Abstract
Background
Favipiravir, an oral, RNA-dependent RNA polymerase inhibitor, has in vitro activity against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Despite limited data, favipiravir is administered to patients with coronavirus disease 2019 (COVID-19) in several countries.
Methods
We conducted a phase 2, double-blind, randomized controlled outpatient trial of favipiravir in asymptomatic or mildly symptomatic adults with a positive SARS-CoV-2 reverse-transcription polymerase chain reaction assay (RT-PCR) within 72 hours of enrollment. Participants were randomized to receive placebo or favipiravir (1800 mg twice daily [BID] day 1, 800 mg BID days 2–10). The primary outcome was SARS-CoV-2 shedding cessation in a modified intention-to-treat (mITT) cohort of participants with positive enrollment RT-PCRs. Using SARS-CoV-2 amplicon-based sequencing, we assessed favipiravir’s impact on mutagenesis.
Results
We randomized 149 participants with 116 included in the mITT cohort. The participants’ mean age was 43 years (standard deviation, 12.5 years) and 57 (49%) were women. We found no difference in time to shedding cessation overall (hazard ratio [HR], 0.76 favoring placebo [95% confidence interval {CI}, .48–1.20]) or in subgroups (age, sex, high-risk comorbidities, seropositivity, or symptom duration at enrollment). We detected no difference in time to symptom resolution (initial: HR, 0.84 [95% CI, .54–1.29]; sustained: HR, 0.87 [95% CI, .52–1.45]) and no difference in transition mutation accumulation in the viral genome during treatment.
Conclusions
Our data do not support favipiravir at commonly used doses in outpatients with uncomplicated COVID-19. Further research is needed to ascertain if higher favipiravir doses are effective and safe for patients with COVID-19.
Clinical Trials Registration
Keywords: COVID-19, SARS-CoV-2, favipiravir, clinical trial
In this phase 2, double-blind, randomized controlled outpatient trial of favipiravir in patients with asymptomatic or uncomplicated COVID-19, we found no difference in time to shedding cessation or time to symptom resolution by treatment arm.
Favipiravir is an oral, RNA-dependent RNA polymerase (RdRp) inhibitor with a wide spectrum of activity, including in vitro activity against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In its active form, favipiravir is incorporated into nascent viral RNA by error-prone viral RdRp disrupting RNA synthesis directly by chain termination or accumulation of deleterious mutations in the SARS-CoV-2 genome [1]. Since 2014, favipiravir has been used in Japan and China for patients with drug-resistant influenza and boasts an established, well-characterized safety profile, making it an attractive potential therapy for coronavirus disease 2019 (COVID-19).
Early data from some open-label trials suggested that favipiravir improved clinical and/or virologic outcomes in patients with COVID-19 [2, 3]. Despite limited data, favipiravir was approved in patients with COVID-19 in some countries. We evaluated favipiravir's efficacy in reducing viral shedding duration and improving symptoms in outpatients with uncomplicated COVID-19.
METHODS
Study Design
We conducted a phase 2, double-blind, randomized, placebo-controlled phase 2 trial at Stanford Healthcare, California. The Stanford University School of Medicine Panel on Human Subjects in Medical Research approved the study protocol. An independent data and safety monitoring board (DSMB) reviewed the study design, trial progress, study integrity, and safety data including interim analysis.
Participants
We enrolled asymptomatic or symptomatic adults without respiratory distress who had a positive SARS-CoV-2 reverse-transcription polymerase chain reaction assay (RT-PCR) collected within 72 hours of enrollment. We excluded individuals who required renal replacement therapy, had liver impairment, were immunocompromised, or were pregnant or breastfeeding (see Supplementary Appendix 1 for full criteria). Participants were randomized 1:1 to favipiravir or placebo using block, Research Electronic Data Capture (REDCap)–implemented, randomization stratified by age (≥50 and <50 years) and sex [4, 5].
Procedures
Participants received placebo or favipiravir 1800 mg twice daily (BID) on day 1, then 800 mg BID on days 2–10. Favipiravir and placebo tablets were identical in appearance to maintain blinding.
