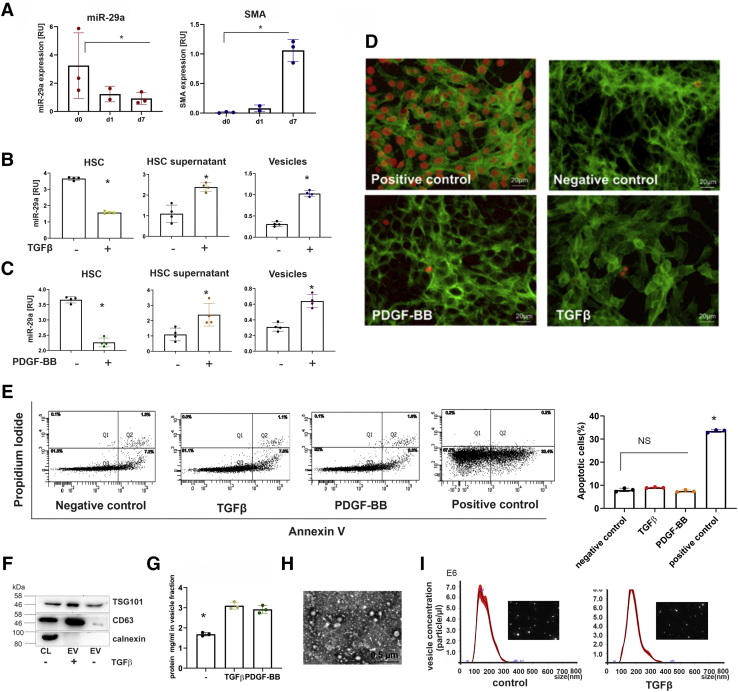
Figure 1.
Intracellular and extracellular miR-29a levels of HSC in response to stimulation with TGFβ or PDGF-BB. MicroRNA-29a levels and αSMA expression freshly isolated HSC (d0) in comparison with levels in HSC of early (day 1, d1) and late primary culture (day 7, d7). Expression profiles were determined in triplicates by quantitative PCR from 3 independent primary HSC preparations (A). In response to TGFβ (B) or PDGF-BB (C) treatment, cellular miR-29a in immortalized HSC (HSC-T6) and extracellular miR-29a in HSC culture supernatants as well as in vesicles were quantified by real-time PCR (B and C). After stimulation by TGFβ and PDGF-BB, apoptosis was determined by TUNEL analysis in HSC; DNase-treated HSCs were used as positive control. HSC were counterstained with fluorescein isothiocyanate (FITC)-conjugated phalloidin. TUNEL-positive cells are shown by red fluorescence (D). HSCs were stained with annexin V-FITC and propidium iodide and analyzed by flow cytometry (E). The percentages of apoptotic cells after TGFβ, PDGF-BB, or H2O2 (positive control) treatment are shown. (∗P value <.05) (E). Vesicular fractions from supernatants of TGFβ- or PDGF-BB-stimulated HSC were characterized by immunoblotting using antibodies against EV markers, TSG101 and CD63. Calnexin was used as control for intracellular component (F). The amounts of secreted vesicles released from control HSC or TGFβ or PDGF-BB treated HSC were estimated by protein determination of the respective extracellular vesicular fractions (G). Moreover, isolated vesicles were imaged by electron microscopy (H) and the nanoparticle tracking method (I).
In an attempt to investigate the mechanism underlying miR-29 release in response to TGFβ and PDGF-BB, vesicles from HSC culture supernatants were purified to quantify the extracellular miR-29 content. Indeed, TGFβ and PDGF-BB stimulation significantly increased the levels of vesicular miR-29a (Figure 1B and C, right panel). By immunoblotting we showed that vesicles were CD63 and TSG101 positive (Figure 1F). Moreover, the increase of extracellular miR-29 in the cell culture supernatant was accompanied by marked elevation of its vesicular fraction (Figure 1G). The vesicle fraction was imaged by electron microscope (Figure 1H) and nanoparticle tracking analysis (Figure I), establishing that their size was 100–200 nm in diameter, which corresponds to the size of small EV.33
Inhibition of Vesicle Transport Leads to Increment of Cellular miR-29a in Response to Profibrogenic Stimulation
Because miR-29a was enclosed in vesicles and secreted into the supernatant upon profibrogenic stimulation, we next investigated which cellular mechanism triggers miR-29a transport into the extracellular environment. It has been previously reported that vesicles require myosin motor proteins.34 Blebbistatin, a myosin II ATPase inhibitor, is a potent inhibitor of both cell contraction and vesicular transport.35 Inhibition of vesicle transport by blebbistatin significantly inhibited TGFβ-induced miR-29a secretion, as evidenced by an increase in cellular miR-29a (Figure 2A, left) on the one hand and a significant decrease in extracellular miR-29a in HSC culture supernatants on the other (Figure 2A, right). Myosin 5a is a versatile non-muscle central motor protein that is crucial for vesicular transport.34 Here we specifically interrupted the vesicular miR-29a export by myosin 5a inhibition. In agreement with the findings after blebbistatin treatment, small interfering RNA (siRNA)-mediated knockdown of myosin 5a led to reduced miR-29a release from primary myofibroblastic HSC (Figure 2B).
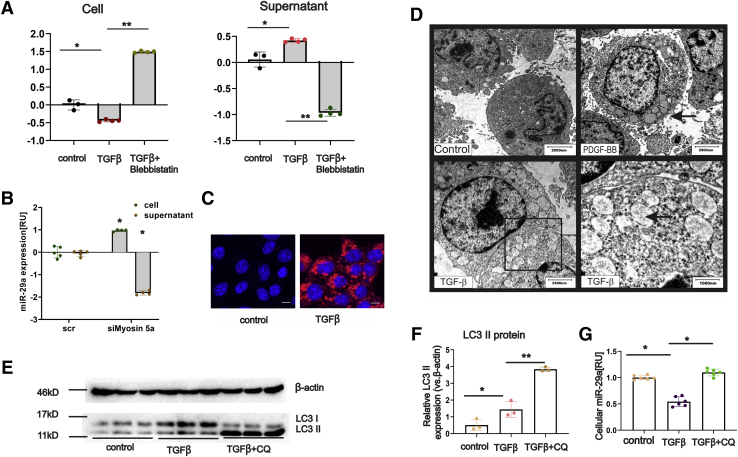
Figure 2.
Stimulation of HSC fibrogenesis by PDGF-BB and TGFβ leads to miR-29a secretion and enhanced autophagy. HSC-T6 cells were treated with TGFβ in combination with or without blebbistatin inhibition. Both the cellular and supernatant RNA was isolated, and real-time PCR was performed to analyze intracellular and extracellular miR-29a levels (A). After treatment of primary HSCs (on day 7 after isolation) with siMyosin 5a, intracellular and extracellular miR-29a levels, quantified by PCR, were shown (B). Cells, nonstimulated or stimulated with TGFβ, were immunostained with LC3 antibodies and subsequently imaged by confocal microscopy (blue = DAPI, red = LC3 protein, scale bar = 15 μm (C). Transmission electron microscopy revealed autophagosome formation in HSCs in response to PDGF-BB and TGFβ treatment (D). LC3 protein expression was shown by immunoblotting using cell lysates from HSCs, nontreated (control), treated with TGFβ, or treated with both TGFβ and chloroquine (CQ) (E and F). Cellular miR-29a levels were determined by qPCR showing that miR-29a decrease upon TGFβ treatment was blocked when HSC were also treated with chloroquine (G). (P value ∗<.05, ∗∗<.001).
Profibrogenic Stimulation Enhances Autophagy-Mediated miR-29 Release From HSC
Myosins play a versatile role in cellular vesicular transport, including the several steps of autophagy pathway including autophagy initiation, phagophore expansion, and fusion of autophagosome and lysosome.36 Moreover, both TGFβ and PDGF-BB can induce autophagy.25,27,28 Hence, we confirmed the formation of autophagosome-like structures in myofibroblastic HSC upon stimulation with TGFβ and PDGF-BB using transmission electron microscopy (Figure 2D). Furthermore, the expression of the autophagosome membrane protein LC3 was highly up-regulated after TGFβ treatment (Figure 2C, E, and F). These results underscore the impact of TGFβ or PDGF-BB stimulation in autophagosome formation and activation of the autophagy.
Chloroquine is a potent autophagy inhibitor, blocking the fusion between autophagosomes and other vesicles.37 In agreement, chloroquine treatment resulted in an increase of the activated LC3 II form (Figure 2E and F). Notably, inhibition of autophagy activity by chloroquine significantly abolished the TGFβ-induced miR-29a secretion (Figure 2G).
