Abstract
Background & Aims
Oncogenic Kirsten Rat Sarcoma virus (KRAS) is the hallmark mutation of human pancreatic cancer and a driver of tumorigenesis in genetically engineered mouse models of the disease. Although the tumor cell–intrinsic effects of oncogenic Kras expression have been widely studied, its role in regulating the extensive pancreatic tumor microenvironment is less understood.
Methods
Using a genetically engineered mouse model of inducible and reversible oncogenic Kras expression and a combination of approaches that include mass cytometry and single-cell RNA sequencing we studied the effect of oncogenic KRAS in the tumor microenvironment.
Results
We have discovered that non–cell autonomous (ie, extrinsic) oncogenic KRAS signaling reprograms pancreatic fibroblasts, activating an inflammatory gene expression program. As a result, fibroblasts become a hub of extracellular signaling, and the main source of cytokines mediating the polarization of protumorigenic macrophages while also preventing tissue repair.
Conclusions
Our study provides fundamental knowledge on the mechanisms underlying the formation of the fibroinflammatory stroma in pancreatic cancer and highlights stromal pathways with the potential to be exploited therapeutically.
Keywords: Pancreatic Cancer, Transformation, Fibroblasts, Macrophages
Abbreviations used in this paper: ADM, acinar-to-ductal metaplasia; ARG1, arginase 1; BSA, bovine serum albumin; CAF, cancer-associated fibroblast; CK19, _; CM, conditioned media; CXCL1, __; DAPI, 4′,6-diamidino-2-phenylindole; DMEM, Dulbecco’s modified Eagle medium; DOX, doxycycline; ESI, Electrospray ionization; iCAF, inflammatory CAFs; IF, immunofluorescence; IL, interleukin; JAK, Janus kinase; JAKi, JAK inhibitor; Kras, Kirsten Rat Sarcoma virus; LC, Liquid Chromatography; MS, mass spectrometer; PanIN, pancreatic intraepithelial neoplasia; Pdx1, Pancreatic and Duodenal Homeobox 1; p-ERK, phosphorylated Extracellular signal-regulated kinase; PME, precursor lesion microenvironment; p-STAT3, phosphorylated signal transducer and activator of transcription 3; Ptf1a, Pancreas Associated Transcription Factor 1a; QqQ, triple quadrupole; SAA, Serum amyloid a; scRNAseq, single-cell RNA sequencing; SMA, smooth muscle actin; STAT3, signal transducer and activator of transcription 3; TAM, tumor-associated macrophage; TBST, Tris buffered saline solution with Tween 20; TME, tumor microenvironment; UMAP, uniform and manifold approximation and projection
Graphical abstract
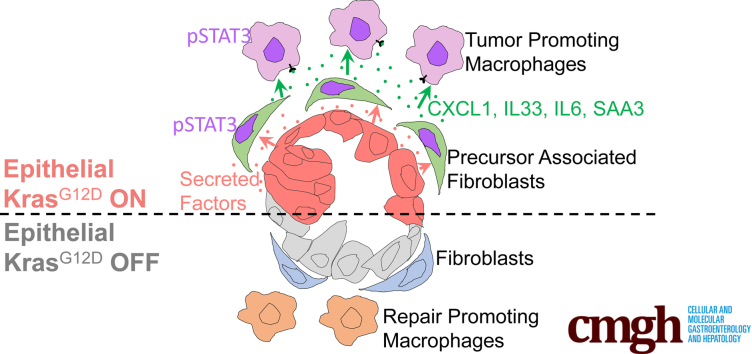
Summary.
Here, we show that epithelial oncogenic Kirsten Rat Sarcoma virus (KRAS) signals in a non–cell autonomous (ie, extrinsic) manner to drive accumulation and maintenance of an immune-suppressive microenvironment during the onset of pancreatic cancer. Furthermore, we show that KRAS-reprogrammed fibroblasts constitute a key mediator of the transformation process.
Pancreatic ductal adenocarcinoma is the third leading cause of cancer-related death in the United States and is among the most lethal malignancies, with an expected 5-year survival of approximately 10%.1 More than 90% of pancreatic ductal adenocarcinoma instances harbor an oncogenic mutation in the KRAS gene, most commonly KRASG12D.2,3 Autopsy studies have shown that more than 75% of the population harbors preneoplastic lesions in the pancreas that are linked to mutations in KRAS, yet pancreatic cancer is relatively rare.4,5 This finding is reproduced in mouse models of the disease, where widespread epithelial expression of oncogenic Kras leads to cancer with long latency. Why some Kirsten Rat Sarcoma virus (KRAS)-mutant lesions maintain an indolent preneoplastic state while others progress to deadly invasive cancer is a fundamental gap in knowledge.
A long-standing question has been the identity of the cell of origin to pancreatic cancer. The most frequently used genetically engineered mouse models of pancreatic cancer express KrasG12D broadly across the pancreas epithelium upon Cre recombination. Two of the most common Cre drivers include the Pancreatic and Duodenal Homeobox 1 (Pdx1) promoter (Pdx1-Cre;KrasLSL-G12D/+) and insertion into the Pancreas Associated Transcription Factor 1a (Ptf1a) locus (Ptf1aCre/+;KrasLSL-G12D/+); both of these models are commonly referred to as KC.6,7 Different precursor lesions to pancreatic cancer have been described in human patients,8 of which pancreatic intraepithelial neoplasia (PanIN) is the most common. KC mice undergo a stepwise carcinogenesis process that mimics the progression of human disease, including PanIN formation.6,7 In mouse models both acinar cells and ductal cells can give rise to pancreatic cancer, however, acinar cell origin is the most frequent.9, 10, 11, 12
Acinar cells are highly plastic: upon tissue damage, they down-regulate expression of digestive enzymes and acquire a duct-like differentiation status through a process known as acinar-to-ductal metaplasia (ADM). Acinar cells that have undergone ADM can be distinguished from ductal cells by their unique expression of acinar progenitor factors.13 In the context of acute injury, ADM is reversible, and the acinar parenchyma is re-established over time. However, expression of oncogenic Kras prevents ADM reversal and the duct-like cells instead undergo neoplastic transformation.14 The cell-intrinsic mechanisms regulating the balance between cellular plasticity and carcinogenesis have been studied extensively. ADM is driven by fundamental changes in the transcriptional regulation of acinar cells, with acinar transcription factors restraining and ductal transcription factors promoting this process (for review see Krah and Murtaugh15). For example, reduction of expression of the acinar transcription factors PTF1a, Muscle,Intestine and Stomach expression 1 (MIST1), or Nuclear Receptor Subfamily 5 Group A Member 2 (NR5A2) promote transformation, while forced expression of MIST1 and PTF1a protect acinar cells from dedifferentiation.16, 17, 18, 19, 20, 21 ADM also is regulated by intracellular signaling, including a requirement for epithelial mitogen-activated protein kinase activation.22,23 Recently, epigenetic reprogramming driven by oncogenic Kras has emerged as a key determinant of progression/redifferentiation of acinar cells upon injury.24
Although the cell-autonomous (ie, intrinsic) effects of oncogenic Kras activation have been studied extensively, the non–cell autonomous effects are less understood. Here, we set out to investigate how oncogenic Kras-expressing cells affect the microenvironment, which we refer to as extrinsic Kras signaling. To understand the role of oncogenic Kras in established PanINs, we take advantage of a unique model of pancreatic cancer, the iKras Add: (p48-Cre;TRE-KrasG12D; R26rtTa/rtTa) mouse, which allows inducible and reversible expression of the oncogene.25,26 PanIN formation is accompanied by accumulation of a precursor lesion microenvironment (PME), with activation of fibroblasts (for review see Helms et al27) and infiltration of immune cells. The latter are largely immune-suppressive,28 and include myeloid cells that both suppress T-cell responses and directly promote pancreatic cancer progression.29,30 Myeloid cells support ADM31 and are required for sustaining ADM and promoting progression to PanIN.32 Conversely, myeloid cells also are required for tissue repair both in the setting of injury and redifferentiation.32,33 Furthermore, experimental induction of acute pancreatitis with its associated inflammation synergizes with oncogenic Kras to accelerate the formation of ADM and PanIN.34,35 Thus, inflammation both accompanies and promotes carcinogenesis, but the relationship between epithelial cells and surrounding stroma in this process remains unclear.
In this study, we dissected the role of oncogenic Kras in driving the formation and maintenance of the PME. We discovered that fibroblast reprogramming occurs during the earliest stages of carcinogenesis in a Janus kinase (JAK)/signal transducer and activator of transcription 3 (STAT3) signaling-dependent manner, resulting in the activation of an inflammatory gene expression program that includes multiple cytokines known to drive the tumor-promoting functional status of myeloid cells infiltrating the pancreas. Consequently, approaches to inhibit fibroblasts reprogramming during carcinogenesis should be explored to prevent, and possibly reverse, KRAS-driven carcinogenesis.
Results
Extrinsic Signaling by Oncogenic Kras Reprograms the Pancreas Microenvironment
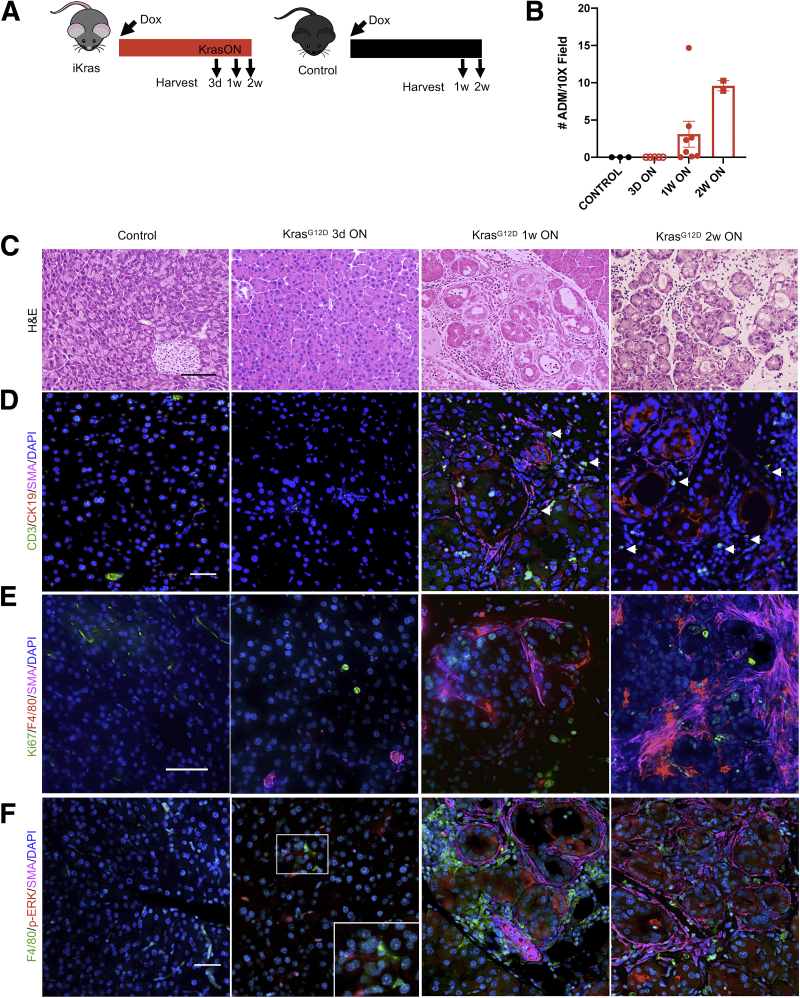
To study the recruitment of immune cells by oncogenic Kras-expressing tumor cells, we used Ptf1aCre/+; R26rtTa-ires-EGFP/rtTa-ires-EGFP;TetO-KrasG12D mice (hereby iKras) that express oncogenic KrasG12D in pancreatic epithelial cells in an inducible and reversible manner upon doxycycline (DOX) administration.25 Initially, we activated KrasG12D in adult mice (age, 6–12 wk) by feeding iKras mice or littermate controls (lacking either Cre or KrasG12D expression) DOX chow for 3 days, 1 week, or 2 weeks (Figure 1A). Pancreata from iKras mice appeared largely histopathologically normal 3 days after KrasG12D activation, with the parenchyma largely composed of acinar cells and Cytokeratin 19 (CK19) expression confined to the ducts (Figure 1B–D). One week after KrasG12D induction, although the pancreas largely was histologically normal, we observed focal ADM accompanied by an increase in the proliferation marker Ki67 that became more widespread by 2 weeks (Figure 1B, C, and E). Three days after KrasG12D induction, we observed patchy epithelial expression of phosphorylated Extracellular signal-regulated kinase (ERK) (p-ERK), indicating activation of the MAPK pathway (Figure 1F) in otherwise histologically normal areas. At later time points, areas of apparent ADM had increased p-ERK staining (Figure 1F), while the remainder of the pancreas had patchy expression of p-ERK. The limited expression of p-ERK even as all pancreas epithelium expresses KrasG12D in this model was consistent with the known requirement of stimulation by various upstream factors to fully activate KrasG12D signaling, including myeloid-derived Epidermal growth factor receptor (EGFR) ligands.36, 37, 38
Figure 1.
Oncogenic Kras drives immune cell recruitment during the onset of tumorigenesis. (A) Experimental design. Control (lacking either the Kras or Ptf1aCre allele) and iKras mice were given DOX chow to activate oncogenic Kras (KrasG12D) and pancreata were harvested 3 days (3d), 1 week (1w), or 2 weeks (2w) after induction of Kras. N = 3–5 mice per group. (B) Quantification of ADM in pancreatic tissue from control or iKras mice that received DOX chow for 3 days, 1 week, or 2 weeks. (C) Representative images of H&E staining of control and iKras pancreata at the indicated time points. N = 3–5 mice per group. Scale bar: 100 μm. (D) Representative images of CD3 (green), CK19 (red), and SMA (magenta) co-immunofluorescent staining in control and iKras pancreata at the indicated time points, arrowheads point to CD3+ T cells. N = 3 mice per group. Scale bar: 50 μm. (E) Immunostaining for Ki67 (green), F4/80 (red), and SMA (magenta). Scale bar: 50 μm. (F) Representative images of F4/80 (green), p-ERK (red), and SMA (magenta) co-immunofluorescent staining in control and iKras pancreata at the indicated time points. N = 3 mice per group. Scale bar: 50 μm.
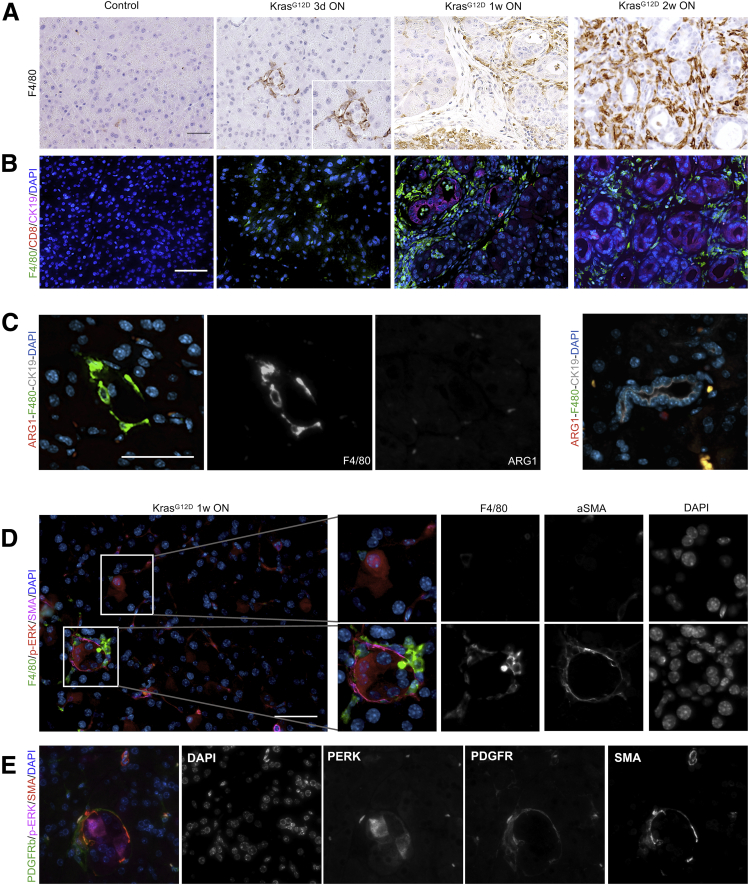
Next, we investigated the dynamics of immune infiltration and the presence of activated fibroblasts (as marked by expression of smooth muscle actin [SMA]). Co-immunofluorescence and immunohistochemistry analysis showed the presence of macrophages surrounding untransformed p-ERK–expressing acinar cells as early as 3 days after KrasG12D activation (Figures 1F and 2A), whereas T cells were rare at this stage (Figures 1D and 2B). Macrophages increased by 1 and 2 weeks after KrasG12D activation, and T cells, including CD8+ T cells, were detected easily at these time points, at least in the ADM areas (Figures 1D and F, and 2A and B). Infiltrating macrophages were largely negative for the immunosuppressive marker arginase 1 (ARG1) (Figure 2C). ADM clusters at this stage also were surrounded by SMA+ fibroblasts (Figure 1D–F). We then examined the areas of the pancreas that were morphologically normal at the 1-week time point (majority of the pancreas) (Figure 2D). Interestingly, high-magnification images showed morphologically normal clusters of acini with increased p-ERK surrounded by both macrophages and a thin layer of SMA+ fibroblasts, indicating that changes in the microenvironment precede acinar transdifferentiation. Careful analysis of the tissue from the 3-day time point also showed some p-ERK–expressing acini with surrounding macrophages but without SMA+ fibroblasts (Figure 1F), but these instances were rare and we cannot exclude that fibroblasts were present outside the plane of the section. Of note, the fibroblasts had no expression of p-ERK at this stage (Figure 2E). Taken together, our findings are consistent with a model whereby epithelial activation of p-ERK, downstream of oncogenic Kras, is accompanied by fibroblast activation and macrophage infiltration, which precede acinar transdifferentiation.