We followed participants for 28 days and performed a clinical assessment (including vital signs and targeted physical examinations) and collected oropharyngeal swabs and blood samples at each visit. Staff-collected oropharyngeal specimens underwent RT-PCR (Viroclinics Biosciences, Rotterdam, The Netherlands). Anti-SARS-CoV-2 serology was performed using a virus plaque reduction neutralization assay (Viroclinics Biosciences, Rotterdam, The Netherlands).
Participants self-collected daily anterior nasal swabs on days 1–10, 14, 21, and 28 and submitted them directly for RT-PCR with an assay that targeted the viral nucleocapsid gene's N1 and N3 regions (Quest Diagnostics, Secaucus, New Jersey).
Participants also completed electronic daily symptom surveys and recorded temperature and oxygen saturation using study-provided devices; data were collected using REDCap Cloud version 1.6 (REDCap Cloud, Encinitas, California).
Outcomes
We defined the primary outcome, SARS-CoV-2 shedding cessation, as time from enrollment to the first of 2 consecutive negative nasal RT-PCRs. We defined time until initial resolution of symptoms as time from randomization until the first of 2 consecutive days without symptoms. We defined time until sustained symptom resolution similarly, with the additional condition that symptoms remain resolved throughout the remainder of the study. Decreased taste/smell, mild fatigue, and mild cough were recorded, but excluded for this analysis (Supplementary Appendix 2) [6]. We censored participants who did not meet the symptom endpoint on their last completed survey. Additional secondary outcomes included incidence of hospitalizations or emergency department visits during the study and adverse events graded for severity [7].
Sample qPCR Testing and Sequencing Protocols
To test whether favipiravir was acting as a mutagen, 1 of its mechanisms of action [1], we sequenced SARS-CoV-2 from residual day 1, 5 and 10 participant nasal swabs using an Illumina MiSeq platform (Supplementary Methods).
Statistical Analysis
We assessed virologic outcomes in a modified intention-to-treat (mITT) cohort, which included all randomized participants whose first available nasal RT-PCR result on days 1–3 was positive. We assessed symptom outcomes in a symptomatic mITT [smITT] cohort, which included all randomized participants who reported ≥1 symptom at enrollment excluding mild cough, mild fatigue, or decreased taste/smell. We assessed safety endpoints in the intention-to-treat (ITT) cohort and adjusted all analyses for age group and sex. Unless otherwise noted, all tests were 2-sided and conducted at an α level of .05. Analyses were performed in R version 4.0.2 software [4, 5].
Primary Analysis
We used a Cox proportional hazards model to compare time until shedding cessation between arms. The final test was performed at the α = .04999 level, allowing for an interim analysis. We censored participants who did not meet the endpoint on the last positive RT-PCR date and verified the proportional hazards assumption by examining Schoenfeld residuals.
Secondary Analyses
We used a Cox proportional hazards model to compare initial and sustained symptom resolution between arms and Fisher's exact test to compare proportions. We evaluated change in cycle threshold (Ct) from day 1 to day 7 or day 10 by treatment arm using generalized linear mixed-effects regression models and defined “reverse Ct” by subtracting the Ct value from 40 (the detection limit, Supplementary Methods).
Post Hoc and Efficacy Sensitivity Analyses
We added a statistical interaction term between arm and these baseline characteristics to the primary efficacy model to test for effect modification: seropositivity; high-risk status; symptom onset within 3, 5, and 7 days of enrollment; age group; and sex. We classified participants as high risk if they met any of these criteria: age ≥65 years, BMI ≥35 kg/m2, chronic kidney disease, diabetes mellitus, or age ≥55 years plus 1 of the following comorbidities: cardiovascular disease, hypertension, or chronic respiratory disease. We added interaction terms to the sustained symptom resolution model for high-risk status and symptom onset within 3 and 5 days of enrollment. We reported P values from a Wald test corresponding to interaction terms and within-subgroup hazard ratios.
