Summary
The regulation of gene expression via protein translation is critical for growth, development, and stress response. While puromycin-based techniques have been used to quantify protein translation in C. elegans, they have been limited to using lysate from whole worms. To achieve tissue-specific quantification of ribosome activity in intact C. elegans, we report the application of O-propargyl-puromycin in a cuticle defective mutant followed by conjugation of an azide fluorophore for detection using fluorescent confocal microscopy. We apply this technique to quantify translation in response to heat shock, cycloheximide, or knockdown of translation factors. Furthermore, we demonstrate that O-propargyl-puromycin can be used to quantify translation between tissues or within a tissue like the germline. This technique is expected to have a broad range of applications in determining how protein translation is altered in different tissues in response to stress or gene knockdowns or with age.
Keywords: translation, puromycin, C. elegans, longevity, tissue specific, fluorescence imaging, germline, cuticle, click chemistry, initiation factors, elongation factors
Graphical abstract
Highlights
-
•
OP-puromycin incorporation is used to measure protein translation in intact C. elegans
-
•
A cuticle defective mutant allows for uptake of OP-puromycin
-
•
Labeling and quantification using “click chemistry” and fluorescence microscopy
-
•
Fluorescently tagged RPS-6 can be used to measure ribosome abundance
Motivation
In C. elegans and other model organisms, many pro-longevity treatments, like caloric restriction or inhibition of the target of rapamycin (TOR) pathway, result in decreased protein translation. There is a growing appreciation of the tissue-specific contributions of these molecular mechanisms to organismal aging, yet quantification of translation in this model organism has traditionally been limited to assaying lysates from pools of worms. Here, we present a method using OP-puromycin, “click chemistry,” and fluorescence microscopy to quantify relative protein translation rates across tissues in intact C. elegans.
Somers et al. present a method that allows protein translation to be quantified in any tissue of interest in C. elegans. The authors use this fluorescence-microscopy-based method to compare protein translation under different stresses, between tissues, and between the distal and proximal portions of the germline.
Introduction
The regulation of mRNA translation is necessary for organismal growth and survival by producing the necessary proteins in the right amount. Rapid growth requires high levels of translation that can consume up to 50% of the energy available (Proud, 2002). Conversely, translation is downregulated under periods of stress to conserve energy resources, maintain proteostasis, and increase survival (Rollins and Rogers, 2017). For example, various forms of caloric restriction extend longevity and decrease global translation rates in C. elegans (Hansen et al., 2007; Rollins et al., 2019). Many pathways that regulate protein translation in response to caloric restriction and other stressors are well conserved among eukaryotes. These pathways include the target of rapamycin pathway, insulin-like signaling, and the integrated stress response. Environmental or genetic perturbation of these pathways can decrease global protein synthesis and extend longevity (Derisbourg et al., 2021; Hansen et al., 2007; Pan et al., 2007). However, there is a growing appreciation that contributions to organismal longevity can arise from tissue-specific responses (Libina et al., 2003; Tain et al., 2021; Srivastava et al., 2020). This phenomenon has led to a mounting interest in quantifying protein translation rates at the tissue and cellular level in response to a variety of stimuli known to modulate resilience and longevity.
The discovery that puromycin incorporation can be used as a proxy for protein translation rates has enabled translation-rate comparisons among cell types under a variety of experimental conditions. A popular variant of this assay consists of treating samples with puromycin at low doses that do not appreciably inhibit protein translation and then quantifying the relative amount of puromycin incorporation using an anti-puromycin antibody (Goodman and Hornberger, 2013; Schmidt et al., 2009). Variations on this technique have been used to quantify protein translation rates between tissues of puromycin-injected mice (Hidalgo San Jose and Signer, 2019). The development of O-propargyl-puromycin (OPP), a puromycin analog containing an alkene group that allows it to be tagged with a suite of “click-chemistry”-compatible azide fluorophores, has enabled the puromycylation of peptides to be quantified using fluorescence microscopy (Liu et al., 2012) or flow cytometry (Hidalgo San Jose and Signer, 2019) and has reduced the sample preparation otherwise needed to use antibodies in permeabilized cells.
In C. elegans, puromycin assays have typically been used on whole animals treated with 1 mM of puromycin to quantify the mean protein translation rate across all cell types and tissues via western blot. This technique has been used to demonstrate no appreciable decrease in global protein synthesis in cey-1;cey-4 double mutants (Arnold et al., 2014) or after RNAi knockdown of rRNA methyltransferases nsun-1 or nsun-5 (Heissenberger et al., 2020). However, it has been used to show that protein translation is reduced upon knockdown of rRNA-processing enzyme fibrillarin (fib-1) (Tiku et al., 2018) and in mutants of the initiation factor iftb-1(wrm53) and target of rapamycin (TOR) pathway kinase rsks-1(sv31) (Derisbourg et al., 2021). While such results are useful for determining whether specific genes play a role in regulating global protein translation rates, they offer no insight as to which tissues and cell types are being affected. To quantify relative protein translation rates across all tissues in intact C. elegans, we present the following methodology in which worms are treated with OPP, fixed, and puromycylated proteins are fluorescently labeled using click chemistry before being quantified using fluorescence microscopy. We then apply this technique to quantify relative protein translation across whole worms treated with heat shock or with RNAi against translation factors ifg-1 and iff-1. To demonstrate the power of this technique in quantifying tissue-specific translation, we use 3D confocal reconstructions of the germline and show that translation is higher in the distal germline compared with the proximal germline. Furthermore, the difference in translation between the distal and proximal germline is maintained even when overall translation is reduced due to knockdown of ifg-1 or iff-1.
Results
Optimization of puromycin uptake into live C. elegans
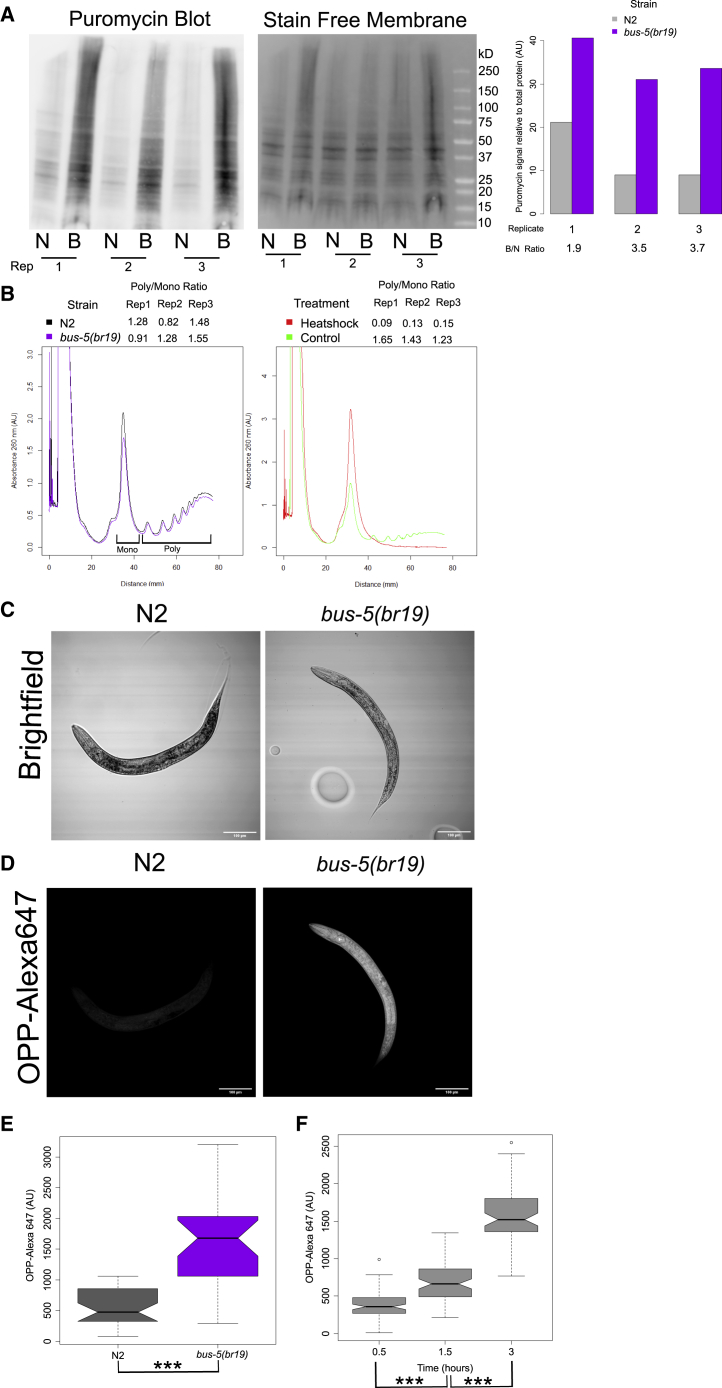
For consistent puromycin labeling across tissue and cell types in C. elegans, we set out to maximize its uptake and detection. To this end, we tested the use of a C. elegans cuticle-defective mutant, different OPP incubation times, different extents of crosslinking, and the effect of live bacteria in the OPP assay. A common mode of entry of drugs into the worm is via ingestion. However, this can be problematic when testing treatments that alter the pumping rate, as the treatment itself may change the uptake of puromycin based on feeding behaviors instead of translation rates. In wild-type C. elegans, the cuticle impedes the uptake of chemicals they are exposed to. Using the mutant strain DC19 (bus-5(br19) X), which was previously shown to have enhanced cuticle permeability (Xiong et al., 2017), we tested whether puromycin incorporation rates are enhanced in this strain. Using a previously established puromycin western blot assay (Arnold et al., 2014), larval stage 4 (L4) worms were incubated in the presence of 1 mM puromycin for 3 h followed by lysis and protein extraction. As expected, puromycin signal on the western blot was greater (mean of 3-fold) in bus-5(br19) than in N2 worms (Figure 1A). However, the possibility remained that this result was due to higher basal translation rates in bus-5(br19) mutants instead of enhanced uptake. To distinguish between these possibilities, we performed polysome profiling on N2 and bus-5(br19) worms (Figure 1B [left]). The ratio of area under the curve of the polysomal mRNA over the monosomal bound mRNA was used to compare the translational activity of bus-5(br19) and N2 worms. The average ratio for bus-5(br19) and N2 was 1.24 and 1.19, respectively. As heat shock inhibits translation, the shift of signal from polysomes to monosomes due to a 1-h heat shock in N2 worms is included as a positive control (Figure 1B [right]) for comparison to our results with N2 and bus-5(br19). The polysome-to-monosome ratio decreased from an average of 1.43 to 0.123 due to the heat shock. Thus, the increased puromycin signal from the puromycin western blot in bus-5(br19) was due to increased uptake and not increased translation rates caused by the bus-5 mutation.
Figure 1.
The bus-5(br19) mutant has increased uptake of OP-puromycin compared with wild-type N2
(A) Western blot of puromycin incorporation in L4 N2 and bus-5(br19) mutants (left). Stain-free total protein measurement served as the loading control (middle). Quantification of puromycin signal corrected by loading control is shown; n = 3 (right). The ratio of total protein to signal is given. B, bus-5(br19); N, N2; Rep, replicate.
