Abstract
Background
Data on the clinical and virological characteristics of the Delta variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are limited. This prospective cohort study compared the characteristics of the Delta variant to other variants.
Methods
Adult patients with mild coronavirus disease 2019 (COVID-19) who agreed to daily saliva sampling at a community isolation facility in South Korea between July and August 2021 were enrolled. Scores of 28 COVID-19-related symptoms were recorded daily. The genomic RNA and subgenomic RNA from saliva samples were measured by real-time reverse-transcription polymerase chain reaction (PCR). Cell cultures were performed on saliva samples with positive genomic RNA results.
Results
A total of 141 patients (Delta group, n = 108 [77%]; non-Delta group, n = 33 [23%]) were enrolled. Myalgia was more common in the Delta group than in the non-Delta group (52% vs 27%, P = .03). Total symptom scores were significantly higher in the Delta group between days 3 and 10 after symptom onset. Initial genomic RNA titers were similar between the 2 groups; however, during the late course of disease, genomic RNA titers were higher in the Delta group. Negative conversion of subgenomic RNA was slower in the Delta group (median 9 vs 5 days; P < .001). The duration of viral shedding in terms of positive viral culture was also longer in the Delta group (median 5 vs 3 days; P = .002).
Conclusions
COVID-19 patients infected with the Delta variant exhibited prolonged viable viral shedding with more severe symptoms than those infected with non-Delta variants.
Keywords: SARS-CoV-2, Delta variant, viral shedding, subgenomic RNA, culture
Symptom score collection, subgenomic RNA measurement and cell cultures showed that patients infected with the Delta variant exhibited more severe symptoms with prolonged symptom duration, higher viral loads, and prolonged viable viral shedding than those infected with non-Delta variants.
The coronavirus disease 2019 (COVID-19) outbreak due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected more than 240 million patients and caused almost 5 million deaths worldwide [1]. Although the development of several vaccines resulted in a temporary decline in the number of cases, hospitalizations, and deaths due to COVID-19 [2, 3], the emergence of variants including the SARS-CoV-2 Pango lineage B.1.617.2, also known as the Delta variant, has caused a new wave of the pandemic [4, 5].
The Delta variant, first identified in India in October 2020, has since become the predominant SARS-CoV-2 variant and currently accounts for more than 99% of the cases in South Korea [6] as well as in the United States [7]. The Delta variant is highly transmissible, with a transmission advantage of 66% over the Alpha variant in England [8] and over 79% in certain regions of France [9]. A previous study reported that compared with other variants, the Delta variant exhibits increased disease severity, higher viral loads, and a longer duration of positive polymerase chain reaction (PCR) results [10]. However, comparative data between the Delta variant with other variants in terms of symptom presentation and the duration of viable virus shedding are limited. The period between July 2021 and August 2021 in South Korea was the transitional time before the Delta era when non-Delta variants and the Delta variant were co-circulating. The proportion of the Delta variant in South Korea was 30% in early July, 62% in early August, and 97% in late August [6]. In this study, we compared the clinical and virological characteristics of the Delta variant to non-Delta variants in adult patients with mild COVID-19.
METHODS
Study Participants
At the time of the study, confirmed asymptomatic or mildly symptomatic COVID-19 patients in South Korea were isolated at non-hospital community facilities. The patients were monitored for at least 10 days after the diagnosis, and those who needed further evaluation or medical care were transferred to nearby hospitals. Following government health policies, patients whose symptoms improved or no longer worsened after 10 days since the diagnosis were discharged from the facility. Between 20 July and 20 August 2021, patients with mild COVID-19 above the age of 18 were recruited at a designated non-hospital community facility in Seoul, South Korea. All participants provided written informed consent. This study was approved by the institutional review board of Asan Medical Center (Seoul, South Korea).
Symptom Score Collection
Patients were instructed to record their symptom scores twice daily (8 AM and 5 PM) in an electronic diary that was accessible to the medical staff. The severity of each of the 28 COVID-19-related symptoms was scored from 0 to 3 points as follows: i) score of 0 (no symptoms), ii) score of 1 (transient or mild discomfort, no interference with daily activities, and no requirement of medical intervention or therapy), iii) score of 2 (mild-to-moderate limitations in daily activities, and symptoms are controlled by medical intervention or therapy), and iv) score of 3 (substantial limitations in daily activities, and symptoms are not well-controlled even with medical intervention or therapy). The questionnaire sheets for the COVID-19-related symptoms are provided in the Supplementary Materials. The mean of the sum of all 28 COVID-19-related symptom scores (“total symptom scores”) per participant per day was used to compare the 2 groups, based on the method used to evaluate the dynamics of symptoms in a previous meta-analysis of human influenza patients [11]. The dynamics of the mean of total symptom scores according to time was drawn for each group, and the mean total symptom scores for each day after symptom onset were compared between the Delta and non-Delta groups.
