Summary
Epichaperomes are disease-associated pathologic scaffolds composed of tightly bound chaperones and co-chaperones. They provide opportunities for precision medicine where aberrant protein-protein interaction networks, rather than a single protein, are detected and targeted. This protocol describes the synthesis and characterization of two 124I-labeled epichaperome probes, [124I]-PU-H71 and [124I]-PU-AD, both which have translated to clinical studies. It shows specific steps in the use of these reagents to image and quantify epichaperome-positivity in tumor bearing mice through positron emission tomography.
For complete details on the use and execution of this protocol, please refer to Bolaender et al. (2021), Inda et al. (2020), and Pillarsetty et al. (2019).
Subject areas: Cell culture, Cancer, Model Organisms, Molecular/Chemical Probes, Chemistry
Graphical abstract

Highlights
-
•
Synthesis and purification of radiolabeled epichaperome probes
-
•
Characterization and validation of the probes in vitro and in vivo
-
•
In vivo PET imaging with option for autoradiography
-
•
Potential uses in cancer and neurodegenerative diseases
Epichaperomes are disease-associated pathologic scaffolds composed of tightly bound chaperones and co-chaperones. They provide opportunities for precision medicine where aberrant protein-protein interaction networks, rather than a single protein, are detected and targeted. This protocol describes the synthesis and characterization of two 124I-labeled epichaperome probes, [124I]-PU-H71 and [124I]-PU-AD, both which have translated to clinical studies. It shows specific steps in the use of these reagents to image and quantify epichaperome-positivity in tumor bearing mice through positron emission tomography.
Before you begin
Institutional permissions
Experiments in this protocol require the handling of radioactive materials, cancer cells, and mice, and therefore need approval according to institutional guidelines. The preclinical experiments using radioactive nuclides were conducted under the non-human use license authorization granted by the Committee on Radiation and Radiation Safety Office of Memorial Sloan Kettering Cancer Center (MSKCC). This protocol also uses cancer cells. In order to use biologicals requiring biosafety level 2 (BSL2), the project was registered with the MSKCC’s Institutional Biosafety Committee (IBC), and the personnel was trained in proper handling and use of hazardous materials. Biohazardous and radioactive materials were handled and disposed of according to applicable state and federal regulations via guidance from both the Radiation Safety Committee, the Environmental Health and Safety Office and the IBC Committee. Experiments also require the use of mice. Ethical approvals are required prior to starting this procedure. Animal studies described in this protocol were performed under an approved Animal Protocol (protocol # 05-11-024) reviewed by the Institutional Animal Care and Use Committee, and conducted in accordance with NIH guidelines. Therefore, before starting experiments described in this protocol, make sure to acquire relevant permissions.
Cell culture for in vitro and in vivo experiments
Timing: 1–2 h
This section describes the steps used to culture, plate and prepare single cell suspensions of the human cancer cell line, MDA-MB-468.
Note: We established this protocol using the human breast cancer MDA-MB-468 cells. The use of another cell line is possible but needs adjustments and further troubleshooting for culture conditions and tumor induction in mice.
Note: Cells should be authenticated using short tandem repeat profiling and tested for mycoplasma before experiments (Dreolini et al., 2020).
Cell culture medium
Timing: 10 min
In a sterile environment, prepare the complete cell culture medium by mixing 445 mL of Dulbecco’s Modified Eagle’s Media (DMEM) with 50 mL of fetal bovine serum and 5 mL of the 100× penicillin-streptomycin solution, as described in materials and equipment.
Note: Culture MDA-MB-468 cancer cells in the above cell culture medium through this study. The cell culture medium can be stored at 4°C for several months without any problem. The cell culture medium should be warmed to 37°C in a water bath prior to cell experiments.
CRITICAL: All solutions employed for cell culture need to be prepared and maintained in sterile conditions by operating under a Class II Biological Safety Cabinet. Sterilize the Class II Biological Safety Cabinet with 70% ethanol before and after using it. All items placed in the Class II Biological safety cabinet must be sterilized with 70% ethanol.
Pause point: Store at 4°C. The cell culture medium can be stored at this temperature for several months without any problem.
Cell culture
Timing: 1–3 days
The human breast cancer MDA-MB-468 (HTB-132; RRID:CVCL_0419) cell line needs to be cultured according to its growth requirements, in complete DMEM cell culture medium.
Note: Perform the cell culture in sterile conditions by operating under a Class II Biological Safety Cabinet.
-
1.
Thaw the frozen vial of MDA-MB-468 purchased from American Type Culture Collection (ATCC) (which was stored in liquid nitrogen) and seed the cells in a 175 cm2 cell culture flask containing 20 mL of cell culture medium.
Note: Check cells for mycoplasma contamination before seeding.
-
2.
Keep the flask in an incubator for 1–3 days at 37°C until cells reach approximately 70–80% confluency.
Note: Maintain physiological O2 concentrations inside the incubator (3% O2, 5% CO2, and 92% N2).
-
3.
For subculturing the cells, remove the cell culture medium from the flask and wash the cells once with 5 mL prewarmed phosphate buffered saline (PBS) to remove all traces of serum that contains trypsin inhibitors.
-
4.
Add 3 mL of prewarmed trypsin-EDTA (0.25%) to the cells and keep the flask in the incubator at 37°C and wait for cells to detach. Generally, it takes 2–3 min to completely detach the cells by trypsin-EDTA (0.25%).
-
5.
Neutralize trypsin-EDTA by adding 5 mL of prewarmed complete culture medium and gently pipette 10–15 times to resuspend the cells and then transfer them to a 15 mL conical tube.
-
6.
Centrifuge the cells at 300 × g for 10 min at 4°C.
-
7.
Carefully remove the cell culture medium from the conical tube and resuspend the cells in fresh culture medium.
Note: The volume of the culture medium will depend on the confluency of the cells, i.e., 5 mL of fresh culture medium for cells at 70% confluency.
-
8.
Count the number of cells in the cell suspension using a cell counter. Use the trypan blue dye exclusion-based method (Strober, 2015) to count viable cells and to calculate the number of cells needed to seed in 6-well plates in a final volume of 2 mL per well. Typically, 0.5 × 106 of MDA-MB-468 cells per well are optimal to get the cells at 70–80% confluency after 24 h.
Note: Culture medium, trypsin, and buffers for the cell culture should be warmed up at 37°C for 30 min before you start.
Note: This protocol uses the commercially available reagents as specified in the key resources table but alternative suppliers or in-house made buffers should work as well.
Note: From one flask we usually obtain 15 million cells which is sufficient to establish approximately 1–2 tumors.
Plating cells
Timing: 30 min
This section describes the steps for plating cells for experiments that test probe’s specificity (see further).
-
9.
Seed the cells into 6-well culture plates at a density of 3 × 106 cells per plate. Add complete culture medium to a final volume of 2 mL/well.
Note: Some cell lines may require higher or lower cell numbers depending on the experimental conditions.
CRITICAL: Do not seed the cells at a density of more than 3 × 106 per plate to avoid over confluent cell cultures during experiment.
CRITICAL: To minimize variation and ensure even distribution between wells, first add the calculated number of required cells to a single 50 mL Falcon tube, and then divide them into the respective wells.
-
10.
Keep the plate in an incubator at 37°C. Approximately 70% confluency will be achieved after 24 h.
Establishment of xenografted tumors in athymic nude mice
Timing: 2–3 weeks
This section describes the steps for preparation and injection of human cancer cells into athymic nude mice (6- to 8-week-old female nude mice).
Note: All animal experiments need to adhere to the Institutional Animal Care and Use Committee (IACUC) guidelines and be performed under an approved protocol. Mice should be acclimatized in the vivarium at least for one week prior to tumor establishment. Mice should be housed in groups of 4–5 mice per individually ventilated cage in a 12 h light/dark cycle (6:00 a.m./6:00 p.m.), with controlled room temperature (22 ± 1°C) and humidity (30–70%). Mice should be provided with food and water ad libitum.
Note: We established this protocol using the human breast cancer MDA-MB-468 cells. The use of another cell line is possible but needs adjustments and further troubleshooting for tumor induction in mice.
Steps 3–8 describes the preparation of cell suspension for xenograft establishment. Use the trypan blue dye exclusion-based method to count the viable cells and to calculate the number of cells needed to be injected in each mouse. Typically, we use 1 × 107 cells per mouse for the MDA-MB-468 cell line. Calculate the total number of cells you need by multiplying 1 × 107 cells with the number of mice to be implanted.
