Abstract
Spoilage in cooked ham is one of the main challenges where microbial contamination can play a fundamental role. This study aimed to characterize pork-cooked ham's microbial community changes among different food production conditions (formulation and processing) using 16S rRNA sequencing and also to investigate the spoilage bacteria in order to elucidate their contamination route. Samples of three pork-cooked ham references with and without post-pasteurization treatment and in contact with the slicing-packaging conveyor belt and slicer and packager surfaces were performed by 16S rRNA gene sequencing. In order to clarify the contamination route, surfaces were sampled by conventional microbiological methods. Results showed that Leuconostoc spp. was the principal genera in spoiled cooked ham and had no relation neither to formulation nor contact with the slicing-packaging conveyor belt. The contamination route found for Leuconostoc spp. was associated with the storage and packaging zone. In addition, the calculated shelf-life decreased to 57.5% independently of the environment interaction minimization when ham casing permeability was changed and linked to contamination of spoilage bacteria during the slicing and packaging process. This research illustrates how the combined approach provides complementary results to implement suggestions in the facility to reduce the cross-contamination with spoilage bacteria. It also generates tools to comprehend and propose transference models understanding the environmental and intrinsic factors related to microbial transfer rate.
Keywords: Cross-contamination, Food spoilage, Shelf-life, Microbiota, Leuconostoc
Graphical abstract
Highlights
-
•
The structure of the bacterial community in cooked ham had no relation to the formulation.
-
•
Genus Leuconostoc dominated the spoilage in cooked ham.
-
•
The methodology allows validating the contamination route for spoilage bacteria.
-
•
Post-pasteurization treatment reduce microbiota diversity.
-
•
The ham shelf lifetime decrease get related to cross-contamination during slicing.
1. Introduction
Elaboration of cooked ham involves multiple steps that modulate the microbial composition and its concentration, affecting the product's shelf-life (Raimondi et al., 2019; Zagdoun et al., 2020). The process of cooked ham comprises grinding, injection, mixing, and stuffing, where raw ingredients have a diverse microbiota, which is principally composed by the genus Pseudomonas, Brochothrix, and lactic acid bacteria (LAB) from Leuconostoc and Lactobacillus genera (Mäkelä et al., 1990; Stoops et al., 2015). The proportion and composition of microbiota vary according to processing, storage, and batches (Cauchie et al., 2020). Nonetheless, cooking will reduce the concentration and diversity of spoilage and pathogen bacteria. On the other hand, the probability of re-contamination with LAB increases due to cross-contamination during cooling, slicing, and packaging (Björkroth and Korkeala, 1997; J Samelis et al., 2006).
The contamination route for spoilage bacteria in cooked ham has been associated with contact to surfaces (Garrett et al., 2008; Kusumaningrum et al., 2003), which generates defects as pH reduction, gas production, atypical flavors, and colors reducing shelf-life (Hamasaki et al., 2003; Pothakos et al., 2015). Spoilage LAB are ubiquitous in facility environments; they are native microbiota in raw meat. Moreover, the contamination routes are variables because LAB are highly adaptable and proliferate from low initial concentrations; these bacteria have multiple stress response mechanisms and low substrates selectivity, increasing the cross-contamination risk (Pothakos et al., 2014). However, spoilage LAB behavior was affected by storage conditions under refrigeration and vacuum atmosphere, which affects competence between species (Björkroth and Korkeala, 1997; Doulgeraki et al., 2012; Raimondi et al., 2019; J Samelis et al., 2006; Vasilopoulos et al., 2015; Zagdoun et al., 2020). Besides that, microbial composition in food processing facilities depends on specific environmental adaptation due to their capacity to form biofilms on equipment surfaces and persistency to cleaning and disinfection practices.
The identification of spoilage bacteria is crucial to design strategies for its control. Consequently, the usage of conventional and molecular techniques is helpful for this aim. Nonetheless, metagenomics has become a popular microbiome mapping technique for typifying resident microbiota that could be transferred to the final food product affecting its safety and quality (Filippis et al., 2021; Stellato et al., 2016). Thus, the molecular methods for determining the microbial composition on food are critical in characterizing the dominant spoilage bacteria and their dynamic under storage, mainly because each product and facility could have unique microbiota according to intrinsic and extrinsic factors (Cauchie et al., 2020). Consequently, some researches are focused on the identification of microbiota composition in meat products by means of 16S rRNA sequencing linked to changes in environmental conditions of facilities, shelf-life, or packaging (Cauchie et al., 2020; Duthoo et al., 2021; Raimondi et al., 2019; Stellato et al., 2016; Zagdoun et al., 2020).
The considerations for process and product development are based on the mitigation of microbial contamination. However, sliced ham spoilage could be attributed to the leading group of bacteria, which comes from raw materials, early stages of processing, survivors to cooking processes, and cross-contamination during slicing and packaging. Consequently, this study aimed to characterize pork-cooked ham's microbial community changes among different food production conditions (formulation and processing) using 16S rRNA sequencing and investigate the spoilage bacteria to elucidate their contamination route by classical microbiology.
2. Materials and methods
2.1. Experiment design
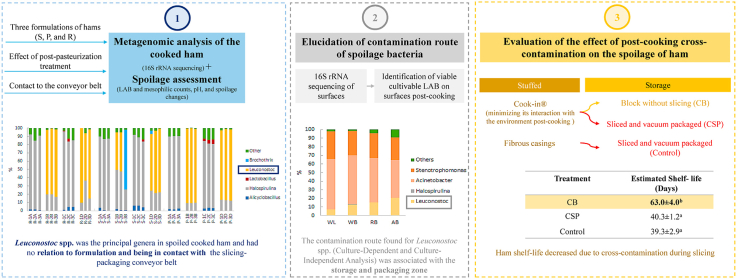
The experiment design had three phases corresponding to the metagenomic analysis of the cooked ham, elucidation of contamination route of spoilage bacteria, and evaluation of the effect of post-cooking cross-contamination on the spoilage of ham.