Saliva Sample Collection and Viral Kinetic Study
Self-collected saliva samples were obtained from the patients starting from the day of study enrollment until the day of discharge. Each day, 2 mL of saliva was collected into an airtight container that was provided after the patient agreed to participate in the study. Patients were instructed to avoid food, water, and teeth brushing for at least 30 minutes prior to sample collection. Saliva samples were picked up within an hour by the medical staff and transported to a designated laboratory where they were aliquoted and stored at –80°C until use. For the viral kinetic study, patients infected with the Delta variant were randomly selected after matching age and sex with the patients infected with a non-Delta variant.
Laboratory Procedure and Identification of Delta Variant
The collected saliva samples were inactivated at 65°C for 30 minutes in a special negative pressure laboratory. Genomic viral RNA was extracted from the specimens using a QIAamp viral RNA Mini kit (Qiagen Inc., Hilden, Germany). The variants were classified by double-multiplex real-time RT-PCR assay using a PowerChek SARS-CoV-2 S-gene Mutation detection kit (Kogene Biotech Co Ltd, Seoul, South Korea) according to the manufacturer’s instructions. The PCR amplifications were performed by AB 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, California, USA), and the Delta variants were identified based on the simultaneous positive results of P681R and L452R mutations. To determine the SARS-CoV-2 genomic viral RNA copy number, multiplex real-time RT-PCR assays targeting the S and N genes were developed; the primer and probe sequences and detailed procedures of the multiplex real-time RT-PCR assay are provided in Supplementary Table 1.
Subgenomic RNA Detection and Cell Culture
Subgenomic RNAs of the SARS-CoV-2 N and S genes were detected by multiplex real-time RT-PCR assays as described in our previous study [12]. The shared forward primer was designed in the 5’ leader sequence, and reverse primers and probes were located in the gene sequences coding the N and S proteins (Supplementary Table 2). Further detailed methods are provided in Supplementary Materials. Culture-based isolation of SARS-CoV-2 from respiratory specimens that revealed positive genomic RNA results was performed by a plaque assay in a Biosafety Level 3 laboratory at Korea University College of Medicine (Seoul, South Korea). Detailed procedures are described in Supplementary Materials.
Statistical Analysis
Categorical variables were compared using χ2 or Fisher exact test, and continuous variables were compared with the Mann–Whitney U test or Kruskal-Wallis test, as appropriate. Time-wise differences in the dynamics of viral shedding and symptom score between patients infected with the Delta variant and those infected with a non-Delta variant were compared using a generalized linear mixed model. Interactions between time and variant groups (Delta vs non-Delta) were also evaluated. Viral shedding values less than the lower limit of quantification (LoQ; 2.6 log10 copies/mL) of the RT-PCR assay but with positive qualitative results were imputed with half of the lower LoQ. Negative RNA values were imputed with 0 log10 copies/mL. In addition, we performed survival analysis to estimate the negative conversion rate of PCR using the Kaplan-Meier plot and log-rank test. Nonparametric maximum-likelihood estimation (NPMLE) was used to estimate the proportion of the viral shedding in terms of subgenomic RNA and viral culture due to interval censoring data. All tests of significance were 2-tailed and P values < .05 were considered significant. Data were analyzed using SPSS Statistics for Windows, version 23.0 (IBM Corp., Armonk, New York, USA) or R version 4.0.4 (R Project for Statistical Computing, Vienna, Austria).