Autophagy Induction Leads to miR-29a Secretion
Next, we studied miR-29a release during the time course of autophagy induction by TGFβ and PDGF-BB. There was a continuous up-regulation of miR-29a secretion and increase of LC3 protein expression in a time-dependent manner during TGFβ (Figure 3A and C) or PDGF-BB stimulation (Figure 3B and D). We also confirmed autophagy induction using cationic amphiphilic tracer dye to measure autophagic vacuoles. Flow cytometry analysis showed that the percentage of autophagic HSCs increased more than 3-fold in response to profibrogenic stimulation (Figure 3E and F). To verify that miR-29 release is related to autophagy, we treated HSC with the autophagy inducers rapamycin or torin-1 (Figure 3E and G). Importantly, an increase in miR-29a secretion into the HSC culture supernatant was confirmed after autophagy induction by rapamycin and torin-1 treatment (Figure 3A and B). Here, we show that induction of autophagy by both profibrogenic mediators and classical autophagy inducers strongly up-regulates extracellular levels of miR-29a (Figure 3A and B).
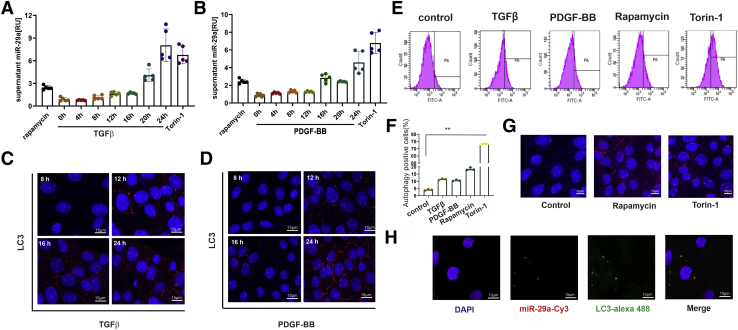
Figure 3.
Time course of miR-29a release from HSC after TGFβ or PDGF-BB stimulation. Extracellular miR-29a levels were analyzed by real-time PCR (A and B), and LC3 protein immunofluorescence staining (C and D) was performed after 0, 4, 8, 12, 16, 20, and 24 hours of TGFβ (A and C) or PDGF-BB (B and D) treatment of HSC-T6 cells. Moreover, autophagosomes were stained by cationic amphiphilic tracer dye followed by fluorescence-activated cell sorter analysis (E). The percentages of autophagy-positive cells are presented in the bar diagram. (∗∗P < .001; data from 3 representative experiments are displayed) (F). Rapamycin and Torin-1 treatment for 24 hours were used as positive controls (G). Cy3-labeled miR-29a (red) was transfected, and colocalization with LC3 protein (green) expression was monitored after LC3 immunocytochemistry (scale bar = 15 μm) (H).
Therefore, miR-29a release is not specific to profibrogenic stimulation but rather to induction of autophagy, which is stimulated either by profibrogenic mediators (TGFβ and PDGF-BB) or by other autophagy-inducing agents. Furthermore, fluorochrome-labeled miR-29a, transfected into HSC-T6, co-localized with cellular LC3 (Figure 3H), indicating that after profibrogenic stimulation vesicular miR-29a release is strongly associated to its recruitment into autophagosomes.
Autophagy Inhibition Leads to Immense Increment of Intracellular miR-29a
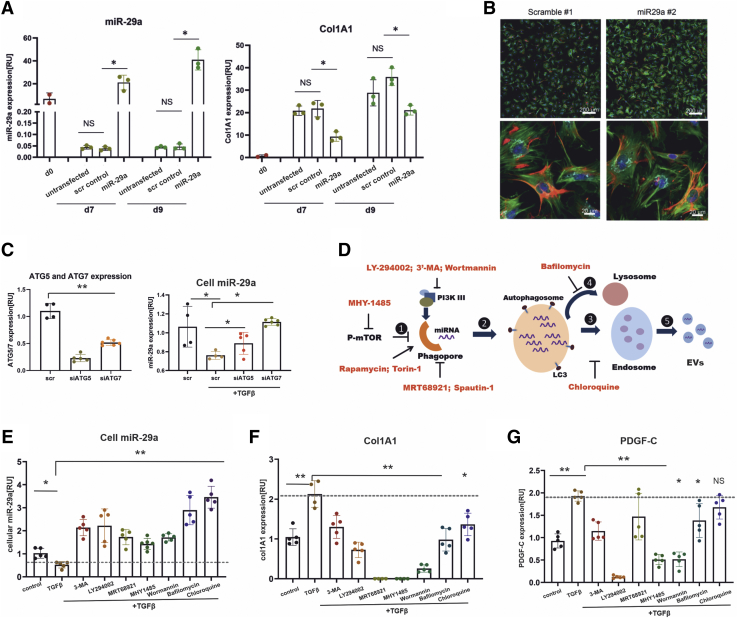
Because miR-29a acts as an antifibrotic miRNA that targets ECM gene expression, we hypothesized that inhibition of autophagy would lead to elevated cellular HSC miR-29a levels, followed by a decrease in miR-29a targeted genes such as the Col1A1 subunit and PDGF-C. In agreement with previous data showing that miR-29 targets ECM synthesis,15, 16, 17, 18 we first proved that miR-29a overexpression leads to a significantly reduced expression of the collagen subunit Col1A1 in primary HSC (Figure 4A) without changing the myofibroblastic HSC morphology (Figure 4B).
Figure 4.
Inhibition of autophagy blocks TGFβ-induced miR-29a secretion and up-regulates miR-29a target gene expression. MicroRNA-29a and Col1A1 gene expression levels were quantified in freshly isolated HSC (d0) or in cultured primary HSC after transgenic overexpression of miR-29a in comparison with non-transfected primary HSC or transfected with scramble RNA (scr). HSC were transduced with miR-29a mimic RNA or scr RNA at days 4 and 6 and harvested 72 hours after transfection. Transgenic miR-29a mimic expression leads to Col1A1 down-regulation (A). Mouse HSC at day 7, fixed 72 hours after transfection with scramble or miR29a, were stained with phalloidin (green) and anti-desmin antibodies (red). Nuclei were stained with DAPI, and collagen1 is shown in aqua blue after immunostaining with anti-collagen 1 antibodies. There were no morphologic changes observed after miR-29 treatment (B). ATG5 and ATG7 expression in HSC-T6 cells treated with siATG5 (50 nmol/L) or siATG7 (50 nmol/L), respectively, versus scramble siRNA was analyzed by quantitative PCR, showing efficient ATG5 and ATG7 inhibition. Quantitative PCR of cellular miR-29a levels proved that ATG5 and ATG7 blockade after siRNA treatment (siATG5, siATG7) restored the TGFβ-stimulated repression in scramble treated HSC-T6 (C). The working mechanisms of autophagy inhibitors are illustrated (D). Briefly, sufficient amino acid supply (or other cytoplasmic changes) sustains the activity of mTOR, the latter of which prevents the induction of autophagy ①. The elongation of phagophores requires the activity of PI3K III and formation of Beclin1/VPS34 and ULK1 complex. After formation of autophagosomes ②, they fuse alternatively with lysosomes to form auto-lysosomes ④, merge with endosomes to form multivesicular bodies ③, or form amphisomes, which ultimately leads to the secretion of EV ⑤. Intracellular miR-29a expression levels were analyzed after dual treatment with TGFβ and different autophagy inhibitors MHY1485, 3-MA, wortmannin, LY294002, MRT68921, bafilomycin A1, chloroquine (E). MicroRNA-29a target genes, Col1A1 (F) and PDGF-C (G), were determined in response to TGFβ stimulation and diverse autophagy inhibition. The induction of Col1A1 and PDGF-C after TGFβ is indicated by the dashed line (F and G). (P value ∗<.05, ∗∗<.001).
The ATGs play an indispensable role in autophagosome formation. ATG5 and ATG7 are crucial for initiation of autophagy.38 To strengthen the link between autophagy and enhanced secretion of miR-29a, we knocked down either ATG5 or ATG7 expression in combination with TGFβ treatment. Strikingly, treatment with small interferingATG5 or small interfering ATG7 significantly neutralized the TGFβ-induced decrease of cellular miR-29a (Figure 4C).
Whereas chloroquine functions mainly by blocking maturation of vesicles and disrupts autophagosome fusion with other vesicles,37 other autophagy inhibitors interfere with autophagy initiation, phagophore expansion, or autophagosome formation. Because autophagy initiation requires the deactivation of mammalian target of rapamycin (mTOR), which is crucial requirement for autophagy, MHY-1485, acting as the mTOR activator, has been widely used to inhibit autophagy. The elongation of phagophores is controlled by kinase activity of PI3K III and formation of beclin1-VPS34 and ULK1 complex. 3-Methyladenine functions as an autophagy inhibitor by targeting PI3K III, whereas bafilomycin inhibits vacuolar H+ ATPase of lysosomes to suppress their acidification (Figure 4D). We used the autophagy inhibitors 3-methyladenine, bafilomycin, chloroquine, MHY-1485, MRT68921, wortmannin, and LY-294002 (Table 1, Figure 4D). Notably, all of the inhibitors blocked miR-29a secretion, showing as significant recovery of the cellular miR-29a levels (Figure 4E). By interfering with lysosomal fusion, chloroquine and bafilomycin especially markedly inhibited cellular miR-29a loss.