Note: This number varies depending on the cell line and should be predetermined.
-
11.
Take the desired number of cells, as calculated above, into a new falcon tube and centrifuge the tube at 4°C for 10 min at 300 × g. Carefully aspirate the cell media. Resuspend the cells in a 1:1 v/v mixture of PBS and Matrigel.
Note: The total number of cells needed for all the mice to be implanted, rather than for individual mice, should be mixed with PBS/Matrigel in a single tube, and then injected. This will minimize variations in injected cells, and in turn in tumor volume, from mouse to mouse.
Note: The Matrigel matrix contains the solubilized extracellular basement membrane proteins that allow cells to remain localized at the injection site. It is always better to prepare a few extra doses due to sample loss in the needle hub.
CRITICAL: In order to reduce cell loss and assure convenient operation, the cell/Matrigel/PBS mix and all items in contact with the mixture (tube, syringe, pipette tip) need to be kept on ice. Matrigel polymerizes above 10°C.
-
12.
Ensure to thoroughly mix the cell/Matrigel/PBS by manually flipping the tube a few times.
-
13.
Fill the syringe with cell/Matrigel/PBS mixture when the mice are ready to be injected.
CRITICAL: To avoid cells settling to the bottom of the tube, and ensure even distribution of cells for each injection, make sure to thoroughly mix the cold cell/Matrigel/PBS mixture immediately before injections.
-
14.
Hold a mouse with one hand and subcutaneously inject 200 μL of cell/Matrigel/PBS mix into the forelimbs of each mouse using a 28G syringe.
Note: If inexperienced with injecting the mice while awake, anesthetize the mice prior to injection. We use isoflurane for this purpose.
CRITICAL: Laboratory mice look identical and require marking by either ear-punching or by marking the tail with permanent marker. Ear-punching is permanent but could be painful and mice could become stressed by weekly tail-marking. For this experiment, each mouse was tail-marked using a non-toxic, permanent marker before cell injection or immediately afterwards to ensure identification.
-
15.
Measure the width and length of each tumor biweekly using digital calipers and calculate the tumor volume using the following formula: volume (mm3) = (width2 × length)/2. When tumor volume of approximately 250–400 mm3 is reached, animals are randomized into study groups.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| BD MatrigelTM | Fisher Scientific | Cat#CB-40234A |
| PU-H71 | He et al. (2006) | N/A |
| PU-HZ151 (also called PU-AD) | Bolaender et al. (2021) | N/A |
| Tris(dibenzylideneacetone)dipalladium(0) [Pd2(dba)3] | Sigma-Aldrich | Cat#328774-1G |
| Hexamethylditin | Sigma-Aldrich | Cat#288020-5G |
| Toluene (anhydrous, 99.8%) | Sigma-Aldrich | Cat#244511-1L |
| Methylene chloride | Fisher Scientific | Cat#D151-4 |
| Ethyl acetate | Fisher Scientific | Cat#E195-4 |
| Hexanes | Fisher Scientific | Cat#H303-4 |
| Ammonia solution (7 N in methanol) | Sigma-Aldrich | Cat#499145-1L |
| Acetic acid | Fisher Scientific | Cat#AC423225000 |
| Chloroform-D (CDCl3) | Fisher Scientific | Cat#NC9754641 |
| Methanol-D4 (CD3OD) | Fisher Scientific | Cat#NC9858470 |
| Ethanol, Absolute, USP | Sigma-Aldrich | Cat#459844-500ML |
| 124I-Sodium Iodide, ≥97% | 3D Imaging | N/A |
| 131I-Sodium Iodide, ≥97% | Nuclear Diagnostic Products | N/A |
| Sodium hydroxide | Fisher Scientific | Cat#S318-500 |
| Chloramine-T | Sigma-Aldrich | Cat#402869-100G |
| Acetonitrile | Fisher Scientific | Cat#A996-4 |
| Trifluoroacetic acid (TFA) | Sigma-Aldrich | Cat#302031-100ML |
| Dimethyl Sulfoxide (DMSO) | Fisher Scientific | Cat#BP231-100 |
| 0.9% Sodium Chloride Injection, USP | Fisher Scientific | Cat#NC9054335 |
| Forane (Isoflurane, USP) | Baxter Healthcare, Deerfield, IL | NDC 10019-360-60 |
| Sterile Water for Injection, USP | N/A | N/A |
| Deionized Water 18.2 MΩ.cm | N/A | N/A |
| Nitrogen gas | N/A | N/A |
| 0.2 μm nylon syringe filter | Fisher Scientific | Cat#03-050-473 |
| 28G syringe | Fisher Scientific | Cat#14-826-79 |
| Thin layer chromatography plates (for preparatory TLC) | Fisher Scientific | Cat#50-465-365 |
| Thin layer chromatography plates (for TLC) | Fisher Scientific | Cat#M1057150001 |
| Silicone oil | Fisher Scientific | Cat#S159-500 |
| ddH2O | N/A | N/A |
| Gibco™ DMEM (supplemented with L-Glutamine) | Fisher Scientific | Cat#11-965-084 |
| Gibco™ Fetal Bovine Serum | Fisher Scientific | Cat#10-082-147 |
| CorningTM Penicillin-Streptomycin Solution (100×) | Fisher Scientific | Cat#MT30002CI |
| Gibco™ PBS, pH 7.4 | Fisher Scientific | Cat#10-010-023 |
| Gibco™ Trypsin-EDTA (0.25%) | Fisher Scientific | Cat#25-200-056 |
| Sep-Pak C18 Light Cartridge | Fisher Scientific | Cat#50-785-175 |
| pH paper (1–12) | Fisher Scientific | Cat#NC9841376 |
| C-18 analytical column | Phenomenex - Luna | Part# 00G-4041-E0 |
| Experimental models: Cell lines | ||
| Human: MDA-MB-468 | ATCC | Cat#HTB-132; RRID:CVCL_0419 |
| Experimental models: Organisms/strains | ||
| Mouse: Hsd:Athymic Nude-Foxn1nu, 6–8 weeks old females | Envigo | Cat#069; RRID:MGI:5652489 |
| Software and algorithms | ||
| PET Acquisition – MicroPET Manager | Siemens Medical Solutions, Inc | Version 2.4 |
| PET Image Analysis – ASIProTM | Siemens Medical Solutions, Inc | N/A |
| Other | ||
| Digital Caliper 6″ | Fisher Scientific | Cat#14-648-17 |
| Genevac centrifugal evaporator | SP Scientific | Model# EZ-2 Mk2 |
| Hotplate stirrer | Fisher Scientific | Cat#NC0649530 |
| Magnetic stirring bar | Fisher Scientific | Cat#22-127100 |
| Rotary evaporator | Büchi | Mfr#23111V000 |
| Water bath | Fisher Scientific | Cat#15-462-20Q |
| Class II Biological Safety Cabinet | NuAire | Cat#NU-407-400 |
| CO2 incubator | NuAire | Cat#NU-8700 |
| Centrifuge | Eppendorf | Product #EP028622180 |
| Cell counter | Beckman Coulter | Product#731196 |
| Mini centrifuge | Fisher Scientific | Cat#05-090-100 |
| 10 mL round-bottom flask | Fisher Scientific | Cat#CG150682 |
| 5 mL round-bottom flask | Fisher Scientific | Cat#CG150680 |
| Rubber balloon | N/A | N/A |
| Rubber septum | Fisher Scientific | Cat#K774261-0014 |
| Oil bath | Fisher Scientific | Cat#08-762-1 |
| Aluminum foil | Fisher Scientific | Cat#01-213-101 |
| 1.5 mL Eppendorf tube | Fisher Scientific | Cat#14-666-319 |
| 1.5 mL brown Eppendorf tube | USA Scientific | Cat#1415-2507 |
| 7 mL glass vial | Fisher Scientific | Cat#03-337-26 |
| 2–20 μL pipette | Fisher Scientific | Cat#14-381-040 |
| 20–200 μL pipette | Fisher Scientific | Cat#14-381-037 |
| 5 mL Serological Pipettes | Fisher Scientific | Cat#13-678-11D |
| 10 mL Serological Pipettes | Fisher Scientific | Cat#13-678-11E |
| Falcon® 15 mL Conical Centrifuge Tubes | Fisher Scientific | Cat#14-959-49B |
| Falcon® 50 mL Conical Centrifuge Tubes | Fisher Scientific | Cat#14-432-22 |
| 175 cm2 cell culture flask | Fisher Scientific | Cat#10-126-13 |
| Polystyrene Petri Dish 6-Well Cell Culture Plate | Fisher Scientific | Cat#08-772-1B |
| 100 mm Cell Culture Dish | Fisher Scientific | Cat#08-772-6 |
| LC-MS system | Waters | Equipped with ELS detector (Model# 2420), Photodiode Array detector (Model# 2998), Sample Manager (Model# 2767) and XBridge C18 5 μm 4.6 × 150 mm Column (Part# 186003116) |
| NMR Spectrometer | Bruker | Avance III UltraShield Plus 500 |
| Dose calibrator | Capintec | CRC-15R |
| Scintillation Counter | PerkinElmer | Wallac 1480 Wizard 3 Automatic Gamma counter |
| Small-animal PET scanner | Siemens Medical Solutions | Focus 120 microPET |
| Bioluminescence Scanner | Xenogen | IVIS-200 |
| LC-MS/MS system | Agilent Technologies | Agilent 6410 Triple Quadrupole LC/MS |
| Shimadzu HPLC system | Shimadzu Scientific Instruments | Equipped with UV detector (SPD-20A), Pump (LC-20AB) and Column (Phenomenex Gemini NX C18, 4.6 × 250 mm, 5 μm) |
| Bioscan Radioactivity detector for HPLC | Eckert & Ziegler | FlowCount |
| Shielded isolator and hot cell | Comecer | FHR1 50 |
Materials and equipment
Complete DMEM cell culture medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Fetal Bovine Serum | 10 % | 50 mL |
| Penicillin Streptomycin Solution (100×) | 100 U/mL (Penicillin), 100 mg/mL (Streptomycin |
5 mL |
| DMEM (High glucose) | N/A | 445 mL |
| Total | N/A | 500 mL |
Note: To prepare 500 mL of complete DMEM cell culture media, add 50 mL of Fetal Bovine Serum and 5 mL of the Penicillin Streptomycin Solution to 445 mL of DMEM. Complete culture media can be stored at 4°C for 1–3 months.