RESULTS
Baseline Clinical Characteristics
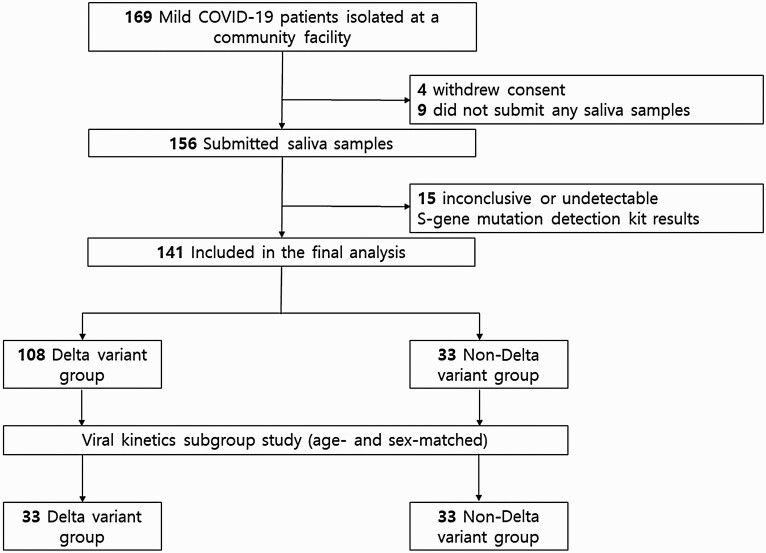
The study patient flow chart is shown in Figure 1. A total of 169 patients were initially enrolled during the study period; of them, 4 patients withdrew consent, and 9 patients who did not participate at least once in saliva sample collection were excluded. Among the 156 patients who submitted saliva samples, the mutation kit results were inconclusive or not detected in 15 patients. Thus, 141 patients were included in the final analysis. Among the 141 patients, 108 (76.6%) were found to have been infected with the Delta variant, whereas 33 (23.4%) were infected with non-Delta variants, including the Alpha variant (n = 3), untypable variants with E484K mutation (n = 26) and L452R mutation (n = 1), and original SARS-CoV-2 (n = 3).
Figure 1.
Flowchart of study participants. Abbreviation: COVID-19, coronavirus disease 2019.
The baseline clinical characteristics of the patients according to the SARS-CoV-2 variant are shown in Table 1. There were no significant differences in terms of age, gender, and the duration from symptom onset to isolation facility admission between the Delta group and the non-Delta group. In addition, there were no significant differences between the Delta group and the non-Delta group in terms of underlying diseases or the proportion of patients with obesity or smoking habits. Nine (8.3%) patients in the Delta group and 7 (21.2%) patients in the non-Delta group were presymptomatic (P = .06). In both groups, the most common symptoms were fever, cough, sore throat, myalgia, and headache, and there were no significant differences in the prevalence of each symptom between the 2 groups (Table 1), with the exception of a higher proportion of myalgia in the Delta group (51.5% vs 26.9%, P = .03) and a higher proportion of loss of taste in the non-Delta group (2.0% vs 26.9%, P < .001). Abnormal infiltrations on chest imaging were seen in approximately 9% of patients in each group (9.3% vs 9.1%, P > .99).
Table 1.
Clinical and Radiologic Characteristics of COVID-19 Patients According to SARS-CoV-2 Variant
| Total (n = 141) | Delta (n = 108) | Non-Delta (n = 33) | P Value | |
|---|---|---|---|---|
| Age, y, median (IQR) | 34.0 (25.0–46.0) | 34.5 (26.5–46.0) | 32.0 (23.0–48.0) | .77 |
| Male sex | 92 (65.2) | 69 (63.9) | 23 (69.7) | .54 |
| Underlying diseases | ||||
| Hypertension | 17 (12.1) | 12 (11.1) | 5 (15.2) | .53 |
| Hyperlipidemia | 6 (4.3) | 4 (3.7) | 2 (6.1) | .63 |
| Diabetes | 12 (8.5) | 7 (6.5) | 5 (15.2) | .12 |
| Psychiatric illness | 2 (1.4) | 1 (0.9) | 1 (3.0) | .42 |
| Asthma/Rhinitis | 2 (1.4) | 1 (0.9) | 1 (3.0) | .42 |
| Cancer | 2 (1.4) | 1 (0.9) | 1 (3.0) | .42 |
| Obesity (BMI ≥ 30) | 16 (11.3) | 12 (11.1) | 4 (12.1) | >.99 |
| Pregnancy | 0 (0) | 0 (0) | 0 (0) | NA |
| Smoking | 44 (31.2) | 37 (34.3) | 7 (21.2) | .16 |
