Abstract
Background
Genetic studies have linked polymorphisms in the interleukin 6 receptor (IL6R) gene to the risk of multiple human diseases and phenotypes, yet have reported inconsistent results. We aimed to synthesize current knowledge of variants in the IL6R gene on the risk of diseases and phenotypes.
Methods
We searched the Medline and Embase databases to identify relevant publications. Meta-analysis was performed utilizing DerSimonian and Laird random-effects model. We also graded cumulative evidence for significant associations. Furthermore, phenome-wide analyses and functional annotations were performed for variants with strong evidence.
Results
We included 155 studies for evaluating the associations between 80 polymorphisms in the IL6R gene and the risk of 102 human diseases and 98 phenotypes. We conducted 58 main meta-analyses, and 41 significant associations were identified. Strong evidence was assigned to 29 associations that investigated ten variants (rs2228145, rs4129267, rs7529229, rs4537545, rs7518199, rs4845625, rs4553185, rs4845618, rs4845371, and rs6667434) related to the risk of four cardiovascular diseases (coronary heart disease, coronary artery disease, atherosclerosis, and abdominal aortic aneurysms), four inflammatory diseases (rheumatoid arthritis, Crohn’s disease, dermatitis, and asthma), and concentration of four phenotypes (C-reactive protein, fibrinogen, IL-6, and sIL-6R). Furthermore, phenome-wide analysis verified that rs2228145 associated with asthma and dermatitis risk. Functional analyses indicated that these polymorphisms fall within exon, enhancer regions.
Conclusions
Our study comprehensively summarizes current data on the genetic architecture of the IL6R gene and highlights the pharmacological targeting potential of IL-6R on cardiovascular and inflammatory diseases.
Keywords: interleukin 6 receptor, variant, cardiovascular diseases, inflammatory diseases, phenotypes
1 Introduction
Interleukin (IL)-6 receptor (IL6R) gene is located in chromosome 1q21.3 (Gene ID: 3570) (1). Its gene production, IL-6R, could specifically bind to its ligand (IL-6) and subsequently recruit two molecules of glycoprotein 130 (gp130) to initiate intercellular signaling (2, 3). IL-6/IL-6R/gp130 signaling is implicated in the development of inflammatory and autoimmune diseases, including novel coronavirus disease 2019 (COVID-19) (4).
Genetic studies have indicated that variants in the IL6R gene are associated with a range of common diseases and phenotypes. In 2003, Kim et al. firstly identified seven single-nucleotide polymorphisms (SNPs) in the IL6R gene in the Korean population (5). One of these variants, Asp358Ala (rs2228145, previously rs8192284), a non-synonymous located in exon 9, has been shown to be associated with the index of obesity in Indians (6) in the same year. Subsequent studies have implicated that Asp358Ala was associated with a series of inflammatory/autoimmune diseases, including inflammatory bowel disease (7), rheumatoid arthritis (8), cardiovascular diseases (CVDs) (9, 10), and type 2 diabetes (11). In 2012, rs2228145 was implicated to play a protective effect in the etiology of CVD diseases, including coronary heart disease (CHD), atrial fibrillation (AF), and abdominal aortic aneurysms (AAA) (12). This effect was verified by findings from a large phenome-wide association analysis among populations from the Million Veteran Program, which showed that rs2228145 was significantly associated with 22 diseases (13). In addition to CVDs, rs2228145 was found to decrease the risk of ulcerative colitis (UC) or Crohn’s disease (CD) by a genome-wide meta-analysis of 38,197 patients with IBD and more than 40,000 controls (7). A genome-wide association study (GWAS) of European ancestry identified that rs2228145 also decreased risk of rheumatoid arthritis (8), and this association has been replicated by subsequent studies (14). Additionally, rs2228145 plays a protective effect in the etiology of type I diabetes (T1D) (12) while positively associated with dermatitis and renal function disorder (13).
Other polymorphisms in the IL6R gene, such as rs4129267 (T allele), rs4845625 (C allele), or rs7529229 (C allele), were also reported to be associated with the risk of many diseases, such as asthma (15) and CVDs (16). A GWAS in Australia identified that rs4129267 (T allele) significantly increased risk of asthma (OR = 1.09, combined p = 2.4×10−8) (15). Then, another GWAS in the Caucasian population indicated that this variant was also associated with an increased risk of AAA. Other large-scale genetic analyses identified that rs4845625 (C allele) and rs7529229 (C allele) were implicated in risk of CVDs, such as atrial fibrillation (17), coronary artery disease (CAD) (16), AAA, and CHD (10).
In addition to diseases, variants in the IL6R gene were also suggested to be related to hematological inflammatory biomarkers and some conventional CVD risk factors. Several publications showed that the C allele of rs2228145 increased circulating soluble IL-6R (sIL-6R) and IL-6 concentrations by 34% and 15%, respectively (7, 9). Furthermore, rs4845371, rs6667434, or rs7529229 was associated with lower levels of circulation of C-reactive protein (CRP), fibrinogen, troponin I, creatine kinase-MB (CK-MB), pro-B-type natriuretic peptide, and systolic blood pressure, which were consistent with a reduced risk of CVDs (9, 13). With the increasing number of genetic studies published since 2003, several meta-analyses have been performed to investigate the role of polymorphisms in IL6R gene on the risk of human diseases and phenotypes. However, existing meta-analyses were limited to a single variant or illness, and the results might not be adequate partly due to the small sample size (18–21). These drawbacks may limit the implementation of the evidence in the clinical practice and disease prevention strategies. Therefore, we aimed to provide a systematic synopsis on the associations between polymorphisms in the IL6R gene and human diseases and phenotypes. Firstly, we conducted a systematic review and meta-analysis to evaluate the epidemiological associations between variants in the IL6R gene and human diseases and phenotypes. Secondly, we graded levels of cumulative evidence for significant associations combining Venice criteria and false-positive report probability (FPRP) tests. Thirdly, we performed a phenome-wide analysis for independent variants with strong evidence using data from UK Biobank to uncover new relationships. Fourthly, we performed functional annotations for the variants in the IL6R gene using data from the Encyclopedia of DNA Elements Project (ENCODE), the 1000 Genomes Project, the Genotype-Tissue Expression (GTEx) Project, and other sources to provide clues for the pathogenesis of these associations.
2 Materials and Methods
The methods used in the present study were based on guidelines proposed by the Human Genome Epidemiology Network (HuGENet) for a systematic review of genetic association studies and the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) (22, 23). This study has been registered to the International Prospective Register of Systematic Reviews (PROSPERO; registration ID: CRD42020201735).
2.1 Literature Search Strategy, Study Eligibility, and Inclusion Criteria
We systematically searched PubMed and Embase using keywords (“interleukin 6 receptor” OR “IL-6R” OR “IL6R” OR “interleukin-6 receptor” OR “IL-6 receptor” OR “rs2228145” OR “Asp358Ala” OR “D358A”) to identify the original genetic association studies of IL6R published in English up to August 1, 2021. We also retrieved published reviews, meta-analyses, pooled analyses, and the included studies to identify additional publications.
Articles were eligible to be included if they met the following inclusion criteria: (i) they were observational studies conducted in humans; (ii) they investigated associations of genetic variants in IL6R gene and human diseases or phenotypes; (iii) they directly provided risk estimates such as odds ratio (OR), relative risk (RR), hazard ratio (HR), and 95% confidence interval (CI), coefficient β and standard error (SE), or provided valid data to calculate these risk estimates under additive genetic model; (iv) or for publications investigating continuous quantitative phenotypes, they should provide mean and standard deviation (SD). When multiple studies use the same population, the most recent or the one with the largest sample size was selected. Studies without full text or sufficient information to calculate effect size were excluded. Two investigators (MZ and YB) independently assessed the eligibility of each publication, and disagreement was settled through discussion with the principal author (DG).
