Abstract
The surface of Gram-positive and Gram-negative bacteria contains long hair-like proteinaceous protrusion known as pili or fimbriae. Historically, pilin proteins were considered to play a major role in the transfer of genetic material during bacterial conjugation. Recent findings however elucidate their importance in virulence, biofilm formation, phage transduction, and motility. Therefore, it is crucial to gain mechanistic insights on the subcellular assembly of pili and the localization patterns of their subunit proteins (major and minor pilins) that aid the macromolecular pilus assembly at the bacterial surface. In this article, we review the current knowledge of pilus assembly mechanisms in a wide range of Gram-positive and Gram-negative bacteria, including subcellular localization patterns of a few pilin subunit proteins and their role in virulence and pathogenesis.
Keywords: Pilin subunits, Pili, Pili assembly, Pili termination, Bacteria
Introduction
Pili are multi-subunit non-flagellar proteinaceous filaments found on the surface of the bacterial cell (Ramirez et al., 2020). Pili acts as an initiator of host-pathogen interaction (Zhou et al., 2021). In particular, pilin subunits proteins (both major and minor) play a significant role in aiding the bacterial attachment and colonization to the host cells and other conditioned surfaces found on implants such as catheters (Flores-Mireles et al., 2015, Govindarajan et al., 2020, Jacobsen et al., 2008, Nallapareddy et al., 2011, Stepanova, 2022). Interaction of pili with host cells takes place in a sequential manner which requires proper subcellular secretion of pilin subunits in the cytoplasmic membrane and translocation to the surface of the bacterial cell to complete the assembly of pilus fiber (Flores-Mireles et al., 2015, Kline et al., 2010, Proft and Baker, 2009).
Gram-positive bacteria express two types of pili, one being the Sortase Assembled (SA) pili (eg. Enterococcus faecalis (E. faecalis)) and the other being type IV pili (eg. Clostridium perfringens (C. perfringens)) that are similar to Gram-negative bacteria (Proft & Baker, 2009). In majority of Gram-positive bacteria, pilus assembly is mediated by membrane-anchored sortase enzyme. Depending upon the species there may be one or more classes of sortase enzymes that can perform housekeeping or polymerizing roles (Hendrickx et al., 2011). In contrast, Gram-negative bacteria (Eg. Escherichia coli (E. coli)) pilus assembly is mediated by Chaperone Usher (CU) pathway (Werneburg et al., 2018). The major elements of the CU system are pilin subunits, periplasmic chaperone, and an outer membrane Usher protein. The principal role of the CU pilus system is the adhesion to the host cell (Du et al., 2021). Both types of bacteria possess well-defined mechanisms for pilus assembly on their surface for effective interaction with the host.
In recent years, a new paradigm has emerged on various pilus assembly mechanisms of both Gram-positive and Gram-negative bacterial species. Therefore, a thorough and collective understanding of the pilus assembly mechanism in a plethora of bacterial species requires a critical review. In doing so, this article elaborates on various pilus assembly mechanisms of both Gram-negative and Gram-positive that was studied to date. In addition, this article critically reviews recent advances in the pilus assembly mechanism and the role of pilin subunits in adherence and biofilm formation. In this review, we aim to provide a clear understanding of how Gram-negative and Gram-positive bacteria assemble their pilin subunits on the cell surface. Furthermore, this article also briefly discusses the localization patterns of virulence proteins and antimicrobial compounds that target them.
Pili in Gram-positive bacteria:
Sortase assembled pili
Sortases are conserved in nearly all Gram-positive bacteria (Hendrickx et al., 2011, Paterson and Mitchell, 2004). Each Gram-positive species has one or more classes of sortase (SrtA to F), which have a specific role, for instance, sortase class A plays a housekeeping role in attaching the target proteins to the cell wall, the sortase class B performs iron acquisition by linking the iron acquisition channel proteins to cell wall, sortase class C catalysis and promotes the pilin polymerization, functions of sortase class D, E, and F was unknown and requires further investigation (Bradshaw et al., 2015, Hendrickx et al., 2011, Mandlik et al., 2008, Marraffini et al., 2006, Spirig et al., 2011, Susmitha et al., 2021). In E. faecalis, SrtA is a housekeeping sortase that initiates the pilus assembly by recognizing its secreted substrate protein that contains C-terminal Cell Wall Sorting Signal (CWSS) sequence (Fig. 1 & Table 1). This recognition helps SrtA to read the remaining components of the substrate’s cell wall sorting signal (CWSS) - a hydrophobic domain and a positively charged tail. SrtA then stalls the substrate protein at the Sec channel to complete the rest of the sorting process (Clancy et al., 2010). Once the substrate protein are recognized, SrtA then cleaves the threonine-glycine bond of the LPXTG motif that are present in CWSS (Hendrickx et al., 2009, Kline et al., 2010).
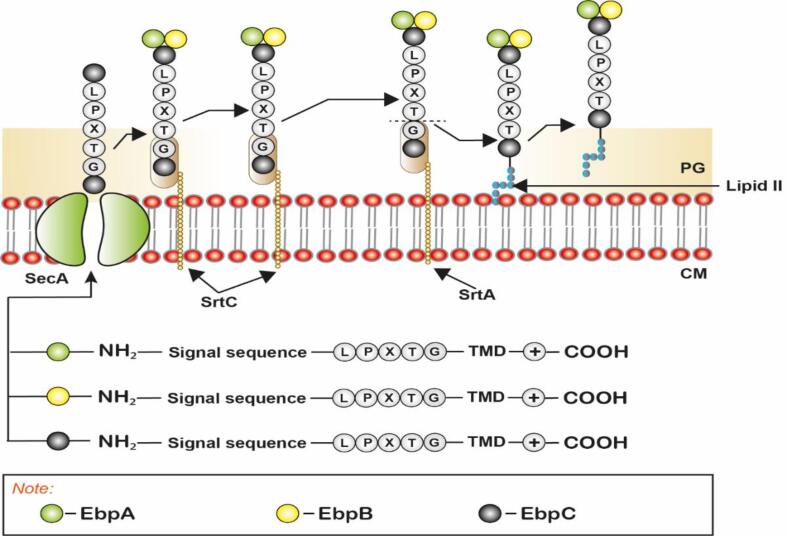
Fig. 1.
SA Pili assembly in Gram-positive bacteria, E. faecalis: Sortase substrate proteins EbpA and B (minor pilin protein), EbpC (Major pilin protein) secreted by the Sec pathway and translocated through SecA. The substrate proteins have a conserved LPXTG motif, Trans-Membrane Domain (TMD), and positively charged tail at their C- terminal and a signal sequence at their N - terminal. SrtC polymerizes pilin subunits, while SrtA recruits the substrate protein by recognizing the signal sequence at their N terminal and cleaves between the Threonine and Glycine of the peptide chain. The entire pilus assembly is then transferred to the lipid II intermediate, which then anchors it to the cell wall. The figure is derived from the previous studies (Hendrickx et al., 2011, Mandlik et al., 2008, Paterson and Mitchell, 2004).
A thioester acyl-enzyme intermediate is formed after the cleavage by SrtA, creating an intermediate complex. The complex is then attached to the peptidoglycan precursor, Lipid II (Fig. 1). In the next step (called nucleophile attack and covalent anchoring), Pentaglycine branched lipid II attacks the amine nucleophile in the intermediate complex, and prepares the complex for covalent anchoring to the cell wall. Lipid II thus plays a role of a middle man receiving the intermediate, attaching it to the growing cell wall and relieving SrtA for further rounds of sorting (Hendrickx et al., 2011, Hendrickx et al., 2009). SrtC, a pilus polymerizing sortase polymerizes the SrtA processed substrates after the substrates are anchored in the cell wall (Fig. 1). The aforementioned process has been well studied in Streptococcus pyogenes (Carlsson et al., 2006), where secretion and translocation of two sortase substrates proteins PrtF and PrtM are also observed, with PrtF localized to the pole and PrtM to the septum (Fig. 1). In addition, both SrtA and SecA (protein of general secretory pathway) were found to be focally colocalized at the septum of E. faecalis (Kandaswamy et al., 2013). Furthermore, it was shown that the SA pili of E. faecalis are of long hairlike extracellular protein fiber also known as the endocarditis- and biofilm-associated (ebp) pilus. SA pili of E. faecalis consist of major (EbpC) and minor subunits (EbpA and EbpB), where EbpA is present at the tip and EbpB is present at the base of an EbpC polymer (Fig. 1) (Nielsen et al., 2013). Studies have also shown that the removal of EbpA or EbpC results in a significant reduction in biofilm formation, while EbpB was found to be dispensable (Sillanpää et al., 2013). Taken together, The spatial positioning of sortase, cleavage of substrates (pilin subunits), and cell-wall anchoring of sortase-specific pilin subunits are extremely important in virulence, efficient pilus assembly, and biofilm formation (Hultgren et al., 2009, Kumari et al., 2020).
Heterotrimeric SpaA pili of Gram-positive bacteria:
The heterotrimeric sortase-mediated pili, SpaA pili was identified in Corynebacterium diphtheriae (C. diphtheriae) which was the major causative bacteria for human diphtheria (Ton-That et al., 2004, Yanagawa and Honda, 1976). Pili confers the virulence factors of C. diphtheriae which causes diphtheritic toxemia in the mouse model (Reardon-Robinson et al., 2015b). These pili exit in heterotrimeric structures made up of the tip adhesin pilin (SpaC) containing LPLTG motif, shaft pilin (SpaA) containing LPLTG motif, and the base pilin (SpaB) containing LAFTG motif (Fig. 2 & Table 1) (Ton-That and Schneewind, 2003). The pilin precursors from the cytoplasm were transferred to the cell membrane by SecA pathway. The membrane-bound thiol-disulfide oxidoreductase (MdbA) protein facilitates the protein folding and disulfide bond formation in the imported pilin subunit to initiate pilus assembly (Reardon-Robinson et al., 2015b). Each subunit contains the cell wall sorting signal with LPLTG or LAFTG motif. Among three subunits, SpaA contains the conserved lysine domain which mediates the crosslinking of pilin monomers, SpaC, and SpaB (Reardon-Robinson et al., 2015b). In the pili assembly mechanism, the pilus specific sortase (SrtA) hydrolysis of the LPLTG motif of the pilin subunit SpaC (tip pilin) and the cleaved threonine residue binds to the lysine residue of the LPLTG motif of the next subunit followed by the addition of several SpaA monomers resulting in pili elongation (McConnell et al., 2018). The pili elongation was terminated by the base pilin, SpaB binds to the sortase (SrtF) and anchored to the peptidoglycan layer (Fig. 2) (Swaminathan et al., 2007).
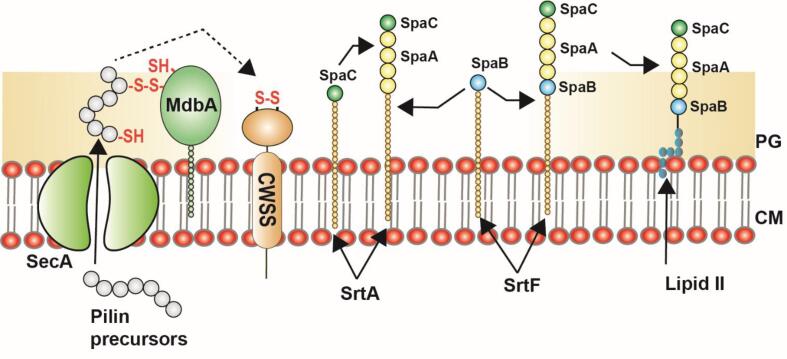
Fig. 2.
Assembly mechanism of heterotrimeric SpaA pili in Gram-positive bacteria, C. diphtheriae: The pilin precursors present in the cytoplasm are transported to the outer membrane through SecA translocon. The MdbA mediates the protein folding and formation of the disulfide bond before insertion into the membrane. The SrtA catalyzes the pilus polymerization by lysine-transpeptidase reaction where the pilin tip (SpaC) is recruited first followed by pilin stalk (SpaA), and pilin base (SpaB). The SpaB was added to the pili by housekeeping sortase (SrtF). Then, the entire pilus assembly is then transferred to the lipid II intermediate, which then anchors it to the cell wall. The figure is derived from previous studies (Siegel et al., 2016a).
Table 1.