Mobile phase for chromatographic purification and characterization (step 5)
| Reagents | Final concentration | Amount |
|---|---|---|
| Methylene chloride | 36.36 % | 40 mL |
| Ethyl acetate | 18.18 % | 20 mL |
| Hexanes | 36.36 % | 40 mL |
| Ammonia solution (7N in methanol) | 9.09 % | 10 mL |
| Total | N/A | 110 mL |
Note: For use in the purification of the trimethylstannyl precursor using preparatory thin layer chromatography, in step 5. Prepare fresh by mixing 40 mL of methylene chloride, 20 mL of ethyl acetate, 40 mL of hexanes and 10 mL of ammonia solution before use. It can be stored for one week at 24 ± 2°C.
Mobile phase for chromatographic purification and characterization (step 21)
| Reagents | Final concentration | Amount |
|---|---|---|
| Acetonitrile | 28 % | 280 mL |
| Water | 72 % | 720 mL |
| TFA | 0.1 % | 1 mL |
| Total | N/A | approx. 1 L |
Note: For use in the characterization of [124I]-PU-H71 using HPLC in step 21. Prepare fresh by mixing 280 mL of acetonitrile, 720 mL of water and 1 mL of TFA. It can be stored for one week at 24 ± 2°C.
Step-by-step method details
Preparation of precursor molecules required for the introduction of the 124I radiolabel
Two epichaperome agents, PU-H71 and PU-AD (also called PU-HZ151) were discovered and translated to clinic as therapeutics for cancer and neurodegenerative diseases (Bolaender et al., 2021; Inda et al., 2020; Jhaveri et al., 2020; Pillarsetty et al., 2019; Rodina et al., 2016; Sugita et al., 2021). Because the agents kinetically select for the epichaperomes over abundant chaperone pools, labeled derivatives of PU-H71 and PU-AD, can be used as epichaperome detection reagents (Bolaender et al., 2021). A favorable feature of these two epichaperome agents is the endogenous presence of iodine on their chemical structure. This naturally occurring stable isotope iodine-127 (127I) can be replaced with the positron emitter iodine-124 (124I) to provide a probe for use in epichaperome detection by positron emission tomography (PET) imaging or with iodine-131 (131I) for detection by autoradiography. These probes could be used to detect, track and quantify epichaperomes, and in turn PPI network dysfunction, at both cellular and organismal level, in either tissues or in live patients (Bolaender et al., 2021; Inda et al., 2020; Jhaveri et al., 2020; Pillarsetty et al., 2019).
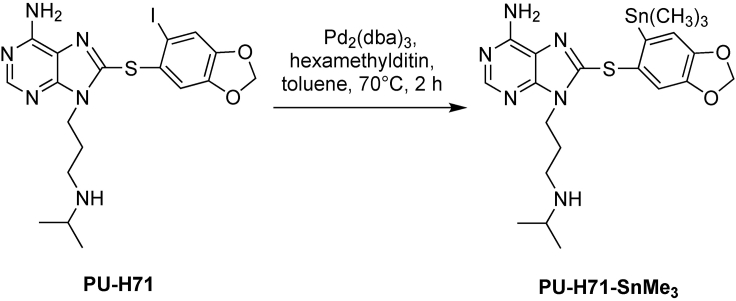
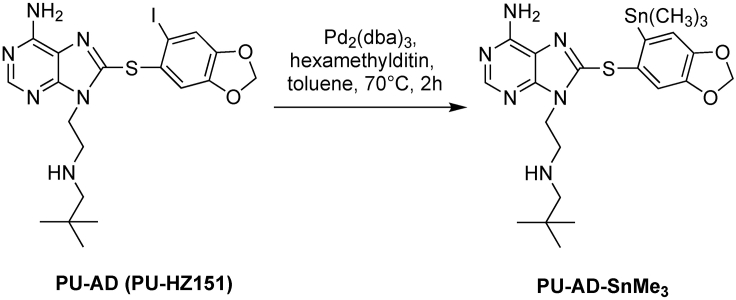
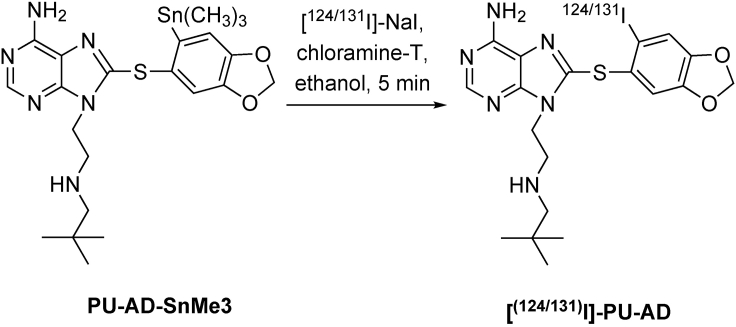
A two-step synthesis was designed for radiolabeling of the epichaperome agents PU-AD and PU-H71. It entails converting the original molecule into the tin precursor (PU-H71-SnMe3 for PU-H71, see Figure 1 and Table 1 and PU-AD-SnMe3 for PU-AD, see Figure 2 and Table 2), which is then radiolabeled using [124I]-NaI or [131I]-NaI (see Figures 3 and 4 and Tables 3 and 4), to yield the epichaperome detection probes.
Figure 1.
Synthetic scheme of PU-H71-SnMe3
Table 1.
Reagents for synthesis of PU-H71-SnMe3
| Reagent | Formula weight | Equivalent | Amount |
|---|---|---|---|
| PU-H71 | 512.37 | 1 | 25 mg (0.05 mmol) |
| Tris(dibenzylideneacetone)dipalladium(0) | 915.72 | 0.1 | 4.5 mg (0.005 mmol) |
| Hexamethylditin | 327.63 | 4 | 42 μL (0.2 mmol) |
Figure 2.
Synthetic scheme of PU-AD-SnMe3
Table 2.
Reagents for synthesis of PU-AD-SnMe3
| Reagent | Formula weight | Equivalent | Amount |
|---|---|---|---|
| PU-AD (PU-HZ151) | 526.39 | 1 | 10 mg (0.02 mmol) |
| Tris(dibenzylideneacetone)dipalladium(0) | 915.72 | 0.1 | 1.75 mg (0.002 mmol) |
| Hexamethylditin | 327.63 | 4 | 17 μL (0.08 mmol) |
Figure 3.