2.2 Data Extraction, Preparation, and Management
Three authors (MZ, YW, and XW) independently extracted the data using a predesigned collection sheet including PMID, first author, publishing year, variants, major and minor alleles, source of population, ethnicity of the study population, study design, sample size of cases and controls, genotype and allele counts, matched information, effects of associations and the corresponding 95% CIs, regression coefficients, SE or p-value (for studies using multiple adjusted models, the most fully adjusted estimates were extracted), mean concentrations of quantitative traits and SD for different genotypes, Hardy–Weinberg equilibrium (HWE) among controls, and genotyping methods.
2.3 Statistical Analysis
In our study, the primary outcome was the risk of disease or categorical phenotypes, measured by OR/RR/HR and 95% CI under an additive genetic model. Due to the relatively low incidence of the events mentioned above, the effect size of OR, HR, and RR was deemed identical in this meta-analysis (24). For continuous quantitative phenotypes, the primary outcome was the mean deviation, measured by standardized mean difference (SMD) and 95% CI after pooling mean concentration and SD under a co-dominant model (mutant-type homozygote vs. wild-type homozygote, heterozygote vs. wild-type homozygote).
Meta-analysis was conducted for variants with at least three independent studies or datasets. We estimated the effects of minor alleles (in Caucasians) on the outcome for each variant. We adopted the methods used in original studies if data were extracted from GWASs or large GWAS meta-analyses from collaborative studies. For heterogeneity assessment, we performed the Cochran’s Q test, and the I 2 statistic was used to evaluate and quantify heterogeneity between studies (25, 26). A random-effects model was performed if heterogeneity was evident (I 2 ≥ 50%), while the fixed effects model was used if I 2 < 50%. For variants that showed significant associations with relevant diseases or phenotypes, sensitivity analyses were conducted to examine whether the associations would be lost when a single study was excluded or when the first positive report or studies with deviation from HWE in controls were excluded. Begg’s test (27) and Egger’s test (28) were conducted to evaluate potential publication bias and small-study bias, respectively.
Statistical analysis was done using STATA, version 15 (Stata, College Station, TX). Tests of heterogeneity and bias were two-tailed, and a p-value of less than 0.10 was considered significant as recommended. All the other statistical tests were two-tailed, and a p-value of < 0.05 was considered statistically significant unless otherwise stated.
2.4 Assessment of Cumulative Evidence
Venice criteria were applied to evaluate the epidemiological credibility of significant associations identified by primary meta-analyses (see Supplementary Notes ) (29). Briefly, credibility was determined by three criteria: (i) amount of evidence, (ii) replication, and (iii) protection from bias. Based on the grade of A, B, or C in these criteria, epidemiological credibility was defined as strong when all three grades were A, moderate when all three grades were A or B, and weak when any grade was C.
We also estimated the noteworthiness of the significant associations by calculating FPRP (30). A prior probability was set at 0.05 to estimate the FPRP value based on the OR obtained from a meta-analysis, and we adopted an FPRP cutoff value of 0.20. Briefly, FPRP value < 0.05, 0.05 < FPRP value < 0.20, and FPRP value > 0.20 were defined as strong, moderate, and weak evidence of true association, respectively. The cumulative epidemiological evidence was adjusted based on the FPRP results: the evidence may be upgraded if FPRP evidence was strong and downgraded if the FPRP result was weak.
2.5 Phenome-Wide Analysis
We performed linkage disequilibrium (LD) analysis for variants significantly associated with risk of diseases and phenotypes in the present meta-analyses, and r 2 > 0.80 was considered as significant using data from the 1000 Genomes Project (31). We then conducted a phenome-wide analysis to estimate associations of the independent variants with 778 phenotypes from UK Biobank in GeneATLAS through querying summary data (32). We considered p-values < 6.43 × 10-5 (0.05/778) as significant, adjusted for multiple comparisons of variants and 778 phenotypes.
2.6 Functional Annotations
We conducted analyses to investigate the potential regulatory effect of polymorphisms significantly associated with relative diseases or phenotypes using the Encyclopedia of DNA Elements (ENCODE) tool HaploReg v4.1 (33) and the University of California, Santa Cruz (UCSC) Genome Browser (34). We examined genome-wide cis-eQTL data in multiple tissues from the Blood eQTL browser (35), and the GTEx Project (36). The significance threshold for these analyses was set to p-values of less than 0.005 accounting for 10 tests. In addition, we searched the published literature for the IL6R gene in relation to diseases from PubMed.
3 Results
3.1 Characteristics of the Included Studies
The literature search strategy is shown in Figure 1 . A total of 155 studies were included for evaluating the associations between 80 polymorphisms in the IL6R gene and the risk of human diseases and phenotypes. Of the 155 publications, 114 regarding the association between 67 variants in the IL6R gene and 102 diseases ( Supplementary Table 1 ) and 67 regarding 50 variants associated with 98 phenotypes ( Supplementary Tables 2, 3 ) were included. A total of 131 articles were cross-sectional or case–control studies, and 24 articles were longitudinal studies. As for the study population, 94 were conducted in populations of European ancestry, 34 were in Asians, four were in the African population, four were in the population of mixed ancestry, and 19 were on other ancestries. The mean number of cases and controls in these articles was 3,855 (range, 17 to 51,441) and 33,415 (range, 20 to 897,488), respectively. Of the 155 studies, 134 were published since 2010 (86.345%).
Figure 1.
Flowchart of the study selection in the present study.
3.2 Relationships Between Variants in IL6R Gene and Human Diseases/Phenotypes by the Meta-Analyses
3.2.1 Relationships Between Variants in IL6R Gene and Human Diseases
We conducted 27 main meta-analyses to estimate associations of six variants in IL6R gene with risk of 20 kinds of diseases under an additive model. Summary findings of meta-analyses are shown in Table 1 . Of the 27 main analyses, 21 associations between six variants and 14 diseases indicated statistical significance (p < 0.05). Specifically, the coronary heart disease risk was negatively associated with minor allele of rs7529229 (OR = 0.954, 95% CI: 0.932–0.977), rs4537545 (OR = 0.940, 95% CI: 0.903–0.978), and rs2228145 (OR = 0.957, 95% CI: 0.948–0.967). Atrial fibrillation risk was inversely associated with minor allele of rs7529229 (OR = 0.900, 95% CI: 0.850–0.960), rs4537545 (OR = 0.901, 95% CI: 0.847–0.952), and rs28638007 (OR = 0.900, 95% CI: 0.850–0.950), but positively associated with rs4845625 (OR = 1.179, 95% CI: 1.044–1.330). The abdominal aortic aneurysms risk was negatively associated with rs7529229 (OR = 0.841, 95% CI: 0.800–0.884), but it was positively associated with rs4129267 (OR = 1.141, 95% CI: 1.101–1.183). As for asthma, rs4129267 (OR = 1.073, 95% CI: 1.035–1.112) and rs2228145 (OR = 1.053, 95% CI: 1.023–1.084) were associated with increased risk of asthma. In addition, the CAD risk was positively associated with rs4845625 (OR = 1.060, 95% CI: 1.042–1.079).
Table 1.