Functions of pili subunits in pili assembly:
| Species | Pili | Pilin subunit | Physiological Functions | Reference |
|---|---|---|---|---|
| Pili in Gram-positive bacteria | ||||
| E. faecalis | SA pili | EbpA, B | Tip pilin mediates the host-pathogen interaction. | (Nielsen et al., 2013, Sillanpää et al., 2013) |
| EbpC | Stalk pilin, Polymerization of pilin subunit along the pilus forms the major pilus rod. | (Nallapareddy et al., 2006) | ||
| C. diphtheriae | Heterotrimeric SpaA | SpaC | Tip pilin mediates the host-pathogen interaction. | (Ton-That et al., 2004) |
| SpaA | Stalk pili, Polymerization of pilin subunit along the pilus forms the major pilus rod. | (Gaspar and Ton-That, 2006, Ton-That et al., 2004) | ||
| SpaB | Base pilin, terminate the pili polymerization. | (Ton-That et al., 2004) | ||
| SrtA | Pilus specific sortase, catalyses the pilus. | (Gaspar & Ton-That, 2006) | ||
| SrtF | The housekeeping sortase facilitates the entire pilus assembly to the lipid II intermediate, which then anchors it to the cell wall. | (Gaspar & Ton-That, 2006) | ||
| L. rhamnosus GG | SpaFED | SpaD | Major Pilin subunit. | (Nishiyama et al., 2016) |
| SpaE | Base pilin subunit. | (Nishiyama et al., 2016) | ||
| SpaF | Tip pilin subunit, adheres to the intestinal cells. | (K. Nishiyama et al., 2016) | ||
| A. oris | HeterodimericType 2 pili | FimB | Tip pilin, mediates the host-pathogen interaction. | (Mishra et al., 2010) |
| CafA | coaggregation pilin subunit. | (Reardon-Robinson et al., 2014) | ||
| FimA | Stalk pili, Polymerization of pilin subunit along the pilus forms the major pilus rod. | (Mishra et al., 2010) | ||
| SrtC2 | Pilus specific sortase, catalyses the pilus. | (Wu et al., 2012) | ||
| SrtA | The housekeeping sortase facilitates the entire pilus assembly to the lipid II intermediate, which then anchors it to the cell wall. | (Siegel et al., 2016a, Siegel et al., 2016b) | ||
| Clostridia | Type IV pili | PilA | Major pilin subunit. | (Varga et al., 2006) |
| PilB | Assembly ATPase, ATP hydrolysis and pushes the filament out of the bacterial cell. | (Varga et al., 2006) | ||
| Tip Pilin | Inner membrane core protein, facilitates the ATPase activity with PilB. | (Hendrick et al., 2017) | ||
| PilD | Prepilin peptidase, cleaves the prepilin and signal peptides then direct to growing pili and PilB respectively. | (Varga et al., 2006) | ||
| PilT | Retraction ATPase, retracts the pilin polymerization. | (Varga et al., 2006) | ||
| Pili in Gram-negative bacteria. | ||||
| E. coli | P pili | PapA | Major pilin subunit along the pili length. | (Jass et al., 2004) |
| PapC | Usher pathway, transfers the pilin subunits from periplasm to outer membrane. | (Kuehn et al., 1992) | ||
| PapD | Chaperone donates its strand to upcoming pili during pili polymerization. | (Lindberg et al., 1989) | ||
| PapE, F, K | The adapter subunit, holds the pilin subunits. | (Tiba et al., 2008) | ||
| PapG | Tip pilin, mediates the host-pathogen interaction. | (Björnham et al., 2009) | ||
| PapH | Base pilin, anchoring the pili to the cell membrane and terminating the pili polymerization. | (Båga et al., 1987) | ||
| Type 1 Pili | FimA | Major pilin subunit along the pili length. | (Walczak et al., 2014) | |
| FimC | Chaperone donates its strand to upcoming pili during pili polymerization. | (Sarowar et al., 2016) | ||
| FimD | Usher pathway, transfers the pilin subunits from periplasm to outer membrane. | (Sarowar et al., 2016) | ||
| FimF, G | The adapter subunit, holds the pilin subunits. | (Busch et al., 2015) | ||
| FimH | Tip pilin, mediates the host-pathogen interaction. | (Sokurenko et al., 1998) | ||
| Curli fibers | CsgA | Major Pilin subunit. | (Evans & Chapman, 2014) | |
| CsgB | Base Pilin subunit. | (Evans & Chapman, 2014) | ||
| CsgC | Chaperone, transfers the pilins to CsgG secretion pore. | (Evans & Chapman, 2014) | ||
| CsgE | Periplasmic protein, direct the CsgA pilins as linear polypeptide to CsgG and prevents the premature amyloid structure formation in periplasm. | (Evans & Chapman, 2014) | ||
| CsgF | Cell wall anchoring protein, anchors the pili to the cell membrane. | (Evans & Chapman, 2014) | ||
| CsgG | Secretion protein, transfers the pilins to outer membrane. | (Evans & Chapman, 2014) | ||
| N. gonorrhoeae | Type IV pili | PilB | Assembly ATPase, ATP hydrolysis and pushes the filament out of the bacterial cell. | (Taha et al., 1988) |
| PilC | Tip pilin, mediates the host-pathogen interaction. | (Burns et al., 1999) | ||
| PilD | Prepilin peptidase, cleaves the prepilin and signal peptides then direct to growing pili and PilB respectively. | (Freitag et al., 1995) | ||
| PilE | Major pilin subunit. | (Freitag et al., 1995) | ||
| PilG | Inner membrane core protein, facilitates the ATPase activity with PilB. | (Craig & Li, 2008) | ||
| PilQ | Outer membrane secretion pore, pushes the pili to the outer membrane. | (Drake and Koomey, 1995) | ||
| P. gingivalis | Type V pili (structural studies in certainly warranted) | Tip pilin | Mediates the host-pathogen interaction. | (Shoji et al., 2020) |
| Stalk pilins | Polymerization of the pilin subunit along the pilus forms the major pilus rod. | (Shoji et al., 2020) | ||
| OM secretion pore | It facilitates the transportation of pilin subunits from the periplasm to the outer membrane. | (Yoshimura et al., 1984) | ||
| Anchor pilin. | Facilitates the entire pilus assembly to anchors in the cell wall. | (Yoshimura et al., 1984) | ||
| Lipoprotein chaperone | Transfers the pilins to OM secretion pore. | (Yoshimura et al., 1984) | ||
Similar heterotrimeric pili were present in the probiotic Lactobacillus rhamnosus GG (LGG) which contains two gene clusters such as spaCBA and spaFED. Previous studies have shown that SpaCBA pili of L. rhamnosus GG are different from heterotrimeric SpaA pili of C. diphtheriae (Segers and Lebeer, 2014). In LGG, the SpaA was the major pilin subunit found in the pilus shaft. SpaC was the tip pilin subunit but also seen in the shaft at a very minimal number when compared to SpaA subunit. Similarly, SpaB was the base pilin present in the pilus base but also found in a few places of the pilus shaft at a very minimal number when compared to SpaA subunit (Reunanen et al., 2012).
The SpaFED pili were found in the intestinal tract, that the environmental factors stimulate the expression of spaFED genes to produce the SpaFED pilin proteins in the host intestinal tract and these genes were not expressed in lab cultures (Kankainen et al., 2009, Lebeer et al., 2012). The SpaFED pili were similar to SpaCAB, the SpaD was the major pilin subunit. SpaF was the tip pilin subunit but also seen in the pilus shaft at a very minimal number when compared to SpaD. Similarly, SpaE was the base pilin present in the pilus shaft at minimal numbers when compared to SpaD subunit. Both tip pilin proteins SpaC, F adheres to the epithelial cells (Susmitha et al., 2021). Being heterotrimeric and non-sequential mixed extracellular pilin protein arrangement, the assembly mechanism of LGG pili is to be warranted (Fig. 3 & Table.1).
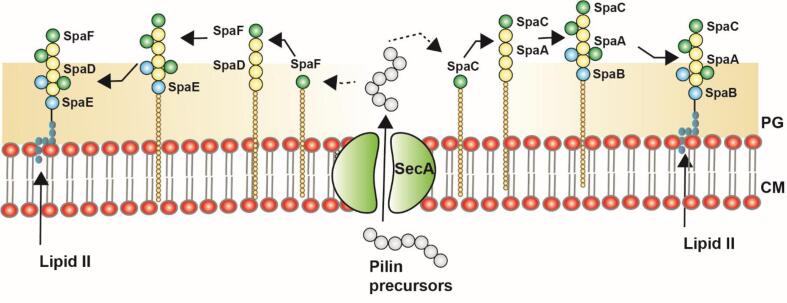
Fig. 3.
Assembly mechanism of heterotrimeric SpaCAB and SpaFED pili in probiotic bacteria, L. rhamnosus GG: The pilin precursors present in the cytoplasm are transported to the outer membrane through SecA translocon. In SpaCAB pili, the SpaC (tip pilins) are transferred to the outer membrane followed by the polymers of SpaA (major pilin), and finally SpaB (basal pilin) attaches to the pili base which lapidates to the cell membrane. The additional pili tip and basal position, SpaC and SpaB also present inbetween SpaA pilin stalk. Similarly, in SpaFED pili, the SpaF (tip pilins) are transferred to the outer membrane followed by the polymers of SpaD (major pilin), and finally SpaE (basal pilin) attaches to the pili base which lapidates to the cell membrane. The additional pili tip and basal position, SpaF and SpaE also present inbetween SpaD pilin stalk. This figure is derived from previous studies. The arrange of the additional tip pilins and basal pilins inbetween the major pilins need to be warranted (K. Nishiyama et al., 2016).
Heterodimeric Type 2 pili in Gram-positive bacteria:
The heterodimeric type 2 fimbriae were found in the Gram-positive bacteria, Actionomyces oris (A. oris) present in the human oral cavity. It causes the dental plague and co-aggregates with the complex biofilms on the oral surfaces and mucosal epithelia (Cisar et al., 1979, Skopek et al., 1993, Zijnge et al., 2010). Heterodimeric Type 2 pili pose binding affinity to the polysaccharides produced by streptococci and host bacterial cells (McIntire et al., 1978, Stathopoulos et al., 2000, Ruhl et al., 1996, Strömberg and Karlsson, 1990). This pilus was exiting in heterodimeric structures made up of the tip adhesin pilin (FimB) containing LPLTG motif, shaft pilin (FimA) containing LPLTG motif, and the co-aggregative pilin (CafA) containing LPXTG motif (Reardon-Robinson et al., 2014). Similar to the assembly mechanism of heterotrimeric SpaA pili, the pilin precursors from the cytoplasm were transferred to the cell membrane by the SecA pathway. The MdbA and Vitamin K epoxide reductase (VKOR) protein facilitates the protein folding and disulfide bond formation in the imported pilin subunit to initiate pilus assembly (Fig. 4 & Table.1) (Reardon-Robinson et al., 2015a, Reardon-Robinson et al., 2015b, Sanchez et al., 2017). Then, the pilus specific sortase (SrtC2) hydrolysis the LPxTG motif of the pilin subunit FimB (tip pilin), and the cleaved threonine residue binds to the lysine residue of LPxTG motif of the next subunit followed by the addition of several FimA monomers resulting in pili elongation (Wu et al., 2012). The housekeeping sortase (SrtA) anchors the pili on the cell membrane (Wu et al., 2014). The pili termination studies of the Type 2 fimbriae in A. oris are warranted.
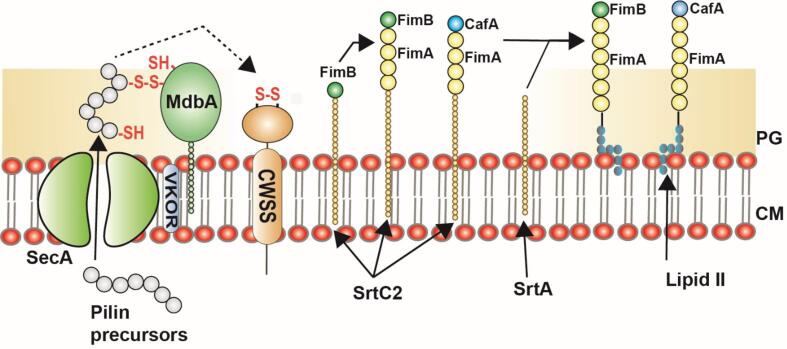
Fig. 4.