| Days from symptom onset to facility admission, median (IQR) | 3 (2–4) | 3 (2–4) | 2 (1–3) | .85 |
| Asymptomatic | 0 (0) | 0 (0) | 0 (0) | NA |
| Presymptomatic | 16 (11.3) | 9 (8.3) | 7 (21.2) | .06 |
| Symptomatic | 125 (88.7) | 99 (91.7) | 26 (78.8) | |
| Symptoms at diagnosis | ||||
| Fever | 89 (71.2) | 73 (73.7) | 16 (61.5) | .22 |
| Cough | 64 (51.2) | 49 (49.5) | 15 (57.7) | .46 |
| Sputum | 25 (20.0) | 18 (18.2) | 7 (26.9) | .32 |
| Sore throat | 60 (48.0) | 43 (43.4) | 17 (65.4) | .05 |
| Dyspnea | 2 (1.6) | 1 (1.0) | 1 (3.8) | .21 |
| Rhinorrhea | 6 (4.8) | 4 (4.0) | 2 (7.7) | .60 |
| Nasal stuffiness | 4 (3.2) | 2 (2.0) | 2 (7.7) | .19 |
| Myalgia | 58 (46.4) | 51 (51.5) | 7 (26.9) | .03 |
| Headache | 43 (34.4) | 34 (34.3) | 9 (34.6) | .98 |
| Chills | 34 (27.2) | 30 (30.3) | 4 (15.4) | .15 |
| Loss of taste | 9 (7.2) | 2 (2.0) | 7 (26.9) | <.001 |
| Loss of smell | 11 (8.8) | 7 (7.1) | 4 (15.4) | .24 |
| Diarrhea | 4 (3.2) | 3 (3.0) | 1 (3.8) | >.99 |
| Vaccination (≥ 1 dose) | 24 (17.0) | 22 (20.4) | 2 (6.1) | .07 |
| mRNA vaccine | 9 (37.5) | 9 (40.9) | 0 (0) | .51 |
| Viral vector vaccine | 15 (62.5) | 13 (59.1) | 2 (100) | |
| Fully vaccinated | 7 (5.0) | 7 (6.5) | 0 (0) | .20 |
| mRNA vaccine | 2 (28.6) | 2 (28.6) | NA | NA |
| Viral vector vaccine | 5 (71.4) | 5 (71.4) | NA | NA |
| Infiltrations on chest X-ray at admissiona | 13 (9.3) | 10 (9.3) | 3 (9.1) | >.99 |
| Clinical course | ||||
| Transfer to hospital | 9 (6.4) | 8 (7.4) | 1 (3.0) | .69 |
| Discharge per protocol | 132 (93.6) | 100 (92.6) | 32 (97.0) |
Data are no. (%) of patients, unless otherwise indicated.
Abbreviations: BMI, body mass index; COVID-19, coronavirus disease 2019; IQR, interquartile range; mRNA, messenger ribonucleic acid; NA, not applicable; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Chest X-ray was not performed in 1 patient in the Delta group due to the possibility of pregnancy.
Dynamics of Symptoms
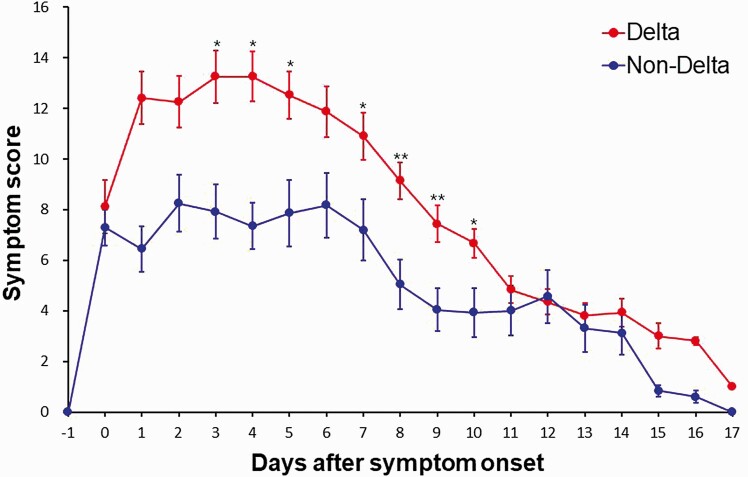
A total of 9 patients were transferred to a hospital facility due to symptom aggravation, among whom 8 were infected with the Delta variant (7.4% vs 3.0%, P = .69). The remaining 132 patients showed stable or reduced symptoms and were discharged from the facility. The dynamics of the mean of total symptom scores in the patients according to the SARS-CoV-2 variant are described in Figure 2 and Supplementary Figure 1. Additional analyses comparing the percentage of patients with more severe symptoms (symptom score 2 or 3) between the two groups are shown in Supplementary Table 3. In both the Delta group and the non-Delta group, the mean of total symptom scores peaked around 2–4 days after symptom onset; the peak of the mean of total symptom scores was significantly higher in the Delta group compared with the non-Delta group (13 vs 8, P = .03). The mean of the total symptom scores were significantly different between the 2 groups on days 3–10 after symptom onset (Figure 2).