Variants in the IL6R gene associated with risk of human diseases in meta-analysis.
| Variant | Allele a | MAF | Diseases | Datasets | Cases | Controls | Risk of human diseases | Heterogeneity | Venice criteria grade | FPRP | Cumulative evidence of association | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95%CI) | p | І 2 | p | ||||||||||
| rs7529229 | T/C | 0.3535 | CHD | 36 | 26,310 | 101,971 | 0.954 (0.932, 0.977) | 1.07×10-4 | 0.0% | 0.686 | AAA | 0.002 | Strong |
| AAA | 5 | 4,529 | 15,734 | 0.841 (0.800, 0.884) | 1.38×10-11 | 8.6% | 0.322 | AAA | <0.001 | Strong | |||
| Atrial fibrillation | 5 | 2,728 | 16,702 | 0.900 (0.850, 0.960) | 0.001 | NA | B-C | 0.026 | Weak | ||||
| rs4845625 | C/T | 0.4495 | CAD | 47 | 63,434 | 110,256 | 1.060 (1.042, 1.079) | 1.12×10-10 | 15.7% | 0.180 | AAA | <0.001 | Strong |
| Atrial fibrillation | 7 | 2,991 | 20,101 | 1.179 (1.044, 1.330) | 0.007 | 52.4% | 0.122 | ACA | 0.187 | Weak | |||
| rs4537545 | C/T | 0.3535 | CHD | 8 | 13,370 | 30,425 | 0.940 (0.903, 0.978) | 0.002 | 27.7% | 0.207 | ABC | 0.040 | Moderate |
| Atrial fibrillation | 5 | 2,728 | 16,702 | 0.901 (0.847, 0.952) | 2.32×10-4 | NA | B-C | 0.004 | Moderate | ||||
| rs4129267 | C/T | 0.3485 | AAA | 7 | 10,204 | 107,766 | 1.141 (1.101, 1.830) | 4.83×10-13 | 0.0% | 0.655 | AAA | <0.001 | Strong |
| Asthma | 7 | 62,596 | 389,460 | 1.073 (1.035, 1.112) | 1.18×10-4 | 61.1% | 0.017 | AAA | <0.001 | Strong | |||
| rs28638007 | T/C | 0.3956 | Atrial fibrillation | 5 | 2,728 | 16,702 | 0.900 (0.850, 0.950) | 1.07×10-4 | NA | B-C | 0.003 | Moderate | |
| rs2228145 | A/C | 0.3485 | Aneurysm | 5 | 27,819 | 352,035 | 0.881 (0.851, 0.912) | 8.68×10-13 | 71.4% | 0.007 | AAA | <0.001 | Strong |
| Atherosclerosis | 4 | 91,520 | 327,474 | 0.929 (0.908, 0.951) | 6.02×10-10 | 63.9% | 0.040 | AAA | <0.001 | Strong | |||
| CHD | 5 | 204,050 | 1,033,873 | 0.957 (0.948, 0.967) | 3.17×10-17 | 0.0% | 0.212 | AAA | <0.001 | Strong | |||
| Crohn’s disease | 3 | 32,880 | 70,025 | 0.946 (0.927, 0.964) | 2.14×10-8 | 0.0% | 0.950 | AAA | <0.001 | Strong | |||
| Dermatitis | 5 | 141,581 | 326,743 | 1.048 (1.026, 1.071) | 2.31×10-5 | 77.0% | 0.002 | AAA | <0.001 | Strong | |||
| Asthma | 5 | 28,762 | 105,138 | 1.053 (1.023, 1.084) | 4.86×10-4 | 2.9% | 0.390 | AAA | 0.009 | Strong | |||
| Ulcerative colitis | 3 | 30,076 | 75,562 | 0.977 (0.957, 0.996) | 0.018 | 0.0% | 0.672 | AAA | 0.254 | Moderate | |||
| RA | 6 | 18,830 | 51,755 | 0.790 (0.693, 0.901) | 4.48×10-4 | 88.0% | 0.000 | AAA | 0.004 | Strong | |||
| Type 1 diabetes | 4 | 38,522 | 780,577 | 0.940 (0.898, 0.984) | 0.009 | 70.1% | 0.018 | AAA | 0.234 | Moderate | |||
| Type 2 diabetes | 5 | 276,906 | 1,441,968 | 0.975 (0.952, 0.998) | 0.033 | 34.1% | 0.194 | AAA | 0.388 | Moderate | |||
| CVD | 4 | 2,335 | 35,900 | 0.726 (0.535, 0.983) | 0.040 | 80.2% | 0.002 | BCC | 0.796 | Weak | |||
| Obesity | 3 | 1,007 | 402 | 1.084 (0.796, 1.476) | 0.609 | 52.0% | 0.124 | ||||||
| Dengue | 4 | 680 | 850 | 0.837 (0.494, 1.417) | 0.507 | 73.4% | 0.010 | ||||||
| Mental disorder | 7 | 1,845 | 8,481 | 1.051 (0.908, 1.216) | 0.505 | 38.1% | 0.138 | ||||||
| Multiple myeloma | 4 | 958 | 433 | 1.129 (0.791, 1.610) | 0.505 | 64.8% | 0.036 | ||||||
| COPD | 6 | 7,519 | 35,653 | 1.019 (0.965, 1.076) | 0.497 | 0.0% | 0.850 | ||||||
| ACL | 3 | 406 | 411 | 0.937 (0.735, 1.194) | 0.597 | 21.9% | 0.278 | ||||||
Major allele/Minor allele. Venice criteria grade was determined by amount of evidence, replication, and protection from bias. CHD, coronary heart disease; AAA, abdominal aortic aneurysms; CAD, coronary artery disease; RA, rheumatoid arthritis; ACL, anterior cruciate ligament injury; COPD, chronic obstructive pulmonary disease; CVD, cardiovascular disease; NA, Not Available.
Furthermore, rs2228145 (C allele) was associated with decreased risk of aneurysm (OR = 0.881, 95% CI: 0.851–0.912), atherosclerosis (OR = 0.929, 95% CI: 0.908–0.951), rheumatoid arthritis (OR = 0.790, 95% CI: 0.693–0.901), type 1 diabetes (OR = 0.940, 95% CI: 0.898–0.984), CVD (OR = 0.726, 95% CI: 0.535–0.983), type 2 diabetes (OR = 0.975, 95% CI: 0.952–0.998), Crohn’s disease (OR = 0.946, 95% CI: 0.927–0.96), and ulcerative colitis (OR = 0.977, 95% CI: 0.957–0.996) while it was positively associated with dermatitis (OR = 1.048, 95% CI: 1.026–1.071). However, we did not find significant associations between rs2228145 and risk of obesity, dengue, mental disorder, multiple myeloma, chronic obstructive pulmonary disease (COPD), and anterior cruciate ligament injury (p > 0.05).
3.2.2 Relationships Between Variants in IL6R Gene and Phenotypes
As shown in Table 2 , we performed 31 meta-analyses for associations between ten variants in IL6R gene and 13 kinds of categorical phenotypes under an additive model. Twenty significant associations were identified, which investigated ten variants associated with five kinds of phenotypes (p < 0.05) ( Table 2 ). Specifically, the minor allele of six variants, rs7529229 (OR = 0.913, 95% CI: 0.904–0.921), rs6667434 (OR = 0.921, 95% CI: 0.897–0.946), rs4845371 (OR = 0.921, 95% CI: 0.897–0.946), rs4537545 (OR = 0.916, 95% CI: 0.882–0.952), rs4129267 (OR = 0.915, 95% CI: 0.903–0.927), and rs2228145 (OR = 0.909, 95% CI: 0.893–0.925), was significantly associated with decreased CRP level while rs4845625 (OR = 1.088, 95% CI: 1.058–1.117) was positively associated with CRP level. Similarly, the minor allele of four variants, rs7518199 (OR = 0.953, 95% CI: 0.936–0.971), rs4537545 (OR = 0.949, 95% CI: 0.933–0.966), rs4129267 (OR = 0.951, 95% CI: 0.933–0.969), and rs2228145 (OR = 0.960, 95% CI: 0.931–0.99), was also significantly associated with decreased fibrinogen level. Furthermore, the minor allele of five polymorphisms, rs7529229 (OR = 1.089, 95% CI: 1.076–1.102), rs7518199 (OR = 1.090, 95% CI: 1.061–1.121), rs4537545 (OR = 1.098, 95% CI: 1.071–1.126), rs4129267 (OR = 1.093, 95% CI: 1.074–1.113), and rs2228145 (OR = 1.130, 95% CI: 1.088–1.174), was associated with increased IL-6 level, whereas two variants rs4845618 (OR = 0.941, 95% CI: 0.927–0.955) and rs4553185 (OR = 0.936, 95% CI: 0.919–0.953) were associated with decreased IL-6 level. In addition, the minor allele of rs4129267 (OR = 1.271, 95% CI: 1.061–1.523) was positively associated with IL-6R level and rs2228145 (OR = 1.346, 95% CI: 1.318–1.374) significantly increased sIL-6R level.