The first phase consisted of evaluating the changes in microbiota composition by 16S rRNA sequencing as well as carried out by a three-factor experiment, where factors corresponded to the references of sliced cooked hams (S, P, and R) according to the formulations shown in Table 1, the effect of post-pasteurization treatment (PP), and contact to the conveyor belt. The metagenomic analysis was complemented with spoilage assessment based on LAB and mesophilic counts, pH, and spoilage changes. Thereby, the three references of pork ham were analyzed by metagenomic of 16S rRNA according to a completely randomized design with three replicates corresponding to three batches; two factors defined as contact of the product to the conveyor belt and PP treatment (85 °C for 2.5 min of holding time, and cooling for 36 min). Four treatments were defined: i) ham in contact to the conveyor belt treated with PP (A), ii) ham in contact to the conveyor belt without treatment PP (B), iii) ham without contact to the conveyor belt treated with PP (C), and iv) ham without contact to the conveyor belt and not PP treatment (D).
Table 1.
Chemical composition (%) of cooked ham.
| Components | Reference 1 (S) | Reference 2 (P) | Reference 3 (R) |
|---|---|---|---|
| Pork meat | 51.60 | 51.50 | 43.00 |
| Water | 34.00 | 32.00 | 41.40 |
| Soy protein and starch | 9.50 | 9.50 | 10.30 |
| NaCl + KCl | 1.80 | 1.80 | 0.90 |
| Flavors, spices, and colorants | 2.01 | 4.92 | 3.61 |
| Sodium fosfate | 0.4 | 0.4 | 0.4 |
| Sodium ascorbate | 0.05 | 0 | 0.06 |
| Sodium nitrite | 0.16 | 0 | 0.33 |
The second phase was focused on elucidating the contamination route. For this reason, two experiments were carried out. The first experiment consisted of the metagenomic analysis (16S rRNA sequencing) of surfaces. For this objective, four samples of the slicing-packaging conveyor belt in the factory were sampled: weber lift (WL), weber blade (WB), rejected belt (RB), and accepted belt (AB). The surfaces samples were taken 1 h after the cleaning and disinfection process with sterile swabs FloQ (COPA, Brescia, Italy). Each sample was taken by rubbing the surface (1 cm2), and then the swab was put in 1 ml of nuclease-free sterile water (Roche, Grenzach, Germany). All samples were transported to the laboratory at 4 °C and stored at −20 °C until their analysis. Based on the results of this experiment, the second one consisted of the identification of viable cultivable LAB on ham contact surfaces post-cooking. For this purpose five surfaces were sampled: ham casing, cooling chamber, cooling shelves, tables for casing removal, and window for ham without casing pass to slicing area. The samples were taken on two different days of production of pork ham after 4 h of the cleaning and disinfection process, with a sterile swab (100 cm2).
For the third phase, where the aim was to evaluate the effect of post-cooking cross-contamination on the spoilage of ham, a completely randomized design was projected on three references of pork ham with three replicates corresponding to different batches. Three treatments were proposed to evaluate the effect on microorganisms that colonize the post-cooking product causing spoilage: i) ham block stuffed in hermetically sealed casings Cook-In ® (Cryovac, Heredia, Costa Rica) without slicing for minimizing its interaction with the environment post-cooking (CB), ii) ham block stuffed in hermetically sealed casings Cook-In ® (Cryovac, Heredia, Costa Rica) sliced and vacuum packaged (CSP), and iii) ham block stuffed in fibrous casings Viskase (Viskase, Lombard, EEUU) sliced and vacuum packaged (Control). For each treatment, spoilage was analyzed based on LAB and mesophilic counts, pH, and spoilage changes.
2.2. Production and sampling procedures
The references of sliced cooked hams (S, P, and R) were prepared in a local meat factory (Medellín, Colombia) according to the formulations shown in Table 1. The raw pork meat was injected with ingredients dissolved in water, mixed and stuffed in a fibrous casing, molded in blocks to form ham bars (0.10 m × 0.1m x 1.5 m), and cooked until the core temperature reached 72 °C. After, the product was cooled and stored from 0 to −2 °C. Subsequently, the casing was removed from hams, and finally, it was sliced and vacuum packaged.
For PP samples, the packaged hams were heated to 85 °C for 2.5 min of holding time by hot water immersion and then cooling for 36 min until 3 °C. UNITHERM and WAPA-6802 (Unitherm Food Systems, Bristow, USA) were used for such procedures. These conditions were determined previously according to the type of packaging, size, and weight of the samples.
2.3. DNA extraction and 16S rRNA sequencing
Total Bacterial DNA collected from ham and surfaces was centrifuged at 14,000 g for 10 min at 4 °C and extracted using the High Pure Template Kit (Roche, Grenzach, Germany) following the manufacturer's protocols. Then, the total DNA was quantified with Nanodrop 2000™ (Thermo Fisher Scientific, Massachusetts, USA). Bacterial identity was determined by NGS techniques using the Miseq Illumina platform (Illumina, San Diego, USA), targeting the V4 region of the 16S rRNA gene. Metagenomic profiles based on amplicons of bacterial communities and their composition analysis were performed under protocols proposed by Caraballo Guzmán et al. (2020). The number of observed OTUs and alpha diversity metrics were calculated as Chao1, Heip, and Shannon indexes for ham treatments. Venn diagrams for description OTUs at gender level in each of the four treatments for hams and four surfaces sampled were performed using Venny 1.0 program (http://bioinfogp.cnb.csic.es/tools/venny/).
2.4. Spoilage assessment
Changes in microbial, pH, and sensory characteristics were analyzed over 45 days of storage at 8 °C for 0, 5, 10, 15, 20, 25, 30, 35, 40, and 45 days. The storage temperature at 8 °C was selected as the most challenging condition, based on the results reported by Jofré et al. (2019), who identified that 30% of the time, the domestic temperature of refrigerators fluctuated between 6 and 8 °C on the position where people usually stored cooked meat products. Moreover, it is the most frequent storage temperature under Colombian distribution and commercialization of cold chains.