Figure 2.
Macrophage expansion precedes acinar transdifferentiation. (A) Immunohistochemistry for F4/80. N = 3 mice per group. (B) Representative images of F4/80 (green), CD8 (red), and CK19 (magenta) co-immunofluorescent staining in control and iKras∗ pancreata. Scale bar: 50 μm. (C) Co-immunostaining for F4/80 (green), ARG1 (red), and CK19 (gray) in iKras∗ pancreata at 3 days ON. Note that macrophages surround CK19- acinar cells. Right: Image shows a CK19+ duct in the same section. Scale bar: 50 μm. (D) Representative images of F4/80 (green), p-ERK (red), and SMA (magenta) co-immunofluorescence staining of an iKras pancreas after activating KrasG12D for 1 week. Regions with high expression of p-ERK are enlarged and single-channel images are included. N = 3 mice. Scale bar: 50 μm. (E) Representative images of PDGFRβ (green), p-ERK (red), and SMA (magenta) co-immunofluorescence staining of an iKras pancreas after activating KrasG12D for 1 week. Single-channel images are included. N = 3 mice.
Epithelial Oncogenic KrasG12D Is Required to Maintain Acinar Transdifferentiation and Fibroblast Accumulation, but Not Immune Infiltration
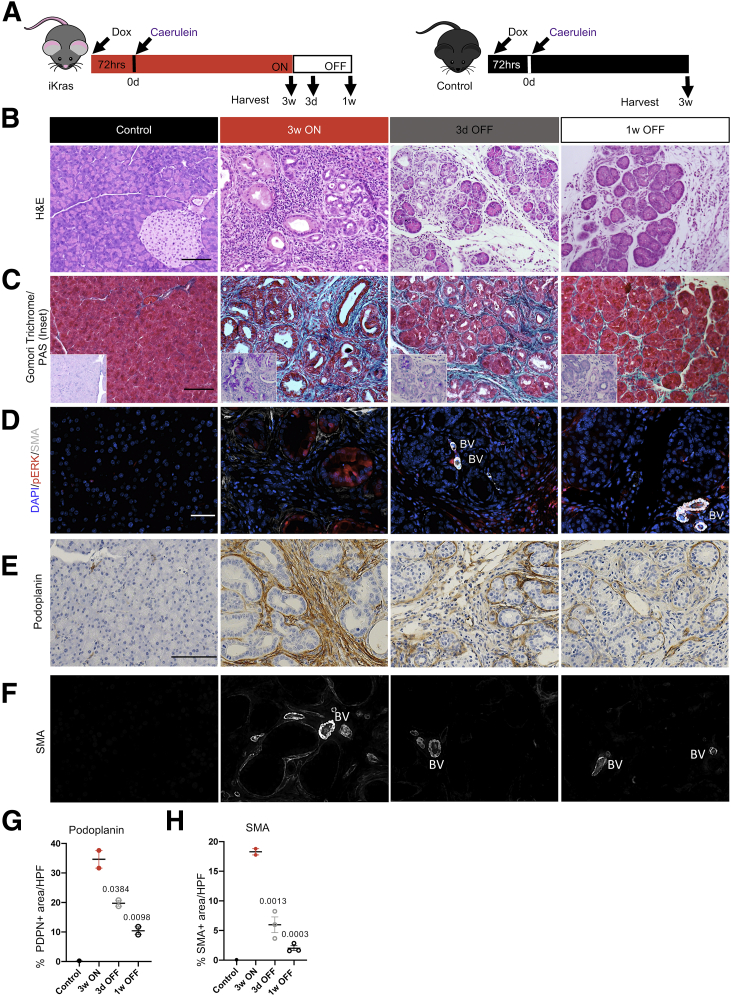
Acute pancreatitis synergizes with oncogenic Kras to drive neoplastic transformation.25,34,35 Previously, we have shown that iKras mice on DOX (KrasG12D ON) develop widespread early PanIN lesions at 3 weeks after pancreatitis induced by caerulein, and withdrawal of DOX (KrasG12D OFF) at this stage leads to PanIN regression and tissue repair.25 Thus, epithelial oncogenic KrasG12D is required to maintain acinar transdifferentiation. Here, using the same experimental strategy, we sought to study the potential role of KrasG12D in regulating the PME. Accordingly, we placed iKras and control mice on DOX and then induced acute pancreatitis and harvested the pancreata 3 weeks after pancreatitis (Figure 3A). Pancreata in iKras mice presented with widespread ADM and low-grade PanINs (with intracellular mucin accumulation as shown by periodic acid–Schiff staining) and accumulation of fibrotic stroma (Figure 3B and C). Expression of p-ERK was confined to the epithelium when KrasG12D was ON, as previously shown (Figure 3D).25 In contrast, control mice treated in parallel had normal pancreas architecture. iKras pancreata also were marked by extensive collagen deposition, fibroblast expansion (as detected by podoplanin), and activation (by SMA expression) (Figure 3C–H). To study the effect of KrasG12D inactivation on the PME, we replaced DOX chow with DOX-free chow to inactivate KrasG12D expression in iKras mice (for 3 days or 1 week) 3 weeks after administration of caerulein (Figure 3A). The 3-day and 1-week time points were chosen to coincide with the early and middle phases of the pancreatic remodeling process.25 As previously described, KrasG12D inactivation led to a drastic reduction in epithelial p-ERK and redifferentiation of acinar cells progressively over time (Figure 3B–D).31,32 We then investigated changes in the stroma after inactivation of KrasG12D. We observed a reduction in both total fibroblasts (podoplanin+ cells) and activated, SMA+ fibroblasts (Figure 3D–F, and quantification in G and H), as well as a reduction in collagen deposition (Figure 3C). These results are consistent with a requirement for continued oncogenic KrasG12D activity to maintain fibroblast expansion and pancreatic fibrosis.
Figure 3.
Sustained expression of KrasG12Dis required to maintain PanIN and fibrosis. (A) Experimental design. Wild-type control and iKras mice were given DOX chow to activate KrasG12D, followed by induction of acute pancreatitis. Mice either remained on DOX chow for 3 weeks and pancreata were harvested (3 weeks [3w] ON), or DOX chow was removed and the pancreata were harvested after 3 days (3d) or 1 week (1w) (labeled 3d OFF or 1w OFF, respectively). N = 8 mice per group. (B) Representative images of H&E staining of control and iKras∗ pancreata at the indicated time points. N = 8 mice per group. Scale bar: 100 μm. (C) Representative images of Gomori trichrome and periodic acid–Schiff Stain (PAS; inset) of control or iKras pancreata at the indicated time points. N = 2–3 mice per group. Scale bar: 100 μm. (D) Immunofluorescent staining for SMA (white) and p-ERK (red). Scale bar: 50 μm. N = 2–3 mice per group. (E) Immunostaining for podoplanin. Scale bar: 100 μm. (F) Immunofluorescent staining for SMA (single channel) used for quantification. Scale bar: 50 μm. N = 2–3 mice per group. Quantification of (G) podoplanin and (H) SMA staining, shown as means ± SEM. Statistical differences were determined by multiple analysis of variance, all compared with the 3w ON group. BV, blood vessels; HPF, high-power field; PDPN, Podoplanin.
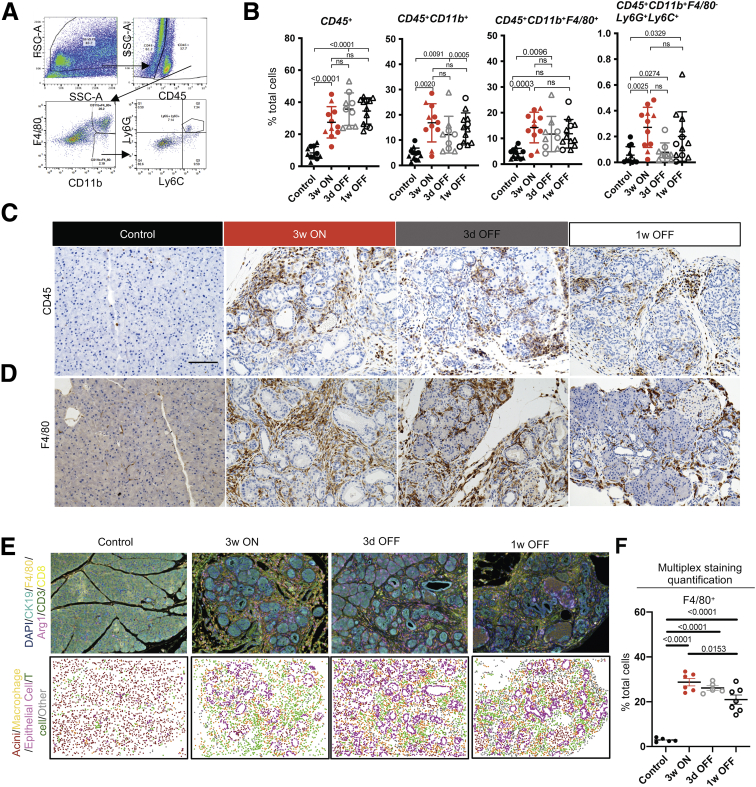
Myeloid cells, including abundant macrophages, infiltrate early during pancreatic carcinogenesis and are required for sustained ADM and progression to PanIN.31,32,39 Therefore, we endeavored to study the effect of KrasG12D inactivation on macrophages and other myeloid cell infiltration in the pancreas. We performed a combination of flow cytometry (gating strategy in Figure 4A) and immunostaining, including multiplex immunofluorescent staining (Opal multiplex immunohistochemistry; Akoya Biosciences, Menlo Park, CA). As expected, we observed an increase in CD45+ immune cells in the iKras mice at 3 weeks ON compared with control mice (Figure 4B and C). This increase included total myeloid cells (CD45+CD11b+) as well as macrophages (CD45+CD11b+F4/80+) and immature myeloid cells (CD45+CD11b+F4/80-Ly6C+Ly6G+, often referred to as Myeloid-derived suppressor cells, MDSCs), with the majority being macrophages (Figure 4B and D–F), consistent with the increase in leukocyte infiltration observed in KC mice.28 Surprisingly, KrasG12D inactivation resulted in little to no reduction in total immune infiltration, total myeloid cells, or macrophages (Figure 4B–F). These results are consistent with the role of myeloid cells during the pancreatic tissue repair process.32
Figure 4.
Immune infiltration persists during tissue repair after epithelial Kras inactivation. (A) Flow cytometry gating strategy for myeloid cells in FlowJo. (B) Quantification of flow cytometry results. Immune cell populations are expressed as the percentage of total cells in control or iKras pancreata at the indicated time points, shown as means ± SD. N = 9–11 mice per group. Statistical analysis by multiple comparison analysis of variance and multiple comparison Kruskal–Wallis. Triangles represent females and circles represent males. Representative immunohistochemistry images for (C) CD45 and (D) F4/80 in the pancreatic tissue of control or iKras∗ mice at the indicated time points. Scale bar: 100 μm. N = 3 mice per group. (E) Top: Representative images of F4/80, Arg1, CD3, CD8, and CK19 immunofluorescent multiplex staining at the indicated time points. N = 5–7 mice per group. Bottom: Identification of cell types using inForm Cell Analysis software. T cell, macrophage, and epithelial cells were selected based on single staining of CD3 (Opal 520), F480 (Opal 540), and CK19 (Opal 690), respectively. Acinar cells were labeled based on the lack of listed fluorophores and by tissue morphology. (F) OPAL staining quantification of the percentage of F4/80-positive cells from total cells in the pancreatic tissue of control or iKras mice at the indicated time points. Data are shown as means ± SEM. N = 5–7 mice per group. Statistical analysis was performed by the 2-tailed unpaired t test. FSC-A, forward scatter area.
Extrinsic Signaling From KrasG12D-Transformed Cells Drives Myeloid Polarization
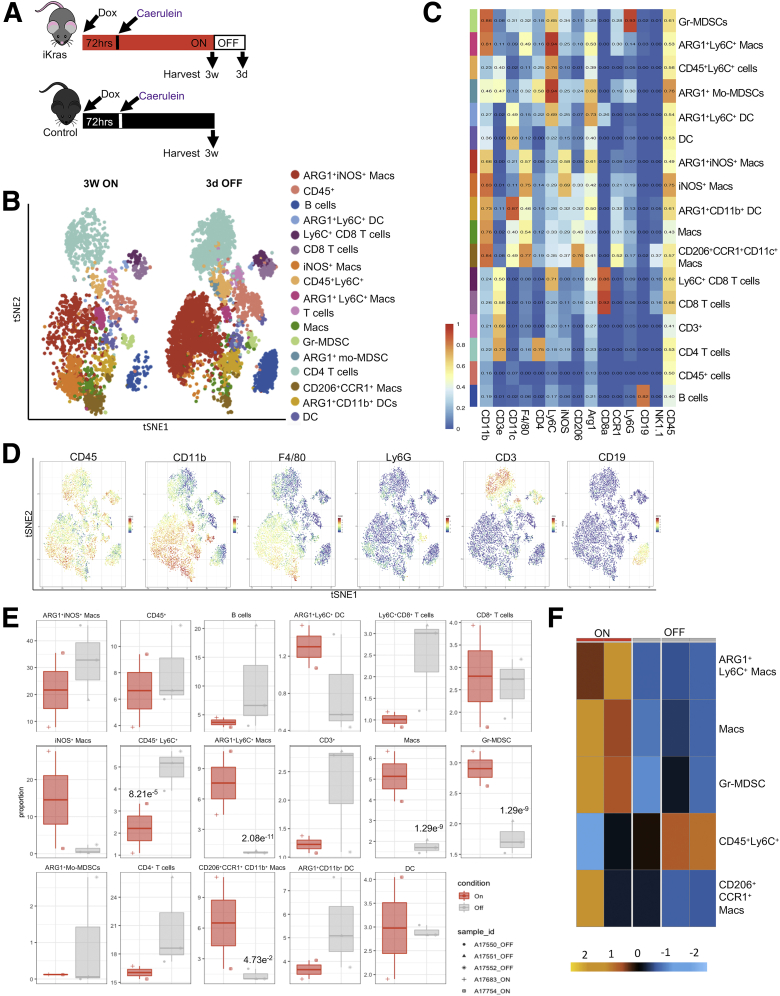
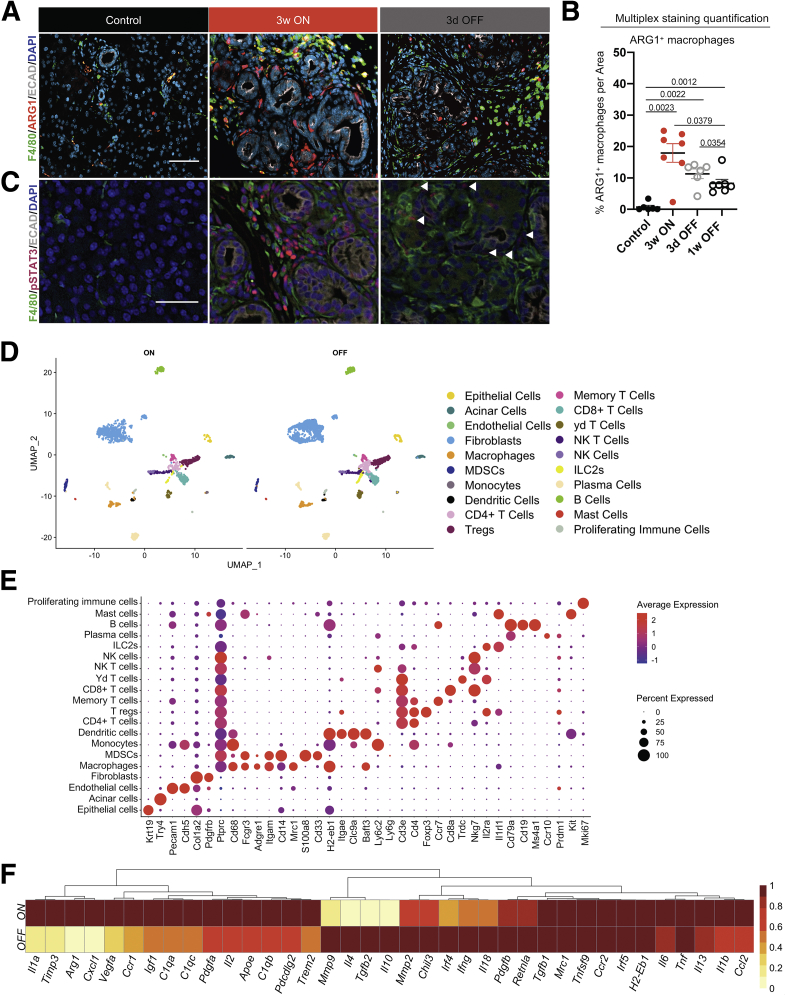
Because myeloid cell numbers did not change upon KrasG12D inactivation, we sought to determine whether their polarization status was affected. For this purpose, we used mass cytometry (CyTOF, Fluidigm, South San Francisco, CA) with a panel of approximately 20 antibodies (Table 1). We limited the scope of the CyTOF experiment to 3 weeks Kras∗ ON (N = 2) and 3 days OFF (N = 3) time points (Figure 5A) to highlight the short-term effect of KrasG12D inactivation, before extensive tissue remodeling. We visualized the multidimensional CyTOF data using t-distributed stochastics neighbor embedding plots and identified 17 distinct immune cell clusters (Figure 5B–D). Among T cells, we observed a trend toward an increase upon KrasG12D inactivation, but no statistical difference (Figure 5E). The myeloid immune landscape was complex, with multiple populations, including heterogeneous macrophage subtypes (Figure 5B, C, and E). Although some populations did not change or trended toward an increase when we compared KrasG12D OFF with KrasG12D ON samples, other populations decreased drastically (Figure 5E). Notably, immunosuppressive ARG1+ Ly6C+ macrophages, granulocytic myeloid-derived suppressor cells (Gr-MDSCs), defined as CD45+CD11b+F4/80-Ly6CloLy6Ghi, and CCR1+CD206+ macrophages, decreased significantly upon KrasG12D inactivation (Figure 5E and F). Down-regulation of ARG1+ macrophages was validated by co-immunofluorescent and multiplex immunofluorescent staining (Figure 6A and B). Furthermore, we discovered that phosphorylated signal transducer and activator of transcription 3 (p-STAT3), indicating activation of JAK/STAT3 signaling and known to promote the immune-suppressive function of macrophages,40 was similarly down-regulated in macrophages and other stromal cells, as well as in epithelial cells, by inactivation of epithelial oncogenic KrasG12D (Figure 6C).