Table 2.
Variants in the IL6R gene associated with relevant categorical phenotypes in meta-analysis.
| Variant | Allelea | MAF | Phenotypes | Datasets | Numbers | Risk of categorical phenotypes | Heterogeneity | Venice criteria grade | FPRP | Cumulative evidence of association | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p | І 2 | p | |||||||||
| rs7529229 | T/C | 0.3535 | CRP level | 5 | 86,998 | 0.913 (0.904, 0.921) | 2.58×10-81 | 0.0% | 0.779 | AAA | <0.001 | Strong |
| IL-6 level | 3 | 38,194 | 1.089 (1.076, 1.102) | 3.88×10-46 | 39.7% | 0.191 | AAA | <0.001 | Strong | |||
| Fibrinogen level | 8 | 82,958 | 0.976 (0.947, 1.006) | 0.121 | 90.1% | <0.001 | ||||||
| rs7518199 | A/C | 0.3600 | Fibrinogen level | 6 | 23,634 | 0.953 (0.936, 0.971) | 2.75×10-7 | 35.5% | – | AAA | <0.001 | Strong |
| IL-6 level | 3 | 12,546 | 1.090 (1.061, 1.121) | 8.79×10-10 | 80.0% | 0.007 | AAA | <0.001 | Strong | |||
| rs6667434 | G/A | 0.4293 | CRP level | 4 | 10,471 | 0.921 (0.897, 0.946) | 1.06×10-9 | 0.0% | 0.967 | AAA | <0.001 | Strong |
| rs4845371 | C/T | 0.4293 | CRP level | 4 | 10,471 | 0.921 (0.897, 0.946) | 1.06×10-9 | 0.0% | 0.967 | AAA | <0.001 | Strong |
| rs4845625 | C/T | 0.4495 | CRP level | 3 | 8,724 | 1.088 (1.058, 1.117) | 1.45×10-9 | 0.0% | 0.371 | AAC | <0.001 | Moderate |
| rs4845618 | T/G | 0.4394 | IL-6 level | 3 | 12,546 | 0.941 (0.927, 0.955) | 8.98×10-16 | 31.4% | 0.233 | AAA | <0.001 | Strong |
| rs4553185 | T/C | 0.4343 | IL-6 level | 3 | 12,546 | 0.936 (0.919, 0.953) | 6.04×10-4 | 52.5% | 0.122 | AAA | <0.001 | Strong |
| rs4537545 | C/T | 0.3535 | CRP level | 8 | 103,289 | 0.916 (0.882, 0.952) | 9.00×10-6 | 91.8% | <0.001 | AAA | <0.001 | Strong |
| Fibrinogen level | 7 | 41,320 | 0.949 (0.933, 0.966) | 3.03×10-9 | 0.0% | 0.409 | AAA | <0.001 | Strong | |||
| IL-6 level | 3 | 12,546 | 1.098 (1.071, 1.126) | 3.10×10-13 | 76.0% | 0.015 | AAA | <0.001 | Strong | |||
| LDL cholesterol level | 4 | 16,251 | 1.080 (0.728, 1.603) | 0.702 | 0.0% | 0.862 | ||||||
| Total cholesterol level | 4 | 16,251 | 0.977 (0.694, 1.375) | 0.893 | 0.0% | 0.887 | ||||||
| Triglyceride level | 3 | 15,256 | 0.973 (0.879, 1.077) | 0.601 | 28.1% | 0.249 | ||||||
| rs4129267 | C/T | 0.3485 | CRP level | 14 | 358,529 | 0.915 (0.903, 0.927) | 6.71×10-43 | 59.1% | 0.003 | AAA | <0.001 | Strong |
| Fibrinogen level | 7 | 41,320 | 0.951 (0.933, 0.969) | 1.70×10-7 | 6.1% | 0.302 | AAA | <0.001 | Strong | |||
| IL-6 level | 3 | 14,271 | 1.093 (1.074, 1.113) | 7.97×10-18 | 48.3% | 0.145 | ABA | <0.001 | Strong | |||
| IL-6R level | 3 | 911 | 1.271 (1.061, 1.523) | 0.009 | 95.0% | <0.001 | BCC | 0.4000 | Weak | |||
| rs2228145 | A/C | 0.3485 | CRP level | 14 | 483,500 | 0.909 (0.893, 0.925) | 3.32×10-26 | 68.6% | <0.001 | AAA | <0.001 | Strong |
| sIL-6R level | 6 | 8,149 | 1.346 (1.318, 1.374) | 4.66×10-171 | 54.6% | 0.051 | ABA | <0.001 | Strong | |||
| IL-6 level | 5 | 42,267 | 1.130 (1.088, 1.174) | 3.17×10-10 | 89.6% | <0.001 | AAA | <0.001 | Strong | |||
| Fibrinogen level | 9 | 98,330 | 0.960 (0.931, 0.990) | 0.009 | 87.8% | <0.001 | AAA | 0.150 | Moderate | |||
| LDL cholesterol level | 3 | 81,202 | 1.000 (0.996, 1.003) | 0.857 | 0.0% | 0.708 | ||||||
| HDL cholesterol level | 5 | 97,754 | 1.076 (0.860, 1.347) | 0.520 | 62.1% | 0.032 | ||||||
| Triglyceride level | 5 | 96,257 | 1.067 (0.855, 1.332) | 0.568 | 59.0% | 0.045 | ||||||
| Systolic blood pressure | 3 | 100,502 | 0.894 (0.713, 1.120) | 0.329 | 44.6% | 0.165 | ||||||
| Fasting glucose | 4 | 185,044 | 1.000 (0.998, 1.003) | 0.799 | 0.0% | 0.668 | ||||||
| Waist circumference | 3 | 69,772 | 1.002 (0.998, 1.005) | 0.283 | 46.7% | 0.131 | ||||||
| BMI | 4 | 895,213 | 1.001 (0.998, 1.004) | 0.383 | 40.7% | 0.168 | ||||||
Venice criteria grade was determined by the amount of evidence, replication, and protection from bias. aMajor allele/Minor allele. CRP, C-reactive protein; IL-6, interleukin 6; IL-6R, interleukin 6 receptor; sIL-6R, soluble IL-6R.
We also conducted meta-analyses for rs2228145 and 12 continuous phenotypes under a co-dominant model ( Supplementary Table 4 ). The results showed that rs2228145 was significantly associated with the levels of IL-6 and CRP.
3.2.3 Heterogeneity, Sensitivity Analysis, and Bias
As shown in Tables 1 and 2 , 24 associations revealed no or little heterogeneity (I 2 < 25%), ten associations revealed moderate heterogeneity (25% > I 2 <50%) and 24 associations revealed strong heterogeneity (I 2 > 50%). The results indicated that publication bias was found in three associations (p < 0.10), and a small study bias was found in nine associations (p < 0.10) ( Supplementary Table 5 ). In addition, sensitivity analysis suggested that most associations did not significantly change when a single or first positive study, small studies, or studies with deviation from HWE in controls were excluded (data not shown).