Thus, 35 unit samples (225 g of ham) for each batch were evaluated for each treatment randomly. The shelf-life time was defined as storage time where at least 25% of samples showed spoilage determined by LAB or mesophilic counts equal or higher than 6 Log CFU/g, pH lower than 5.8, or samples with milkiness or swelling according to procedures by the standard Guide for Sensory Evaluation Methods to Determine Sensory Shelf Life of Consumer Products (ASTM, 2020). The LAB, mesophilic counts, and pH thresholds were previously identified based on consumers' rejection.
LAB were counted using TEMPO LAB® kit (BioMéreux Industry, l'Étoile, France), and TEMPO AC® kit (BioMéreux Industry, l'Étoile, France) was used for mesophilic counts, according to ISO 966.23 method (For the Determination of Aerobic Plate Count, Most Probable Number of Coliform Bacteria, and Escherichia coli and Staphylococcus spp. in Products, 1989).
The pH was determined using a pH meter (Oakton ION 2700 Benchtop Meter, Columbus, USA). pH data correspond to a mean of four measurements per sample at central points. Milkiness (milky exudates) and swelling conditions were qualified using a scale of four set levels based on the sensory analysis carried out quantitative descriptive analysis (QDA) during storage assisted by 5 trained judges, where 0 corresponds to the standard product, 1 to onset of spoilage, 2 to spoiled product, and 3 to advanced spoilage.
2.5. Identification of viable cultivable LAB on ham contact surfaces post-cooking
The samples of five surfaces were taken with a sterile swab (100 cm2). Then, the sterilized swab was placed in 10 ml of Man, Rogosa and Sharpe (MRS) broth (Scharlau Chemie S.A., Barcelona, Spain) and incubated for 4 day at 30 °C. This procedure was performed in duplicates.
All tubes of incubated MRS broth were harvested on MRS agar (Scharlau Chemie S.A., Barcelona, Spain) and incubated for 48 h at 30 °C and 25 °C under aerobic and anaerobic conditions. Isolated bacterial colonies were purified in Columbia Agar with 5% lamb Blood (BioMerieux, Paris, France) for later biochemical identification by VITEK® 2 (BioMerieux, Paris, France). Colonies were suspended in a salt solution, and the turbidity was adjusted to the 0.5 McFarland index. According to the manufacturer, the prepared bacterial cultures were analyzed using the Gram-Positive identification card (GP) or Bacillus card (BCL) and the VITEK® 2 system (BioMerieux, Paris, France). Results were expressed as defined by the manufacturer (96%–100%, excellent identification; 93%–95%, very good identification; 89%–92%, good identification; 85%–88%, acceptable identification; below 85%, no identification) (Pincus, 2006).
2.6. Statistical analysis
Parameters obtained on the diversity index from the metagenomic analysis were compared for each treatment and analyzed using the F-test, assuming the quantitative approach planned test focused on variance analysis (p < 0.05). For comparison of times of shelf-life from spoilage assessment, Duncan's test (α = 0.05) was applied. The values of pH, mesophilic, and LAB counts were compared to each storage time for control, CSP, and CB treatments by the F-test, assuming the quantitative planned test approach with Duncan's test. Moreover, these kinetics were fitted to the smoothed lines based on the second-order polynomial model and moving average for microbial counts of CSP treatment. The statistical software SAS University was used for the analyses (SAS Institute, Inc., Cary, NC).
3. Results and discussion
3.1. Metagenomic analysis in cooked ham by 16S rRNA sequencing
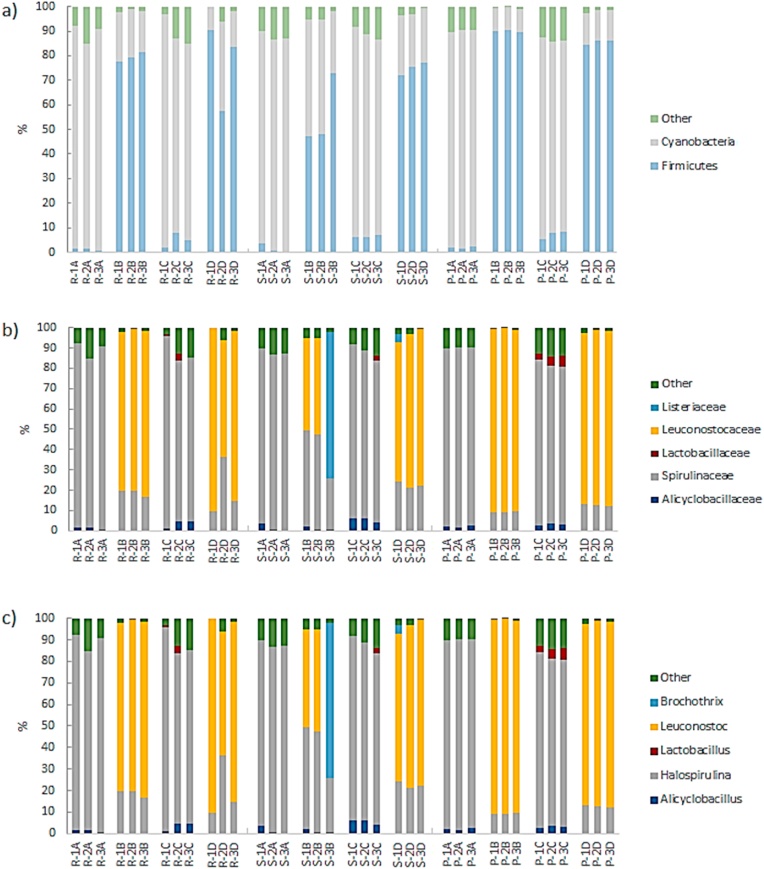
The results of the metagenomic analysis are shown in Fig. 1 for all treatments, formulations (references of ham), and batches. Treatments without PP (B and D) had the genus Leuconostoc in relative abundance more significant than 50%, independently of their contact with the slicing-packaging conveyor belt. In addition, this behavior was consistent among batches and three commercial ham references despite the formulation changes, which were based on pork meat quantity, ascorbate, and sodium nitrite. Moreover, these treatments without PP showed swelling at 28 days of storage for two ham references; but 77% of the samples without conveyor belt contact showed this spoilage at 40 days. Additionally, 44% of the samples showed milkiness at 35 days of storage for all ham references. However, the treatments with PP (A and C) did not present swelling in neither of the samples stored for 40 days, which are concordant with the absence of the genus Leuconostoc independently of their contact with the slicing-packaging conveyor belt.
