Abstract
Enterotoxigenic Bacteroides fragilis (ETBF) has received significant attention for a possible association with, or causal role in, colorectal cancer (CRC). The goal of this review was to assess the status of the published evidence supporting (i) the association between ETBF and CRC and (ii) the causal role of ETBF in CRC. PubMed and Scopus searches were performed in August 2021 to identify human, animal, and cell studies pertaining to the role of ETBF in CRC. Inclusion criteria included the use of cell lines, mice, exposure to BFT or ETBF, and detection of bft. Review studies were excluded, and studies were limited to the English language. Quality of study design and risk of bias analysis was performed on the cell, animal, and human studies using ToxRTools, SYRCLE, and NOS, respectively. Ninety-five eligible studies were identified, this included 22 human studies, 24 animal studies, 43 cell studies, and 6 studies that included both cells and mice studies. We found that a large majority of studies supported an association or causal role of ETBF in CRC, as well as high levels of study bias was detected in the in vitro and in vivo studies. The high-level heterogeneity in study design and reporting made it difficult to synthesize these findings into a unified conclusion, suggesting that the need for future studies that include improved mechanistic models, longitudinal in vitro and in vivo evidence, and appropriate control of confounding factors will be required to confirm whether ETBF has a direct role in CRC etiopathogenesis.
Keywords: Colorectal cancer (CRC), Enterotoxigenic Bacteroides fragilis (ETBF), B. fragilis toxin (BFT), Etiology
Introduction
Colorectal cancer (CRC) is the third most diagnosed cancer in men and women around the world [1]. The vast majority of the 1.9 million annual CRC cases are sporadic and can be attributed to a variety of environmental factors [2]. The environmental influence of the gastrointestinal microbiome has become an important research consideration in the etiology of CRC, including the role of microbes and microbially-produced metabolites and toxins as causal agent in the initiation and progression of CRC. Numerous studies of CRC in humans, animal models, and cell models have provided data supporting the role of microbes as a causative agent of CRC. This review focuses on one area generating strong interest, the role of enterotoxigenic Bacteroides fragilis (ETBF) as a causal agent of CRC [3], [4], [5].
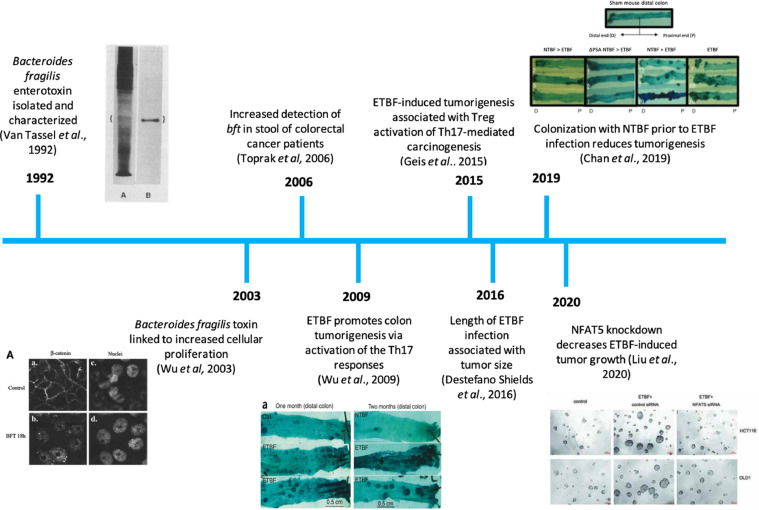
A brief overview of the historical evidence for the role of ETBF in the etiology of CRC is given by Fig. 1. Briefly, the B. fragilis toxin (BFT) is a metalloprotease located on a pathogenicity island, BfPAI, which encodes both the bft gene and mpII, a second metalloprotease [6,7]. The association between certain strains of B. fragilis and secretory diarrhea in farm animals and humans was initially reported in the 1980s and BFT was subsequently isolated and characterized [8], [9], [10]. A review of the literature shows that ETBF has been associated with both colitis and CRC [11,12]. For example, Toprak et al. report a 38% carriage rate of bft in CRC patients using stool samples [12] and Boleij et al. report an ETBF colonization rate of 85.5% in mucosal tissue samples taken from CRC patients [60]. Mechanistic studies carried out using human cell lines and animal models also provide evidence for the ability of ETBF to enhance tumorigenesis, including immune-mediated inflammation. This review summarizes existing evidence for association of ETBF and CRC as well as the current state of knowledge regarding molecular mechanisms by which BFT influences the etiology of CRC.
Fig. 1.
A timeline of some of the key discoveries concerning the potential causal relationship between ETBF colonization and colorectal cancer.
Methods
Search strategy and paper selection
A systematic literature search was performed in August 2021 using SCOPUS and PubMed to identify human observational studies that investigated an association between ETBF colonization and a CRC diagnosis, and in vitro cell studies and in vivo mouse studies that explored a causal relationship between BFT/ ETBF colonization and CRC pathology.
Potential human observational studies were identified using search terms “colo* cancer”, “Bacteroides fragilis”, “bft”, “enterotoxigenic Bacteroides”, “enterotoxigenic”, “ETBF”, “fragilysin”, “metagenome*”, “microbiota*; in vitro studies were identified using search terms “Bacteroides fragilis enterotoxin", “ETBF”, “BFT”, "enterotoxigenic Bacteroides", “fragilysin”, "in vitro", and “cell”; and in vivo studies were identified using search terms "Bacteroides fragilis", “ETBF”, “BFT”, "enterotoxigenic Bacteroides”, “fragilysin”, “mouse”, and studies were limited to the English language.
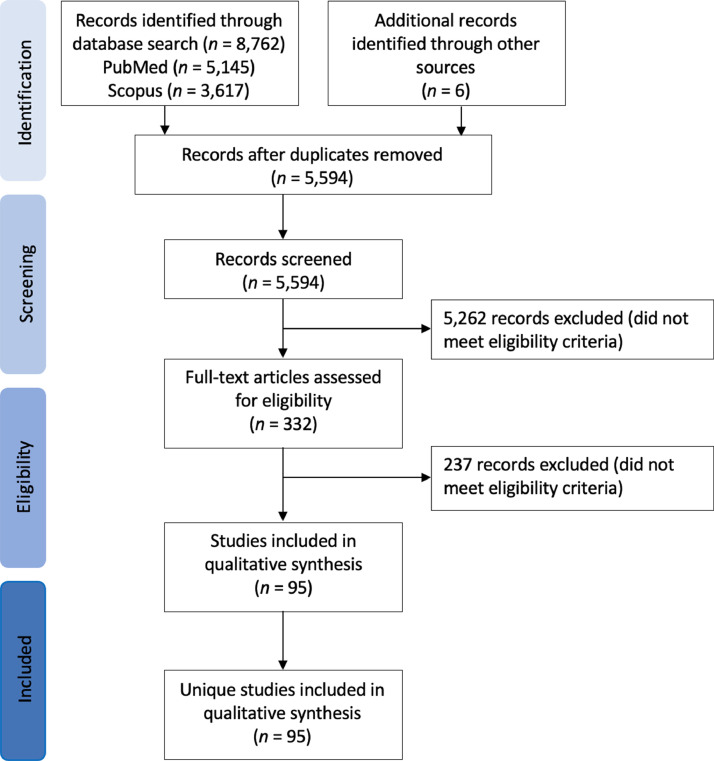
Following an initial screen of titles and abstracts, full text papers were screened for eligibility using pre-defined criteria (Fig. 2, Supplementary Materials S1 Fig. 1). Inclusion criteria for the human observational studies included the detection of ETBF carriage and/ or bft (gene or transcript) in CRC patients using colonic washings, stool samples, mucosal samples, and tissue biopsies of colorectal neoplasms (polyps, adenomas, carcinomas). Inclusion criteria for the in vivo studies included use of mouse models and exposure to ETBF and/ or purified or recombinant BFT, and inclusion criteria for the in vitro studies involved use of cell lines, primary cells, or tissues biopsies and exposure to ETBF and/ or BFT. Studies were excluded if they were review papers, did not present original data, presented protocol development only, and/or performed toxin characterization only. Additionally, 3 studies were removed following concerns regarding how the qPCR analysis was performed, and 1 study was removed for relying on aerobic culture only.
Fig. 2.
PRISMA flow diagram for identification, screening, eligibility, and inclusion of human observational studies, mouse in vitro studies and cell in vivo studies included in this systematic review.
Assessment of methodology quality and risk of bias
The Newcastle-Ottawa Quality Assessment (NOS) tool adapted to human cross-sectional studies, as described by Modesti et al. [13], was performed to assess study design and bias in the human observational studies, the Systematic Review Center for Laboratory animal Experimentation (SYRCLE) tool was used to assess the in vivo mouse studies [14], and ToxRTools used to analyze the in vitro cell studies [15]. Difference assessment tools were used to account for differences in study design across the three study groups.
The NOS and SYRCLE tools involved a set of criteria designed to assess potential bias with regards to selection of study subjects, comparability between case and control subjects, and assessment of outcome. Additionally, SYRCLE also included a list of criteria designed to detect potential bias as a consequence of flaws in the experimental design (see Supplementary Material S2 for a full list of criteria). A ‘yes’ score indicates that the criteria has been achieved and that there is low risk of bias, a ‘no’ score indicates that the criteria was not achieved and that there is a high risk of bias, and an ‘unclear’ score is recorded when the paper has not provided sufficient information to determine whether the criteria has been achieved or not.
The ToxRTool differed in that it involved five criteria groups designed to assess the reliability of toxicological data by evaluating how the test substance was identified (total possible score of 4), how the test system was characterized (score ≤ 3), the description of the study design (score ≤ 6), how the results were documented and presented (score ≤ 3), and the plausibility of the study design and data (score ≤ 2). For each criteria a score of one is achieved when the criteria is met and a score of zero is achieved when the criteria has not been met. The total score is quantified and used to assess how reliable the study is. For in vitro studies, a total score of 15–18 indicates the study is reliable without restrictions (reliability category 1), a score of 11-–14 indicates the study is reliable with restrictions (reliability category 2), and a score <11 indicates that the study is not reliable (reliability category 3). Additionally, six criteria, one in Group I, four in Group 2, and one in Group V, are deemed essential. A failure to meet all six criteria results in the study being categorized as not reliable (see Supplementary Material S2 for the full list of ToxRTool criteria).
Results
The goal of this review was to provide a systematic overview of the current state of evidence for the causal role of ETBF in CRC using standardized guidelines. A meta-analysis of guidelines for meta-analyses show broad agreement on the need to carefully access a few key characteristics across studies, including eligibility criteria (100%), method of data extraction (100%), risk of bias (100%), and heterogeneity in study design (100%) [16]. We highlight the study design, methodologies, risk of bias, and provide quantitative and qualitative descriptions of heterogeneity in our discussion of the study results to highlight the difficulty of synthesizing the existing evidence.
Observational human studies
Study designs
Twenty-two human observational studies were included in the analysis [12,[17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33], [34], [35], [36], [37] (Table 1). Study formats were cross-sectional, case-control or meta-analysis. Nine studies examined samples taken from CRC patients [19,20,23,[28], [29], [30],34,37] and 13 studies compared sample results from CRC patients to healthy controls [12,17,21,22,[24], [25], [26], [27],[31], [32], [33],35,36].
Table 1.
