Abstract
There are racial disparities in the outcome of triple negative breast cancer (TNBC) patients between women of African ancestry and women of European ancestry, even after accounting for lifestyle, socioeconomic and clinical factors. MicroRNA (miRNA) are non-coding molecules whose level of expression is associated with cancer suppression, proliferation and drug resistance; therefore, these have potential for biomarker applications in cancers including TNBC. Historically, miRNAs up-regulated in African American (AA) patients have received less attention than for patients of European ancestry. Using laser capture microdissection (LCM) to acquire ultrapure tumor cell samples, miRNA expression was evaluated in 15 AA and 15 European American (EA) TNBC patients. Tumor sections were evaluated using RNA extraction followed by miRNA analysis and profiling. Results were compared based on ethnicity and method of tissue fixation. miRNAs that showed high differential expression in AA TNBC patients compared to EA included: miR-19a, miR-192, miR-302a, miR-302b, miR-302c, miR-335, miR-520b, miR-520f and miR-645. LCM is a useful technique for isolation of tumor cells. We found a greater abundance of RNA in frozen samples compared to formalin fixed, paraffin embedded samples. miRNA appears to be a useful biomarker for TNBC to improve diagnosis and treatment.
Keywords: African American, European American, Caucasian, human, laser-capture microdissection, microRNA, racial disparity, triple negative breast cancer
Although women of African ancestry exhibit a lower incidence of breast cancer than European American (EA) women, African American (AA) women experience significantly poorer survival (Danforth 2013; Thomas et al. 2019). Even accounting for the fact that AA are more likely to be diagnosed at more advanced stages (Banegas and Li 2012), have a more aggressive subtype of cancer (Carey et al. 2006; Howlader et al. 2014) and are less likely to undergo recommended treatment (Ooi et al. 2011), the trend persists. Factors including incidence, higher tumor growth index (Morris and Mitchell 2008), tumor location (Han et al. 2019) and microenvironment (Deshmukh et al. 2017) have all been proposed as potential reasons for racial disparities in breast cancer. Translation of predisposing information into alterations of treatment and attitudes, however, has not yet followed. Characterization and discovery of biomarkers that are expressed differentially in races could elicit information useful for the diagnosis, treatment and overall prognosis to narrow racial disparities and lower breast cancer mortality.
MicroRNA (miRNA) is a small non-coding RNA molecule that binds to target mRNA to prevent subsequent protein production (MacFarlane and Murphy 2010; McNally et al. 2013). Although specifics concerning the function of miRNA molecules are relatively unknown, miRNAs are important factors for regulating gene expression (Esau et al. 2006; Garzon et al. 2006; Naguibneva et al. 2006); they are an intriguing possibility for predicting disease risk. miRNAs are useful biomarkers; they are stable in vitro and exhibit long duration in vivo, two significant advantages over mRNA (Lim et al. 2005; Lu et al. 2005). A significant number of miRNAs could have oncogenic potential and could signal discrepancies in outcomes due to race. For example, miRNA-19a is related to malignancy by regulating drug resistance (Wang et al. 2013) and cell proliferation (Xu et al. 2012) as well as having anti-inflammatory characteristics (Sonkoly and Pivarcsi 2009).
Other miRNAs that regulate inflammatory responses, miRNAs 192 and 335, also have been implicated in oncogenic activity (Braun et al. 2008; Sonkoly and Pivarcsi 2009; Zhu et al. 2014; Arner and Kulyté 2015; Dong et al. 2018; Ali Ahmed et al. 2020). Controlling inflammatory responses is critical for many diseases; these responses participate in cancer, particularly onset of carcinogenesis and difficulty of treatment (Greten and Grivennikov 2019). Prior to an oncogenic event, normal inflammatory responses often are regulated incorrectly, which results in chronic inflammation, immunosuppression and ultimately cancer (Okabe and Medzhitov 2016; Greten and Grivennikov 2019). Reduction of inflammation using nonsteroidal, anti- inflammatory drugs such as aspirin is an effective preventative (Castelao et al. 2000; Baron et al. 2003) and co-therapy approach (Todoric et al. 2016; Ritter and Greten 2019). Although miRNA levels and their association with inflammation and cancers have been reported, there is significant under-representation of patients who are not of European ancestry. Few reports have addressed the disproportionate levels of miRNA in AA patients compared to EA patients. It is possible that lack of representation in clinical studies is among the reasons that AA patients experience worse outcomes despite lower incidence.
Studies of miRNA expression often are concerned with the entire cancer tissue and use techniques that do not isolate cancer cells completely, which results in inclusion of stromal cells, vasculature and immune cells that can be confounding factors in efforts to determine true expression of miRNA in cancer cells (Wang et al. 2010). To prevent sample contamination by extracellular matrix/stromal structures, laser capture microdissection (LCM) can be used to isolate tumor cells (Wang et al. 2010; Hackler et al. 2012).
Establishing racial differences in miRNA expression in triple negative breast cancer (TNBC) patients could improve diagnosis and treatment to facilitate determination of outcome associated with race. We investigated differential miRNA expression in TNBC in AA and EA patients using LCM.
Material and methods
Sample collection
We used 30 TNBC specimens acquired from University of Alabama at Birmingham under an approved IRB for specimens collected as part of the tissue repository at the University. To determine potential differences in TNBC miRNA from either AA or EA female patients, the samples were matched for breast cancer subtype, stage (IIB–IIIA), age (+/− 5 years). Both frozen and paraffin embedded specimens were prepared. All 30 patients were classified by race, sex, age, stage (IIIA or IIB) and diagnosis. Two samples were obtained for each specimen, one for frozen section and the other for paraffin embedding (60 total specimens). Fifteen patients were self-identified as AA and 15 were self-identified as EA with a diagnosis of TNBC at stages IIB or IIIA. Patients were 40−55 years old and had a confirmed diagnosis of TBNC at either stage IIB or IIIA. AA samples were selected based on availability of samples large enough for two samples each at least 7 mm. Table 1 is a summary of sample characteristics. Prior to LCM, sections of either frozen or formalin fixed, paraffin embedded (FFPE) were cut at 8 µm from each specimen. Frozen sections remained on dry ice until LCM. Initial sections were stained with hematoxylin and eosin (H & E) (Kerr et al. 2013; Garza-Morales et al. 2018; MacCuaig et al. 2021). A board-certified pathologist (WEG) identified areas of tissues with sufficient malignancy for LCM.
Table 1.