Fig. 1.
Relative abundance by phyla (a), family (b), and genera (c) on cooked pork ham with different formulation for reference 1 (R-), reference 2 (S-), and reference 3 (P-) for treatments: ham in contact to the conveyor belt treated with PP (A), ii) ham in contact to the conveyor belt without treatment PP (B), iii) ham without contact to transportation band treated with PP (C), and iv) ham without contact to the conveyor belt and did not treat with PP (D). The numbers 1–3 indicate different repetitions correspond to different samples from different batches.
This behavior agrees with Pothakos et al. (2014a) results, which associate the presence of L. gelidum as the main spoilage bacteria in sliced turkey in Belgium with relative abundances greater than 80% on 16S rRNA metagenomic analysis. Hu et al. (2009) related the presence of Leuconostoc mesenteroides subsp. mesenteroides and Lactobacillus sakei subsp. carnosus with spoilage of sliced vacuum-packed cooked ham, where the presence of culturable and non-culturable species of Leuconostoc was associated with spoilage in early stages of cooked products shelf-life. These bacteria were coming from raw meats and they are classified as native microbiota of meat factories.
The genus Brochothrix spp. was identified in one sample (S–3B) with 72.3% relative abundance (Fig. 1c). It is coherent with reports of this genus where it was classified as spoilage bacteria due to the undesirable aroma produced by glucose fermentation under aerobic conditions, non-proteolytic spoilage, and ethanol production under glucose starvation under anaerobic conditions (Holley, 2014; Pellissery et al., 2019; Raimondi et al., 2019).
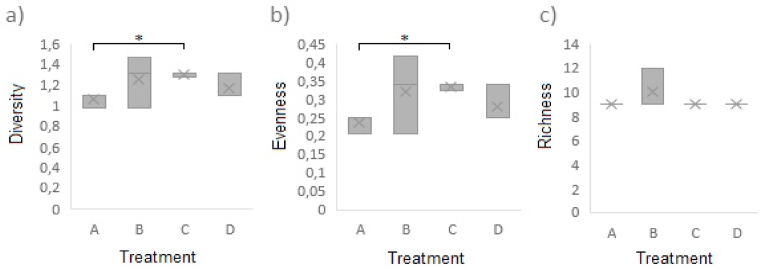
Shannon diversity index (Fig. 2a), Heip (Fig. 2b), and Chao1 richness (Fig. 2c) have the highest values for treatment B (Fig. 3a) without significant differences regarding other treatments (p > 0.05). The genus Halospirulina was founded in all treatments and samples. Furthermore, the genus Alyciclobacillus was found in all treatments except treatment (D) (Fig. 3a), which corresponded to ham without contact with the conveyor belt and without PP treatment. Halospirulina and Alyciclobacillus were found as genera with high relative abundances without attributable effects to spoilage. Perri et al. (2020) reported the co-occurrence of Halospirulina spp. with Leuconostoc under environmental conditions. Therefore, an amensalism relation may occur between these genera. Moreover, the antagonistic effect of Halospirulina spp. against Lactobacillus spp. has been reported on the anaerobic culture of chicken feces (Sohail and Hume, 2019), legumes, and pseudocereals (Perri et al., 2020). This effect could be related to the low relative abundance of the genus Lactobacillus (<3%). Additionally, the genus Alyciclobacillus is composed of thermo-acidophilic, Gram-positive bacteria described as spoilage in fruit juices (Smit et al., 2011) with inhibitory effect against S. aureus, Acinetobacter baumani, and Pseudomonas aureginosa (Yang et al., 2016).
Fig. 2.
Diversity index for each treatment for cooked ham. (a) Shannon diversity index, (b) Heip evenness, and (c) Chao1 richness. For treatments: ham in contact to transportation band treated with PP (A), ii) ham in contact to transportation band without treatment PP (B), iii) ham without contact to transportation band treated with PP (C), and iv) ham without contact to transportation band and did not treat with PP (D).
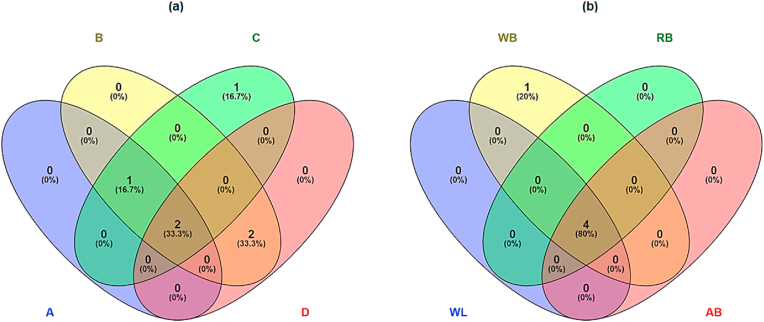
Fig. 3.
Venn diagrams of Genera identified. The letter and number together outside the ovals and their colors represent each treatment for cooked ham (a) and sampled surface (b). Where, treatments for cooked ham: ham in contact to transportation band treated by PP (A), ii) ham in contact to transportation band without PP treatment (B), iii) ham without contact to transportation band and treated by PP (C), and iv) ham without contact to transportation band and without PP treatment (D). For sampled surfaces: weber lift (WL), weber blade (WB), the rejected band (RB), and accepted band (AB). The numbers outside of the intersections are the genera found only in those cooked ham or surfaces. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
On the other hand, Lactobacillus was isolated only from ham without contact to the conveyor belt and treated with PP (C) (Fig. 3a); consequently, this treatment has the most significant number of unique genera (Fig. 1). Furthermore, PP-A has the lowest Shannon diversity index with significant differences against B (p < 0.05), that could be attributable to PP treatment which reduces the microbial count, maximizing transfer effect from the slicing-packaging band. Furthermore, the variation on non-classified microorganisms in PP treatments is related to the Heip's index, which detects significant changes in the relative abundance of the infrequent genus.