Observational studies (n = 22) included in review. CRC = colorectal cancer.
| Author [reference] | Number of Subjects | Method of detection | Major findings |
|---|---|---|---|
| Toprak et al. 2006 [12] | 132 (73 CRC, 59 control) | qPCR of stool culture isolates | bft detected at higher rates in CRC compared to controls (38% v 12%, P = 0.009) |
| Van et al. 2012 [17] | 99 (49 polyps, 50 without polyps) | Cytotoxicity assay, qPCR of colonic washing culture isolates | ETBF carriage did not positively correlate to polyp incidence. |
| Dutilh et al. 2013 [28] | 12 (12 CRC) | Metatranscriptomics of DNA extracted from tissue biopsy | No significant bft expression in tumor or matched normal sections |
| Zeller et al. 2014 [31] | 491 (114 CRC, 41 adenoma, 335 control) | Metagenomics of DNA extracted from stool samples | B. fragilis was not detected |
| Boleij et al. 2015 [32] | 98* (49* CRC/adenoma, 49** control)*23 antibiotic-treated cases excluded from comparison to controls due to poor culture recovery**including 11 adenomas | qPCR of bacterial colonies isolated anaerobically from mucosal colon tissue | The bft gene was associated with colorectal neoplasia, especially in late-stage CRC. Detection of bft occurred more often in the right tumor |
| Nakatsu et al. 2015 [33] | 276 (102 CRC, 88 adenoma, 86 control) | Characterized the colorectal mucosal microbiome using 16S rRNA sequencing | B. fragilis was enriched in the adenoma-carcinoma sequence |
| Viljoen et al. 2015 [34] | 73 (73 CRC*)*55 fresh-frozen, 18 FFPE | Quantified ETBF in paired tumor and normal tissue samples from 55 CRC patients using qPCR | ETBF was enriched in late stage (III/V) colorectal cancers |
| Keenan et al. 2016 [35] | 142 (71 CRC, 71 control) | Screened stool samples for bft gene using PCR or qPCR of DNA or cultured colonies isolated from stool. | qPCR was more sensitive than standard PCR for bft detection. bft was detected at an increased rate in CRC patients |
| Lennard et al. 2016 [37] | 19 (19 CRC) | Transcriptomics (microarray) of DNA extracted from tissue biopsy | Found no differential expression between ETBF-positive and negative tumor, and no differential expression between ETBF-positive and negative adjacent normal |
| Purcell et al.2016 [18] | 19 (19 CRC) | Standard PCR, qPCR, digital PCR to detect bft gene from matched luminal and stool samples from 19 CRC patients | SYBR qPCR under-detected bft in clinical samples |
| Snezhkina et al. 2016 [19] | 36 (36 CRC) | qRT-PCR was used to quantify SMOX gene and ETBF colonization in 50 paired specimens of stages I-IV CRC tumors and adjacent morphologically normal tissues from CRC patients | Found no association between ETBF colonization and SMOX expression |
| Zhou et al. 2016 [36] | 135 (87 CRC, 48 control) | Quantified ETBF present in resected tumors and adjacent normal tissues from 97 CRC patients using qPCR | ETBF was detected significantly higher in the tumor tissues compared to normal tissue and healthy controls |
| Purcell et al. 2017 [30] | 150 (77 CRC/adenoma/dysplasia, 73 without lesions) | qPCR was used to quantified bft gene present in mucosal tissue from up to four different colonic sites obtained from a consecutive series from 150 patients referred for colonoscopy | ETBF positivity was associated with the presence of low-grade dysplasia, tubular adenomas, and serrated polyps. Increased ETBF and abundance was also associated with left-sided biopsies. |
| Hale et al. 2018 [20] | 83 (83 CRC) | 16S rRNA sequencing was performed on paired colon tumor and normal-adjacent tissue and mucosa samples from patients who underwent partial or total colectomies for CRC | B. fragilis was enriched in deficient MMR CRC but not proficient MMR CRC |
| Bao et al. 2019 [23] | 96 (96 CRC) | qPCR was used to quantify ETBF, mRNA, and microRNAs present in CRC tissue samples | Increased expression of BFAL1 and high abundance of ETBF in CRC tissues predicted poor outcome in CRC patients |
| Haghi et al. 2019 [21] | 120 (60 CRC, 60 control) | Stool samples were screened for B. fragilis using PCR targeting the marker genes neu and bft | B. fragilis was detected at a higher frequency in the CRC patients. Detection of bft was greater in stage III samples compared to stages I and II. |
| Saffarian et al. 2019 [22] | 67 (58 CRC, 9 control) | Characterized the microbiome from crypts and associated adjacent mucosal surfaces from CRC patients and controls using 16S rRNA gene sequencing, qPCR, and FISH analysis | B. fragilis was more abundant in the right-side tumors. B. fragilis abundance was increased in tumor samples compared to the controls |
| Wirbel et al. 2019 [24] | 768 (386 CRC, 382 control) | Metagenomic meta-analysis of DNA extracted from stool samples | No significant difference in bft between cases and controls |
| Jasemi et al. 2020 [25] | 62 (31 CRC, 31 control) | Phenotypic tests and PCR were performed on bacterial isolates cultured from colorectal tissue | bft gene was detected with a greater frequency in the CRC samples compared to the controls. ETBF had an increased ability to form biofilms |
| Zamani et al. 2020 [26] | 120 (68 CRC, 52 control) | qPCR was used to detect B. fragilis and bft gene from bacterial isolates cultured from mucosal biopsies from patients with precancerous and cancerous legions and healthy controls | B. fragilis and bft was detected with increased frequency in the patient samples compared to the controls. ETBF was associated with serrated lesions and adenoma with low-grade dysplasia |
| Shen et al. 2021 [27] | 24 [8 colorectal adenoma, 11 laterally spreading tumor (LST), 5 control] – mucosal475 (113 CRC, 208 adenoma, 109 LST, 113 control) - stool | 16S rRNA sequencing was performed in mucosal samples and qPCR was performed on fecal samples to characterize microbial signature | High abundance of ETBF was associated with LST and CRC groups. ETBF also had strong diagnostic power and was associated with malignant LST and IL-6. |
| Shariati et al. 2021 [29] | 30 (30 CRC) | qPCR was used to quantify B. fragilis present in paired tumors and normal tissue specimens from CRC patients | B. fragilis was detected in equal levels in the tumor and control samples. 15% of B. fragilis patients were infected with ETBF in both adenocarcinoma and matched adjacent normal samples |
Method of ETBF assessment
Methods of detection included direct PCR or qPCR of ETBF marker genes (bft, neu) (13 studies) [18], [19], [20], [21], [22], [23],26,27,29,30,33,34,36], selective culture followed by PCR (four studies) [12,17,25,32], comparison of direct PCR to culture with PCR (one study) [35], transcriptomic analysis (one study) [37], or detection via metagenomic or metatranscriptomic data (three studies) [24,28,31]. Nine studies examined tissue biopsy samples for bft and/ or ETBF colonization [19,20,23,25,28,29,34,36,37], six examined stool samples [12,18,21,24,31,35], five looked at mucosal samples [22,26,30,32,33], one study used colonic washings [17], and one study examined both stool and mucosal samples [27] (Table 1).
Study results
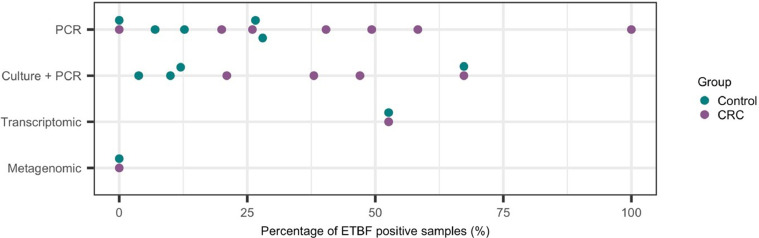
In nine studies that examined only CRC subjects, the prevalence of ETBF ranged from 0 to 100% [18], [19], [20],22,23,28,29,34,37] (Fig. 3). Most studies used qPCR for detection of bft, but specific methods, including primers, vary between studies. In patients positive for bft in tissue by qPCR, Bao et al. [23] found greater ETBF in tumor samples, but two other studies found no difference between tumor and adjacent normal [29,34]. In addition, Hale et al. [20] identified bft in only six of 75 subjects by qPCR. Dutilh et al. [28] used metatranscriptomic data to search for bacterial toxin gene expression. While they identified many reads mapping to the B. fragilis genome, there was no significant expression of bft. Lennard et al. [37] found no difference in host gene expression between ETBF positive and negative samples, for either tumor or normal samples.
Fig. 3.
Comparison of enterotoxin Bacteroides fragilis (ETBF) prevalence using different methods of detection. The percentage of colorectal cancer (CRC) and healthy control subjects that tested positive for the bft gene are plotted. Each data point represents the results reported from a different study, and only studies that clearly reported the abundance of bft are plotted.
In studies examining CRC and controls, the association between ETBF and CRC was unclear (Fig. 3). Six of eight case-control studies found differences in ETBF prevalence between cases and controls [12,21,25,26,31,32,35,36]. Boleij et al. [32] performed selective Bacteroides culture followed by touch-down PCR of selected Bacteroides colonies and identified a statistically significant difference between cases and controls, but only after excluding almost half of the cases and not controls for poor culture recovery, potentially biasing the results. Three cross-sectional studies identified an association between ETBF and CRC, but involved different or unreported inclusion criteria, or explicitly did not address irritable bowel disease (IBD) as a potential confounder [27,30,33]. Unfortunately, systematic comparison across studies with so much heterogeneity is not straightforward. An attempt to perform a meta-analysis of fecal metagenomics data across 768 subjects found clearly detectable bft in deeply sequenced fecal metagenomes but no significant difference between cases and controls [24]. It was reported that bft levels differed broadly with respect to abundance, significant and cross-study consistency of enrichment [24], and thus lack of significance may be due to variability in sequencing depth across the different studies included in the analysis, and thus further metagenomic studies will be required to confirm these findings. Conflicting results may be a consequence of study design. It was observed that studies that reported a significant difference in bft/ ETBF often involved the use of selective bacterial culturing prior to performing bft PCR, while studies that found no significant difference were typically studies that performed PCR/ sequencing directly on DNA samples obtained from the clinical samples. It may be that in samples with low levels of B. fragilis, performing PCR and/ or sequencing directly on DNA extracted from the clinical samples may not detect as significant difference as a result of low levels of B. fragilis DNA, and that improved B. fragilis DNA concentrations as a consequence of selective culturing may improve statistical power. However, it should be noted that the metagenomics studies included significantly higher numbers of subjects, and thus future studies should aim to increase total number of subjects included in order to determine if DNA concentrations and/ or number of included subjects influences the overall findings.
The ability to identify association between ETBF and precursor lesions would be consistent with the hypothesis that ETBF plays a causal role in CRC. Conflicting results between the seven studies and heterogeneity in reporting [17,26,27,[30], [31], [32], [33] has made it difficult to meaningfully compare the results from different studies. We note some examples to highlight some of the difficulties in making broad assessments. In a cross-sectional study of colonoscopy patients using qPCR Van et al. [17] did not find a difference in ETBF prevalence between patients with polyps or without polyps. Boleij et al. [32] defined subjects with tubular adenomas (TA) as either cases or controls based on the type of medical procedure they underwent (surgical resection or colonoscopy). Nakatsu et al. [33] identified a significant percentage of bft positive cases in neoplasms relative to controls in a discovery cohort but not validation cohort. We refer the interested reader to the studies listed in Table 1.
Study limitations and bias analysis
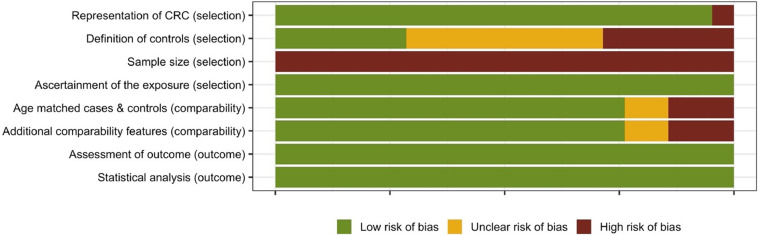
Observational studies were analyzed for bias using the NOS bias tool adapted to cross-sectional studies and the criteria of selection, comparability and outcome. The included observational human studies displayed a relatively low risk of bias (Fig. 4). All studies, however, failed to perform a power analysis to determine the minimum number of subjects and/ or samples required to successfully demonstrate an association between ETBF colonization and CRC. This, therefore, makes it difficult to determine whether the results presented are complete. Observational studies without the engineered use of a toxin/microbial exposure are relatively simple protocols with little risk presented to the individuals, and it may be this simplicity in design that enabled the studies to score a low risk of bias.
Fig. 4.
Quality of reporting and risk of bias assessment using the NOS bias tool adapted to cross-sectional studies. Assessment of the selection, comparability, detection, and outcome is presented as a percentage across all included observational human studies.