Patient information.
| Patient race | Histology | Stage | Age |
|---|---|---|---|
| AA | TNBC | IIB | 42 |
| AA | TNBC | IIB | 56 |
| AA | TNBC | IIIA | 52 |
| AA | TNBC | IIB | 48 |
| AA | TNBC | IIB | 41 |
| AA | TNBC | IIIA | 50 |
| AA | TNBC | IIB | 48 |
| AA | TNBC | IIB | 46 |
| AA | TNBC | IIIA | 43 |
| AA | TNBC | IIB | 50 |
| AA | TNBC | IIB | 49 |
| AA | TNBC | IIB | 41 |
| AA | TNBC | IIB | 55 |
| AA | TNBC | IIIA | 52 |
| AA | TNBC | IIB | 43 |
| EA | TNBC | IIB | 44 |
| EA | TNBC | IIIA | 46 |
| EA | TNBC | IIB | 50 |
| EA | TNBC | IIIA | 51 |
| EA | TNBC | IIB | 40 |
| EA | TNBC | IIB | 58 |
| EA | TNBC | IIB | 50 |
| EA | TNBC | IIB | 43 |
| EA | TNBC | IIIA | 50 |
| EA | TNBC | IIB | 45 |
| EA | TNBC | IIB | 48 |
| EA | TNBC | IIB | 50 |
| EA | TNBC | IIIA | 49 |
| EA | TNBC | IIB | 47 |
| EA | TNBC | IIIA | 45 |
AA, African American; EA, European American; TBNC, triple negative breast cancer.
LCM
To prepare for LCM, frozen or paraffin tissue sections were dehydrated and stained as follows: 100% EtOH, 1 min; 95% EtOH, 30 sec; 70% EtOH, 30 sec; 50% EtOH, 30 sec; 0.5% crystal violet, 1 min; 50% EtOH, 30 sec; 70% EtOH 30 sec; 95% EtOH, 30 sec; and 100% EtOH, 1 min. Samples were dried completely using a desiccator. Individual tumor cells were selected based upon morphology and collected from the slide using a high energy UV laser pulse to catapult tissue into 10 µl of RNAse-free PCR caps that contained cell lysis buffer. At least a 500,000 µm2 area of cells were collected using LCM. Cell pellets were collected by centrifugation at 100 x g for 5 min at 27 °C and placed on dry ice until extraction of RNA.
RNA extraction from frozen tissue samples
We used the RNAqueous-Micro kit (Ambion Ltd., Cambridgeshire, UK) to extract RNA from frozen samples. The protocol for miRNA isolation has been described by Huang et al. (2013). Briefly, frozen pelleted cells were washed with phosphate buffered saline (PBS), then re-suspended and mixed with 100 µl lysis solution and 50 µl 100% EtOH. The solution was filtered using a microfilter cartridge at 21,330 x g for 1 min. Wash solutions 1, 2 and 3, 180 µl each, were used to wash the sample three times. The filtrate was discarded and the sample was collected by centrifugation at 21,330 x g for 1 min; 10 µl eluates were obtained in micro-elution tubes. The LCM sample was placed in 30 µl lysis solution and incubated for approximately 30 min. The cartridge assembly filter was moistened with 30 µl lysis solution for 5 min and 3 µl LCM additive was introduced and mixed. Then 1.25 volume of 100% EtOH was added and the mixture passed through another microfilter assembly at 10,000 x g for 1 min. Three washes with each of the three solutions, 180 µl each, produced 10 µl of eluted RNA.
RNA extraction from FFPE samples
We used the RecoverAll Total Nucleic Acid Isolation kit (Ambion Ltd., Sydney, NSW) to isolated RNA from FFPE cells as described by Yang et al. (2015) except for the tube type. Instead, a smaller filter cartridge and tube was used (AM1066G; Invitrogen, Green Island, NY) for concentrating a smaller quantity of sample. Briefly, 100 µl digestion buffer and 4 µl protease were mixed with each sample followed by incubation for 15 min at 50 or 80 ºC. Then, 100 µl isolation additive and 275 µl 100% EtOH were added to the filter cartridge followed by centrifugation at 10,000 x g for 30 sec. Samples were washed with 500 µl wash 1 and 200 µl each washes 2 and 3. Solutions were incubated for 30 min at 50 ºC with 60 µl DNAse. Samples were washed again, i.e., 500 µl, wash 1; 200 µl, wash 2; 200 µl, wash 3 and eluted into 5 µl 75 ºC nuclease-free water. Total volume was increased to 10 µl by adding 5 µl nuclease-free water. RNA isolation RIN (RNA Integrity Number) values were consistent with expected results.
MiRNA expression
The TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA) was used to perform reverse transcription with MegaPlex RT primer as described earlier (Mestdagh et al. 2008). Samples,15 µl, were incubated for 40 cycles as follows: 16 ºC for 2 min, 42 ºC for 1 min, 50 ºC for 1 sec, 85 ºC for 5 min and maintaining subsequently at 4 ºC. Pre-amplification used Megaplex Preamp Primer Pools A and B and the following cycle: 95 ºC for 10 min, 55 ºC for 2 min; 72 ºC for 2 min, 12 cycles at 95 ºC for 15 sec, 60 ºC for 4 min, 99.9 ºC for 10 min, then maintained at 4 ºC. The 7300 Real-time PCR System (Applied Biosystems) was used to quantify pre- amplified product in a 384-well Taqman Open Array Real-time PCR plate. We detected 750 miRNA targets in three samples for each run. SDS 1.4 (Applied Biosystems) was used to calculate cycle threshold values. miRNA expression levels in samples were calculated using a standard concentration curve constructed from synthetic miRNA calibration; 2-ΔΔCt was used to determine changes in gene expression.
Statistical analysis
Sex, age, tumor type, stage and pathology were factors in case matching. Log-linear generalized linear models (GLMs) were used to calculate differential expression between FFPE and frozen samples between AA and EA patients. Poisson distribution was used to read counts and the EdgeR package was used to estimate of the gene-wise dispersion factor using the empirical Bayes procedure. A significant difference in linear models was determined by a likelihood ratio test. Heatmaps and PC plots were used to confirm roles in tumor classification. Two-sample t-test for differences in read counts between FFPE vs. frozen and AA or EA was used to determine sample size. Differential miRNA expression in the FFPE and frozen samples from the two patient types were calculated using linear model fitting. Specifically, the linear model was fit to the ΔCt values derived from Taq-Man low density arrays. Controlling the false discovery rate (FDR) to 0.05 was performed to account for multiple comparisons. Values for p ≤ 0.05 were considered significant.