The results of microbial composition are concordant with those reported in sliced vacuum-packed ham by Raimondi et al. (2019), who found the highest diversity index in early stages of shelf-life, which could be related to the DNA presence of non-viable bacteria inactivated by cooking and cross-contamination during slicing and packaging. Concerning later stages of shelf-life and spoiled product have similar values on diversity index, it was evident that higher participation on the microbiota of Firmicutes was LAB from genera Carnobacterium, Lactobacillus, Leuconostoc, and Weissella with relative abundances higher than 17% in 7 from 19 samples, while the genus Brochothrix was found in 3 from 19 samples.
However, different factors affect the composition and abundance of food microbiota. Zagdoun et al. (2020) found that the changes in cooked ham microbiota depend on pre-cooking parameters, slicing lines and variability of seasons. On the other hand, Woods et al. (2019) did not find significant differences in the number of OTUs in marinated meat analyzed in 0, 20, and 40 days of storage. Besides that, the authors found the highest relative abundance of genera Marinilactibacillus, Carnobacterium, Leuconostoc, Vibrio, Pseudoalteromonas, and Marinomonas, where Marinilactibacillus, Carnobacterium, Leuconostoc, and Vibrio dominated the microbial composition over two years of production.
3.2. Elucidation of contamination route of spoilage bacteria
Results of microbiota composition on slicing-packaging surfaces by 16S rRNA profiles are shown in Fig. 4, where Proteobacteria was the most frequent phylum (70.5–90.8%). The genus Halospirulina was found only from WB; concerning other surfaces' microbiota composition (WL, RB, and AB), the same genus composition was found (Fig. 3b), proving the resident microbiome of the specific genus on facility surfaces. Moreover, the genus Leuconostoc was found in the four sampled surfaces, validating the cross-contamination product-surfaces-product. These results are concordant with the study on meat products factories reported by Caraballo Guzmán et al. (2020), where Proteobacteria represent 51.7% and 45.4% of OTUs of two factories. Along with that, the family Alyciclobacillus was found in occurrences of 66.6% and 33.3% for both meat factories, and the genus Acinetobacter had the highest occurrences due to persistence and survival ability on facility environments. Furthermore, genera as Acinetobacter and Pseudomonas have been associated with spoilage microbiota which is predominant under temperatures abuse conditions of overcooling (Doulgeraki et al., 2012).
Fig. 4.
Relative abundance by phyla (a), family (b), and genera (c) on surfaces of weber lift (WL), weber blade (WB), the rejected band (RB), and accepted band (AB).
The results on viable cultivable LAB isolated from surfaces before slicing (casings, cooling chamber, and the window where ham passes to slicing area without casing) showed the presence of L. citreum with percentages of identity >91%. Furthermore, samples taken from surfaces of tables used for casing removal did not show growth of bacteria in MRS broth; this could be related to a higher frequency of operative cleaning and disinfection process. In addition, other bacteria were identified in the cooling shelves, such as Aerococo viridians, L. pseudomesenteroides, K. rosea, L. citreum, and K. varians, where Kocuria spp. are classified as environmental bacteria and have been found in pork carcasses by 16S rRNA gene profiles. However, their presence and abundance were related to bacteria adaptive response to specific conditions of the slaughterhouse (Peruzy et al., 2021).
Consequently, the processing of cooked ham contributes to microbiota composition where spoilage bacteria of the genus Leuconostoc contaminates the product by different surfaces and environmental contact, where their abundance, survival, and adaptation are associated with cleaning and disinfection practices. Thus, L. carnosum had been reported as omnipresent bacteria on the production chain of ham. This bacterium has two subgroups whose prevalence and participation depend on temperature. The first subgroup is commonly found below 12 °C and the second subgroup is present in products storage above 12 °C. However, their isolation in raw materials or products in the early stages of their shelf-life is difficult, perhaps because their concentration is low until the food reaches abuse temperatures (>12 °C) which are considered as favorable growth conditions (Vasilopoulos et al., 2010).
3.3. Effect of post-cooking cross-contamination on the spoilage of ham
The assessment of ham spoilage was based on the kinetics of pH, mesophilic, and LAB counts and appearance of the main defects (milkiness and swelling) for shelf-life time.
3.3.1. Mesophilic and LAB counts
The growth of mesophilic and LAB are presented in Fig. 5a and b, respectively. Although bacteria behavior was highly variable, it reached a maximum concentration of 6 Log CFU/g for total plate count and LAB from 2 to 5 Log CFU/g between 20 and 50 days of storage. The higher bacterial concentration was reached at day 40, corresponding to the end of the shelf-life time for treatments Control and CSP with significant differences for mesophilic and LAB counts against CB (p < 0.05). On the other hand, the smoothed trend lines show a lag phase for Control treatment near 22 days for mesophilic and LAB counts and exponential phase until to 40 days; while for CB, the shown lag phase was higher, corresponding to 28 days. For CSP treatment, microbial counts remained constant in approximately 4 Log CFU/g between 7 and 24 days of storage, followed by the decreased trend of a bacteria population that could be related to pH reduction at a high rate.
Fig. 5.
Kinetic of behavior of pH (a), mesophilic count (b), and LAB count (c) in cooked pork ham, for treatments: control (▲), CSP ( ), and CB (o). Non-continuous lines were showed the general smoothing of the behavior of spoilage variables during storage: control (--), CSP (- -), and CB (- -).
), and CB (o). Non-continuous lines were showed the general smoothing of the behavior of spoilage variables during storage: control (--), CSP (- -), and CB (- -).
Furthermore, the changes in LAB counts did not show any trend in the dynamic of population related to the appearance of early spoilage phenomena that could be attributed to limitations of methodology based on fluorescence for microbial recovery. Therefore, the bacterial growth curve showed high fluctuation between counts in each batch and wide variability in initial concentration, which could be related to contamination by cross-contamination under facility process conditions. Thus, the ISO method for mesophilic count could have limited selectivity, especially for psychotropic bacteria generally underestimated (Pothakos et al., 2014).