Sample Size Determination
Assuming 1:1 randomization and a 2-sided log-rank test at the α = .04999 level for the final analysis, we anticipated 79 shedding cessation events, which provided 80% power to detect a hazard ratio of 2.03. We additionally assumed a median of 14 and 7 days to shedding cessation in the control and treatment arms, respectively; 3-month accrual period; 4-week follow-up period after randomization of the last patient; and 10% dropout in the control arm. This enabled an interim analysis conducted at α = .00001 to assess overwhelming efficacy after 50% of participants completed 24 hours of follow-up. We estimated that the total sample size required to achieve 79 events was 120 (60 participants per arm).
At interim review, the DSMB recommended increasing the sample size with the goal of 120 participants in the mITT cohort.
Variant Identification
We used the nfcore/viralrecon v.2.3dev bioinformatic pipeline to perform variant calling and to generate consensus sequences from raw reads (Supplementary Methods) [8]. We predicted that favipiravir would impact viral diversity by study day 5 and result in a higher transition mutation rate [1, 9].
To assess favipiravir's impact on SARS-CoV-2 within-host diversity, we tested if the number of intrahost single nucleotide variants (iSNVs), transitions, and/or either iSNVs and transitions standardized by the total number of bases sequenced in a sample differed between the treatment arms on day 5 using 2-sided t tests with R package rstatix [10]. To standardize by sequencing effort, we divided the number of iSNVs identified by the number of sequenced base-pairs, the product of read-length and number of mapped reads, for each sample. We fit independent linear models for number of iSNVs, standardized number of iSNVs, number of transitions, and standardized number of transitions with study day and treatment group as predictor variables in the R package stats [11]. We used a P value threshold of .05 to identify predictors significantly associated with within-host viral diversity.
RESULTS
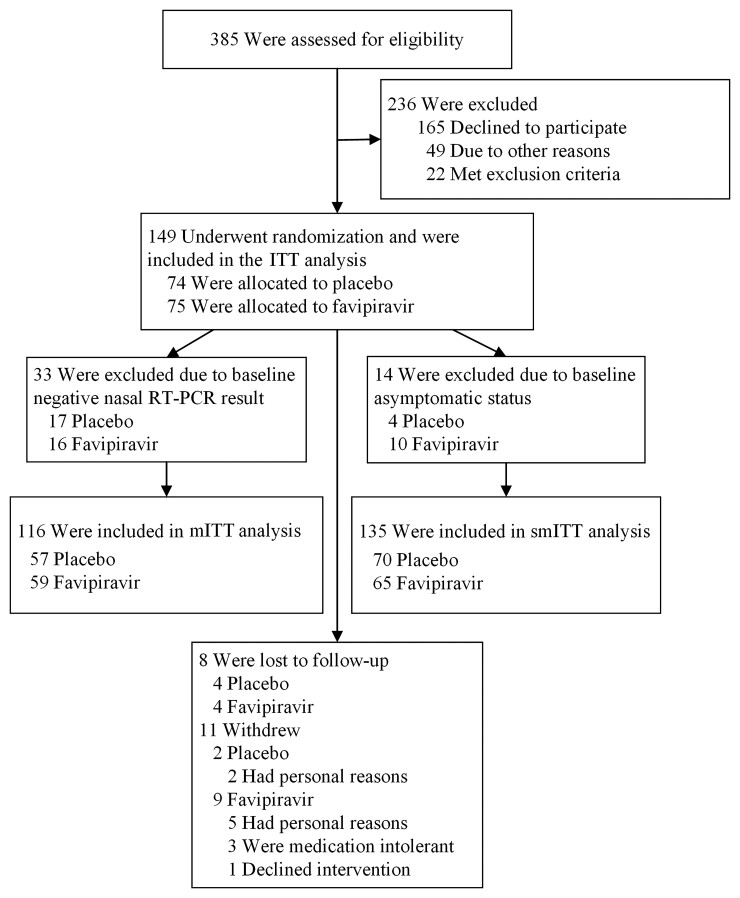
From 8 July 2020 to 23 March 2021, we screened 385 patients and randomized 149 who were included in the ITT cohort (74 placebo, 75 favipiravir; Figure 1). Of these, 116 and 135 were included in the mITT and smITT cohorts, respectively; 112 participants were included in all 3 analytic cohorts (Supplementary Figure 1).