In addition to the examples already discussed, it is worth noting some of the limitations in the remaining studies included in Table 1. For example, while Shen et al. [27] found a difference between lateral spreading tumors compared to controls or adenomas, results were only provided as part of a figure, and explicit values were not reported. Purcell et al. [30] reported a significant association of ETBF colonization with the left side of the colon, however, they also reported a within-subject concordance rate of 86% (e.g., a subject's samples were all negative or all positive for ETBF for 129 of 150 subjects). Presumably the reported association is then limited to the 21 cases with discordant results between anatomic sites, raising questions regarding the significance of this result. Additionally, the study identified an association with dysplasia, but did not differentiate between dysplasia arising in adenoma or dysplasia within the setting of IBD. Given that there is a well-established association between IBD and CRC, and that ETBF has also been associated with IBD, this is an important potential confounder and limitation of the study. Nakatsu et al. [33] examined two geographically distinct cohorts and results were reported as percentages only, making it difficult to compare results even within their one study.
In vivo studies
Study designs
Thirty in vivo mouse studies were identified and included in the analysis [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68]. 87% of these studies used the C57BL6 mouse strain (26 studies [38,[40], [41], [42], [43], [44], [45], [46], [47], [48], [49],[51], [52], [53],[57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69]), 10% used the BALB/c mouse strain (3 studies [54], [55], [56]), and 3% used a germ free (GF) NIH mouse strain (1 study [39]) to investigate the role of ETBF in colitis and tumorigenesis (Table 2). The mouse strains used had varying genetic backgrounds (see Supplementary Materials S1, Table 1) and 47% of the studies involved knockout (KO) mouse strains, this included 2 studies that used just the C57BL/6J-ApcMin (Min) mouse strain [41,51], six studies that compared wild type (WT) C5BL6 mice to Min mice [49,53,62,65,67,68], two studies that compared WT C5BL6 mice to various KO mouse strains [45,60], and four studies that compared WT C5BL6, MIN, and KO mouse strains [47,49,64,66] (for additional details on KO strains used, see Supplementary Materials Table 1).
Table 2.
Mouse studies (n = 30) included in review.
| Author [reference] | Study design | Method of analysis | Major findings |
|---|---|---|---|
| Kim et al. 2005 [38] | SPF C57BL6Cr were injected with BFT | Histopathological examination | Inhibition of p38 prevented BFT-induced enteritis |
| Kim et al. 2006 [50] | SPF C57BL6Cr mice were treated with either buffer or a COX-2 inhibitor and injected with BFT | cAMP assay, Histopathological examination, ELISA | Suppression of COX-2 activity prevented BFT-induced fluid secretion |
| Nakano et al. 2006 [39] | GF NIH mice were treated with ETBF or NTBF | Histopathological examination, multiplex-PCR | ETBF induced ulceration, edema, and inflammatory infiltration in the intestine. NTBF was not associated with histological alterations. |
| Rabizadeh et al. 2007 [61] | SPF C57BL/6 mice were inoculated with buffer, NTBF, or ETBF | PCR, hematoxylin and eosin staining | ETBF alone stimulated colitis and significantly enhanced colonic inflammation |
| Rhee et al. 2009 [63] | SPF C57BL/6J or GF 129S6/SvEv mice were orally inoculated with WT ETBF, WT NTBF, WT NTBF overexpressing bft (rNTBF), or WT NTBF overexpressing a biologically inactive mutated bft | Colonic histopathology, Western blot, ex vivo E-cadherin cleavage | ETBF and rNTBF caused colitis in both SPF and GF mice but was lethal only in GF mice.Colonic neoplasms were not observed in mice persistently colonized with ETBF or rNTBF (up to 16 months) |
| Wu et al. 2009 [64] | SPF multiple intestinal neoplasmia (Min)Apc716+/− mice, C57BL/6 mice, and conditional CD4 Stat3–KO mice were colonized with ETBF or NTBF | Histopathology, flow cytometry, depletion of T lymphocytes, cytokine blockade, RT-PCR, Western blotting | Only ETBF triggered colitis and strongly induced colonic tumors. This was associated with Stat3 activation and a selected Th17 response |
| Goodwin et al. 2011 [65] | SPF C57BL/6 and Min mice were treated with ETBF | Immunohistochemical staining, Western blotting, qRT-PCR | ETBF treatment induced colitis that was associated with increased SMO expression. Treatment with MDL 72527 reduced ETBF-induced chronic intestinal inflammation and proliferation, and reduced ETBF-induced colon tumorigenesis in the Min mouse mode |
| Wick et al. 2014 [66] | C57BL/6 WT, C57BL/6Stat3ΔIEC , and Rag-1 mice were inoculated with NTBF or ETBF | Immunohistochemistry (hematoxylin and eosin staining), Western blot, EMSA, mucosal permeability, flow cytometry | ETBF increased mucosal permeability and induced rapid-onset colitis that persisted for up to a year. Stat3 activity was increased. |
| Geis et al. 2015 [67] | C57BL/6 and Min mice were inoculated with ETBF | Flow cytometry, quantitative RT-PCR, histology and microadenoma counts | Tregs initiate IL17-mediated carcinogenesis. Depletion of Tregs in ETBF-colonized C57BL/6 FOXP3DTR mice enhanced colitis but diminished tumorigenesis |
| Destefano Shields et al. 2016 [68] | SPF Min mice and SPF C57BL/6 mice were colonized with ETBF | Mucosal colonization, TaqMan qPCR analysis | Median colon tumor numbers increased with duration of ETBF colonization. ETBF clearance associated with decreased IL-17 expression |
| Hecht et al. 2016 [40] | SPF C57BL/6J mice were co-colonized with NTBF and ETBF | Histological staining, ELISA, quantitative reverse transcriptase PCR, sequential colonization | Competitive exclusion of ETBF by NTBF limited toxin exposure and protected against ETBF-induced colitis. |
| Housseau et al. 2016 [41] | MinApc+/− mice were colonized with ETBF | Tumor counting, histopathology, flow cytometry, RT-PCR, | Ablation of Th17 cells delayed but did not eliminate ETBF-induced tumorigenesis. IL17 blockade significantly attenuated tumorigenesis |
| Wagner et al. 2016 [42] | GF C57BL/6 mice were colonized with human fecal microbiota containing NTBF or ETBF | bft PCR assay, histological analysis, mass spectrometry, immune cell isolation and characterization, microbial RNA-seq, cytokine quantification | ETBF caused weight loss and NTBF reduced BFT expression |
| Casterline et al. 2017 [44] | SPF C57BL/6 mice were inoculated with NTBF and then challenged with ETBF | Sequential colonization, Western blot, quantitative reverse transcriptase PCR, | In sequential B. fragilis colonization, secondary colonization in SPF mice was strain-specific. Bfpai is neither necessary nor sufficient for secondary colonization but was demonstrated to provide an advantage to one strain of ETBF in successful secondary colonization. |
| Hecht et al. 2017 [45] | SPF C57BL/6 Muc2+/+ and SPF C57BL/6 Muc2-/- mice were inoculated with various ETBF clones | Protein overexpression, bacterial mutants, Western blot, qRT-PCR, EMSA | Muc2-deficient mice succumbed to lethal disease from ETBF colonization in a BFT- dependent manner. BFT expression was suppressed by RprXY. Overexpression of RprXY was sufficient to prevent lethal disease in Muc2-deficient mice. |
| Lv et al. 2017 [46] | SPF C57BL/6J mice were treated with AOM/DSS and BFT | Histopathological examination, immunohistochemical examination, tumor examination | BFT blocked formation of adenocarcinoma and size of tumors. BFT treatment was associated with increased adenoma counts. |
| Thiele Orberg et al. 2017 [47] | C57BL/6 (WT), CD45.1 C57BL/6, MinApc716/+ (Min), and OT-1 T cell receptor transgenic RAG−/− mice were colonized with ETBF | Flow cytometry, cell sorting. | ETBF-triggered colon tumorigenesis was associated with an IL-17 driven myeloid signature characterized by subversion of steady-state myelopoiesis in favor of the generation of pro-tumoral monocytic-MDSCs (MO-MDSCs) |
| Chung et al. 2018 [48] | Mice with a C57BL/6 background were colonized with ETBF | Tumor and microadenoma counts, flow cytometry and cell sorting, gene expression, immunohistochemistry, immunofluorescence, Western blot, immunoblotting | ETBF-induced tumorigenesis requires BFT, epithelial IL-17 and Stat3 signaling. |
| Dejea et al. 2018 [49] | Apc+/Δ716 Min mice and SPF C57BL/6J mice treated with AOM were colonized with ETBF | Flow cytometry, qRT-PCR, ELISA, immunohistochemistry | Tumor-prone mice co-colonized with E. coli and ETBF showed increased IL-17 in the colon and DNA damage in colonic epithelium with faster tumor onset and greater mortality, compared mono-colonized mice. |
| Chan et al. 2019 [53] | SPF C57BL/6 WT and MinApc716+/− mice were treated with NTBF and ETBF | qRT-PCR, histology, microadenoma & macroadenoma counts, immunofluorescence and FISH staining, flow cytometry | Sequential treatment with NTBF followed by ETBF diminished ETBF-induced colitis and tumorigenesis |
| Gu et al. 2019 [51] | SPF C57BL/6J-ApcMin mice were colonized with ETBF | Flow cytometry, cytospin analysis, IFN β neutralization, qRT-PCR | Expansion of Treg in the colon of ETBF-colonized mice was driven by CX3CR1+ tissue-resident macrophages in a IFN β-dependent manner.Knockout or suppression of CX3CR1+ myeloid cells reduced tumors |
| Hwang et al. 2019 [52] | SPF C57BL/6 mice was colonized with ETBF and treated with zerumbone | Hematoxylin and eosin staining, Western blot, qRT-PCR, ELISA, nitric oxide assay, immunohistochemistry | Zerumbone did not affect ETBF colonization or BFT-mediated E-cadherin cleavage. Zerumbone did prevent weight loss, splenomegaly, decrease macrophage infiltration, and suppress BFT-induced NF-kB signaling and aIL-8 secretion |
| Cho et al. 2020 [54] | SPF ETBF-colonized BALB/c mice were treated with AOM/DSS and zerumbone | V3-V4 16S MiSeq sequencing, microbiome taxonomic profiling | B. fragilis could be activated by zerumbone. ETBF significantly decreased microbial diversity |
| Hwang et al. 2020 [55] | ETBF-colonized BALB/c mice with AOM/DSS-induced tumorigenesis were treated with zerumbone | Tumor enumeration, histopathology | Oral treatment with zerumbone inhibited colonic polyp numbers and macroadenoma progression |
| Hwang et al. 2020 [56] | SPF BALC/c mice were colonized with ETBF or NTBF | Histology, quantitative reverse transcriptase PCR, ELISA, | ETBF colonization resulted in formation of numerous, larger-sized polyps in the colon. Polyp formation was associated with bft expression |
| Hwang et al. 2020 [57] | SPF C57BL/6 mice were colonized with ETBF and fed a normal salt diet (NSD) or high salt diet (HSD) | qPCR, nitric oxide assay, histology, ELISA | HSD decreased ETBF-induced tumorigenesis through suppression of IL-17A and iNOS expression |
| Liu et al. 2020 [58] | SPF C57BL/6 mice were treated with AOM and colonized with ETBF or ETBF.SPF BALB/c nude mice were injected with ETBF-treated or untreated cancer stem cells | RT-PCR, Western blotting, RNA interference, ChIP assays, immunohistology | ETBF increased the number and volume of intestinal tumors and enhanced expression of NANOG and SOX2. NFAT5 and TLR4 knockdowns decreased tumor growth |
| Patterson et al. 2020 [59] | SPF C57BL/6 mice were colonized with ETBF | Lipidomic analysis, confocal microscopy, qRT-PCR, Western blot, flow cytometry | BFT increases glucosylceramide levels |
| Boleij et al. 2021 [60] | SPF WT C57BL6 and GPR35−/− (KO) mice were colonized with ETBF | qPCR, Histopathology, | Choice of antibiotic pre-treatment influenced severity of ETBF-colitis.GPR35 knockdown resulted in reduced ETBF-induced weight loss, less severe colitis, increased survival rate, and reduced expression of IL-22, Cxcl5, and Mt2 |
| Destefano Shields et al. 2021 [62] | C57BL/6J and BRAFV600ELgr5CreMin (BLM) mice were colonized with ETBF | Flow cytometry, histology and immunohistology, immunohistochemistry, MBD-Seq, RNA-seq, anti-PD-L1 therapy | BRAF mutation drove right-sided ETBF-induced colon tumorigenesis and resulted in disruption of the mucus layer and significant changes in myeloid populations in ETBF-colonized mice |
Study methods
Study methods include inoculation of specific pathogen free (SPF) mice following antibiotic treatment, mono-colonization of GF mice, colonization in conjunction with chemical instigation of colitis, and exposure to purified or recombinant toxin. The two most commonly used murine models of CRC was the azoxymethane/dextran sodium sulfate (AOM/DSS) model of colitis-associated carcinoma [70] and the Min model of multiple intestinal neoplasia [71]. The AOM/DSS model used a pro-carcinogen (AOM) as initiator followed by repeated cycles of DSS-induced colitis, while the Min model involved the use of an engineered APC mutation in the murine genome that results in the formation of multiple small intestine adenomas. Investigation of the potential causal association between ETBF colonization and CRC was achieved by colonizing mice with ETBF in 53% of studies [41,45,[47], [48], [49],51,52,54,55,57,59,60,62,65,67,68], colonizing mice with ETBF or NTBF in 23% of studies [39,56,58,61,63,64,66], colonizing mice with both ETBF and NTBF (either concurrently or subsequential) in 10% of studies [40,44,53], and inoculating mice with BFT in 10% of studies [38,46,50]. Additionally, mice were inoculated with human fecal microbiota (either ETBF positive of NTBF positive) in one study [42], and in another study the mice were inoculated with ETBF-treated or untreated stem cells [58]. Studies that involved colonizing the mice with ETBF and/ or NTBF typically utilized antibiotics to disrupt the gastrointestinal microbiota prior to bacterial inoculation in order to encourage successful colonization. In total 10% of studies utilized a GF mouse model [39,42,63], 67% of studies utilized antibiotics to generate SPF mouse models [38,40,45,46,[49], [50], [51], [52], [53], [54],[56], [57], [58], [59], [60], [61],64,65,68,72], 3% of studies utilized both GF and SPF mice [63], and 23% of studies involved WT mice with no disruptions to the gastrointestinal microbiota [41,47,48,55,62,66,67].