3.3.2. pH kinetics
pH kinetics are shown in Fig. 5c, where Control and CSP have similar behavior in pH values under 5.8 for 39 days without significant differences between treatments for each storage time (p > 0.05). Furthermore, the pH decline trend changes from day 41, with an accelerated reduction attributable to the metabolic behavior of the leading group of spoilage bacteria. Thus, pH for CB remained stable until 63 days with an average value of 6.05 with significant differences against Control and CSP (p < 0.05); in addition, the smoothed trend lines were similar for the pH reduction in Control and CSP treatments, which could be related to similar contamination routes for spoilage bacteria during casing removal, slicing, and packaging processes.
Moreover, a decrease in the pH (5.64 ± 0.20) on slicing treatments (Control and CSP) coincides with high LAB counts. These results are concordant with results reported by Hu et al. (2009) on sliced vacuum-packed ham storage at 4 °C where pH is reduced from 6.54 to 5.24 between 7 and 15 days related to increases on mesophilic and LAB counts from 106 to 108 CFU/g between 3 and 35 days. Likewise, decreases in pH kinetic were linked to the metabolic activity of L. gelidum, L. carnosum, and L. mesenterorides (Raimondi et al., 2019; John Samelis et al., 2000). Comi et al. (2016) reported the pH of cooked bacon without spoilage remained at 5.6 while spoiled product was reduced to 5.3 as well as related to increases on LAB and mesophilic counts of 8.7 and 6.3 Log CFU/g, respectively.
Additionally, L. mesenteroides was identified as a principal spoilage microorganism. This bacterium causes a decrease in pH due to their fast growth and milkiness, and off-flavors attributed to hexanal and acetic acid production on morcilla de Burgos vacuum-packaged refrigerated (Diez et al., 2009). Moreover, L. mesenteroides subsp. mesenteroides, L. mesenteroides subsp. dextranicum and L. citreum produce dextrans linked to milkiness and the survival of bacteria in acidic matrices and biofilm formation ability (Holland and Liu, 2011). These phenomena could explain that low pH values did not inhibit the LAB growth. In addition to this, Jääskeläinen et al. (2013) reported that Leuconostoc gasicomitatum grows faster under the anaerobic condition with exogenous iron, typically for meat products. However, its metabolic activity did not affect pH behavior in vacuum packaged pork meat; in fact, meat quality was acceptable during 13–18 days, although L. gasicomitatum reached its maximal population density.
3.3.3. Shelf-life
Based on changes in pH, bacterial behavior, and appearance of milkiness and swelling, the calculated shelf-life time of cooked ham storage at 8 °C was 39 ± 3 days for Control treatment without significant differences (p > 0.05) against CSP treatment (40 ± 1 days). However, the shelf-life time of CB increased with significant differences (p < 0.05) to 63 ± 4 days, which indicates the high impact of spoilage bacteria contamination during slicing and packaging linked to decreasing shelf-life time (57.5%) independently of the environment interaction minimization when casing permeability was changed. On the other hand, Dušková et al. (2016) found that cooking at 70 °C for 10 min of holding time in ham reduces between 4 and 5 Log CFU/g of spoilage bacteria from raw materials generating concentrations below 10 CFU/g of spoilage LAB of the genus Leuconostoc and Lactobacillus.
Consequently, it is possible to elucidate the main contamination route for Leuconostoc from the results that showed the presence of this genus in cooling areas. Thereby, the surface of the casing may be contaminated by contact with shelves and the environment, where its distribution may respond to ecological reasons of growing and persist in the meat matrix. For example, high relative humidity in the processing areas could increase the probability of cross-contamination from casing to ham because contaminated water steam could permeate the casing of ham and introduce Leuconostoc because this bacterium is not motile. Thus, contaminated ham could contaminate the slicing and packaging area, where the transference process would be from product to surfaces and vice-versa.
4. Conclusions
The presence of Leuconostoc was found in relative abundances above 50%, and it was linked to spoilage (milkiness and swelling) of cooked vacuum-packed pork ham independently of their formulations. Besides, results demonstrate that cooked ham shelf-life time decrease was related to cross-contamination during slicing. Furthermore, the LAB growth curve showed wide fluctuation on counts for the different batches and the initial concentration, which could be related to contamination by cross-contamination under facility process conditions.
For practical applications, it is crucial to identify culturable and non-culturable spoilage bacteria in food products and their potential contamination route under industrial conditions. Furthermore, future research is needed to consider the effect of intrinsic, extrinsic, and implicit factors on bacterial behavior and possible changes on the microbiota for proposing mathematical models to describe cross-contamination phenomena understanding factors governing bacterial transfer to ham and linked to shelf-life alterations.
CRediT authorship contribution statement
Carla María Blanco – Lizarazo: conceptualized the study, analyzed data and wrote the manuscript. Andrea Sierra-Cadavid: conceptualized the study, edited and review the manuscript and supervised the study. Alejandra M. Montoya R: performed laboratory work and supervised the study. Juan Camilo Ospina-E: performed laboratory work, conceptualized the study, review the manuscript and supervised the study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The authors acknowledge the financial support of the Ministry of Science, Technology, and Innovation (Minciencias) and Industria de Alimentos Zenú S.A.S, RC No. 843283969237 and Postdoctoral Grant 80740-277-2020 of National Science Found “Francisco José de Caldas” (Announcement 848-2019). Also, the laboratory team at Industria de Alimentos Zenú S.A.S with Luz Amparo Montoya, Verónica Bedoya and Alejandra Rozo who participated in this research, and Ernesto López Guerrero for English review.