Figure 1.
Consolidated Standards of Reporting Trials (CONSORT) diagram. Trial schematic showing participants screened, randomized, and followed through study completion. Two of the 3 participants randomized to receive favipiravir withdrew due to nausea and dizziness. Abbreviations: ITT, intention-to-treat; mITT, modified intention-to-treat; RT-PCR, reverse-transcription polymerase chain reaction; smITT, symptomatic modified intention-to-treat.
Baseline demographic and disease characteristics were balanced between the 2 groups in all analytic cohorts (Table 1). In the mITT cohort, 31% of participants had ≥1 comorbidity of interest, and 37% had a body mass index ≥30 kg/m2. Of those with a positive RT-PCR upon enrollment, the median Ct was 24 (interquartile range, 21–28) for the N1 target, and only 10 participants had detectable antibodies (placebo 4, favipiravir 6).
Table 1.
Baseline Characteristics
| Characteristic | mITT (n = 116) | smITT (n = 135) | ||||
|---|---|---|---|---|---|---|
| Placebo (n = 57) | Favipiravir (n = 59) | SMD | Placebo (n = 70) | Favipiravir (n = 65) | SMD | |
| Age at randomization, y, mean (SD) | 43.4 (12.8) | 42.9 (12.3) | 0.04 | 42.8 (12.6) | 42.5 (12.0) | 0.03 |
| Female sex | 29 (50.9) | 28 (47.5) | 0.07 | 37 (52.9) | 32 (49.2) | 0.07 |
| Race/ethnicity | 0.14 | 0.20 | ||||
| Latinx | 24 (42.1) | 26 (44.1) | 29 (41.4) | 28 (43.1) | ||
| White | 21 (36.8) | 19 (32.2) | 26 (37.1) | 22 (33.8) | ||
| Asian | 5 (8.8) | 6 (10.2) | 7 (10.0) | 6 (9.2) | ||
| Native Hawaiian/Pacific Islander | 1 (1.8) | 2 (3.4) | 1 (1.4) | 3 (4.6) | ||
| Other/unknown | 6 (10.5) | 6 (10.2) | 7 (10.0) | 6 (9.2) | ||
| Mean BMI, kg/m2 (SD) | 29.3 (6.0) | 27.8 (5.7) | 0.25 | 28.9 (5.9) | 28.0 (5.8) | 0.15 |
| BMI ≥30 kg/m2 | 25 (43.9) | 18 (30.5) | 0.33 | 29 (41.4) | 21 (32.3) | 0.19 |
| Comorbid conditions | ||||||
| None | 39 (68.4) | 41 (69.5) | 0.02 | 48 (68.6) | 47 (72.3) | 0.08 |
| Diabetes mellitus | 3 (5.3) | 7 (11.9) | 0.24 | 4 (5.7) | 8 (12.3) | 0.23 |
| Hypertension | 5 (8.8) | 5 (8.5) | 0.01 | 8 (11.4) | 6 (9.2) | 0.07 |
| Chronic lung disease | 3 (5.3) | 2 (3.4) | 0.09 | 3 (4.3) | 2 (3.1) | 0.06 |
| Asymptomatic | 1 (1.8) | 3 (5.1) | 0.18 | 0 | 0 | <0.01 |
| Days from symptom onset to randomization, median (IQR) | 5 (4–6) | 5 (3–7) | 0.01 | 5 (4–7) | 5 (3–7) | 0.08 |
| No. of symptoms reported at randomization, median (IQR) | 6 (4–9) | 6 (4–8.5) | 0.28 | 6 (4–9) | 6 (4–8) | 0.16 |
| Symptoms at randomization | ||||||
| Fever | 2 (3.5) | 1 (1.7) | 0.11 | 3 (4.3) | 1 (1.5) | 0.16 |