3.3 Cumulative Evidence of Significant Associations
Using Venice criteria and FPRP tests, we graded cumulative evidence level for the 41 significant associations, which evaluated associations between 11 variants in the IL6R gene and 14 diseases and five phenotypes ( Tables 1 , 2 and Supplementary Table 5 ). According to Venice criteria, strong, moderate, and weak evidence were assigned to 29, four, and 12 associations, respectively. The FPRP test suggested that 34 of them had an FPRP value less than 0.05, two associations had an FPRP value between 0.05 and 0.20, and five associations had an FPRP value greater than 0.20. Combining Venice Criteria and FPRP results, strong, moderate, and weak evidence were finally assigned to 29, 8, and four associations, respectively. The 29 strong associations evaluated ten polymorphisms related to nine diseases and four phenotypes.
3.4 Phenome-Wide Profile
Linkage disequilibrium analysis ( Supplementary Table 6 ) showed that five variants (rs2228145, rs7518199, rs4537545, rs7529229, and rs4129267) were highly correlated with each other, while another five SNPs (rs4845625, rs4845618, rs4845371, rs6667434, and rs4553185) were also highly correlated with each other according to r 2 (r 2 > 0.8) in the European population.
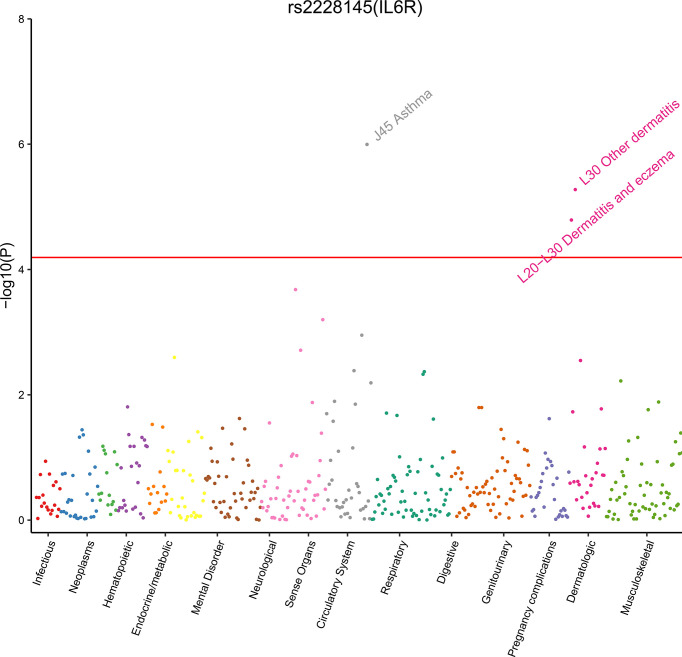
Therefore, phenome-wide association analysis was just conducted for two SNPs (rs2228145 and rs4845625) using data from UK Biobank. The results are shown in Table 3 and Figure 2 . Variant rs2228145 and its highly correlated SNPs were found to be significantly associated with three binary phenotypes (asthma, eczema/dermatitis, and other dermatitis) and 11 non-binary phenotypes (monocyte percentage, monocyte count, mean corpuscular hemoglobin, mean corpuscular volume, mean platelet volume, platelet count, red blood cell distribution width, hemoglobin concentration, platelet distribution width, lymphocyte count, and mean sphered cell volume) at p < 6.43 × 10-6. Variant rs4845625 and its highly correlated SNPs were significantly associated with four non-binary phenotypes of mean platelet (thrombocyte) volume, platelet count, mean corpuscular hemoglobin, and monocyte count.
Table 3.
Significant associations in phenome-wide analysis of the two independent variants using data from UK Biobank.
| SNP | Position | Allelea | MAF | Phenotype (ICD10 code) | UK Biobank (n = 452,264) | ||
|---|---|---|---|---|---|---|---|
| Cases | OR (95% CI) | p | |||||
| rs2228145 | 154426970 | A/C | 0.4094 | Asthma (J45) | 28,628 | 1.003 (1.002, 1.004) | 1.29×10-6 |
| Eczema/dermatitis | 11,552 | 1.002 (1.001, 1.003) | 7.96×10-9 | ||||
| Other dermatitis (L30) | 1,654 | 1.001 (1.000, 1.001) | 5.26×10-6 | ||||
| Monocyte percentage | 14,307 | 1.040 (1.032, 1.048) | 1.09×10-24 | ||||
| Monocyte count | 14,322 | 1.003 (1.002, 1.003) | 2.66×10-19 | ||||
| Mean corpuscular hemoglobin | 13,433 | 1.024 (1.018, 1.030) | 2.19×10-16 | ||||
| Mean corpuscular volume | 13,219 | 1.060 (1.045, 1.075) | 1.07×10-15 | ||||
| Mean platelet (thrombocyte) volume | 13,220 | 0.988 (0.985, 0.991) | 9.81×10-14 | ||||
| Platelet count | 13,258 | 1.761 (1.467, 2.113) | 1.21×10-9 | ||||
| Red blood cell (erythrocyte) distribution width | 13,429 | 0.991 (0.988, 0.994) | 1.30×10-7 | ||||
| Hemoglobin concentration | 13,226 | 1.009 (1.005, 1.012) | 4.85×10-7 | ||||
| Platelet distribution width | 13,220 | 0.996 (0.994, 0.998) | 2.10×10-6 | ||||
| Lymphocyte count | 14,209 | 0.995 (0.993, 0.997) | 4.39×10-6 | ||||
| Mean sphered cell volume | 20,355 | 0.961 (0.945, 0.978) | 6.01×10-6 | ||||
| rs4845625 | 154422067 | C/T | 0.4221 | Mean platelet (thrombocyte) volume | 13,220 | 1.011 (1.008, 1.015) | 2.79×10-12 |
| Platelet count | 13,258 | 0.570 (0.475, 0.685) | 1.62×10-9 | ||||
| Mean corpuscular hemoglobin | 13,433 | 0.986 (0.980, 0.992) | 1.63×10-6 | ||||
| Monocyte count | 14,322 | 0.999 (0.998, 0.999) | 6.08×10-6 | ||||
MAF in UB Biobank. aMajor allele/Minor allele.
Figure 2.
Plot of phenome-wide analysis for rs2228145 using data from UB Biobank. We estimated rs2228145 associated with 778 phenotypes in UK Biobank cohort. Phenotypes are grouped along the x-axis by categorization within the PheWAS code hierarchy. We considered a p-value < 6.43 × 10−5 (the red line) as significant.
3.5 Functional Annotation of Variants With Strong Evidence
Bioinformatics analyses suggested that the ten variants with strong evidence and variants highly correlated with them might fall within an exon, a DNase I hypersensitivity site, a strong prompter, and an enhancer activity region ( Table 4 and Supplementary Figure 1 ). Of those SNPs, rs2228145 is a nonsynonymous variant located in exon 9 of the IL6R gene. In variant rs2228145, A to C transition is observed at position 358 in the IL6R gene, resulting in the alteration of amino acid from alanine to aspartate or valine. The C allele of rs2228145 is associated with inflammatory/autoimmune diseases as well as quantitative traits (7, 9), and the mechanisms might be involved in regulating the balance of transmembrane IL-6R (mIL-6R) and sIL-6R concentrations and responding to IL-6 (12). Additionally, rs2228145 is in LD with four intronic SNPs (rs4129267, rs4537545, rs7529229, and rs7518199) in Europeans (r 2 > 0.85 for all tests), indicating similar functional mechanisms in this ethnic group. According to LD analysis in the European population, cis-eQTL analysis was conducted for rs2228145 and rs4845625 ( Supplementary Table 7 ). The findings indicate that rs2228145 and rs4845625 are associated with a decrease in IL-6R in an artery or heart tissue. In addition, rs2228145 is an eQTL for SHE, TDRD10, and UBE2Q1 genes, and rs4845625 is an eQTL for PSMD8P1, TDRD10, UBAP2L, and AQP10 genes.
Table 4.