References
- Björkroth K., Korkeala H. Use of rRNA gene restriction patterns to evaluate lactic acid bacterium contamination of vacuum-packaged sliced cooked whole-meat product in a meat processing plant. Appl. Environ. Microbiol. 1997;63(2):448–453. doi: 10.1128/AEM.63.2.448-453.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraballo Guzmán A., González Hurtado M.I., Cuesta-Astroz Y., Torres G. Metagenomic characterization of bacterial biofilm in four food processing plants in Colombia. Braz. J. Microbiol. 2020;51(3):1259–1267. doi: 10.1007/s42770-020-00260-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchie E., Delhalle L., Taminiau B., Tahiri A., Korsak N., Burteau S., Fall P.A., Farnir F., Baré G., Daube G. Assessment of spoilage bacterial communities in food wrap and modified atmospheres-packed minced pork meat samples by 16S rDNA metagenetic analysis. Front. Microbiol. 2020;10:3074. doi: 10.3389/FMICB.2019.03074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comi G., Andyanto D., Manzano M., Iacumin L. Lactococcus lactis and Lactobacillus sakei as bio-protective culture to eliminate Leuconostoc mesenteroides spoilage and improve the shelf life and sensorial characteristics of commercial cooked bacon. Food Microbiol. 2016;58:16–22. doi: 10.1016/j.fm.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Diez A., Björkroth J., Jaime I., Rovira J. Microbial, sensory and volatile changes during the anaerobic cold storage of morcilla de Burgos previously inoculated with Weissella viridescens and Leuconostoc mesenteroides. Int. J. Food Microbiol. 2009;131(2–3):168–177. doi: 10.1016/J.IJFOODMICRO.2009.02.019. [DOI] [PubMed] [Google Scholar]
- Doulgeraki A.I., Ercolini D., Villani F., Nychas G.-J.E. Spoilage microbiota associated to the storage of raw meat in different conditions. Int. J. Food Microbiol. 2012;157:130–141. doi: 10.1016/j.ijfoodmicro.2012.05.020. [DOI] [PubMed] [Google Scholar]
- Dušková M., Kameník J., Lačanin I., Šedo O., Zdráhal Z. Lactic acid bacteria in cooked hams - sources of contamination and chances of survival in the product. Food Control. 2016;61:1–5. doi: 10.1016/j.foodcont.2015.09.019. [DOI] [Google Scholar]
- Duthoo E., Rasschaert G., Leroy F., Weckx S., Heyndrickx M., De Reu K. The microbiota of modified-atmosphere-packaged cooked charcuterie products throughout their shelf-life period, as revealed by a complementary combination of culture-dependent and culture-independent analysis. Microorganisms. 2021;9(6) doi: 10.3390/MICROORGANISMS9061223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippis F. De, Valentino V., Cotter P.D., Alvarez-ordo A., Ercolini D. Environmental microbiome mapping as a strategy to improve quality and safety in the food industry. Curr. Opin. Food Sci. 2021;38:168–176. [Google Scholar]
- For the determination of aerobic plate count . Association of Official Analytical Chemist (AOAC); 1989. Most Probable Number of Coliform Bacteria, and Escherichia coli and Staphylococcus in Products.https://www.edgeanalytical.com/wp-content/uploads/Food_AOAC-966.23.pdf Pub. L. No. 966.23. [Google Scholar]
- Garrett T.R., Bhakoo M., Zhang Z. Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 2008;18(9):1049–1056. doi: 10.1016/J.PNSC.2008.04.001. [DOI] [Google Scholar]
- Hamasaki Y., Ayaki M., Fuchu H., Sugiyama M., Morita H. Behavior of psychrotrophic lactic acid bacteria isolated from spoiling cooked meat products. Appl. Environ. Microbiol. 2003;69(6):3668. doi: 10.1128/AEM.69.6.3668-3671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland R., Liu S.Q. Lactic acid bacteria | Leuconostoc spp. encyclopedia of dairy sciences. 2011. second ed.,138-142. [DOI]
- Holley R.A. Brochothrix. Encyclop. Food Microbiol. 2014;1:331–334. doi: 10.1016/B978-0-12-384730-0.00048-3. [DOI] [Google Scholar]
- Hu P., Zhou G., Xu X., Li C., Han Y. Characterization of the predominant spoilage bacteria in sliced vacuum-packed cooked ham based on 16S rDNA-DGGE. Food Control. 2009;20(2):99–104. doi: 10.1016/j.foodcont.2008.02.007. [DOI] [Google Scholar]
- Jääskeläinen E., Johansson P., Kostiainen O., Nieminen T., Schmidt G., Somervuo P., Mohsina M., Vanninen P., Auvinen P., Björkroth J. Significance of heme-based respiration in meat spoilage caused by Leuconostoc gasicomitatum. Appl. Environ. Microbiol. 2013;79(4):1078–1085. doi: 10.1128/AEM.02943-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofré A., Latorre-Moratalla M.L., Garriga M., Bover-Cid S. Domestic refrigerator temperatures in Spain: assessment of its impact on the safety and shelf-life of cooked meat products. Food Res. Int. 2019;126(July):108578. doi: 10.1016/j.foodres.2019.108578. [DOI] [PubMed] [Google Scholar]
- Kusumaningrum H., Riboldi G., Hazeleger W., Beumer R. Survival of foodborne pathogens on stainless steel surfaces and cross-contamination to foods. Int. J. Food Microbiol. 2003;85(3):227–236. doi: 10.1016/S0168-1605(02)00540-8. [DOI] [PubMed] [Google Scholar]
- Mäkelä P., Korkeala H., Laine J. Raw materials of cooked ring sausages as a source of spoilage lactic acid bacteria. J. Food Protect. 1990;53(11):965–968. doi: 10.4315/0362-028X-53.11.965. [DOI] [PubMed] [Google Scholar]
- Pellissery A.J., Vinayamohan P.G., Amalaradjou M.A.R., Venkitanarayanan K. Meat Quality Analysis: Advanced Evaluation Methods, Techniques, and Technologies. Elsevier Inc; 2019. Spoilage bacteria and meat quality. [DOI] [Google Scholar]