Table 1.
CyTOF Antibodies
| Antibodies | Clone | Isotope | Dilution | |
|---|---|---|---|---|
| CD45 | Fluidigm | 30-F11 | Y89 | 1:200 |
| Ly6G | Fluidigm | 1AB | Pr141 | 1:400 |
| CD11b | Fluidigm | M1/70 | Nd143 | 1:300 |
| CD4 | Fluidigm | RM4/5 | Nd145 | 1:200 |
| F480 | Fluidigm | BM8 | Nd146 | 1:100 |
| CD140a | Fluidigm | APA5 | Nd148 | 1:100 |
| CD19 | Fluidigm | 6D5 | Nd149 | 1:200 |
| Ly6C | Fluidigm | HK1.4 | Nd150 | 1:400 |
| CD3e | Fluidigm | 145-2C11 | Sm152 | 1:100 |
| CD105 | Custom | MJ7/18 | Dy164 | 1:100 |
| TCR gd | Fluidigm | GL3 | Tb159 | 1:100 |
| CD191 CCR1 | Custom | S15040E | Gd160 | 1:100 |
| iNOS | Fluidigm | CXNFT | Dy161 | 1:100 |
| Arginase1 | Custom | Polyclonal | Er166 | 1:400 |
| CD8a | Fluidigm | 53-6.7 | Er168 | 1:200 |
| CD206 | Fluidigm | C068C2 | Tm169 | 1:200 |
| CD161 NK1.1 | Fluidigm | PK136 | Er170 | 1:100 |
| CD11c | Fluidigm | N418 | Bi209 | 1:100 |
Figure 5.
Oncogenic Kras regulates myeloid cell polarization status in the PME. (A) Experimental design. N = 8–9 mice per group. (B) CyTOF analysis of CD45+ cells from iKras pancreata visualized by t-distributed stochastics neighbor embedding (tSNE) plot for 3 weeks (3w) ON and 3 days (3d) OFF time points. Nineteen distinct cell clusters were identified by FlowSOM (Bioconductor, Buffalo, NY) using 18 markers and 1061 randomly selected cells per group. N = 2–3 mice per group. (C) Heatmap of the median marker intensity generated with FlowSOM of the CyTOF samples (iKras 3w ON and 3d OFF combined). Colors on the left represent the clusters shown in the t-SNE plot. N = 2–3 per group. All population were gated from CD45+ cells. (D) tSNE plots showing the expression of different immune lineage markers in the clusters from the CyTOF data. (E) Quantification of identified clusters proportion by CyTOF analysis at the indicated time points. Statistical analysis by Wrapper function. (F) Heatmap showing differential abundance of clusters from CyTOF analysis of iKras pancreata for 3w ON and 3d OFF time points. N = 2–3 mice per group.
Figure 6.
Oncogenic Kras inactivation results in transcriptional reprogramming of infiltrating macrophages. (A) Immunostaining for F4/80 (green), ARG1 (red), and E-cadherin (Ecad) (white) in control and iKras pancreata at the indicated time points. N = 3 mice per group. Scale bar: 50 μm. (B) OPAL staining quantification of ARG1+ macrophages in pancreatic tissue of control or iKras mice at the indicated time points. Data are shown as means ± SEM. Statistical analysis by the 2-tailed unpaired t test. (C) Immunostaining for F4/80 (green), p-STAT3 (magenta), and Ecad (white). N = 3 mice per group. Scale bar: 50 μm. (D) UMAP visualization of scRNAseq data showing unsupervised clustering of cells from iKras pancreatic samples (3 weeks [3w] ON, N = 2; 3 days [3d] OFF, N = 3). Each color represents a distinct cellular cluster. (E) Dot plot of the key genes used to identify different cell populations shown in the UMAP, identified by unsupervised clustering of the scRNAseq samples (3w ON and 3d OFF samples combined). Size of the dots reflects the proportion of cells expressing a determined gene and color represents the average expression level. (F) Heatmap showing the averaged scRNAseq expression data (relative to the highest expressor) for genes in macrophages selected from a curated list of macrophage polarization and functional markers.
Because CyTOF and immunostaining are limited by available antibodies, we performed single-cell RNA sequencing (scRNAseq) to elucidate how epithelial KrasG12D expression alters the microenvironment, and, in particular, macrophage polarization. We harvested pancreata from mice with KrasG12D ON for 3 weeks plus 3 days (N = 2, pooled for submission) or ON for 3 weeks and then OFF for 3 days (N = 3, pooled for submission). The experiment was designed such that pancreata were harvested and processed at the same time to avoid batch effects. The multidimensional scRNAseq data included a total of 5073 cells overall, with 1984 cells from the 3 weeks ON sample and 3089 cells from the 3 days OFF sample. The data were analyzed using the Seurat (Satija Lab, New York Genome Center, New York, NY) package in R (version 3.2.2) and visualized by uniform and manifold approximation and projection (UMAP) (Figure 6D). By comparing the transcriptome of each cluster with known markers of epithelial, immune, and stromal cell types (Figure 6E), we identified acinar cells, CK19+ epithelial cells (PanIN or ducts), fibroblasts, endothelial cells, and 16 distinct immune cell populations. Similar cell populations were present in both the ON and OFF condition, allowing us to compare gene expression changes upon inactivation of KrasG12D within each. We first assessed gene expression changes in macrophages (Figure 6F). Notably, tumor-associated macrophage (TAM) markers such as Arg1 and Ccr1 were down-regulated upon KrasG12D inactivation. We also observed down-regulation of apolipoprotein E (Apoe), a secreted protein that our group recently described as highly expressed in TAMs and as promoting immune suppression in invasive pancreatic cancer.41 In addition, we observed down-regulation of the complement genes C1qa, C1qb, C1qc, and the transcription factor Trem2, which together define TAMs in several malignancies including, as described by our group, primary and metastatic pancreatic cancer.42,43 These findings are consistent with KrasG12D being required to maintain TAM-like macrophage polarization. Conversely, inactivation of KrasG12D induced a repair-associated gene expression program, including Tgfb, Il10, Il4, Pdgfb, and Mmps (Figure 6F). Although oncogenic Kras is not required to maintain immune infiltration, it extrinsically (non–cell autonomously) regulates macrophage gene expression in PanIN.
Epithelial KrasG12D Drives Inflammatory Reprogramming of Pancreatic Fibroblasts
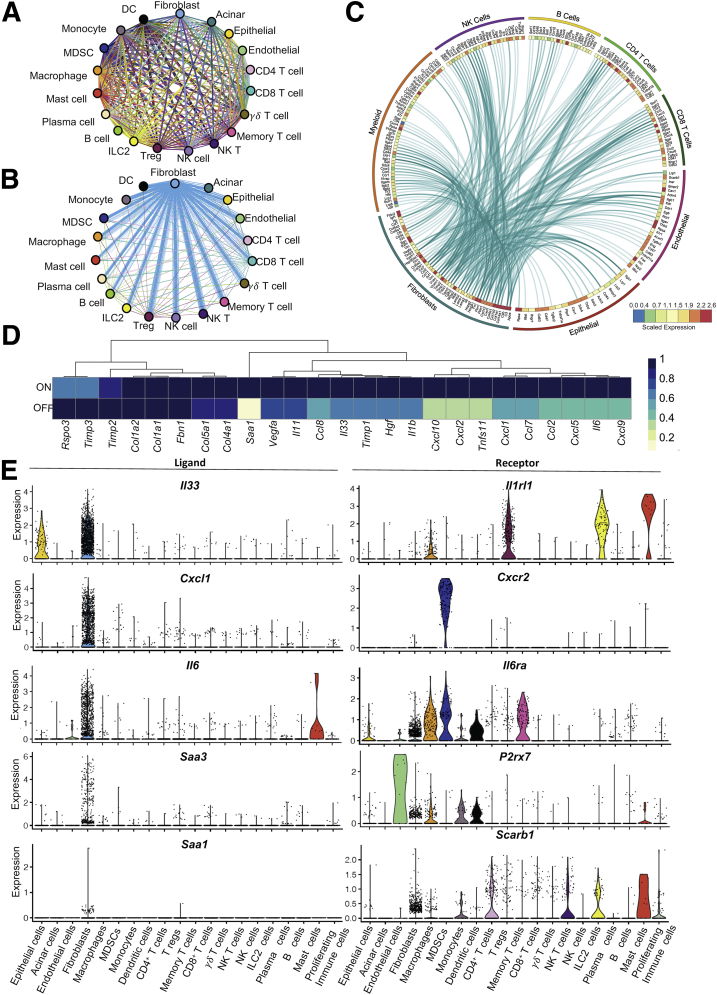
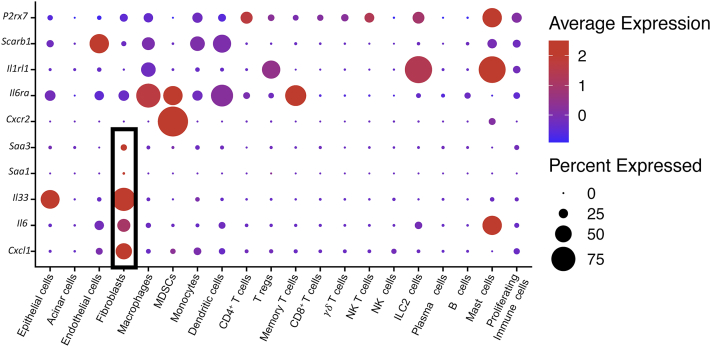
We next sought to understand how KrasG12D expression in epithelial cells directed cellular crosstalk to account for the changes in macrophage polarization. To achieve this, we filtered the scRNAseq data for expression of known ligand and receptor pairs using a published list further curated in our laboratory.44,45 We thus identified potential predicted interactions within the tissue (Figure 7A). We then plotted interactions that were induced by oncogenic Kras (hence down-regulated upon KrasG12D inactivation). Strikingly, most Kras-driven interactions connected fibroblast-expressed ligands with receptors on epithelial cells and immune cells (Figure 7B and C). We then queried the fibroblast gene expression data to identify cytokines specifically expressed in an epithelial KrasG12D-dependent manner (Figure 7D). Finally, we selected from that list the genes encoding for cytokines expressed mainly, or exclusively, by fibroblasts, including Il33, Il6, C-X-C motif chemokine ligand 1 (Cxcl1), and the inflammatory mediators Serum amyloid a1 (Saa1) and Saa3 (the ortholog of human SAA1) (Figures 7E and 8). Intriguingly, these aforementioned cytokines all have known roles in pancreatic cancer.46, 47, 48, 49, 50 Conversely, the respective receptors for these secreted factors were expressed by immune cells, including Cxcr2 (CXCL1 receptor) on immature myeloid cells, Il6r (interleukin [IL]6 receptor), P2rx7 and Scarb1 (SAA3 and SAA1 receptors) on several myeloid populations, and Il1rl1 (IL33 receptor also known as Suppression Of Tumorigenicity 2, ST2) on macrophages, regulatory T cells, mast cells, and innate lymphoid cells type 2. Thus, fibroblasts are extrinsically reprogrammed by KrasG12D epithelial cells to activate a gene expression profile that includes cytokines with cognate receptors expressed by myeloid cells, and with a known role in driving myeloid cell/macrophage-mediated immune suppression.
Figure 7.
Fibroblasts express inflammatory cytokines in response to epithelial oncogenic Kras. (A) Interactome analysis showing all predicted ligand-receptor interactions between different cell populations identified in iKras pancreatic scRNAseq analysis. Each line represents a ligand-receptor pair, color-coded by the cell type expressing the ligand. (B) Differential interactions from panel A positively regulated by oncogenic Kras (higher in 3 weeks [3w] ON compared with 3 days [3d] OFF) (adjusted P value < .05). (C) Circos plot showing the average expression of fibroblast ligands connected to their predicted receptors on various cell populations as measured in the pancreatic scRNAseq analysis. Ligands shown are from panel B (adjusted P value < .05). (D) Heatmap showing averaged scRNAseq expression data (relative to the highest expressor) for genes in fibroblasts from a curated list of immunomodulatory factors. (E) Violin plots showing expression of Il33, Cxcl1, Il6, Saa3, and Saa1 and their respective receptors Il1rl1, Cxcr2, Il6ra, P2rx7, and Scarb1 across all identified cell populations in both Kras∗ ON and OFF samples combined. DC, Dendritic cell; NK T, Natural killer T cell.
Figure 8.
Fibroblasts secrete immunomodulatory factors with receptors in myeloid cells. Dot plot showing average expression of Il33, Il6, Cxcl1, Saa1, and Saa3 and their respective receptors Il1rl1, Il6ra, Cxcr2, P2rx7, and Scarb1 across cell populations. Size of the dots reflects the proportion of cells expressing a determined gene and color represents the average expression level. NK T, Natural killer T cell.