Synthetic scheme of [124/131I]-PU-H71
Figure 4.
Synthetic scheme of [124/131I]-PU-AD
Table 3.
Reagents for synthesis of [124/131I]-PU-H71
| Reagent | Formula weight | Equivalent | Amount |
|---|---|---|---|
| PU-H71-SnMe3 | 549.28 | 1 | 25 μg (0.046 μmol) |
| [124/131I]-NaI in 0.1 N sodium hydroxide | – | – | 100 μL |
| Chloramine-T solution (2 mg/mL in acetic acid) | 227.64 | 1 | 5 μL [10 μg] (0.045 μmol) |
Table 4.
Reagents for synthesis of [124/131I]-PU-AD
| Reagent | Formula weight | Equivalent | Amount |
|---|---|---|---|
| PU-AD-SnMe3 | 563.31 | 1 | 20 μg (0.035 μmol) |
| [124/131I]-NaI in 0.1 N sodium hydroxide | – | – | 200 μL |
| Chloramine-T solution (2 mg/mL in acetic acid) | 227.64 | 1.3 | 5 μL [10 μg] (0.045 μmol) |
The steps below describe the first part of the protocol, which is to prepare, purify and characterize the tin precursor molecules.
Note: All reagents were purchased from commercial suppliers and used without further purification. PU-H71 and PU-AD, which we use below to prepare the tin precursors, were prepared as previously reported (Bolaender et al., 2021; He et al., 2006). Alternatively, both chemicals are available and can be purchased from several commercial suppliers.
Synthesis of 9-(3-(isopropylamino)propyl)-8-((6-(trimethylstannyl)benzo[d][1,3]dioxol-5-yl)thio)-9H-purin-6-amine (PU-H71-SnMe3) and of 9-(2-(Neopentylamino)ethyl)-8-((6-(trimethylstannyl)benzo[d][1,3]dioxol-5-yl)thio)-9H-purin-6-amine (PU-AD-SnMe3)
Timing: 2 h for each precursor
These steps describe how to prepare PU-H71-SnMe3 (See Figure 1) and PU-AD-SnMe3 (See Figure 2). Please refer to Tables 1 and 2 for the amounts of individual reagents to be added at each of the steps.
-
1.
Weigh PU-H71 (or PU-AD) and Tris(dibenzylideneacetone)dipalladium(0) [Pd2(dba)3] in a 10 mL round-bottom flask containing a magnetic stirring bar. Troubleshooting 1.
-
2.
Stopper the flask with a rubber septum. Insert through the septum a syringe attached to a nitrogen-filled balloon to replace the air in the reaction vessel with an inert atmosphere.
CRITICAL: It is important to carry out these reactions in the dark (eg. wrap the reaction vessel in aluminum foil) under an inert atmosphere and using dry solvents to obtain the maximum yield.
-
3.
Mix the hexamethylditin in 3 mL of dry toluene and add this mixture through the septum using a syringe.
-
4.
Place the reaction flask in an oil bath containing silicone oil kept on a hotplate stirrer and stir the reaction solution at 500 rpm for 2 h at 70°C.
-
5.Concentrate the reaction mixture using a rotary evaporator and purify the resulting residue by preparatory thin layer chromatography (PTLC) as mentioned below:
-
a.Dissolve the residue in 0.5 mL of CH2Cl2:MeOH (1:1) and load it onto the PTLC as a uniform band.
-
b.Develop the PTLC using a solvent mixture (mobile phase) composed of CH2Cl2:EtOAc:Hexane:NH3/MeOH (7N) at 2:1:2:0.5 (see materials and equipment for mobile phase recipe).
-
c.Scrap off the silica band containing the desired product (Rf∼0.45). The deiodinated byproduct band is observed at Rf∼0.33.
-
d.Dissolve the desired product in 50 mL of CH2Cl2:MeOH (10:1) and filter this suspension using a sintered glass funnel. Dry the filtrate under vacuum to afford the tin precursor molecules. Troubleshooting 2.
-
a.
-
6.
Characterize each product by NMR spectroscopy (1H NMR and 13C NMR) and mass spectrometry (MS) (see the expected outcomes section).
Note: PU-H71 and PU-AD contain a secondary alkyl amine which could theoretically interfere during the synthesis of the corresponding trimethylstannyl derivative using palladium (0) catalyst resulting in unwanted side reactions. Former synthetic protocols, employing tetrakis(triphenylphosphine)Pd(0) as a catalyst, used Boc to protect the free amino group (Taldone et al., 2016). In the current protocol we use tris(dibenzylideneacetone)dipalladium(0) as catalyst and have confirmed using LC-MS that the secondary alkyl amine is tolerant to this synthetic protocol and does not need Boc protection.
Note: Using 25 mg of PU-H71 and 10 mg of PU-AD as the starting materials, yields approximately 19 mg of PU-H71-SnMe3 and 6.8 mg PU-AD-SnMe3, respectively (∼70% and 64% yield, respectively). The synthesis has been repeated at least three times, and the yield of the reaction has been stable. A 2.5 mg stock of PU-H71-SnMe3 (or 2 mg PU-AD-SnMe3) is sufficient for 100 labeling reactions. This amount of product can be aliquoted for individual reactions after dissolving it first in 1 mL of methylene chloride:methanol 1:1 v/v and then transferring 10 μL aliquots into brown Eppendorf tubes. Upon drying under vacuum in the Genevac for 2–3 h, the vials are then stored at −20°C. The remaining product can be stored at −20°C as solid powder in an aluminum wrapped glass vial.
Note: It is recommended to check precursor stability every 6 months by reconstituting the content of 1 vial in 20 μL of methylene chloride:methanol 1:1 v/v and applying the mix onto a LC-MS instrument.
Pause point: The products can be stored at −20°C for at least a year.
Synthesis of the radiolabeled probes
Timing: 30 min
The steps below describe the second part of the protocol, which is to prepare, purify and characterize the radiolabeled epichaperome probes, specifically [124I]-PU-H71 (See Figure 3 and Table 3) or [131I]-PU-H71. Similar steps can be used to prepare [124/131I]-PU-AD (See Figure 4 and Table 4).
Note: The following steps use and yield radioactive compounds, and therefore, require special laboratories and permission to handle radioactivity.
CRITICAL: Radioactive iodine isotopes (iodine-124, iodine-131) that we use for radiolabeling emit high energy gamma rays and are considered potent radiation sources. Appropriate training of personnel and the use of proper facilities are required to handle radioactive substances. Regular monitoring of workers exposure, including thyroid monitoring, is necessary.
CRITICAL: This protocol requires the handling of radioactive materials and therefore needs radiation safety and appropriate biosafety approvals according to institutional guidelines.
-
7.
Pipette 20 μL of ethanol into the brown Eppendorf tube containing 25 μg of PU-H71-SnMe3 precursor.
-
8.
Pipette ≤100 μL of 124I-Sodium Iodide solution (0.5–20 mCi) into the vial.
-
9.
Pipette 5 μL of chloramine-T solution (2 mg/mL in acetic acid) into the vial and allow the reaction mixture to incubate for 5 min at 24 ± 2°C.
Note: After the addition of the above reagents, it is recommended to centrifuge the samples in a minifuge (approximately 12 × g) for 15 s. This is to ensure that all the reagents are at the bottom of the Eppendorf, and thus able to react. This is to minimize deposition of the reagents on the sides of the Eppendorf which can lead to low/no yields.
Note: The reaction should be shielded from light. Performing the reaction in the brown Eppendorf tube is typically sufficient for this purpose.
-
10.
Assay the amount of activity in the reaction vial by placing in the dose calibrator chamber. Select iodine-124 as the radionuclide in the isotope selector. Troubleshooting 3.
Note: The dose measurement is sensitive to placement of the reaction vial within the chamber. Please ensure that the measurement vial is suspended from the plastic dipper to be placed near the center of the chamber. Capintec recommends using a calibration setting number of 570 for 124I on the CRC-15R dose calibrator.
-
11.
Pre-condition the C18 cartridge with 10 mL of ethanol followed by 10 mL water. Troubleshooting 4.
-
12.