Table 1.
Autophagy Inhibitors and Inducers
| Chemical reagent | Working concentration | Function (references) | Sites of target in autophagy signaling |
|---|---|---|---|
| Chloroquine | 50 μmol/L | Autophagy inhibitor | Autophagosome maturation |
| Bafilomycin | 50 nmol/L | Autophagy inhibitor | V-type ATPase inhibitor |
| MRT68921 | 1 μmol/L | Autophagy inhibitor | ULK1 complex |
| LY-294002 | 10 mmol/L | Autophagy inhibitor | Class I and Class III PI3K |
| 3′-MA | 5 mmol/L | Autophagy inhibitor | Class I and Class III PI3K |
| Wortmannin | 50 nmol/L | Autophagy inhibitor | Class I and Class III PI3K |
| MHY1485 | 2 μmol/L | Autophagy inhibitor | mTOR |
| Torin-1 | 5 μmol/L | Autophagy inducer | mTOR |
| Rapamycin | 100 ng/mL | Autophagy inducer | mTOR |
MicroRNA-29 acts as an antifibrotic miRNA because it targets transcripts that encode ECM proteins such as different collagens but also profibrogenic mediators such as PDGF-C.15,16,30,31,39 The enhanced cellular miR-29a levels, caused by autophagy inhibition (Figure 4E), resulted in a significant down-regulation of the miR-29a target genes, Col1A1 and PDGF-C (Figure 4F and G). However, the inhibition of the TGFβ-mediated effects on the two miR-29 targets differed between autophagy inhibitors and was less pronounced on PDGF-C expression than on Col1A1 expression. Interestingly, chloroquine showed the strongest restoration of cellular miR-29a, but in contrast to bafilomycin and the PI3K inhibitors, it did not markedly influence Col1A1 or PDGF-C expression (Figure 4E–G). Although besides the miR-29a control, the regulation of miR-29a targeted genes is suggested to underlie additional mechanistic links, our findings definitively show that autophagy inhibition repressed the Col1A and PDGF-C expression by restoring miR-29a function.
Increased Circulating miR-29a Levels During Liver Fibrosis
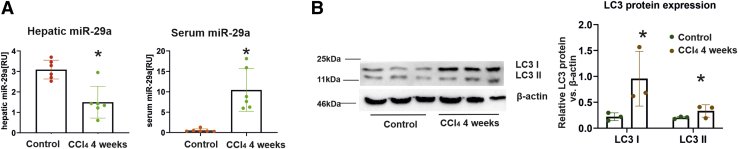
To determine whether autophagic flux-mediated miR-29 release is increased during fibrogenesis, we quantified miR-29a levels in liver and serum from a mouse CCl4 fibrosis model and in clinical samples from patients with chronic HCV. Hepatic miR-29a levels were down-regulated in mice treated with CCl4 for 4 weeks, whereas miR-29a in corresponding serum samples were up-regulated more than 5-fold (Figure 5A). In addition, we observed a significant overexpression of LC3 protein in livers of CCl4-treated mice, indicating that autophagic activity was strongly intensified during CCl4-induced fibrogenesis (Figure 5B).
Figure 5.
Hepatic LC3 overexpression in murine fibrosis is associated with miR-29a release in the serum. After 4 weeks of CCl4 intoxication fibrosis was developed, and total RNA was isolated from both liver and serum samples of CCl4-treated mice (n = 6) or control mice (n = 6). Liver and serum miR-29a levels were quantified by real-time PCR (A). LC3 protein expression levels were determined by immunoblotting and subsequent analysis using quantitative chemiluminescent imaging (B). (P value ∗<.05, ∗∗<.001).
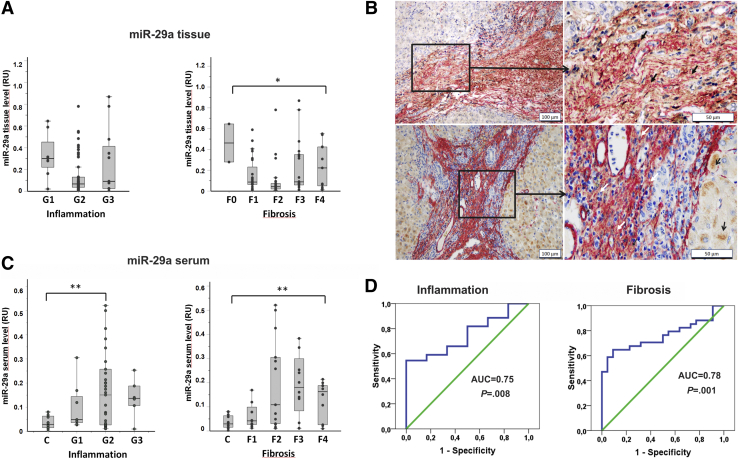
Next, we analyzed 84 human biopsies representing different stages of fibrosis during chronic HCV infection (NF0 = 2, NF1 = 23, NF2 = 26, NF3 = 22, NF4 = 11, and one not specified sample). Patients’ clinical datasets were previously described by Trebicka et al40 and Wedemeyer et al.41 In agreement with the data in murine fibrosis, there was a loss of miR-29a in liver biopsies, showing inflammation and fibrosis (Figure 6A). Notably, by αSMA and LC3 double staining, we demonstrated active autophagy within myofibroblastic HSC in the fibrotic septa of liver biopsies with chronic hepatitis C (Figure 6B).
Figure 6.
Autophagy-associated miR-29a secretion into circulation is a biomarker of fibrosis. MicroRNA-29a levels in 85 biopsy specimens that represent different stages of inflammation (G1–G3) and fibrosis (F0–F4) (A). Localization of the LC3 protein (brown) in SMA-positive myofibroblastic cells (red) was found mainly in fibrotic septa (closed arrows). LC3-positive hepatocytes are shown by open arrows (B). MicroRNA-29a levels in 56 serum samples including 12 healthy volunteers and 44 patients with different grades of inflammation (G1–G3) or different stages of fibrosis (F1–F4) (C). Whereas hepatic miR-29a levels were significantly reduced in biopsies of fibrotic livers (P = .041) (A), the circulating miR-29a was distinctly increased in serum samples of patients with chronic hepatitis C (P = .004), depending on the severity of fibrosis (F2–F4) (C). Receiver operating characteristic curve analysis for serum miR-29a as the continuous variable and inflammation (G1–3) or progression of fibrosis (F1/F2–F4) as target variables is shown in (D). (P value ∗<.05, ∗∗<.001).
For circulating miR-29 quantification, 44 matching serum samples (NF1 = 10, NF2 = 13, NF3 = 11, NF4 = 10) were collected. Serum miR-29a levels in patients with hepatitis C were highly elevated compared with healthy controls (N = 12) but did not correlate with alanine aminotransferase values (data not shown). Importantly, significantly elevated miR-29a levels were strongly associated with the severity of fibrosis (Figure 6B). Receiver operating characteristic analysis reinforced the potential of fibrosis-associated miR-29 release as a biomarker of chronic liver disease progression and fibrosis (Figure 6C).