(B) Polysome profiles comparing N2 with bus-5(br19) mutants (left) or comparing heat shocked N2 worms with unstressed controls (right). The ratio of the area under the curve for polysomes and monosomes (P/M) is given above; n = 3. The areas quantified for monosomes (mono) and polysomes (poly) are indicated underneath.
(C and D) Bright-field (C) and confocal (D) images of Alexa-Fluor-647-labeled OPP in N2 (left) and bus-5(br19) (right).
(E) Quantification of OPP fluorescence intensity from treated N2 and bus-5(br19). n = 30 from three independent replications.
(F) Quantification of OPP fluorescence intensity from bus-5(br19) mutant at the times indicated. n = 69 summed across three independent replicates.
Notches in box plots indicate the estimated 95% confidence interval of the median value (black line). ∗∗∗p < 0.001. Wilcoxon rank sum test.
We next compared the uptake of 10 μM OPP between N2 and bus-5(br19) strains after 3 h using click chemistry and fluorescence microscopy (Figures 1C–1E; additional representative images in Figure S2A). Quantification of the OPP-Alexa Fluor 647 signal was performed on a single plane transecting the dorsoventral axis of the worm to give a readout of global translation. The lower concentration of 10 μM OPP was tested because concentrations in this range are commonly used for labeling in mammalian cell culture (Barrett et al., 2016; Henrich, 2016). Quantification of the mean fluorescence intensity showed that bus-5(br19) worms had 2.9-fold significantly higher OPP incorporation after 3 h compared with wild-type N2 worms (Figure 1E; p = 9.5e−9; Wilcoxon rank sum test), like the results from the puromycin western blot assay (Figure 1A). Furthermore, the presence of florescence was more evenly distributed in bus-5(br19) compared with N2 (Figures 1C and 1D). To quantify this, the ratio of signal intensity from the midsection of the worm over the signal from either the head or tail regions was compared between N2 and bus-5(br19) (Figure S1A). The difference between the midsection and head signal in the bus-5 mutant was 1.12, significantly lower than the 1.4 ratio in N2 (Figure S1B; p = 0.002; one-way ANOVA). In a similar manner, the ratio between the midsection and the tail was also significantly lower (Figure S1C; p = 0.002; one-way ANOVA) in bus-5 mutant (1.3) as compared with N2 (1.8). Thus, bus-5(br19) worms have increased permeability to OPP and puromycin compared with N2 worms.
Having determined that the bus-5(br19) mutant had increased uptake of OPP, we wanted to find the optimal incubation time in 10 μM OPP. Keeping the incubation time and dose of OPP as low as possible is imperative to minimize the impact of puromycin on global translation in C. elegans. We tested incubation times of 0.5, 1.5, and 3 h. At all three time points, fluorescently labeled puromycin was detectable above background and the overall intensity significantly increased between 0.5 and 1.5 h (Figure 1F; p = 2.71e−14; Wilcoxon rank sum test; representative images in Figure S2B) and between the 1.5- and 3-h incubations (Figure 1F; p = 2.20e−16; Wilcoxon rank sum test). To determine whether the OPP incorporation was evenly distributed at each time point, the ratio of signal between the midsection of the worm compared with the extremities was quantified (Figure S1D). OPP incorporation was the most variable after 0.5 h in both the head and tail sections (Figures S1D and S1F). After 1.5 h, OPP incorporation was more even between the midsection and the extremities but with some outliers, with twice as much OPP in the midsection compared with the head or tail regions (Figures S1E and S1F). After 3 h, OPP was evenly incorporated between the midsection and the head and tail regions (Figures S1E and S1F; additional images in Figure S6B). Based on the higher signal and even incorporation of OPP-Alexa Fluor 647, we determined that 3 h was an optimal incubation time with OPP for this assay.
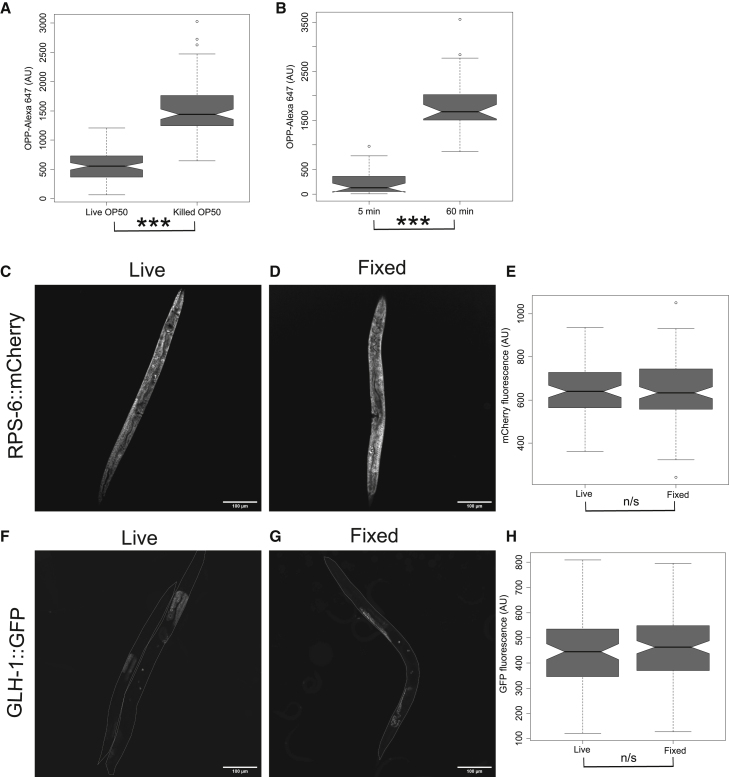
The typical food source for C. elegans in research is the E. coli strain OP50. As puromycin inhibits translation in both eukaryotes and prokaryotes, the presence of live bacteria may compete with the worms in the uptake of puromycin. To test this, we performed the OPP assay in bus-5(br19) in the presence of live OP50 or paraformaldehyde (PFA)-killed OP50 (Beydoun et al., 2021). Compared with PFA-killed OP50, live OP50 had 63% lower puromycin signal (Figure 2A; p = 3.0e−28; Wilcoxon rank sum test; representative images in Figure S2C). Thus, the presence of live bacteria interferes with the uptake of puromycin into worms, and we recommend the use of killed bacteria when conducting this assay.
Figure 2.
Killed OP50 food source during OPP incubation and PFA fixation after OPP treatment enhanced detection
(A) Quantification of OPP fluorescence intensity in bus-5(br19) using live or PFA-killed OP50 E. coli as a food source during the incubation. n = 171 summed across three independent replicates.
(B) Effect of 5-min and 60-min fixation in 4% PFA on retainment of OPP fluorescence intensity in bus-5(br19). n = 84 summed across three independent replicates.
(C and D) Confocal image of RPS-6::mCherry in (C) live and (D) PFA-fixed worms with the bus-5(br19) background.
(E) Quantification of RPS-6::mCherry fluorescence in live worms or with PFA fixation. n = 84 summed across three independent replicates.
(F and G) Confocal images of GLH-1::GFP in (F) live and (G) PFA-fixed worms in the bus-5(br19) background.
(H) Quantification of GLH-1::mCherry fluorescence in live worms or with PFA fixation. n = 87 summed across three independent replicates.
Notches in box plots indicate the estimated 95% confidence interval of the median value (black line). n.s., not significant p > 0.05 and ∗∗∗p < 0.001. Wilcoxon rank sum test is shown.
Incorporation of OPP into nascent peptides triggers the release of the puromycylated peptide from the ribosome (Enam et al., 2020; Hobson et al., 2020). To prevent these puromycylated peptides from being washed away from their cells of origin during sample preparation, PFA fixation was used to crosslink puromycylated peptides in place for subsequent imaging. Fixation with 4% PFA for 1 h at 10°C resulted in a >7-fold increase in the signal retained compared with a 5-min fixation that was sufficient to kill the worms (Figure 2B; p = 2.6e−30; Wilcoxon rank sum test; quantified representative images are found in Figure S3A). Therefore, crosslinking greatly enhances the retainment of puromycylated peptides within the sample. To be able to colocalize the labeled OPP signal with fluorescently tagged proteins, we additionally wanted to ensure that the PFA fixation did not cause an appreciable loss of fluorescence, as has been reported (Schnell et al., 2012). Using a strain expressing an endogenously mCherry-tagged ribosomal 40S subunit (rps-6::mCherry) in the bus-5 background, we compared fluorescence intensity between live (Figure 2C; additional representative images in Figure S3B) and PFA-treated samples (Figure 2D; additional representative images in Figure S3B). There was no appreciable change in mCherry fluorescence between live and fixed worms (Figure 2E; p = 0.98; Wilcoxon rank sum test). In addition, we used a strain expressing a germline-specific marker fused to GFP (glh-1::GFP; Andralojc et al., 2017) in the bus-5 background to compare GFP fluorescence between live (Figure 2F; additional representative images in Figure S3C) and fixed conditions (Figure 2G; additional representative images in Figure S3C). As with mCherry, there was no appreciable difference in GFP intensity between the two conditions (Figure 2H; p = 0.33; Wilcoxon rank sum test). Thus, our fixation step for this assay is compatible for use with strains expressing mCherry, GFP, and likely other fluorescent tags.
Validation of OPP fluorescent signal as measure of relative translation rates
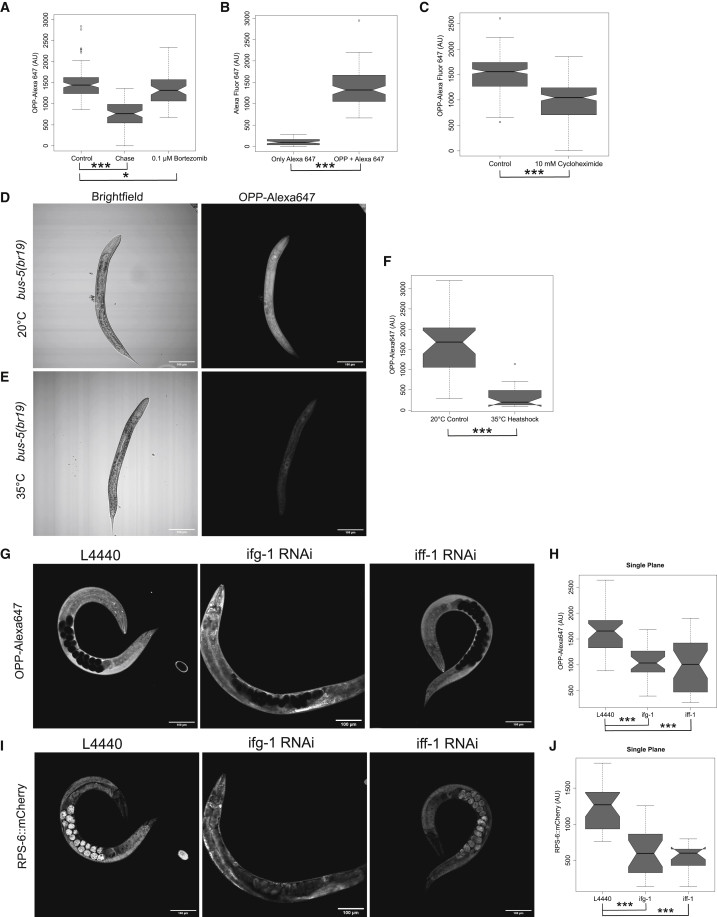
Having optimized the conditions for efficient uptake and retention of puromycin into C. elegans tissues, we set out to validate that the fluorescent signal was indicative of puromycin incorporation and thus of relative protein translation rates. Since puromycylated peptides resulting from premature termination of translation are degraded by the proteasome (Liu et al., 2012), we would expect the signal to decrease over time after the worms are removed from the presence of OPP. Furthermore, we would expect the OPP signal to be maintained in the presence of the proteasomal inhibitor bortezomib. In line with these expectations, the mean puromycin signal decreased significatively by 51% 1 h after removal of OPP relative to controls that were immediately fixed (Figure 3A; p = 4.80e−24; Wilcoxon rank sum test; quantified representative images are found in Figure S4A). In comparison, the presence of bortezomib during the 1 h incubation without OPP resulted in only a 0.7 % decrease in signal (Figure 3A; p = 0.035; Wilcoxon rank sum test). As there was still residual fluorescence remaining after the OPP had been removed for 1 h (Figure 3A), it is possible that the fluorescence was due to unreacted Alexa Fluor 647-azide that had not been washed out or due to unspecific click chemistry. However, performing the assay in the absence of OPP showed only minimally detectable Alexa Fluor 647 fluorescence (Figure 3B; p = 2.2e−16; Wilcoxon rank sum test; quantified representative images in Figure S6A), demonstrating that our washing is sufficient to remove any excess fluorophore and that the fluorophore azide does not appreciable react with endogenous macromolecules in the worm. Therefore, we conclude that the fluorescent signal quantified is specific to puromycylated peptides.