Results
LCM and RNA extraction
Two specimensof RNA were extracted from each of 15 AA and 15 EA patients: 1 FFPE and 1 frozen fixed for each patient, for a total of 60 specimens. Following drying and staining, tumor cells were isolated using LCM as illustrated in Figures 1 and 2. Because TNBC tumors are heterogeneous, separation of tumor cells from inflammatory cells enables assessment of the tumor cell population for specific miRNAs that may be important for tumor initiation and progression. Figures 1 and 2 show H & E staining of a TNBC tissue section containing tumor cells (red circle), inflammatory cells (blue circles) and stroma. Figure 2 shows the process of collecting a homogenous TNBC tumor cell population using LCM without potential contamination from immune cells. Isolated homogenous TNBC tumor cells were collected in PCR caps following LCM. RNA was extracted using FFPE/frozen extraction methods (Huang et al. 2013; Yang et al. 2015).
Figure 1.
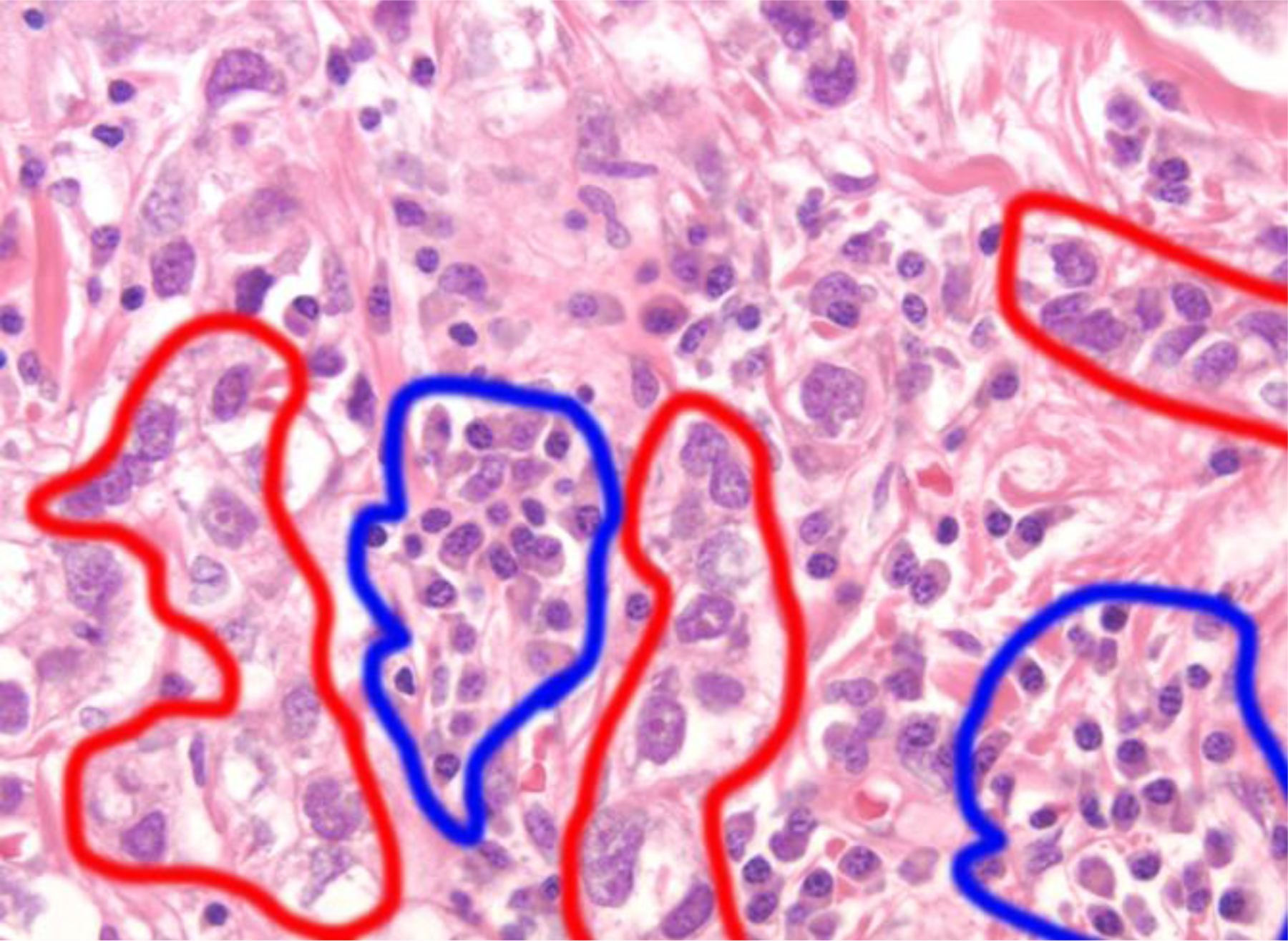
H & E staining of TNBC tissue isolated using laser capture microdissection. Cells outlined in blue are inflammatory cells; cells outlined in red are cancer cells as indicated by enlarged nuclei. 40X magnification.
Figure 2.
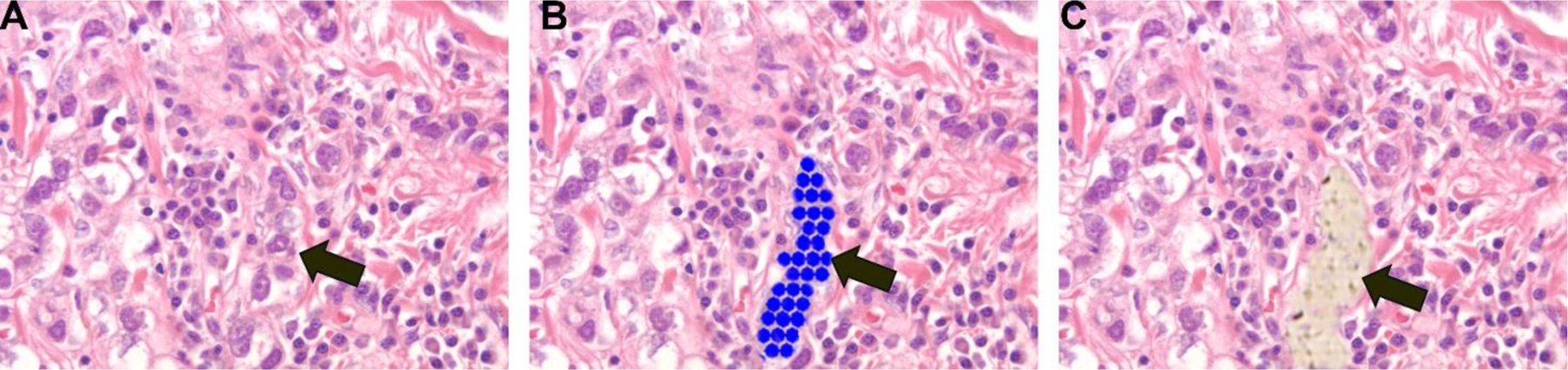
TNBC tumor cells were identified and captured using laser capture microdissection. A) Visualization of initial TNBC tissue. B) Identification of TNBC tumor cells to be captured using LCM. Blue dots denote the area to be collected using LCM and identified with an arrow. C) Arrow shows the complete removal of the target area following microdissection. 40X magnification.
miRNA expression
We assessed 756 miRNAs using open array plates. miRNA expression was evaluated by comparing ethnicity, i.e., AA or EA, and type of fixation, i.e., frozen or FFPE. Significant differences in miRNA expression in FFPE samples of AA and EA TNBC patients were observed (Figure 3A). Specifically, the following miRNAs showed racial disparities: 19a, 30a-30, 95, 192, 206, 211, 302a, 302b, 302c, 335, 367, 372, 375, 520b, 520f 601, 548d, 367c, let7g, and 645. LCM isolated tumor cells also exhibited differential miRNA expression depending on whether the sections were frozen or embedded in paraffin (Figure 3B).