Assembly mechanism of Heterodimeric Type 2 pili in Gram-positive bacteria, A. oris: Similar to the SpaA pili, the pilin precursors present in the cytoplasm are transported to the outer membrane through SecA translocon. The translocated pilin subunits undergo folding and bond formation which was mediated by MdbA and vitamin K epoxide reductase (VKOR). The pilus specific sortase (SrtC2) catalyzes the pilus polymerization in the order of FimB (pilin tip), CafA (coaggregation factor), and FimA (major pilin). The housekeeping sortase facilitates the entire pilus assembly to the lipid II intermediate, which then anchors it to the cell wall. The figure is derived from the previous studies (Sanchez et al., 2017).
Type IV pili of Gram-positive bacteria:
The Type IV Pili (T4P) are surface fibers that facilitate several functions in the bacteria such as host cell adhesion, protein secretion, and locomotion (twitching and gliding motility) which was flagella independent (Burrows, 2012). The T4P are composed of pili subunits such as PilA, B, C, D, T, M, N, and O which were initially studied in several Gram-negative species and recent findings identified the type IV pili in Gram-positive species, such as Clostridium acetobutylicum (Nölling et al., 2001), C. perfringens (Myers et al., 2006, Shimizu et al., 2002), Clostridium difficile (C. difficile) (Borriello et al., 1990), and Clostridium beijerinckii (C. beijerinckii) (Varga et al., 2006). The Gram-negative and Gram-positive species have different cell wall architecture like a thick peptidoglycan layer with the cell membrane in Gram-positive bacteria and thin peptidoglycan layer with the outer and inner cell membrane in Gram-negative bacteria, however, they both possess a similar T4P assembly mechanism such as assembly/retraction of ATPase, PilB, and PilT, respectively.
In C. perfringens, the expression of pilC and pilT genes in T4P facilitates the gliding motility in the absence of flagella and chemotaxis genes (Varga et al., 2006). Similarly, C. beijerinckii co-express both flagella and type IV pili genes which provide both flagella-mediated movement and T4P mediated gliding motility (Varga et al., 2006).
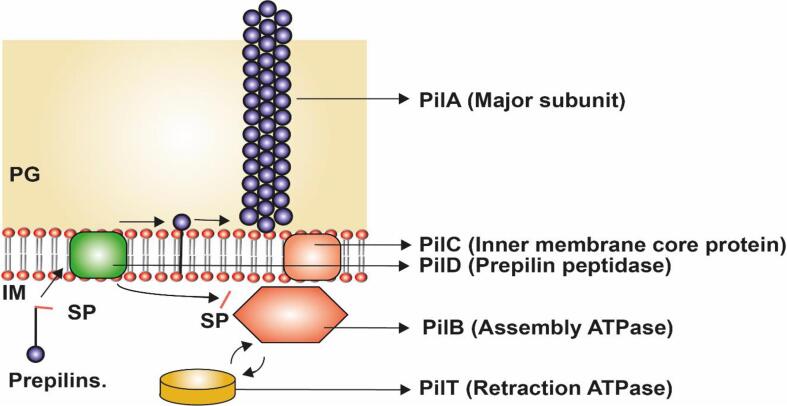
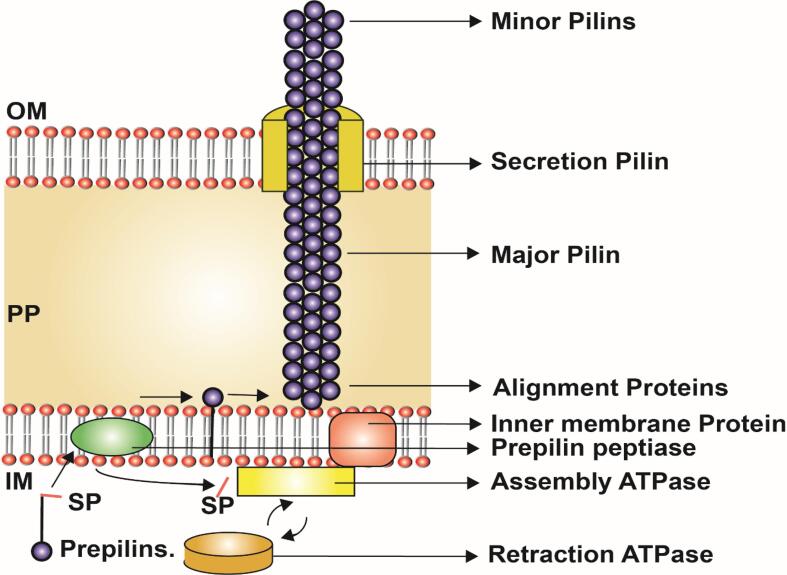
PilA was the major subunit recruited first to the outer membrane during the pilus assembly. The PilA was present inside the inner membrane as a prepilin coupled with a signal peptide which was recruited by the PilD subunit in the inner membrane (Varga et al., 2006) (Fig. 5 & Table 1). The PilD cleaves the signal peptides from PilA (prepilin) and directs the signal peptides to PilB and PilA. The transferred PilA in the outer peptidoglycan layer anchors to the cell membrane (Varga et al., 2006). The inner membrane core protein, PilC links the anchoring pili subunit PilA in the peptidoglycan layer to the inner membrane pilin subunit PilB to continue the addition of the next subunit to the growing pili (Varga et al., 2006). The polymerization and pili assembly of the PilA in the peptidoglycan was controlled by the assembly/retraction ATPase hydrolysis reaction i.e., the ATP was hydrolyzed to form ADP + inorganic phosphate (Pi), the Pi then binds to the next ADP to form ATP which results in the addition of one PilA monomer to the growing pili (Crowther et al., 2004, Megli and Taylor, 2013). This ATPase mechanism promotes the assembly of the PilA polymers along the pili length. (Fig. 5 & Table.2) (Wolfgang et al., 1998a, Wolfgang et al., 1998b).
Fig. 5.
Assembly mechanism of Type IV Pili in Gram-positive bacteria, Clostridia: the subunits are synthesized as prepilin (PilA, major subunit) whose signal peptide (SP) is cleaved by prepilin peptidase (PilD) and added to the growing fiber emerging out from inner membrane and periplasm. PilC forms the inner membrane protein to which PilB (assembly ATPase), together undergoes a conformational change during ATP hydrolysis and pushes the pilus filament out of the bacterial cell. Besides, the pili contain retraction ATPase (PilT) that can remove the pilin subunits at the pilus base ensuring twitching motility. The figure is derived from the previous studies (Varga et al., 2006).
Table 2.
Type IV pili subunits in pili assembly.
| Pilin Category/Bacterial strains | Major Pilin | Minor Pilins | Assembly ATPase | Inner membrane core protein | Prepilin peptidase | Outer membrane secretion protein | Retraction ATPase | Inner membrane accessory proteins | Reference |
|---|---|---|---|---|---|---|---|---|---|
| aClostridia Sp. | PilA | Various proteins | PilB | PilC | PilD | NA | PilT | PilM, N, O | (Nölling et al., 2001, Varga et al., 2006) |
| bP. aeruginosa | PilE, X, W, V, U | PilQ | PilM, N, O, P | (Beaussart et al., 2014, Craig et al., 2019, Giltner et al., 2010) | |||||
| bM. Xanthus | PilX1, V1, W1, FimU1 | (Craig et al., 2019, Hu et al., 2016, Li et al., 2003) | |||||||
| bN. gonorrhoeae | PilE | PilH, I, J, K, L | PilF | PilG | (Burdman et al., 2011, Craig et al., 2019, Proft and Baker, 2009) | ||||
| bV. cholerae | TcpA | TcpB | TcpT | TcpE | TcpJ | TcpC | TctT | TcpR, D, S | (Craig et al., 2019, Lim et al., 2010) |
Gram-positive bacteria, bGram-negative bacteria.
Pili in Gram-negative bacteria
Gram-negative bacteria contain several non-flagellar structures on the outer cell membrane called fimbriae or pili. These pili are composed of several protein monomers and they pose diverse functions during pathogenesis (Thanassi et al., 2012). The Pilus assembly was mediated by several pilin subunit proteins present in both the outer and inner cell membrane or only in the outer membrane (Costa et al., 2015). These pili confer the bacteria-host interaction, bacterial adhesin to the abiotic surfaces, bacterial interaction within the bacterial community, adaptation to the external environment, pathogenicity, bacterial motility, and formation of biofilm (Berry and Pelicic, 2015, Fronzes et al., 2008, Govindarajan and Kandaswamy, 2022, Van Gerven et al., 2015). Pili in Gram-negative bacteria are classified into five types based on their assembly mechanism and are Chaperone Usher (CU) Pili which includes Pap pili and Fim pili, Curli, Type IV pili, and Type V pili. Here we review some of those well-studied pili types of Gram-negative bacteria and discuss how they differ in their respective assembly mechanism.
Pap pili (P Pili) and Fim pili (F pili)
The P pili and F pili were found in the uropathogenic E. coli (UPEC) that was assembled via the Chaperone Usher (CU) pathway. Among the five pili types in Gram-negative bacterial, P pili and F pili were mostly found in the UPEC strains which confers the recurrent Urinary Tract Infection (UTI) such as pyelonephritis by P pili (Lane and Mobley, 2007) and cystitis by F pili (Mobley et al., 2009). The CU pili consist of a periplasmic chaperone, Outer Membrane (OM) pore pilus assembly, and major pili subunits (Table 1, Fig. 6, Fig. 7). The P pili subunits are PapA (major subunit), PapC (usher), PapD (periplasmic chaperone), PapE, F, K (adapter protein), PapG (pili tip), and PapH (terminator). The F pili subunits are FimA (major subunit), FimC (chaperone), FimD (usher), FimF, G (adapter proteins), FimH (pili tip). The aforementioned subunits play a key role in pilus assembly initiated after the translocation of pili subunits from the inner membrane to the periplasm via the Sec pathway (Stathopoulos et al., 2000). The pilus initiation is followed by a donor strand complementation and donor strand exchange mechanisms drive the assembly and bond formation between the pili subunits.
Fig. 6.
Schematic representation of the donor strand complementation and donor strand exchange of pili assembly on P pili and F pili: (a) the chaperone donates its G strand parallel to the pili subunit called Donor Strand Complementation (b) the G strand residues accommodates the pocket region present in the F strand, and (C) the G strand was exchanged between the Nte of the upcoming pili subunit during the pili assembly, this cycle repeats along the pili length and the pili length, this mechanism was termed as Donor Strand Exchange. The tip pilin does not undergo strand exchange and this pilus polymerization was terminated by the base pilin.
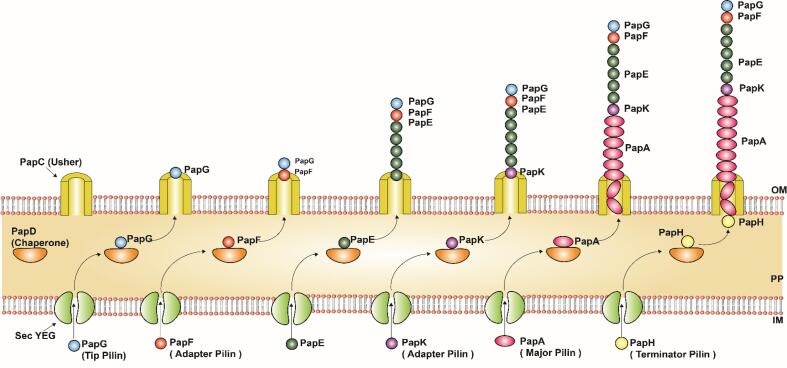
Fig. 7.
Assembly mechanism of P pili in Gram negative bacteria, E. coli: P Pili subunits PapG (Tip fibrillum), PapF (Adapter protein), PapE, PapK (Adapter protein), PapA (Major subunit) and PapH (Terminator) are secreted by Sec pathway and translocated to the inner membrane (IM). The periplasmic (PP) chaperone (PapD) then traps the subunits and helps in their proper folding and finally directs the subunits to the outer membrane (OM) Usher protein (PapC) where the subunits are assembled in ordered sequence. The figure is derived from the previous studies (Waksman, 2017).