Discussion
The miR-29a acts as an antifibrotic miRNA in fibroblasts and fibroblast-related cell types and is crucial in suppressing ECM synthesis.16,19 During HSC activation, hepatic miR-29a is gradually decreased in parallel with enhanced production of ECM and autocrine TGFβ stimulation.19,31 TGFβ is a prominent stimulus that induces ECM production and provokes loss of miR-29 from fibroblasts as shown in different organs.16,19
In this study, we provide compelling evidence that the profibrogenic mediators TGFβ and PDGF-BB strongly stimulate vesicular miR-29a secretion from HSC, the principal collagen-producing cells in liver injury. Because miR-29a targets mRNA transcripts of a wide spectrum of ECM members such as different collagen subunits, elastin, and fibrillin 1, resulting in less ECM accumulation and fibrosis,11,15, 16, 17, 18, 19 increase of cellular miR-29a in HSC by inhibition of TGFβ and PDGF-BB mediated release of cellular miR-29a is of high therapeutic relevance. Indeed treatment of HSC with miR-29a mimics and subsequent miR-29a overexpression leads to less ECM expression and less fibrosis.42
Vesicles released after profibrogenic stimulation of HSC with TGFβ and PDGF-BB had the size of small EV32,33 and contained the exosome markers TSG101 and CD63, which are part of tetraspanin protein family43 and endosome sorting complex required for transport,44 respectively. Recently EV release has been demonstrated as a consequence of cellular autophagy, although the mechanistic links are not yet fully understood.45, 46, 47 Conventional autophagy is regarded as a degradation process in which autophagosomes fuse with lysosomes, leading to the turnover of their contents. However, unconventional autophagy processes activate secretory pathways, resulting in cytokine secretion that does not involve the Golgi apparatus.48,49 TGFβ and PDGF-BB are both potent autophagy inducers, which is consistent with the finding that autophagy induces HSC activation, the initiation of myofibroblast transdifferentiation, and subsequently liver fibrosis.25,26,50 Thus, inhibition of autophagy by rapamycin treatment abrogated the marked increase of procollagen synthesis during murine CCl4-induced fibrosis, supporting our results of an autophagy-miRNA secretion that mediates miR-29a loss and enhanced ECM synthesis. Furthermore, autophagy inhibition in HSC in vivo by stellate cell–specific knockout of the ATG7 gene resulted in alleviated fibrosis.26 Autophagy induction by TGFβ is based on its interaction with insulin-like growth factor binding protein-associated protein 1 and activation of the PI3K/AKT/mTOR pathway.51 In addition, autophagy is induced by hypoxic and endoplasmic reticulum stress in response to metabolic starvation during fibrogenesis.52 Although HSC-specific inhibition of autophagy retards fibrosis progression,26 its therapeutic application is challenging because of the different influences on the different hepatic cell types. Thus, autophagy is beneficial in hepatocytes such that global autophagy inhibition in liver could be deleterious.21,22 Moreover, inhibition of autophagy in hepatocytes leads to development of hepatocellular carcinoma.23,24 Therefore, the therapeutic impact of inhibition of miR-29a release from HSC to the bloodstream should be ideally shown on experimental fibrosis in HSC-specific, autophagy-deficient mice. After demonstration in vitro that the interruption of the autophagy pathway in HSC leads to cellular miR-29 accumulation, followed by retardation of ECM synthesis, in vivo studies on the miR-29a/autophagy axis will be of high relevance for therapeutic strategies of fibrosis prevention.
In conclusion, autophagy-regulated processes are multifaceted, including nonconventional autophagy pathways that link autophagy-related signalling to the endocytotic and exocytotic pathway and EV secretion. Hence, the autophagy marker protein LC3 was also identified in late endosomes and multivesicular bodies.53 Importantly, PDGF has been discovered to promote exosomal protein secretion due to the mTOR pathway activation.54 In addition, the recent findings of Gao et al54 show that PDGF-induced vesicular release may be blocked by mTOR pathway inhibition, followed by canonical autophagy activation. These findings led us to suspect that the autophagy-related EV secretion involves endosomes and multivesicular bodies of the endocytotic and exocytotic network and that it competes with the canonical autophagy progress, which results in lysosomal degradation. Indeed, in cancers, the release of EVs is negatively regulated by induction of canonical autophagy pathways.55 In our study, we now prove that the interaction of autophagy processes with the endocytotic and exocytotic pathway is involved in miR-29a release.
It is noteworthy that autophagy-regulated pathways leading to fibrosis-associated vesicular release of miR-29a were illustrated in clinical samples of HCV patients with injury and fibrosis. In those tissue and serum samples, miR-29 reduction in the tissue corresponds to increase of that at serum levels, which is consistent with the progression of liver fibrosis.19 In summary, our findings point to an autophagy-related secretory pathway that controls the release of miR-29a, similar to its effects on cytokine secretion.48,49 Importantly, the controlled miRNA release is regulated by autophagy, linking this process to endocytotic and exocytotic flux that culminates in vesicular secretion.
Materials and Methods
Cell Culture and Stimulation of HSC
Primary mouse HSC were isolated from male BALB/c mice and retired breeder rats after liver perfusion and Nycodenz density centrifugation as previously described.56 Primary HSC were incubated in Dulbecco modified Eagle medium (DMEM) with 20% fetal calf serum (FCS) and supplemented with 2% penicillin for the first 2 days after isolation. Then, HSC were sustained in DMEM with 10% FCS. Myofibroblastic transition was characterized by αSMA induction. HSC at 36 hours were considered as quiescent compared with HSC on day 7 when cells express high levels of αSMA, therefore considering them as myofibroblastic cells.
HSC-T6 cells were cultured in DMEM containing 10% fetal bovine serum as previously described.57 HSC were stimulated with 30 ng/mL PDGF-BB (Sigma-Aldrich, Taufkirchen, Germany) or 5 ng/mL TGFβ (R&D Systems, Wiesbaden-Nordenstadt, Germany). For additional stimulation, cells were incubated for 2 hours with medium containing 50 μmol/L blebbistatin (Sigma-Aldrich Chemie GmbH, Munich, Germany) and then with TGFβ for additional 24 hours. For autophagy induction and inhibition, reagents are summarized in Table 1. Autophagy was monitored by fluorescence microscopy or quantified using the cationic amphiphilic tracer dye followed by flow cytometry.
Autophagy Induction and Inhibition
Cells were seeded in 6-well plates in DMEM containing 10% FCS. The next day, the medium was changed to DMEM containing 0.5% FCS. Cells were incubated in DMEM without FCS before stimulating with 5 ng/mL TGFβ (R&D Systems) or 30 ng /mL PDGF-BB (Sigma-Aldrich) for 24 hours. As positive control for autophagy activation, rapamycin or torin1 treatment was performed according to the concentrations listed in Table 1. For autophagy inhibition, 4 hours after TGFβ stimulation, agents listed in Table 1 were added.
For autophagy inhibition using different siRNAs (Table 2), cells were transfected with 50 nmol/L siRNA using lipofectamine (Thermo Fisher, Germany) according to the manufacturer’s instructions. After 24 hours, HSC were stimulated with TGFβ or PDGF-BB for another 24 hours and then lysed either for RNA isolation or for protein expression analysis.
Table 2.
mRNA Sequences Targeted by siRNA
| siRNA | Company | Target mRNA sequences |
|---|---|---|
| Rat siATG5 | Dharmacon, USA | (a) AGUCAGGUGAUCAACGAAA |
| (b) CGUGUGGUUUGGACGGAUU | ||
| (c) GGAUGAGAUAACUGAACGA | ||
| (d) ACCGGAAACUCAUGGAAUA | ||
| Rat siATG7 | Dharmacon, USA | (a) CAAAGUUAACAGUCGGUGU |
| (b) AGUGAAUGCCAGAGGGUUC | ||
| (c) CUGGAGGAACUCAUUGAUA | ||
| (d) CCCAGAAGAAGUUGAACGA | ||
| Rat Myosin 5a | Dharmacon, USA | - CGCUACAAGAAGCUCCAUA |
Autophagy Analysis by Flow Cytometry
After profibrogenic stimulation, cells were stained with cationic amphiphilic tracer dye using CYTO-ID autophagy detection kit (Enzo Life Sciences, Lörrach, Germany). Subsequently, fluorescence was analyzed by flow cytometry using FACS Aria III (Becton Dickinson, Heidelberg, Germany).
Vesicle Preparations, Negative Staining, and Electron Microscopy
Supernatant of HSC was collected after stimulation for 24 hours, and 250 μL was taken for vesicle preparation using the Exosome Precipitation Solution (SBI, Mountain View, CA) according to the manufacturer’s instructions. The vesicle pellets were dissolved in 25 μL phosphate-buffered saline and used for miRNA quantification, protein determination by the BCA protein assay (Pierce, Thermo Scientific, Rockford, IL), or negative staining.
For negative staining, 20 μL of vesicle suspension was placed on carbon-coated grids and blotted on filter paper after washing with an aqueous solution of 2% uranyl acetate. Transmission electron microscopy of HSC upon TGFβ and PDGF-BB stimulation was performed as previously described.58 Then, cells were trypsinized and centrifuged after stimulation. Cell pellets were fixed by glutaraldehyde solution and incubated in osmium tetroxide (OsO4) solution. After dehydration through different ethanol concentrations, cell pellets were embedded by a tissue-embedding machine (Leica, Wetzler, Germany; EM TP). Ultrathin sections were stained with lead citrate and uranyl citrate. Uranyl acetate stained vesicles and ultrathin sections were monitored by electron microscopy using the EM902A (Zeiss, Oberkochen, Germany).
Nanoparticle Tracking Analysis
Cell culture supernatants were collected after 24 hours with or without TGFβ stimulation. A series of centrifugation was performed to remove dead cells and debris as described previously.59 After 0.22 μm filtration, the supernatants were taken for vesicle isolation exoEasy Maxi Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Directly after isolation, vesicles were diluted 1:1000 in phosphate-buffered saline and imaged by Nanosight Range NS300 (Malvern, Kassel, Germany).
Detection of Cell Death and Apoptosis
HSC were starved for 24 hours before stimulation. After 24 hours of starvation, cells were centrifuged and washed twice with cold phosphate-buffered saline and stained with annexin V-FITC and propidium iodide for 15 minutes, followed by flow cytometry analysis (FACS Aria III; Becton Dickinson). Cells treated with 100 μmol/L hydrogen peroxide were used as positive control.