Figure 3.
Validation of OPP fluorescence signal as measure of relative translation rates in whole C. elegans
(A) Quantification of OPP fluorescence intensity in bus-5(br19) after 3 h incubation with 10 μM OPP only (control), with a 1-h incubation chase in PBS without OPP after the initial incubation or 1-h PBS chase with 0.1 μM bortezomib; n = 57 summed across three independent replicates.
(B) Comparison of fluorescence intensity between worms treated with both OPP followed and Alexa Fluor 647 azide or treated with only Alexa Fluor 647 azide without prior incubation with OPP. n = 51 summed across three independent replicates.
(C) Quantification of OPP fluorescence intensity from control and cycloheximide-treated bus-5(br19); n = 180 summed across three independent replicates.
(D and E) Confocal bright-field (left) and fluorescent (right) images of OPP-treated bus-5(br19) (D) incubated at 20°C for 1 h or (E) heat shocked at 35°C for 1 h.
(F) Quantification of OPP fluorescence intensity from control and heat-shocked worms. n = 57 summed across three independent replicates.
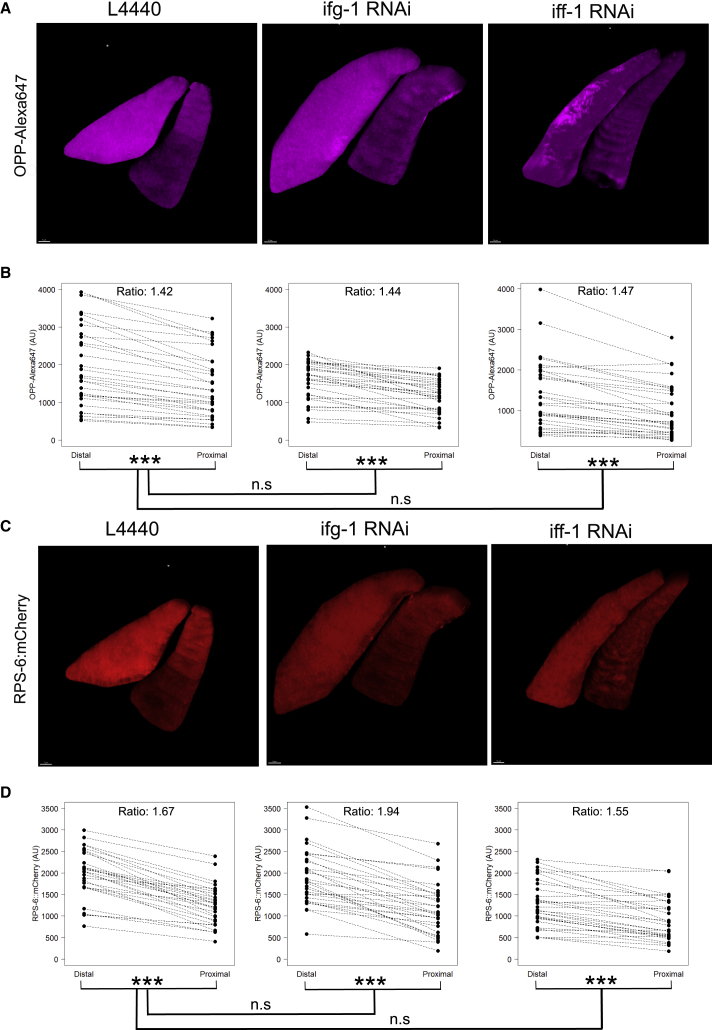
(G) Single-plane confocal images of OPP-Alexa Fluor 647 in adult bus-5(br19):rps-6::mCherry mutants after 48 h treatment with RNAi control vector L4440 (left), ifg-1 RNAi (middle), or iff-1 RNAi (right).
(H) Quantification of OPP fluorescence from a single plane in worms treated with RNAi as described in (E); n = 30 from three independent replicates.
(I) Confocal images of RPS-6:mCherry in adult bus-5(br19):rps-6::mCherry mutants after 48 h treatment with RNAi control vector L4440 (left), ifg-1 RNAi (middle), or iff-1 RNAi.
(J) Quantification of RPS-6::mCherry fluorescence from a single plane in worms treated with RNAi as described in (G); n = 30 from three independent replicates.
Notches in box plots indicate the estimated 95% confidence interval of the median value (black line). n.s. p > 0.05, ∗p < 0.05, and ∗∗∗p < 0.001. Wilcoxon rank sum test is shown.
Cycloheximide is a protein synthesis inhibitor that reduces ribosomal elongation rates. As incorporation of OPP into nascent peptides requires elongation, exposure to cycloheximide should reduce OPP incorporation in treated worms. Prior to addition of OPP to the assay, bus-5 mutants were treated with 10 mM cycloheximide for 1 h. Incorporation of OPP was significantly reduced by 37% (Figure 3C; p = 2.2e−16; Wilcoxon rank sum test; quantified representative images are found in Figure S4C) due to treatment with cycloheximide.
To demonstrate that OPP incorporation is indicative of translation rates when C. elegans is subject to stress, we quantified OPP fluorescence intensity in bus-5(br19) mutants at the control temperature of 20°C (Figure 3D; additional representative images in Figure S4B) and after 1 h heat shock at 35°C (Figure 3E; additional representative images in Figure S4B). Heat shock is a known inhibitor of translation, and using polysome profiling, we have previously shown that polysomes are reduced after 1 h at 35°C compared with controls (Figure 1B; Shaffer and Rollins, 2020). Like the assays conducted above, quantification of the OPP signal was performed on a single plane transecting the dorsoventral axis of the worm to give a readout of global translation. Compared with controls, heat shock resulted in a 78% decrease in fluorescent OPP signal (Figure 3F; p = 7.30e−09; Wilcoxon rank sum test). Thus, OPP can be used to quantify relative changes in global translation rates in C. elegans in response to environmental stressors, such as heat shock.
As further proof of principle that OPP incorporation is indicative of translation rates, we treated bus-5(br19);rps-6::mCherry mutants with RNAi targeting iff-1, ifg-1, or control RNAi vector L4440. The introduction of endogenously tagged rps-6::mCherry into the bus-5 background allows for ribosome abundance to be quantified in addition to translation rates. Knockdown of iff-1 was chosen, as it is a germline-specific translation and elongation factor (Hanazawa et al., 2004; Schuller et al., 2017) and because it was previously determined by us and others that its knockdown promotes longevity (Rollins et al., 2019). Reduction of the human ortholog of iff-1, eiF5A, results in global defects in elongation (Schuller et al., 2017). Thus, knockdown of iff-1 in worm is expected to reduce protein translation specifically in the germline. Knockdown of ifg-1 was chosen, as it is an ortholog of the translation initiation factor eiF4G and is broadly expressed across most tissues in C. elegans (Mounsey et al., 2002). Furthermore, RNAi of ifg-1 has been previously shown to reduce abundance of its protein by 80% after 48 h (Howard et al., 2016). Reduction of ifg-1 in adult worms also promotes longevity (Pan et al., 2007; Rogers et al., 2011). As with heat shock, we quantified the effect of these knockdowns across the whole worm in a single plane. Incorporation of OPP upon knockdown of ifg-1 or iff-1 (Figure 3G; additional representative images are found in Figure S5A) decreased by 37% (Figure 3H; p = 2.0e−06; Wilcoxon rank sum test) and 41% (Figure 3H; p = 7.7e−06; Wilcoxon rank sum test), respectively, across the whole worm after 48 h of RNAi treatment, compared with control vector L4440. The intensity of RPS-6::mCherry upon knockdown of ifg-1 or iff-1 (Figure 3I; additional representative images in Figure S5B) decreased by 46% (Figure 3J; p = 2.8e−08; Wilcoxon rank sum test) and 45% (Figure 3J; p = 3.4e−09; Wilcoxon rank sum test), respectively, compared with control vector L4440. Thus, reduction of translation factors ifg-1 or iff-1 both caused similar reductions in protein synthesis and in ribosome abundance when quantified across whole worms.
Quantification of tissue-specific translation
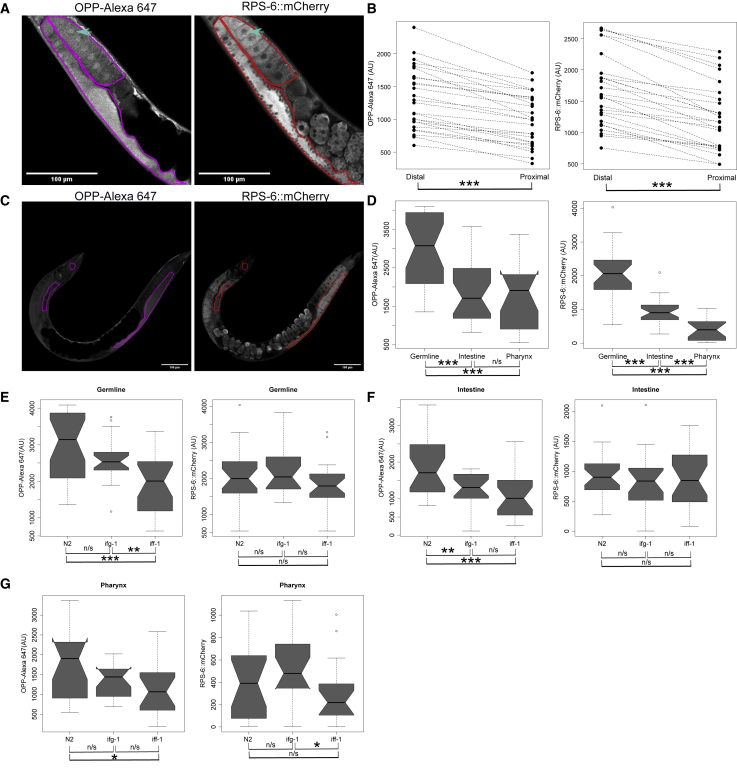
To demonstrate the utility of this OPP assay in the quantification of tissue-specific translation, we compared OPP incorporation in the proximal gonad arm, which mostly contains maturing oocytes, with the distal gonad, which predominantly contains the mitotic cells and meiotic cells in a syncytium (Figure 4A [left]; additional representative images in Figure S6C). The distal and proximal regions were defined using the bend in the germline. The abundance of the ribosome was also compared between the arms of the germline using endogenously tagged ribosomal protein RPS-6::mCherry (Figure 4A [right]; additional representative images in Figure S6C). As selective translation of specific transcripts is critical for the gene expression changes necessary in oocyte development (Nousch and Eckmann, 2013), we sought to establish how bulk translation rates change during this process and whether it is dependent on ribosome levels. In addition, we quantified and compared OPP incorporation and RPS-6::mCherry abundance in the distal germline, intestine, and the metacorpus of the pharynx to further demonstrate that this method can be used to quantify relative tissue-specific translation rates. We then compared incorporation rates and ribosome abundance in the each of these tissues after knockdown of ifg-1 or iff-1 as described above.