Donor strand complementation
The pilin subunits sequentially reach the periplasm. For instance, PapH monomer (Pilin tip) first reaches the periplasm followed by PapA, PapK PapE, PapF and ends with PapG monomer that forms the base (Fig. 7). Similarly, the sequential order of the Fim pili subunits that reach the periplasm are several FimA monomers, FimF monomer, FimG monomer, and FimH monomer (Fig. 8). After reaching periplasm, the pili proteins form disulfide bonds between their β-strands A and B which were catalyzed by periplasmic oxidoreductase DsbA (Totsika et al., 2009). The pilus subunits with disulfide bonds between their β-strands A and B in the N terminal (Nte) with incomplete Ig-like folds at the C terminal (Cte) which leads to a deep hydrophobic channel on the pilin surface (Totsika et al., 2009). To complete the stable strand, the chaperone recognizes the deep hydrophobic channels present in the unstable pilin subunit and confers the early stage of stability by offering the chaperone's strand to the hydrophobic channel in parallel to the F strand of the unstable pili subunit, this process was termed as donor strand complementation (DSC) (Crespo et al., 2012). However, forms a complete strand with a non-canonical Ig fold. The chaperone’s donor strand holds five conserved hydrophobic residues (P1 to P4 residues except for P5 which is a free pocket) which binds to the pocket region (P1 to P4 pockets) present in the unstable pili subunit which in turn forms a chaperone-subunit complex. The P5 pocket initiates the donor strand exchange between the chaperone-subunit complex and the incoming pili subunit during the pili assembly (Fig. 6) (Waksman, 2017). Finally, the chaperone-subunit complex targets the usher protein to initiate the pili elongation (Barnhart et al., 2000). Overall, DSC promotes the stabilization of pilus subunits, proper folding, and prevents the subunit's premature self-polymerization (Busch and Waksman, 2012). DSC is followed by Donar Strand Exchange (DSE) for Subunit-subunit interaction (Sauer et al., 1999).
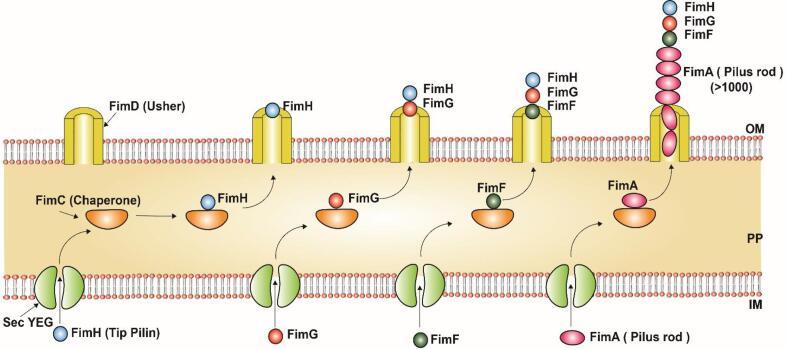
Fig. 8.
Assembly mechanism of Type 1 Pili in Gram-negative bacteria, E. coli: Type 1 Pili subunits FimH (Tip pilin), FimG (Minor tip pilin), FimF (Minor tip pilin), and FimA (Major subunit) are secreted by Sec pathway and translocated to the inner membrane (IM). The periplasmic (PP) Chaperone (FimC) then traps the subunits and helps in their proper folding and finally directs the subunits to the Outer membrane (OM) Usher protein (FimD) where the subunits are assembled in an ordered sequence. The figure is derived from the previous studies (Barnhart et al., 2000).
Donar strand exchange
The chaperone (FimC_and PapD) was not a part of the pilus in the outer membrane thus the chaperone strand was replaced by the strand of the next subunit during the assembly mechanism of pili (Waksman, 2017). This was achieved by the exchange of the G1 strand with the (Nte) of the next subunit was termed DSE (Remaut et al., 2006). During the strand exchange, the G1 strand pockets (P1 to P5 pockets) of the chaperone-subunit complex accommodate (P1 to P5 residues) of Nte of the incoming subunit. Accommodation of Nte with G1 strand drives the G1′s β-strands opposite to F strand forms a canonical Ig-fold which in turn promotes the DSE. The Nte replaces the chaperone strand with its strand in a zip-in-zip-out mechanism (Rose et al., 2008, Zavialov et al., 2003). The repetitive cycle in replacing the chaperone strand with Nte takes place during the assembly of pili length. Interestingly the pilus tip subunit does not have P5 pockets to undergo DSE and the pili extension ends (Fig. 6) (Verger et al., 2006).
Interaction of the chaperone with the pili subunits leads to the conformational change to the usher binding site of the chaperone (Busch & Waksman, 2012). The conformational change was based on the incoming pili subunit and their binding affinity to the chaperone (Kline et al., 2010, Verger et al., 2008). The pili tip has (PapH in pap pili and FimA in Fim pili) the highest affinity and hence it is recruited first while other subunits have comparatively fewer affinities followed by the pili subunit (Busch et al., 2015). The affinity is also determined by the DSE rate between the subunits (Allen et al., 2013, Nishiyama et al., 2008). Also, the pilin subunit recruitment was based on their concentration (Busch et al., 2015). In P pili, PapG was the first subunit to be recruited by PapD and the PapD-PapG complex binds to PapC followed by PapD-PapF which initiates the tip fibrillin growth. This process takes place by donor strand exchange (Busch et al., 2015, Busch and Waksman, 2012). Once the PapG subunit is linked to PapF it is followed by the addition of PapE subunits. PapF has the capability of correctly linking PapG to PapE subunits (Båga et al., 1987). It was followed by the incorporation of PapK (adapter protein) which terminates the growth of the pili tip and creates binding sites for PapD- PapA which results in the polymer of PapA subunits (major subunit) which forms the pilus rod (Fig. 7 & Table 1) (Busch & Waksman, 2012). Finally, the inclusion of PapH subunit terminates the pilus growth and anchors the pilus on the cell membrane (Kline et al., 2009b). The terminator subunit PapH also lacks a P5 pocket, hence it cannot undergo DSE and pilus assembly (Busch et al., 2015, Verger et al., 2006) (Fig. 6). The interaction between the P pili and the host cell was facilitated by the PapG which targets the gallibiose moieties in glycolipids of host epithelial cells (Busch et al., 2015, Kline et al., 2010).
In type I pili, FimH was the first subunit recruited by FimC and the FimC-FimH complex binds to FimD followed by FimC-FimG which initiates the growth of the pili tip followed by the addition of FimF subunit and then form a polymer of FimA subunits (major subunit) which forms the pilus rod (Busch et al., 2015) (Fig. 8). Unlike PapH subunit in P pili, the termination pili subunit was missing in Type 1 pilus assembly to control the pili length hence the termination mechanism remains unclear (Busch & Waksman, 2012). The interaction between the F pili to the host cell was facilitated by FimH, the pili tip subunit, which binds to the mannose portion in the host cell (Fig. 8 & Table 1) (Busch et al., 2015, Kline et al., 2010).
Usher’s role
Outer membrane Usher protein (FimD in fim pili and PapC in pap pili) (Fig. 7,8 & Table 1) contains 5 domains – N terminal periplasmic domain, translocation pore domain, plug domain, two C terminal periplasmic domain (CTD1, CTD2) (Busch et al., 2015, Capitani et al., 2006, Nishiyama et al., 2003, Thanassi et al., 2002). In the translocation pore domain when stopped by the plug domain, the usher protein remains in an inactive state (apo form) (Busch et al., 2015). This prevents the passage of pilus subunits through usher proteins (Kuehn et al., 1993). When the usher protein is triggered by the chaperone subunit complex, the plug domain in the usher swings out and the usher turns into an active state (open form) and thereby it initiates the translocation of pilus subunits (Busch et al., 2015).
The Chaperone-pili subunit complex interacts initially with the Nte domain of the usher because the chaperone alone without pili subunits does not bind to usher protein. The chaperone contains conserved proline residues on its binding surface to usher which blocks the chaperone-usher interaction (Busch et al., 2015, Di Yu et al., 2012). When the chaperone binds to a pili subunit, the proline lock was rotated away making the chaperone subunit complex bind to the N terminal domain of the usher protein (Di Yu et al., 2012). Then the pilus assembly begins in the outer membrane.
Curli fibers:
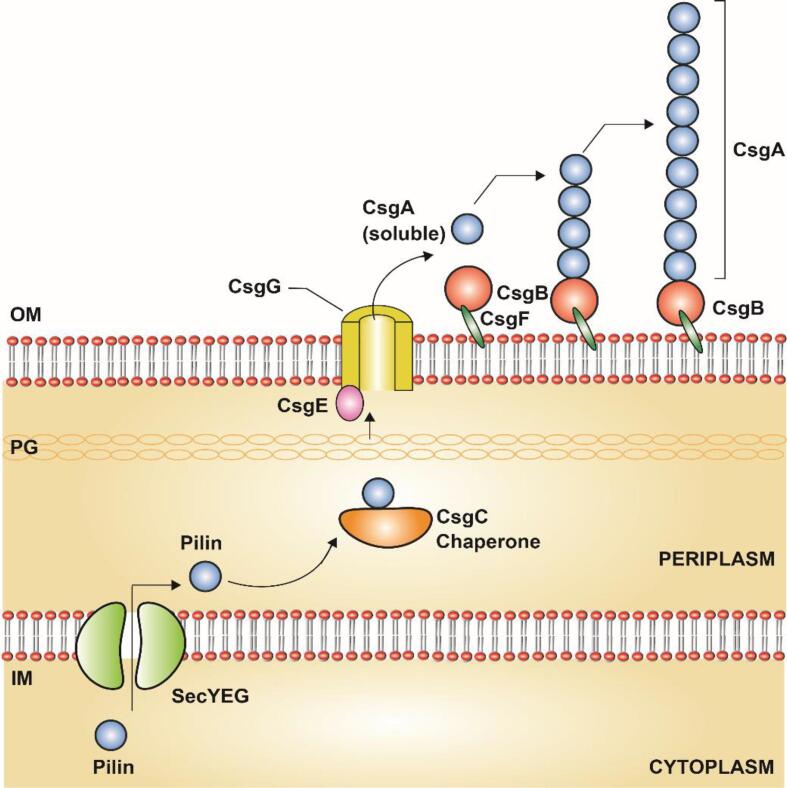
Curli are first identified as amyloid extracellular protein fibers present in E. coli and Salmonella enterica (Olsén et al., 1989, Zogaj et al., 2001). Curli fibers mediate bacterial colonization, host-pathogen interaction, immune evasion in the host, and biofilm formation (Kai-Larsen et al., 2010, McCrate et al., 2013, Pawar et al., 2005). The curli-mediated rugose biofilms are localized in the air-colony interface regions which protect the biofilms from oxidative stress compared to planktonic cells (DePas et al., 2013). Successful curli-mediated biofilm formation relies on the coordination of the curli gene (csg) expression and extracellular protein production (Hammar et al., 1995). Initially, the pilins CsgA, B, and F were transferred from the cytoplasm to the periplasm through the Sec pathway. The periplasmic chaperone, CsgC traps the pilin subunits and directs them to the CsgG, an outer membrane secretion pore (Hammer et al., 2007). The periplasmic protein CsgE facilitates the pilin subunits through the CsgG pore (Nenninger et al., 2009). The CsgB and F were transferred to the outer membrane where the CsgB (minor subunit) was anchored to the cell wall by the CsgF, anchoring protein. In periplasm, the secreted CsgA occurs as a linear polypeptide, the CsgE (periplasmic protein) prevents the CsgA to undergo protein folding and transfer the CsgA to the outer membrane (Andersson et al., 2013). Once CsgA is secreted, it occurs in the soluble linear polypeptide chain with the imperfect repeated amyloid core domain at the C-terminal consists of SxQxGxGNxAxQ forms the cross-β structures. (Fig. 9 & Table.1) The repeated Glycine and Alanine residues stack the polymers of amyloid fibers of CsgA along the curli fiber length (Shewmaker et al., 2009).
Fig. 9.
Assembly mechanism of Curli fibers in Gram-negative bacteria, E. coli: Curli fiber subunits, CsgA (Major pilin), CsgB (Minor pilin), CsgF (Cell wall anchoring pilins) are secreted by the SecYEG and translocated to the inner membrane (IM). The CsgC (Periplasmic Chaperone) then traps the subunits (CsgB and CsgF) and directs the subunits to the outer membrane (OM) through the CsgG (OM secretion pore). The CsgF anchors the CsgB to the cell wall. The CsgE (Periplasmic protein) direct the CsgA pilins as linear polypeptide to CsgG and prevents the premature amyloid structure formation in periplasm. Once the CsgA transferred to OM as a soluble linear polypeptide interacts with the CsgB which initiates the amyloid structure formation on soluble linear polypeptide, CsgA. In the following steps, CsgA forms the major pilin subunit along the pili length. This figure is derived from the previous studies (Evans and Chapman, 2014).