In addition, cells were analyzed for apoptosis by TUNEL assay using the cell death detection kit from Roche Diagnostics (Mannheim, Germany) following the recommendation of the manufacturer. DNAse treatment (1 U/mL) was taken as positive control. TUNEL-positive cells are shown by red fluorescence. After TUNEL staining, cells were counterstained by FITC-conjugated phalloidin (Sigma-Aldrich, Heidelberg, Germany) and monitored by a Nikon (Tokyo, Japan) Eclipse T2 microscope.
Induction of Acute Liver Injury and Fibrosis
In this study, 12 BALB/c male mice (25–35 g) were used, from which 6 were subjected to CCl4 injury and 6 control mice were treated only with mineral oil as described earlier.25,39 All experiments were performed under the supervision of the National Health and Medical Research Committee Guidelines for Animal Experimentation. Liver tissues were taken and snap-frozen in liquid nitrogen for RNA isolation and hydroxyproline determination as described before. Liver tissue chosen from liver segment IV was fixed in 4% buffered paraformaldehyde and embedded in paraffin for histopathologic evaluation.
Patients, Human Biopsies, and Serum Specimens
A total of 85 patients with well-documented chronic hepatitis C infection were selected from the database of HepNet, the German Competence Network on Viral Hepatitis (www.kompetenznetz-hepatitis.de). All patients gave their written advised consent before undergoing liver biopsy. Clinical characteristics of the cohort were previously reported by Wedemeyer et al.41 Briefly, the cohort included 57.14% men and 42.86% women; 78% of the patients were positive for HCV genotype 1 and 19% for genotype 3.
For grading of inflammation (G1–G4) and staging of fibrosis (F0–F4), histologic assessment was performed independently by 2 experienced pathologists (UD, HPD) according to the method of Desmet et al.60 From a total of 85 biopsies (Ntotal = 85) representing different stages of fibrosis (NF0 = 2, NF1 = 23, NF2 = 26, NF3 = 22, NF4 = 11, and one not specified sample), RNA was extracted and used in this study.
For circulating miR-29 quantification, 56 serum specimens including 12 samples of healthy blood donors and 44 samples of chronic hepatitis C patients with fibrosis (NF1 = 10, NF2 = 13, NF3 = 11, NF4 = 10) were available and used for RNA isolation as described above.
Total RNA Extraction
Intracellular total RNA from snap-frozen tissues or cells was isolated using the Qiazol reagent following the instructions of the supplier (Qiagen, Hilden, Germany).
Extracellular RNA was isolated from cell culture supernatants, vesicle fractions, or serum samples of patients with chronic hepatitis C liver disease. Cell culture supernatants were collected from HSC with or without stimulation, centrifuged at 12,000g for 20 minutes, and filtered through 0.22 μm filters. Then, 200 μL of filtered supernatants, vesicle fractions, or serum samples from patients or mice were applied to RNA isolation using Qiazol. For quality control and later normalization, we added 2 pmol spike SV-40 RNA to all extracellular specimens before RNA isolation as previously described.40
The quantity of RNA obtained from cells and tissues was determined by A260-measurement using the ND-1000 NanoDrop spectrophotometer (NanoDrop, Wilmington, DE), and the quality was assessed by microcapillary electrophoresis (2100 BioAnalyser; Agilent Technologies, Waldbronn, Germany).
Total RNA preparations from human formalin-fixed and paraffin-embedded biopsies were performed according to previous reports.61,62
Intracellular and Extracellular miRNA Quantification by Quantitative Real-Time Polymerase Chain Reaction
RNA was polyadenylated and reverse transcribed by means of miScript II reagents from Qiagen. Subsequently, miRNA was analyzed by quantitative polymerase chain reaction (PCR) using the miRNA-SYBR Green PCR Kit (Qiagen). Primers used for cDNA synthesis and real-time PCR were selected and purchased from the GeneGlobe Search Center (Qiagen) (Table 3). All steps were performed in triplicate and in agreement with the supplier’s guidelines.
Table 3.
Primers Used for Quantitative PCR
| Oligonucleotide | Sequence | Company |
|---|---|---|
| rno Atg5-F | CTG TTC GAT CTT CTTGCATCA | Eurofins MWG, GER |
| rno Atg5-R | TCC TTT TCT GGA AAA CTC TTG AA | Eurofins MWG, GER |
| rno Atg7-F | CTT GGC TGC TAC TTC TGC AA | Eurofins MWG, GER |
| rno Atg7-R | AGT CCG GTC TCT GGT GGA A | Eurofins MWG, GER |
| rno mTOR-F | ATC AAG CAA GCG ACA TCT CA | Eurofins MWG, GER |
| rno mTOR-R | CAG GCC TTG GTT ACC AGA A | Eurofins MWG, GER |
| rno coL1A1A-F | CAT GTT CAG CTT TGT GGA CCT | Eurofins MWG, GER |
| rno coL1A1A-R | GCA GCT GAC TTC AGG GAT GT | Eurofins MWG, GER |
| rno PDGF-C-F | GCG GAA GCG CAT CTA TAT CT | Eurofins MWG, GER |
| rno PDGF-C-R | AAT GAA TAG GTC CTC AGA GTC CA | Eurofins MWG, GER |
| rno HPRT-F | GAC CGG TTC TGT CAT GTC G | Eurofins MWG, GER |
| rno HPRT-F | ACC TGG TTC ATC ATC ACT AAT CAC | Eurofins MWG, GER |
| miR-29a | Qiagen, GER | |
| RNU6 | Qiagen, GER | |
| Universal primer | Qiagen, GER | |
| SV40-miRNA | Qiagen, GER |
GER, Germany.
Cellular miRNA levels were normalized using RNU6 as reference. For normalization of extracellular miRNA levels, SV40-miRNA (Qiagen), which has been added to serum samples, vesicle fractions, and cell culture supernatants before the RNA isolation procedure, was used.
Analysis of mRNA Expression by Real-Time PCR
After polyadenylation and reverse transcription using the Qiagen miScript reverse transcription kit as described above, 2 ng cDNA was applied to real-time PCR assays using the GoTaq qPCR Master Mix (Promega, Walldorf, Germany) and primer sets listed in Table 3. Real-time PCR was performed in triplicates on a Bio-Rad (Hercules, CA) C1000M thermal cycler. After demonstration that primer sets exert equal and high efficiencies, relative expression was calculated by the ΔΔCt method using the transcript levels of hypoxanthine-guanine phosphoribosyl transferase for normalization.
siRNA Transfection
Cells were seeded in 6-well plates in DMEM containing 10% FCS. After reaching 30% confluency, cells were transfected using the X-tremeGENE transfection reagent (Sigma-Aldrich, Taufkirchen, Germany). A mix of siRNAs (Dharmacon Inc, Lafayette, CO) targeting the respective transcript of interest (Table 1) was used for transfection following the manufacturer’s instructions.
Immunoblotting
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed by means of the Bio-Rad Mini protein gel system. Briefly, protein samples were loaded with Laemmli sample buffer, heat-denatured, and directly loaded onto the gels. After electrophoresis and protein transfer, PCDF membranes were blocked in 10% milk powder solution. After incubation with the first antibodies (Table 4), membranes were incubated with the respective peroxidase-conjugated secondary antibodies, and then membranes were developed using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific, Germany) according to the manufacturer’s instructions. Signals were evaluated using the ChemiDoc Imaging System (Bio-Rad).
Table 4.
Antibodies Used in This Study
| Targeted antigen | Company | Dilution (immunoblotting) | Dilution (immunochemistry) |
|---|---|---|---|
| LC3B | Abcam, USA | 1:1000 | 1:1000 |
| SQSTM1/p62 | Abcam, USA | 1:1000 | --- |
| β-actin | Sigma-Aldrich, GER | 1:1000 | --- |
| ATG7 | Abcam, USA | 1:1000 | --- |
| α-SMA | DAKO, GER | — | 1:400 |
| Phalloidin | Thermo Scientific | — | 1:500 |
| Collagen1 | Southern Biotech | — | 1:50 |
| Desmin | Thermo Scientific | — | 1:200 |
Immunofluorescence Staining and Immunohistochemistry
HSC were plated in chamber slides, and after 24 hours of starvation without FCS, they were treated with either TGFβ or PDGF-BB. After stimulation, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (pH 8.0) and stained with rabbit LC3 antibody (Abcam, Cambridge, MA), secondary antibody, goat anti-rabbit immunoglobulin G conjugated with Alexa 488 (Abcam), according to manufacturer’s instructions. In addition, primary HSC were immunostained with anti-αSMA, anti-desmin, and anti-collagen I antibodies listed in Table 4. Cells were analyzed by confocal laser scanning microscopy (Leica TCS SP8; Leica Microsystems).