- Perri G., Calabrese F.M., Rizzello C.G., De Angelis M., Gobbetti M., Calasso M. Sprouting process affects the lactic acid bacteria and yeasts of cereal, pseudocereal and legume flours. LWT (Lebensm.-Wiss. & Technol.) 2020;126(February):109314. doi: 10.1016/j.lwt.2020.109314. [DOI] [Google Scholar]
- Peruzy M.F., Houf K., Joossens M., Yu Z., Proroga Y.T.R., Murru N. Evaluation of microbial contamination of different pork carcass areas through culture-dependent and independent methods in small-scale slaughterhouses. Int. J. Food Microbiol. 2021;336:108902. doi: 10.1016/J.IJFOODMICRO.2020.108902. [DOI] [PubMed] [Google Scholar]
- Pincus D.H. Encyclopedia of Rapid Microbiological Methods. 2006. Microbial identification using the bioMérieux Vitek® 2 system; pp. 1–32.www.pda.org/bookstore Biomerieux. [Google Scholar]
- Pothakos V., Snauwaert C., De Vos P., Huys G., Devlieghere F. Psychrotrophic members of Leuconostoc gasicomitatum, Leuconostoc gelidum and Lactococcus piscium dominate at the end of shelf-life in packaged and chilled-stored food products in Belgium. Food Microbiol. 2014;39:61–67. doi: 10.1016/j.fm.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Pothakos V., Taminiau B., Huys G., Nezer C., Daube G., Devlieghere F. Psychrotrophic lactic acid bacteria associated with production batch recalls and sporadic cases of early spoilage in Belgium between 2010 and 2014. Int. J. Food Microbiol. 2014;191:157–163. doi: 10.1016/j.ijfoodmicro.2014.09.013. [DOI] [PubMed] [Google Scholar]
- Pothakos V., Stellato G., Ercolini D., Devlieghere F. Processing environment and ingredients are both sources of Leuconostoc gelidum, which emerges as a major spoiler in ready-to-eat meals. Appl. Environ. Microbiol. 2015;81(10):3529–3541. doi: 10.1128/AEM.03941-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi S., Luciani R., Sirangelo T.M., Amaretti A., Leonardi A., Ulrici A., Foca G., D'Auria G., Moya A., Zuliani V., Seibert T.M., Søltoft-Jensen J., Rossi M. Microbiota of sliced cooked ham packaged in modified atmosphere throughout the shelf life: microbiota of sliced cooked ham in MAP. Int. J. Food Microbiol. 2019;289:200–208. doi: 10.1016/j.ijfoodmicro.2018.09.017. July 2018. [DOI] [PubMed] [Google Scholar]
- Samelis John, Kakouri A., Rementzis J. The spoilage microflora of cured, cooked Turkey breasts prepared commercially with or without smoking. Int. J. Food Microbiol. 2000;56(2–3):133–143. doi: 10.1016/S0168-1605(99)00190-7. [DOI] [PubMed] [Google Scholar]
- Samelis J., Björkroth J., Kakouri A., Rementzis J. Leuconostoc carnosum associated with spoilage of refrigerated whole cooked hams in Greece. J. Food Protect. 2006;69(9):2268–2273. doi: 10.4315/0362-028X-69.9.2268. [DOI] [PubMed] [Google Scholar]
- Standard Guide for Sensory Evaluation Methods to Determine Sensory Shelf Life of Consumer Products. vol. 1. Pub. L.; 2020. https://standards.globalspec.com/std/14182192/astm-e2454-20 No. ASTM E2454-20, ASTM International. [Google Scholar]
- Smit Y., Cameron M., Venter P., Witthuhn R.C. Alicyclobacillus spoilage and isolation - a review. Food Microbiol. 2011;28(3):331–349. doi: 10.1016/j.fm.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Sohail M.U., Hume M.E. Evaluation of antimicrobial action of chitosan and acetic acid on broiler cecal bacterial profiles in anaerobic cultures inoculated with Salmonella typhimurium. J. Appl. Poultry Res. 2019;28(1):176–183. doi: 10.3382/japr/pfy061. [DOI] [Google Scholar]
- Stellato G., La Storia A., De Filippis F., Borriello G., Villani F., Ercolini D. Overlap of spoilage-associated microbiota between meat and the meat processing environment in small-scale and large-scale retail distributions. Appl. Environ. Microbiol. 2016;82(13):4045–4054. doi: 10.1128/AEM.00793-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoops J., Ruyters S., Busschaert P., Spaepen R., Verreth C., Claes J., Lievens B., Van Campenhout L. Bacterial community dynamics during cold storage of minced meat packaged under modified atmosphere and supplemented with different preservatives. Food Microbiol. 2015;48:192–199. doi: 10.1016/J.FM.2014.12.012. [DOI] [PubMed] [Google Scholar]
- Vasilopoulos C., De Maere H., De Mey E., Paelinck H., De Vuyst L., Leroy F. Technology-induced selection towards the spoilage microbiota of artisan-type cooked ham packed under modified atmosphere. Food Microbiol. 2010;27(1):77–84. doi: 10.1016/J.FM.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Vasilopoulos C., De Vuyst L., Leroy F. Shelf-life reduction as an emerging problem in cooked hams underlines the need for improved preservation strategies. Crit. Rev. Food Sci. Nutr. 2015;55(10):1425–1443. doi: 10.1080/10408398.2012.695413. [DOI] [PubMed] [Google Scholar]
- Woods D.F., Kozak I.M., Flynn S., O'Gara F. The microbiome of an active meat curing brine. Front. Microbiol. 2019;10(JAN):1–11. doi: 10.3389/fmicb.2018.03346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.F., Yang C.H., Liang M.T., Gao Z.J., Wu Y.W., Chuang L.Y. Chemical composition, antioxidant, and antibacterial activity of wood vinegar from litchi chinensis. Molecules. 2016;21(9):1–10. doi: 10.3390/molecules21091150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagdoun M., Coeuret G., N'Dione M., Champomier-Vergès M.C., Chaillou S. Large microbiota survey reveals how the microbial ecology of cooked ham is shaped by different processing steps. Food Microbiol. 2020;91:103547. doi: 10.1016/J.FM.2020.103547. [DOI] [PubMed] [Google Scholar]