Functional annotation of the ten variants showing strong evidence using data from the ENCODE project.
| SNP | Position a | Allele b | RAF in CEU/ASN/AFR | Annotation | Promoter histone marks c | Enhancer histone marks d | DNase e | Proteins bound f | Motifs changed g |
|---|---|---|---|---|---|---|---|---|---|
| rs2228145 | 154454494 | A/C,T | 0.36/0.36/0.09 | Missense | 5 tissues | 16 tissues | |||
| rs4129267 | 154453788 | C/T | 0.36/0.36/0.09 | Intronic | 8 tissues | 20 tissues | 24 tissues | STAT3, KAP1 | EBF, Elf3, Pou1f1 |
| rs4537545 | 154446403 | C/T | 0.37/0.35/0.68 | Intronic | 4 tissues | 16 tissues | 18 tissues | MAFK | |
| rs7529229 | 154448302 | T/C | 0.37/0.36/0.69 | Intronic | GI | 10 tissues | MUS | ZEB1 | |
| rs4553185 | 154438479 | C/T | 0.54/0.46/0.46 | Intronic | BLD, SKIN, GI | 12 tissues | Pax-6, Pou3f4, RORalpha1 | ||
| rs4845371 | 154435864 | T/C | 0.55/0.46/0.68 | Intronic | 4 tissues | 17 tissues | MUS | 4 altered motifs | |
| rs4845618 | 154427539 | G/T | 0.53/0.48/0.48 | Intronic | 8 tissues | 23 tissues | 5 tissues | HDAC2, Irf, STAT | |
| rs4845625 | 154449591 | T/C | 0.54/0.47/0.72 | Intronic | 8 tissues | PLCNT | Pax-2, Pax-5 | ||
| rs6667434 | 154436624 | A/G | 0.55/0.46/0.67 | Intronic | ESDR, SKIN, GI | 13 tissues | 4 tissues | BCL, NRSF | |
| rs7518199 | 154434943 | A/C | 0.36/0.37/0.2 | Intronic | 10 tissues | 19 tissues | 10 tissues | NRSF |
The chromosome position (bp) is based on NCBI, Build 38.
Reference allele/Risk allele.
Evidence of local H3K4Me1 and H3K27Ac modification (cell lines/types: if >3, only the number is included).
Evidence of local H3K4Me3 modification (cell lines/types: if >3, only the number is included).
Evidence of chromatin hypersensitivity to DNase (cell lines/types: if >3, only the number is included).
ChIP-seq experiments indicate alteration in binding of transcription factor (if >3, only the number is included).
Evidence of alteration in regulatory motif (if >3, only the number is included).
RAF, risk allele frequency; CEU, Utah residents with Northern and Western European ancestry from the CEPH collection; ASN, Asian; AFR, African.
4 Discussion
Our study summarizes the published data thus far on relationships between variants in the IL6R gene and the risk of human diseases and phenotypes. The meta-analyses results show that 12 variants in the IL6R gene are significantly associated with risk of CVDs (CAD, coronary heart disease, atrial fibrillation, aneurysms, and atherosclerosis), inflammatory or autoimmune diseases (rheumatoid arthritis, asthma, Crohn’s disease, ulcerative colitis, dermatitis, and type 1 diabetes), and levels of inflammatory biomarkers (CRP, fibrinogen, IL-6, IL-6R, and sIL-6R). We then assigned strong cumulative evidence to 29 significant associations, which assessed ten variants in this gene related to the risk of five cardiovascular diseases, four inflammatory diseases, and the concentration of four phenotypes. Further analysis of functional annotation indicates that the strong variants might fall in putative functional regions.
IL6R gene encodes a subunit of an IL-6R protein complex that exists in two forms, an mIL-6R and sIL-6R, resulting in different signal transduction mechanisms known as IL-6/IL-6R classic signaling and trans-signaling, respectively. These two models of signaling can mediate the anti-inflammatory (classic signaling) and pro-inflammatory (trans-signaling) response of IL-6 (37, 38). By specifically binding to IL-6, IL-6/IL-6R signaling plays an essential role in the pathogenesis of many diseases, including COVID-2019 (4, 7, 9, 10).
Previous studies have suggested that rs2228145 has similar synergistic effects as tocilizumab on a series of cardiovascular and inflammatory biomarkers (10, 12, 19). Consistent with this, our meta-analysis found that this variant was significantly related to a decreased serum concentration of CRP and fibrinogen. There were also consistent associations with strong cumulative evidence between rs2228145 and a reduced risk of CHD, atherosclerosis, AAA, rheumatoid arthritis, and inflammatory bowel disease identified by this meta-analysis, findings that suggest targeting the IL-6R is a plausible strategy in cardiovascular and inflammatory diseases. On the contrary, the C allele of rs2228145 is strongly related to higher levels of sIL-6R and IL-6, which seems paradoxical. This apparent paradox might be interpreted that the C allele reduces mIL-6R level either by alternative splicing or increasing the membrane-bound form shedding rather than by regulating the production of IL-6R or IL-6 (9). Such mechanisms could account for the accumulation of both sIL-6R and IL-6 in the circulation of individuals carrying the C allele of rs2228145.
The mutation of rs2228145 promotes the proteolysis of the IL-6R, which increases the level of sIL-6R and decreases the level of mIL-6R (39). Thus, this variant exerts its functional mechanism by regulating the balance between mIL-6R and sIL-6R (12). The minor allele of rs2228145 strongly increases the concentration of sIL-6R, whereas it reduces the surface expression of mIL-6R in individual immune cells. Reducing mIL-6R expression leads to impaired IL-6 responsiveness, as measured by decreased phosphorylation of the transcription factors STAT3 and STAT1 following stimulation with IL-6 (12, 19). Therefore, it is speculated that rs2228145 impairs classical IL-6/IL-6R signaling and reduces the inflammatory response, thus leading to a lower risk of CVDs, inflammatory, or autoimmune diseases (19). Our findings also link rs2228145 with an increased risk of asthma and dermatitis, but the exact mechanisms for these associations remain unclear. Experimental evidence indicates that IL-6/sIL-6R trans-signaling might play an essential role in the development of asthma (40, 41) and dermatitis (42). Further studies are warranted to elucidate these mechanisms.
LD analysis using data from the 1000 Genomes Project reveals that four variants (rs7518199, rs4537545, rs7529229, and rs4129267) are highly correlated with rs2228145 in the European population (r 2 > 0.90 for all tests), indicating that the functional mechanisms of these SNPs associated with diseases and phenotypes might be similar. In addition, the variant rs4845625 (T allele) strongly associated with CAD is in weak or moderate LD with rs2228145 (r 2 = 0.04, 0.462, and 0.63 in African, European, and Asian populations, respectively), reflecting that functional mechanisms of IL6R variants associated with CVD risk might be distinct. Additional studies are needed to investigate rs4845625 and its related SNPs affecting the development of CVD.
Clinically, targeting the IL-6/IL-6R pathway is a plausible therapeutic strategy in several cardiovascular and inflammatory diseases, including COVID-2019 (37, 43–45). For example, clinical trials suggest that tocilizumab, a humanized monoclonal biologic drug that blocks both sIL-6R and mIL-6R, is an effective treatment for rheumatoid arthritis (44). In addition, sgp130Fc, a protein that specifically blocks trans-signaling, is an effective therapy for various preclinical chronic and autoimmune disease models (37, 42). Therefore, our findings have clinical implications and may provide tools for identifying patients who can benefit from the therapeutic intervention in the IL-6/IL-6R pathway.