Serial sections of formalin-fixed and paraffin-embedded liver biopsies from patients with chronic hepatitis C infection were applied to LC3 and αSMA immunocytochemistry (Table 4) using BOND IHC/ISH Staining Instruments (Leica Biosystems, Wetzlar, Germany) according to the manufacturer’s instructions. For double staining, LC3 immunodetection was developed using peroxidase-polymer conjugated secondary antibodies and diaminobenzidine as substrate, whereas SMA was detected by means of phosphatase-linked anti-mouse antibodies and new fuchsine (pararosaniline) as substrate, using the BOND Polymer Refine Detection kit (Leica Biosystems).
Acknowledgments
The authors thank the German Liver Foundation and the German Competence Network for Viral Hepatitis (HepNet), initiated and coordinated by Michael Manns (Medical School Hannover, Hannover, Germany), for having the opportunity to obtain biopsy and serum specimens from the central tissue and serum bank, respectively. Furthermore, they thank Monika Kwiecinski and Hannah Eischeidt-Scholz for their support in cell culture and miR-29 analysis and Melanie Ruhrländer for RNA extraction from FFPE biopsies. They thank Claudia Köhne for her support in electron microscopy and Silke Bellinghausen for the excellent technical assistance in primary HSC isolation.
CRediT Authorship Contributions
Margarete Odenthal, PhD (Conceptualization: Lead; Funding acquisition: Lead; Investigation: Lead; Project administration: Equal; Resources: Equal; Writing – original draft: Lead)
Xiaojie Yu (Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Supporting; Project administration: Supporting; Validation: Lead; Visualization: Lead; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Inge Mannaerts (Investigation: Lead; Project administration: Lead; Data curation: supporting)
Natalia Elfimova (Data curation: Supporting)
Marion Mueller (Methodology: Supporting)
Daniel Bachurski (Methodology: Supporting; Data curation: supporting)
Ulrike Koitzsch (Methodology: Supporting)
Uta Drebber (Resources: Supporting)
Esther Mahabir (Resources: Supporting; Writing – review & editing: Supporting)
Hinrich P. Hansen (Methodology: Supporting)
Scott Laurence Friedman (Resources: Supporting; Writing – review & editing: Supporting)
Sabine Klein (Methodology: Supporting)
Hans Peter Dienes (Resources: Supporting)
Marianna Hoesel (Resources: Supporting; Writing – review & editing: Supporting)
Reinhard Buettner (Funding acquisition: Equal; Resources: Equal)
Jonel Trebicka (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting)
Vangelis Kondylis (Resources: Supporting; Writing – review & editing: Supporting)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by German Liver Foundation (MO), the Heinrich Hertz Foundation (MO), the Center for Molecular Medicine Cologne (CMMC) to RB, MO, and the German Research Foundation (DFG OD/6-1) to MO, (SFB TRR57/ P18) to JT, (DFG KO 5827/2-1) to VK, and (DFG HA2432/3-2) to HH. In addition, financial support was received from the Research and Education programme of the Medical Faculty of the University of Cologne and the German Competence Network for Viral Hepatitis (HepNet), funded by the German Federal Ministry of Education and Research (BMBF), Grant No 01KI0601 (HPD/MO). Jonel Trebicka is supported by the European Union’s Horizon 2020 Research and Innovation Program (Galaxy, No. 668031, MICROB-PREDICT, No. 825694 and DECISION No.84794), and Societal Challenges - Health, Demographic Change and Wellbeing (No. 731875), and Cellex Foundation (PREDICT). S.L.F. is supported by NIH grant R01 DK128289 and I.M. by a FWO–V post-doctoral fellowship (12N5419 N). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Inge Mannaerts, Email: inge.mannaerts@vub.be.
Margarete Odenthal, Email: m.odenthal@uni-koeln.de.
References
- 1.Altamirano-Barrera A., Barranco-Fragoso B., Mendez-Sanchez N. Management strategies for liver fibrosis. Ann Hepatol. 2017;16:48–56. doi: 10.5604/16652681.1226814. [DOI] [PubMed] [Google Scholar]
- 2.Hernandez-Gea V., Friedman S.L. Pathogenesis of liver fibrosis. Annu Rev Pathol. 2011;6:425–456. doi: 10.1146/annurev-pathol-011110-130246. [DOI] [PubMed] [Google Scholar]
- 3.Gabele E., Brenner D.A., Rippe R.A. Liver fibrosis: signals leading to the amplification of the fibrogenic hepatic stellate cell. Front Biosci. 2003;8:d69–d77. doi: 10.2741/887. [DOI] [PubMed] [Google Scholar]
- 4.Meng X.M., Nikolic-Paterson D.J., Lan H.Y. TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol. 2016;12:325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y.E. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009;19:128–139. doi: 10.1038/cr.2008.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Annoni G., Weiner F.R., Zern M.A. Increased transforming growth factor-beta 1 gene expression in human liver disease. J Hepatol. 1992;14:259–264. doi: 10.1016/0168-8278(92)90168-o. [DOI] [PubMed] [Google Scholar]
- 7.Czaja M.J., Weiner F.R., Flanders K.C., Giambrone M.A., Wind R., Biempica L., Zern M.A. In vitro and in vivo association of transforming growth factor-beta 1 with hepatic fibrosis. J Cell Biol. 1989;108:2477–2482. doi: 10.1083/jcb.108.6.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida K., Matsuzaki K. Differential regulation of TGF-beta/Smad signaling in hepatic stellate cells between acute and chronic liver injuries. Front Physiol. 2012;3:53. doi: 10.3389/fphys.2012.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pinzani M., Gesualdo L., Sabbah G.M., Abboud H.E. Effects of platelet-derived growth factor and other polypeptide mitogens on DNA synthesis and growth of cultured rat liver fat-storing cells. J Clin Invest. 1989;84:1786–1793. doi: 10.1172/JCI114363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman S.L., Arthur M.J. Activation of cultured rat hepatic lipocytes by Kupffer cell conditioned medium: direct enhancement of matrix synthesis and stimulation of cell proliferation via induction of platelet-derived growth factor receptors. J Clin Invest. 1989;84:1780–1785. doi: 10.1172/JCI114362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sekiya Y., Ogawa T., Yoshizato K., Ikeda K., Kawada N. Suppression of hepatic stellate cell activation by microRNA-29b. Biochem Biophys Res Commun. 2011;412:74–79. doi: 10.1016/j.bbrc.2011.07.041. [DOI] [PubMed] [Google Scholar]
- 12.Jiang Y., Zhao Y., He F., Wang H. Artificial microRNA-mediated Tgfbr2 and Pdgfrb co-silencing ameliorates carbon tetrachloride-induced hepatic fibrosis in mice. Hum Gene Ther. 2019;30:179–196. doi: 10.1089/hum.2018.047. [DOI] [PubMed] [Google Scholar]
- 13.Patel D.J., Ma J.B., Yuan Y.R., Ye K., Pei Y., Kuryavyi V., Malinina L., Meister G., Tuschl T. Structural biology of RNA silencing and its functional implications. Cold Spring Harb Symp Quant Biol. 2006;71:81–93. doi: 10.1101/sqb.2006.71.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aravin A., Tuschl T. Identification and characterization of small RNAs involved in RNA silencing. FEBS Lett. 2005;579:5830–5840. doi: 10.1016/j.febslet.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 15.van Rooij E., Sutherland L.B., Thatcher J.E., DiMaio J.M., Naseem R.H., Marshall W.S., Hill J.A., Olson E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwiecinski M., Noetel A., Elfimova N., Trebicka J., Schievenbusch S., Strack I., Molnar L., von Brandenstein M., Tox U., Nischt R., Coutelle O., Dienes H.P., Odenthal M. Hepatocyte growth factor (HGF) inhibits collagen I and IV synthesis in hepatic stellate cells by miRNA-29 induction. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurer B., Stanczyk J., Jungel A., Akhmetshina A., Trenkmann M., Brock M., Kowal-Bielecka O., Gay R.E., Michel B.A., Distler J.H., Gay S., Distler O. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum. 2010;62:1733–1743. doi: 10.1002/art.27443. [DOI] [PubMed] [Google Scholar]