Draw 10 mL of water into a syringe. Eject 1 mL of the water from the syringe into the reaction mixture, then draw the water/reaction mixture back into the same syringe.
-
13.
Load the reaction mixture onto the preconditioned C18 cartridge and elute the leftover radioactive iodine with water. Discard the water fraction as waste.
Note: The water fraction will contain some amount of radioactivity. Ensure that the water fraction is stored as radioactive waste and can be disposed as non-radioactive waste only after complete decay.
-
14.
Rinse the C18 cartridge with an additional 10 mL of water and discard the eluted water as waste.
Note: See note above.
-
15.
Use ≤3 mL of ethanol to elute the product from the C18 Sep-Pak into individual Eppendorf tubes. Record the eluted activity and time.
Note: To minimize ethanol content, it is recommended that users elute the compound in 0.3 mL fractions and combine fractions with highest activities.
-
16.
Working at a temperature ≤45°C, use inert gas flow to reduce the ethanol volume to approximately 0.1–0.3 mL. Record the reduced ethanol volume.
Note: The inert gas flow should be highly regulated at a flow rate of <100 mL/min to minimize creating vortexes that can lead to spilling of the evaporating liquid to the sides or out of the vial leading to loss of radioactivity. Complete drying of solvent is not recommended because redissolving will require high volumes.
-
17.
Formulate the reaction mixture in 1–10 mL of saline and pass through a 0.2 μm sterile filter into the final product vial.
-
18.
Record the final product information.
-
19.
Remove 0.05 mL of the final product using sterile and pyrogen free pipettes or tuberculin syringe for QC test (see below steps 20–24).
Quality control of the radiolabeled epichaperome probes
Timing: 60 min
The steps below describe the method employed for the analysis and characterization of [124I]-PU-H71. Similar steps can be performed for [124I]-PU-AD or the I-131 labeled probes.
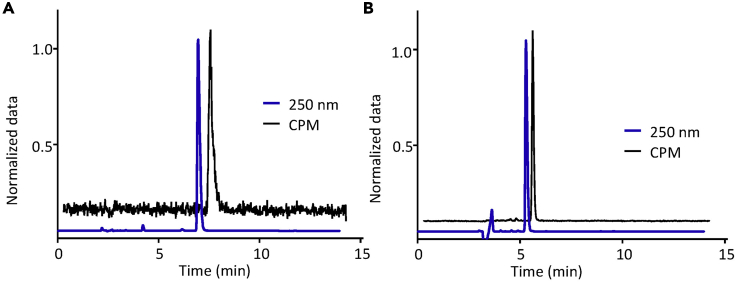
Because the mass of radiopharmaceutical being synthesized is in the sub nanogram range, traditional analytical methods such as NMR or mass spectrometry cannot be used. Therefore, the most common way to ascertain the identity of the radiolabeled probe is using HPLC wherein the retention time of the radioactive sample is matched with the retention time of non-radioactive (cold) standard (Figure 5). The radioactive HPLC chromatogram can be used to evaluate the % purity of the sample by integrating the area under the curve of desired compound and of observed impurities. If the compound is administered to animals or humans, it needs to be formulated in a saline and/or buffered solution with minimal amounts of excipients. Ethanol is commonly used as an excipient to increase solubility, but its concentration should be lower than <5% v/v in saline or buffer. Usually, gas chromatography is employed to determine residual solvent content. The injectable should be clear and particle free upon visual inspection (Table 5).
-
20.
Prepare the blank solution by adding 0.6 mL of ethanol to a 10 mL vial of saline.
-
21.PU-H71 analytical run is done using isocratic condition (28% acetonitrile/ 72% water with added 0.1% TFA, see materials and equipment for mobile phase recipe) and running the samples in the following sequence:
-
a.Blank, 20 μL.
-
b.Reference Standard Solution, 20 μL.
-
c.Blank, 20 μL.
-
d.[124I]-PU-H71, 20 μL.
-
a.
-
22.
The radiochemical identity of the compound is confirmed by matching its retention time with that of the cold standard.
Note: Instead of sequential runs, both cold standard and radioactive standard can be co-injected to confirm the identical retention time.
-
23.
The radiochemical purity of the radioactive product is calculated by integrating the areas under the curve (in the radioactivity channel) of radioactive compound and the rest of the peaks.
-
24.
Take 2 μL of the final formulation and drop it onto a pH paper to measure the pH. The pH should be between 5.5 and 8.0 for the radiopharmaceutical.
Note: Ideally the pH should be near 7.4, but depending on the nature of the compound, the injectate can be buffered within a pre-defined acceptable range to improve solubility. This also minimizes residualization of radioactivity on the syringe used for injections.
Figure 5.
Quality control analysis of radiolabeled probes using HPLC
(A) HPLC chromatogram of cold PU-AD (UV channel @ 250 nm, blue line) and [124I]-PU-AD (radioactive counts channel, black line) from two separate runs. The UV absorbance data and count data has been normalized and radioactive chromatogram is offset (on x axis) to aid clear visualization. CPM, counts per minute.
(B) Same as (A) for PU-H71 and [124I]-PU-H71.
Table 5.
QC pre-release test results for [124I]-PU-H71
| Test | Acceptance criteria |
|---|---|
| pH | 5.5–8.0 |
| Visual Inspection | Clear and particle free |
| Radiochemical Purity | ≥95% |
| Radiochemical Identity | Matches the retention time of the non-radioactive standard |
In vitro blocking experiment to validate the specificity of the radiolabeled probe
Timing: 2–3 h
This section describes the steps performed to validate the specific binding of the [124I]-PU-AD probe. Similar steps can be used for the other radiolabeled epichaperome probes.
Note: This experiment uses radioactive compounds, and therefore, requires special facilities and the permission to handle radioactivity.
CRITICAL: Radioactive iodine isotopes (iodine-124) emit high energy gamma rays and are considered potent radiation sources. Appropriate training of personnel and the use of proper facilities are required to handle radioactive substances. Regular monitoring of workers exposure, including thyroid monitoring, is necessary.
CRITICAL: This protocol requires the handling of radioactive particles and therefore needs biosafety approval according to institutional guidelines.
-
25.
Steps 8–10 in ‘before you begin’ section described the process of plating cells. Use the trypan blue dye exclusion-based method to count viable cells and to calculate the number of cells needed to seed in 6-well plates for each condition in a final volume of 2 mL per well. Typically, 0.5 × 106 of MDA-MB-468 cells per well are optimal for this experiment to get the cells at 70% confluence the next day.
Note: Some cell lines may require higher or lower cell numbers depending on the experimental conditions.
CRITICAL: To minimize variation and ensure even distribution between wells, add the calculated number of required cells in a single 50 mL Falcon tube first and then distribute to the respective wells.
-
26.
Treat the cells with Vehicle, 1 μM of PU-AD and 1 μM of PU-H71 in triplicates.
Note: Use a 10 mM stock solution of PU-H71 or PU-AD in DMSO. To obtain the 1 μM final concentration, add 2 μL of the stock solution to each well containing 2 mL of media.
-
27.
Add 18.5–37 kBq (0.5–1 μCi) of [124I]-PU-AD per well and incubate for 1 h at 37°C.
-
28.
Add the same amount of sample in separate Eppendorf tubes to serve as controls for the added total activity. This sample will be used for counting in the gamma counter.
-
29.
From the 6 well plates, carefully aspirate the media from the wells and wash cells three times with the cold 1× PBS to remove unbound radioactivity.
-
30.
Detach the cells using 0.25% trypsin for 5 min and collect them into Eppendorf tubes.
-
31.
Use a gamma counter to measure the radioactivity associated with each experimental condition. Record the raw values in CPM (counts per minute) for each sample. Measure the radioactivity in controls (step 26).
-
32.
To obtain % uptake (normalized to each experimental condition), divide the CPM values from each sample to CPM values from the control and multiply by 100 (Figure 6). In general, we present the data as % uptake/million cells.
Note: The number of cells in each well is determined prior to plating (see step 25). Alternatively, cells can be plated in control wells (in triplicate) and counted on the day of the radioactivity measurements to determine the average cell number per well.