Figure 4.
OPP assay in whole worms allows for tissue-specific quantification in the germline
(A) Confocal image of OPP-treated adult bus-5(br19);rps-6:mCherry germline from OPP-Alexa Fluor 647 fluorescence (left) and from RPS-6::mCherry (right). Arrow indicates a nucleus within an oocyte in the proximal germline. The regions quantified for the distal (left) and proximal (right) germline are outlined in red or magenta.
(B) Interaction plots comparing OPP-Alexa Fluor 647 (left) and RPS-6::mCherry (right) mean signal intensity between the distal and proximal germline as quantified from single-plane images. n = 26 summed across three independent replicates.
(C) Representative images indicating the regions considered for quantification of the distal germline, intestine, and terminal bulb of the pharynx for OPP (left) and mCherry (right).
(D) Quantification of OPP-Alexa Fluor 647 fluorescence (left) and from RPS-6::mCherry (right) in the distal germline, intestine, and terminal bulb of the pharynx. n = 54 summed from three independent replicates.
(E–G) Quantification of OPP-Alexa Fluor 647 fluorescence (left) and from RPS-6::mCherry (right) after treatment with control vector L4440, ifg-1, or iff-1 RNAi in the (E) distal germline, (F) intestine, and (G) terminal bulb of the pharynx. n = 54 summed across three independent replicates.
Notches in box plots indicate the estimated 95% confidence interval of the median value (black line). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Wilcoxon matched-pairs signed rank test was used in (B). Wilcoxon rank sum test was used in (D)–(G).
Quantification of confocal images transecting the center of the gonad revealed that OPP incorporation in the distal gonad was 1.28-fold higher than the proximal gonad (Figure 4B [left]; p = 2.98e−08; Wilcoxon matched-pairs signed rank test). In support of this observation, we saw a similar significant increase in RPS-6 intensity between the distal and proximal gonad of 1.36-fold (Figure 4B [right]; p = 2.98e−08; Wilcoxon matched-pairs signed rank test) in the distal arm compared with the proximal gonad. Therefore, the intensity of puromycylated peptides in the germline tracks well with the presence of ribosomes on the cellular level. The overlap of OPP with RPS-6::mCherry was not restricted to the germline, as seen in the overlay of the two signals in whole worms (Figure S1G). However, we note that, on the subcellular level, puromycylated peptides accumulated in the nucleus (indicated by arrows in Figure 4A), whereas RPS-6::mCherry was largely absent there.
Having shown that OPP labeling can be used to compare relative translation rates between the distal and proximal arms of the germline, we extended this technique to quantify translation rates between the germline, the intestine, and the metacorpus of the pharynx in 2-day-old adults. Representative images showing the areas used to quantify the germline, the intestine, and the metacorpus of the pharynx from the OPP-Alexa Fluor 647 signal (Figure 4C [left]; additional representative images in Figure S7A) and the RPS-6::mCherry signal (Figure 4C [right]; additional representative images in Figure S7A) are given. Based on the mean intensity of the OPP-Alexa Fluor 647 signal, the germline had higher translation than the pharynx and the intestine (Figure 4D, left; p = 7.282e−4 and p = 5.592e−4, respectively; Wilcoxon rank sum test), while translation was similar between pharynx versus intestine (p = 0.437; Wilcoxon rank sum test). Based on the RPS-6::mCherry signal, ribosome abundance was highest in the germline (germline versus pharynx: p = 6.36e−08; germline versus intestine: p = 3.113e−07; Wilcoxon rank sum test), intermediate in the intestine (intestine versus pharynx: p = 8.458e−06; Wilcoxon rank sum test), and lowest in the pharynx. Thus, both ribosome abundance and translational activity appear to differ between the germline and the somatic tissues.
RNAi knockdown of the translation factors iff-1 and ifg-1 resulted in similar decreases in OPP and RPS-6 when quantified across whole worms (Figures 3H and 3J). To determine whether there were tissue-specific effects on translation, we quantified OPP incorporation in the germline, intestine, and in the terminal bulb of the pharynx. In the distal germline, iff-1 RNAi led to a significant decrease in OPP incorporation compared with the empty vector control (Figure 4E [left]; p = 2.081e−4; Wilcoxon rank sum test), while ifg-1 did not (p = 0.1759). However, RPS-6::mCherry levels in the germline were similar between the three treatments (Figure 4E [right]; L4440 versus ifg-1: p = 0.1759; L4440 versus iff-1: p = 0.1984; iff-1 versus ifg-1: p = 0.06005; Wilcoxon rank sum test). In the intestine, OPP incorporation was lower than controls in both iff-1 and ifg-1 RNAi-treated animals (Figure 4F [left]; iff-1 versus L4440: p = 3.936e−4; ifg-1 versus L4440: p = 0.005579; Wilcoxon rank sum test). RPS-6::mCherry levels in the intestine were similar between the three treatments (Figure 4F [right]; L4440 versus ifg-1: p = 0.3861; L4440 versus iff-1: p = 0.6481; ifg-1 versus iff-1: p = 0.6121; Wilcoxon rank sum test). In the metacorpus of the pharynx, iff-1 RNAi led to a decrease in OPP incorporation compared with controls (Figure 4G [left]; p = 0.02586; Wilcoxon rank sum test) that was not as pronounced in the ifg-1 RNAi treatment (Figure 4G [left]; L4440 versus ifg-1: p = 0.104; Wilcoxon rank sum test). RPS-6::mCherry levels in the pharynx were lowest due to the iff-1 RNAi, which was significantly lower than ifg-1-treated worms, but not controls (Figure 4G [right]; iff-1 versus ifg-1: p = 0.009043; L4440 versus iff-1: p = 0.2184; Wilcoxon rank sum test). In summary, iff-1 knockdown resulted in significantly decreased OPP incorporation in the germline, intestine, and pharynx, while ifg-1 knockdown led to a significant decrease in the germline only. However, these RNAi treatments did not significantly affect ribosome abundance in these tissues as quantified by RPS-6::mCherry intensity.
Quantification of translation in distal and proximal arms of the germline using 3D volumetric reconstructions
As single-plane images may not accurately reflect translational regulation occurring in the entire volume of the tissue, 3D reconstructions of confocal image z stacks were also used to quantify the OPP and RPS-6 signal in the whole volume of the germline. To limit quantification to the germline specifically, an endogenously GFP-tagged germline reporter glh-1(sam24[glh-1::GFP::3xFLAG]) (Andralojc et al., 2017) was used to guide image mask creation in 3D. Therefore, reconstructions were performed on bus-5(br19);rps-6::mCherry;glh-1::GFP mutants treated with control RNAi (Figure 5A [left]; additional representative images in Figure S7B) or RNAi targeting ifg-1 (Figure 5A [middle]; additional representative images in Figure S7B) or iff-1 (Figure 5A [right]; additional representative images in Figure S7B). An example 3D projection is provided in Video S1, showing stained nuclei, the OPP-Alexa Fluor 647 signal, RPS-6::mCherry, and GLH-1::GFP in the germline. Quantification of the 3D reconstructed germlines in RNAi controls revealed that translation rates in the proximal gonad were 1.42-fold higher than the distal gonad (Figure 5B [left]; p = 1.863e−09; paired Wilcoxon signed rank test). OPP incorporation was also significantly higher in the proximal germline after knockdown of ifg-1 (Figure 5B [middle]; p = 3.725e−09; paired Wilcoxon signed rank test) or iff-1 (Figure 5B [right]; p = 4.657e−08; paired Wilcoxon signed rank test) with ratios of 1.44 and 1.47, respectively. The ratio of OPP signal between the distal and proximal germline was similar between the three treatments (Figure 5B; L4440 versus ifg-1: p = 0.150; L4440 versus iff-1: p = 0.982; ifg-1 versus iff-1: p = 0.218; Wilcoxon rank sum test), despite iff-1 and ifg-1 RNAi decreasing translation in whole-worm measurements (Figure 3H). However, we note that, given a significance level of 0.05 and power of 0.8, our statistical power to detect differences in the mean OPP ratio between L4440 and iff-1 or ifg-1 treatment is 0.28 and 0.22, respectively.
Figure 5.
Quantification of translation in distal and proximal arms of the germline using 3D volumetric reconstructions
(A) Representative 3D reconstructions of OPP-Alexa Fluor 647 signal in adult bus-5(br19);rps-6:mCherry;glh-1::GFP mutant germlines after 48 h treatment with RNAi control vector L4440 (left), ifg-1 RNAi (middle), or iff-1 RNAi (right). In each image, the distal gonad is on the left and proximal on the right.
(B) Interaction plots comparing OPP-Alexa Fluor 647 mean signal intensity between the distal and proximal germline after treatment with RNAi described in (A). n = 30 from three independent replicates.
(C) Representative 3D reconstructions of RPS-6::mCherry signal in adult bus-5(br19);rps-6:mCherry;glh-1::GFP mutant germline after 48 h treatment with RNAi control vector L4440 (left), ifg-1 RNAi (middle), or iff-1 RNAi (right).
(D) Interaction plots comparing RPS-6::mCherry mean signal intensity between the distal and proximal germline after treatment with RNAi described in (C). n = 30 from three independent replicates.
Notches in box plots indicate the estimated 95% confidence interval of the median value (black line). ∗∗∗p < 0.001. Wilcoxon matched-pairs signed rank test was used to compare signal between distal and proximal germline. Wilcoxon rank sum test was used to compare the average ratio between RNAi treatments.
The initial view shows all the fluorophores present, NuclearMask Blue Stain (cyan), GLH-1:GFP (green), RPS-6::mCherry (red), and OPP (magenta). The surfaces created with the Imaris software are displayed (gray), overlaying OPP and GLH::1-GFP. The germline including distal and proximal regions are displayed, demonstrating the 3D regions that were measured from the OPP signal, as well as RPS-6::mCherry.