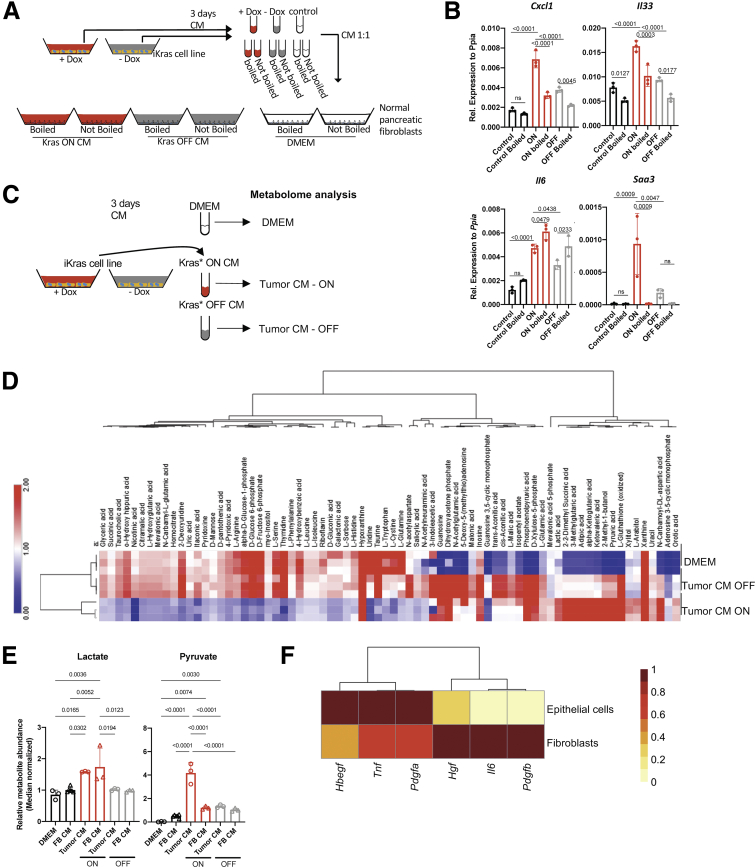
To investigate the mechanism of fibroblast reprogramming, we used an in vitro approach. We harvested conditioned media (CM) from iKras cells derived from a Ptf1aCre/+; R26rtTa-ires-EGFP/rtTa-ires-EGFP;TetO-KrasG12D;Trp53R172H/+ (iKras;P53) mouse tumor51; the inclusion of mutant Trp53 being required for growth in cell culture. In brief, iKras cells51 were grown in the presence of DOX, thus expressing KrasG12D. Then, media was changed to either DOX-containing or DOX-free media, thus maintaining KrasG12D expression or inactivating it, respectively. After 48–72 hours, we harvested CM from each condition and boiled half of it to denature heat-labile components. CM then was administered to cultured primary mouse fibroblasts (CD1WT and B6318) derived from healthy adult pancreata as previously described41,52 (Figure 9A). Compared with control Dulbecco’s modified Eagle medium (DMEM) media, CM from iKras cells induced expression of Cxcl1, Il6, Il33, and Saa3 in fibroblasts in a KrasG12D-dependent manner (Figure 9B), recapitulating the in vivo results. To distinguish between a KrasG12D-dependent protein factor or metabolite signaling to fibroblasts, we compared intact CM with boiled CM, and found that the latter induced Il6 but none of the genes encoding the other cytokines of interest (Figure 9B). Thus, a combination of cancer cell–derived, KrasG12D-dependent heat-labile factors such as proteins and metabolites mediated fibroblast reprogramming. Oncogenic Kras regulates the intracellular metabolome of pancreatic cancer cells.26 We inquired whether the extracellular metabolome is similarly dependent on expression of oncogenic Kras. Indeed, mass spectrometry–based metabolomic profiling showed numerous extracellular metabolites whose abundance changed depending on KrasG12D expression (Figure 9C and D). Among these, several released metabolites have been reported to act as direct extracellular signaling molecules (eg, pyruvate, lactate, α-ketoglutarate) while, alternatively, the decreased abundance of other metabolites may impact signaling downstream of Mammalian target of rapamycin (mTOR) (eg, arginine, isoleucine, cystine, glutamine) (Figure 9D).53, 54, 55, 56 We then investigated the extracellular metabolite composition after incubation of the iKrasG12D-conditioned medium from the various conditions described earlier. We observed that the lactate concentration was not affected while pyruvate was depleted upon culturing with fibroblasts (Figure 9E). Thus, it appears that fibroblasts consume pyruvate from the medium, an intriguing finding given that fibroblasts serve as a source of pyruvate in breast cancer.57
Figure 9.
Fibroblasts are reprogrammed through epithelial KrasG12D-induced secreted molecules. (A) Experimental design. CM was collected from iKras∗ cancer cells cultured with +DOX to activate KrasG12D expression (Kras ON) or without -DOX (Kras OFF). The media samples then were boiled to denature protein factors, or left intact and used to culture pancreatic fibroblasts. DMEM was used as control. (B) Quantitative reverse-transcription polymerase chain reaction for Cxcl1, Il33, Il6, and Saa3 expression in fibroblasts (CD1WT) that were cultured with CM either from Kras ON, Kras OFF, or DMEM, either boiled or not boiled. Gene expression was normalized to Peptidylprolyl Isomerase A (Ppia). Data are shown as means ± SD. N = 3 per group. The statistical differences were determined by multiple-comparison analysis of variance. (C) Experimental design. CM was collected from iKras cancer cells cultured with +DOX to activate KrasG12D expression (Kras∗ ON) or without -DOX (KrasG12D OFF), and used for metabolomic analysis. (D) Heatmap showing levels of extracellular metabolites in iKras cancer cell CM with KrasG12D ON or OFF. Both conditions are compared with regular DMEM media. (E) Fibroblasts were exposed to +DOX CM and -DOX CM for 48 hours. The resulting CM was analyzed for metabolites, in parallel with iKras cell CM and DMEM. Relative metabolite abundance of lactate and pyruvate from all conditions are shown. (F) Heatmap showing averaged scRNAseq expression data (relative to the highest expressor) for genes encoding for ligands that are secreted by fibroblasts and epithelial cells. FB, Fibroblast; Rel., relative.
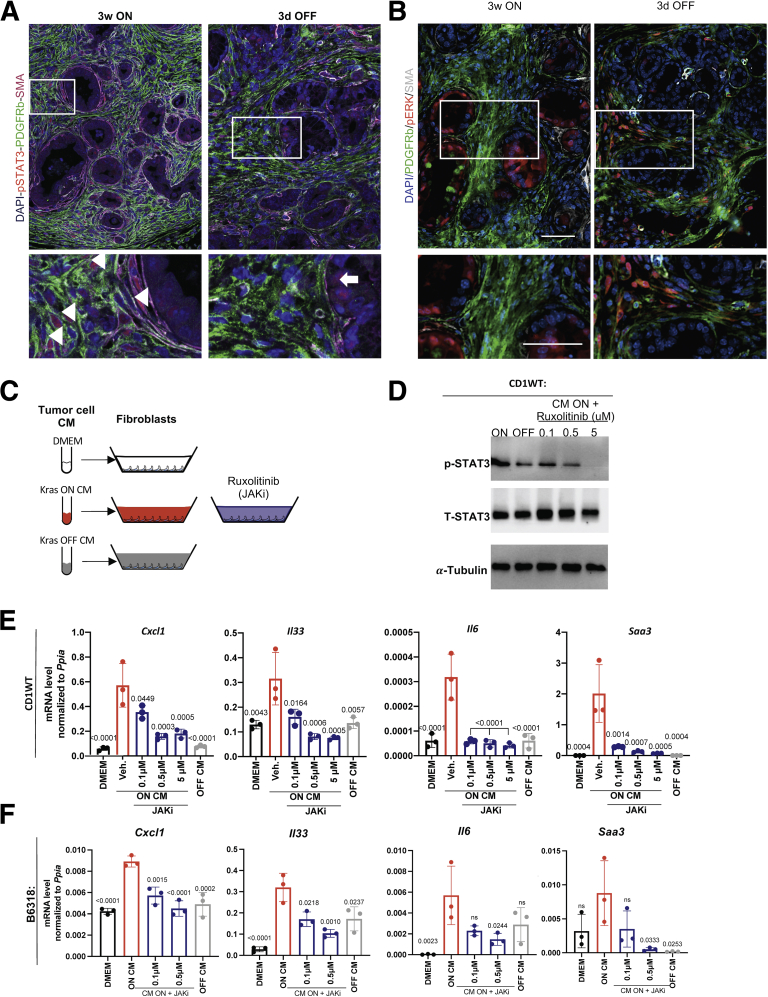
We then investigated the list of secreted factors expressed by KrasG12D-expressing epithelial cells in the single-cell sequencing data. We noted, among others, expression of Hbegf, Tnf, Pdgfa, Pdgfb, Hgf, and Il6 (Figure 9F). Expression of Hbegf, Tnf, and Pdgfa was higher in epithelial cells, whereas Hgf, Il6, and Pdgfb were higher in fibroblasts. Given the multitude of factors, we reasoned that we were unlikely to identify a single factor driving fibroblast reprogramming. We focused instead on signaling pathways that might be activated in fibroblasts. In vivo, we observed expression of p-STAT3, but not p-ERK, in Platelet-derived growth factor receptors (PDGFR)+ or SMA+ fibroblasts when KrasG12D is ON in iKras mice (Figure 10A and B). Similar to the p-STAT3 expression within macrophages (Figure 6C), p-STAT3 expression in fibroblasts was down-regulated upon KrasG12D inactivation, identifying it as a potential mediator of fibroblast reprogramming (Figure 10A). In contrast, p-ERK was up-regulated in fibroblasts upon KrasG12D inactivation (Figure 10B), consistent with its role during tissue repair.22,32 We then repeated the in vitro experiment described earlier, this time inhibiting JAK/STAT signaling using ruxolitinib (JAK inhibitor, JAKi) (Figure 10C and D). CM-induced fibroblast expression of Cxcl1, Il33, Saa3, and Il6 was inhibited by JAKi (Figure 10E and F). Thus, in vitro activation of fibroblast JAK/STAT3 signaling is required for epithelial cell–induced reprogramming.
Figure 10.
Fibroblast reprogramming requires JAK/STAT signaling. (A) Immunofluorescent staining of p-STAT3 (red), Platelet-derived growth factor receptor beta (PDGFRβ) (green), and SMA (gray). N = 2–3 mice per group. Arrowheads point to p-STAT3–positive fibroblasts and arrow points to positive epithelial cells. (B) Immunofluorescent staining for PDGFRβ (green), SMA (white), and p-ERK (red). Scale bar: 50 μm. N = 2–3 mice per group. (C) Experimental design. Fibroblasts were exposed to iKras∖tumor cell CM either from KrasG12D ON, KrasG12D OFF, or DMEM. Fibroblasts cultured with KrasG12D ON CM media were treated with a JAK/STAT2/3 inhibitor (ruxolitinib) or vehicle. (D) Western blot showing protein levels of total STAT3 or phospho-STAT3 from CD1WT fibroblasts treated as indicated. α-tubulin was used as a loading control. (E) Quantitative reverse-transcription polymerase chain reaction results for Cxcl1, Il33, Il6, and Saa3 expression in fibroblasts (CD1WT) treated with CM as described in panel C. Data are shown as means ± SD. N = 3 per group. The statistical differences were determined by multiple-comparison analysis of variance. (F) Quantitative reverse-transcription polymerase chain reaction for Cxcl1, Il33, Il6, and Saa3 expression in fibroblasts (B6318) treated with iKras cell CM either from KrasG12D ON, KrasG12D OFF, or DMEM. Fibroblasts cultured with KrasG12D ON CM medium were treated with a JAK/STAT2/3 inhibitor (ruxolitinib) or vehicle. Data are shown as means ± SD. The statistical differences were determined by multiple-comparison analysis of variance. mRNA, messenger RNA.
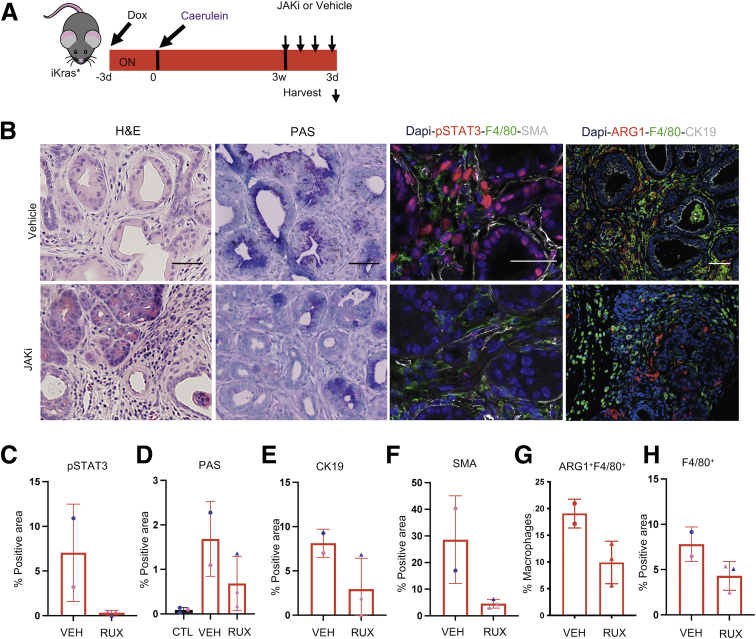
We then studied the effect of JAK/STAT inhibition in vivo. We administered DOX to iKras mice and induced acute pancreatitis; 3 weeks later, the mice were randomized to ruxolitinib (JAKi, 180 mg/kg) or vehicle daily for 3 days (Figure 11A). As expected, p-STAT3 was reduced upon JAKi treatment (Figure 11B and C). Intriguingly, histology analysis showed that the pancreata of mice treated with JAKi had fewer PanIN lesions and more acini areas when compared with vehicle-treated mice (Figure 11B, D, and E). We also observed a reduction in SMA+ fibroblasts (Figure 11F). Although the number of F4/80+ macrophages did not change in JAKi-treated mice, the number of ARG1+ macrophages tended toward decreasing (Figure 11G and H). Thus, inhibition of JAK/STAT in early PanINs partially reversed tumorigenesis and drove stromal remodeling.
Figure 11.
Inhibition of the JAK/STAT pathway mimics KrasG12Dinactivation-induced tissue repair. (A) Experimental design. iKras mice were given DOX and after 3 days acute pancreatitis was induced. After pancreatitis, KrasG12D was left ON for 3 weeks and at 3 weeks mice either received ruxolitinib (180 mg/kg) daily for 3 days or were treated with vehicle. N = 2–3 mice per group. (B) H&E, periodic acid–Schiff (PAS), and immunofluorescent staining for p-STAT3 (red), F4/80 (green), and SMA (white), or ARG1 (red), F4/80 (green), and CK19 (white). Scale bar: 50 μm. N = 2–3 mice per group. (C) Quantification for the p-STAT3 staining. (D) Quantification for the PAS staining. (E) Quantification of CK19-positive area. (F) Quantification of SMA-positive area. (G) Quantification of ARG1+-positive area within the F4/80+ macrophages. (H) Quantification of F4/80+-positive area. All data were analyzed using a t test. Blue, male; pink, females. Rux, ruxolitinib; Veh, vehicle.
Discussion
In summary, our results showed that oncogenic KrasG12D in epithelial cells drives the recruitment of macrophages and activates fibroblasts before ADM formation. Furthermore, continuous KrasG12D expression is required for fibroblast reprogramming, including expression of an inflammatory gene expression panel with cytokines known to be key mediators of macrophage polarization in pancreatic cancer. Thus, KrasG12D signaling drives formation and maintenance of an immune-suppressive stroma from early stages of carcinogenesis in an extrinsic manner.
Mutations in the KRAS gene are present in most human pancreatic cancers58 and have a high prevalence in PanINs.59 To date, the only clinically available KRAS inhibitor targets the KRASG12C mutant form,60,61 which is exceedingly rare in pancreatic cancer.8 Thus, understanding the downstream effects of oncogenic KRAS signaling is essential to design alternative targeting strategies. Furthermore, even when/if inhibitors of the more common KRASG12D and KRADG12V mutations become available, resistance to single-agent targeting is to be expected, and understanding which agents might synergize with KRAS inhibition is essential. Oncogenic Kras expression in pancreatic cancer has been associated with immune evasion62; in lung cancer, KRAS and Myelocytomatosis oncogene (MYC) cooperate to shape the immune microenvironment.63
Pancreatic cancer is characterized by an extensive tumor microenvironment (TME) rich in fibroblasts and suppressive immune cells. PanIN formation similarly is accompanied by accumulation of a fibroinflammatory stroma, which we refer to as precursor lesion microenvironment (PME).28 The mechanisms underlying the formation of the PME are poorly understood. In the current study, we set out to understand how the interplay between epithelial cells expressing oncogenic Kras and the surrounding components of the pancreatic microenvironment regulate the balance between tissue repair and carcinogenesis. We exploited the iKras genetically engineered mouse model25 in which oncogenic Kras can be activated and inactivated at will, to understand how inactivation of oncogenic Kras affects the precursor lesion microenvironment. Our results show that immune infiltration is not reduced upon KrasG12D inactivation, but polarization of macrophages and other myeloid cells is, with epithelial Kras inducing genes expressed in TAMs and inhibiting expression of repair promoting factors, consistent with previous findings by our group.32 When we investigated the mechanism of this polarity change, we discovered that cytokines known to regulate macrophage polarization are expressed prevalently by fibroblasts and regulated extrinsically by oncogenic Kras. Thus, our work supports the notion to target fibroblasts to reverse their reprogramming and block the activation of inflammatory pathways in the pancreas. Mechanistically, we show that a combination of epithelial factors drives fibroblast reprogramming, and that reprogramming can be inhibited by blocking the JAK/STAT3 signaling pathway.