Some limitations should be noted in this study. Firstly, although we have tried our best to perform a comprehensive meta-analysis to include all the relevant literature, a small part of articles may still be missed. Secondly, several studies were omitted without valid data for extraction. For example, some articles that just provide a histogram of the average level of phenotypes corresponding to different genotypes were excluded. Thirdly, meta-analysis was not performed for some diseases and phenotypes, such as breast cancer, colorectal cancer, stroke, and systemic lupus erythematosus because fewer than three datasets were included. Fourthly, we could not define a unified analytical standard across studies since we could not obtain raw data from the original studies. To minimize bias, we adopted age, sex, or multivariable-adjusted risk estimates. Fifthly, moderate and weak evidence of significant associations needs to be explained with caution. Future studies with a larger sample size are required to refute or confirm these associations.
In conclusion, our study provides comprehensive evidence that variants in the IL6R gene are associated with the risk of inflammatory or autoimmune diseases and levels of inflammatory biomarkers. Also, it highlights the significant role of the IL-6/IL-6R pathway in the pathogenesis of cardiovascular and inflammatory diseases. Phenome-wide association analysis confirmed that variants in the IL6R gene are associated with asthma and dermatitis. Bioinformatics analyses showed that most of these variants fall in putative functional regions. Those findings highlight that the variations in the IL6R gene may be considered a useful genetic tool for investigating the pharmacological targeting potential of IL-6R. More efforts should be made to understand the biological pathways further and apply these lines of evidences for clinical practice and public health for risk evaluation and management.
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.
Ethics Statement
Ethical review and approval were not required for the study on human participants in accordance with the local legislation and institutional requirements. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
DG initiated and designed this study and approved the final draft manuscript. MZ and YB performed study selection, data extraction, and statistical analysis. LW, HC, and MT performed the cumulative evidence assessments and phenome-wide analysis. YW and XW conducted the functional analyses. MZ also drafted the first manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Natural Science Foundation of China (81903393 and 81903398). The funding agencies of this study had no role in study design, data collection, data management, data analysis, data interpretation, writing of the manuscript, or the decision for submission.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.860703/full#supplementary-material
References
- 1. van Dongen J, Jansen R, Smit D, Hottenga JJ, Mbarek H, Willemsen G, et al. The Contribution of the Functional IL6R Polymorphism Rs2228145, eQTLs and Other Genome-Wide SNPs to the Heritability of Plasma sIL-6R Levels. Behav Genet (2014) 44:368–82. doi: 10.1007/s10519-014-9656-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang X, Guo Y, Wen C, Lv M, Gan N, Zhou H, et al. Molecular Characterization of Grass Carp Interleukin-6 Receptor and the Agonistic Activity of Its Soluble Form in Head Kidney Leucocytes. Fish Shellfish Immunol (2019) 86:1072–80. doi: 10.1016/j.fsi.2018.12.031 [DOI] [PubMed] [Google Scholar]
- 3. Mihara M, Hashizume M, Yoshida H, Suzuki M, Shiina M. IL-6/IL-6 Receptor System and its Role in Physiological and Pathological Conditions. Clin Sci (Lond) (2012) 122:143–59. doi: 10.1042/cs20110340 [DOI] [PubMed] [Google Scholar]
- 4. Karcioglu Batur L, Hekim N. Correlation Between Interleukin Gene Polymorphisms and Current Prevalence and Mortality Rates Due to Novel Coronavirus Disease 2019 (COVID-2019) in 23 Countries. J Med Virol (2021) 93:5853–63. doi: 10.1002/jmv.27127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim LH, Lee HS, Kim YJ, Jung JH, Kim JY, Park BL, et al. Identification of Novel SNPs in the Interleukin 6 Receptor Gene (IL6R). Hum Mutat (2003) 21:450–1. doi: 10.1002/humu.9130 [DOI] [PubMed] [Google Scholar]
- 6. Wolford JK, Colligan PB, Gruber JD, Bogardus C. Variants in the Interleukin 6 Receptor Gene Are Associated With Obesity in Pima Indians. Mol Genet Metab (2003) 80:338–43. doi: 10.1016/j.ymgme.2003.07.003 [DOI] [PubMed] [Google Scholar]
- 7. Parisinos CA, Serghiou S, Katsoulis M, George MJ, Patel RS, Hemingway H, et al. Variation in Interleukin 6 Receptor Gene Associates With Risk of Crohn’s Disease and Ulcerative Colitis. Gastroenterology (2018) 155:303–306.e2. doi: 10.1053/j.gastro.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Eyre S, Bowes J, Diogo D, Lee A, Barton A, Martin P, et al. High-Density Genetic Mapping Identifies New Susceptibility Loci for Rheumatoid Arthritis. Nat Genet (2012) 44:1336–40. doi: 10.1038/ng.2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Collaboration IRGCERF. Sarwar N, Butterworth AS, Freitag DF, Gregson J, Willeit P, et al. Interleukin-6 Receptor Pathways in Coronary Heart Disease: A Collaborative Meta-Analysis of 82 Studies. Lancet (2012) 379:1205–13. doi: 10.1016/S0140-6736(11)61931-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Interleukin-6 Receptor Mendelian Randomisation Analysis Consortium. Swerdlow DI, Holmes MV, Kuchenbaecker KB, Engmann JE, Shah T, et al. The Interleukin-6 Receptor as a Target for Prevention of Coronary Heart Disease: A Mendelian Randomisation Analysis. Lancet (2012) 379:1214–24. doi: 10.1016/S0140-6736(12)60110-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hamid YH, Urhammer SA, Jensen DP, Glümer C, Borch-Johnsen K, Jørgensen T, et al. Variation in the Interleukin-6 Receptor Gene Associates With Type 2 Diabetes in Danish Whites. Diabetes (2004) 53:3342–5. doi: 10.2337/diabetes.53.12.3342 [DOI] [PubMed] [Google Scholar]
- 12. Ferreira RC, Freitag DF, Cutler AJ, Howson JM, Rainbow DB, Smyth DJ, et al. Functional IL6R 358Ala Allele Impairs Classical IL-6 Receptor Signaling and Influences Risk of Diverse Inflammatory Diseases. PloS Genet (2013) 9:e1003444. doi: 10.1371/journal.pgen.1003444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cai T, Zhang Y, Ho YL, Link N, Sun J, Huang J, et al. Association of Interleukin 6 Receptor Variant With Cardiovascular Disease Effects of Interleukin 6 Receptor Blocking Therapy: A Phenome-Wide Association Study. JAMA Cardiol (2018) 3:849–57. doi: 10.1001/jamacardio.2018.2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ahmed S, Hussain S, Ammar A, Jahan S, Khaliq S, Kaul H. Interleukin 6 Receptor (IL6-R) Gene Polymorphisms Underlie Susceptibility to Rheumatoid Arthritis. Clin Lab (2017) 63:1365–9. doi: 10.7754/Clin.Lab.2017.170216 [DOI] [PubMed] [Google Scholar]