The RPS-6::mCherry signal was significantly 1.67-fold higher in the distal germline than the proximal germline (Figure 5C [left]; additional representative images in Figure S7B) when quantified in 3D (Figure 5D [left]; p = 1.863e−09; paired Wilcoxon signed rank test) in empty vector controls. After knockdown of ifg-1 or iff-1 (Figure 5C [middle and right]), the ratio of RPS-6::mCherry signal was still significantly higher in the proximal germline with ratios of 1.94 (Figure 5D; p = 1.863e−09; paired Wilcoxon signed rank test) and 1.55 (Figure 5D; p = 3.148e−07; paired Wilcoxon signed rank test), respectively. These ratios were similar to each other (Figure 5D; p = 0.3581; Wilcoxon rank sum test) and to empty vector controls (L4440 versus ifg-1: p = 0.9124; L4440 versus iff-1: p = 0.4761; Wilcoxon rank sum test). The authors note that, given a significance level of 0.05 and power of 0.8, our estimated ability to detect differences in the mean RPS-6 ratio between L4440 and iff-1 or ifg-1 treatment is 0.60 and 0.30, respectively. Thus, the difference in protein translation and ribosome abundance is maintained in the distal and proximal germline, even when the overall translation rates are decreased by loss of the translation factors iff-1 and ifg-1.
Discussion
The increased cuticle permeability of bus-5(br19) greatly enhances the sensitivity and reproducibility of this technique, showing increased and better distributed uptake of puromycin in tissues compared with wild-type worms (Figures 1E, S1E, and S1F). Using the OPP assay to quantify global changes in translation, we show that heat shock caused a 50% decrease after 1 h (Figure 3E), in line with reductions in polysome profiles in heat-shocked samples (Figure 1B) shown here and shown previously (Shaffer and Rollins, 2020). Comparison of OPP incorporation between the distal and proximal germline revealed a 1.42-fold increase in translation in the distal germline compared with the oocytes in the proximal germline (Figure 5B). We additionally show that RNAi knockdown of the protein initiation factor ifg-1 or the elongation factor iff-1 results in a decrease in protein translation across the whole worm by 37% and 41%, respectively (Figures 3G and 3I). Furthermore, we demonstrate tissue-specific changes in showing that OPP incorporation and ribosome abundance is higher in the germline than the intestine (Figure 4D). Thus, this technique is suitable for the quantification of protein translation between worms under different conditions as well as between tissues within a single worm.
This technique offers several advantages over quantification of puromycin incorporation by western blot. Instead of relying on bulk populations, single worms are quantified so that variation between individuals can be captured. More importantly, imaging individual intact worms allows translation rates of tissues and cells to be quantified and compared. Our assay uses a much lower dose of OPP (10 μM) than the 1 mM dose of puromycin typically used for detection via western blot in C. elegans (Arnold et al., 2014; Heissenberger et al., 2020). As high doses of puromycin and OPP appreciably inhibit protein translation (Liu et al., 2012), the very phenomenon we are trying to measure, lower doses are less prone to giving spurious results.
A key component to this assay is the use of the bus-5 mutation to increase puromycin uptake to penetrate tissues within 3 h. Polysome profiling of bus-5(br19) was unable to detect any differences in polysome-bound mRNA compared with N2s (Figure 1B), suggesting that translation rates are not appreciably affected by the bus-5 mutation. The bus-5 mutant was originally selected for use in toxicity screens, as it offered increased cuticle permeability with minimal effects on development and fecundity compared with N2 (Xiong et al., 2017). The gene bus-5 (also known as rml-2) encodes for a deoxythymidine diphosphate (dTDP)-glucose 4,6-dehydratase, which is required for biosynthesis of dTDP-rhamnose, a component of bacterial cell walls and a likely component of the C. elegans cuticle (Feng et al., 2016). The likelihood of this mutation impacting global translation rates in C. elegans is low, as expression of a bus-5::GFP reporter was restricted to the hypodermis and absent in adult worms (Feng et al., 2016). While knockdown of bus-5 may recapitulate the enhanced cuticle phenotype needed for the OPP assay, RNAi by feeding has variable knockdown efficiencies (Simmer et al., 2003; Zugasti et al., 2016) that could lead to inconsistent cuticle permeability between experiments that would hinder the interpretation of results. We therefore recommended to cross the bus-5(br19) allele into any strain that would be used with OPP imaging.
The quantification of OPP intensity in the germline was guided by imaging in a strain expressing the germline-specific marker glh-1::GFP. This allowed accurate determination of the borders of the germline in three dimensions. As shown in Figures 2E and 2H, our fixation step does not appreciably affect the fluorescence of mCherry or GFP. This is important, as future applications of this technique will likely rely on the expression of fluorophores-driven, tissue-specific promoters to accurately quantify OPP incorporation into tissues of interest in C. elegans. Using a tissue-specific fluorescent signal as a mask can be used to limit quantification of OPP within the worm to that area. The discovery that translation rates are lower in the proximal germline than the distal germline in this study further exemplifies the importance of translational regulation in this tissue and in oocyte maturation and the need for tissue-specific translation measurements. Several RNA-binding proteins have been implicated in the translational promotion or repression of mRNAs necessary in this process (Huelgas-Morales and Greenstein, 2018; Kaymak and Ryder, 2013; Tsukamoto et al., 2017). However, our data suggest that selective repression of mRNA translation might be the predominant net activity of these RNA-binding proteins in oocyte maturation, as translation among oocytes was lower than in the distal germline.
RNAi knockdown of iff-1 caused appreciable loss of translation across the entire worm, despite being purported as germline specific (Hanazawa et al., 2004). This is possibly due to the similarity between the coding sequences of iff-1 and its somatic ortholog iff-2, resulting in the RNAi targeting both transcripts. It is also possible that reduction of translation in the germline has cell non-autonomous effects to signal reduction of translation in other tissues (Lan et al., 2019; Robinson-Thiewes et al., 2021). Knockdown of either iff-1 or ifg-1 resulted in similar decreases in OPP incorporation across the whole worm (Figure 3H), despite having different roles promoting translation initiation and translation elongation, respectively. Interestingly, the reduction of either iff-1 or ifg-1 also led to a reduction in RPS-6::mCherry across the whole worm (Figure 3J), signifying that overall ribosome abundance was reduced by these knockdowns. However, after knockdown of iff-1, ribosome abundance was not significantly decreased in the germline, intestine, or pharynx (Figures 4E–4G), despite OPP incorporation being decreased here. Reduction in ribosome abundance has been seen before upon the reduction of initiation factor eIF3 or depletion of individual ribosomal proteins (Juli et al., 2016; Smekalova et al., 2020). Thus, it is possible that reduced translation observed in some tissues from these RNAi treatments after 48 h is due to reduced availability of ribosomes as opposed to reduced initiation rates or elongation rates directly. However, this does not appear to be the case in the germline, pharynx, or intestine. This distinction is important in understanding whether knockdowns of these translation initiation factors are pro-longevity due to changes in selective translation, energetics, and/or enhanced proteostasis (Rollins and Rogers, 2017). While reduction of ifg-1 has been shown to increase the selective translation of genes associated with stress resistance (Rogers et al., 2011), the reduction in ribosome biosynthesis described here raises the question to which changes in selective translation are specific to loss of ifg-1 activity and which are common to any treatment that reduced translation.
In the present study, we demonstrated that differences in translation exist between tissues under control conditions. For example, OPP incorporation was 33% lower in the intestine compared with the germline (Figure 4D). Furthermore, RNAi treatments that inhibit translation had tissue-specific effects (Figures 4E–4G). There is a growing appreciation of the relationship between protein translation rates and longevity, with the focus turning to the contribution of individual tissues. For example, translation elongation rates were shown to differ between 3-month-old mouse kidney, liver, and skeletal muscle using a form of ribosome profiling (Gerashchenko et al., 2020). In the same study, elongation rates were shown to decrease with age in the liver by ∼20%, in line with other observations that bulk protein synthesis declines with age (Rattan, 1996; Ward and Richardson, 1991). However, as other tissues were not examined, presumably due to the complexity of executing such experiments, it raises the question of whether the decline in translation with age is equal among all tissues or variable and dependent on tissue type. The use of OPP and click chemistry to quantify protein translation rates in intact, whole worms allows for a rapid assessment of tissue-specific changes with age or in response to anti-aging treatments. We anticipate its application will broaden our understanding of which tissues are impacted the most by pro-longevity treatments that affect translation like stress, gene knockdown, or drug therapies.
Limitations of the study
There are a few limitations to this technique pertaining to sample permeability, subcellular localization of the OPP signal, and proteasomal activity. The use of a defective cuticle mutant to increase puromycin incorporation highlights an important limitation of the OPP assay in C. elegans; that conditions or genetic backgrounds that change the cuticle composition or pharynx pumping rates can lead to changes in puromycin incorporation unrelated to translational activity. Thus, it is recommended to confirm changes in global translation by independent methods, such as polysome profiling. In addition, the total protein content between N2 and bus-5(br19) mutants have not been compared, and thus, we cannot eliminate the possibility that there are physiological differences between the two strains other than cuticle permeability that might be relevant to this assay.