The most commonly used methods of characterizing the effects of ETBF/ BFT exposure in the mouse studies included histopathological examination (utilized by 73%) to assess changes to intestinal inflammation, the formation of tumors, polyps and neoplasms, and the development of ulceration, edema, colitis; PCR (70% of studies) to confirm the presence of ETBF/ BFT and to quantify mRNA levels for inflammatory genes of interest; flow cytometry (40% of studies) for cell quantification; and Western blot (30% of studies) and ELISA (23% of studies) for quantification of BFT, inflammatory proteins (chemokines, cytokines) and other proteins of interest (antibodies, Stat3, Casp3, E-Cadherin) (Table 2).
Study results
Of the 26 studies that colonized mice with ETBF and/ or NTBF, 24 studies reported induced pathogenic traits associated with CRC, this included tumorigenesis (reported in 11 studies) [41,[46], [47], [48], [49],51,53,57,58,62,64,65,67,68], intestinal inflammation (reported in six studies) [39,47,51,52,61,65], colitis (reported in six studies) [40,53,60,61,[63], [64], [65], [66], [67], polyp formation (reported in two studies) [55,56], and ulceration [39], edema [39], splenomegaly [52], and macroademona progression [55] (all reported in one study each). Of the studies that colonized mice with NTBF, colonization was found to be non-pathogenic [39,56,61,63,64], and colonization with NTBF prior to colonization with ETBF was observed to reduce the toxic effects of ETBF colonization [40,42,53] (Table 2). Inoculation with just BFT, however, produced more variable results, Kim et al. [38] and Kim et al. [50] reported BFT-induced enteritis and fluid secretion, respectively, while Ly et al. reported that inoculation with BFT blocked the formation of CRC and reduce the number and size of tumors [46], indicating that BFT was in fact protective against CRC.
Study limitations and bias analysis
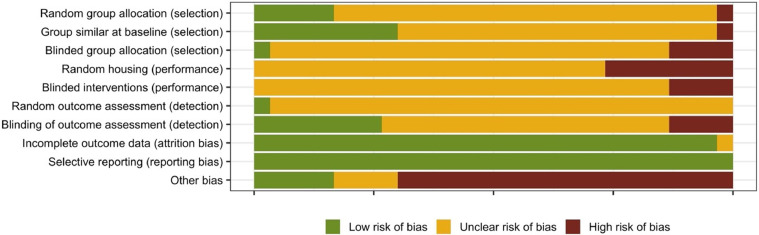
In the majority of the in vivo studies, poorly reported methodology led to an unclear risk of bias for the selection, performance, and detection sections of the bias assessment tool. The authors frequently failed to discuss how the mice were assigned to the treatment group, whether the caregivers, intervenors, or assessors were blinded to which treatment the mice received, whether mice for each treatment group were housed separately or together, and whether results from all the mice were included in the outcome assessment (Fig. 5). Additionally, a significantly number of the in vivo studies used pre-treatment antibiotics but failed to address how this might impact outcome, resulting in a high risk of other bias (Fig. 5).
Fig. 5.
Quality of reporting and risk of bias assessment using SYRCLE'S risk of bias tool. Assessment of the selection, performance, detection, attrition, reporting, and other bias is presented as a percentage across all included in vivo mouse studies.
Analysis of the study design revealed a number of limitations associated with the in vivo studies. Firstly, despite widespread use of SPF mouse models, excluded pathogens vary by vendor and institution and were not reported by investigators [73]. Extremely limited data was available about gastrointestinal microbiota community composition prior to antibiotic treatment or following ETBF colonization This was of significant concern given that Boleij et al. reported that the choice of antibiotic used to generate SPF mice prior to ETBF colonization significantly influences the severity of ETBF-induced colitis [60]. Gut microbiome dysbiosis has been associated with colitis [74], [75], [76], [77], and thus the use of SPF mice makes it difficult to determine whether the development of colitis is a consequence of microbial dysbiosis as a result of non-specific antibiotic targeting of gut commensals or if its caused by ETBF colonization. Moreover, the failure of most studies to determine whether B. fragilis strains were present in the gastrointestinal microbiota prior to treatment means that it is impossible to determine whether treatment response was due to ETBF, NTBF, or BFT inoculation or if it was the results of native species of B, fragilis or other microbial species undergoing expansion due to reduced competition as a result of antibiotic treatment [40,72].
Another concern was the lack of consistent results within the same model system. ETBF inoculation was found to enhance tumorigenesis in AOM/DSS-induced tumorigenesis model when reported by Hwang et al. [56], yet Lv et al. reported that BFT treatment reduced adenocarcinoma, as evidenced by the reduced number and size of tumors in AOM/DSS mice treated with recombinant BFT compared to AOM/DSS mice not exposed to BFT [46]. However, BFT treatment was associated with increased adenoma counts and when the total number of adenocarcinomas and adenomas were combined the number of neoplasms detected were similar across the BFT-treated and non-treated groups [46].
The interaction between NTBF and ETBF has been shown to be important, but the lack of follow-up studies on these questions make the role of ETBF in carcinogenesis more uncertain. Hecht et al. [40] reported that co-colonization of NTBF and ETBF prevented the exacerbation of DSS-induced colitis caused by ETBF colonization alone. Wagner et al. [42] reported that NTBF reduced expression of ETBF and prevented weight loss in a human microbiome associated (HMA) mouse model of childhood undernourishment . Inoculation of the Min (APC +/−) mouse model with ETBF promotes rapid development of colonic tumors [64]. However, wild-type strains have not demonstrated tumor development in response to ETBF colonization alone, despite development of chronic colitis [63]. Results of co-colonization of ETBF and NTBF in Min (APC +/−) mice have not been reported, but this is an important question in light of the Wagner and Hecht studies.
In vitro studies
Study characteristics
Forty-nine in vitro studies were included in this analysis [6,7,10,38,50,[58], [59], [60],65,[78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102], [103], [104], [105], [106], [107], [108], [109], [110], [111], [112], [113], [114], [115], [116], [117] (Table 3, see also Supplementary Materials S1, Table 2). At least thirty cell lines have been studied (Table 3, Supplementary Materials S1, Table 2), of which the colon carcinoma HT-29 cell line was the most frequently utilized (74% of studies), followed by the human intestinal epithelial cell lines T84 and Caco-2, which were utilized in 10% of the studies each (Table 3). Additionally, several of the studies used human primary colon cells [38,50,86,88,91,102], rat primary cells [83,100], and mouse primary cells [58,59,105] (Table 3, Supplementary Materials S1, Table 2).
Table 3.
In vitro studies (n = 49) included in review.