Figure 3.
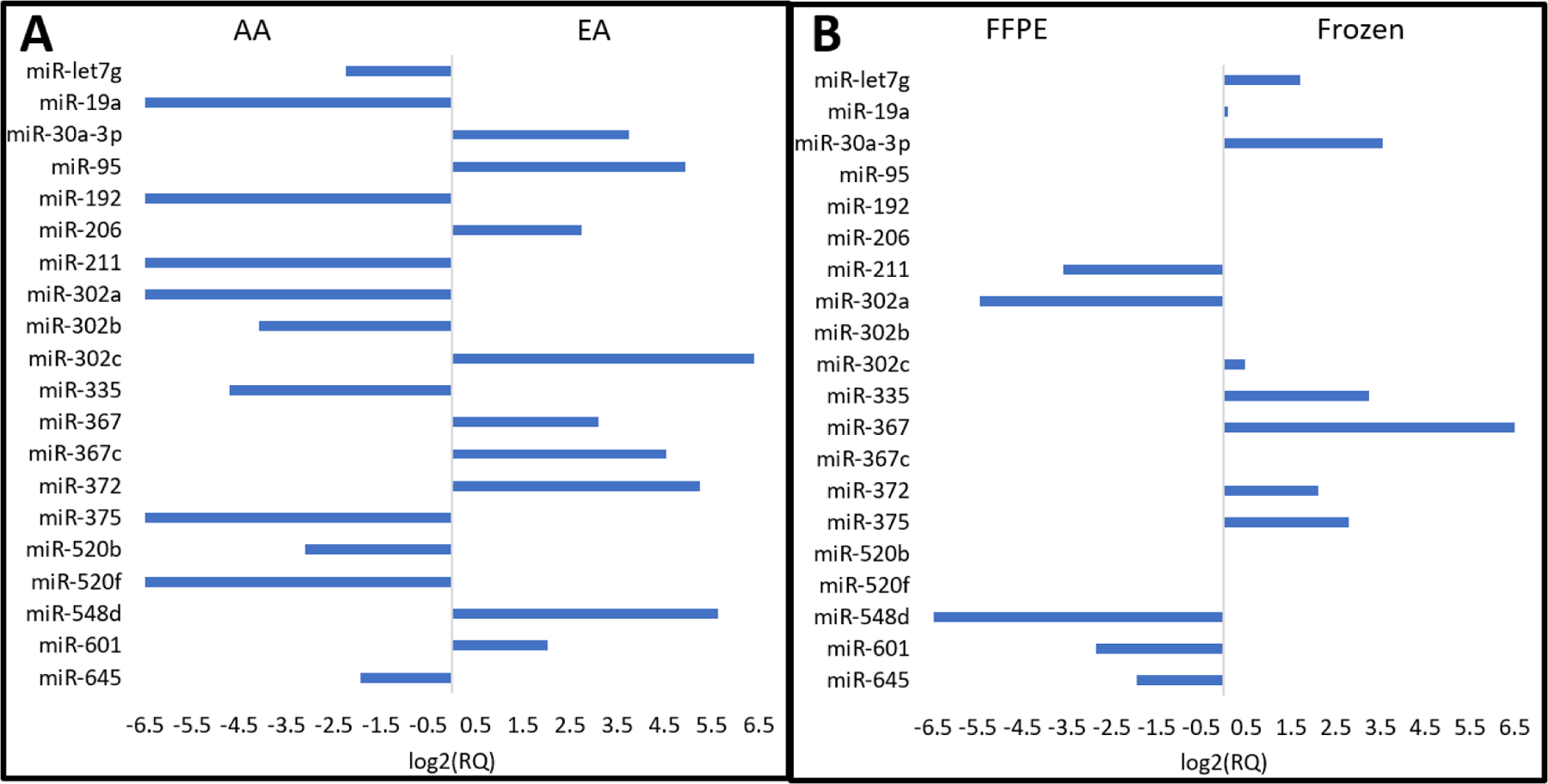
Differential miRNA expression in tumor cells. A) AA patients compared to EA TNBC tumor cells that were preserved using FFPE. B) TNBC tumor cells preserved using formalin fixed, paraffin embedded sections were compared to frozen sections. Data compares method of fixation and contains both AA and EA samples. Selected miRNAs are listed from top to bottom: let7g, 19a, 30a-3p, 95, 192, 206, 211, 302a, 302b, 302c, 335, 367, 367c, 372, 275, 520b, 520f, 548d, 601 and 645. The length of the bar represents increased quantity of difference in miRNA expression.
Discussion
LCM techniques enabled precise isolation of tumor cells while excluding cellular matrix components such as stromal and immune cells. Exclusion of the latter cells enables collection of RNA that is specific to tumor cells, and miRNA extraction and profiling, which improves interpretation concerning miRNA expression. LCM is an important technique for establishing relations between miRNA expression and cancer progression and proliferation (Shi et al. 2008). We found that precise and accurate interpretation of miRNA expression facilitated determination of racial disparities in TNBC. Figure 3A shows results of differential miRNA expression of 20 miRNAs based on patient race: miRNAs 19a, 30a-30, 95, 192, 206, 211, 302a, 302b, 302c, 335, 367, 372, 375, 520b, 520f, 601, 548d, 367c, let7g and 645. The bars in Figure 2A represent increased quantity of specific miRNA in the respective group. Some of these miRNAs have been linked to tumor progression and we report here that they were implicated in racial disparities between AA and EA TNBC patients. Many of the miRNAs implicated in AA, including miRNAs19a, 192, and 335, are linked to immune/inflammatory regulation processes and have been much less documented in relation to breast cancer than other miRNAs that are highly expressed in EA patients. Specifically, we found that miRNA-19a, a member of the miRNA-17–92 cluster, was expressed significantly less in AA patients compared to EA patients. Although this miRNA has not been linked to racial disparities in cancer previously, it has been established as both a critical component in anti-inflammatory response and cancer progression. miRNA-19a expression is significantly greater in drug-resistant gastric and prostate cancer cell lines compared to normal prostate tissue, and transfection with specific miRNA-19a leads to decreased drug sensitivity and inhibition of apoptosis (Wang et al. 2013; Lu et al. 2015). Further, miRNA-19a targets CUL5 and up-regulation of miRNA-19a promotes cell growth and invasion in a cervical carcinoma model (Xu et al. 2012). Because miRNA-19a serum levels are significantly decreased after chemotherapy for breast cancer, miRNA-19a could prove to be a novel cancer specific agent for detecting reduction in tumor following treatment in the future (Sochor et al. 2014). miRNA-19a, together with the remainder of the miRNA-17–92 cluster, participates in B cell development and T cell function; under-expression suggests depression of the immune system, which is conducive to carcinogenesis (Sonkoly and Pivarcsi 2009; Greten and Grivennikov 2019).