Type IV pili (Type 4a and b):
Type IV pili (T4P) in bacteria are hair-like multi-subunit compounds that acts a key role in cellular adhesion, colonization, virulence, twitching motility, and biofilm formation (Carbonnelle et al., 2005, Jain et al., 2012, Persat et al., 2015, Wolfgang et al., 2000). The T4P poses unique features like pilin polymerization and depolymerization which in turn extends and retracts the pili length. It consists of polymer copies of one or more pilin subunits assembled in helix structures (Piepenbrink and Sundberg, 2016). These pilin polymers are secreted from the inner membrane pilin reservoir which was transported to the cell membrane to display the T4P in the bacterial cell wall, reversibly retracts the pilin subunits. This uniqueness in polymerization/de-polymerization makes T4P a dynamic pilus. T4P was identified in Gram-negative bacterial species like enteropathogenic E. coli, Pseudomonas aeruginosa (P. aeruginosa), Neisseria gonorrhoeae (N. gonorrhoeae), Neisseria meningitidis (Shi and Sun, 2002, Burns et al., 1999).
The T4P was composed of several pilin subunits with different functions such as major pilin subunit, minor pilin subunits, a pre-pilin peptidase that breaks the pilin signal sequence from prepilins, specific ATPase which acts as a motor and provides energy for pilus assembly, inner membrane protein recruits ATPase from the cytoplasm, outer membrane secretory protein for the secretion of pili proteins, inner membrane accessory proteins, and the retraction ATPase promotes the depolymerization of pilus fiber called twitching motility (Proft & Baker, 2009).
The T4P were differentiated into two T4a and T4b pili depending on their differences in the length of the signal peptide and major pilin subunits (Burdman et al., 2011). The T4a subunits consist of shorter signal peptides (<10aa) where a typical length of about 150-160aa (Craig et al., 2004). T4a pilins are present in P. aeruginosa, N. gonorrhoeae, and Myxococcus xanthus (M. xanthus). T4b pilins are present in enteric pathogens such as Vibrio cholera (V. cholera) (Craig and Li, 2008).
The T4a present in P. aeruginosa and N. gonorrhoeae have similar pili assembly. The T4a present in V. cholera, and M. xanthus vary in the pilin subunits but undergoes similar pili assembly mechanism (Craig et al., 2019). In T4P of P. aeruginosa, the major pilin subunit was PilA, outer membrane secretion pilin (PilQ), inner membrane accessory proteins (PilM, N, O, and P), inner membrane protein (PilC), retraction of ATPase pilin (PilT) was similar in all T4a, the assembly ATPase pilin (PilB), and the minor pilins (PilE, X, W, V, and U). Similar pilin subunits were found in T4a of M. xanthus (Craig et al., 2019).
In T4b of V. cholerae, the major pilin subunit was TcpA, outer membrane secretion pilin (TcpC), inner membrane accessory proteins (TcpR, D, and S), inner membrane protein (TcpE), retraction of ATPase were yet to be warranted, the assembly ATPase pilin (TcpT), and the minor pilins (TcpB) (Craig et al., 2019).
Assembly mechanism
In the pili assembly mechanism of T4P, the pilin subunits were initially synthesized as prepilin (prepilin + Signal proteins). The signal proteins were recognized by pre pilin peptidase which cleaves the N-terminal signal peptide and methylates the N-terminal phenylalanine of mature pilin (Carbonnelle et al., 2005, Kline et al., 2009a). The synthesized pilin subunits (prepilin) from the cytoplasm were transferred to the inner membrane through the Sec pathway (Mandlik et al., 2008). The transferred prepilin present in negative charge attracts the positively charged alignment protein subunits in the growing pilus length (Proft & Baker, 2009). Following interactions with the globular pili domains, accommodate the incoming subunit to assemble in the existing gap of the pilus base (Proft & Baker, 2009). Then, the major pilin subunit is added to the pilus chain, emerging outside the cell via the outer membrane secretion protein (Burdman et al., 2011, Collins et al., 2001, Wolfgang et al., 2000). The Pili assembly was carried out in a molecular platform of inner membrane platform proteins, assembly ATPase, and retraction ATPase (Proft & Baker, 2009). The inner membrane platform proteins push the prepilin to the growing pili length and the pili length was controlled by assembly/retraction ATPase (Proft and Baker, 2009, Tønjum et al., 1995, Wolfgang et al., 2000). Followed by the transfer of the tip pilin subunit to the pili terminal end. In addition to the major pilins, minor pilin monomers were added along the pilin length (Kline et al., 2009a, Rudel et al., 1995) (Fig. 10 & Table 1), the different T4P pilin subunits present in Gram-negative bacteria was tabulated (Table 2).
Fig. 10.
Assembly mechanism of Type 4 Pilus in Gram-negative bacteria, N. gonorrhoeae: The pili subunits are synthesized as prepilin (Pile, major subunit) whose signal peptide (SP) is cleaved by prepilin peptidase and added to the growing fiber emerging out via outer membrane (OM) secretion. The inner membrane protein links the growing fiber to assembly ATPase which undergoes a conformational change during ATP hydrolysis and pushes the pilins out of the membrane to the gap in growing fibert. Besides, type 4 pili contain retraction ATPase which can remove the pilin subunits at the pilus base ensuring twitching motility. The figure is derived from the previous studies (Kline et al., 2009a, Kline et al., 2009b, Proft and Baker, 2009).
Type V pili:
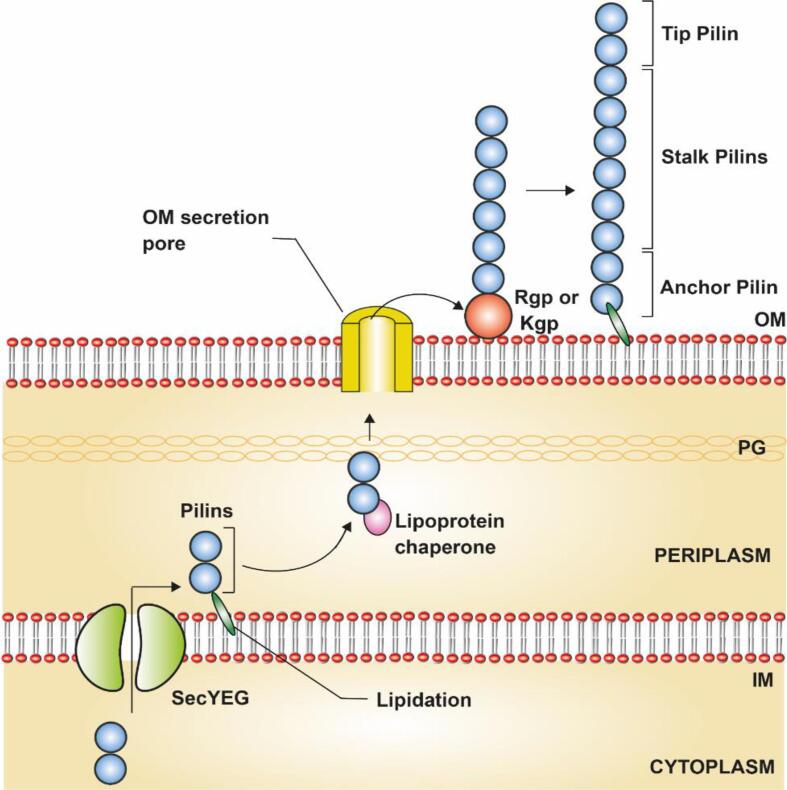
The type V pili are present in the outer membrane of Gram-negative oral pathogen, Porphyromonas gingivalis (P. gingivalis), which facilitates the bacterial adhesin, aggregation in internal bacterial communities, and formation of biofilm. The P. gingivalis causes severe periodontitis and gingivitis (Yoshimura et al., 2009). In P. gingivalis, two distinct types of V pili were present: major pili and minor pili where the major pili were long of about 0.3–1.6 μm in length (Yoshimura et al., 1984), and the minor pili were short of about 80–120 μm in length (Hamada et al., 1996). The Major and minor pili differ only in length but the pili are encoded with similar operons (Yoshimura et al., 2009). The pili synthesis was initiated in the cytoplasm as a lipoprotein precursor with a Nte signal peptide which was termed ‘lipobox’. Then these pili proteins are transferred to the periplasm through SecYEG, then the cysteine at the N-terminal of the pili protein was lipidated to cleave the signal residues. Next, the lipoprotein chaperone binds and transfers the pili subunits to the outer membrane and this pathway protein remains unknown (Shoji et al., 2004, Xu et al., 2016). In the outer membrane, gingipain K (Kgp) or gingipain R (Rgp), mediates the production of the mature pili by polymerization (Shoji et al., 2004) (Fig. 11 & Table 1). The C-terminal of the mature pili anchors on the outer membrane and the pili tip, Nte might be capped by another protein that remains unknown that prevents further polymerization of the pili elongation and also promotes the interaction with the suitable ligands. The complete assembly mechanism of pili subunits and their functions are yet to be studied.
Fig. 11.
Assembly mechanism of Type V pili in Gram-negative bacteria, P. gingivalis: The type V pili assembly was initiated as prepilins in the cytoplasm and transferred to the periplasm across the inner membrane (IM) via SecYEG pathway. The imported pilins are transferred to the outer membrane (OM) which was facilitated by the chaperone and the OM secretory proteins were not studied so far. In OM, Rgp/Kgp proteins promote the pili polymerization and the mature pili were anchored to the cell membrane (anchoring mechanism yet to be studied). The figure is derived from the previous studies (Yoshimura et al., 2009).
Concluding and perspectives
Collective studies and reviews on the pilus assembly mechanism of Gram-positive and Gram-negative bacteria and their role in bacterial species created a path for drug and vaccine development (Govindarajan et al., 2020, Kline et al., 2010, Ramirez et al., 2020). This article aims to provide a clear and concise review of the pilus assembly mechanism by elaborating on the role of individual pilin subunits along with their physiological functions. The most common features found in the pilus assembly are pilus-specific substances such as sortases of Gram-positive bacteria and chaperones of the Gram-negative bacteria that induce the pili polymerization followed by the cell was anchoring by the base pilins. Both Gram-positive and Gram-negative bacteria possess unique pili initiation mechanisms via transpeptidation and transglycosylation (Gram-positive) reaction and DSC/DSE (Gram-negative (Hendrickx et al., 2009, Marraffini et al., 2006, Waksman, 2017, Walczak et al., 2014)). In addition, the majority of Gram-positive species contain class A sortase that performs a housekeeping role by recruiting and cleaving the substrate protein while class C sortase performs pili polymerization (Kandaswamy et al., 2013, Kline et al., 2009b, Spirig et al., 2011). Recently, it was discovered that class B sortase performs the role of iron acquisition. The functions of other classes of sortases such as D, E, and F remains unknown and requires further investigation (Spirig et al., 2011).
Similarly, the CU pathway of pilus assembly is well studied in gram-negative species, however, there are other pilus assembly mechanisms such as curli, type IV, and type V pili that are either studied only in a few representative species and in some cases, there is a significant variation in assembly mechanism from one species to other. In moving forward, researchers should focus on identifying the pilin protein that is responsible for both the initiation and termination of pilus assembly. Furthermore, it was observed that there is a significant variation in the length of pili across bacterial species and there is no information available on what determines the pilus length and how bacterial species terminate the pilus assembly. Taken together, it is crucial to identify the structure and function of the entire family of pilin proteins of both Gram-positive and Gram-negative species, as this would provide more leads for the discovery of a novel antimicrobial agent that can selectively target bacterial surface proteins.
CRediT authorship contribution statement
Tamilarasi Shanmugasundarasamy: Conceptualization, Data curation, Methodology, Writing – original draft, Formal analysis. Deenadayalan Karaiyagowder Govindarajan: Conceptualization, Data curation, Methodology, Writing – original draft, Formal analysis, Writing – review & editing, Validation, Visualization. Kumaravel Kandaswamy: Writing – review & editing, Validation, Visualization, Funding acquisition, Investigation, Project administration, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
The authors gratefully acknowledge the DST SERB Start-Up Research Grant (File Number: SRG/2019/000094) from the Ministry of Science and Technology, Government of India.