Our work is complementary with studies in later stages of carcinogenesis, in which oncogenic KRAS blocks antitumor immune responses.62 Furthermore, changes in the microenvironment can mediate escape from oncogenic Kras dependence.64 Here, we identify fibroblasts as a key mediator of the extrinsic role of oncogenic KRAS and identify JAK/STAT3 signaling as required for fibroblast reprogramming and a potential target to reverse this process.
Based on our data, fibroblasts are a source of multiple cytokines that are dependent on epithelial KrasG12D expression: some are expressed by other cell types (Il33 and Il6), while others are uniquely fibroblast-specific (Cxcl1, Saa1, and Saa3). Recently, IL33 has been described as expressed by epithelial cells during the early stages of carcinogenesis and noted to promote neoplastic progression24; interestingly, we found that IL33 also is highly expressed by fibroblasts in early tumorigenesis stages, while its receptor is expressed by multiple immune cell populations. Fibroblasts are also a source of IL6, with a known role in pancreatic cancer progression46,48,65; Saa3 (ortholog to human SAA1), an apolipoprotein required for neoplastic progression49 and CXCL1, has been studied in epithelial cells, and shown to be a key moderator of T-cell exclusion in malignant disease.50 In our study, fibroblasts emerged as mediators of epithelial/immune cell crosstalk during the onset of pancreatic carcinogenesis. We found that fibroblasts are reprogrammed when exposed to CM from oncogenic Kras-expressing epithelial cells, supporting direct communication between these cell types. Interestingly, although the majority of cytokines we assessed in fibroblasts required a heat-labile component of the conditioned media, Il6 did not, implying that a combination of KrasG12D-dependent signals are required for establishing a complete reprogramming of pancreatic fibroblasts, potentially including both peptide and metabolic secreted factors (Figure 6).
The role and origin of fibroblasts in pancreatic cancer remains poorly understood. Fibroblasts secrete a number of cytokines that influence the immune milieu (eg, CXCL12, which has been shown to reduce T-cell infiltration in pancreatic cancer), and as such they promote cancer growth.66 However, in other models, fibroblast depletion promotes carcinogenesis.67 These contradictory findings might be explained by the heterogeneity of fibroblast populations. In recent years, the nature and origin of fibroblasts in advanced disease have been addressed by several studies, and previous assumptions regarding these cell populations have been challenged. Cancer-associated fibroblasts (CAFs) have long been assumed to derive from a pancreatic stellate cell population in the pancreas (similar to hepatic stellate cells), but recent lineage tracing work by our group has shown that a substantial proportion of CAFs derive from perivascular fibroblasts present in the normal pancreas.68 Accordingly, a lineage tracing study following pancreatic stellate cells in pancreatic cancer showed that they only contribute to a small subset of CAFs.69 In vitro characterization and, more recently, scRNAseq studies have identified CAF subsets with specific transcriptional signatures and functional roles.70, 71, 72
In advanced disease, fibroblasts have been classified as myofibroblastic CAFs (myCAFs), expressing high levels of SMA and genes encoding for extracellular matrix components, and inflammatory CAFs (iCAFs), inflammatory fibroblasts secreting cytokines such as IL6.72,73 Formation of iCAFs is dependent on IL1 and JAK/STAT3 signaling.70 Although fibroblast heterogeneity has been described in precursor lesions,74 whether the biological function of different fibroblast subsets is similar in early and advanced disease is not known. Here, we show that fibroblasts activated during the earliest stages of carcinogenesis have increased SMA, but also are a source of inflammatory cytokines such as IL6; furthermore, fibroblast activation, including SMA expression, requires JAK/STAT3 signaling. This suggests that the reported iCAF/myCAF dichotomy in established tumors is not as distinct in premalignant lesions. Whether other populations, such as the antigen-presenting CAFs,71 are recapitulated in early lesions remains to be determined. Analysis of the myeloid cells showed other differences in the PME compared with the TME. In the earliest stages of Kras activation, macrophages have low/no expression of ARG1, while expression is increased in established PanINs and in pancreatic cancer. This observation recapitulates earlier functional studies on the role of macrophages in pancreatic carcinogenesis. In the PME, macrophages promote PanIN formation/progression independently from their ability to inhibit CD8+ T-cell responses, likely through direct signaling to epithelial cells.32 In contrast, in advanced disease, the main role of macrophages is to inhibit anti–tumor immunity.29,30,75 Our findings thus highlight profound differences between the PME and the TME, and raise the question as to whether fibroblasts are equally required to regulate production of inflammatory cytokines in the TME, or whether they become dispensable at later stages. It also is possible that different tumor types, such as basal and classic, have a different reliance on fibroblasts, an area of future exploration.
In this study, we close the loop on communication between epithelial cells, fibroblasts, and immune cells. Reprogramming of myeloid cells to alleviate the profound immune suppression of pancreatic cancer is a key concept in potential therapeutic approaches.76,77 Unfortunately, the initial results from clinical trial testing of a CD40 agonist administered to reprogram myeloid cells combined with immune checkpoint inhibition and chemotherapy did not benefit the majority of patients,78 pointing to the need for yet additional avenues by which to target the microenvironment. Our current work suggests that fibroblast reprogramming should be considered in high-risk patients to prevent malignant progression and also could be explored to prevent tumor relapse in surgical patients. Finally, our study provides another piece of the puzzle in our understanding of the events that lead to the onset and progression of pancreatic cancer, and as such contribute to our fundamental knowledge of this deadly disease.
Methods
Mice
Mice were housed in the specific pathogen-free animal facility at the Rogel Cancer Center at the University of Michigan and overseen by the Unit for Laboratory Animal Medicine. Ptf1a(p48)-Cre;Rosa26rtTa/rtTa mice were crossed to TRE-KrasG12D;R26rtTa/rtTa to generate the p48-Cre;TRE-KrasG12D;R26rtTa/rtTa (iKras∗), as described.25 Expression of KrasG12D was induced in adult mice (age, 8–14 wk) by replacing regular chow with doxycycline chow (1 g/kg; Bio-Serv, Flemington, NJ). Acute pancreatitis was induced by administering 8 hourly doses of caerulein (75 μg/kg; Sigma-Aldrich, St. Louis, MO) intraperitoneally for 2 consecutive days. For JAK/STAT pathway inhibition, the JAK1/2 inhibitor ruxolitinib (INCB18424; MedChemExpress, Monmouth Junction, NJ) was administered orally once a day for a total of 4 doses at 180 mg/kg in 100 μL of 10% dimethyl sulfoxide in corn oil. Control mice received vehicle in parallel. All animal studies were conducted in compliance with the guidelines of the Institutional Animal Care and Use Committee at the University of Michigan.
Histology and Immunohistochemistry
Pancreatic tissue samples from experimental and control mice were fixed in 10% neutral-buffered formalin (FisherBrand, Fisher Scientific, Pittsburgh, PA) overnight and then embedded in paraffin and sectioned into slides. H&E, Gomori trichome, periodic acid–Schiff, and immunofluorescence (IF) staining were performed as previously described.25 For immunohistochemistry, fresh cut paraffin sections were rehydrated using 2 series of xylene, 2 series of 100% ethanol, and then 2 series of 95% ethanol. Water was used to wash all residues from previous washes. Antigen retrieval was performed using Antigen Retrieval CITRA Plus (BioGenex, Fremont, CA) and microwaved for a total of 8 minutes. Upon cool down, sections were blocked using 1% bovine serum albumin (BSA) in phosphate-buffered saline for 30 minutes and then primary antibodies (Table 2) were used at their corresponding dilutions. Primary antibody incubation was performed at 4°C overnight. Biotinylated secondary antibodies were used in 1:300 dilution and applied to sections for 45 minutes at room temperature. After secondary antibody incubation, sections were incubated for 30 minutes with the ABC reagent from the Vectastain Elite ABC Kit (peroxidase), followed by 3,3′-diaminobenzidine tetra hydrochloride (Vector, Burlingame, CA). For IF, Alexa Fluor secondary antibodies (Invitrogen) were used, then slides were mounted with Prolong Diamond Antifade Mountant with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen, Waltham, MA). The TSA Plus Fluorescein system (PerkinElmer, Waltham, MA) was used in IF for mouse primary antibodies. An Olympus BX53F microscope, Olympus DP80 digital camera (Olympus, Tokyo, Japan), and CellSens Standard software (Olympus, Tokyo, Japan) were used for imaging. Quantification of positive cell number or area was performed using ImageJ (National Institutes of Health, Bethesda, MD) using 3–5 images/slide (magnification, 200× or 400×) taken from 2–4 samples per group. We also used the Leica STELLARIS 8 FALCON Confocal Microscopy System and the LAS X software to acquire and visualize images (Leica Microsystems, Wetzlar, Germany).
Table 2.
Immunohistochemistry, IF, and Western Blot Antibodies
| Antibody | Supplier | Catalog number | Immunohistochemistry dilution | IF dilution | WB dilution |
|---|---|---|---|---|---|
| CD45 | BD Pharmingen, San Diego, CA | 553076 | 1:200 | ||
| F4/80 | Cell Signaling, Danvers, MA | 70076S | 1:250 | 1:250 | |
| CD4 | Cell Signaling | 25229S | 1:00 | ||
| CD8 | Cell Signaling | 98941S | 1:400 | 1:100 | |
| Foxp3 | Cell Signaling | 12653S | 1:100 | ||
| Arginase 1 | Cell Signaling | 93668S | 1:250 | ||
| αSMA | Sigma Aldrich, Burlington, MA | A2547 | 1:1000 | ||
| CK19 (Troma III) | Developmental Studies Hybridoma Bank, Iowa City, IA | - | 1:50 | ||
| Perk1/2 | Cell Signaling | 4370L | 1:100 | ||
| Ki67 | Abcam, Cambridge, UK | ab15580 | 1:100 | ||
| CC3 | Cell Signaling | 9661L | |||
| E-cadherin | Cell Signaling | 14472S | 1:100 | ||
| CD3 | Abcam | ab5690 | 1:100 | ||
| p-Stat3 Y705 | Cell Signaling | 9145S | 1:500 | ||
| Total STAT3 | Cell Signaling | 9139S | 1:1000 | ||
| p-ERK (T202/Y204) | Cell Signaling | 470L | 1:1000 | ||
| Total ERK | Cell Signaling | 4695S | 1:1000 | ||
| Vinculin | Cell Signaling | 13901 | 1:2000 | ||
| -tubulin | Cell Signaling | 3873S | 1:2000 | ||
| PDGFRβ | Abcam | ab32570 | 1:100 |
WB, Western blot.
Multiplex Immunohistochemistry Staining and Analysis
Multiplex immunohistochemistry staining was performed on paraffin-embedded pancreatic tissue sections as follows. Slides were baked in a hybridization oven for 1 hour at 60ºC, cooled for 10 minutes at room temperature, then dipped sequentially (×3) into xylene for 10 minutes each for removal of paraffin. Slides then were rehydrated in alcohol with dilutions of 100%, 95%, and then 70% for 10 minutes each, followed by a wash in deionized water for 2 minutes. Slides then were placed in neutral buffered formalin for 30 minutes. The slides then were washed for 2 minutes in deionized water, and then microwaved at 100% power in Rodent Decloaker (Biocare Medical, Pacheco, CA) for 30 seconds, the power level was reduced to 20% and microwaving continued for an additional 10 minutes, followed by a resting step of 15 minutes at room temperature. Microwaving continued at 10% power for an additional 10 minutes. Before microwaving with Rodent Decloaker, plastic wrap was secured on top of the microwave-proof slide box with rubber bands, with a partial opening for steam escape to prevent loss of solution. After the last microwaving step, slides were left to cool until slides and solution achieved room temperature. The multiplex staining was performed for each primary color combination. Slides were placed in a deionized water wash for 2 minutes followed by a Tris buffered saline solution with Tween 20 (TBST) wash for 2 minutes. Slides were placed in a slide incubation chamber and Bloxall (Vector, Burlingame, CA) was applied for 10 minutes followed by an additional blocking step of 1% BSA (in TBST) for 20 minutes, primary antibody was applied after tapping the slide to remove the primary antibody and was left to incubate for 1 hour, slides were washed in TBST (×3) for 2 minutes each, secondary antibody was applied, followed by TBST wash (×3) for 2 minutes each, Opal color was applied for 10 minutes, and a TBST wash (×3) for 2 minutes each was performed. The slides then were microwaved with either AR6 or AR9 for 45 seconds at 100% followed by 15 minutes at 20%. The previous steps then were repeated for each of the following antibodies and Opal colors in exact listed order: (1) F4/80 at 1:600 (ab6640; Abcam, Cambridge, UK); (2) CD3 at 1:400 (A0452; Dako, Agilent, Santa Clara, CA) TSA 520; (3) CD8 at 1:400 (98941; Cell Signaling, Danvers, MA); (4) ARG1 at 1:100 (93668; Cell Signaling, Danvers, MA); and (5) CK19 at 1:400 (Troma III, Developmental Studies Hybridoma Bank, Iowa City, IA). After the last application of multiplex was completed, slides were washed as described previously and placed in AR6, and then microwaved. After cooling, the slides were washed in deionized water followed by TBST for 2 minutes each. Opal spectral DAPI solution was applied (3 drops diluted in 1 mL TBST for 10 minutes followed by a wash in TBST for 30 seconds). Coverslips were mounted with Prolong Diamond, and slides were left to lie flat overnight away from light. If the entire multiplex could not be completed without interruption, the slides were left in AR6 or AR9 after a microwaving step, and covered from light until the next day. All primary antibodies were diluted in 1% BSA and all TSA Opal colors were diluted in TSA diluent at 1:50.
The Mantra Quantitative Pathology Work Station was used to image sections of each of the slides. One to 3 images per slide was captured at a magnification of ×20. Cube filters (DAPI, CY3, CY5, CY7, Texas Red, Qdot, Thermo Fisher Scientific, Waltham, MA) were used in taking each image capture. All images were analyzed using inForm Cell Analysis software (Akoya Biosciences, Marlborough, MA). Sixty-three images encompassing the 26 different control and experimental slides were batch analyzed after using a mouse formulated library consisting of each single TSA fluorophore listed previously. Unmixing was performed and no spectral overlap was found between fluorophores. Using inForm version 2.3.0 training software, cell compartments were segmented into the nucleus, cytoplasm, and membrane. DAPI was used to identify the nucleus of the cells and to determine their shape and size. The cytoplasm was segmented using ARG1 (Opal 650), F4/80 (Opal 540), and CD3 (Opal 520). The inner distance to the nucleus was set at 0 pixels and the outer distance of the nucleus was set at 6 pixels. The membrane was segmented using CK19 (Opal 690) with the maximum cell size set at 25 pixels from the nucleus to the membrane, and each pixel is 0.496 μm. X and Y coordinates were assigned to each identified cell in each image. Cell phenotypes (T cell, macrophage, epithelial cell, acinar cell, other cell) were selected manually at random throughout the 63 images. T cell, macrophage, and epithelial cells were selected based on single staining of CD3 (Opal 520), F4/80 (Opal 540), and CK19 (Opal 690), respectively. Other cells were selected manually based on the lack of staining of the earlier-listed fluorophores, and acinar cells were labeled based on the lack of listed fluorophores and tissue morphology. A fluorescent intensity of ARG1 (Opal 650) in the nucleus at a maximum threshold of 200 and a positivity threshold level of 13 was used to identify ARG1+ cells. R programs were used to make complex phenotypes by combining the primary cell phenotypes (T cell, macrophage, epithelial cell, acinar cell, and other cell) and the positivity thresholds for CD8+ and ARG1+ (Table 3).
Table 3.