| Cough/dyspnea | 44 (77.2) | 42 (71.2) | 0.14 | 48 (68.6) | 47 (72.3) | 0.08 |
| Fatigue | 41 (71.9) | 40 (67.8) | 0.09 | 51 (72.9) | 47 (72.3) | 0.01 |
| Joint pain | 18 (31.6) | 20 (33.9) | 0.05 | 20 (28.6) | 22 (33.8) | 0.11 |
| Myalgias | 36 (63.2) | 36 (61.0) | 0.04 | 42 (60.0) | 38 (58.5) | 0.03 |
| Headache | 37 (64.9) | 40 (67.8) | 0.06 | 45 (64.3) | 43 (66.2) | 0.04 |
| Received at least 1 dose of COVID-19 vaccine | 2 (3.5) | 0 (0.0) | 0.27 | 2 (2.9) | 0 (0.0) | 0.24 |
| Baseline seropositivity | 4 (7.0) | 6 (10.2) | 0.30 | 11 (15.7) | 9 (13.8) | 0.14 |
| Baseline anterior nares RT-PCR Ct, median, (IQR) | 25.1 (22.2–28.9) | 22.2 (19.7–27.2) | 0.30 | 28.3 (23.2–38.4) | 24.3 (20.7–31.9) | 0.38 |
| Baseline oropharyngeal RT-PCR positivity | 50 (87.7) | 54 (91.5) | 0.18 | 52 (74.3) | 53 (81.5) | 0.24 |
| Baseline laboratory values, median (IQR) | ||||||
| AST, U/L | 32.0 (26.0–42.5) | 29.0 (25.0–34.0) | 0.39 | 29.5 (25.8–39.3) | 29.0 (25.0–34.0) | 0.31 |
| ALT, U/L | 29.0 (20.0–48.0) | 25.0 (19.5–38.0) | 0.18 | 24.5 (18.8–46.5) | 25.0 (19.0–37.0) | 0.16 |
| Creatinine, mg/dL | 0.8 (0.6–1.0) | 0.8 (0.6–1.0) | 0.09 | 0.8 (0.7–1.0) | 0.8 (0.6–1.0) | 0.12 |
| Uric acid, mg/dL | 4.5 (3.5–5.8) | 4.4 (3.9–5.3) | <0.01 | 4.5 (3.5–5.6) | 4.4 (3.9–5.3) | 0.02 |
Data are presented as No. (%) unless otherwise indicated.
Abbreviations: ALT, alanine aminotransferase; AST, aspartate aminotransferase; BMI, body mass index; Ct, cycle threshold; COVID-19, coronavirus disease 2019; IQR, interquartile range; mITT, modified intention-to-treat population; RT-PCR, reverse-transcription polymerase chain reaction; SD, standard deviation; SMD, standardized mean difference; smITT, symptomatic modified intention-to-treat population.
Primary Analysis
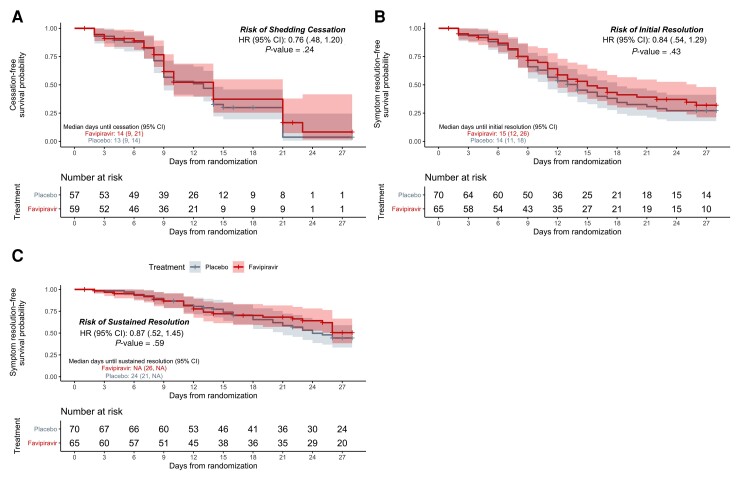
Of the mITT population, 79 participants met the primary endpoint (44/57 [77%] placebo vs 35/59 [59%] favipiravir). Although the likelihood of shedding cessation favored placebo, we found no statistically significant difference in time to shedding cessation by treatment arm (hazard ratio [HR], 0.76 [95% confidence interval {CI}, .48–1.20], P = .24; Figure 2). We detected no difference in median time to shedding cessation between groups (13 days [95% CI, 9–14 days] vs 14 [95% CI, 9–21 days] for placebo vs favipiravir, respectively; Table 2). Of the 37 participants who did not meet the primary outcome, 18 had at least 1 negative RT-PCR during the study (8 placebo, 10 favipiravir).