Authors Contribution
Tamilarasi Shanmugasundarasamy, Deenadayalan Karaiyagowder Govindarajan, and Kumaravel Kandaswamy: Conceptualization, and design of the study. Tamilarasi Shanmugasundarasamy and Deenadayalan Karaiyagowder Govindarajan: Data curation, methodology, writing - original draft, and format analysis. Deenadayalan Karaiyagowder Govindarajan, and Kumaravel Kandaswamy: writing - review & editing, validation, and visualization. Kumaravel Kandaswamy: Funding acquisition, investigation, project administration, and supervision.
Data availability
Data will be made available on request.
References
- Allen W.J., Phan G., Hultgren S.J., Waksman G. Dissection of pilus tip assembly by the FimD usher monomer. J. Mol. Biol. 2013;425(5):958–967. doi: 10.1016/j.jmb.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson E., Bengtsson C., Evans M., Chorell E., Sellstedt M., Lindgren A.G., Hufnagel D., Bhattacharya M., Tessier P., Wittung-Stafshede P., Almqvist F., Chapman M. Modulation of curli assembly and pellicle biofilm formation by chemical and protein chaperones. Chem. Biol. 2013;20(10):1245–1254. doi: 10.1016/j.chembiol.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Båga M., Norgren M., Normark S. Biogenesis of E. coli Pap pili: PapH, a minor pilin subunit involved in cell anchoring and length modulation. Cell. 1987;49(2):241–251. doi: 10.1016/0092-8674(87)90565-4. [DOI] [PubMed] [Google Scholar]
- Barnhart M.M., Pinkner J.S., Soto G.E., Sauer F.G., Langermann S., Waksman G., Frieden C., Hultgren S.J. PapD-like chaperones provide the missing information for folding of pilin proteins. Proc. Natl. Acad. Sci. 2000;97(14):7709–7714. doi: 10.1073/pnas.130183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaussart A., Baker A.E., Kuchma S.L., El-Kirat-Chatel S., O’Toole G.A., Dufrêne Y.F. Nanoscale adhesion forces of Pseudomonas aeruginosa type IV pili. ACS Nano. 2014;8(10):10723–10733. doi: 10.1021/nn5044383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry J.-L., Pelicic V. Exceptionally widespread nanomachines composed of type IV pilins: the prokaryotic Swiss Army knives. FEMS Microbiol. Rev. 2015;39(1):134–154. doi: 10.1093/femsre/fuu001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björnham O., Nilsson H., Andersson M., Schedin S. Physical properties of the specific PapG–galabiose binding in E. coli P pili-mediated adhesion. Eur. Biophys. J. 2009;38(2):245–254. doi: 10.1007/s00249-008-0376-y. [DOI] [PubMed] [Google Scholar]
- Borriello S., Davies H., Kamiya S., Reed P., Seddon S. Virulence factors of Clostridium difficile. Rev. Infect. Dis. 1990;12(Supplement_2):S185–S191. doi: 10.1093/clinids/12.supplement_2.s185. [DOI] [PubMed] [Google Scholar]
- Bradshaw W.J., Davies A.H., Chambers C.J., Roberts A.K., Shone C.C., Acharya K.R. Molecular features of the sortase enzyme family. FEBS J. 2015;282(11):2097–2114. doi: 10.1111/febs.13288. [DOI] [PubMed] [Google Scholar]
- Burdman S., Bahar O., Parker J.K., De La Fuente L. Involvement of type IV pili in pathogenicity of plant pathogenic bacteria. Genes. 2011;2(4):706–735. doi: 10.3390/genes2040706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns D.L., Scheuerpflug I., Rudel T., Ryll R., Pandit J., Meyer T.F. Roles of PilC and PilE proteins in pilus-mediated adherence of Neisseria gonorrhoeae and Neisseria meningitidis to human erythrocytes and endothelial and epithelial cells. Infect. Immun. 1999;67(2):834–843. doi: 10.1128/iai.67.2.834-843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows L.L. Prime time for minor subunits of the type II secretion and type IV pilus systems. Mol. Microbiol. 2012;86(4):765–769. doi: 10.1111/mmi.12034. [DOI] [PubMed] [Google Scholar]
- Busch A., Waksman G. Chaperone–usher pathways: diversity and pilus assembly mechanism. Philos. Trans. R. Soc. B Mathemat. Phys. Eng. Sci. 2012;367(1592):1112–1122. doi: 10.1098/rstb.2011.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A., Phan G., Waksman G. Molecular mechanism of bacterial type 1 and P pili assembly. Philos. Trans. R. Soc. A Mathemat. Phys. Eng. Sci. 2015;373(2036):20130153. doi: 10.1098/rsta.2013.0153. [DOI] [PubMed] [Google Scholar]
- Capitani G., Eidam O., Grütter M.G. Evidence for a novel domain of bacterial outer membrane ushers. Proteins Struct. Funct. Bioinf. 2006;65(4):816–823. doi: 10.1002/prot.21147. [DOI] [PubMed] [Google Scholar]
- Carbonnelle E., Hélaine S., Prouvensier L., Nassif X., Pelicic V. Type IV pilus biogenesis in Neisseria meningitidis: PilW is involved in a step occurring after pilus assembly, essential for fibre stability and function. Mol. Microbiol. 2005;55(1):54–64. doi: 10.1111/j.1365-2958.2004.04364.x. [DOI] [PubMed] [Google Scholar]
- Carlsson F., Stålhammar-Carlemalm M., Flärdh K., Sandin C., Carlemalm E., Lindahl G. Signal sequence directs localized secretion of bacterial surface proteins. Nature. 2006;442(7105):943–946. doi: 10.1038/nature05021. [DOI] [PubMed] [Google Scholar]
- Cisar J.O., Kolenbrander P., McIntire F. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 1979;24(3):742–752. doi: 10.1128/iai.24.3.742-752.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy K.W., Melvin J.A., McCafferty D.G. Sortase transpeptidases: insights into mechanism, substrate specificity, and inhibition. Pept. Sci. 2010;94(4):385–396. doi: 10.1002/bip.21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R.F., Davidsen L., Derrick J.P., Ford R.C., Tønjum T. Analysis of the PilQ secretin from Neisseria meningitidis by transmission electron microscopy reveals a dodecameric quaternary structure. J. Bacteriol. 2001;183(13):3825–3832. doi: 10.1128/JB.183.13.3825-3832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa T.R., Felisberto-Rodrigues C., Meir A., Prevost M.S., Redzej A., Trokter M., Waksman G. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 2015;13(6):343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- Craig L., Li J. Type IV pili: paradoxes in form and function. Curr. Opin. Struct. Biol. 2008;18(2):267–277. doi: 10.1016/j.sbi.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L., Pique M.E., Tainer J.A. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2004;2(5):363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- Craig L., Forest K.T., Maier B. Type IV pili: dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 2019;17(7):429–440. doi: 10.1038/s41579-019-0195-4. [DOI] [PubMed] [Google Scholar]
- Crespo M.D., Puorger C., Schärer M.A., Eidam O., Grütter M.G., Capitani G., Glockshuber R. Quality control of disulfide bond formation in pilus subunits by the chaperone FimC. Nat. Chem. Biol. 2012;8(8):707–713. doi: 10.1038/nchembio.1019. [DOI] [PubMed] [Google Scholar]
- Crowther L.J., Anantha R.P., Donnenberg M.S. The inner membrane subassembly of the enteropathogenic Escherichia coli bundle-forming pilus machine. Mol. Microbiol. 2004;52(1):67–79. doi: 10.1111/j.1365-2958.2003.03963.x. [DOI] [PubMed] [Google Scholar]
- DePas W.H., Hufnagel D.A., Lee J.S., Blanco L.P., Bernstein H.C., Fisher S.T., James G.A., Stewart P.S., Chapman M.R. Iron induces bimodal population development by Escherichia coli. Proc. Natl. Acad. Sci. 2013;110(7):2629–2634. doi: 10.1073/pnas.1218703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Yu X., Dubnovitsky A., Pudney A.F., MacIntyre S., Knight S.D., Zavialov A.V. Allosteric mechanism controls traffic in the chaperone/usher pathway. Structure. 2012;20(11):1861–1871. doi: 10.1016/j.str.2012.08.016. [DOI] [PubMed] [Google Scholar]
- Drake S.L., Koomey M. The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol. Microbiol. 1995;18(5):975–986. doi: 10.1111/j.1365-2958.1995.18050975.x. [DOI] [PubMed] [Google Scholar]
- Du M., Yuan Z., Werneburg G.T., Henderson N.S., Chauhan H., Kovach A., Zhao G., Johl J., Li H., Thanassi D.G. Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis. Nat. Commun. 2021;12(1) doi: 10.1038/s41467-021-25522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M.L., Chapman M.R. Curli biogenesis: order out of disorder. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2014;1843(8):1551–1558. doi: 10.1016/j.bbamcr.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores-Mireles A.L., Walker J.N., Caparon M., Hultgren S.J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015;13(5):269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag N.E., Seifert H.S., Koomey M. Characterization of the pilF—pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol. Microbiol. 1995;16(3):575–586. doi: 10.1111/j.1365-2958.1995.tb02420.x. [DOI] [PubMed] [Google Scholar]
- Fronzes R., Remaut H., Waksman G. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 2008;27(17):2271–2280. doi: 10.1038/emboj.2008.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar A.H., Ton-That H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J. Bacteriol. 2006;188(4):1526–1533. doi: 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giltner C.L., Habash M., Burrows L.L. Pseudomonas aeruginosa minor pilins are incorporated into type IV pili. J. Mol. Biol. 2010;398(3):444–461. doi: 10.1016/j.jmb.2010.03.028. [DOI] [PubMed] [Google Scholar]
- Govindarajan D.K., Kandaswamy K. Virulence factors of uropathogens and their role in host pathogen interactions. Cell Surf. 2022;8:100075. doi: 10.1016/j.tcsw.2022.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindarajan D.K., Viswalingam N., Meganathan Y., Kandaswamy K. Adherence patterns of Escherichia coli in the intestine and its role in pathogenesis. Med. Microecol. 2020;5:100025. [Google Scholar]
- Hamada N., Sojar H.T., Cho M.-I., Genco R.J. Isolation and characterization of a minor fimbria from Porphyromonas gingivalis. Infect. Immun. 1996;64(11):4788–4794. doi: 10.1128/iai.64.11.4788-4794.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammar M., Arnqvist A., Bian Z., Olsén A., Normark S. Expression of two csg operons is required for production of fibronectin-and congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 1995;18(4):661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- Hammer N.D., Schmidt J.C., Chapman M.R. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc. Natl. Acad. Sci. 2007;104(30):12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrick W.A., Orr M.W., Murray S.R., Lee V.T., Melville S.B. Cyclic di-GMP binding by an assembly ATPase (PilB2) and control of type IV pilin polymerization in the Gram-positive pathogen Clostridium perfringens. J. Bacteriol. 2017;199(10):e00034–00017. doi: 10.1128/JB.00034-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx A., Budzik J.M., Oh S.-Y., Schneewind O. Architects at the bacterial surface—sortases and the assembly of pili with isopeptide bonds. Nat. Rev. Microbiol. 2011;9(3):166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- Hendrickx A.P., Willems R.J., Bonten M.J., van Schaik W. LPxTG surface proteins of enterococci. Trends Microbiol. 2009;17(9):423–430. doi: 10.1016/j.tim.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Hu W., Gibiansky M.L., Wang J., Wang C., Lux R., Li Y., Shi W. Interplay between type IV pili activity and exopolysaccharides secretion controls motility patterns in single cells of Myxococcus xanthus. Sci. Rep. 2016;6(1):1–10. doi: 10.1038/srep17790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultgren S.J., Henriques-Normark W.B., Caparon M.G., Nallapareddy B.E.M., Lim B., Pinkner J.S., Sreedhar R. Mechanism for Sortase Localization and the. J. Bacteriol. 2009;191(10):3237. doi: 10.1128/JB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen S., Stickler D., Mobley H., Shirtliff M. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin. Microbiol. Rev. 2008;21(1):26–59. doi: 10.1128/CMR.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Behrens A.-J., Kaever V., Kazmierczak B.I. Type IV pilus assembly in Pseudomonas aeruginosa over a broad range of cyclic di-GMP concentrations. J. Bacteriol. 2012;194(16):4285–4294. doi: 10.1128/JB.00803-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jass J., Schedin S., Fällman E., Ohlsson J., Nilsson U.J., Uhlin B.E., Axner O. Physical properties of Escherichia coli P pili measured by optical tweezers. Biophys. J. 2004;87(6):4271–4283. doi: 10.1529/biophysj.104.044867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai-Larsen Y., Lüthje P., Chromek M., Peters V., Wang X., Holm Å., Kádas L., Hedlund K.