Figure 2.
Dynamics of the mean of total symptom scores in the Delta group and non-Delta group. Summary curves for the Delta group (n = 108) and non-Delta group (n = 33) were compared using mean ± standard error of the mean. *P < .05; **P < .01.
Viral Shedding Kinetics
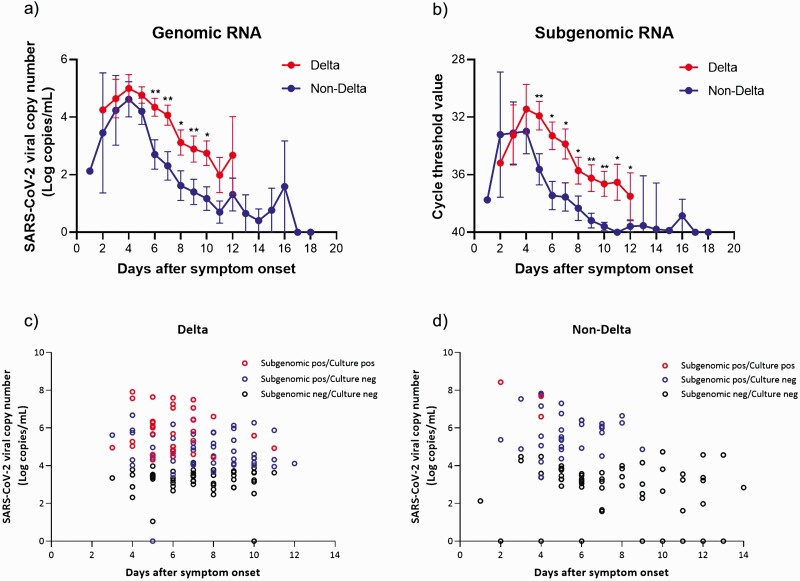
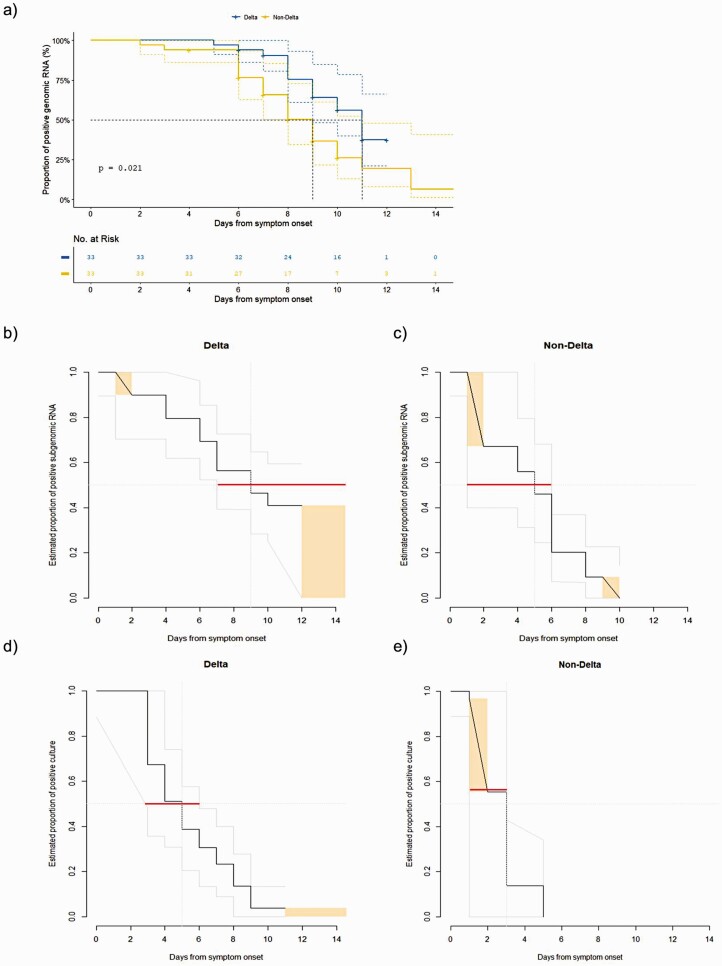
Of the 108 patients infected with the Delta variant, 33 were matched with those in the non-Delta group according to age and sex (Figure 1). Baseline clinical characteristics of these matched patients are shown in Supplementary Table 4. There were no significant differences in terms of age, gender, underlying diseases, and the duration from symptom onset to isolation facility admission between the Delta group and the non-Delta group. A total of 366 samples from the 66 patients were analyzed using genomic RNA assay and their viral shedding kinetics are shown in Figure 3. The initial viral loads (log10 copies/mL) were not significantly different between the Delta group and the non-Delta group; however, during the late course of disease (day 3 to day 10 after symptom onset), the genomic RNA titers were significantly higher in the Delta group than in the non-Delta group (Figure 3). In all patients, viral loads decreased according to time (P for time effect < .001), and the viral loads were significantly different between the 2 groups over time (P for group effect = .046). No significant group-by-time interaction effect was observed (P for interaction = .41). However, the degree of declines in the viral loads from the peak values was more rapid in the non-Delta group than in the Delta group, as the median number of days from symptom onset to negative conversion of genomic RNA detection was 9 days (95% confidence interval [CI]: 8–11) in the non-Delta group compared with 11 days (lower limit of the 95% CI: 9 days; upper limit of the 95% CI: not available in our data) in the Delta group (P by log-rank test = .02, Figure 4A).
Figure 3.
SARS CoV-2 viral loads (Log10 copies/mL), subgenomic RNA, and virus culture as a function of days after symptom onset. A, Comparison of SARS CoV-2 viral genomic copy number (Log10 copies/mL) between the Delta group (red circles) and the non-Delta group (blue circles). B, Comparison of cycle threshold values detecting N gene subgenomic RNA between Delta (red circle) and non-Delta (blue circle) variants. C, Distribution of saliva samples identified as Delta variants. D, Distribution of saliva samples identified as wild type or non-Delta variants; red dot, positive subgenomic RNA and positive virus culture; blue dot, positive subgenomic RNA and negative virus culture; black dot, negative subgenomic RNA and negative virus culture; *P < .05; **P < .01. Abbreviation: SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Figure 4.