Figure 2.
Kaplan-Meier analyses of the primary and key secondary outcomes in the modified intention-to-treat population. Time until shedding cessation of SARS-CoV-2 in RT-PCR from nasal swabs (A), initial symptom resolution (B), and sustained symptom resolution (C), stratified by treatment arm: favipiravir (red) vs placebo (gray). Participants who did not experience the endpoint were censored (+ symbols) at their last positive swab for the primary outcome or at the last completed symptom questionnaire for the key secondary outcomes. Solid lines represent Kaplan-Meier survival probability; shading represents 95% confidence intervals. Abbreviations: CI, confidence interval; HR, hazard ratio; NA, not applicable.
Table 2.
Primary and Secondary Outcomes
| Outcome | Treatment Arm | Measure of Association | ||
|---|---|---|---|---|
| Placebo | Favipiravir | aHR (95% CI) | P Value | |
| Primary outcomea | ||||
| Days until viral shedding cessation, median (IQR) | 13 (9–14) | 14 (9–21) | 0.76 (.48–1.20) | .24 |
| Secondary clinical outcomes | ||||
| Hospitalizations by day 28b, No. (%) of participants | 4/74 (5) | 0 | … | .06 |
| ED visits by day 28b, No. (%) of participants | 7/74 (10) | 5/75 (7) | … | .56 |
| Days until initial resolution of symptomsc, median (IQR) | 14 (11–18) | 15 (12–26) | 0.84 (.54–1.29) | .43 |
| Days until sustained resolution of symptomsc, median (IQR) | 24 (21–NA) | NA (26–NA) | 0.87 (.52–1.45) | .59 |
| Secondary virologic outcomesa | Δ Reverse Ctd (95% CI) |
P Value | ||
| Change in reverse Ctd from day 1 to 7, mean (SD) | −7.0 (5.6) | −9.2 (5.0) | −2.06 (−4.34 to .22) | .08 |
| Change in reverse Ctd from day 1 to 10, mean (SD) | −10.5 (5.1) | −12.9 (5.9) | −1.83 (−4.19 to .53) | .13 |
| Negative by RT-PCR on day 7, No. (%) of participants | 10/47 (21) | 10/42 (24) | … | .80 |
| Negative by RT-PCR on day 10, No. (%) of participants | 23/45 (51) | 20/35 (57) | … | .65 |
| Safety outcomesb | ||||
| Serious AEs, No. (%) of events | 1 (1.4) | 0 | … | … |
| Resulting in death | 0 | 0 | … | … |
| Resulting in hospitalization | 1 (100.0) | 0 | … | … |
| AEs, No. of events | 15 | 27 | … | … |
| AEs, No. (%) of participants | 10 (13.5) | 19 (25.3) | … | .11 |
| Grade 3 AEs, No. (%) | 2 (13.3) | 2 (7.4) | … | … |
| Most common AEs, No. (%) of participants | ||||
| Dizziness | 2 (2.7) | 3 (4.0) | … | … |
| Nausea | 3 (4.1) | 1 (1.3) | … | … |
| Day 10 uric acid, mg/dL, median (IQR) | 4.9 (4.1–6.0) | 7.4 (6.3–9.0) | … | … |
All virologic endpoints use anterior nares swab results unless otherwise noted.