-O., Johansson J., Chapman M.R., Jacobson S.H., Römling U., Agerberth B., Brauner A., Mabbott N. Uropathogenic Escherichia coli modulates immune responses and its curli fimbriae interact with the antimicrobial peptide LL-37. PLoS Pathog. 2010;6(7):e1001010. doi: 10.1371/journal.ppat.1001010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandaswamy K., Liew T.H., Wang C.Y., Huston-Warren E., Meyer-Hoffert U., Hultenby K., Schröder J.M., Caparon M.G., Normark S., Henriques-Normark B., Hultgren S.J., Kline K.A. Focal targeting by human β-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc. Natl. Acad. Sci. 2013;110(50):20230–20235. doi: 10.1073/pnas.1319066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kankainen M., Paulin L., Tynkkynen S., von Ossowski I., Reunanen J., Partanen P., Satokari R., Vesterlund S., Hendrickx A.P.A., Lebeer S., De Keersmaecker S.C.J., Vanderleyden J., Hämäläinen T., Laukkanen S., Salovuori N., Ritari J., Alatalo E., Korpela R., Mattila-Sandholm T., Lassig A., Hatakka K., Kinnunen K.T., Karjalainen H., Saxelin M., Laakso K., Surakka A., Palva A., Salusjärvi T., Auvinen P., de Vos W.M. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc. Natl. Acad. Sci. 2009;106(40):17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline K.A., Fälker S., Dahlberg S., Normark S., Henriques-Normark B. Bacterial adhesins in host-microbe interactions. Cell Host Microbe. 2009;5(6):580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- Kline K.A., Kau A.L., Chen S.L., Lim A., Pinkner J.S., Rosch J., Nallapareddy S.R., Murray B.E., Henriques-Normark B., Beatty W., Caparon M.G., Hultgren S.J. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J. Bacteriol. 2009;191(10):3237–3247. doi: 10.1128/JB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline K.A., Dodson K.W., Caparon M.G., Hultgren S.J. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 2010;18(5):224–232. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn M.J., Heuser J., Normark S., Hultgren S.J. P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature. 1992;356(6366):252–255. doi: 10.1038/356252a0. [DOI] [PubMed] [Google Scholar]
- Kuehn M.J., Ogg D.J., Kihlberg J., Slonim L.N., Flemmer K., Bergfors T., Hultgren S.J. Structural basis of pilus subunit recognition by the PapD chaperone. Science. 1993;262(5137):1234–1241. doi: 10.1126/science.7901913. [DOI] [PubMed] [Google Scholar]
- Kumari, P., Nath, Y., Murty, U. S., Ravichandiran, V., & Mohan, U. (2020). Sortase A mediated bioconjugation of common epitopes decreases biofilm formation in Staphylococcus aureus. Frontiers in microbiology, 1702. [DOI] [PMC free article] [PubMed]
- Lane M., Mobley H. Role of P-fimbrial-mediated adherence in pyelonephritis and persistence of uropathogenic Escherichia coli (UPEC) in the mammalian kidney. Kidney Int. 2007;72(1):19–25. doi: 10.1038/sj.ki.5002230. [DOI] [PubMed] [Google Scholar]
- Lebeer S., Claes I., Tytgat H.L.P., Verhoeven T.L.A., Marien E., von Ossowski I., Reunanen J., Palva A., de Vos W.M., De Keersmaecker S.C.J., Vanderleyden J. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 2012;78(1):185–193. doi: 10.1128/AEM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Sun H., Ma X., Lu A., Lux R., Zusman D., Shi W. Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. 2003;100(9):5443–5448. doi: 10.1073/pnas.0836639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim M.S., Ng D., Zong Z., Arvai A.S., Taylor R.K., Tainer J.A., Craig L. Vibrio cholerae El Tor TcpA crystal structure and mechanism for pilus-mediated microcolony formation. Mol. Microbiol. 2010;77(3):755–770. doi: 10.1111/j.1365-2958.2010.07244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg F., Tennent J., Hultgren S., Lund B., Normark S. PapD, a periplasmic transport protein in P-pilus biogenesis. J. Bacteriol. 1989;171(11):6052–6058. doi: 10.1128/jb.171.11.6052-6058.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A., Swierczynski A., Das A., Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16(1):33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini L.A., DeDent A.C., Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2006;70(1):192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell S.A., Amer B.R., Muroski J., Fu J., Chang C., Ogorzalek Loo R.R., Loo J.A., Osipiuk J., Ton-That H., Clubb R.T. Protein labeling via a specific lysine-isopeptide bond using the pilin polymerizing sortase from Corynebacterium diphtheriae. J. Am. Chem. Soc. 2018;140(27):8420–8423. doi: 10.1021/jacs.8b05200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCrate O.A., Zhou X., Reichhardt C., Cegelski L. Sum of the parts: composition and architecture of the bacterial extracellular matrix. J. Mol. Biol. 2013;425(22):4286–4294. doi: 10.1016/j.jmb.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire F., Vatter A., Baros J., Arnold J. Mechanism of coaggregation between Actinomyces viscosus T14V and Streptococcus sanguis 34. Infect. Immun. 1978;21(3):978–988. doi: 10.1128/iai.21.3.978-988.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megli C.J., Taylor R.K. Secretion of TcpF by the Vibrio cholerae toxin-coregulated pilus biogenesis apparatus requires an N-terminal determinant. J. Bacteriol. 2013;195(12):2718–2727. doi: 10.1128/JB.01122-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A., Wu C., Yang J., Cisar J.O., Das A., Ton-That H. The Actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol. Microbiol. 2010;77(4):841–854. doi: 10.1111/j.1365-2958.2010.07252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley H.L., Donnenberg M.S., Hagan E.C. Uropathogenic Escherichia coli. EcoSal Plus. 2009;3(2) doi: 10.1128/ecosalplus.8.6.1.3. [DOI] [PubMed] [Google Scholar]
- Myers G.S.A., Rasko D.A., Cheung J.K., Ravel J., Seshadri R., DeBoy R.T., Ren Q., Varga J., Awad M.M., Brinkac L.M., Daugherty S.C., Haft D.H., Dodson R.J., Madupu R., Nelson W.C., Rosovitz M.J., Sullivan S.A., Khouri H., Dimitrov G.I., Watkins K.L., Mulligan S., Benton J., Radune D., Fisher D.J., Atkins H.S., Hiscox T., Jost B.H., Billington S.J., Songer J.G., McClane B.A., Titball R.W., Rood J.I., Melville S.B., Paulsen I.T. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 2006;16(8):1031–1040. doi: 10.1101/gr.5238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy S.R., Singh K.V., Sillanpää J., Garsin D.A., Höök M., Erlandsen S.L., Murray B.E. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J. Clin. Investig. 2006;116(10):2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy S.R., Singh K.V., Sillanpää J., Zhao M., Murray B.E., Camilli A. Relative contributions of Ebp Pili and the collagen adhesin ace to host extracellular matrix protein adherence and experimental urinary tract infection by Enterococcus faecalis OG1RF. Infect. Immun. 2011;79(7):2901–2910. doi: 10.1128/IAI.00038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenninger A.A., Robinson L.S., Hultgren S.J. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc. Natl. Acad. Sci. 2009;106(3):900–905. doi: 10.1073/pnas.0812143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H.V., Flores-Mireles A.L., Kau A.L., Kline K.A., Pinkner J.S., Neiers F., Normark S., Henriques-Normark B., Caparon M.G., Hultgren S.J. Pilin and sortase residues critical for endocarditis-and biofilm-associated pilus biogenesis in Enterococcus faecalis. J. Bacteriol. 2013;195(19):4484–4495. doi: 10.1128/JB.00451-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M., Vetsch M., Puorger C., Jelesarov I., Glockshuber R. Identification and characterization of the chaperone-subunit complex-binding domain from the type 1 pilus assembly platform FimD. J. Mol. Biol. 2003;330(3):513–525. doi: 10.1016/s0022-2836(03)00591-6. [DOI] [PubMed] [Google Scholar]
- Nishiyama M., Ishikawa T., Rechsteiner H., Glockshuber R. Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science. 2008;320(5874):376–379. doi: 10.1126/science.1154994. [DOI] [PubMed] [Google Scholar]
- Nishiyama K., Sugiyama M., Mukai T. Adhesion properties of lactic acid bacteria on intestinal mucin. Microorganisms. 2016;4(3):34. doi: 10.3390/microorganisms4030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nölling Jörk, Breton G., Omelchenko M.V., Makarova K.S., Zeng Q., Gibson R., Lee H.M., Dubois JoAnn, Qiu D., Hitti J., Wolf Y.I., Tatusov R.L., Sabathe F., Doucette-Stamm L., Soucaille P., Daly M.J., Bennett G.N., Koonin E.V., Smith D.R. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J. Bacteriol. 2001;183(16):4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsén A., Jonsson A., Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338(6217):652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- Paterson G.K., Mitchell T.J. The biology of Gram-positive sortase enzymes. Trends Microbiol. 2004;12(2):89–95. doi: 10.1016/j.tim.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Pawar D., Rossman M., Chen J. Role of curli fimbriae in mediating the cells of enterohaemorrhagic Escherichia coli to attach to abiotic surfaces. J. Appl. Microbiol. 2005;99(2):418–425. doi: 10.1111/j.1365-2672.2005.02499.x. [DOI] [PubMed] [Google Scholar]
- Persat A., Inclan Y.F., Engel J.N., Stone H.A., Gitai Z. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 2015;112(24):7563–7568. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenbrink K.H., Sundberg E.J. Motility and adhesion through type IV pili in Gram-positive bacteria. Biochem. Soc. Trans. 2016;44(6):1659–1666. doi: 10.1042/BST20160221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft T., Baker E. Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell. Mol. Life Sci. 2009;66(4):613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez N.A., Das A., Ton-That H. New paradigms of pilus assembly mechanisms in gram-positive actinobacteria. Trends Microbiol. 2020;28(12):999–1009. doi: 10.1016/j.tim.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon-Robinson M.E., Wu C., Mishra A., Chang C., Bier N., Das A., Ton-That H. Pilus hijacking by a bacterial coaggregation factor critical for oral biofilm development. Proc. Natl. Acad. Sci. 2014;111(10):3835–3840. doi: 10.1073/pnas.1321417111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon-Robinson M.E., Osipiuk J., Chang C., Wu C., Jooya N., Joachimiak A., Das A., Ton-That H. A disulfide bond-forming machine is linked to the sortase-mediated pilus assembly pathway in the Gram-positive bacterium Actinomyces oris. J. Biol. Chem. 2015;290(35):21393–21405. doi: 10.1074/jbc.M115.672253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon-Robinson M.E., Osipiuk J., Jooya N., Chang C., Joachimiak A., Das A., Ton-That H. A thiol-disulfide oxidoreductase of the G ram-positive pathogen C orynebacterium diphtheriae is essential for viability, pilus assembly, toxin production and virulence. Mol. Microbiol. 2015;98(6):1037–1050. doi: 10.1111/mmi.13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaut H., Rose R.J., Hannan T.J., Hultgren S.J., Radford S.E., Ashcroft A.E., Waksman G. Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted β strand displacement mechanism. Mol. Cell. 2006;22(6):831–842. doi: 10.1016/j.molcel.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Reunanen J., von Ossowski I., Hendrickx A.P., Palva A., de Vos W.M. Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 2012;78(7):2337–2344. doi: 10.1128/AEM.07047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose R.J., Welsh T.S., Waksman G., Ashcroft A.E., Radford S.E., Paci E. Donor-strand exchange in chaperone-assisted pilus assembly revealed in atomic detail by molecular dynamics. J. Mol. Biol. 2008;375(4):908–919. doi: 10.1016/j.jmb.2007.10.077. [DOI] [PubMed] [Google Scholar]
- Rudel T., Scheuerpflug I., Meyer T.F. Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature. 1995;373(6512):357–359. doi: 10.1038/373357a0. [DOI] [PubMed] [Google Scholar]