Previous staining of dissected C. elegans germlines with 20 μM OPP for 5 min resulted in more pronounced labeling of the nuclei than the cytoplasm (Enam et al., 2020), despite the paucity of ribosomes there. In the present study, in which live intact worms were assayed, a greater accumulation of OPP was also seen in the nuclei (Figure 4A [left]). Imaging of endogenously tagged ribosomal 40S subunit RPS-6::mCherry confirmed that ribosomes were largely absent from these OPP-labeled nuclei (Figure 4A [right]). Thus, we stress caution in using this assay to quantify subcellular localization of translation, as puromycylated peptides are possibly prone to diffusion and trafficking (Enam et al., 2020), especially after a 3-h incubation, as used here. Finally, since it takes time for OPP to infiltrate all tissues, some cells are presumably exposed to puromycin longer than others. Therefore, this assay assumes that a steady state between OPP incorporation and degradation of puromycylated peptides is reached. As shown with treatment with bortezomib (Figure 3A), inhibition of the proteosome results in retention of the OPP signal. Thus, mutations or treatments that alter proteasomal activity could invalidate the accuracy of this assay in quantifying relative translation rates.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Puromycin Antibody, clone 12D10 | Sigma Aldrich | Cat# MABE343 |
| Goat Anti-Rabbit IgG H&L (HRP) preadsorbed | Abcam | Cat# ab97080 |
| Bacterial and virus strains | ||
| Escherichia coli OP50 | Caenorhabditis Genetics Center | Cat# OP50 |
| Escherichia coli HT115 (L4440) | Caenorhabditis Genetics Center | Cat# GC363 |
| Escherichia coli HT115 (iff-1) | Ahringer feeding library | III-5A04 |
| Escherichia coli HT115 (ifg-1) | Ahringer feeding library | II-6E05 |
| Chemicals, peptides, and recombinant proteins | ||
| Agar, Granulated, High Gel Strength | Research Products International | Cat# A20250-1000.0 |
| Peptone, Granulated | Research Products International | Cat# P20250-1000.0 |
| Sodium Chloride | Research Products International | Cat# S23020-1000.0 |
| Potassium Chloride | Research Products International | Cat# P41000-500.0 |
| Potassium Phosphate, Monobasic, ACS Grade | Research Products International | Cat# P41200-500.0 |
| Sodium Phosphate Dibasic | Sigma Aldrich | Cat# 7558-79-4 |
| TRIS Hydrochloride [Tris(hydroxymethyl) aminomethane HCl] | Research Products International | Cat# T60050-500.0 |
| OPP (O-propargyl-puromycin) | Thermo Fisher Scientific | Cat# C10459 |
| Paraformaldehyde reagent grade, crystalline | Sigma Aldrich | Cat# P6148-500G |
| Cycloheximide | Research Products International | Cat# C81040-1.0 |
| Bortezomib, 99% | Fisher Scientific | Cat# AAJ60378MA |
| Magnesium Chloride Hexahydrate (Crystalline/Certified ACS) | Fisher Scientific | Cat# M33-500 |
| Triton X-100 Surfactant | Fisher Scientific | Cat# MTX15681 |
| Tween-20 | Sigma Aldrich | Cat# P1379 |
| IPTG [Isopropyl-β-D-thiogalactopyranoside], Dioxane Free | Research Products International | Cat# I56000-50.0 |
| Carbenicillin, Disodium Salt | Research Products International | Cat# C46000-25.0 |
| EGTA [Ethylene glycol-bis (2-aminoethylether)-N,N,N',N'- tetraacetic acid] |
Research Products International | Cat# E57060-25.0 |
| Sodium Deoxycholate | Thermo Fisher Scientific | Cat# 89904 |
| Sucrose | Research Products International | Cat# S24060-1000.0 |
| Critical commercial assays | ||
| DC™ Protein Assay Kit I | Bio-Rad | Cat# 5000111 |
| O- Click-iT™ Plus OPP Alexa Fluor™ 647 Protein Synthesis Assay Kit | Thermo Fisher Scientific | Cat# C10458 |
| EveryBlot Blocking Buffer | Bio-Rad | Cat# 12010947 |
| 4–20% Mini-PROTEAN® TGX Stain-Free™ Protein Gels | Bio-Rad | Cat# 4568095 |
| cOmplete(TM), Mini, EDTA-free Protease Inhibitor Cocktail | Thomas Scientific | Cat# C852A34 |
| Experimental models: Organisms/strains | ||
| Caenorhabditis elegans strain: N2 wildtype | Caenorhabditis Genetics Center | WormBase ID: WBStrain00000004 |
| Caenorhabditis elegans strain: DC19 bus-5(br19) X. | Caenorhabditis Genetics Center | WormBase ID: WBStrain00005604 |
| Caenorhabditis elegans strain: JAR016 rps-6(rns6[rps-6::mCherry]) I | This paper | Rollins Lab at MDIBL |
| Caenorhabditis elegans strain: DUP64 glh-1(sam24[glh-1::gfp::3xFLAG]) I | Andralojc et al., 2017 | WormBase ID: WBStrain00048695 |
| Caenorhabditis elegans strain: JAR030 bus-5(br19) X ; rps-6(rns6[rps-6::mCherry]) I | This paper | WormBase ID: WBStrain00005604 |
| Caenorhabditis elegans strain: JAR033 bus-5(br19) X ; rps-6(rns6[rps-6::mCherry]) I ; glh-1::GFP I | This paper | WormBase ID: WBStrain00005604 |
| Oligonucleotides | ||
| crRNA - RPS-6::mCherry CAGCAGTATTTACTTCTTGC |
This paper | n/a |
| Recombinant DNA | ||
| Homologous recombination template for rps-6::mCherry: CCTCGCCAAGTACTCC AAGGAAGAGCACGACGCCAAGATCGCC CGCAGACGCTCTTCGGCTTCCCATCAC TCCGAGTCCGAGGTCAAGAAGACaAGC AAGAAGGGAGCATCGGGAGCCTCAGGA GCATCGATGGTCTCAAAGGGTGAAGAAG ATAACATGGCAATTATCAAGGAGTTCATG CGTTTCAAGGTCCACATGGAGGGATCC GTCAACGGACACGAGTTCGAGATCGAG GGAGAGGGAGAGGGACGTCCATACGAG GGAACCCAAACCGCCAAGCTCAAGGTA AGTTTAAACATATATATACTAACTAACCC TGATTATTTAAATTTTCAGGTCACCAAGG GAGGACCACTCCCATTCGCCTGGGACA TCCTCTCCCCACAATTCATGTACGGATC CAAGGCCTACGTCAAGCACCCAGCCG ACATCCCAGACTACCTCAAGCTCTCCT TCCCAGAGGGATTCAAGTGGGAGCGT GTCATGAACTTCGAGGACGGAGGAGT CGTCACCGTCACCCAAGACTCCTCCC TCCAAGACGGAGAGTTCATCTACAAGG TCAAGCTCCGTGGAACCAACTTCCCAT CCGACGGACCAGTCATGCAAAAGAAG ACCATGGGATGGGAGGCCTCCTCCGA GCGTATGTACCCAGAGGACGGAGCCC TCAAGGGAGAGATCAAGCAACGTCTC AAGCTCAAGGACGGAGGACACTACG ACGCCGAGGTCAAGACCACCTACAA GGCCAAGAAGCCAGTCCAACTCCCA GGAGCCTACAACGTCAACATCAAGC TCGACATCACCTCCCACAACGAGGAC TACACCATCGTCGAGCAATACGAGCG TGCCGAGGGACGTCACTCCACCGGA GGAATGGACGAGCTCTACAAGTAAAT ACTGCTGCTCGTTATTGTTTCCTAATG AAATTGTTGTTAAACTTAAATCTTTTTT TTTTGTTGTTCACTCCATTCGTCTC |
This paper | n/a |
| Software and algorithms | ||
| Fiji is just ImageJ (Fiji) v1.53i | Schindelin et al., 2012 | https://imagej.net/software/fiji/ |
| Imaris v.9.5 | Oxford Instruments | https://imaris.oxinst.com/ |
| Zen Blue software (Zen Pro 3.1) | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html |
| Olympus Fluoview software (v4.2.3.6) | Olympus | https://www.olympus-lifescience.com/en/software/ |
| R Studio (version 3.6.1) | This paper | https://www.rstudio.com/products/rstudio/ |
| Other | ||
| Zeiss LSM 980 | Zeiss | https://www.zeiss.com/microscopy/us/products/confocal-microscopes/lsm-980.html |
| Olympus FV 1000 | Olympus | https://www.olympus-lifescience.com/en/technology/museum/micro/2004/ |
| BioComp Gradient Fractionator and Gradient Master | BioComp Instruments | https://biocompinstruments.com/ |
Resource availability
Lead contact
Information regarding resources or reagents can be requested should be directed to and will be fulfilled by the lead contact, Jarod Rollins (jrollins@mdibl.org).
Materials availability
-
•
This study did not generate new unique reagents.
-
•
Strains used in this paper are available upon request from the Rollins lab at MDI Biological Laboratory.
Experimental model and subject details
Strains
N2 wildtype (Bristol), DC19 bus-5(br19) X, JAR030 bus-5(br19) X; rps-6(rns6[rps-6::mCherry]) I, and JAR033 bus-5(br19) X; rps-6(rns6[rps-6::mCherry]) I ; glh-1::GFP I. The DC19 bus-5(br19) mutant was obtained from the CGC. The JAR030 strain was creating by crossing the bus-5(br19), and JAR016 rps-6(rns6[rps-6::mCherry]) I. The JAR033 strain was created by crossing the JAR030 strain with DUP64 glh-1::GFP I.
Culturing C. elegans
Worms were grown at 20°C on nematode growth media (NGM) agar plates seeded with OP50 bacteria at 2 × 1010 CFU. Hermaphrodites were utilized for all assays. To age synchronize populations, adult worms were transferred to a NGM plate for 1 h to lay eggs, and then were removed. Worms were harvested from NGM plates by washing with liquid NGM or PBS buffer (pH 7.4) containing 0.01% triton-X.
RNAi interference by feeding
The RNAi bacterial strains were grown in LB broth containing 25 μg/mL carbenicillin at 37°C overnight and concentrated to 2 × 1010 CFU. NGM plates containing 1 mM IPTG and 25 μg/mL carbenicillin were spotted with 100 μL RNAi bacteria and allowed to dry at room temperature for 18 h before worms were transferred to them. Worms were fed RNAi starting at day 1 of adulthood. RNAi used for this assay included: Empty vector control L4440, ifg-1, and iff-1 from the Ahringer feeding library (Kamath, 2003). The worms were treated with RNAi for 48 h prior to puromycin incubation.
CRISPR strain construction
A co-CRISPR technique was used in the generation of the JAR016 rps-6(rns6[rps-6::mCherry]) I allele as described (Paix et al., 2017). The guide RNA sequences, and repair templates used are listed in key resources. Proper insertion of the mCherry sequence was confirmed using sanger sequencing.
Method details
Biochemical assays
Western Blot analysis
For gel electrophoresis, 30 μg of total protein as determined from DC protein assay (Bio-Rad) was separated on 4-20% mini-Protean TGX stain-free gels (Bio-Rad). The resulting gels were exposed to UV for 5 min to allow fluorescent detection of total protein present before electroblotting samples to PVDF. Blots were then imaged under UV to image total protein transferred to them. Anti-puromycin antibody (clone 12D10, Millipore) was diluted 1:3000 in Everyblot (Bio-Rad) and incubated at RT for 1 h with shaking. After washing, the blot was incubated with anti-mouse HRP conjugated antibody diluted 1:10000 in Everyblot. Three washes of Everyblot were used after each incubation step. Images of total protein and of chemiluminescence were background subtracted and quantified using imageJ.
Polysome profiling
Profiles were conducted and analyzed as described previously (Shaffer and Rollins, 2020). Briefly, for each profile, 100 μL of pelleted worms were homogenized on ice in 350 μL of solubilization buffer (300 mM NaCl, 50 mM Tris–HCl [pH 8.0], 10 mM MgCl2, 1 mM EGTA, 400 U RNAsin/mL, 0.2 mg cycloheximide/mL, 1% Triton X-100, and 0.1% sodium deoxycholate). After homogenization, an additional 200 μL of solubilization buffer was added and the sample was incubated at 4°C. Debris was pelleted by centrifugation at 14,000 × g for 15 min at 4°C. Of the resulting supernatant, 300 μL was applied to the top of a 5–50% sucrose gradient in high salt resolving buffer (140 mM NaCl, 25 mM Tris–HCl [pH 8.0], and 10 mM MgCl2) and centrifuged in a Sorvall TH-641 rotor at 38,000 rpm for 2 h @ 4°C. Gradients were fractionated using a Piston Gradient Station equipped with a Triax flow cell (BioComp Instruments) with continuous monitoring of absorbance at 260 nm.
O-propargyl-puromycin translation assay
Preparation
Prepare OP50 prior to OPP assay by growing OP50 to approximately 2 × 109 CFU/mL. Incubate the OP50 for 1 h with 1% PFA with shaking. Pellet the OP50 with centrifugation and resuspend the pellet with M9 to wash out the PFA. Repeat for a total of three washes and the resuspend the PFA killed OP50 to 2 × 108 CFU/mL.