- 18.Cushing L., Kuang P.P., Qian J., Shao F., Wu J., Little F., Thannickal V.J., Cardoso W.V., Lu J. MiR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roderburg C., Urban G.W., Bettermann K., Vucur M., Zimmermann H., Schmidt S., Janssen J., Koppe C., Knolle P., Castoldi M., Tacke F., Trautwein C., Luedde T. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 20.Roderburg C., Luedde M., Vargas Cardenas D., Vucur M., Mollnow T., Zimmermann H.W., Koch A., Hellerbrand C., Weiskirchen R., Frey N., Tacke F., Trautwein C., Luedde T. MiR-133a mediates TGF-beta-dependent derepression of collagen synthesis in hepatic stellate cells during liver fibrosis. J Hepatol. 2013;58:736–742. doi: 10.1016/j.jhep.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 21.Weiskirchen R., Tacke F. Relevance of autophagy in parenchymal and non-parenchymal liver cells for health and disease. Cells. 2019;8:16. doi: 10.3390/cells8010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gracia-Sancho J., Guixe-Muntet S. The many-faced role of autophagy in liver diseases. J Hepatol. 2018;68:593–594. doi: 10.1016/j.jhep.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 23.Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathew R., Karp C.M., Beaudoin B., Vuong N., Chen G., Chen H.Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R.S., Karantza-Wadsworth V., White E. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thoen L.F., Guimaraes E.L., Dolle L., Mannaerts I., Najimi M., Sokal E., van Grunsven L.A. A role for autophagy during hepatic stellate cell activation. J Hepatol. 2011;55:1353–1360. doi: 10.1016/j.jhep.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Hernandez-Gea V., Ghiassi-Nejad Z., Rozenfeld R., Gordon R., Fiel M.I., Yue Z., Czaja M.J., Friedman S.L. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dewidar B., Meyer C., Dooley S., Meindl-Beinker A.N. TGF-beta in hepatic stellate cell activation and liver fibrogenesis-updated 2019. Cells. 2019;8:1419. doi: 10.3390/cells8111419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghavami S., Cunnington R.H., Gupta S., Yeganeh B., Filomeno K.L., Freed D.H., Chen S., Klonisch T., Halayko A.J., Ambrose E., Singal R., Dixon I.M. Autophagy is a regulator of TGF-beta1-induced fibrogenesis in primary human atrial myofibroblasts. Cell Death Dis. 2015;6 doi: 10.1038/cddis.2015.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye H.L., Zhang J.W., Chen X.Z., Wu P.B., Chen L., Zhang G. Ursodeoxycholic acid alleviates experimental liver fibrosis involving inhibition of autophagy. Life Sci. 2019;242 doi: 10.1016/j.lfs.2019.117175. [DOI] [PubMed] [Google Scholar]
- 30.Zhou L., Wang L., Lu L., Jiang P., Sun H., Wang H. Inhibition of miR-29 by TGF-beta-Smad3 signaling through dual mechanisms promotes transdifferentiation of mouse myoblasts into myofibroblasts. PLoS One. 2012;7 doi: 10.1371/journal.pone.0033766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwiecinski M., Elfimova N., Noetel A., Tox U., Steffen H.M., Hacker U., Nischt R., Dienes H.P., Odenthal M. Expression of platelet-derived growth factor-C and insulin-like growth factor I in hepatic stellate cells is inhibited by miR-29. Lab Invest. 2012;92:978–987. doi: 10.1038/labinvest.2012.70. [DOI] [PubMed] [Google Scholar]
- 32.Maji S., Matsuda A., Yan I.K., Parasramka M., Patel T. Extracellular vesicles in liver diseases. Am J Physiol Gastrointest Liver Physiol. 2017;312:G194–G200. doi: 10.1152/ajpgi.00216.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thery C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., Ayre D.C., Bach J.M., Bachurski D., Baharvand H., Balaj L., Baldacchino S., Bauer N.N., Baxter A.A., Bebawy M., Beckham C., Bedina Zavec A., Benmoussa A., Berardi A.C., Bergese P., Bielska E., Blenkiron C., Bobis-Wozowicz S., Boilard E., Boireau W., Bongiovanni A., Borras F.E., Bosch S., Boulanger C.M., Breakefield X., Breglio A.M., Brennan M.A., Brigstock D.R., Brisson A., Broekman M.L., Bromberg J.F., Bryl-Gorecka P., Buch S., Buck A.H., Burger D., Busatto S., Buschmann D., Bussolati B., Buzas E.I., Byrd J.B., Camussi G., Carter D.R., Caruso S., Chamley L.W., Chang Y.T., Chen C., Chen S., Cheng L., Chin A.R., Clayton A., Clerici S.P., Cocks A., Cocucci E., Coffey R.J., Cordeiro-da-Silva A., Couch Y., Coumans F.A., Coyle B., Crescitelli R., Criado M.F., D’Souza-Schorey C., Das S., Datta Chaudhuri A., de Candia P., De Santana E.F., De Wever O., Del Portillo H.A., Demaret T., Deville S., Devitt A., Dhondt B., Di Vizio D., Dieterich L.C., Dolo V., Dominguez Rubio A.P., Dominici M., Dourado M.R., Driedonks T.A., Duarte F.V., Duncan H.M., Eichenberger R.M., Ekstrom K., El Andaloussi S., Elie-Caille C., Erdbrugger U., Falcon-Perez J.M., Fatima F., Fish J.E., Flores-Bellver M., Forsonits A., Frelet-Barrand A., Fricke F., Fuhrmann G., Gabrielsson S., Gamez-Valero A., Gardiner C., Gartner K., Gaudin R., Gho Y.S., Giebel B., Gilbert C., Gimona M., Giusti I., Goberdhan D.C., Gorgens A., Gorski S.M., Greening D.W., Gross J.C., Gualerzi A., Gupta G.N., Gustafson D., Handberg A., Haraszti R.A., Harrison P., Hegyesi H., Hendrix A., Hill A.F., Hochberg F.H., Hoffmann K.F., Holder B., Holthofer H., Hosseinkhani B., Hu G., Huang Y., Huber V., Hunt S., Ibrahim A.G., Ikezu T., Inal J.M., Isin M., Ivanova A., Jackson H.K., Jacobsen S., Jay S.M., Jayachandran M., Jenster G., Jiang L., Johnson S.M., Jones J.C., Jong A., Jovanovic-Talisman T., Jung S., Kalluri R., Kano S.I., Kaur S., Kawamura Y., Keller E.T., Khamari D., Khomyakova E., Khvorova A., Kierulf P., Kim K.P., Kislinger T., Klingeborn M., Klinke D.J., 2nd, Kornek M., Kosanovic M.M., Kovacs A.F., Kramer-Albers E.M., Krasemann S., Krause M., Kurochkin I.V., Kusuma G.D., Kuypers S., Laitinen S., Langevin S.M., Languino L.R., Lannigan J., Lasser C., Laurent L.C., Lavieu G., Lazaro-Ibanez E., Le Lay S., Lee M.S., Lee Y.X.F., Lemos D.S., Lenassi M., Leszczynska A., Li I.T., Liao K., Libregts S.F., Ligeti E., Lim R., Lim S.K., Line A., Linnemannstons K., Llorente A., Lombard C.A., Lorenowicz M.J., Lorincz A.M., Lotvall J., Lovett J., Lowry M.C., Loyer X., Lu Q., Lukomska B., Lunavat T.R., Maas S.L., Malhi H., Marcilla A., Mariani J., Mariscal J., Martens-Uzunova E.S., Martin-Jaular L., Martinez M.C., Martins V.R., Mathieu M., Mathivanan S., Maugeri M., McGinnis L.K., McVey M.J., Meckes D.G., Jr., Meehan K.L., Mertens I., Minciacchi V.R., Moller A., Moller Jorgensen M., Morales-Kastresana A., Morhayim J., Mullier F., Muraca M., Musante L., Mussack V., Muth D.C., Myburgh K.H., Najrana T., Nawaz M., Nazarenko I., Nejsum P., Neri C., Neri T., Nieuwland R., Nimrichter L., Nolan J.P., Nolte-‘t Hoen E.N., Noren Hooten N., O’Driscoll L., O’Grady T., O’Loghlen A., Ochiya T., Olivier M., Ortiz A., Ortiz L.A., Osteikoetxea X., Ostergaard O., Ostrowski M., Park J., Pegtel D.M., Peinado H., Perut F., Pfaffl M.W., Phinney D.G., Pieters B.C., Pink R.C., Pisetsky D.S., Pogge von Strandmann E., Polakovicova I., Poon I.K., Powell B.H., Prada I., Pulliam L., Quesenberry P., Radeghieri A., Raffai R.L., Raimondo S., Rak J., Ramirez M.I., Raposo G., Rayyan M.S., Regev-Rudzki N., Ricklefs F.L., Robbins P.D., Roberts D.D., Rodrigues S.C., Rohde E., Rome S., Rouschop K.M., Rughetti A., Russell A.E., Saa P., Sahoo S., Salas-Huenuleo E., Sanchez C., Saugstad J.A., Saul M.J., Schiffelers R.M., Schneider R., Schoyen T.H., Scott A., Shahaj E., Sharma S., Shatnyeva O., Shekari F., Shelke G.V., Shetty A.K., Shiba K., Siljander P.R., Silva A.M., Skowronek A., Snyder O.L., 2nd, Soares R.P., Sodar B.W., Soekmadji C., Sotillo J., Stahl P.D., Stoorvogel W., Stott S.L., Strasser E.F., Swift S., Tahara H., Tewari M., Timms K., Tiwari S., Tixeira R., Tkach M., Toh W.S., Tomasini R., Torrecilhas A.C., Tosar J.P., Toxavidis V., Urbanelli L., Vader P., van Balkom B.W., van der Grein S.G., Van Deun J., van Herwijnen M.J., Van Keuren-Jensen K., van Niel G., van Royen M.E., van Wijnen A.J., Vasconcelos M.H., Vechetti I.J., Jr., Veit T.D., Vella L.J., Velot E., Verweij F.J., Vestad B., Vinas J.L., Visnovitz T., Vukman K.V., Wahlgren J., Watson D.C., Wauben M.H., Weaver A., Webber J.P., Weber V., Wehman A.M., Weiss D.J., Welsh J.A., Wendt S., Wheelock A.M., Wiener Z., Witte L., Wolfram J., Xagorari A., Xander P., Xu J., Yan X., Yanez-Mo M., Yin H., Yuana Y., Zappulli V., Zarubova J., Zekas V., Zhang J.Y., Zhao Z., Zheng L., Zheutlin A.R., Zickler A.M., Zimmermann P., Zivkovic A.M., Zocco D., Zuba-Surma E.K. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7 doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Langford G.M. Myosin-V, a versatile motor for short-range vesicle transport. Traffic. 2002;3:859–865. doi: 10.1034/j.1600-0854.2002.31202.x. [DOI] [PubMed] [Google Scholar]