| Reference | Study design | Method of analysis | Major findings |
|---|---|---|---|
| Van Tassell et al. 1992 [10] | Colon carcinoma HT-29 cells were treated with BFT | Cytotoxicity assay | BFT induced cytotoxic response (cell rounding) |
| Weikel et al. 1992 [117] | Human intestinal epithelial cells T84, Caco-2, HT-29 cell lines were co-cultured with ETBF and NTBF cultures | Cell morphology (bright-field light microscopy) | Exposure to ETBF induced morphological changes |
| Pantosti et al. 1994 [120] | 146 B fragilis strains and 64 Bacteroides isolates were tested for ability to produce BFT | Anti-serum testing, Cytotoxicity assay | 16 strains of ETBF were identified (11% of B. fragilis strains examined)Clinical isolates were associated with tissue destruction |
| Moncrief et al. 1995 [115] | HT29 cells were treated with BFT | Cytotoxicity, SSP-PCT, protein assays, ELISA, PAGE, Western blot | BFT exhibited cytotoxic activity that was inhibited by pretreatment with a metal chelator |
| Donelli et al. 1996 [79] | HT-29 cells were treated with BFT | Fluorescence and electron microscopy | BFT induces morphological cell changes by reversibly modifying the actin cytoskeleton |
| Koshy et al. 1996 [80] | Cloned human colonic epithelial cells (HT29/C1) were treated with BFT | Fluorescent phallicidin staining. Cell volume | BFT exposure resulted in distribution of F-actin with loss of stress fibers and cellular membrane blebbing |
| Saidi and Sears 1996 [81] | HT29/C1 cells were treated with BFT | Cytotoxicity assay | BFT rapidly and irreversibly intoxicates HT29/C1 cells in a concentration- and temperature-dependent manner |
| Wells et al. 1996 [82] | HT-29 enterocytes were treated with BFT and then co-cultured with enteric bacteria | Viability, transepithelial electrical resistance (TEER), Light and electron microscopy, bacterial internalization | BFT treatment decreased transepithelial electrical resistance, decreased Listeria monocytogenes internalization, increased internalization of other enteric species |
| Obiso Jr. et al. 1997 [83] | HT-29, rat lung type II, and canine kidney epithelium cells were treated with BFT | Mannitol flux assay, Tight junction resistance recovery assay, epifluorescence microscopy | BFT increased permeability of the paracellular barrier of epithelial cells |
| Chambers et al. 1997 [84] | T84 cells were treated with BFT | Light and electron microscopy, Cell viability, F-actin staining, Ussing chambers | BFT treated induced morphological changes, loss of cellular microvilli, and complete dissolution of some tight junctions |
| Saidi et al. 1997 [85] | HT29/C1 cells were treated with BFT | Spectrofluorimetry, Confocal microscopy, Western blot | BFT alters the F and G-actin cytoskeletal architecture of HT29/C1 cells without direct proteolysis of actin or decrease in F-actin content |
| Sanfilippo et al. 1998 [86] | Human primary colon cells were treated with BFT | Cytotoxicity, Electron microscopy | BFT treatment induced morphological changes (cell rounding, separation from adjacent cells, detachment from basement membrane) and cell cytotoxicity |
| Wu et al. 1998 [87] | HT29/C1 cells were treated with BFT | Western blot, Immunofluorescent, confocal microscopy, Northern blot, Reverse transcription PCR | BFT cleaves the extracellular domain of E-cadherin |
| Chung et al. 1999 [116] | 89 B. fragilis strains were tested for BFT productionHT29/C1 cells were co-cultured with NTBF and ETBF | Colony blot hybridization, PCR, Western blot | 38% of B. fragilis strains examined were ETBF, BFT cleaved E-cadherin |
| Riegler et al. 1999 [88] | Treated colonic mucosa with BFT | Ussing chambers, confocal microscopy | BFT treatment increased cell permeability and damaged crypt and surface colonocytes |
| Sanfilippo et al. 2000 [89] | Intestinal epithelial cell lines HT29, T84, Caco-2, and IEC-6 were treated with BFT | Transmission electron microscopy, reverse transcription PCR, sandwich ELISA | BFT exposure increased expression of IL-8 and secretion of TGF-β (T84), induced morphology changes (HT29), loss of tight junctions (T84), and detachment (T84) |
| Kim et al. 2001 [90] | HT29 and Caco-2 cells were treated with BFT | Quantitative real-time (qRT)-PCR, ELISA | BFT exposure increased expression of neutrophil chemoattractant and activators (ENA-78, GRO-α, IL-8) |
| Kim et al. 2002 [91] | HT29, T84, and primary human colon epithelial cells were treated with BFT | Supershift EMSA, Western blot, qRT-PCR, ELISA | BFT induced NF-κB activation and IκB degradation |
| Franco et al. 2002 [6] | HT29/C1 cells were treated with BFT | Reverse transcription PCR | The B. fragilis pathogenicity island and its flanking regions modulate bft expression |
| Wu et al. 2003 [92] | HT29/C1 cells were treated with BFT | Western blot, Immunofluorescent confocal microscopy, Reverse transcription PCR | BFT activates T-cell factor-dependent transcriptional activation and promotes cell proliferation |
| Wu et al. 2004 [93] | HT29/C1 cell were treated with BFT | Western blot, ELISA, reverse transcription PCR | BFT stimulates IL-8 secretion |
| Kim et al. 2005 [38] | HT29 cells were treated with BFT | qRT-PCR, ELISA, EMSA, Western blot | BFT activated three major MAPK cascades (p38, JNK, ERK1/2) and AP-1 signals composed of c-Jun/c-Fos heterodimers |
| Kim et al. 2006 [50] | HT29 cells were treated with BFT | qRT-PCR, Western blot, Luciferase assay | BFT exposure increased expression of COX-2 and prostaglandin E2 |
| Sears et al. 2006 [94] | HT29/C1 cells were treated with BFT | Western blot, reverse transcription PCR | The deletion of 2 amino acids in the C terminus of BFT reduced biological activity |
| Wu et al. 2006 [95] | HT29/C1 cells were treated with BFT | Confocal microscopy, flow cytometry, acid wash | BFT binds irreversibly to intestinal epithelial cells in a polarized, metalloprotease-dependent manner |
| Wu et al. 2007 [96] | HT29/C1 cells were treated with BFT | Western blot, RNA interference, immunostaining | BFT mediated shedding of cell membrane proteins. Cleavage of E-cadherin was dependent on toxin metalloprotease and γ-secretase. |
| Kim et al. 2008 [97] | HT29 cells were treated with BFT | Cell Death detection ELISA, flow cytometry, qRT-PCR, Western blot, luciferase assay | BFT induced apoptosis and activated the phosphorylation of ERK1/2, p38, and JNK |
| Kim et al. 2009 [98] | HT29 cells were treated with BFT | Quantitative reverse transcription PCR, RT-PCR, ELISA, Western blot | BFT-induced phosphorylation of both IκBα and IκB kinase (IKK) signals was prevented in eupatilin-pretreated HT29 cells |
| Yoon et al. 2010 [99] | HT-29 and Caco-2 cells were treated with BFT | qRT-PCR, ELISA, EMSA, Western blot | BFT induced human ß-defensin 2 in a dose- and time-dependent manner that could be regulated by a MAPK, IKK-, and NF-kB-dependent signaling pathway. BFT also activated ERK1/2, p38, and JNK |
| Goodwin et al. 2011 [65] | HT29/C1 and T84 cells were treated with BFT | qRT-PCR, Western blot, enzyme activity assays, | BFT upregulates spermine oxidase (SMO), resulting in SMO-dependent generation of ROS and induction of a DNA damage marker (γ-H2A.x) |
| Roh et al. 2011 [100] | HUVECs and rat aortic endothelial cells were treated with BFT | qRT-PCR, flow cytometry, immunofluorescence assay, EMSA, ELISA | BFT induced ICAM-1 expression. Upregulation of ICAM-1 was dependent on the activation of IkB and NF-kB signaling pathways. |
| Hwang et al.2013 [101] | HT29/C1 wells were treated with BFT | ELSA and Western blot | BFT induced E-cadherin degradation and IL-8 secretion |
| Yoo et al. 2013 [102] | HT29 cells were treated with BFT | Quantitative reverse transcriptase PCR, ELISA, EMSA, luciferase assay, Western blot | BFT induced upregulation of lipocalin 2 in an AP-1 signaling dependent manner that was regulated by MAPKs (ERK, p38) |
| Remacle et al. 2014 [103] | Human colorectal carcinoma cell lines (HTC116, HT29, HT29/C1) were treated with BFT | Immunofluorescence microscopy, immunoprecipitation of E-cadherin | BFT cleaved E-cadherin, |
| Shiryaev et al. 2014 [7] | HT29 cells were treated with BFT | Immunoprecipitation of E-cadherin, cell aggregation assay | BFT repressed cell aggregation |
| Kharlampieva et al. 2015 [104] | HT29 cells were treated with BFT | Site-directed mutagenesis, recombination, Western blot | BFT induced endogenous E-cadherin cleavage. Cleavage activity required the native structure of zinc-binding motif |
| Ko et al. 2016 [105] | Murine intestinal epithelial cells were treated with BFT | Quantitative reverse transcriptase PCR, EMSA, transfection assay, Western blot, ELISA immunofluorescence, apoptosis assay | BFT upregulated expression of heme oxygenase-1 (HO-1) in a p38 and IKK-NF-xB dependent manner |
| Ko et al. 2017 [106] | HUVECs were treated with BFT | Western blot, ELISA, immunofluorescence assay, EMSAs, transfection assay | BFT increased light chain 3 protein II (LC3-II) conversion from LC3-I and protein expression of p62, Atg5, and Atg12. BFT increased indices of autophagosomal fusion with lysosomes, activated ATP-1, and upregulated expression of C/EBP |
| Jeon et al. 2019 [107] | Human colon epithelial cells (HCT 116) were treated with BFT | Quantitative reverse transcriptase PCR, ELISA, Western blot | BFT reduced expression of β-catenin. Suppression of β-catenin resulted in increased NF-kB activity and IL-8 expression. |
| Metz et al. 2019 [109] | Ht29/C1 cells were treated with BFT | Morphological assay, thermal shift assay | Chenodeoxycholic acid inhibits BFT |
| Allen et al. 2019 [110] | HT29/C1 cells were treated with BFT | Quantitative PCR, RNA-seq assay | BFT induced differential expression of genes related to bacterial interactions with colon epithelial cells. Ceacam1 was increased and Muc2 was decreased |
| Jeon et al. 2020 [108] | HCT 116 cells were treated with BFT | Western blot, ELISA, EMSA, Cell death detection ELISA | BFT increased expression of sulfiredoxin 1 (Srx-1) in a time-dependent manner. BFT also activated transcriptional signals (Nrf2, AP-1, and NF-kB). Srx-1 induction was dependent on the activation of Nrf2 signals. Overexpression of Srx-1 attenuated apoptosis |
| Ko et al. 2020 [111] | Murine dendritic cells were exposed to BFT | Quantitative reverse transcriptase-PCR, EMSA, transfection assays, Western blot, ELISA, ROS assay | BFT upregulated HO-1expression and activated transcription factors (NF-kB, AP-1, Nrf2). Upregulation of HO-1 was dependent on Nrf2 activation and regulated by ERK and p38. BFT also increased production of ROS. |
| Liu et al. 2020 [58] | Murine colonoids were co-cultured with ETBF | qRT- PCR, sphere forming assay, Western blot, immunohistochemistry | ETBF increased cell stemness and enhanced expression of core stemness transcription factors (NANOG, SOX2). ETBF also activated the Toll-Like 4 pathway |
| Patterson et al. 2020 [59] | Colon organoids and HT29/C1 cells were treated with BFT | Lipidomic analysis, confocal microscopy, q RT-PCR, Western blot, flow cytometry | BFT increased glucosylceramide levels and decreased colonoid permeability and bursting. |
| Becker et al. 2021 [112] | Caco-2 cells were exposed to bft-positive and -negative strains of B. fragilis | TEER, Real-time qPCR, whole genome sequencing, NMR spectroscopy. | BFT increased intestinal barrier function |
| Cao et al. 2021 [113] | Human CRC cell lines (HCT116, SW480) were co-cultured with NTBF and ETBF | microRNA sequencing, semiquantitative reverse-transcription PCR, RT-PCR | ETBF promoted CRC cell proliferation by down-regulating miR-149-3p |
| Xie et al. 2021 [114] | Human CRC cell line SW620 and normal colon cell line NCM460 were cultured/ treated with Recombinant BFT (rBFT) | ELISA, Western blot, cell proliferation assays, | rBFT promoted CRC cell proliferation and accelerated tumor growth. This was associated with upregulation of CCL3, CCR5, NF-kB, and TRAF-6 |
| Boleij et al. 2021 [60] | HT29/C1 cells were treated with BFT | CRISPRcas GPR35-knockout, Western blot, ELISA, immunofluorescence, confocal imaging, RT-PCR | GPR35 identified as a signaling molecule for BFT |
Study methods
The most common method of investigation was direct exposure to the BFT protein (80% of studies) [6,7,10,38,50,59,60,65,[80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102], [103], [104], [105], [106], [107], [108],110,111,114,115,118], followed by co-culture with both ETBF and NTBF (4% of studies, includes studies where identification of ETBF/ NTBF was determination after co-culture) [112,113,116,117], co-culture with ETBF (2% of studies) [58], co-culture with either ETBF or NTBF (2% of studies) [78] (Table 3). The effect of BFT/ ETBF exposure on the cultured cells was determined by a variety of methods (see Table 3), but the most common included Western blotting, utilized by 57% of studies to detect BFT [59,60,94], assess the ability of BFT to bind and / or degrade to proteins of interest [85,91], to cleave E-cadherin [60,87,96,104,116], determine protein expression/ levels [50,58,59,[97], [98], [99],[104], [105], [106], [107],111,114], assess electrophoretic mobility [116] and protein activation [60,108]; PCR (reverse transcription, real-time) was used in 49% of studies to detect and quantify expression of bft [6,65,94,112,116], E-cadherin [87], Cox-2 [50,98], beta-defensin, ICAM-1 [100] cytokines [60,89,90,93,97,98,107,113], chemokines [38,91], Muc-2 [110], heme oxygenase [105,111], and transcription factors [58,92]; microscopy (bright-field, fluorescence, electron, confocal, immunofluorescence) was used in 33% of the studies to assess changes in morphology [59,60,79,[82], [83], [84], [85], [86], [87], [88], [89],92,103,117], proliferation [92], and permeability [59,[82], [83], [84],88] in the exposed cells; and 12% of studies performed cytotoxicity assays to cellular sensitivity to BFT [10,78,81,86,115] (Table 3). The cytotoxicity were typically semi-quantitative and relied on morphologic changes such as cell rounding or detachment following incubation with purified toxin [16]. Quantification of purified toxin was reported as a concentration such as picomolar or ng/mL, or as titers of cytotoxic activity (highest dilution causing at least 50% cell rounding after 4-hour incubation).
Study results
Exposure to BFT was found to induce a number of cellular changes that are associated with CRC pathogenesis. This included morphological changes [79,80,84,85,89,117], cell permeability [83,84,88,89], cytotoxicity response [10,78,81,86,115], tissue damage [78,88], gene expression (cytokines, transcriptional factors, Cox-2, ICAM-1, B-catenin, Heme oxygenase) [38,50,65,[89], [90], [91], [92], [93],97,[99], [100], [101],105,107,108,110,111], cell proliferation [92,114], tumorigenesis [65], and reduced apoptosis [108]. Co-culture with ETBF also resulted in changes to morphology [117], CRC cell proliferation [113] and tumorigenesis [65], increased tissues destruction, increased expression of core stemness transcriptional factors [58], and activation of the TLR4 pathway [58].