- 15. Ferreira MA, Matheson MC, Duffy DL, Marks GB, Hui J, Le Souef P, et al. Identification of IL6R and Chromosome 11q13.5 as Risk Loci for Asthma. Lancet (2011) 378:1006–14. doi: 10.1016/S0140-6736(11)60874-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Consortium CAD. Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, et al. Large-Scale Association Analysis Identifies New Risk Loci for Coronary Artery Disease. Nat Genet (2013) 45:25–33. doi: 10.1038/ng.2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schnabel RB, Kerr KF, Lubitz SA, Alkylbekova EL, Marcus GM, Sinner MF, et al. Large-Scale Candidate Gene Analysis in Whites and African Americans Identifies IL6R Polymorphism in Relation to Atrial Fibrillation: The National Heart, Lung, and Blood Institute’s Candidate Gene Association Resource (CARe) Project. Circ Cardiovasc Genet (2011) 4:557–64. doi: 10.1161/CIRCGENETICS.110.959197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y, Du Z, Wang X, Wang G, Li W. Association of IL-6 Promoter and Receptor Polymorphisms With Multiple Myeloma Risk: A Systematic Review and Meta-Analysis. Genet Test Mol Biomarkers (2016) 20:587–96. doi: 10.1089/gtmb.2015.0169 [DOI] [PubMed] [Google Scholar]
- 19. Harrison SC, Smith AJ, Jones GT, Swerdlow DI, Rampuri R, Bown MJ, et al. Interleukin-6 Receptor Pathways in Abdominal Aortic Aneurysm. Eur Heart J (2013) 34:3707–16. doi: 10.1093/eurheartj/ehs354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou J, Chen X, Ye H, Peng P, Ba Y, Yang X, et al. An Association Study Between Genetic Polymorphism in the Interleukin-6 Receptor Gene and Coronary Heart Disease. BioMed Res Int (2014) 2014:504727. doi: 10.1155/2014/504727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hudson ZD, Miller BJ. Meta-Analysis of Cytokine and Chemokine Genes in Schizophrenia. Clin Schizophr Relat Psychoses (2018) 12:121–9B. doi: 10.3371/CSRP.HUMI.070516 [DOI] [PubMed] [Google Scholar]
- 22. Sagoo GS, Little J, Higgins JP. Systematic Reviews of Genetic Association Studies. Human Genome Epidemiology Network. PloS Med (2009) 6:e28. doi: 10.1371/journal.pmed.1000028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shamseer L, Moher D, Clarke M, Ghersi D, Liberati A, Petticrew M, et al. Preferred Reporting Items for Systematic Review and Meta-Analysis Protocols (PRISMA-P) 2015: Elaboration and Explanation. BMJ (2015) 350:g7647. doi: 10.1136/bmj.g7647 [DOI] [PubMed] [Google Scholar]
- 24. Lu D, Pan C, Ye C, Duan H, Xu F, Yin L, et al. Meta-Analysis of Soy Consumption and Gastrointestinal Cancer Risk. Sci Rep (2017) 7:4048. doi: 10.1038/s41598-017-03692-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lau J, Ioannidis JP, Schmid CH. Quantitative Synthesis in Systematic Reviews. Ann Intern Med (1997) 127:820–6. doi: 10.7326/0003-4819-127-9-199711010-00008 [DOI] [PubMed] [Google Scholar]
- 26. Higgins JP, Thompson SG. Quantifying Heterogeneity in a Meta-Analysis. Stat Med (2002) 21:1539–58. doi: 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 27. Begg CB, Mazumdar M. Operating Characteristics of a Rank Correlation Test for Publication Bias. Biometrics (1994) 50:1088–101. doi: 10.2307/2533446 [DOI] [PubMed] [Google Scholar]
- 28. Egger M, Davey Smith G, Schneider M, Minder C. Bias in Meta-Analysis Detected by a Simple, Graphical Test. BMJ (1997) 315:629–34. doi: 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ioannidis JP, Boffetta P, Little J, O’Brien TR, Uitterlinden AG, Vineis P, et al. Assessment of Cumulative Evidence on Genetic Associations: Interim Guidelines. Int J Epidemiol (2008) 37:120–32. doi: 10.1093/ije/dym159 [DOI] [PubMed] [Google Scholar]
- 30. Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the Probability That a Positive Report Is False: An Approach for Molecular Epidemiology Studies. J Natl Cancer Inst (2004) 96:434–42. doi: 10.1093/jnci/djh075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Genomes Project C. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An Integrated Map of Genetic Variation From 1,092 Human Genomes. Nature (2012) 491:56–65. doi: 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Canela-Xandri O, Rawlik K, Tenesa A. An Atlas of Genetic Associations in UK Biobank. Nat Genet (2018) 50:1593–9. doi: 10.1038/s41588-018-0248-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ward LD, Kellis M. HaploReg: A Resource for Exploring Chromatin States, Conservation, and Regulatory Motif Alterations Within Sets of Genetically Linked Variants. Nucleic Acids Res (2012) 40:D930–4. doi: 10.1093/nar/gkr917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, et al. The UCSC Genome Browser Database: 2018 Update. Nucleic Acids Res (2018) 46:D762–9. doi: 10.1093/nar/gkx1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic Identification of Trans eQTLs as Putative Drivers of Known Disease Associations. Nat Genet (2013) 45:1238–43. doi: 10.1038/ng.2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Consortium GT . Human Genomics. The Genotype-Tissue Expression (GTEx) Pilot Analysis: Multitissue Gene Regulation in Humans. Science (2015) 348:648–60. doi: 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rose-John S. The Soluble Interleukin 6 Receptor: Advanced Therapeutic Options in Inflammation. Clin Pharmacol Ther (2017) 102:591–8. doi: 10.1002/cpt.782 [DOI] [PubMed] [Google Scholar]
- 38. Zegeye MM, Lindkvist M, Fälker K, Kumawat AK, Paramel G, Grenegård M, et al. Activation of the JAK/STAT3 and PI3K/AKT Pathways Are Crucial for IL-6 Trans-Signaling-Mediated Pro-Inflammatory Response in Human Vascular Endothelial Cells. Cell Commun Signal (2018) 16:55. doi: 10.1186/s12964-018-0268-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garbers C, Monhasery N, Aparicio-Siegmund S, Lokau J, Baran P, Nowell MA, et al. The Interleukin-6 Receptor Asp358Ala Single Nucleotide Polymorphism Rs2228145 Confers Increased Proteolytic Conversion Rates by ADAM Proteases. Biochim Biophys Acta (2014) 1842:1485–94. doi: 10.1016/j.bbadis.2014.05.018 [DOI] [PubMed] [Google Scholar]
- 40. Ruwanpura SM, McLeod L, Dousha LF, Seow HJ, Alhayyani S, Tate MD, et al. Therapeutic Targeting of the IL-6 Trans-Signaling/Mechanistic Target of Rapamycin Complex 1 Axis in Pulmonary Emphysema. Am J Respir Crit Care Med (2016) 194:1494–505. doi: 10.1164/rccm.201512-2368OC [DOI] [PubMed] [Google Scholar]
- 41. Wang Y, Hu H, Wu J, Zhao X, Zhen Y, Wang S, et al. The IL6R Gene Polymorphisms Are Associated With sIL-6r, IgE and Lung Function in Chinese Patients With Asthma. Gene (2016) 585:51–7. doi: 10.1016/j.gene.2016.03.026 [DOI] [PubMed] [Google Scholar]
- 42. Kano H, Kita J, Makino S, Ikegami S, Itoh H. Oral Administration of Lactobacillus Delbrueckii Subspecies Bulgaricus OLL1073R-1 Suppresses Inflammation by Decreasing Interleukin-6 Responses in a Murine Model of Atopic Dermatitis. J Dairy Sci (2013) 96:3525–34. doi: 10.3168/jds.2012-6514 [DOI] [PubMed] [Google Scholar]
- 43. Biggioggero M, Crotti C, Becciolini A, Favalli EG. Tocilizumab in the Treatment of Rheumatoid Arthritis: An Evidence-Based Review and Patient Selection. Drug Des Devel Ther (2019) 13:57–70. doi: 10.2147/dddt.S150580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yao X, Huang J, Zhong H, Shen N, Faggioni R, Fung M, et al. Targeting Interleukin-6 in Inflammatory Autoimmune Diseases and Cancers. Pharmacol Ther (2014) 141:125–39. doi: 10.1016/j.pharmthera.2013.09.004 [DOI] [PubMed] [Google Scholar]
- 45. Toniati P, Piva S, Cattalini M, Garrafa E, Regola F, Castelli F, et al. Tocilizumab for the Treatment of Severe COVID-19 Pneumonia With Hyperinflammatory Syndrome and Acute Respiratory Failure: A Single Center Study of 100 Patients in Brescia, Italy. Autoimmun Rev (2020) 19:102568. doi: 10.1016/j.autrev.2020.102568 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding author.