Proportion of the viral shedding. A, Genomic RNA. B, Subgenomic RNA in the Delta group. C, Subgenomic RNA in the non-Delta group. D, Viral culture in the Delta group. E, Viral culture in the non-Delta group. Orange rectangles indicate the proportions of censored data, and gray lines represent the 95% CI of the fitted distributions. Red horizontal lines indicate the 95% CI of the time-to-negative viral culture or subgenomic RNA for the 50 percentile of the patients. Abbreviation: CI, confidence interval.
Subgenomic RNA Viral Shedding and Isolation of SARS-CoV-2 by Culture
We further analyzed subgenomic RNA from all saliva samples to infer the duration and prevalence of viable virus shedding. As shown in Figure 3B–3D, subgenomic RNAs were detected for a considerably longer duration in the Delta group than in the non-Delta group. In the Delta group, subgenomic RNA was positive up to 12 days after symptom onset, with the rate of subgenomic RNA positivity gradually decreasing over time (Figure 3C and Supplementary Table 5). In the non-Delta group, subgenomic RNA was positive up to 9 days after diagnosis and not thereafter (Figure 3D and Supplementary Table 5).
In addition, we performed cell cultures by using 231 saliva samples with positive genomic RNA results. As shown in Figure 3C and 3D, viable viruses were detected for a considerably longer duration in the Delta group than in the non-Delta group. In the Delta group, cell culture revealed positive results up to 11 days after symptom onset, with the rate of cell culture positivity gradually decreasing over time (Figure 3C and Supplementary Table 6). In the non-Delta group, cell culture exhibited positive results only until day 4 after diagnosis and not thereafter (Figure 3D and Supplementary Table 6). Moreover, the negative conversion of subgenomic RNA was slower in the Delta group at a median of 9 days from symptom onset (lower limit of the 95% CI: 6 days; upper limit of the 95% CI: not available in our data) compared with the non-Delta group at a median of 5 days (95% CI: 1–6; P < .001; Figure 4B and 4C). The time to negative conversion of viral culture was also longer in the Delta group (median, 5 days; 95% CI: 3–6) compared with the non-Delta group (median, 3 days; 95% CI: 1–3; P = .002; Figure 4D and 4E).