Study limitations and bias analysis
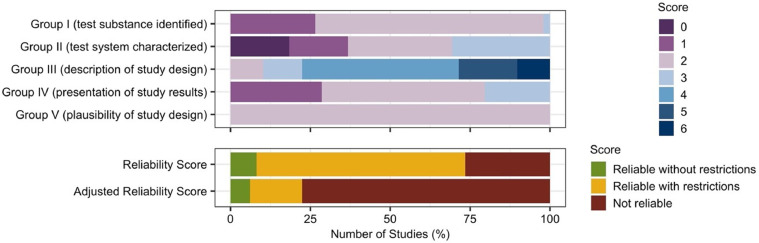
The reliability of the toxicological data generated by the in vitro studies was assessed by the ToxR Tool (Fig. 6). The average score for Group criteria 1: test substance identified was 1.8 (range = 1–3, total possible score = 3), the average score for Group II: test system characterization was 1.8 (range = 0–3, total possible score = 3), the average score for Group III: description of study design was 4.1 (range = 2–6, total possible score = 6), the average score for Group IV: presentation of study results was 1.9 (range = 1–3, total possible score = 3, and the average score for Group V: plausibility of study design was 2.0 (range = 2–2, total possible score = 2) (Fig. 6). Overall, the average total score was 11.6, resulting in an average reliability score of 2, reliable but with restrictions (Fig. 6). However, 38 studies (78% of the studies) failed to meet the six critical criteria, resulting in 13 studies scoring 3 on the reliability scale (data is not reliable) and 25 studies being downgraded from a reliability score of 2 to a reliability score of 3. This led to an average adjusted reliability score of 2.7 (Fig. 6) (Supplementary Materials S2).
Fig. 6.
Quality of reporting and the risk of bias assessment using ToxRTool. The reliability of the in vitro cell studies was determined by scoring the test substance identified, test system characterized, description of study design, presentation of study results, and plausibility of the study design. Total score is quantified and both reliability score and adjusted reliability score were quantified. Bias score is presented as a percentage across all included in vitro cell studies.
Of the six essential criteria, it was the essential criteria included in Group III: study design description where the in vitro studies analyzed failed to meet all essential criteria. The included studies frequently failed to disclose the concentration of BFT used, how the cells were exposed to BFT/ ETBF, the frequency and time points of exposure, and if a positive control had been included (Supplementary Materials S2). Studies which referred the reader to previous papers were score ‘no’ for these criteria, and often the studies the reader is recommended to refer to also failed to achieve these essential criteria. This meant that the in vitro studies were highly unreliable as the studies could not be replicated and potential influencing factors that might bias the results could not be determined.
The in vitro studies were found to have several flaws in methodology and presented with conflicting evidence. An early diagnostic test for the presence of BFT took advantage of the “exquisitely sensitive” response of the HT-29/C1 colon adenocarcinoma cell line [84,121] and since the development of this diagnostic tool the vast majority of in vitro studies investigating a potential causal relationship between ETBF and CRC have utilized the HT-29 cell line. However, Van Tassell et al. [10] also exposed 14 mammalian cell lines in addition to HT-29 to BFT, including the CCD-3CO cell line (human colon fibroblasts), NCI-H508 (human cecal adenocarcinoma), LS174T (human colon adenoma), Caco-2 (human colon carcinoma), and T-84 (human colon carcinoma). The authors found that BFT only induced a cytotoxic response on the HT-29 cells [10], suggesting that only the HT-29 cell line is sensitive to the toxin, and that this unique property may mean that that the HT-29 model may not be the most appropriate cell line to investigate the relationship between ETBF colonization and CRC. However, it should be noted that other groups have subsequently demonstrated T84 responsiveness to BFT [84].
There is also conflicting evidence for the effects of BFT on barrier function. While Chambers et al. [84] reported decreased monolayer resistance in T84 cells and Riegler et al. [88] identified increased permeability in primary human colonic mucosal strips, Becker et al. [112] reported increased barrier function in Caco-2 cells and human colonic organoids [84,88]. Differences in results across the three studies is likely due to the cell lines used in the different studies having different responses to BFT. Additionally, Becker et al. used live cultures while Chambers and Riegler exposed the cells to purified BFT. It has been previously reported that inoculation of B. fragilis corrects gut permeability in a maternal immune activation (MIA) mouse model [122], suggesting that the improved barrier function reported by Becker et al may be due to additional activities of B. fragilis. Further investigations will be required to determine whether differences in findings is a consequence of different concentrations of BFT used, different cell lines used, or the use of BFT compared to co-culture with ETBF.
Discussion
This systemic review lays out the evidence for the association between ETBF and human CRC as well as ETBF's role in causing CRC. Though the initial reports have been promising, important biological questions remain as part of future studies. One of the key challenges for any broader attempt at synthesizing evidence is the heterogeneity of the studies and the potential for bias. Future epidemiological studies of CRC could improve the status of our knowledge through stratification of normal, adenomas and carcinomas, and matched controls to assess known confounding risk factors such as colitis. On a technical level, newer sequencing technology can potentially reduce the variation in detection methods and the variability apparent in PCR and culture. There is opportunity to move from exploratory studies to more concrete assessments that more consistently report power calculations and detect ETBF using multiple testing methodologies to enhance reproducibility and reduce bias. This would help us better understand the nature of any geographic variation in ETBF prevalence.
Mechanistic studies have been extremely promising but reducing the risk of bias by expanding the animal models used for testing will be a key additional piece of evidence in support of the causal role of ETBF in CRC. It is worth noting that many of the in vitro studies do not provide key details essential for reproducibility including identification of the cell line being used, source of material for cell lines, and concentration of toxin being used in experiments. Reproducing these results would be a key first step to understanding the robustness of the overall findings from in vitro testing. Reporting concentrations of toxin exposures would be particularly helpful for understanding the physiological applicability to the human colon. Additional future areas of interest for understanding the broader relationship between ETBF and human CRC include assessing the amount of toxin production in an asymptomatic human carriers; what drives heterogeneity of colonocyte response to toxin; and how the context of the gastrointestinal microbial community, including the presence of other B. fragilis strains, modulates ETBF behavior.
Conclusion
The role of individual microbes and the gastrointestinal microbiome as a whole in the initiation and progression of CRC is an important area of active research. While the initial studies have brought to light an intriguing potential relationship between ETBF and CRC, a combination of multiple lines of high-quality evidence will be important to further this hypothesis. Future studies should seek to reduce heterogeneity and bias by employing appropriate controls for key confounding factors. In addition, reducing risk of bias in experimental testing by diversifying the models used as well as reporting key data such as cell line or toxin concentrations used would greatly improve the ability of synthesis findings into a broader understanding of the role of ETBF in CRC. This review of the literature supports the International Cancer Microbiome Consortia's 2019 statement that “there is currently no direct evidence that the human commensal microbiome is a key determinant in the etiopathogenesis of cancer” [123], at least with regards to ETBF, and identifies specific areas where additional evidence is needed.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100797.
Appendix. Supplementary materials
References
- 1.Sung H., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Islami F., et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J Clin. 2018;68:31–54. doi: 10.3322/caac.21440. [DOI] [PubMed] [Google Scholar]
- 3.Park C.H., Eun C.S., Han D.S. Intestinal microbiota, chronic inflammation, and colorectal cancer. Intest Res. 2018;16:338. doi: 10.5217/ir.2018.16.3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng Y., Ling Z., Li L. The intestinal microbiota and colorectal cancer. Front Immunol. 2020;11:3100. doi: 10.3389/fimmu.2020.615056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez L.R., Bleich R.M., Arthur J.C. Annual review of medicine microbiota effects on carcinogenesis: initiation, promotion, and progression. Annu Rev Med. 2021;72:2020. doi: 10.1146/annurev-med-080719-091604. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franco A.A., Cheng R.K., Goodman A., Sears C.L. Modulation of bft expression by the Bacteroides fragilis pathogenicity island and its flanking region. Mol Microbiol. 2002;45:1067–1077. doi: 10.1046/j.1365-2958.2002.03077.x. [DOI] [PubMed] [Google Scholar]
- 7.Shiryaev S.A., Remacle A.G., Cieplak P., Strongin A.Y. Peptide sequence region that is essential for the interactions of the enterotoxigenic Bacteroides fragilis metalloproteinase II with E-cadherin. J Proteolysis. 2014;1:3. [PMC free article] [PubMed] [Google Scholar]
- 8.Myers L.L., Shoop D.S., Firehammer B.D., Border M.M. Association of enterotoxigenic Bacteroides fragilis with diarrheal disease in calves. J Infect Dis. 1985;152:1344–1347. doi: 10.1093/infdis/152.6.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers L.L., et al. Isolation of enterotoxigenic Bacteroides fragilis from humans with diarrhea. J Clin Microbiol. 1987;25:2330–2333. doi: 10.1128/jcm.25.12.2330-2333.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Tassell R.L., Lyerly D.M., Wilkins T.D. Purification and characterization of an enterotoxin from Bacteroides fragilis. Infect Immun. 1992;60:1343–1350. doi: 10.1128/iai.60.4.1343-1350.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basset C., Holton J., Bazeos A., Vaira D., Bloom S. Are helicobacter species and enterotoxigenic Bacteroides fragilis involved in inflammatory bowel disease? Dig Dis Sci. 2004;49:1425–1432. doi: 10.1023/b:ddas.0000042241.13489.88. [DOI] [PubMed] [Google Scholar]
- 12.Ulger Toprak N., et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 13.Modesti P.A., et al. Panethnic differences in blood pressure in Europe: a systematic review and meta-analysis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0147601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hooijmans C.R., et al. SYRCLE's risk of bias tool for animal studies. BMC Med Res Methodol. 2014;14:43. doi: 10.1186/1471-2288-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schneider K., et al. ToxRTool”, a new tool to assess the reliability of toxicological data. Toxicol Lett. 2009;189:138–144. doi: 10.1016/j.toxlet.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Mueller M., et al. Methods to systematically review and meta-analyse observational studies: a systematic scoping review of recommendations. BMC Med Res Methodol. 2018;18:1–18. doi: 10.1186/s12874-018-0495-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van N., et al. Evaluation of enterotoxigenic Bacteroides fragilis from colonic washings from patients undergoing colonoscopy. Biomed Sci. 2012;18(4):362–368. [Google Scholar]
- 18.Purcell R.V., Pearson J., Frizelle F.A., Keenan J.I. Comparison of standard, quantitative and digital PCR in the detection of enterotoxigenic Bacteroides fragilis. Sci Rep. 2016;6:1–8. doi: 10.1038/srep34554. 2016 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snezhkina A.V., et al. The dysregulation of polyamine metabolism in colorectal cancer is associated with overexpression of c-Myc and C/EBPβ rather than enterotoxigenic Bacteroides fragilis infection. Oxid Med Cell Longev. 2016;2016 doi: 10.1155/2016/2353560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hale V.L., et al. Distinct microbes, metabolites, and ecologies define the microbiome in deficient and proficient mismatch repair colorectal cancers. Genome Med. 2018;10:78. doi: 10.1186/s13073-018-0586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haghi F., Goli E., Mirzaei B., Zeighami H. The association between fecal enterotoxigenic B. fragilis with colorectal cancer. BMC Cancer. 2019;19:879. doi: 10.1186/s12885-019-6115-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saffarian A., et al. Crypt- and mucosa-associated core microbiotas in humans and their alteration in colon cancer patients. MBio. 2019;10:pp.e01315–19. doi: 10.1128/mBio.01315-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bao Y., et al. Long noncoding RNA BFAL1 mediates enterotoxigenic Bacteroides fragilis-related carcinogenesis in colorectal cancer via the RHEB/mTOR pathway. Cell Death Dis. 2019;10:1–14. doi: 10.1038/s41419-019-1925-2. 2019 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wirbel, J. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed]