Multiplex Immunohistochemistry Antibodies and Cell Phenotyping
| Complex phenotype | Opal | Primary phenotype | Antibody scoring | Cell segment | Mean signal intensity |
|---|---|---|---|---|---|
| Macrophage | F4/80-Opal 540 | Macrophage | None | N/A | N/A |
| Arg1+ macrophage | Arg1-Opal 650 | Macrophage | Arg1 | Nucleus | 13 and above |
| Arg1- macrophage | – | Macrophage | Arg1 | Nucleus | <13 |
| Epithelial cell | CK19-Opal 690 | Epithelial | None | N/A | N/A |
| Acinar cell | – | Acinar cell | None | N/A | N/A |
N/A, not applicable.
Flow Cytometry
Pancreata were harvested and dissociated to single cells by mincing the tissue finely using scissors followed by collagenase IV (1 mg/mL; Sigma) digestion for 30 minutes at 37°C while shaking. A 40-um mesh strainer was used to separate single cells. Red blood cell lysis buffer (eBioscience, San Diego, CA) was used to lyse all the red blood cells. Live cells were stained for surface markers using the antibodies listed in Table 4. The cells were either fixed after the primary antibody staining and used for analysis or the cells were fixed and permeabilized before intracellular staining using the antibodies listed in Table 4. Flow-cytometric analysis was performed on the Cyan ADP analyzer (Beckman Coulter, Brea, CA) and the ZE5 analyzer (Bio-Rad, Hercules, CA). Data were analyzed using FlowJo v10 software (Flowjo, LCC; Ashland, OR).
Table 4.
Flow Cytometry Antibodies
| Antibodies | Supplier | Catalog number | Clone | Dilution |
|---|---|---|---|---|
| CD45 | Invitrogen, Waltham, MA | MCD4530 | 30-F11 | 1:100 |
| CD45 | BD Horizon, Franklin Lakes, NJ | 563891 | 30-F11 | 1:100 |
| CD11b | BD Pharmingen | 557657 | M1/70 | 1:100 |
| F4/80 | Invitrogen | 15-4801-82 | BM8 | 1:100 |
| Ly6G | BD Pharmingen | 551460 | 1A8 | 1:100 |
| Ly6C | BD Pharmingen | 560592 | AL-21 | 1:100 |
| Arg1 | R&D Systems, Minneapolis, MN | IC5868F | Polyclonal | 1:50 |
| iNOS | Invitrogen | 12-5920-82 | CXNFT | 1:100 |
| CD206 | BD Pharmingen | 565250 | MR5D3 | 1:100 |
CyTOF
Pancreas was harvested and disrupted to single cells as described earlier. Three mesh strainers were used: 500 um, 100 um, and 40 um to separate single cells. Cells were washed twice in phosphate-buffered saline and incubated with Cell-ID cisplatin (1.67 umol/L) for 5 minutes at room temperature as a viability marker. Surface and intracellular staining was performed as detailed in the manufacturer’s instructions (Fluidigm) with the antibodies listed in Table 1. Cells were shipped in intercalator buffer on ice overnight to the Flow Cytometry Core at the University of Rochester Medical Center at Rochester, NY, where sample preparation was finalized, and CyTOF2 Mass Cytometer (Helios) analysis was performed. Data analysis was performed using the Premium CytoBank Software (cytobank.org) and R studio using the CyTOF workflow from Nowicka et al.79
scRNAseq
Pancreatic tissues were mechanically minced and enzymatically digested with collagenase IV (1 mg/mL in Roswell Park Memorial Institute (RPMI) 1640). Cell suspensions subsequently were filtered through 500-μm, 100-μm, and 40-μm mesh to obtain single cells. Dead cells were removed using the MACS Dead Cell Removal Kit (Miltenyi Biotec, Bergish Gladbas, DE)). The resulting single-cell suspensions were pooled by experimental group (Kras ON/Kras OFF). Single-cell complementary DNA libraries were prepared and sequenced at the University of Michigan Advanced Genomics Core using the 10× Genomics Platform (raw and processed data are available at GSM4175981 and GSE179846). Samples were run using 50-cycle, paired-end reads on the HiSeq 4000 (Illumina, San Diego, CA) to a depth of 100,000 reads. The raw data were processed and aligned by the University of Michigan DNA Sequencing Core. Cell Ranger count version 3.0.0 (10× Genomics, Pleasanton, CA) was used with default settings, with an initial expected cell count of 10,000. R version 3.6.2, RStudio version 1.2.5033, and R package Seurat version 3.2.2 were used for scRNAseq data analysis (RStudio Team RStudio: Integrated Development for R, RStudio, 2015; http://www.rstudio.com; R Core Development Team R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing, 2017; https://www.R-project.org80,81). Data initially were filtered to include only cells with at least 100 genes and genes that appeared in more than 3 cells. Data were normalized using the NormalizeData function with a scale factor of 10,000 and the LogNormalize normalization method. Data then were filtered manually to include only cells with 1000–60,000 transcripts and <15% mitochondrial genes. Variable genes were identified using the FindVariableFeatures function. Data were scaled and centered using linear regression of transcript counts. PCA was run with the RunPCA function using the previously defined variable genes. Cell clusters were identified via the FindNeighbors and FindClusters functions, using dimensions corresponding to approximately 90% variance as defined by PCA. UMAP clustering algorithms were performed with RunUMAP. Clusters were defined by user-defined criteria. The complete R script including figure-specific visualization methods is publicly available on GitHub (https://github.com/PascaDiMagliano-Lab).
For interactome analysis, ligand-receptor pairs were defined based on a curated literature-supported list in Ramilowski et al,44 further curated as described in Steele et al.45 The average expression values of ligands and receptors in each cell population for both experimental groups (Kras ON/Kras OFF) were calculated individually, and ligands and receptors expressed below a user-defined threshold (median average expression) were removed from analysis. Ligand-receptor pairs also were excluded if the ligands and receptors were not expressed in both experimental groups. Differences in the ligands and receptors between groups were determined using the Wilcoxon ranked test, and P values were adjusted for multiple comparisons using the Bonferroni correction method. Ligands and receptors were considered significantly different if P < .05. The resulting data table was visualized using Cytoscape (version 3.7.2) software (https://cytoscape.org/).82 The complete R script is publicly available on GitHub (https://github.com/PascaDiMagliano-Lab). The Circos plot was built with Circos software (version 0.69-9) http://circos.ca/ and the heatmap values within the circus plot show the average expression of each fibroblast ligand and their corresponding receptor across the iKras pancreatic tissue.
Cell Culture
All cell lines were cultured in DMEM with 10% fetal bovine serum and 1% penicillin streptomycin. The tumor cell line (9805) was derived from an iKras G12DP53R172H (ptf1a-Cre; TetO-KrasG12D;Rosa26rtTa/+; p53R172H/+) pancreatic cancer.51 The fibroblast cell lines, CD1WT and B6318, were derived from wild-type pancreata of a mixed or C57BL/6J background, respectively. The 9805 cell line was maintained in DOX-containing (1 ug/mL) medium. To generate conditioned media, fresh medium containing DOX (KrasG12D ON) or without DOX (KrasG12D OFF) was replaced and harvested after 2–3 days based on cell confluency. The media then were centrifuged (300 × g, 10 minutes at 4ºC) to remove contaminating cancer cells. The CM was used to culture fibroblasts with a 1:1 ratio of CM to normal DMEM for 2–3 days. An aliquot of the CM before and after culturing fibroblasts was collected for metabolomics analysis (described later). To test the effect of heat labile factors, media were boiled for 10 minutes to denature secondary and tertiary peptide structures. For inhibitor experiments, ruxolitinib (INCB018424; Selleckchem, Houston, TX) at 0.1 μmol/L, 0.5 μmol/L, and 5 μmol/L, or trametinib (GSK1120212; Selleckchem) at 0.1 μmol/L, 0.5 μmol/L, and 2 μmol/L or vehicle control were added to the medium.
Quantitative Reverse-Transcription Polymerase Chain Reaction
RNA was extracted using the QIAGEN kit (Hilden, Germany). RNA samples went through reverse-transcription polymerase chain reaction using the complementary DNA kit (Applied Biosystems, Waltham, MA). Complementary DNA samples for quantitative real-time polymerase chain reaction were prepared using a mix of 1× Fast-SYBR Green PCR Master Mix (Applied Biosystems) and the primers listed in Table 5. The reaction conditions were used as previously described (Collins et al. 201225). Cyclophilin A (Ppia) was used as housekeeping control.
Table 5.
Primers for Quantitative Reverse-Transcription Polymerase Chain Reaction
| Genes | Forward primer | Reverse primer |
|---|---|---|
| Cxcl1 | 5’-CTGGGATTCACCTCAAGAACATC-3’ | 5’-CAGGGTCAAGGCAAGCCTC-3’ |
| Il6 | 5’-TTCCATCCAGTTGCCTTCTTGG-3’ | 5’-TTCTCATTTCCACGATTTCCCAG-3’ |
| Il33 | 5’-TGAGACTCCGTTCTGGCCTC-3’ | 5’-CTCTTCATGCTTGGTACCCGA T-3’ |
| Saa3 | 5’-TGCCATCATTCTTTGCATCTTGA-3’ | 5’-CCGTGAACTTCTGAACACCCT-3’ |
Western Blot
Cells were lysed using RIPA buffer (Sigma-Aldrich) with protease and phosphatase inhibitors (Sigma-Aldrich). Protein levels were quantified and the same amount of protein was loaded to the wells in a 4%–15% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (Bio-Rad). Protein was transferred to a Polyvinylidene fluoride or polyvinylidene difluoride (PVDF) membrane (Bio-Rad) that was blocked with milk and then incubated with primary antibodies listed in Table 2 overnight. Horseradish-peroxidase–conjugated secondary anti-rabbit and anti-mouse (1:5000) were used and detected by using the enhance chemiluminescent substrate (PerkinElmer). The bands were visualized using the ChemiDoc Imaging System (Bio-Rad).
Metabolomics Analysis
The conditioned medium from tumor cells and fibroblasts (described earlier) was aspirated and 1 mL of 80% methanol—cooled in dry ice—was added per well. The plates were incubated on dry ice for 10 minutes, then scraped and transferred to sample tubes. The samples were vortexed and centrifuged for 10 minutes at 13,000 × g at 4°C. Then, 1 mL of the supernatant was aspirated from each tube, transferred to a tightly capped sample tube, and stored at -80°C until analysis.
Samples were run on an Agilent 1290 Infinity II LC-6470 triple quadrupole (QqQ) tandem mass spectrometer (MS/MS) system with the following components: Agilent Technologies Triple Quad 6470 LC-MS/MS system with a 1290 Infinity II LC Flexible Pump (Quaternary Pump), multisampler, and multicolumn thermostat with 6 port valves. Agilent Masshunter Workstation Software Liquid Chromatography Mas Spectometry (LC/MS) Data Acquisition for 6400 Series Triple Quadrupole MS with version B.08.02 was used for compound optimization, calibration, and data acquisition.
The following solvents were used for LC analysis. Solvent A (97% water, 3% methanol, 15 mmol/L acetic acid, and 10 mmol/L tributylamine, pH 5), solvent B (15 mmol/L acetic acid and 10 mmol/L tributylamine in methanol), and washing solvent C (acetonitrile). LC system seal washing solvent (90% water and 10% isopropanol) and needle wash solvent (75% methanol and 25% water) were used. Solvents were purchased from the following vendors: GC grade tributylamine 99% (ACROS ORGANICS, Geel, Belgium), LC/MS grade acetic acid Optima (Fisher Chemical, Waltham, MA), InfinityLab (Dallas, TX) deactivator additive, Electrospray ionization (ESI)–L low concentration tuning mix (Agilent Technologies), LC-MS grade water, acetonitrile, methanol (Millipore, Burlington, MA), and isopropanol (Fisher Chemical).
For LC analysis, 2 μL sample was injected into an Agilent ZORBAX RRHD Extend-C18 column (2.1 × 150 mm, 1.8 μm) with ZORBAX Extend Fast Guards. The LC gradient profile was as follows: flow rate, 0.25 mL/min, 0–2.5 minutes, 100% A; 2.5–7.5 minutes, 80% A and 20% B; 7.5–13 minutes 55% A and 45% B; 13–24 minutes, 1% A and 99% B; 24–27 minutes, 1% A and 99% C; and 27–27.5 minutes, 1% A and 99% C. Then, at 0.8 mL/min, 27.5–31.5 minutes, 1% A and 99% C; at 0.6 mL/min, 31.5–32.25 minutes, 1% A and 99% C; at 0.4 mL/min, 32.25–39.9 minutes, 100% A; and at 0.25 mL/min, 40 minutes, 100% A. Column temperature was maintained at 35ºC while the samples were at 4ºC.
For MS analysis, a 6470 Triple Quad MS calibrated with the Agilent ESI-L low concentration tuning mix was used. Source parameters: gas temperature, 150ºC; gas flow, 10 L/min; nebulizer, 45 psi; sheath gas temperature, 325ºC; sheath gas flow, 12 L/min; capillary, -2000 V; and delta electron multiplier voltage (EMV), -200 V. Dynamic MRM scan type was used with 0.07-minute peak width and 24-minute acquisition time. Delta retention time of ±1 minute, fragmentor of 40 eV, and cell accelerator of 5 eV were incorporated in the method.
The MassHunter (Agilent) Metabolomics Dynamic multiple reaction monitoring (MRM) Database and Method was used for target identification. Key parameters of AJS ESI were as follows: gas temperature, 150ºC; gas flow, 13 L/min; nebulizer, 45 psi; sheath gas temperature, 325ºC; sheath gas flow, 12 L/min; capillary, 2000 V; and nozzle, 500 V. Detector delta EMV, (-) 200. The QqQ data were preprocessed with the Agilent MassHunter Workstation QqQ Quantitative Analysis Software (B0700). The abundance level of each metabolite in every sample was divided by the median of all abundance levels across all samples for proper comparisons, statistical analyses, and visualization. The statistical significance test was performed by a 2-tailed t test with a significance threshold level of 0.05.
Heatmaps were generated with Morpheus (https://software.broadinstitute.org/morpheus).
Statistics
We used GraphPad Prism (San Diego, CA) version 8 software for most of our analysis. The normality was checked in all data sets and either a t test or the Mann–Whitney test was performed for statistical analysis, with statistical significance at P < .05. Quantitative reverse-transcription polymerase chain reaction data were analyzed using multiple comparison analysis of variance and considered statistically significant when P < .05.
Acknowledgment
The authors thank Daniel Long for histologic services.
CRediT Authorship Contributions
Ashley Velez-Delgado (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Funding acquisition: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Katelyn L. Donahue (Data curation: Lead; Formal analysis: Lead; Software: Lead; Visualization: Lead; Writing – review & editing: Supporting)
Kristee L. Brown (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Validation: Supporting)
Wenting Du (Formal analysis: Equal; Investigation: Equal; Validation: Equal; Writing – review & editing: Equal)
Valerie Irizarry-Negron (Formal analysis: Equal; Investigation: Equal; Validation: Equal)
Rosa E. Menjivar (Formal analysis: Supporting; Investigation: Supporting; Writing – review & editing: Equal)
Emily L. Lasse Opsahl (Investigation: Supporting; Validation: Supporting)
Nina Steele (Formal analysis: Supporting; Writing – review & editing: Equal)
Stephanie The (Software: Lead)
Jenny Lazarus (Data curation: Lead; Methodology: Lead; Software: Lead; Visualization: Lead; Writing – review & editing: Supporting)
Veerin R. Sirihorachai (Software: Lead; Visualization: Supporting)
Wei Yan (Investigation: Supporting; Validation: Supporting)
Samantha B. Kemp (Investigation: Supporting)
Samuel A. Kerk (Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Visualization: Equal)
Murali Bollampally (Investigation: Supporting)
Sion Yang (Investigation: Supporting)
Michael K. Scales (Formal analysis: Supporting)
Faith R. Avritt (Formal analysis: Supporting)
Fatima Lima (Formal analysis: Supporting; Investigation: Supporting)
Costas A. Lyssiotis (Resources: Supporting; Supervision: Supporting)
Arvind Rao (Conceptualization: Supporting; Resources: Supporting; Software: Supporting; Supervision: Supporting)
Howard C. Crawford (Resources: Supporting; Writing – review & editing: Supporting)
Filip Bednar (Conceptualization: Supporting; Software: Supporting; Visualization: Supporting; Writing – review & editing: Supporting)
Timothy L. Frankel (Conceptualization: Supporting; Resources: Supporting; Supervision: Supporting; Writing – review & editing: Supporting)
Benjamin L. Allen (Resources: Equal; Supervision: Supporting)
Yaqing Zhang (Conceptualization: Lead; Supervision: Lead; Visualization: Supporting; Writing – review & editing: Supporting)
Marina Pasca di Magliano (Conceptualization: Lead; Funding acquisition: Lead; Methodology: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – review & editing: Lead)
Footnotes
Current affiliation of Stephanie The: Cancer Data Science Shared Resource, University of Michigan, Ann Arbor, Michigan; current affiliation of Jenny Lazarus: Lazarus Medical Corporation, Stonyford, California; and current affiliation of Howard C. Crawford: Henry Ford Pancreatic Cancer Center, Henry Ford Health System, Detroit, Michigan.