Incubation
Each assay started with washing approximately 100 worms from NGM plates using liquid NGM. Tubes containing worms were briefly centrifuged for 30 s on a tabletop centrifuge at 1500 × g to create a loose pellet allowing the supernatant to be removed. The worm pellets were then washed with liquid NGM to remove OP50. Again, the tubes were centrifuged, and the supernatant was removed. Each wash step was conducted this way throughout the assay. The OPP assay was based on the Click-iT® Plus kit (Molecular Probes) manufacturer’s instructions with slight alterations for use in C. elegans. Worms were incubated for 3 h at 20°C with gentle shaking in 10 μM O-propargyl-puromycin (Invitrogen) diluted in liquid NGM containing 2 × 108 CFU/mL of PFA killed OP50 in a total volume of 1 mL in a 1.5 mL microcentrifuge tube. Before PFA fixation worm pellets were washed with PBS.
Fixation
Incubate 0.4 g of paraformaldehyde in 8 mL DEPC-treated H2O, treated with 2 drops of 10 M NaOH, at 65°C, mixing occasionally until the powder is completely dissolved. Chill the solution on ice and then add the 10X PBS. Adjust the volume to 10 mL with DEPC-treated H2O. Prepare fresh before use. For fixation, 100 μL of freshly prepared 4% paraformaldehyde (pH 7) in PBS was added for 1 h at 10°C with shaking. After fixation, the worm pellet was washed three times with PBS.
Conjugation of fluorescent azide
Conjugation of a fluorophore to the puromycylated peptides was performed using the Click-iT® Plus Alexa 647 Fluor® Picolyl Azide Tool Kit (Molecular Probes). The fixed worms were incubated in 870-880 μL of Click-iT® reaction buffer containing 2.5–10 μL of 500 μM Alexa Fluor® picolyl azide, 20 μL 100 mM copper protectant, and 100 μL of the reaction buffer additive, in 1 mL total solution and incubated overnight (16 h) at 10°C in shaking incubator at 900 rpm. The worm pellets were then washed 3 times in PBS with shaking for 30 min each wash to remove unconjugated Alexa Fluor® picolyl azide.
Nucleus staining
HCS NuclearMask™ Blue Stain (Invitrogen) was used based on the manufacturer instructions. Click-iT® Plus protocol. 5 μL of the blue stain 2000× concentrate was added to 995 μL of PBS buffer. Each worm pellet was incubated with 1 mL of the blue stain solution. The blue stain was added during the second wash step after addition of azide. The worms were incubated with the blue stain for 1 h. The blue stain solution was then washed out a final time for 30 min, along with any excess Alexa Fluor 647® Picolyl Azide.
C. elegans treatments
Heat shock treatment
Prior to OPP incubation the N2 and bus-5(br19) strains were grown to the L4 larval stage on NGM plates, and then heat shocked for 1 h at 35°C. Following the heat shock the worms were allowed to recover for 30 min and were then incubated in OP-puromycin as described above.
Bortezomib treatment
The bus-5(br19) strain was utilized for this assay at the L4 stage. The worms were incubated in 0.1 μM bortezomib in DMSO for 1 h at 20°C in M9 following incubation in OPP. As a control another tube of worms was incubated in M9 with DMSO at 0.1 μM for 1 h. The worms were then treated as described above during the PFA fixation and staining.
Cycloheximide treatment
The bus-5(br19) strain was utilized for this assay at the L4 stage. Once the worms were L4 they were transferred to a solution of 1 mL of 10 mM cycloheximide in M9 for 1 h in a 1.5 mL Eppendorf tube shaking at 900 rpm at 20°C on a table-top shaking incubator. The control worms were incubated in M9 for 1 h simultaneously. After 1 h the control worms were treated as normal. The cycloheximide treated worms were incubated in a solution of OPP with 10 mM cycloheximide for the normal 3 h incubation and were treated as normal following this 3 h incubation.
Fluorescent microscopy
2D image acquisition
All 2D images for the OP-Puromycin assay were collected with an Olympus FV 1000 confocal microscope on an inverted Olympus IX81 stand equipped with an Olympus Plan Fluo 20X/0.5NA objective. HCS NuclearMask™ Blue Stain fluorescence (excitation ⁄emission maxima of 355⁄460 nm, similar to DAPI) was excited with a 405 nm line from a 50-mW laser diode at 20% laser intensity. EGFP fluorescence was excited with a 473 nm line from a 15-mW laser diode, 5% laser intensity. mCherry fluorescence was excited with a 559 nm line from a 15-mW laser diode, 15% laser intensity. Alexa 647 fluorescence was excited with a 635 nm line from a 20-mW laser diode, 16% laser intensity. Transmitted light imaging was performed using the 473 nm line from a 15-mW laser diode, 5% laser intensity. Laser beam attenuation was achieved using acousto-optic tunable filters.
To image samples for DAPI, brightfield, and Alexa 647, the excitation Dichroic Mirror used was DM405/473/559/635 nm. Transmitted light images were collected with the trans PMT. DAPI fluorescence was filtered using a beam splitter SDM560 and a barrier filter of 425-475 nm. Alexa 647 fluorescence was filtered using a barrier filter of 650-750 nm. For all channels pinhole diameter was set to automatic.
To image experiments for DAPI, brightfield, EGFP, mCherry, and Alexa 647 fluorescence, the virtual channel scan was used as following: DAPI and mCherry were imaged together using the excitation Dichroic Mirror DM 405/473/559 nm and the fluorescence was filtered, respectively, using a beam splitter SDM 560 nm with a barrier filter of 425-475 nm and with a barrier filter of 570-670 nm. EGFP and Alexa 647 fluorescence were imaged together using the excitation Dichroic Mirror DM 405/473/559/635 nm and the signal was filtered, respectively, using a beam splitter SDM 560 and with a barrier filter of 485-585 nm and with a barrier filter of 650-750 nm. Brightfield image was collected with the trans PMT. The pinhole diameter was set on automatic.
For all acquisitions, images were sequentially acquired at zoom 1, with a line average of 4, a resolution of 1024 × 1024 pixels, a pixel time of 4 μs, 16-bit, in Unidirectional mode and controlled with Olympus Fluoview software (v4.2.3.6). All imaging parameters were identical between experiments for compared image sets and saved in OIF image file output.
3D image acquisition
All 3D images for the OP-Puromycin assay were collected with a Zeiss LSM 980 confocal microscope (Carl Zeiss Microscopy) on an upright Zeiss Axio Examiner stand equipped with a Zeiss Plan Apo 20X/0.8NA objective (Carl Zeiss Microscopy).
DAPI fluorescence was excited with the 405 nm line with 1% laser intensity, from a 30-mW laser diode (Laser beam attenuation via direct modulation). EGFP fluorescence was excited with the 488 nm line, 1% laser intensity from a 30-mW laser diode. mCherry fluorescence was excited with the 594 nm line, 1% laser intensity from a 8-mW laser diode. Alexa 647 fluorescence was excited with a 639 nm line, 1% laser intensity from a 25-mW laser diode. Laser beam attenuation for all channels other than DAPI was by using visible range acousto-optic tunable filters. Images were captured using Airyscan 2 with the following detection wavelengths: DAPI from 422 to 497 nm, EGFP from 499 to 557 nm and 659 to 720 nm, mCherry from 422 to 477 nm and 573 to 627 nm, Alexa 647 from 499 to 557 nm and 659 to 720 nm.
Images were sequentially acquired in Super Resolution mode (SR) at zoom 1.7, with a line average of 1, a resolution of 512 × 512 pixels, a pixel time of 0.85 μs, in 8-bit and in bidirectional mode. Z-stack images were collected with a step size of 0.31 μm with the Motorized Scanning Stage 130 × 85 PIEZO (Carl Zeiss Microscopy) mounted on Z-piezo stage insert WSB 500 (Carl Zeiss Microscopy). Microscope was controlled using Zen Blue software (Zen Pro 3.1), Airy scan images were processed manually in 3D with a strength of the deconvolution set to 4.1 and saved in CZI file format. All imaging parameters were identical between experiments for compared images.
Quantification and statistical analysis
Image analysis
2D image quantification
Fiji is just ImageJ (Fiji) v1.53i was utilized for image analysis. To measure the OPP-Alexa 647 fluorescence, the worms or tissue in each image were individually identified and circled using the Freehand selection tool. The mean fluorescence intensity of each area was recorded using the measurement tool.
3D image quantification
The image analyses were done on the entire germline in 3D, using Imaris v.9.5. Raw data were segmented by manually outlining the germline using the glh-1::GFP signal by navigation through the Z-stack. The segmented rendered surface was used to mask the raw data and create a subset on which OPP-Alexa 647 and mCherry intensity measurements were performed on the selected distal and proximal germline using the surfaces statistics tool for each sample. The automatic Imaris detection was first validated manually, by examining individual z-planes using the Oblique slicer tool and a combination of 2 Clipping plane tools to isolate and examine a specific tissue region. Threshold settings were manually adjusted for each sample to yield the best possible detection.
Statistical analysis
All statistical tests were conducted using R (version 3.6.1). ANOVAs were run using the aov() function. Wilcoxon ranked sum tests and Wilcoxon signed rank test were run using the wilcox.test() function. Power tests were run using the power.t.test() function from the stats package (version 3.6.1). The exact p-values for each test are reported in the main text and thresholds for significance are given in the figure legends.
Acknowledgments
The authors would like to thank the Updike lab for providing the DUP64 strain. We would also like to thank both Dr. Dustin Updike and Dr. Brett Kieper for thoughtful discussions. In addition, some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Images were acquired using equipment of the Light Microscopy Facility at the MDIBL, which is supported by the Maine INBRE grant (GM103423) from the National Institute of General Medical Sciences at the National Institutes of Health. Research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant numbers P20GM103423 and P20GM104318. This work was supported by the Morris Discovery Fund at MDI Biological Laboratory.
Author contributions
Conceptualization, J.A.R.; methodology, J.A.R., H.M.S., and F.X.A.B.; validation, H.M.S.; formal analysis, J.A.R., H.M.S., and F.X.A.B.; investigation, H.M.S. and J.H.F.; resources, J.H.F.; writing – original draft, J.A.R. and H.M.S.; writing – review & editing, J.A.R. and H.M.S.; visualization, H.M.S.; supervision, J.A.R.; project administration, J.A.R.; funding acquisition, J.A.R.
Declaration of interests
J.A.R. is a paid consultant for Biocomp Instruments (Fredericton, Canada).
Published: April 25, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2022.100203.
Supplemental information
Data and code availability
-
•
Any additional information regarding data sets and acquisition is available from the lead contact by request.
-
•
This paper does not report original code.
-
•
Any additional information required for reanalysis is available from the lead contact.