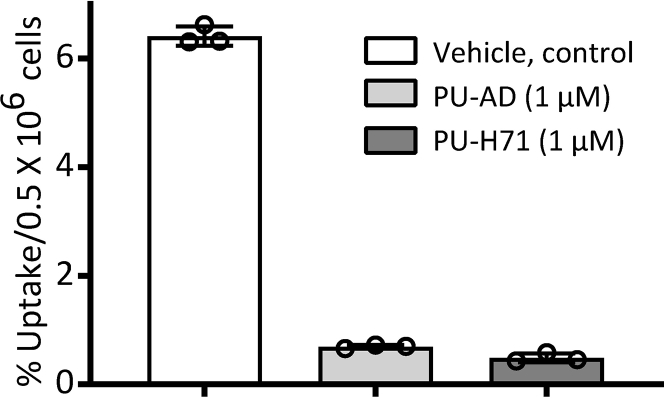
Figure 6.
Representative bar graph indicating the percentage uptake of the radiolabeled PU-AD
Graphed is the specific uptake in MDA-MB-468 cells after 1 h of concurrent treatment of cells with Vehicle (DMSO), PU-AD or PU-H71 (1 μM) and a tracer amount of radiolabeled PU-AD (1 μCi, approximately 1 pmole). Graph, mean±SD, n = 3 replicates.
PET imaging in mice
This section describes the steps to inject the radiolabeled epichaperome probe, to acquire data through non-invasive positron emission tomography imaging and to process the acquired data. It provides an example in the use of the probes for detecting and profiling epichaperome-positive tissues in vivo and demonstrates the specificity of the probe for epichaperomes in the context of a whole organism (e.g., mouse).
Note: All animal experiments must follow the Institutional Animal Care policies and Use Committee (IACUC) guidelines under an approved protocol.
PET scanning
Timing: 1 h – 72 h (depending on how many imaging time-points are planned)
Tail vein injections: 2–5 min per animal.
Anesthesia: 5 min per animal.
Imaging time: Depends on the scanning time, approximately 10 min at early time points and up to 30 min at later time points.
Steps 11–15 in ‘before you begin’ section described the process of tumor implantation. We suggest performing the PET imaging experiment when the size of the tumors reaches 250–400 mm3. For this experiment female athymic nude mice bearing single or bilateral tumor xenografts on forelimbs were used. The iodine-124 has a long life (t1/2 = 4.2 days), which permits the study of the drug kinetics non-invasively for a prolonged time.
CRITICAL: Make sure the mouse cages are clearly labeled with radioactive cage cards.
CRITICAL: Starting 48 h prior to tracer administration, add 0.01% potassium iodide solution to the drinking water to minimize the thyroid uptake of free iodide arising from metabolism of I-124 labeled probe.
-
33.
Using a 1.0 mL insulin syringe, inject intravenously via the tail vein, 5.55–9.25 MBq (150–250 mCi) of [124I]-PU-H71 or [124I]-PU-AD formulated in 200 μL of 5% ethanol in saline. Troubleshooting 5.
Note: Place the mouse under a heat lamp for 2–4 min and rub the area with an alcohol swab to induce vasodilation for intravenous administration. If necessary, to confirm placement of needle in the tail vein, pull the syringe’s plunger outward to observe the blood flow.
Note: Syringe withdrawal from the tail vein can lead to minor bleeding. If necessary, using a sterile alcohol wipe, apply mild pressure at the injection site for 5–15 s to control the bleeding. Ensure that the tail is wiped clean after the injection using the sterile alcohol wipe. If tail markings are used for mouse identification, then remark them as necessary.
-
34.
At designated time points post probe administration, place the animal in an induction chamber for anesthesia using 2% isoflurane in oxygen at a flow rate of 2 L/min.
Note: Anesthesia takes about 5 min to be effective so consider scheduling your experiments accordingly.
-
35.Prepare the PET scanner.
-
a.Create a new file in the MicroPET 2.0. program with appropriate acquisition time parameters (10 min for early time points and 20 min for time points after 16 h).
-
b.Input the necessary information regarding the mouse and the administered radioactive dose, including the physical half-life of 124I, the remaining activity in the used syringe, and the time-point of the measurement.
-
a.
-
36.Place the animal in the scanner to be at the center of the field of view of the PET scanner.
-
a.Place the mouse in a prone and headfirst position.
-
b.Maintain the animal under 2% isoflurane anesthesia in oxygen at a flow rate of 2 L/min during the entire scanning period.
-
a.
-
37.
Initiate a 30 min PET scan.
-
38.
Obtain list-mode data (10–30 min acquisitions) for each animal at various time points to obtain a minimum of 5 million coincident counts. Troubleshooting 6.
Note: Use an energy window of 420–580 keV and a coincidence timing window of 6 ns.
-
39.
Take the mouse out of the scanner when the acquisition is finished and move it back to the mouse cage displaying a radioactive cage card. Mice administered a radioactive compound must be housed separately from other mice. The bedding, cage, food, water, waste is considered as radioactive waste and should be disposed accordingly.
Analyzing data from PET imaging
Timing: 2 h (varies depending on the number of animals)
-
40.
Sort the resulting list-mode data into 2-dimensional histograms by Fourier rebinding and reconstruct transverse images by filtered back projection (FBP).
-
41.
Correct the image data for dead-time count losses and physical decay to the time of injection (no correction applied for attenuation, scatter, or partial-volume averaging).
Note: The measured reconstructed spatial resolution of the Focus 120 is 1.6-mm FWHM at the center of the field of view.
Note: The transmission scan is performed for clinical PET/CT to correct for attenuation, which can be significant for humans. For mice no such corrections are applied.
-
42.
Open the corresponding data file using the ASIPro VMTM (Siemens Medical Solutions, USA), or other software. Perform ROI analysis of the reconstructed images using ASIPro VMTM (Siemens Medical Solutions, USA), and record the maximum pixel value for each tissue/organ ROI.
-
43.
Reconstructed microPET images, including 124I images, are initially parameterized in terms of count rate (cps/voxel). These images are then quantitated, that is, parameterized in terms of activity concentration (μCi 124I/mL) using a system calibration factor (i.e., μCi/mL/cps/voxel), derived from reconstructed images of a mouse-sized water-filled cylinder containing a known concentration (μCi /mL) of 18F. The 18F calibration factor is converted to the 124I calibration factor by adjusting for the difference in positron branching ratio between 18F (0.967) and 124I (0.228) to convert the 124I count rates per voxel to activity concentrations.
-
44.
Normalize the resulting image data to the administered activity to parameterize the microPET images in terms of %ID/g (corrected for decay of 124I to the time of injection).
-
45.
Post-reconstruction smoothing can be applied to improve the quality of images. Smoothened images/frames should not be used for obtaining parametric data (%ID/g). Troubleshooting 7.
Note: When analyzing data from PET imaging, %ID/g values are more commonly used for mice and other small animal studies, whereas for humans, the standardized uptake value (SUV) is preferred. The standardized uptake value (SUV) can be calculated using the formula:
Note: Micro-CT can be used in conjunction with epichaperome PET functional imaging to provide high-resolution anatomic information, if needed. For relevant protocols please refer to (Bolaender et al., 2021; Clark and Badea, 2014; Laperre et al., 2011).
Expected outcomes
In summary, this protocol will allow the synthesis and characterization of [124/131I]-PU-H71 and [124/131I]-PU-AD, reagents with use in detecting, quantifying and tracing epichaperome localization in vivo, in live mice or human patients, through non-invasive imaging. To our knowledge, no other reagents (i.e., antibodies or other small molecules) are available for such use. An example is provided in the use of one such reagent to identify epichaperome positivity and its anatomical localization in a mouse model of triple-negative breast cancer (Figure 7). For detecting and quantifying epichaperome positivity at single-cell level through flow cytometry, please refer to a published protocol (Merugu et al., 2020).
Figure 7.
Representative biological applications for the PU-H71 and PU-AD radiolabeled epichaperome probes
(A) Experimental design to image epichaperome positivity in tumors. Mice bearing MDA-MB-468 tumor xenografts on forelimbs (as per this protocol) or intracranially (see (Bolaender et al., 2021)) were injected with a single tracer dose of [124I]-PU-H71 or [124I]-PU-AD and imaged as indicated. The radiolabeled PU-AD should be used for CNS applications instead of PU-H71 (Bolaender et al., 2021).