miRNAs 192 and 335 exhibited significantly less expression in AA TNBC patients compared to EA patients. Like miRNA-19a, these miRNAs are related to inflammatory processes, which affect development of cancer and its microenvironment both directly and indirectly (Sonkoly and Pivarcsi 2009; Zhu et al. 2014; Arner and Kulyté 2015). For example, miRNA-192 inhibits proliferation and metastasis in colon (Braun et al. 2008) and liver cancers (Yan-Chun et al. 2017) while also regulating inflammation levels in the gastrointestinal system (Sonkoly and Pivarcsi 2009). By targeting chemokine MIP-2α, miRNA-192 greatly affects inflammatory gastrointestinal diseases, e.g., active ulcerative colitis (Wu et al. 2008). Chronic inflammatory diseases related to ulcerative colitis are responsible for many gastrointestinal cancers including colorectal, stomach and liver cancers (Grivennikov et al. 2010; Trinchieri 2012).
miRNA-335 exhibits tumor invasion and metastasis suppressor activity in breast cancer (Heyn et al. 2011; Chen et al. 2016; Dong et al. 2018). Expression of miRNA-335 is decreased significantly in cases of breast cancer (Ali Ahmed et al. 2020); it permits unregulated tumor growth that is exacerbated in AA patients. miRNA-335 also has been implicated in suppression of inflammation; therefore, expression of miRNA-335 may further amplify complications of cancer and obesity by high levels of inflammation (Zhu et al. 2014; Gao et al. 2018; Miller et al. 2021). Inclusion of these miRNAs should be considered for future therapeutic TNBC strategies, because each may participate in progression of cancer both directly and indirectly through inflammatory pathways. The evidence suggests that miRNA expression and subsequent biological events have a disproportionate impact on AA TNBC patients.
We found that the miRNA-302 class (a, b, c) is implicated in racial disparities in TNBC patients. Specifically, miRNA-302a and 302b were down-regulated in AA patients, while miRNA-302c was up-regulated. Up-regulated miRNA-302a is associated with suppression of proliferation and metastasis in colorectal (Wei et al. 2015), prostate (Zhang et al. 2015) and breast (Liang et al. 2014) cancers by regulation of MAPK and AKT signaling pathways and CXC4 expression. These investigators also reported that reduction of miRNA-302a expression resulted in cancer growth and metastasis (Liang et al. 2014; Wei et al. 2015; Zhang et al. 2015). Also, cooperative action of the miRNA-302 class sensitizes breast cancer to adriamycin via the ERK pathway (Zhao et al. 2016). Sensitization is achieved by directly binding to and down-regulating levels of MAP/ERK kinase 1 (MEKK1), which decreases drug resistance of cells due to over-expression of P- glycoprotein (Zhao et al. 2016; Lee et al. 2020). Also, miRNA-302b regulates inflammatory responses (Zhou et al. 2014). Weak expression of inflammatory regulators causes chronic inflammation, and increased risk and difficulty of cancer treatment (Grivennikov et al. 2010; Greten and Grivennikov 2019). Because we observed lower miRNA-302a/b expression in AA patients, we conclude that lack of miRNA- 302a/b expression could be a factor in the poor outcome of AA TNBC patients compared to EA patients due to rapid proliferation, metastasis and complications due to inflammation.
We also found that the class of miRNA-520 (b, f) was less expressed in AA TNBC patients compared to EA patients. miRNA-520b has been identified as a tumor suppressor in several types of malignancies. This function includes inhibition of growth and proliferation in glioblastoma (Liu et al. 2016), hepatoma (Zhang et al. 2012) and colorectal cancers (Xiao et al. 2018) as well as sensitizing breast cancer to drugs (Cui et al. 2010; Keklikoglou et al. 2012). Also, miRNA-520f inhibits cancer cell proliferation and reverses epithelial to mesenchymal transition in several cell models (van Kampen et al. 2017; Du et al. 2018). In addition to direct cancer effects, the miRNA-502 class is associated with insulin insensitivity and inflammation, which are indirect risk factors for cancer (Arner and Kulyté 2015; Guo et al. 2016). Because we observed less expression of both miRNA-502b and 502f in AA TNBC patients, it is possible that these miRNAs participate in the poorer outlook for AA patients with TNBC.
In addition to the miRNAs that are under-expressed in AA TNBC patients, others exhibited increased expression, particularly, miRNA-95 and 548d in EA patients compared to AA patients. miRNA-95 influences radiation sensitivity (Ma et al. 2016). miRNA-95 causes resistance to radiation and over-expression causes a highly proliferative, invasive phenotype (Huang et al. 2013). Although miRNA-95 generally is up-regulated in breast cancer, the racial disparity in breast cancer suggests the need for further exploration.
miRNA-548d also is associated with AA TNBC patients. Strong expression of miRNA-548d promotes proliferation and suppresses apoptosis both in vitro and in vivo (Song et al. 2016; Zhao et al. 2016). Little is known about miRNA-548d expression, but it is expressed strongly in AA compared to EA TNBC patients. Inclusion of miRNAs such as miRNA-95 and 548d must be considered together with other differentially expressed miRNAs as potential diagnostic and therapeutic tools for TNBC that could decrease differences in racial outcomes.
Prostate cancer exhibits differences in miRNA expression between AA and EA patients. Over-expression of miRNA-26a (Theodore et al. 2010) and IL-16 miRNA (Hughes et al. 2013) in AA prostate cancer patients has been implicated in accelerated onset of carcinogenesis as well as specific miRNA-mRNA pairings (Wang et al. 2015), epigenetic factors (Theodore et al. 2014) and increased inflammation in the tumor microenvironment (Wallace et al. 2008). miRNA-182 has been implicated in lower survival of AA colon cancer patients compared to EA colon cancer patients (Li et al. 2014). Although miRNA and other tumor microenvironmental factors have been linked to inflammation and overall breast cancer development (Iorio et al. 2005; Martin et al. 2009; Dietze et al. 2015), the need remains to characterize further the differences in AA TNBC compared to other populations. We found differential miRNA expression levels in TNBC patients, but more studies are required to incorporate our findings into diagnostic and therapeutic techniques to improve outcomes.