- Ruhl S., Sandberg A., Cole M., Cisar J. Recognition of immunoglobulin A1 by oral actinomyces and streptococcal lectins. Infect. Immun. 1996;64(12):5421–5424. doi: 10.1128/iai.64.12.5421-5424.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez B.C., Chang C., Wu C., Tran B., Ton-That H. Electron transport chain is biochemically linked to pilus assembly required for polymicrobial interactions and biofilm formation in the Gram-positive actinobacterium Actinomyces oris. MBio. 2017;8(3):e00399–00317. doi: 10.1128/mBio.00399-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarowar S., Hu O.J., Werneburg G.T., Thanassi D.G., Li H., Christie P.J. The Escherichia coli P and type 1 pilus assembly chaperones PapD and FimC are monomeric in solution. J. Bacteriol. 2016;198(17):2360–2369. doi: 10.1128/JB.00366-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer F.G., Futterer K., Pinkner J.S., Dodson K.W., Hultgren S.J., Waksman G. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285(5430):1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- Segers M.E., Lebeer S. Towards a better understanding of Lactobacillus rhamnosus GG-host interactions. Microb. Cell Fact. 2014;13(1):1–16. doi: 10.1186/1475-2859-13-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewmaker F., McGlinchey R.P., Thurber K.R., McPhie P., Dyda F., Tycko R., Wickner R.B. The functional curli amyloid is not based on in-register parallel β-sheet structure. J. Biol. Chem. 2009;284(37):25065–25076. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W., Sun H. Type IV pilus-dependent motility and its possible role in bacterial pathogenesis. Infect. Immun. 2002;70(1):1–4. doi: 10.1128/IAI.70.1.1-4.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Ohtani K., Hirakawa H., Ohshima K., Yamashita A., Shiba T., Ogasawara N., Hattori M., Kuhara S., Hayashi H. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. 2002;99(2):996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji M., Naito M., Yukitake H., Sato K., Sakai E., Ohara N., Nakayama K. The major structural components of two cell surface filaments of Porphyromonas gingivalis are matured through lipoprotein precursors. Mol. Microbiol. 2004;52(5):1513–1525. doi: 10.1111/j.1365-2958.2004.04105.x. [DOI] [PubMed] [Google Scholar]
- Shoji M., Shibata S., Sueyoshi T., Naito M., Nakayama K. Biogenesis of type V pili. Microbiol. Immunol. 2020;64(10):643–656. doi: 10.1111/1348-0421.12838. [DOI] [PubMed] [Google Scholar]
- Siegel S.D., Liu J., Ton-That H. Biogenesis of the Gram-positive bacterial cell envelope. Curr. Opin. Microbiol. 2016;34:31–37. doi: 10.1016/j.mib.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel S.D., Wu C., Ton-That H., Schneewind O. A type I signal peptidase is required for pilus assembly in the Gram-positive, biofilm-forming bacterium Actinomyces oris. J. Bacteriol. 2016;198(15):2064–2073. doi: 10.1128/JB.00353-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillanpää J., Chang C., Singh K.V., Montealegre M.C., Nallapareddy S.R., Harvey B.R., Ton-That H., Murray B.E., Hancock L. Contribution of individual Ebp pilus subunits of Enterococcus faecalis OG1RF to pilus biogenesis, biofilm formation and urinary tract infection. PLoS ONE. 2013;8(7):e68813. doi: 10.1371/journal.pone.0068813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopek R., Liljemark W.F., Bloomquist C., Rudney J.D. Dental plaque development on defined streptococcal surfaces. Oral Microbiol. Immunol. 1993;8(1):16–23. doi: 10.1111/j.1399-302x.1993.tb00537.x. [DOI] [PubMed] [Google Scholar]
- Sokurenko E.V., Chesnokova V., Dykhuizen D.E., Ofek I., Wu X.-R., Krogfelt K.A., Struve C., Schembri M.A., Hasty D.L. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc. Natl. Acad. Sci. 1998;95(15):8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirig T., Weiner E.M., Clubb R.T. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 2011;82(5):1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos C., Hendrixson D.R., Thanassi D.G., Hultgren S.J., Geme J.W.S., III, Curtiss R., III Secretion of virulence determinants by the general secretory pathway in gram-negative pathogens: an evolving story. Microbes Infect. 2000;2(9):1061–1072. doi: 10.1016/s1286-4579(00)01260-0. [DOI] [PubMed] [Google Scholar]
- Stepanova N. How Advanced Is Our Understanding of the Role of Intestinal Barrier Dysfunction in the Pathogenesis of Recurrent Urinary Tract Infections. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.780122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strömberg N., Karlsson K.-A. Characterization of the binding of Actinomyces naeslundii (ATCC 12104) and Actinomyces viscosus (ATCC 19246) to glycosphingolipids, using a solid-phase overlay approach. J. Biol. Chem. 1990;265(19):11251–11258. [PubMed] [Google Scholar]
- Susmitha A., Bajaj H., Madhavan Nampoothiri K. The divergent roles of sortase in the biology of Gram-positive bacteria. The Cell Surface. 2021;7:100055. doi: 10.1016/j.tcsw.2021.100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan A., Mandlik A., Swierczynski A., Gaspar A., Das A., Ton-That H. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol. Microbiol. 2007;66(4):961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taha M., So M., Seifert H., Billyard E., Marchal C. Pilin expression in Neisseria gonorrhoeae is under both positive and negative transcriptional control. The EMBO journal. 1988;7(13):4367–4378. doi: 10.1002/j.1460-2075.1988.tb03335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi D.G., Stathopoulos C., Dodson K., Geiger D., Hultgren S.J. Bacterial outer membrane ushers contain distinct targeting and assembly domains for pilus biogenesis. J. Bacteriol. 2002;184(22):6260–6269. doi: 10.1128/JB.184.22.6260-6269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanassi D.G., Bliska J.B., Christie P.J. Surface organelles assembled by secretion systems of Gram-negative bacteria: diversity in structure and function. FEMS Microbiol. Rev. 2012;36(6):1046–1082. doi: 10.1111/j.1574-6976.2012.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiba M.R., Yano T., Leite D., d. S. Genotypic characterization of virulence factors in Escherichia coli strains from patients with cystitis. Revista do Instituto de Medicina Tropical de São Paulo. 2008;50(5):255–260. doi: 10.1590/s0036-46652008000500001. [DOI] [PubMed] [Google Scholar]
- Tønjum T., Freitag N.E., Namork E., Koomey M. Identification and characterization of pilG, a highly conserved pilus-assembly gene in pathogenic Neisseria. Mol. Microbiol. 1995;16(3):451–464. doi: 10.1111/j.1365-2958.1995.tb02410.x. [DOI] [PubMed] [Google Scholar]
- Ton-That H., Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 2003;50(4):1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- Ton-That H., Marraffini L.A., Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 2004;53(1):251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- Totsika M., Heras B., Wurpel D.J., Schembri M.A. Characterization of two homologous disulfide bond systems involved in virulence factor biogenesis in uropathogenic Escherichia coli CFT073. J. Bacteriol. 2009;191(12):3901–3908. doi: 10.1128/JB.00143-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gerven N., Klein R.D., Hultgren S.J., Remaut H. Bacterial amyloid formation: structural insights into curli biogensis. Trends Microbiol. 2015;23(11):693–706. doi: 10.1016/j.tim.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J.J., Nguyen V., O'Brien D.K., Rodgers K., Walker R.A., Melville S.B. Type IV pili-dependent gliding motility in the Gram-positive pathogen Clostridium perfringens and other Clostridia. Mol. Microbiol. 2006;62(3):680–694. doi: 10.1111/j.1365-2958.2006.05414.x. [DOI] [PubMed] [Google Scholar]
- Verger D., Miller E., Remaut H., Waksman G., Hultgren S. Molecular mechanism of P pilus termination in uropathogenic Escherichia coli. EMBO Rep. 2006;7(12):1228–1232. doi: 10.1038/sj.embor.7400833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger D., Rose R.J., Paci E., Costakes G., Daviter T., Hultgren S., Remaut H., Ashcroft A.E., Radford S.E., Waksman G. Structural determinants of polymerization reactivity of the P pilus adaptor subunit PapF. Structure. 2008;16(11):1724–1731. doi: 10.1016/j.str.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Waksman G. Structural and molecular biology of a protein-polymerizing nanomachine for pilus biogenesis. J. Mol. Biol. 2017;429(17):2654–2666. doi: 10.1016/j.jmb.2017.05.016. [DOI] [PubMed] [Google Scholar]
- Walczak M.J., Puorger C., Glockshuber R., Wider G. Intramolecular donor strand complementation in the E. coli type 1 pilus subunit FimA explains the existence of FimA monomers as off-pathway products of pilus assembly that inhibit host cell apoptosis. J. Mol. Biol. 2014;426(3):542–549. doi: 10.1016/j.jmb.2013.10.029. [DOI] [PubMed] [Google Scholar]
- Werneburg G.T., Thanassi D.G., Donnenberg M.S. Pili assembled by the chaperone/usher pathway in Escherichia coli and Salmonella. EcoSal Plus. 2018;8(1) doi: 10.1128/ecosalplus.esp-0007-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M., Lauer P., Park H.S., Brossay L., Hébert J., Koomey M. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 1998;29(1):321–330. doi: 10.1046/j.1365-2958.1998.00935.x. [DOI] [PubMed] [Google Scholar]
- Wolfgang M., Park H.-S., Hayes S.F., Van Putten J.P., Koomey M. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. 1998;95(25):14973–14978. doi: 10.1073/pnas.95.25.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfgang M., van Putten J.P., Hayes S.F., Dorward D., Koomey M. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 2000;19(23):6408–6418. doi: 10.1093/emboj/19.23.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Mishra A., Reardon M.E., Huang I.-H., Counts S.C., Das A., Ton-That H. Structural determinants of Actinomyces sortase SrtC2 required for membrane localization and assembly of type 2 fimbriae for interbacterial coaggregation and oral biofilm formation. J. Bacteriol. 2012;194(10):2531–2539. doi: 10.1128/JB.00093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Huang I.H., Chang C., Reardon-Robinson M.E., Das A., Ton-That H. Lethality of sortase depletion in Actinomyces oris caused by excessive membrane accumulation of a surface glycoprotein. Mol. Microbiol. 2014;94(6):1227–1241. doi: 10.1111/mmi.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., Shoji M., Shibata S., Naito M., Sato K., Elsliger M.-A., Grant J., Axelrod H., Chiu H.-J., Farr C., Jaroszewski L., Knuth M., Deacon A., Godzik A., Lesley S., Curtis M., Nakayama K., Wilson I. A distinct type of pilus from the human microbiome. Cell. 2016;165(3):690–703. doi: 10.1016/j.cell.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa R., Honda E. Presence of pili in species of human and animal parasites and pathogens of the genuscorynebacterium. Infect. Immun. 1976;13(4):1293–1295. doi: 10.1128/iai.13.4.1293-1295.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura F., Takahashi K., Nodasaka Y., Suzuki T. Purification and characterization of a novel type of fimbriae from the oral anaerobe Bacteroides gingivalis. J. Bacteriol. 1984;160(3):949–957. doi: 10.1128/jb.160.3.949-957.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura F., Murakami Y., Nishikawa K., Hasegawa Y., Kawaminami S. Surface components of Porphyromonas gingivalis. J. Periodontal Res. 2009;44(1):1–12. doi: 10.1111/j.1600-0765.2008.01135.x. [DOI] [PubMed] [Google Scholar]
- Zavialov A.V., Berglund J., Pudney A.F., Fooks L.J., Ibrahim T.M., MacIntyre S., Knight S.D. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell. 2003;113(5):587–596. doi: 10.1016/s0092-8674(03)00351-9. [DOI] [PubMed] [Google Scholar]
- Zhou X., Li Y., Peng X., Ren B., Xiao L., Li M., Guo Q. From Healthy Microflora to Disease; Atlas of Oral Microbiology: 2021. Basic Biology of Oral Microbes; pp. 1–24. [Google Scholar]
- Zijnge V., van Leeuwen M.B.M., Degener J.E., Abbas F., Thurnheer T., Gmür R., Harmsen M., H. j. Oral biofilm architecture on natural teeth. PLoS ONE. 2010;5(2) doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X., Nimtz M., Rohde M., Bokranz W., Römling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 2001;39(6):1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.