(B) As in (A). The epichaperome probes have a slow off-rate from epichaperomes but clear rapidly from normal tissues (i.e., epichaperome non-expressors) and from plasma providing an optimal signal to noise ratio for imaging. Full-body imaging of [124I]-PU-H71 shows its distribution in the mouse body (1 h post-probe injection imaging time point) with clear labeling of epichaperome positive MDA-MB-468 tumors (48 h, imaging time point). The signal seen in the gastrointestinal tract represents clearance of the probe and/or metabolites; thyroid signal, thyroid uptake of free iodide arising from metabolism of I-124 labeled probe. Adapted from (Pillarsetty et al., 2019).
(C) As in (A) for radiolabeled PU-AD. PET/CT image obtained at 48 h post-[124I]-PU-AD injection (microdose of 15 μCi/g), representative digital autoradiography ([131I]-PU-AD, 15 μCi/g iv, 6 h post-injection) and H&E staining of brain cryosections are shown. Inset shows the heterogeneity of the tumor with clusters of cancer cells surrounded by stroma. Adapted from (Bolaender et al., 2021).
(D) Experimental design and representative image to map epichaperome localization via autoradiography using [131I]-PU-AD in the brain of PS19 transgenic mice (mouse model of tauopathies, such as is Alzheimer’s disease, AD). Dark field microscopy images were taken to visualize brain anatomy. Representative coronal brain section of a 7 months of age female PS19 mouse is shown. Scale bar, 2 mm. Affected in AD: Hippocampal formation: entorhinal cortex (ENT), hippocampus (HIP), and subiculum (SUB). HIP regions: dentate gyrus (DG), cornu ammonis CA1 and CA3 layers. Adapted from (Inda et al., 2020).
[124/131I]-PU-H71 and [124/131I]-PU-AD are obtained by applying a two-step synthetic protocol, where a trimethyl tin-derivative is generated as a precursor molecule for radiolabel introduction. Synthetic schemes are shown in Figure 1, Figure 2, Figure 3, Figure 4. NMR spectra and LC-MS chromatograms for the precursors are given in supplemental information (Figures S1–S4), and the analytical data are summarized in Table 6. HPLC chromatograms and characterization data for the radiolabeled probes are summarized in Table 6 and presented in Figure 5.
Table 6.
Analytical data
| Compound | IUPAC name and yield | 1H NMR (500 MHz, CDCl3) δ [ppm] | 13C NMR (125 MHz, CDCl3/CD3OD) δ [ppm] | MS (m/z) |
|---|---|---|---|---|
| PU-H71-SnMe3 | 9-(3-(isopropylamino) propyl)-8-((6-(trimethylstannyl) benzo[d][1,3]dioxol-5-yl)thio)-9H-purin-6-amine and 70% | 8.24 (s, 1H), 7.01 (s, 1H), 6.98 (s, 1H), 5.96 (s, 2H), 5.83 (br s, 2H), 4.28 (t, J = 6.0 Hz, 2H), 2.78 (spt, J = 6.0 Hz, 1H), 2.60 (t, J = 6.0 Hz, 2H), 2.02–2.07 (m, 2H), 1.09 (d, J = 6.0 Hz, 6H), 0.28 (s, 9H) | 153.93, 152.07, 152.02, 149.18, 149. 05, 148.96, 143.12, 127.54, 119.66, 115.58, 115.21, 101.46, 48.91, 43.40, 41.02, 29.57, 22.45, -7.49 | [M + H]+ ion at m/z 551.2 |
| [124/131I]-PU-H71 | Identity confirmation using HPLC by matching retention time with non-radioactive standard (see also Figure 5) | |||
| PU-AD-SnMe3 | 9-(2-(Neopentylamino) ethyl)-8-((6-(trimethylstannyl)benzo[d][1,3]dioxol-5-yl)thio)-9H-purin-6-amine and 64% | 8.28 (s, 1H), 6.98 (s, 1H), 6.97 (s, 1H), 5.97 (s, 2H), 5.48 (br s, 2H) 4.29 (t, J = 6.0 Hz, 2H), 3.02 (t, J = 6.0 Hz, 2H), 2.36 (s, 2H), 0.85 (s, 9H), 0.31 (s, 9H) | 154.00, 151.84, 151.63, 150.67, 149.66, 144.65, 134.36, 126.35, 119.22, 119.19, 115.88, 101.81, 61.97, 49.40, 43.33, 31.53, 27.65, -7.63 | [M + H]+ ion at m/z 565.2 |
| [124/131I]-PU-AD | Identity confirmation using HPLC by matching retention time with non-radioactive standard (see also Figure 5) | |||
A single injection of trace (250 pmoles, considering average specific activity of 1 Ci/μmole) amount of the radiolabeled epichaperome probe is sufficient for epichaperome detection using these reagents. This small amount is completely non-perturbing biologically and allows serial PET imaging for monitoring non-invasively epichaperome changes for multiple days. This application is of use when changes in disease localization or intensity are of interest or when changes during disease progression and/or as induced by drug treatment need to be monitored in real-time, in the same individual, non-invasively. Furthermore, these probes have a potential as diagnostic tools, as epichaperome levels positively correlate with cancer sensitivity to epichaperome-targeting compounds, such as PU-H71 and PU-AD (Jhaveri et al., 2020; Joshi et al., 2018; Rodina et al., 2016). For example, Pillarsetty and colleagues used these probes combined with PET imaging to demonstrate detection of epichaperome-positive tumors is feasible in the clinical setting (Pillarsetty et al., 2019). They also showed that these probes can be used to monitor target engagement by PU-H71 in patients’ tumors at single-lesion resolution in real time, and illustrated that quantitative evaluation at the level of individual tumors can be used to optimize dose and schedule selection (Pillarsetty et al., 2019). Jhaveri and colleagues used the [124I]-PU-H71 probe and PET imaging to record the baseline epichaperome expression in the tumors of metastatic breast cancer patients that then received PU-H71 therapy (Jhaveri et al., 2020). They reported a positive significant correlation between baseline epichaperome levels (defined by the PET signal value at 24 h after [124I]-PU-H71 injection) and time-to-progression on therapy, as patients with highest baseline epichaperome levels benefitted longest. Bolaender and colleagues used [124I]-PU-AD to show a positive correlation between epichaperome levels determined by PET and the vulnerability of gliomas to epichaperome therapy (Bolaender et al., 2021). These tumors were derived from glioma-stem cells stereotactically implanted into the mouse brain, which is reported to result in tumors that mimic the features of the parental (i.e., human patient) tumor.
Limitations
For applications in the use of these probes to track the regional and temporal formation of epichaperomes in the mouse brain or other organs and tissues, we recommend signal detection via autoradiography instead of PET imaging. Autoradiography may provide a better regional accuracy and overcome limitations of small animal PET imaging which arise due to the relatively small size of mouse brain regions and partial volume effects that can lead to inaccurate quantification (Herfert et al., 2020). Please refer to published protocols for the use of these probes in detecting epichaperomes in intracranial tumors (e.g., glioblastoma and metastatic breast cancer (Bolaender et al., 2021) and mouse models of tauopathies (Inda et al., 2020) (see Figure 7 for examples).
Iodine-124 decays via positron (β+) emission 23% of the time, while the remaining 77% decay occurs via electron capture, yielding a lower signal than traditional PET isotopes such as 11C and 18F. Additionally, there is prompt gamma emission at 602 keV (100 %) that results in lowering the image quality by reducing signal to noise ratio. For the current study no correction factors for prompt gamma and partial volume effects were applied for image analysis. For human PET scanners, correction factors are applied routinely to account for such potential interferences.
Another important challenge to overcome is in the production of 124I, which requires solid targetry on the cyclotron. Most hospital- or academic-based cyclotrons do not have solid target stations and therefore production of this isotope is possible only at specialized centers. Nonetheless, 124I is now commercially available, and the 4-day half-life of 124I ensures that the radiolabeled probe (e.g., [124I]-PU-H71 or [124I]-PU-AD) can be distributed worldwide.
Troubleshooting
Problem 1
Low reaction yield (step_1).
Potential solution
Tris(dibenzylideneacetone)dipalladium(0) and hexamethylditin are air and moisture sensitive reagents. Make sure to use new or properly stored reagent for the reaction.
Problem 2
The deiodinated side product is majorly formed (step_5).
Potential solution
Deiodination occurs when the reaction is carried out under light. Be careful to properly cover the reaction vessel during reaction and when evaporating the solvent.
Problem 3
Variable radioactivity readings (step_10).