Conflicts of interest This author discloses the following: Costas A. Lyssiotis has received consulting fees from Astellas Pharmaceuticals and is an inventor on patents pertaining to Kirsten Rat Sarcoma virus (KRAS) regulated metabolic pathways, redox control pathways in pancreatic cancer, and targeting the Glutamic-Oxaloacetic Transaminase 1 (GOT1) pathway as a therapeutic approach. The remaining authors disclose no conflicts.
Funding This project was supported by National Institutes of Health/National Cancer Institute grants R01CA151588 and R01CA198074, the University of Michigan Cancer Center Support Grant (NCI P30CA046592), and the American Cancer Society (M.P.M.); and the National Cancer Institute U01CA-224145 (M.P.M. and H.C.C.). Funded by NCI-R50CA232985 (Y.Z.). Supported by the Rackham Merit Fellowship, the Cellular Biotechnology Training Program (T32GM008353), and National Cancer Institute grant F31-CA247037 (A.V.D.). Funded by the Cancer Biology Training Program T32-CA009676 (K.L.D., N.G.S., and V.R.S.). Supported by American Cancer Society Postdoctoral Award PF-19-096-01 and the Michigan Institute for Clinical and Healthy Research Postdoctoral Translational Scholar Program fellowship award (N.G.S.). WD was supported by University of Michigan Training Program in Organogenesis (NIH T32-HD007505). Supported by the National Institutes of Health Cellular and Molecular Biology Training Grant T32-GM007315 and the Center for Organogenesis Training Program (National Institutes of Health T32 HD007505) (R.E.M.). Supported by institutional start-up funds from the University of Michigan, National Cancer Institute grants R37CA214955 and P30CA046592, and a Research Scholar Grant from the American Cancer Society (RSG-16-005-01) (S.T. and A.R.). Supported by National Institutes of Health grant T32-GM113900 and National Cancer Institute grant F31-CA247076 (S.B.K.). Supported by F31CA24745701 (S.A.K.). Supported by National Cancer Institute grants the National Cancer Institute R37CA237421 and R01CA248160 and University of Michigan Comprehensive Cancer Center (UMCCC) Core Grant P30CA046592 (C.A.L.). Funded by the Association of Academic Surgery Joel Roslyn Award (F.B.). Supported by the national institute of health grant K08CA201581 (T.F.). Metabolomics studies performed at the University of Michigan were supported by National Institutes of Health grant DK097153, the Charles Woodson Research Fund, and the University of Michigan (UM) Pediatric Brain Tumor Initiative. This project also was supported by the Tissue and Molecular Pathology and Flow Cytometry Shared Resources at the Rogel Cancer Center and the University of Michigan DNA Sequencing Core. CyTOF (Fluidigm, South San Francisco, CA) was performed at the University of Rochester Medical Center Flow Cytometry Shared Resource and at the Indiana University Simon Cancer Center Flow Cytometry Service. This work used a Leica STELLARIS 8 FALCON Confocal Microscopy System that was purchased with funds from a National Institutes of Health SIG grant S10OD28612-01-A1. The study sponsors had no role in the study design, collection, analysis, or interpretation of data.
Data transparency All the data and study material will be available. Codes can be found at: https://github.com/PascaDiMagliano-Lab/Extrinsic-KRAS-signaling-shapes-the-pancreatic-microenvironment-through-fibroblast-reprogramming
iKras∗ ON: GEO accession number: GSM4175981, iKras∗ OFF: GEO accession number: GSE179846.
Contributor Information
Yaqing Zhang, Email: yaqingzh@umich.edu.
Marina Pasca di Magliano, Email: marinapa@umich.edu.
References
- 1.American Cancer Society . American Cancer Society; Atlanta: 2020. Cancer facts & figures. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smit V.T., Boot A.J., Smits A.M., Fleuren G.J., Cornelisse C.J., Bos J.L. KRAS codon 12 mutations occur very frequently in pancreatic adenocarcinomas. Nucleic Acids Res. 1988;16:7773–7782. doi: 10.1093/nar/16.16.7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almoguera C., Shibata D., Forrester K., Martin J., Arnheim N., Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 4.Pour P.M., Sayed S., Sayed G. Hyperplastic, preneoplastic and neoplastic lesions found in 83 human pancreases. Am J Clin Pathol. 1982;77:137–152. doi: 10.1093/ajcp/77.2.137. [DOI] [PubMed] [Google Scholar]
- 5.Matsuda Y., Furukawa T., Yachida S., Nishimura M., Seki A., Nonaka K., Aida J., Takubo K., Ishiwata T., Kimura W., Arai T., Mino-Kenudson M. The prevalence and clinicopathological characteristics of high-grade pancreatic intraepithelial neoplasia: autopsy study evaluating the entire pancreatic parenchyma. Pancreas. 2017;46:658–664. doi: 10.1097/MPA.0000000000000786. [DOI] [PubMed] [Google Scholar]
- 6.Hingorani S.R., Petricoin E.F., Maitra A., Rajapakse V., King C., Jacobetz M.A., Ross S., Conrads T.P., Veenstra T.D., Hitt B.A., Kawaguchi Y., Johann D., Liotta L.A., Crawford H.C., Putt M.E., Jacks T., Wright C.V., Hruban R.H., Lowy A.M., Tuveson D.A. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 7.Aguirre A.J., Bardeesy N., Sinha M., Lopez L., Tuveson D.A., Horner J., Redston M.S., DePinho R.A. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ying H., Dey P., Yao W., Kimmelman A.C., Draetta G.F., Maitra A., DePinho R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2016;30:355–385. doi: 10.1101/gad.275776.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De La O.J., Emerson L.L., Goodman J.L., Froebe S.C., Illum B.E., Curtis A.B., Murtaugh L.C. Notch and Kras reprogram pancreatic acinar cells to ductal intraepithelial neoplasia. Proc Natl Acad Sci U S A. 2008;105:18907–18912. doi: 10.1073/pnas.0810111105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopp J.L., von Figura G., Mayes E., Liu F.F., Dubois C.L. Morris JPt, Pan FC, Akiyama H, Wright CV, Jensen K, Hebrok M, Sander M. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Habbe N., Shi G., Meguid R.A., Fendrich V., Esni F., Chen H., Feldmann G., Stoffers D.A., Konieczny S.F., Leach S.D., Maitra A. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad Sci U S A. 2008;105:18913–18918. doi: 10.1073/pnas.0810097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Figura G., Fukuda A., Roy N., Liku M.E., Morris Iv J.P., Kim G.E., Russ H.A., Firpo M.A., Mulvihill S.J., Dawson D.W., Ferrer J., Mueller W.F., Busch A., Hertel K.J., Hebrok M. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol. 2014;16:255–267. doi: 10.1038/ncb2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy N., Takeuchi K.K., Ruggeri J.M., Bailey P., Chang D., Li J., Leonhardt L., Puri S., Hoffman M.T., Gao S., Halbrook C.J., Song Y., Ljungman M., Malik S., Wright C.V., Dawson D.W., Biankin A.V., Hebrok M., Crawford H.C. PDX1 dynamically regulates pancreatic ductal adenocarcinoma initiation and maintenance. Genes Dev. 2016;30:2669–2683. doi: 10.1101/gad.291021.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morris J.P.t., Wang S.C., Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krah N.M., Murtaugh L.C. Differentiation and inflammation: ‘best enemies’ in gastrointestinal carcinogenesis. Trends Cancer. 2016;2:723–735. doi: 10.1016/j.trecan.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krah N.M., De La O.J., Swift G.H., Hoang C.Q., Willet S.G., Chen Pan F., Cash G.M., Bronner M.P., Wright C.V., MacDonald R.J., Murtaugh L.C. The acinar differentiation determinant PTF1A inhibits initiation of pancreatic ductal adenocarcinoma. Elife. 2015;4 doi: 10.7554/eLife.07125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krah N.M., Narayanan S.M., Yugawa D.E., Straley J.A., Wright C.V.E., MacDonald R.J., Murtaugh L.C. Prevention and reversion of pancreatic tumorigenesis through a differentiation-based mechanism. Dev Cell. 2019;50:744–754 e4. doi: 10.1016/j.devcel.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi G., DiRenzo D., Qu C., Barney D., Miley D., Konieczny S.F. Maintenance of acinar cell organization is critical to preventing Kras-induced acinar-ductal metaplasia. Oncogene. 2013;32:1950–1958. doi: 10.1038/onc.2012.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuveson D.A., Zhu L., Gopinathan A., Willis N.A., Kachatrian L., Grochow R., Pin C.L., Mitin N.Y., Taparowsky E.J., Gimotty P.A., Hruban R.H., Jacks T., Konieczny S.F. Mist1-KrasG12D knock-in mice develop mixed differentiation metastatic exocrine pancreatic carcinoma and hepatocellular carcinoma. Cancer Res. 2006;66:242–247. doi: 10.1158/0008-5472.CAN-05-2305. [DOI] [PubMed] [Google Scholar]
- 20.von Figura G., Morris JPt, Wright C.V., Hebrok M. Nr5a2 maintains acinar cell differentiation and constrains oncogenic Kras-mediated pancreatic neoplastic initiation. Gut. 2014;63:656–664. doi: 10.1136/gutjnl-2012-304287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cobo I., Martinelli P., Flandez M., Bakiri L., Zhang M., Carrillo-de-Santa-Pau E., Jia J., Sanchez-Arevalo Lobo V.J., Megias D., Felipe I., Del Pozo N., Millan I., Thommesen L., Bruland T., Olson S.H., Smith J., Schoonjans K., Bamlet W.R., Petersen G.M., Malats N., Amundadottir L.T., Wagner E.F., Real F.X. Transcriptional regulation by NR5A2 links differentiation and inflammation in the pancreas. Nature. 2018;554:533–537. doi: 10.1038/nature25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collins M.A., Yan W., Sebolt-Leopold J.S., Pasca di Magliano M. MAPK signaling is required for dedifferentiation of acinar cells and development of pancreatic intraepithelial neoplasia in mice. Gastroenterology. 2014;146:822–834 e7. doi: 10.1053/j.gastro.2013.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collisson E.A., Trejo C.L., Silva J.M., Gu S., Korkola J.E., Heiser L.M., Charles R.P., Rabinovich B.A., Hann B., Dankort D., Spellman P.T., Phillips W.A., Gray J.W., McMahon M. A central role for RAF-->MEK-->ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2012;2:685–693. doi: 10.1158/2159-8290.CD-11-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alonso-Curbelo D., Ho Y.J., Burdziak C., Maag J.L.V., Morris JPt, Chandwani R., Chen H.A., Tsanov K.M., Barriga F.M., Luan W., Tasdemir N., Livshits G., Azizi E., Chun J., Wilkinson J.E., Mazutis L., Leach S.D., Koche R., Pe'er D., Lowe S.W. A gene-environment-induced epigenetic program initiates tumorigenesis. Nature. 2021;590:642–648. doi: 10.1038/s41586-020-03147-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collins M.A., Bednar F., Zhang Y., Brisset J.C., Galban S., Galban C.J., Rakshit S., Flannagan K.S., Adsay N.V., Pasca di Magliano M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122:639–653. doi: 10.1172/JCI59227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ying H., Kimmelman A.C., Lyssiotis C.A., Hua S., Chu G.C., Fletcher-Sananikone E., Locasale J.W., Son J., Zhang H., Coloff J.L., Yan H., Wang W., Chen S., Viale A., Zheng H., Paik J.H., Lim C., Guimaraes A.R., Martin E.S., Chang J., Hezel A.F., Perry S.R., Hu J., Gan B., Xiao Y., Asara J.M., Weissleder R., Wang Y.A., Chin L., Cantley L.C., DePinho R.A. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helms E., Onate M.K., Sherman M.H. Fibroblast heterogeneity in the pancreatic tumor microenvironment. Cancer Discov. 2020;10:648–656. doi: 10.1158/2159-8290.CD-19-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark C.E., Hingorani S.R., Mick R., Combs C., Tuveson D.A., Vonderheide R.H. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 2007;67:9518–9527. doi: 10.1158/0008-5472.CAN-07-0175. [DOI] [PubMed] [Google Scholar]
- 29.Mitchem J.B., Brennan D.J., Knolhoff B.L., Belt B.A., Zhu Y., Sanford D.E., Belaygorod L., Carpenter D., Collins L., Piwnica-Worms D., Hewitt S., Udupi G.M., Gallagher W.M., Wegner C., West B.L., Wang-Gillam A., Goedegebuure P., Linehan D.C., DeNardo D.G. Targeting tumor-infiltrating macrophages decreases tumor-initiating cells, relieves immunosuppression, and improves chemotherapeutic responses. Cancer Res. 2013;73:1128–1141. doi: 10.1158/0008-5472.CAN-12-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Velez-Delgado A., Mathew E., Li D., Mendez F.M., Flannagan K., Rhim A.D., Simeone D.M., Beatty G.L., Pasca di Magliano M. Myeloid cells are required for PD-1/PD-L1 checkpoint activation and the establishment of an immunosuppressive environment in pancreatic cancer. Gut. 2017;66:124–136. doi: 10.1136/gutjnl-2016-312078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liou G.Y., Doppler H., Necela B., Krishna M., Crawford H.C., Raimondo M., Storz P. Macrophage-secreted cytokines drive pancreatic acinar-to-ductal metaplasia through NF-kappaB and MMPs. J Cell Biol. 2013;202:563–577. doi: 10.1083/jcb.201301001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Yan W., Mathew E., Kane K.T., Brannon A., 3rd, Adoumie M., Vinta A., Crawford H.C., Pasca di Magliano M. Epithelial-myeloid cell crosstalk regulates acinar cell plasticity and pancreatic remodeling in mice. Elife. 2017;6 doi: 10.7554/eLife.27388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Criscimanna A., Coudriet G.M., Gittes G.K., Piganelli J.D., Esni F. Activated macrophages create lineage-specific microenvironments for pancreatic acinar- and beta-cell regeneration in mice. Gastroenterology. 2014;147:1106–1118 e11. doi: 10.1053/j.gastro.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 34.Carriere C., Young A.L., Gunn J.R., Longnecker D.S., Korc M. Acute pancreatitis markedly accelerates pancreatic cancer progression in mice expressing oncogenic Kras. Biochem Biophys Res Commun. 2009;382:561–565. doi: 10.1016/j.bbrc.2009.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guerra C., Collado M., Navas C., Schuhmacher A.J., Hernandez-Porras I., Canamero M., Rodriguez-Justo M., Serrano M., Barbacid M. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19:728–739. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wen H.J., Gao S., Wang Y., Ray M., Magnuson M.A., Wright C.V.E., Di Magliano M.P., Frankel T.L., Crawford H.C. Myeloid cell-derived HB-EGF drives tissue recovery after pancreatitis. Cell Mol Gastroenterol Hepatol. 2019;8:173–192. doi: 10.1016/j.jcmgh.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ardito C.M., Gruner B.M., Takeuchi K.K., Lubeseder-Martellato C., Teichmann N., Mazur P.K., Delgiorno K.E., Carpenter E.S., Halbrook C.J., Hall J.C., Pal D., Briel T., Herner A., Trajkovic-Arsic M., Sipos B., Liou G.Y., Storz P., Murray N.R., Threadgill D.W., Sibilia M., Washington M.K., Wilson C.L., Schmid R.M., Raines E.W., Crawford H.C., Siveke J.T. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer Cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Navas C., Hernandez-Porras I., Schuhmacher A.J., Sibilia M., Guerra C., Barbacid M. EGF receptor signaling is essential for k-ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:318–330. doi: 10.1016/j.ccr.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol. 2017;14:296–304. doi: 10.1038/nrgastro.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu H., Kortylewski M., Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 41.Kemp S.B., Carpenter E.S., Steele N.G., Donahue K.L., Nwosu Z.C., Pacheco A., Velez-Delgado A., Menjivar R.E., Lima F., The S., Espinoza C.E., Brown K., Long D., Lyssiotis C.A., Rao A., Zhang Y., Pasca di Magliano M., Crawford H.C. Apolipoprotein E promotes immune suppression in pancreatic cancer through NF-kB-mediated production of CXCL1. Cancer Res. 2021;81:4305–4318. doi: 10.1158/0008-5472.CAN-20-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Katzenelenbogen Y., Sheban F., Yalin A., Yofe I., Svetlichnyy D., Jaitin D.A., Bornstein C., Moshe A., Keren-Shaul H., Cohen M., Wang S.Y., Li B., David E., Salame T.M., Weiner A., Amit I. Coupled scRNA-seq and intracellular protein activity reveal an immunosuppressive role of TREM2 in cancer. Cell. 2020;182:872–885 e19. doi: 10.1016/j.cell.2020.06.032. [DOI] [PubMed] [Google Scholar]