DISCUSSION
A previous study reported that the Delta variant exhibited increased disease severity and had higher and longer viral loads than other variants [10], suggesting a higher transmission potential. However, comparative data on the kinetics of viable viral shedding between the Delta variant and non-Delta variants are limited. Our study clearly demonstrated that patients with the Delta variant had prolonged viable viral shedding by subgenomic RNA assay and cell culture when compared with other variants. Previous studies reported that in most patients with mild-to-moderate COVID-19, viable virus was not isolated after 10 days following symptom onset [13, 14]. In contrast, in some patients with severe COVID-19, viable virus was isolated between 10 and 20 days after symptom onset [15, 16]. Our previous study on asymptomatic or mild COVID-19 patients before the emergence of the Delta variant also demonstrated that the duration of viable virus shedding of symptomatic COVID-19 patients was longer than that of asymptomatic patients with SARS-CoV-2 infection, and that the time-wise changes in viral shedding kinetics after symptom onset overlapped the changes in symptom scores [17]. In this context, our findings on the higher symptom scores and higher viral load with more prolonged viable viral shedding in the Delta group than in the non-Delta group are consistent with the previous studies [13–17].
Currently, the Centers for Disease Control and Prevention (CDC) recommends that isolation and precautions may be discontinued 10 days after symptom onset for most symptomatic patients with COVID-19, although there are few studies on the viable viral shedding of the Delta variant. The present study showed that viable virus was detected up to 11 days after symptom onset in patients with mild COVID-19 caused by the Delta variant (Figure 3, Supplementary Tables 5 and 6). Therefore, our findings support the CDC recommendation in the era of the Delta variant. However, the kinetics of viable viral shedding in elderly patients, hospitalized patients, or immunocompromised patients with the Delta variant are largely unknown. Thus, further studies are urgently needed in this area to provide evidence-based recommendations on the isolation of patients.
This study has several limitations. First, there may be unmeasured confounding variables between the Delta variant and non-Delta variant groups. However, as randomized trials are neither feasible nor ethical in this setting, alternative study designs are needed to answer important policy questions. Our study has strengths in terms of the similar distribution of baseline characteristics between the 2 groups. In addition, we had a unique chance to compare patients who were infected with the Delta strain and those who were infected with non-Delta strains during the same period, which may reduce the biases that may entail comparisons between contemporary and historical cohorts. Second, Lechien et al. developed the COVID-19 symptom index (CSI) [18] and demonstrated the internal consistency of the CSI by assessing the test-retest reliability and the external validity by a correlation analysis between the CSI and other scoring systems such as the sinonasal outcome test (SNOT)-22. The weakness of our study is the lack of internal and external validation of the symptom scoring system used in our study. However, our previous study conducted in the same isolation facility between 10 January and 22 February 2021, when the non-Delta variant was dominant in South Korea [17] showed similar symptom scores compared to those in patients with the non-Delta variant between 20 July and 20 August 2021 in this study (Supplementary Figure 2). Finally, during the study period, only a limited number of fully vaccinated patients with breakthrough Delta variant infection were included. Previous studies revealed that vaccination reduced the risk of onward transmission by accelerating viral clearance [19, 20]. Therefore, our findings may not be generalized in the setting of a highly vaccinated population.
In conclusion, COVID-19 patients infected with the Delta variant had more systemic symptoms with more prolonged symptom duration than those infected with non-Delta variants. In addition, patients with the Delta variant exhibited higher viral loads and more prolonged viable viral shedding than those with non-Delta variants.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare, South Korea (grant number HW20C2062); and the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT, Republic of Korea (grant number NRF-2017M3A9E4061995).
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
Contributor Information
Sunghee Park, Department of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea.
So Yun Lim, Department of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea.
Ji Yeun Kim, Department of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea.
Heedo Park, BK21 Graduate Program, Department of Biomedical Sciences, Korea University College of Medicine, Seoul, Republic of Korea; Department of Microbiology, Institute for Viral Diseases, Biosafety Center, College of Medicine, Korea University, Seoul, South Korea.
Joon Seo Lim, Clinical Research Center, Asan Institute for Life Sciences, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea.
Seongman Bae, Department of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea.