Potential solution
In measuring iodine-124 radioactivity, there may be variability depending on the volume and container material used in the measurement (e.g., glass versus plastic). Some groups recommend the use of a copper filter and a calibration setting number of 494 (for the Capintec CRC-15R calibrator) to minimize such variations (Beattie et al., 2014). For our application we used only plastic containers and the total volume of reagents was < 1 mL.
Problem 4
Low product yield coming off the column (step_11).
Potential solution
Make sure to properly prep the C18 cartridge. Incomplete washing of the C18 cartridge with water post ethanol treatment can lead to inefficient capture of reactants, resulting in loss of product during water washes after loading.
Problem 5
Intravenous vascular access could be technically challenging in mice (step_33).
Potential solution
Under such scenarios, intraperitoneal injection (i.p.) can be used to administer the drug. Drugs delivered intraperitoneally are absorbed typically much slower and cannot provide peak concentrations achieved using i.v. administration but have extended circulation times when compared to intravenous injection. Administering radiotracer intramuscularly can lead to high non-specific signal from the injection site.
Problem 6
Low signal to noise ratio for the radiolabeled probes (step_38).
Potential solution
Longer acquisition times and applying prompt gamma compensation to acquired images can improve the signal to noise ratios for the radiolabeled PET iodine probes.
Problem 7
In vivo studies with radioiodinated probes can result in high thyroid uptake (step_45).
Potential solution
This is the result of natural metabolism of iodinated compounds in animals. The iodine (released as iodide, I-) is taken up by thyroid and organified to thyroxine derivatives. This can be minimized by administering 100 μL of 1 mg/mL solution of potassium iodide in saline (0.9%) i.p. in mice 30 min before administering radiotracer and adding 100 μL of saturated potassium iodide solution in feeding water of mice starting 24–48 h prior to administering the tracer.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Gabriela Chiosis (chiosisg@mskcc.org).
Materials availability
This study did not generate new unique reagents.
Acknowledgments
This work was supported in part by the US National Institutes of Health (NIH) (R01 CA172546, R56 AG061869, R01 CA155226, P01 CA186866, R01 AG067598, R56 AG072599, R01 AG071805, U01 AG032969, R01 AG074004, and P50 CA192937), Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, and the Experimental Therapeutics Center of the Memorial Sloan Kettering Cancer Center. Technical services were provided by MKSCC’s Radiochemistry and Molecular Imaging Probes Core and Small-Animal Imaging Core Facility supported in part by R24 CA83084 and P30 CA08748 are gratefully acknowledged.
Author contributions
S.S., T.K., S.J., C.S.D., P.P., E.B., and S.G.L. performed the experiments. S.S., N.P., and G.C. wrote the manuscript. All authors contributed to the manuscript and approved it for publication.
Declaration of interests
Memorial Sloan Kettering Cancer Center holds the intellectual rights to the epichaperome imaging probe portfolio. Samus Therapeutics Inc., of which G.C. is a founder, has partial ownership, and is a member of its board of directors, has licensed this portfolio. G.C. and N.P. are inventors on the licensed intellectual property. All other authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2022.101318.
Contributor Information
Nagavarakishore Pillarsetty, Email: pillarsn@mskcc.org.
Gabriela Chiosis, Email: chiosisg@mskcc.org.
Supplemental information
Data and code availability
This study did not generate unique code or new data.
References
- Beattie B.J., Pentlow K.S., O'Donoghue J., Humm J.L. A recommendation for revised dose calibrator measurement procedures for 89Zr and 124I. PLoS One. 2014;9:e106868. doi: 10.1371/journal.pone.0106868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolaender A., Zatorska D., He H., Joshi S., Sharma S., Digwal C.S., Patel H.J., Sun W., Imber B.S., Ochiana S.O., et al. Chemical tools for epichaperome-mediated interactome dysfunctions of the central nervous system. Nat. Commun. 2021;12:4669. doi: 10.1038/s41467-021-24821-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D.P., Badea C.T. Micro-CT of rodents: state-of-the-art and future perspectives. Phys. Med. 2014;30:619–634. doi: 10.1016/j.ejmp.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreolini L., Cullen M., Yung E., Laird L., Webb J.R., Nelson B.H., Hay K.A., Balasundaram M., Kekre N., Holt R.A. A rapid and sensitive nucleic acid amplification technique for mycoplasma screening of cell therapy products. Mol. Ther. Methods Clin. Dev. 2020;17:393–399. doi: 10.1016/j.omtm.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H., Zatorska D., Kim J., Aguirre J., Llauger L., She Y., Wu N., Immormino R.M., Gewirth D.T., Chiosis G. Identification of potent water soluble purine-scaffold inhibitors of the heat shock protein 90. J. Med. Chem. 2006;49:381–390. doi: 10.1021/jm0508078. [DOI] [PubMed] [Google Scholar]
- Herfert K., Mannheim J.G., Kuebler L., Marciano S., Amend M., Parl C., Napieczynska H., Maier F.M., Vega S.C., Pichler B.J. Quantitative rodent brain receptor imaging. Mol. Imaging Biol. 2020;22:223–244. doi: 10.1007/s11307-019-01368-9. [DOI] [PubMed] [Google Scholar]
- Inda M.C., Joshi S., Wang T., Bolaender A., Gandu S., Koren J., III, Che A.Y., Taldone T., Yan P., Sun W., et al. The epichaperome is a mediator of toxic hippocampal stress and leads to protein connectivity-based dysfunction. Nat. Commun. 2020;11:319. doi: 10.1038/s41467-019-14082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhaveri K.L., Dos Anjos C.H., Taldone T., Wang R., Comen E., Fornier M., Bromberg J.F., Ma W., Patil S., Rodina A., et al. Measuring tumor epichaperome expression using [(124)I] PU-H71 positron emission tomography as a biomarker of response for PU-H71 plus nab-paclitaxel in HER2-negative metastatic breast cancer. JCO Precis Oncol. 2020;4:1414–1424. doi: 10.1200/PO.20.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S., Wang T., Araujo T.L.S., Sharma S., Brodsky J.L., Chiosis G. Adapting to stress - chaperome networks in cancer. Nat. Rev. Cancer. 2018;18:562–575. doi: 10.1038/s41568-018-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laperre K., Depypere M., van Gastel N., Torrekens S., Moermans K., Bogaerts R., Maes F., Carmeliet G. Development of micro-CT protocols for in vivo follow-up of mouse bone architecture without major radiation side effects. Bone. 2011;49:613–622. doi: 10.1016/j.bone.2011.06.031. [DOI] [PubMed] [Google Scholar]
- Merugu S., Sharma S., Kaner J., Digwal C., Sugita M., Joshi S., Taldone T., Guzman M.L., Chiosis G. Chemical probes and methods for single-cell detection and quantification of epichaperomes in hematologic malignancies. Methods Enzymol. 2020;639:289–311. doi: 10.1016/bs.mie.2020.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillarsetty N., Jhaveri K., Taldone T., Caldas-Lopes E., Punzalan B., Joshi S., Bolaender A., Uddin M.M., Rodina A., Yan P., et al. Paradigms for precision medicine in epichaperome cancer therapy. Cancer Cell. 2019;36:559–573.e557. doi: 10.1016/j.ccell.2019.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodina A., Wang T., Yan P., Gomes E.D., Dunphy M.P., Pillarsetty N., Koren J., Gerecitano J.F., Taldone T., Zong H., et al. The epichaperome is an integrated chaperome network that facilitates tumour survival. Nature. 2016;538:397–401. doi: 10.1038/nature19807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strober W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2015;111:A3 B 1–A3 B 3. doi: 10.1002/0471142735.ima03bs111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita M., Wilkes D.C., Bareja R., Eng K.W., Nataraj S., Jimenez-Flores R.A., Yan L., De Leon J.P., Croyle J.A., Kaner J., et al. Targeting the epichaperome as an effective precision medicine approach in a novel PML-SYK fusion acute myeloid leukemia. NPJ Precis Oncol. 2021;5:44. doi: 10.1038/s41698-021-00183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taldone T., Zatorska D., Ochiana S.O., Smith-Jones P., Koziorowski J., Dunphy M.P., Zanzonico P., Bolaender A., Lewis J.S., Larson S.M., et al. Radiosynthesis of the iodine-124 labeled Hsp90 inhibitor PU-H71. J. Labelled Comp. Radiopharm. 2016;59:129–132. doi: 10.1002/jlcr.3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate unique code or new data.