- 25.Jasemi S., et al. Toxigenic and non-toxigenic patterns I, II and III and biofilm-forming ability in Bacteroides fragilis strains isolated from patients diagnosed with colorectal cancer. Gut Pathog. 2020;12:1–7. doi: 10.1186/s13099-020-00366-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zamani S., et al. Enterotoxigenic Bacteroides fragilis: a possible etiological candidate for bacterially-induced colorectal precancerous and cancerous lesions. Front Cell Infect Microbiol. 2020;9:449. doi: 10.3389/fcimb.2019.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen X., et al. Fecal enterotoxigenic Bacteroides fragilis–peptostreptococcus stomatis–parvimonas micra biomarker for noninvasive diagnosis and prognosis of colorectal laterally spreading tumor. Front Oncol. 2021;11:1. doi: 10.3389/fonc.2021.661048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dutilh B.E., Backus L., Van Hijum S.A.F.T., Tjalsma H. Screening metatranscriptomes for toxin genes as functional drivers of human colorectal cancer. Best Pract Res Clin Gastroenterol. 2013;27:85–99. doi: 10.1016/j.bpg.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Shariati A., et al. Association between colorectal cancer and Fusobacterium nucleatum and Bacteroides fragilis bacteria in Iranian patients: a preliminary study. Infect Agent Cancer. 2021;16:1–10. doi: 10.1186/s13027-021-00381-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Purcell R.V., et al. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One. 2017;12 doi: 10.1371/journal.pone.0171602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeller G., et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol Syst Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boleij A., et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60:208–215. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakatsu G., et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun. 2015;6:1–9. doi: 10.1038/ncomms9727. 2015 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viljoen K.S., Dakshinamurthy A., Goldberg P., Blackburn J.M. Quantitative profiling of colorectal cancer-associated bacteria reveals associations between fusobacterium spp., enterotoxigenic Bacteroides fragilis (ETBF) and clinicopathological features of colorectal cancer. PLoS One. 2015;10 doi: 10.1371/journal.pone.0119462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keenan J.I., et al. Screening for enterotoxigenic Bacteroides fragilis in stool samples. Anaerobe. 2016;40:50–53. doi: 10.1016/j.anaerobe.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 36.Zhou Y., et al. Association of oncogenic bacteria with colorectal cancer in South China. Oncotarget. 2016;7:80794. doi: 10.18632/oncotarget.13094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lennard K.S., Goosen R.W., Blackburn J.M. Bacterially-associated transcriptional remodelling in a distinct genomic subtype of colorectal cancer provides a plausible molecular basis for disease development. PLoS One. 2016;11 doi: 10.1371/journal.pone.0166282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J.M., et al. Mitogen-activated protein kinase and activator protein-1 dependent signals are essential for Bacteroides fragilis enterotoxin-induced enteritis. Eur J Immunol. 2005;35:2648–2657. doi: 10.1002/eji.200526321. [DOI] [PubMed] [Google Scholar]
- 39.Nakano V., Gomes D.A., Arantes R.M.E., Nicoli J.R., Avila-Campos M.J. Evaluation of the pathogenicity of the Bacteroides fragilis toxin gene subtypes in gnotobiotic mice. Curr Microbiol. 2006;53:113–117. doi: 10.1007/s00284-005-0321-6. 2006 532. [DOI] [PubMed] [Google Scholar]
- 40.Hecht A.L., et al. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep. 2016;17:1281–1291. doi: 10.15252/embr.201642282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Housseau F., et al. Microenvironment and immunology redundant innate and adaptive sources of IL17 production drive colon tumorigenesis. Cancer Res. 2016;76:2115–2124. doi: 10.1158/0008-5472.CAN-15-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner V.E., et al. Effects of a gut pathobiont in a gnotobiotic mouse model of childhood undernutrition. Sci Transl Med. 2016;8 doi: 10.1126/scitranslmed.aah4669. 366ra164-366ra164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wickens K., Pearce N., Crane J., Beasley R. Antibiotic use in early childhood and the development of asthma. Clin Exp Allergy. 1999;29:766–771. doi: 10.1046/j.1365-2222.1999.00536.x. [DOI] [PubMed] [Google Scholar]
- 44.Casterline B.W., Hecht A.L., Choi V.M., Bubeck Wardenburg J. The Bacteroides fragilis pathogenicity island links virulence and strain competition. Gut Microbes. 2017;8:1–10. doi: 10.1080/19490976.2017.1290758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hecht A.L., Casterline B.W., Choi V.M., Bubeck Wardenburg J. A two-component system regulates Bacteroides fragilis toxin to maintain intestinal homeostasis and prevent lethal disease. Cell Host Microbe. 2017;22:443–448. doi: 10.1016/j.chom.2017.08.007. .e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lv Y., et al. Suppression of colorectal tumorigenesis by recombinant Bacteroides fragilis enterotoxin-2 in vivo. World J Gastroenterol. 2017;23:603. doi: 10.3748/wjg.v23.i4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thiele Orberg E., et al. The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 2016;10:421–433. doi: 10.1038/mi.2016.53. 2017 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung L., et al. Bacteroides fragilis toxin coordinates a pro-carcinogenic inflammatory cascade via targeting of colonic epithelial cells. Cell Host Microbe. 2018;23:203–214. doi: 10.1016/j.chom.2018.01.007. .e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dejea C.M., et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science. 2018;359:592–597. doi: 10.1126/science.aah3648. (80-.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J.M., et al. Bacteroides fragilis enterotoxin induces cyclooxygenase-2 and fluid secretion in intestinal epithelial cells through NF-kappa B activation. Eur J Immunol. 2006;36:2446–2456. doi: 10.1002/eji.200535808. [DOI] [PubMed] [Google Scholar]
- 51.Gu T., Li Q., Egilmez N.K. IFNβ-producing CX3CR1+ macrophages promote T-regulatory cell expansion and tumor growth in the APCmin/+ /Bacteroides fragilis colon cancer model. Oncoimmunology. 2019;8:e1665975. doi: 10.1080/2162402X.2019.1665975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hwang S., et al. Zerumbone suppresses enterotoxigenic Bacteroides fragilis infection-induced colonic inflammation through inhibition of NF-κΒ. Int J Mol Sci. 2019;20:4560. doi: 10.3390/ijms20184560. 2019, Vol. 20, Page 4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan J.L., et al. Non-toxigenic Bacteroides fragilis (NTBF) administration reduces bacteria-driven chronic colitis and tumor development independent of polysaccharide A. Mucosal Immunol. 2019;12:164. doi: 10.1038/s41385-018-0085-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cho H.W., Rhee K.J., Eom Y.B. Zerumbone restores gut microbiota composition in ETBF colonized AOM/DSS mice. J Microbiol Biotechnol. 2020;30:1640–1650. doi: 10.4014/jmb.2006.06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwang S., et al. Protective effects of zerumbone on colonic tumorigenesis in enterotoxigenic Bacteroides fragilis (ETBF)-colonized AOM/DSS BALB/c mice. Int J Mol Sci. 2020;21:857. doi: 10.3390/ijms21030857. 2020, Vol. 21, Page 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hwang S., et al. Enterotoxigenic Bacteroides fragilis infection exacerbates tumorigenesis in AOM/DSS mouse model. Int J Med Sci. 2020;17:145. doi: 10.7150/ijms.38371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hwang S., Yi H.C., Hwang S., Jo M., Rhee K.J. Dietary salt administration decreases enterotoxigenic Bacteroides fragilis (ETBF)-promoted tumorigenesis via inhibition of colonic inflammation. Int J Mol Sci. 2020;21:8034. doi: 10.3390/ijms21218034. 2020, Vol. 21, Page 8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Q.Q., et al. Enterotoxigenic Bacteroides fragilis induces the stemness in colorectal cancer via upregulating histone demethylase JMJD2B. Gut Microbes. 2020;12(1):1–19. doi: 10.1080/19490976.2020.1788900. 10.1080/19490976.2020.1788900/SUPPL_FILE/KGMI_A_1788900_SM0276.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patterson L., et al. Glucosylceramide production maintains colon integrity in response to Bacteroides fragilis toxin-induced colon epithelial cell signaling. FASEB J. 2020;34:15922–15945. doi: 10.1096/fj.202001669R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boleij A., et al. G-protein coupled receptor 35 (GPR35) regulates the colonic epithelial cell response to enterotoxigenic Bacteroides fragilis. Commun Biol. 2021;4:1–14. doi: 10.1038/s42003-021-02014-3. 2021 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rabizadeh S., et al. Enterotoxigenic Bacteroides fragilis: a potential instigator of colitis. Inflamm Bowel Dis. 2007;13:1475–1483. doi: 10.1002/ibd.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shields D.S., et al. Bacterial-driven inflammation and mutant braf expression combine to promote murine colon tumorigenesis that is sensitive to immune checkpoint therapy. Cancer Discov. 2021;11:1792–1807. doi: 10.1158/2159-8290.CD-20-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rhee K.J., et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect Immun. 2009;77:1708–1718. doi: 10.1128/IAI.00814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu S., et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009 doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goodwin A.C., et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc Natl Acad Sci U S A. 2011;108:15354–15359. doi: 10.1073/pnas.1010203108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wick, E.C. et al. Stat3 activation in murine colitis induced by enterotoxigenic Bacteroides fragilis. (2014) doi: 10.1097/MIB.0000000000000019. [DOI] [PMC free article] [PubMed]
- 67.Geis A.L., et al. Regulatory T-cell response to enterotoxigenic Bacteroides fragilis colonization triggers IL17-dependent colon carcinogenesis. Cancer Discov. 2015;5:1098–1109. doi: 10.1158/2159-8290.CD-15-0447. 1099 Cancer Discov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Destefano Shields C.E., et al. Reduction of murine colon tumorigenesis driven by enterotoxigenic Bacteroides fragilis using cefoxitin treatment. J Infect Dis. 2016;214:122–129. doi: 10.1093/infdis/jiw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee K.S., et al. Modulation of airway remodeling and airway inflammation by peroxisome proliferator-activated receptor gamma in a murine model of toluene diisocyanate-induced asthma. J Immunol. 2006;177:5248–5257. doi: 10.4049/jimmunol.177.8.5248. [DOI] [PubMed] [Google Scholar]
- 70.De Robertis M., et al. The AOM/DSS murine model for the study of colon carcinogenesis: from pathways to diagnosis and therapy studies. J Carcinog. 2011;10 doi: 10.4103/1477-3163.78279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moser A.R., Dove W.F., Roth K.A., Gordon J.I. The min (multiple intestinal Neoplasia) mutation: its effect on gut epithelial cell differentiation and interaction with a modifier system. J Cell Biol. 1992;116:1517–1526. doi: 10.1083/jcb.116.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Casterline, B.W., Hecht, A.L., Choi, V.M. & Bubeck Wardenburg, J. The Bacteroides fragilis pathogenicity island links virulence and strain competition. 10.1080/19490976.2017.1290758 1–10 (2017). [DOI] [PMC free article] [PubMed]
- 73.Dobson G.P., Letson H.L., Biros E., Morris J. Specific pathogen-free (SPF) animal status as a variable in biomedical research: have we come full circle? EBioMedicine. 2019;41:42–43. doi: 10.1016/j.ebiom.2019.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wlodarska M., et al. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect Immun. 2011;79:1536–1545. doi: 10.1128/IAI.01104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Duranti S., et al. Elucidating the gut microbiome of ulcerative colitis: bifidobacteria as novel microbial biomarkers. FEMS Microbiol Ecol. 2016;92 doi: 10.1093/femsec/fiw191. [DOI] [PubMed] [Google Scholar]