Figure 3B compares the quantity of miRNA measured in FFPE tissue vs. frozen tissue regardless of race. These data may be used to determine the location of miRNA. Historically, frozen samples were used for determination of mRNA, because FFPE tissue samples produced poor quality extracted RNA due to rapid degradation and chemical modification (Masuda et al. 1999; Srinivasan et al. 2002). miRNAs, however, are a class of small RNAs that have the potential for increased survivability and resistance to alterations when prepared as FFPE samples (Li et al. 2007). Our findings suggest that the quantity of miRNAs is greater when prepared as FFPE samples compared to frozen samples, but this not consistent for all miRNAs measured. Fenestrations in cellular membranes due to freezing samples could explain the decreased miRNA l in the cytosol. For example, miRNA 548d was less common in frozen samples than in FFPE samples, which suggests that miRNA 548d is a cytosol miRNA and that it leaked from the cytosol through the cell membrane. By contrast, miRNA 367 may be a nuclear miRNA, because large amounts were found in frozen samples. This suggests that both frozen and FFPE techniques are useful for interpreting miRNA abundance.
We identified racial disparities in miRNA expression in AA and EA TNBC patients with emphasis on miRNAs that have been shown to be related to cancer and related inflammatory regulation pathways. Our findings could be exploited to improve diagnosis and therapeutic targets to decrease disparities in TNBC outcomes associated with race. We also provide evidence for the critical need to include patients of a variety of ethnic backgrounds in studies to characterize biomarker association with cancers.
Funding
This research was funded by the National Institute of Health grants R01CA205941 and R01CA212350.
Footnotes
Declaration of interest
The authors report no conflict of interest.
References
- Ali Ahmed E, Abd El-Basit SA, Mohamed MA, Swellam M. 2020. Clinical role of MiRNA 29a and MiRNA 335 on breast cancer management: their relevance to MMP2 protein level. Arch Physiol Biochem 1–8. [DOI] [PubMed] [Google Scholar]
- Arner P, Kulyté A. 2015. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol 11:276–2015. [DOI] [PubMed] [Google Scholar]
- Banegas MP, Li CI. 2012. Breast cancer characteristics and outcomes among Hispanic Black and Hispanic White women. Breast Cancer Res Treat 134:1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron JA, Cole BF, Sandler RS, Haile RW, Ahnen D, Bresalier R, McKeown-Eyssen G, Summers RW, Rothstein R, Burke CA. 2003. A randomized trial of aspirin to prevent colorectal adenomas. N Engl J Med 348:891–899. [DOI] [PubMed] [Google Scholar]
- Braun CJ, Zhang X, Savelyeva I, Wolff S, Moll UM, Schepeler T, Ørntoft TF, Andersen CL, Dobbelstein M. 2008. p53-Responsive microRNAs 192 and 215 are capable of inducing cell cycle arrest. Cancer Res 68:10094–10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S. 2006. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. J Am Med Assoc 295:2492–2502. [DOI] [PubMed] [Google Scholar]
- Castelao J, Yuan J, Gago-Dominguez M, Yu M, Ross R. 2000. Non-steroidal anti-inflammatory drugs and bladder cancer prevention. Br J Cancer 82:1364–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Song Y, Lu Y, Zheng W, Ma W, Zhang C. 2016. miR-335 inhibits cell proliferation, migration and invasion in HeLa cervical cancer cells. Int J Clin Exp Pathol 9:10351–10362. [Google Scholar]
- Cui W, Zhang Y, Hu N, Shan C, Zhang S, Zhang W, Zhang X, Ye L. 2010. miRNA-520b and miR-520e sensitize breast cancer cells to complement attack via directly targeting 3′ UTR of CD46. Cancer Biol Ther 10:232–241. [DOI] [PubMed] [Google Scholar]
- Danforth DN Jr. 2013. Disparities in breast cancer outcomes between Caucasian and African American women: a model for describing the relationship of biological and nonbiological factors. Breast Cancer Res 15:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh SK, Srivastava SK, Tyagi N, Ahmad A, Singh AP, Ghadhban AA, Dyess DL, Carter JE, Dugger K, Singh S. 2017. Emerging evidence for the role of differential tumor microenvironment in breast cancer racial disparity: a closer look at the surroundings. Carcinogenesis 38:757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietze EC, Sistrunk C, Miranda-Carboni G, O’regan R, Seewaldt VL. 2015. Triple-negative breast cancer in African American women: disparities versus biology. Nat Rev Cancer 15:248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Liu Y, Jiang A, Li R, Yin M, Wang Y. 2018. MicroRNA-335 suppresses the proliferation, migration, and invasion of breast cancer cells by targeting EphA4. Mol Cell Biochem 439:95–104. [DOI] [PubMed] [Google Scholar]
- Du X, Fan W, Chen Y. 2018. microRNA-520f inhibits hepatocellular carcinoma cell proliferation and invasion by targeting TM4SF1. Gene 657:30–38. [DOI] [PubMed] [Google Scholar]
- Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3:87–98. [DOI] [PubMed] [Google Scholar]
- Gao X-L, Li J-Q, Dong Y-T, Cheng E-J, Gong J-N, Qin Y-L, Huang Y-Q, Yang J-J, Wang S-J, An D-D. 2018. Upregulation of microRNA-335–5p reduces inflammatory responses by inhibiting FASN through the activation of AMPK/ULK1 signaling pathway in a septic mouse model. Cytokine 110:466–478. [DOI] [PubMed] [Google Scholar]
- Garza-Morales R, Gonzalez-Ramos R, Chiba A, de Oca-Luna RM, McNally L, McMasters K, Gomez-Gutierrez J. 2018. Temozolomide enhances triple-negative breast cancer virotherapy in vitro. Cancers 10:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, Volinia S, Bhatt D, Alder H, Marcucci G. 2006. MicroRNA fingerprints during human megakaryocytopoiesis. Proc Natl Acad Sci. USA 103:5078–5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greten FR, Grivennikov SI. 2019. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity 51:27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. 2010. Immunity, inflammation, and cancer. Cell 140:883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Zhang Y, Zhang L, Huang F, Li J, Wang S. 2016. MicroRNAs, TGF-β signaling, and the inflammatory microenvironment in cancer. Tumor Biol 37:115–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackler L, Masuda T, Oliver VF, Merbs SL, Zack DJ. 2012. Use of laser capture microdissection for analysis of retinal mRNA/miRNA expression and DNA methylation. Retinal Development Springer; Humana Press, Totowa, NJ. p. 289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Moore JX, Langston M, Fuzzell L, Khan S, Lewis MW, Colditz GA, Liu Y. 2019. Do breast quadrants explain racial disparities in breast cancer outcomes? Cancer Causes Contr 30:1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Engelmann M, Schreek S, Ahrens P, Lehmann U, Kreipe H, Schlegelberger B, Beger C. 2011. MicroRNA miR‐335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int J Cancer 129:2797–2806. [DOI] [PubMed] [Google Scholar]