Jeonghun Kim, BK21 Graduate Program, Department of Biomedical Sciences, Korea University College of Medicine, Seoul, Republic of Korea; Department of Microbiology, Institute for Viral Diseases, Biosafety Center, College of Medicine, Korea University, Seoul, South Korea.
Jiwon Jung, Department of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea.
Min Jae Kim, Department of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea.
Yong Pil Chong, Department of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea.
Sang-Ho Choi, Department of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea.
Sang-Oh Lee, Department of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea.
Yang Soo Kim, Department of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea.
Man-Seong Park, BK21 Graduate Program, Department of Biomedical Sciences, Korea University College of Medicine, Seoul, Republic of Korea; Department of Microbiology, Institute for Viral Diseases, Biosafety Center, College of Medicine, Korea University, Seoul, South Korea.
Sung-Han Kim, Department of Infectious Diseases, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea.
References
- 1. Johns Hopkins University and Medicine. Coronavirus resource center. COVID-19 dashboard. Available at: https://coronavirus.jhu.edu/map.html. Accessed 19 October 2021.
- 2. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 2020; 383:2603–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dagan N, Barda N, Kepten E, et al. BNT162b2 mRNA Covid-19 vaccine in a nationwide mass vaccination setting. N Engl J Med 2021; 384:1412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. del Rio C, Malani PN, Omer SB. Confronting the Delta variant of SARS-CoV-2, summer 2021. JAMA 2021; 326:1001–2. [DOI] [PubMed] [Google Scholar]
- 5. Levine-Tiefenbrun M, Yelin I, Alapi H, et al. Viral loads of Delta-variant SARS-CoV-2 breakthrough infections after vaccination and booster with BNT162b2. Nat Med 2021; 27:2108–10. [DOI] [PubMed] [Google Scholar]
- 6. Korea Disease Control and Prevention Agency. COVID-19 variant surveillance. Available at: https://nih.go.kr/contents.es?mid=a20107040000. Accessed 19 October 2021.
- 7. Centers for Disease Control and Prevention. COVID data tracker. Available at: https://covid.cdc.gov/covid-data-tracker/#variant-proportions. Accessed 19 October 2021.
- 8. Mahase E. Delta variant: what is happening with transmission, hospital admissions, and restrictions? BMJ 2021; 373:n1513. [DOI] [PubMed] [Google Scholar]
- 9. Alizon S, Haim-Boukobza S, Foulongne V, et al. Rapid spread of the SARS-CoV-2 Delta variant in some French regions, June 2021. Euro Surveill 2021; 26:2100573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ong SWX, Chiew CJ, Ang LW, et al. Clinical and virological features of SARS-CoV-2 variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta). Clin Infect Dis 2021; ciab721. doi: 10.1093/cid/ciab721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carrat F, Vergu E, Ferguson NM, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol 2008; 167:775–85. [DOI] [PubMed] [Google Scholar]
- 12. Kim JY, Bae J-Y, Bae S, et al. Diagnostic usefulness of subgenomic RNA detection of viable SARS-CoV-2 in patients with COVID-19. Clin Microbiol Infect 2022; 28:101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Owusu D, Pomeroy MA, Lewis NM, et al. Persistent SARS-CoV-2 RNA shedding without evidence of infectiousness: a cohort study of individuals with COVID-19. J Infect Dis 2021; 224:1362–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020; 581:465–9. [DOI] [PubMed] [Google Scholar]
- 15. Baang JH, Smith C, Mirabelli C, et al. Prolonged severe acute respiratory syndrome coronavirus 2 replication in an immunocompromised patient. J Infect Dis 2021; 223:23–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Avanzato VA, Matson MJ, Seifert SN, et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell 2020; 183:1901–1912.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bae S, Kim JY, Lim SY, et al. Dynamics of viral shedding and symptoms in patients with asymptomatic or mild COVID-19. Viruses 2021; 13:2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lechien JR, Chiesa-Estomba CM, Hans S, et al. Validity and reliability of the COVID-19 symptom index, an instrument evaluating severity of general and otolaryngological symptoms. Acta Otolaryngol 2021; 141:615–20. [DOI] [PubMed] [Google Scholar]
- 19. Singanayagam A, Hakki S, Dunning J, et al. Community transmission and viral load kinetics of the SARS-CoV-2 Delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Infect Dis 2022; 22:183–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eyre DW, Taylor D, Purver M, et al. Effect of Covid-19 vaccination on transmission of Alpha and Delta variants. N Engl J Med 2022; 386:744–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.