References
- Andralojc K.M., Campbell A.C., Kelly A.L., Terrey M., Tanner P.C., Gans I.M., Senter-Zapata M.J., Khokhar E.S., Updike D.L. ELLI-1, a novel germline protein, modulates RNAi activity and P-granule accumulation in Caenorhabditis elegans. PLoS Genet. 2017;13:e1006611. doi: 10.1371/journal.pgen.1006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold A., Rahman M.M., Lee M.C., Muehlhaeusser S., Katic I., Gaidatzis D., Hess D., Scheckel C., Wright J.E., Stetak A., et al. Functional characterization of C. elegans Y-box-binding proteins reveals tissue-specific functions and a critical role in the formation of polysomes. Nucleic Acids Res. 2014;42:13353–13369. doi: 10.1093/nar/gku1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett R.M., Liu H., Jin H., Goodman R.H., Cohen M.S. Cell-specific profiling of nascent proteomes using orthogonal enzyme-mediated puromycin incorporation. ACS Chem. Biol. 2016;11:1532–1536. doi: 10.1021/acschembio.5b01076. [DOI] [PubMed] [Google Scholar]
- Beydoun S., Choi H.S., Dela-Cruz G., Kruempel J., Huang S., Bazopoulou D., Miller H.A., Schaller M.L., Evans C.R., Leiser S.F. An alternative food source for metabolism and longevity studies in Caenorhabditis elegans. Commun. Biol. 2021;4:1–9. doi: 10.1038/s42003-021-01764-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derisbourg M.J., Wester L.E., Baddi R., Denzel M.S. Mutagenesis screen uncovers lifespan extension through integrated stress response inhibition without reduced mRNA translation. Nat. Commun. 2021;12:1678. doi: 10.1038/s41467-021-21743-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enam S.U., Zinshteyn B., Goldman D.H., Cassani M., Livingston N.M., Seydoux G., Green R. Puromycin reactivity does not accurately localize translation at the subcellular level. ELife. 2020;9:e60303. doi: 10.7554/eLife.60303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L., Shou Q., Butcher R.A. Identification of a dTDP-rhamnose biosynthetic pathway that oscillates with the molting cycle in Caenorhabditis elegans. Biochem. J. 2016;473:1507–1521. doi: 10.1042/BCJ20160142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko M.V., Peterfi Z., Yim S.H., Gladyshev V.N. Translation elongation rate varies among organs and decreases with age. Nucleic Acids Res. 2020;49:e9. doi: 10.1093/nar/gkaa1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman C.A., Hornberger T.A. Measuring protein synthesis with SUnSET: a valid alternative to traditional techniques? Exerc. Sport Sci. Rev. 2013;41:107–115. doi: 10.1097/JES.0b013e3182798a95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanazawa M., Kawasaki I., Kunitomo H., Gengyo-Ando K., Bennett K.L., Mitani S., Iino Y. The Caenorhabditis elegans eukaryotic initiation factor 5A homologue, IFF-1, is required for germ cell proliferation, gametogenesis and localization of the P-granule component PGL-1. Mech. Development. 2004;121:213–224. doi: 10.1016/j.mod.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Hansen M., Taubert S., Crawford D., Libina N., Lee S.-J., Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- Heissenberger C., Rollins J.A., Krammer T.L., Nagelreiter F., Stocker I., Wacheul L., Shpylovyi A., Tav K., Snow S., Grillari J., et al. The ribosomal RNA m5C methyltransferase NSUN-1 modulates healthspan and oogenesis in Caenorhabditis elegans. ELife. 2020;9:e56205. doi: 10.7554/eLife.56205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrich C.J. A microplate-based nonradioactive protein synthesis assay: application to TRAIL sensitization by protein synthesis inhibitors. PLoS One. 2016;11:e0165192. doi: 10.1371/journal.pone.0165192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidalgo San Jose L., Signer R.A.J. Cell-type-specific quantification of protein synthesis in vivo. Nat. Protoc. 2019;14:441–460. doi: 10.1038/s41596-018-0100-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson B.D., Kong L., Hartwick E.W., Gonzalez R.L., Sims P.A. Elongation inhibitors do not prevent the release of puromycylated nascent polypeptide chains from ribosomes. ELife. 2020;9:e60048. doi: 10.7554/eLife.60048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard A.C., Rollins J., Snow S., Castor S., Rogers A.N. Reducing translation through eIF4G/IFG-1 improves survival under ER stress that depends on heat shock factor HSF-1 in Caenorhabditis elegans. Aging Cell. 2016;15:1027–1038. doi: 10.1111/acel.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelgas-Morales G., Greenstein D. Control of oocyte meiotic maturation in C. elegans. Semin. Cell Dev. Biol. 2018;84:90–99. doi: 10.1016/j.semcdb.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juli G., Gismondi A., Monteleone V., Caldarola S., Iadevaia V., Aspesi A., Dianzani I., Proud C.G., Loreni F. Depletion of ribosomal protein S19 causes a reduction of rRNA synthesis. Sci. Rep. 2016;6:35026. doi: 10.1038/srep35026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath R. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kaymak E., Ryder S.P. RNA recognition by the Caenorhabditis elegans oocyte maturation determinant OMA-1. J. Biol. Chem. 2013;288:30463–30472. doi: 10.1074/jbc.M113.496547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Rollins J.A., Zang X., Wu D., Zou L., Wang Z., Ye C., Wu Z., Kapahi P., Rogers A.N., et al. Translational regulation of non-autonomous mitochondrial stress response promotes longevity. Cell Rep. 2019;28:1050–1062.e6. doi: 10.1016/j.celrep.2019.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N., Berman J.R., Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Liu J., Xu Y., Stoleru D., Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. PNAS. 2012;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounsey A., Bauer P., Hope I.A. Evidence suggesting that a fifth of annotated Caenorhabditis elegans genes may be pseudogenes. Genome Res. 2002;12:770–775. doi: 10.1101/gr208802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousch M., Eckmann C.R. In: Germ Cell Development in C. Elegans. Schedl T., editor. Springer New York; 2013. Translational control in the Caenorhabditis elegans germ line; pp. 205–247. [DOI] [PubMed] [Google Scholar]
- Paix A., Folkmann A., Seydoux G. Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods. 2017;121–122:86–93. doi: 10.1016/j.ymeth.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan K.Z., Palter J.E., Rogers A.N., Olsen A., Chen D., Lithgow G.J., Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–119. doi: 10.1111/j.1474-9726.2006.00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud C.G. Regulation of mammalian translation factors by nutrients: control of translation factors by nutrients. Eur. J. Biochem. 2002;269:5338–5349. doi: 10.1046/j.1432-1033.2002.03292.x. [DOI] [PubMed] [Google Scholar]
- Rattan S.I. Synthesis, modifications, and turnover of proteins during aging. Exp. Gerontol. 1996;31:33–47. doi: 10.1016/0531-5565(95)02022-5. [DOI] [PubMed] [Google Scholar]
- Robinson-Thiewes S., Dufour B., Martel P.-O., Lechasseur X., Brou A.A.D., Roy V., Chen Y., Kimble J., Narbonne P. Non-autonomous regulation of germline stem cell proliferation by somatic MPK-1/MAPK activity in C. elegans. Cell Rep. 2021;35:109162. doi: 10.1016/j.celrep.2021.109162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers A.N., Chen D., McColl G., Czerwieniec G., Felkey K., Gibson B.W., Hubbard A., Melov S., Lithgow G.J., Kapahi P. Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab. 2011;14:55–66. doi: 10.1016/j.cmet.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins J., Rogers A. In: Ageing: Lessons from C. Elegans. Olsen A., Gill M.S., editors. Springer International Publishing; 2017. Translational control of longevity; pp. 285–305. [Google Scholar]
- Rollins J.A., Shaffer D., Snow S.S., Kapahi P., Rogers A.N. Dietary restriction induces posttranscriptional regulation of longevity genes. Life Sci. Alliance. 2019;2:e201800281. doi: 10.26508/lsa.201800281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E.K., Clavarino G., Ceppi M., Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- Schnell U., Dijk F., Sjollema K.A., Giepmans B.N.G. Immunolabeling artifacts and the need for live-cell imaging. Nat. Methods. 2012;9:152–158. doi: 10.1038/nmeth.1855. [DOI] [PubMed] [Google Scholar]
- Schuller A.P., Wu C.C.-C., Dever T.E., Buskirk A.R., Green R. eIF5A functions globally in translation elongation and termination. Mol. Cell. 2017;66:194–205.e5. doi: 10.1016/j.molcel.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer D., Rollins J. Fluorescent polysome profiling in Caenorhabditis elegans. Bio-protocol. 2020;10 doi: 10.21769/BioProtoc.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer F., Moorman C., van der Linden A.M., Kuijk E., van den Berghe P.V.E., Kamath R.S., Fraser A.G., Ahringer J., Plasterk R.H.A. Genome-Wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:e12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smekalova E.M., Gerashchenko M.V., O’Connor P.B.F., Whittaker C.A., Kauffman K.J., Fefilova A.S., Zatsepin T.S., Bogorad R.L., Baranov P.V., Langer R., et al. Vivo RNAi-mediated eIF3m knockdown affects ribosome biogenesis and transcription but has limited impact on mRNA-specific translation. Mol. Ther. Nucleic Acids. 2020;19:252–266. doi: 10.1016/j.omtn.2019.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A., Barth E., Ermolaeva M.A., Guenther M., Frahm C., Marz M., Witte O.W. Tissue-specific gene expression changes are associated with aging in mice. Genomics Proteomics Bioinformatics. 2020;18:430–442. doi: 10.1016/j.gpb.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tain L.S., Sehlke R., Meilenbrock R.L., Leech T., Paulitz J., Chokkalingam M., Nagaraj N., Grönke S., Fröhlich J., Atanassov I., et al. Tissue-specific modulation of gene expression in response to lowered insulin signalling in Drosophila. ELife. 2021;10:e67275. doi: 10.7554/eLife.67275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiku V., Kew C., Mehrotra P., Ganesan R., Robinson N., Antebi A. Nucleolar fibrillarin is an evolutionarily conserved regulator of bacterial pathogen resistance. Nat. Commun. 2018;9:3607. doi: 10.1038/s41467-018-06051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T., Gearhart M.D., Spike C.A., Huelgas-Morales G., Mews M., Boag P.R., Beilharz T.H., Greenstein D. LIN-41 and OMA ribonucleoprotein complexes mediate a translational repression-to-activation switch controlling oocyte meiotic maturation and the oocyte-to-embryo transition in Caenorhabditis elegans. Genetics. 2017;206:2007–2039. doi: 10.1534/genetics.117.203174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward W., Richardson A. Effect of age on liver protein synthesis and degradation. Hepatology. 1991;14:935–948. doi: 10.1002/hep.1840140529. [DOI] [PubMed] [Google Scholar]
- Xiong H., Pears C., Woollard A. An enhanced C. elegans based platform for toxicity assessment. Sci. Rep. 2017;7:1–11. doi: 10.1038/s41598-017-10454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zugasti O., Thakur N., Belougne J., Squiban B., Kurz C.L., Soulé J., Omi S., Tichit L., Pujol N., Ewbank J.J. A quantitative genome-wide RNAi screen in C. elegans for antifungal innate immunity genes. BMC Biol. 2016;14:35. doi: 10.1186/s12915-016-0256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The initial view shows all the fluorophores present, NuclearMask Blue Stain (cyan), GLH-1:GFP (green), RPS-6::mCherry (red), and OPP (magenta). The surfaces created with the Imaris software are displayed (gray), overlaying OPP and GLH::1-GFP. The germline including distal and proximal regions are displayed, demonstrating the 3D regions that were measured from the OPP signal, as well as RPS-6::mCherry.
Data Availability Statement
-
•
Any additional information regarding data sets and acquisition is available from the lead contact by request.
-
•
This paper does not report original code.
-
•
Any additional information required for reanalysis is available from the lead contact.