- 35.Liu Z., van Grunsven L.A., Van Rossen E., Schroyen B., Timmermans J.P., Geerts A., Reynaert H. Blebbistatin inhibits contraction and accelerates migration in mouse hepatic stellate cells. Br J Pharmacol. 2010;159:304–315. doi: 10.1111/j.1476-5381.2009.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruppa A.J., Kendrick-Jones J., Buss F. Myosins, actin and autophagy. Traffic. 2016;17:878–890. doi: 10.1111/tra.12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.J., Coppes R.P., Engedal N., Mari M., Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 39.Mannaerts I., Eysackers N., Onyema O.O., Van Beneden K., Valente S., Mai A., Odenthal M., van Grunsven L.A. Class II HDAC inhibition hampers hepatic stellate cell activation by induction of microRNA-29. PLoS One. 2013;8 doi: 10.1371/journal.pone.0055786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trebicka J., Anadol E., Elfimova N., Strack I., Roggendorf M., Viazov S., Wedemeyer I., Drebber U., Rockstroh J., Sauerbruch T., Dienes H.P., Odenthal M. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J Hepatol. 2013;58:234–239. doi: 10.1016/j.jhep.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 41.Wedemeyer I., Bechmann L.P., Odenthal M., Jochum C., Marquitan G., Drebber U., Gerken G., Gieseler R.K., Dienes H.P., Canbay A. Adiponectin inhibits steatotic CD95/Fas up-regulation by hepatocytes: therapeutic implications for hepatitis C. J Hepatol. 2009;50:140–149. doi: 10.1016/j.jhep.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 42.Matsumoto Y., Itami S., Kuroda M., Yoshizato K., Kawada N., Murakami Y. MiR-29a assists in preventing the activation of human stellate cells and promotes recovery from liver fibrosis in mice. Mol Ther. 2016;24:1848–1859. doi: 10.1038/mt.2016.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreu Z., Yanez-Mo M. Tetraspanins in extracellular vesicle formation and function. Front Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lotvall J., Hill A.F., Hochberg F., Buzas E.I., Di Vizio D., Gardiner C., Gho Y.S., Kurochkin I.V., Mathivanan S., Quesenberry P., Sahoo S., Tahara H., Wauben M.H., Witwer K.W., Thery C. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles. 2014;3 doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fader C.M., Colombo M.I. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 2009;16:70–78. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- 46.Buratta S., Tancini B., Sagini K., Delo F., Chiaradia E., Urbanelli L., Emiliani C. Lysosomal exocytosis, exosome release and secretory autophagy: the autophagic- and endo-lysosomal systems go extracellular. Int J Mol Sci. 2020;21:2576. doi: 10.3390/ijms21072576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baixauli F., Lopez-Otin C., Mittelbrunn M. Exosomes and autophagy: coordinated mechanisms for the maintenance of cellular fitness. Front Immunol. 2014;5:403. doi: 10.3389/fimmu.2014.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang M., Kenny S.J., Ge L., Xu K., Schekman R. Translocation of interleukin-1beta into a vesicle intermediate in autophagy-mediated secretion. Elife. 2015;4 doi: 10.7554/eLife.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nuchel J., Ghatak S., Zuk A.V., Illerhaus A., Morgelin M., Schonborn K., Blumbach K., Wickstrom S.A., Krieg T., Sengle G., Plomann M., Eckes B. TGFB1 is secreted through an unconventional pathway dependent on the autophagic machinery and cytoskeletal regulators. Autophagy. 2018;14:465–486. doi: 10.1080/15548627.2017.1422850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thoen L.F., Guimaraes E.L., Grunsven L.A. Autophagy: a new player in hepatic stellate cell activation. Autophagy. 2012;8:126–128. doi: 10.4161/auto.8.1.18105. [DOI] [PubMed] [Google Scholar]
- 51.Huang T.J., Ren J.J., Zhang Q.Q., Kong Y.Y., Zhang H.Y., Guo X.H., Fan H.Q., Liu L.X. IGFBPrP1 accelerates autophagy and activation of hepatic stellate cells via mutual regulation between H19 and PI3K/AKT/mTOR pathway. Biomed Pharmacother. 2019;116 doi: 10.1016/j.biopha.2019.109034. [DOI] [PubMed] [Google Scholar]
- 52.Hernandez-Gea V., Hilscher M., Rozenfeld R., Lim M.P., Nieto N., Werner S., Devi L.A., Friedman S.L. Endoplasmic reticulum stress induces fibrogenic activity in hepatic stellate cells through autophagy. J Hepatol. 2013;59:98–104. doi: 10.1016/j.jhep.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jacquin E., Leclerc-Mercier S., Judon C., Blanchard E., Fraitag S., Florey O. Pharmacological modulators of autophagy activate a parallel noncanonical pathway driving unconventional LC3 lipidation. Autophagy. 2017;13:854–867. doi: 10.1080/15548627.2017.1287653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao J., Wei B., de Assuncao T.M., Liu Z., Hu X., Ibrahim S., Cooper S.A., Cao S., Shah V.H., Kostallari E. Hepatic stellate cell autophagy inhibits extracellular vesicle release to attenuate liver fibrosis. J Hepatol. 2020;73:1144–1154. doi: 10.1016/j.jhep.2020.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Villarroya-Beltri C., Baixauli F., Mittelbrunn M., Fernandez-Delgado I., Torralba D., Moreno-Gonzalo O., Baldanta S., Enrich C., Guerra S., Sanchez-Madrid F. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun. 2016;7:13588. doi: 10.1038/ncomms13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guimaraes E.L., Empsen C., Geerts A., van Grunsven L.A. Advanced glycation end products induce production of reactive oxygen species via the activation of NADPH oxidase in murine hepatic stellate cells. J Hepatol. 2010;52:389–397. doi: 10.1016/j.jhep.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 57.Vogel S., Piantedosi R., Frank J., Lalazar A., Rockey D.C., Friedman S.L., Blaner W.S. An immortalized rat liver stellate cell line (HSC-T6): a new cell model for the study of retinoid metabolism in vitro. J Lipid Res. 2000;41:882–893. [PubMed] [Google Scholar]
- 58.Martinet W., Timmermans J.P., De Meyer G.R. Methods to assess autophagy in situ: transmission electron microscopy versus immunohistochemistry. Methods Enzymol. 2014;543:89–114. doi: 10.1016/B978-0-12-801329-8.00005-2. [DOI] [PubMed] [Google Scholar]
- 59.Szatanek R., Baran J., Siedlar M., Baj-Krzyworzeka M. Isolation of extracellular vesicles: determining the correct approach (review) Int J Mol Med. 2015;36:11–17. doi: 10.3892/ijmm.2015.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Desmet V.J., Gerber M., Hoofnagle J.H., Manns M., Scheuer P.J. Classification of chronic hepatitis: diagnosis, grading and staging. Hepatology. 1994;19:1513–1520. [PubMed] [Google Scholar]
- 61.Siebolts U., Varnholt H., Drebber U., Dienes H.P., Wickenhauser C., Odenthal M. Tissues from routine pathology archives are suitable for microRNA analyses by quantitative PCR. J Clin Pathol. 2009;62:84–88. doi: 10.1136/jcp.2008.058339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elfimova N., Sievers E., Eischeid H., Kwiecinski M., Noetel A., Hunt H., Becker D., Frommolt P., Quasdorff M., Steffen H.M., Nurnberg P., Buttner R., Teufel A., Dienes H.P., Drebber U., Odenthal M. Control of mitogenic and motogenic pathways by miR-198, diminishing hepatoma cell growth and migration. Biochim Biophys Acta. 2013;1833:1190–1198. doi: 10.1016/j.bbamcr.2013.01.023. [DOI] [PubMed] [Google Scholar]