- Howlader N, Altekruse SF, Li CI, Chen VW, Clarke CA, Ries LA, Cronin KA. 2014. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J Natl Cancer Inst 106: dju055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Egger M, Grizzle W, McNally L. 2013. MicroRNA-100 regulates IGF1-receptor expression in metastatic pancreatic cancer cells. Biotech Histochem 88:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Taeb S, Jahangiri S, Emmenegger U, Tran E, Bruce J, Mesci A, Korpela E, Vesprini D, Wong CS. 2013. miRNA-95 mediates radio-resistance in tumors by targeting the sphingolipid phosphatase SGPP1. Cancer Res 73:6972–6986. [DOI] [PubMed] [Google Scholar]
- Hughes L, Ruth K, Rebbeck TR, Giri VN. 2013. Genetic variation in IL-16 miRNA target site and time to prostate cancer diagnosis in African American men. Prost Cancer Prost Dis 16:308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu C-G, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M. 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65:7065–7070. [DOI] [PubMed] [Google Scholar]
- Keklikoglou I, Koerner C, Schmidt C, Zhang J, Heckmann D, Shavinskaya A, Allgayer H, Gückel B, Fehm T, Schneeweiss A. 2012. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene 31:4150–4163. [DOI] [PubMed] [Google Scholar]
- Kerr EH, Frederick PJ, Egger ME, Stockard CR, Sellers J, DellaManna D, Oelschlager DK, Amm HM, Eltoum I-E, Straughn JM, Buchsbaum DJ, Grizzle WE, McNally LR. 2013. Lung resistance-related protein (LRP) expression in malignant ascitic cells as a prognostic marker for advanced ovarian serous carcinoma. Ann Surg. Onc 20:3059–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Rauch J, Kolch W. 2020. Targeting MAPK signaling in cancer: mechanisms of drug resistance and sensitivity. Int J Mol Sci 21:1102–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Ji P, Ouyang N, Zhang Y, Wang XY, Rubin DC, Davidson NO, Bergamaschi R, Shroyer KR, Burke S. 2014. Differential expression of miRNAs in colon cancer between African and Caucasian Americans: implications for cancer racial health disparities. Int J Oncol 45:587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Smyth P, Flavin R, Cahill S, Denning K, Aherne S, Guenther SM, O’Leary JJ, Sheils O. 2007. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol 7:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, Bian X, Shim H. 2014. Inhibition of breast cancer metastasis with microRNA-302a by downregulation of CXCR4 expression. Breast Cancer Res Treat 146:535–542. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433:769–773. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang F, Tian L, Wang T, Zhang W, Li B, Bai Y-a. 2016. MicroRNA-520b affects the proliferation of human glioblastoma cells by directly targeting cyclin D1. Tumor Biol 37:7921–7928. [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. 2005. MicroRNA expression profiles classify human cancers. Nature 435:834–838. [DOI] [PubMed] [Google Scholar]
- Lu K, Liu C, Tao T, Zhang X, Zhang L, Sun C, Wang Y, Chen S, Xu B, Chen M. 2015. MicroRNA- 19a regulates proliferation and apoptosis of castration-resistant prostate cancer cells by targeting BTG1. FEBS Lett 589:1485–1490. [DOI] [PubMed] [Google Scholar]
- Ma W, Ma C, Li X, Zhang Y. 2016. Examining the effect of gene reduction in miR-95 and enhanced radiosensitivity in non-small cell lung cancer. Cancer Gene Ther 23:66–71. [DOI] [PubMed] [Google Scholar]
- MacCuaig WM, Fouts BL, McNally MW, Grizzle WE, Chuong P, Samykutty A, Mukherjee P, Li M, Jasinski JB, Behkam B, McNally LR. 2021. Active targeting significantly outperforms nanoparticle size in facilitating tumor-specific uptake in orthotopic pancreatic cancer. ACS Appl. Mater. Interf 13: 49614–49630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane L-A, R Murphy P. 2010. MicroRNA: biogenesis, function and role in cancer. Curr Genom 11:537–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DN, Boersma BJ, Yi M, Reimers M, Howe TM, Yfantis HG, Tsai YC, Williams EH, Lee DH, Stephens RM. 2009. Differences in the tumor microenvironment between African American and European American breast cancer patients. PloS One 4(2):e4531–e4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. 1999. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucl Acids Res 27:4436–4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally LR, Manne U, Grizzle WE. 2013. Post-transcriptional processing of genetic information and its relation to cancer. Biotech Histochem 88:365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh P, Feys T, Bernard N, Guenther S, Chen C, Speleman F, Vandesompele J. 2008. High- throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucl Acids Res 36:e143–e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller B, Chalfant H, Thomas A, Wellberg E, Henson C, McNally MW, Grizzle WE, Jain A, McNally LR. 2021. Diabetes, obesity, and inflammation: impact on clinical and radiographic features of breast cancer. Int J Mol Sci 22:2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GJ, Mitchell EP. 2008. Higher incidence of aggressive breast cancers in African American women: a review. J Natl Med Assoc 100:698–702. [DOI] [PubMed] [Google Scholar]
- Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. 2006. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol 8:278–284. [DOI] [PubMed] [Google Scholar]
- Okabe Y, Medzhitov R. 2016. Tissue biology perspective on macrophages. Nat Immunol 17:9–17. [DOI] [PubMed] [Google Scholar]