- 76.Schneditz G., et al. Enterotoxicity of a nonribosomal peptide causes antibiotic-associated colitis. Proc Natl Acad Sci U S A. 2014;111:13181–13187. doi: 10.1073/pnas.1403274111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Noor S.O., et al. Ulcerative colitis and irritable bowel patients exhibit distinct abnormalities of the gut microbiota. BMC Gastroenterol. 2010;10:1–9. doi: 10.1186/1471-230X-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pantosti A., Cerquetti M., Colangeli R., D'Ambrosio F. Detection of intestinal and extra-intestinal strains of enterotoxigenic Bacteroides fragilis by the HT-29 cytotoxicity assay. J Med Microbiol. 1994;41:191–196. doi: 10.1099/00222615-41-3-191. [DOI] [PubMed] [Google Scholar]
- 79.Donelli G., Fabbri A., Fiorentini C. Bacteroides fragilis enterotoxin induces cytoskeletal changes and surface blebbing in HT-29 cells. Infect Immun. 1996;64:113–119. doi: 10.1128/iai.64.1.113-119.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koshy S., Montrose M.H., Sears C.L. Human intestinal epithelial cells swell and demonstrate actin rearrangement in response to the metalloprotease toxin of Bacteroides fragilis. Infect Immun. 1996;64:5022–5028. doi: 10.1128/iai.64.12.5022-5028.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saidi R.F., Sears C.L. Bacteroides fragilis toxin rapidly intoxicates human intestinal epithelial cells (HT29/C1) in vitro. Infect Immun. 1996;64:5029–5034. doi: 10.1128/iai.64.12.5029-5034.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wells C.L., et al. Bacteroides fragilis enterotoxin modulates epithelial permeability and bacterial internalization by HT-29 enterocytes. Gastroenterology. 1996;110:1429–1437. doi: 10.1053/gast.1996.v110.pm8613048. [DOI] [PubMed] [Google Scholar]
- 83.Obiso R.J., Azghani A.O., Wilkins T.D. The Bacteroides fragilis toxin fragilysin disrupts the paracellular barrier of epithelial cells. Infect Immun. 1997;65:1431–1439. doi: 10.1128/iai.65.4.1431-1439.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chambers F.G., et al. Bacteroides fragilis toxin exhibits polar activity on monolayers of human intestinal epithelial cells (T84 cells) in vitro. Infect Immun. 1997;65:3561–3570. doi: 10.1128/iai.65.9.3561-3570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saidi R.F., Jaeger K., Montrose M.H., Wu S., Sears C.L. Bacteroides fragilis toxin rearranges the actin cytoskeleton of HT29/C1 cells without direct proteolysis of actin or decrease in F-actin content. Cell Motil. 1997;37:159–165. doi: 10.1002/(SICI)1097-0169(1997)37:2<159::AID-CM8>3.0.CO;2-3. Cytoskeleton. [DOI] [PubMed] [Google Scholar]
- 86.Sanfilippo L., Baldwin T.J., Menozzi M.G., Borriello S.P., Mahida Y.R. Heterogeneity in responses by primary adult human colonic epithelial cells to purified enterotoxin of Bacteroides fragilis. Gut. 1998;43:651–655. doi: 10.1136/gut.43.5.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu S., Lim K.C., Huang J., Saidi R.F., Sears C.L. Vol. 95. 1998. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Riegler M., et al. Bacteroides fragilis toxin 2 damages human colonic mucosa in vitro. Gut. 1999;44:504–510. doi: 10.1136/gut.44.4.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sanfilippo L., et al. Bacteroides fragilis enterotoxin induces the expression of IL-8 and transforming growth factor-beta (TGF-β) by human colonic epithelial cells. Clin Exp Immunol. 2001;119:456–463. doi: 10.1046/j.1365-2249.2000.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim J.M., Oh Y.K., Kim Y.J., Oh H.B. Polarized secretion of CXC chemokines by human intestinal epithelial cells in response to Bacteroides fragilis enterotoxin: NF-κB plays a major role in the regulation of IL-8 expression. Clin Exp Immunol. 2001;123:421–427. doi: 10.1046/j.1365-2249.2001.01462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim J.M., et al. Nuclear factor-kappa B activation pathway in intestinal epithelial cells is a major regulator of chemokine gene expression and neutrophil migration induced by Bacteroides fragilis enterotoxin. Clin Exp Immunol. 2002;130:59–66. doi: 10.1046/j.1365-2249.2002.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu, S., Morin, P.J., Maouyo, D. & Sears, C.L. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. (2003) doi: 10.1053/gast.2003.50047. [DOI] [PubMed]
- 93.Wu S., et al. Bacteroides fragilis enterotoxin induces intestinal epithelial cell secretion of interleukin-8 through mitogen-activated protein kinases and a tyrosine kinase-regulated nuclear factor-κB pathway. Infect Immun. 2004;72:5832–5839. doi: 10.1128/IAI.72.10.5832-5839.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sears C.L., Buckwold S.L., Shin J.W., Franco A.A. The C-terminal region of Bacteroides fragilis toxin is essential to its biological activity. Infect Immun. 2006;74:5595–5601. doi: 10.1128/IAI.00135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wu S., et al. The Bacteroides fragilis toxin binds to a specific intestinal epithelial cell receptor. Infect Immun. 2006;74:5382–5390. doi: 10.1128/IAI.00060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu S., Rhee K.J., Zhang M., Franco A., Sears C.L. Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and γ-secretase-dependent E-cadherin cleavage. J Cell Sci. 2007;120:1944–1952. doi: 10.1242/jcs.03455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim J.M., Lee J.Y., Kim Y.J. Inhibition of apoptosis in Bacteroides fragilis enterotoxin-stimulated intestinal epithelial cells through the induction of c-IAP-2. Eur J Immunol. 2008;38:2190–2199. doi: 10.1002/eji.200838191. [DOI] [PubMed] [Google Scholar]
- 98.Kim J.M., et al. 5,7-dihydroxy-3,4,6-trimethoxyflavone inhibits the inflammatory effects induced by Bacteroides fragilis enterotoxin via dissociating the complex of heat shock protein 90 and IκBα and IκB kinase-γ in intestinal epithelial cell culture. Clin Exp Immunol. 2009;155:541–551. doi: 10.1111/j.1365-2249.2008.03849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yoon Y.M., et al. Bacteroides fragilis enterotoxin induces human β-defensin-2 expression in intestinal epithelial cells via a mitogen-activated protein kinase/IκB kinase/NF-κB-dependent pathway. Infect Immun. 2010;78:2024–2033. doi: 10.1128/IAI.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Roh H.C., Yoo D.Y., Ko S.H., Kim Y.J., Kim J.M. Bacteroides fragilis enterotoxin upregulates intercellular adhesion molecule-1 in endothelial cells via an aldose reductase-, MAPK-, and NF-κB–dependent pathway, leading to monocyte adhesion to endothelial cells. J Immunol. 2011;187:1931–1941. doi: 10.4049/jimmunol.1101226. [DOI] [PubMed] [Google Scholar]
- 101.Hwang S., Gwon S.Y., Kim M.S., Lee S., Rhee K.J. Bacteroides fragilis toxin induces IL-8 secretion in HT29/C1 cells through disruption of E-cadherin junctions. Immune Netw. 2013;13:213–217. doi: 10.4110/in.2013.13.5.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yoo D.Y., et al. Bacteroides fragilis enterotoxin upregulates lipocalin-2 expression in intestinal epithelial cells. Lab Investig. 2013;93:384–396. doi: 10.1038/labinvest.2013.1. 2013 934. [DOI] [PubMed] [Google Scholar]
- 103.Remacle A.G., Shiryaev S.A., Strongin A.Y. Distinct interactions with cellular E-cadherin of the two virulent metalloproteinases encoded by a Bacteroides fragilis pathogenicity island. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kharlampieva, D. et al. Recombinant fragilysin isoforms cause E-cadherin cleavage of intact cells and do not cleave isolated E-cadherin. (2015) doi: 10.1016/j.micpath.2015.05.003. [DOI] [PubMed]
- 105.Ko S.H., et al. Bacteroides fragilis enterotoxin upregulates heme oxygenase-1 in intestinal epithelial cells via a mitogen-activated protein kinase- and NF-κB-dependent pathway, leading to modulation of apoptosis. Infect Immun. 2016;84:2541–2554. doi: 10.1128/IAI.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ko S.H., Jeon J.I., Myung H.S., Kim Y.J., Kim J.M. Bacteroides fragilis enterotoxin induces formation of autophagosomes in endothelial cells but interferes with fusion with lysosomes for complete autophagic flux through a mitogen-activated protein kinase-, AP-1-, and C/EBP homologous proteindependent pathway. Infect Immun. 2017;85:e00420–17. doi: 10.1128/IAI.00420-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jeon J.I., Ko S.H., Kim J.M. Intestinal epithelial cells exposed to bacteroides fragilis enterotoxin regulate NF-ĸB activation and inflammatory responses through β-catenin expression. Infect Immun. 2019;87:e00312–19. doi: 10.1128/IAI.00312-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jeon J.I., Choi J.H., Lee K.H., Kim J.M. Bacteroides fragilis enterotoxin induces sulfiredoxin-1 expression in intestinal epithelial cell lines through a mitogen-activated protein kinases- and Nrf2-dependent pathway, leading to the suppression of apoptosis. Int J Mol Sci. 2020;21:5383. doi: 10.3390/ijms21155383. 2020, Vol. 21, Page 5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Metz P., et al. Drug discovery and repurposing inhibits a major gut pathogen-derived oncogenic toxin. Front Cell Infect Microbiol. 2019;9:364. doi: 10.3389/fcimb.2019.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Allen J., Hao S., Sears C.L., Timp W. Epigenetic changes induced by Bacteroides fragilis toxin. Infect Immun. 2019;87:e00447–18. doi: 10.1128/IAI.00447-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ko S.H., Jeon J.I., Woo H.A., Kim J.M. Bacteroides fragilis enterotoxin upregulates heme oxygenase-1 in dendritic cells via reactive oxygen species-, mitogen-activated protein kinase-, and Nrf2-dependent pathway. World J Gastroenterol. 2020;26:291–306. doi: 10.3748/wjg.v26.i3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Becker H.E.F., et al. Higher prevalence of Bacteroides fragilis in Crohn's disease exacerbations and strain-dependent increase of epithelial resistance. Front Microbiol. 2021;12:1305. doi: 10.3389/fmicb.2021.598232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cao Y., et al. Enterotoxigenic Bacteroides fragilis promotes intestinal inflammation and malignancy by inhibiting exosome-packaged miR-149-3p. Gastroenterology. 2021;161:1552–1566. doi: 10.1053/j.gastro.2021.08.003. .e12. [DOI] [PubMed] [Google Scholar]
- 114.Xie X., et al. Recombinant Bacteroides fragilis enterotoxin-1 (rBFT-1) promotes proliferation of colorectal cancer via CCL3-related molecular pathways. Open Life Sci. 2021;16:408–418. doi: 10.1515/biol-2021-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Moncrief J.S., et al. The enterotoxin of Bacteroides fragilis is a metalloprotease. Infect Immun. 1995;63:175–181. doi: 10.1128/iai.63.1.175-181.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chung G.T., et al. Identification of a third metalloprotease toxin gene in extraintestinal isolates of Bacteroides fragilis. Infect Immun. 1999;67:4945–4949. doi: 10.1128/iai.67.9.4945-4949.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Weikel C.S., Grieco F.D., Reuben J., Myers L.L., Sack R.B. Human colonic epithelial cells, HT29/C1, treated with crude Bacteroides fragilis enterotoxin dramatically alter their morphology. Infect Immun. 1992;60:321–327. doi: 10.1128/iai.60.2.321-327.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Donelli G., Fabbri A., Fiorentini C. Bacteroides fragilis enterotoxin induces cytoskeletal changes and surface blebbing in HT-29 cells. Infect Immun. 1996;64:113–119. doi: 10.1128/iai.64.1.113-119.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pantosti A., Cerquetti M., Colangeli R., D'Ambrosio F. Detection of intestinal and extra-intestinal strains of enterotoxigenic Bacteroides fragilis by the HT-29 cytotoxicity assay. J Med Microbiol. 1994;41:191–196. doi: 10.1099/00222615-41-3-191. [DOI] [PubMed] [Google Scholar]
- 121.Mundy L.M., Sears C.L. Detection of toxin production by Bacteroides fragilis: assay development and screening of extraintestinal clinical isolates. Clin Infect Dis. 1996;23:269–276. doi: 10.1093/clinids/23.2.269. [DOI] [PubMed] [Google Scholar]
- 122.Hsiao E.Y., et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Scott A.J., et al. International cancer microbiome consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut. 2019;68:1624–1632. doi: 10.1136/gutjnl-2019-318556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.