- 43.Kemp S.B., Steele N.G., Carpenter E.S., Donahue K.L., Bushnell G.G., Morris A.H., The S., Orbach S.M., Sirihorachai V.R., Nwosu Z.C., Espinoza C., Lima F., Brown K., Girgis A.A., Gunchick V., Zhang Y., Lyssiotis C.A., Frankel T.L., Bednar F., Rao A., Sahai V., Shea L.D., Crawford H.C., Pasca di Magliano M. Pancreatic cancer is marked by complement-high blood monocytes and tumor-associated macrophages. Life Sci Alliance. 2021;4 doi: 10.26508/lsa.202000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramilowski J.A., Goldberg T., Harshbarger J., Kloppmann E., Lizio M., Satagopam V.P., Itoh M., Kawaji H., Carninci P., Rost B., Forrest A.R. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Steele N.G., Carpenter E.S., Kemp S.B., Sirihorachai V.R., The S., Delrosario L., Lazarus J., E-aD Amir, Gunchick V., Espinoza C., Bell S., Harris L., Lima F., Irizarry-Negron V., Paglia D., Macchia J., Chu A.K.Y., Schofield H., Wamsteker E.-J., Kwon R., Schulman A., Prabhu A., Law R., Sondhi A., Yu J., Patel A., Donahue K., Nathan H., Cho C., Anderson M.A., Sahai V., Lyssiotis C.A., Zou W., Allen B.L., Rao A., Crawford H.C., Bednar F., Frankel T.L., Pasca di Magliano M. Multimodal mapping of the tumor and peripheral blood immune landscape in human pancreatic cancer. Nat Cancer. 2020;1:1097–1112. doi: 10.1038/s43018-020-00121-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lesina M., Kurkowski M.U., Ludes K., Rose-John S., Treiber M., Kloppel G., Yoshimura A., Reindl W., Sipos B., Akira S., Schmid R.M., Algul H. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–469. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 47.Mace T.A., Shakya R., Pitarresi J.R., Swanson B., McQuinn C.W., Loftus S., Nordquist E., Cruz-Monserrate Z., Yu L., Young G., Zhong X., Zimmers T.A., Ostrowski M.C., Ludwig T., Bloomston M., Bekaii-Saab T., Lesinski G.B. IL-6 and PD-L1 antibody blockade combination therapy reduces tumour progression in murine models of pancreatic cancer. Gut. 2018;67:320–332. doi: 10.1136/gutjnl-2016-311585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y., Yan W., Collins M.A., Bednar F., Rakshit S., Zetter B.R., Stanger B.Z., Chung I., Rhim A.D., di Magliano M.P. Interleukin-6 is required for pancreatic cancer progression by promoting MAPK signaling activation and oxidative stress resistance. Cancer Res. 2013;73:6359–6374. doi: 10.1158/0008-5472.CAN-13-1558-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Djurec M., Grana O., Lee A., Troule K., Espinet E., Cabras L., Navas C., Blasco M.T., Martin-Diaz L., Burdiel M., Li J., Liu Z., Vallespinos M., Sanchez-Bueno F., Sprick M.R., Trumpp A., Sainz B., Jr., Al-Shahrour F., Rabadan R., Guerra C., Barbacid M. Saa3 is a key mediator of the protumorigenic properties of cancer-associated fibroblasts in pancreatic tumors. Proc Natl Acad Sci U S A. 2018;115:E1147–E1156. doi: 10.1073/pnas.1717802115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li J., Byrne K.T., Yan F., Yamazoe T., Chen Z., Baslan T., Richman L.P., Lin J.H., Sun Y.H., Rech A.J., Balli D., Hay C.A., Sela Y., Merrell A.J., Liudahl S.M., Gordon N., Norgard R.J., Yuan S., Yu S., Chao T., Ye S., Eisinger-Mathason T.S.K., Faryabi R.B., Tobias J.W., Lowe S.W., Coussens L.M., Wherry E.J., Vonderheide R.H., Stanger B.Z. Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity. 2018;49:178–193 e7. doi: 10.1016/j.immuni.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins M.A., Brisset J.C., Zhang Y., Bednar F., Pierre J., Heist K.A., Galban C.J., Galban S., di Magliano M.P. Metastatic pancreatic cancer is dependent on oncogenic Kras in mice. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mathew E., Collins M.A., Fernandez-Barrena M.G., Holtz A.M., Yan W., Hogan J.O., Tata Z., Allen B.L., Fernandez-Zapico M.E., di Magliano M.P. The transcription factor GLI1 modulates the inflammatory response during pancreatic tissue remodeling. J Biol Chem. 2014;289:27727–27743. doi: 10.1074/jbc.M114.556563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colegio O.R., Chu N.Q., Szabo A.L., Chu T., Rhebergen A.M., Jairam V., Cyrus N., Brokowski C.E., Eisenbarth S.C., Phillips G.M., Cline G.W., Phillips A.J., Medzhitov R. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu J.Y., Huang T.W., Hsieh Y.T., Wang Y.F., Yen C.C., Lee G.L., Yeh C.C., Peng Y.J., Kuo Y.Y., Wen H.T., Lin H.C., Hsiao C.W., Wu K.K., Kung H.J., Hsu Y.J., Kuo C.C. Cancer-derived succinate promotes macrophage polarization and cancer metastasis via succinate receptor. Mol Cell. 2020;77:213–227 e5. doi: 10.1016/j.molcel.2019.10.023. [DOI] [PubMed] [Google Scholar]
- 55.Saxton R.A., Sabatini D.M. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 56.Hooftman A., O'Neill L.A.J. The immunomodulatory potential of the metabolite itaconate. Trends Immunol. 2019;40:687–698. doi: 10.1016/j.it.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 57.Becker L.M., O'Connell J.T., Vo A.P., Cain M.P., Tampe D., Bizarro L., Sugimoto H., McGow A.K., Asara J.M., Lovisa S., McAndrews K.M., Zielinski R., Lorenzi P.L., Zeisberg M., Raza S., LeBleu V.S., Kalluri R. Epigenetic reprogramming of cancer-associated fibroblasts deregulates glucose metabolism and facilitates progression of breast cancer. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Biankin A.V., Waddell N., Kassahn K.S., Gingras M.C., Muthuswamy L.B., Johns A.L., Miller D.K., Wilson P.J., Patch A.M., Wu J., Chang D.K., Cowley M.J., Gardiner B.B., Song S., Harliwong I., Idrisoglu S., Nourse C., Nourbakhsh E., Manning S., Wani S., Gongora M., Pajic M., Scarlett C.J., Gill A.J., Pinho A.V., Rooman I., Anderson M., Holmes O., Leonard C., Taylor D., Wood S., Xu Q., Nones K., Fink J.L., Christ A., Bruxner T., Cloonan N., Kolle G., Newell F., Pinese M., Mead R.S., Humphris J.L., Kaplan W., Jones M.D., Colvin E.K., Nagrial A.M., Humphrey E.S., Chou A., Chin V.T., Chantrill L.A., Mawson A., Samra J.S., Kench J.G., Lovell J.A., Daly R.J., Merrett N.D., Toon C., Epari K., Nguyen N.Q., Barbour A., Zeps N., Australian Pancreatic Cancer Genome I. Kakkar N., Zhao F., Wu Y.Q., Wang M., Muzny D.M., Fisher W.E., Brunicardi F.C., Hodges S.E., Reid J.G., Drummond J., Chang K., Han Y., Lewis L.R., Dinh H., Buhay C.J., Beck T., Timms L., Sam M., Begley K., Brown A., Pai D., Panchal A., Buchner N., De Borja R., Denroche R.E., Yung C.K., Serra S., Onetto N., Mukhopadhyay D., Tsao M.S., Shaw P.A., Petersen G.M., Gallinger S., Hruban R.H., Maitra A., Iacobuzio-Donahue C.A., Schulick R.D., Wolfgang C.L., Morgan R.A., Lawlor R.T., Capelli P., Corbo V., Scardoni M., Tortora G., Tempero M.A., Mann K.M., Jenkins N.A., Perez-Mancera P.A., Adams D.J., Largaespada D.A., Wessels L.F., Rust A.G., Stein L.D., Tuveson D.A., Copeland N.G., Musgrove E.A., Scarpa A., Eshleman J.R., Hudson T.J., Sutherland R.L., Wheeler D.A., Pearson J.V., McPherson J.D., Gibbs R.A., Grimmond S.M. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanda M., Matthaei H., Wu J., Hong S.M., Yu J., Borges M., Hruban R.H., Maitra A., Kinzler K., Vogelstein B., Goggins M. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142:730–733 e9. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Canon J., Rex K., Saiki A.Y., Mohr C., Cooke K., Bagal D., Gaida K., Holt T., Knutson C.G., Koppada N., Lanman B.A., Werner J., Rapaport A.S., San Miguel T., Ortiz R., Osgood T., Sun J.-R., Zhu X., McCarter J.D., Volak L.P., Houk B.E., Fakih M.G., O’Neil B.H., Price T.J., Falchook G.S., Desai J., Kuo J., Govindan R., Hong D.S., Ouyang W., Henary H., Arvedson T., Cee V.J., Lipford J.R. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 61.Wilhelm S., Carter C., Lynch M., Lowinger T., Dumas J., Smith R.A., Schwartz B., Simantov R., Kelley S. Discovery and development of sorafenib: a multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5:835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- 62.Ischenko I., D’Amico S., Rao M., Li J., Hayman M.J., Powers S., Petrenko O., Reich N.C. KRAS drives immune evasion in a genetic model of pancreatic cancer. Nat Commun. 2021;12:1482. doi: 10.1038/s41467-021-21736-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kortlever R.M., Sodir N.M., Wilson C.H., Burkhart D.L., Pellegrinet L. Brown Swigart L, Littlewood TD, Evan GI. Myc cooperates with Ras by programming inflammation and immune suppression. Cell. 2017;171:1301–1315 e14. doi: 10.1016/j.cell.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hou P., Kapoor A., Zhang Q., Li J., Wu C.J., Li J., Lan Z., Tang M., Ma X., Ackroyd J.J., Kalluri R., Zhang J., Jiang S., Spring D.J., Wang Y.A., DePinho R.A. Tumor microenvironment remodeling enables bypass of oncogenic KRAS dependency in pancreatic cancer. Cancer Discov. 2020;10:1058–1077. doi: 10.1158/2159-8290.CD-19-0597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fukuda A., Wang S.C., Morris J.P., Folias A.E., Liou A., Kim G.E., Akira S., Boucher K.M., Firpo M.A., Mulvihill S.J., Hebrok M. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feig C., Jones J.O., Kraman M., Wells R.J., Deonarine A., Chan D.S., Connell C.M., Roberts E.W., Zhao Q., Caballero O.L., Teichmann S.A., Janowitz T., Jodrell D.I., Tuveson D.A., Fearon D.T. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110:20212–22027. doi: 10.1073/pnas.1320318110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ozdemir B.C., Pentcheva-Hoang T., Carstens J.L., Zheng X., Wu C.C., Simpson T.R., Laklai H., Sugimoto H., Kahlert C., Novitskiy S.V., De Jesus-Acosta A., Sharma P., Heidari P., Mahmood U., Chin L., Moses H.L., Weaver V.M., Maitra A., Allison J.P., LeBleu V.S., Kalluri R. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2014;25:719–734. doi: 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garcia P.E., Adoumie M., Kim E.C., Zhang Y., Scales M.K., El-Tawil Y.S., Shaikh A.Z., Wen H.J., Bednar F., Allen B.L., Wellik D.M., Crawford H.C., Pasca di Magliano M. Differential contribution of pancreatic fibroblast subsets to the pancreatic cancer stroma. Cell Mol Gastroenterol Hepatol. 2020;10:581–599. doi: 10.1016/j.jcmgh.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Helms E.J., Berry M.W., Chaw R.C., DuFort C.C., Sun D., Onate M.K., Oon C., Bhattacharyya S., Sanford-Crane H., Horton W., Finan J.M., Sattler A., Makar R., Dawson D.W., Xia Z., Hingorani S.R., Sherman M.H. Mesenchymal lineage heterogeneity underlies non-redundant functions of pancreatic cancer-associated fibroblasts. bioRxiv. 2021 doi: 10.1158/2159-8290.CD-21-0601. 2021.05.01.442252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Biffi G., Oni T.E., Spielman B., Hao Y., Elyada E., Park Y., Preall J., Tuveson D.A. IL1-Induced JAK/STAT signaling is antagonized by TGFbeta to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 2019;9:282–301. doi: 10.1158/2159-8290.CD-18-0710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elyada E., Bolisetty M., Laise P., Flynn W.F., Courtois E.T., Burkhart R.A., Teinor J.A., Belleau P., Biffi G., Lucito M.S., Sivajothi S., Armstrong T.D., Engle D.D., Yu K.H., Hao Y., Wolfgang C.L., Park Y., Preall J., Jaffee E.M., Califano A., Robson P., Tuveson D.A. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9:1102–1123. doi: 10.1158/2159-8290.CD-19-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ohlund D., Handly-Santana A., Biffi G., Elyada E., Almeida A.S., Ponz-Sarvise M., Corbo V., Oni T.E., Hearn S.A., Lee E.J., Chio, Hwang C.I., Tiriac H., Baker L.A., Engle D.D., Feig C., Kultti A., Egeblad M., Fearon D.T., Crawford J.M., Clevers H., Park Y., Tuveson D.A. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214:579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Biffi G., Tuveson D.A. Diversity and biology of cancer-associated fibroblasts. Physiol Rev. 2021;101:147–176. doi: 10.1152/physrev.00048.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hosein A.N., Huang H., Wang Z., Parmar K., Du W., Huang J., Maitra A., Olson E., Verma U., Brekken R.A. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single-cell resolution. JCI Insight. 2019;5 doi: 10.1172/jci.insight.129212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Steele C.W., Karim S.A., Leach J.D., Bailey P., Upstill-Goddard R., Rishi L., Foth M., Bryson S., McDaid K., Wilson Z., Eberlein C., Candido J.B., Clarke M., Nixon C., Connelly J., Jamieson N., Carter C.R., Balkwill F., Chang D.K., Evans T.R., Strathdee D., Biankin A.V., Nibbs R.J., Barry S.T., Sansom O.J., Morton J.P. CXCR2 inhibition profoundly suppresses metastases and augments immunotherapy in pancreatic ductal adenocarcinoma. Cancer Cell. 2016;29:832–845. doi: 10.1016/j.ccell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhu Y., Knolhoff B.L., Meyer M.A., Nywening T.M., West B.L., Luo J., Wang-Gillam A., Goedegebuure S.P., Linehan D.C., DeNardo D.G. CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res. 2014;74:5057–5069. doi: 10.1158/0008-5472.CAN-13-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beatty G.L., Chiorean E.G., Fishman M.P., Saboury B., Teitelbaum U.R., Sun W., Huhn R.D., Song W., Li D., Sharp L.L., Torigian D.A., O'Dwyer P.J., Vonderheide R.H. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331:1612–1616. doi: 10.1126/science.1198443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O'Hara M.H., O'Reilly E.M., Wolff R.A., Wainberg Z.A., Ko A.H., Rahma O.E., Fisher G.A., Lyman J.P., Cabanski C.R., Karakunnel J.J., Gherardini P.F., Kitch L.J., Bucktrout S., Christopher E., Mick R., Chen R., Trifan O.C., Salvador L., O'Donnell-Tormey J., Vonderheide R.H. Gemcitabine (Gem) and nab-paclitaxel (NP) ± nivolumab (nivo) ± CD40 agonistic monoclonal antibody APX005M (sotigalimab), in patients (Pts) with untreated metastatic pancreatic adenocarcinoma (mPDAC): phase (Ph) 2 final results. J Clin Oncol. 2021;39(Suppl):4019. [Google Scholar]
- 79.Nowicka M., Krieg C., Crowell H.L., Weber L.M., Hartmann F.J., Guglietta S., Becher B., Levesque M.P., Robinson M.D. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Res. 2017;6:748. doi: 10.12688/f1000research.11622.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902 e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]