- Ooi SL, Martinez ME, Li CI. 2011. Disparities in breast cancer characteristics and outcomes by race/ethnicity. Breast Cancer Res Treat 127:729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter B, Greten FR. 2019. Modulating inflammation for cancer therapy. J Exp Med 216:1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi XB, Tepper CG, White RW. 2008. MicroRNAs and prostate cancer. J Cell Mol Med 12:1456–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sochor M, Basova P, Pesta M, Dusilkova N, Bartos J, Burda P, Pospisil V, Stopka T. 2014. Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BioMed Cent Cancer 14:448–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Song J, Wang Q, Ma Y, Sun N, Ma J, Chen Q, Xia G, Huo Y, Yang L. 2016. miR‐548d‐ 3p/TP 53 BP 2 axis regulates the proliferation and apoptosis of breast cancer cells. Cancer Med 5:315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E, Pivarcsi A. 2009. MicroRNAs in inflammation. Int Rev Immunol 28:535–561. [DOI] [PubMed] [Google Scholar]
- Theodore SC, Rhim JS, Turner T, Yates C. 2010. MiRNA 26a expression in a novel panel of African American prostate cancer cell lines. Ethn Dis 20(Suppl. 1):96–100. [PMC free article] [PubMed] [Google Scholar]
- Srinivasan M, Sedmak D, Jewell S. 2002. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol 161:1961–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodore SC, Davis M, Zhao F, Wang H, Chen D, Rhim J, Dean-Colomb W, Turner T, Ji W, Zeng G. 2014. MicroRNA profiling of novel African American and Caucasian prostate cancer cell lines reveals a reciprocal regulatory relationship of miR-152 and DNA methyltranferase 1. Oncotarget 5:3512–3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Rhoads A, Pinkerton E, Schroeder MC, Conway KM, Hundley WG, McNally LR, Oleson J, Lynch CF, Romitti PA. 2019. Incidence and survival among young women with stage I–III breast cancer: SEER 2000–2015. J Natl Cancer Inst Cancer Spect 3:pkz040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todoric J, Antonucci L, Karin M. 2016. Targeting inflammation in cancer prevention and therapy. Cancer Prevent Res 9:895–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinchieri G. 2012. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Ann Rev Immunol 30:677–706. [DOI] [PubMed] [Google Scholar]
- van Kampen JG, van Hooij O, Jansen CF, Smit FP, van Noort PI, Schultz I, Schaapveld RQ, Schalken JA, Verhaegh GW. 2017. miRNA-520f reverses epithelial-to-mesenchymal transition by targeting ADAM9 and TGFBR2. Cancer Res 77:2008–2017. [DOI] [PubMed] [Google Scholar]
- Wallace TA, Prueitt RL, Yi M, Howe TM, Gillespie JW, Yfantis HG, Stephens RM, Caporaso NE, Loffredo CA, Ambs S. 2008. Tumor immunobiological differences in prostate cancer between African American and European American men. Cancer Res 68:927–936. [DOI] [PubMed] [Google Scholar]
- Wang B-D, Ceniccola K, Yang Q, Andrawis R, Patel V, Ji Y, Rhim J, Olender J, Popratiloff A, Latham P. 2015. Identification and functional validation of reciprocal microRNA-mRNA pairings in African American prostate cancer disparities. Clin Cancer Res 21:4970–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Li T, Zhang B, Li H, Wu Q, Yang L, Nie Y, Wu K, Shi Y, Fan D. 2013. MicroRNA-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochem Biophys Res Commun 434:688–694. [DOI] [PubMed] [Google Scholar]
- Wang S, Wang L, Zhu T, Gao X, Li J, Wu Y, Zhu H. 2010. Improvement of tissue preparation for laser capture microdissection: application for cell type-specific miRNA expression profiling in colorectal tumors. BioMed Cent Genom 11:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z-J, Tao M-L, Zhang W, Han G-D, Zhu Z-C, Miao Z-G, Li J-Y, Qiao Z-B. 2015. Up- regulation of microRNA-302a inhibited the proliferation and invasion of colorectal cancer cells by regulation of the MAPK and PI3K/Akt signaling pathways. Int J Clin Exp Pathol 8:4481–4491. [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. 2008. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2α. Gastroenterology 135:1624–1635. [DOI] [PubMed] [Google Scholar]
- Xiao J, Li G, Zhou J, Wang S, Liu D, Shu G, Zhou J, Ren F. 2018. MicroRNA-520b functions as a tumor suppressor in colorectal cancer by inhibiting defective in cullin neddylation 1 domain containing 1 (DCUN1D1). Oncol Res Preclin Clin Cancer Ther 26:593–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X-M, Wang X-B, Chen M-M, Liu T, Li Y-X, Jia W-H, Liu M, Li X, Tang H. 2012. MicroRNA- 19a and-19b regulate cervical carcinoma cell proliferation and invasion by targeting CUL5. Cancer Lett 322:148–158. [DOI] [PubMed] [Google Scholar]
- Yan-Chun L, Hong-Mei Y, Zhi-Hong C, Qing H, Yan-Hong Z, Ji-Fang W. 2017. MicroRNA-192– 5p promote the proliferation and metastasis of hepatocellular carcinoma cell by targeting SEMA3A. Appl Immunohistochem Mol Morphol 25:251–260. [DOI] [PubMed] [Google Scholar]
- Yang X-D, Xu X-H, Zhang S-Y, Wu Y, Xing C-G, Ru G, Xu H-T, Cao J-P. 2015. Role of miR-100 in the radio resistance of colorectal cancer cells. Am J Cancer Res 5:545–559. [PMC free article] [PubMed] [Google Scholar]
- Zhang G-M, Bao C-Y, Wan F-N, Cao D-L, Qin X-J, Zhang H-L, Zhu Y, Dai B, Shi G-H, Ye D-W. 2015. MicroRNA-302a suppresses tumor cell proliferation by inhibiting AKT in prostate cancer. PLoS One 10(4): e0124410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Kong G, Zhang J, Wang T, Ye L, Zhang X. 2012. MicroRNA-520b inhibits growth of hepatoma cells by targeting MEKK2 and cyclin D1. PloS One 7(2): e31450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Wang T, Huang Q-K, Pu M, Sun W, Zhang Z-C, Ling R, Tao K-S. 2016. MicroRNA- 548a-5p promotes proliferation and inhibits apoptosis in hepatocellular carcinoma cells by targeting Tg737. World J Gastroenterol 22:5364–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Wang Y, Jiang L, He M, Bai X, Yu L, Wei M. 2016. MiR-302a/b/c/d cooperatively sensitizes breast cancer cells to adriamycin via suppressing P-glycoprotein (P-gp) by targeting MAP/ERK kinase 1 (MEKK1). J Exp Clin Cancer Res 35:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Li X, Ye Y, Zhao K, Zhuang Y, Li Y, Wei Y, Wu M. 2014. MicroRNA-302b augments host defense to bacteria by regulating inflammatory responses via feedback to TLR/IRAK4 circuits. Nat Commun 5:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Chen L, Shi C-M, Xu G-F, Xu L-L, Zhu L-L, Guo X-R, Ni Y, Cui Y, Ji C. 2014. MiR-335, an adipogenesis-related microRNA, is involved in adipose tissue inflammation. Cell Biochem Biophys 68:283–290. [DOI] [PubMed] [Google Scholar]