Abbreviations: AE, adverse event; aHR, adjusted hazard ratio (adjusted for age ≥50 years and sex); CI, confidence interval; Ct, cycle threshold; ED, emergency department; IQR, interquartile range; NA, undefined; RT-PCR, reverse-transcription polymerase chain reaction; SD, standard deviation.
Among the modified intention-to-treat (mITT) population.
Among the ITT population.
Among the symptomatic mITT (smITT) population.
Reverse Ct was defined by subtracting the Ct value from 40 (the limit of detection; see Supplementary Methods for details).
In prespecified and post hoc analyses, we found no difference in time to shedding cessation by subgroup including age group, sex, high-risk comorbid conditions, seropositivity, or duration of symptoms at enrollment (Supplementary Table 1).
In a sensitivity analysis using the ITT cohort, the median time to shedding cessation decreased to 9 days for both arms.
Secondary Analyses
In the smITT cohort, both groups reported a median of 5 days of symptoms at enrollment (range, 1–21 days vs 1–14 days for placebo vs favipiravir, respectively; Table 1). The most common symptoms included cough/dyspnea, fatigue, myalgias, and headache.
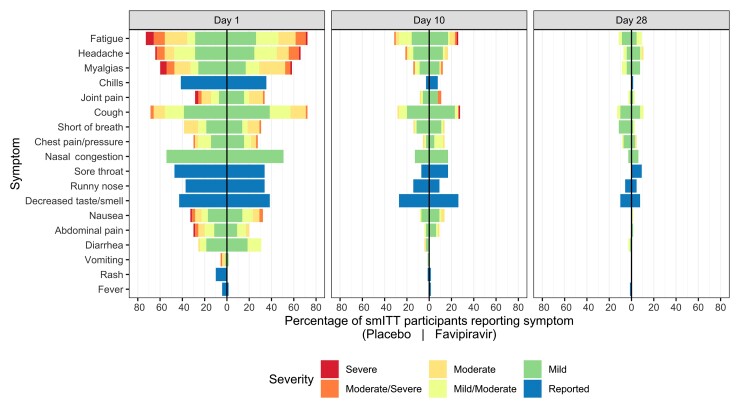
We found no statistically significant difference in time to initial or sustained symptom resolution by treatment arm (initial: HR, 0.84 [95% CI, .54–1.29]; sustained: HR, 0.87 [95% CI, .52–1.45]; Table 2, Figure 3). The median time to initial symptom resolution was 1 day shorter in the placebo arm than the favipiravir arm (14 days [95% CI, 11–18 days] vs 15 days [95% CI, 12–26 days], respectively). Although participants reported fewer and milder symptoms over time, 30 (18 placebo, 12 favipiravir) continued to report ≥1 symptom on day 28 (Figure 3, Supplementary Figures 3 and 4).
Figure 3.
Symptom prevalence in the symptomatic modified intention-to-treat (smITT) population. Mirrored bar plots of percentage of smITT participants reporting symptoms by treatment arm and study day, colored by symptom severity. Numerator is the number of participants reporting the symptom severity per study day and treatment arm; denominator is the number of overall participants in the treatment arm (n = 70 in placebo and n = 65 in favipiravir). Symptoms are ordered by day 1 relative frequency within their respective organ systems (lower respiratory, upper respiratory, systemic, gastrointestinal, other). Bars to the right of the centered black line represent favipiravir symptom distributions, while those on the left are representative of placebo.
In the ITT cohort, 12 participants reported ≥1 emergency department visit during the study (7 [9.5%] placebo vs 5 [6.7%] favipiravir, P = .56). Four were hospitalized and all 4 received placebo (Table 2).
Of the 124 randomized participants who did not have detectable antibodies at baseline, 71 (57%) were seropositive at day 28 (Supplementary Table 2).
Virologic Analyses
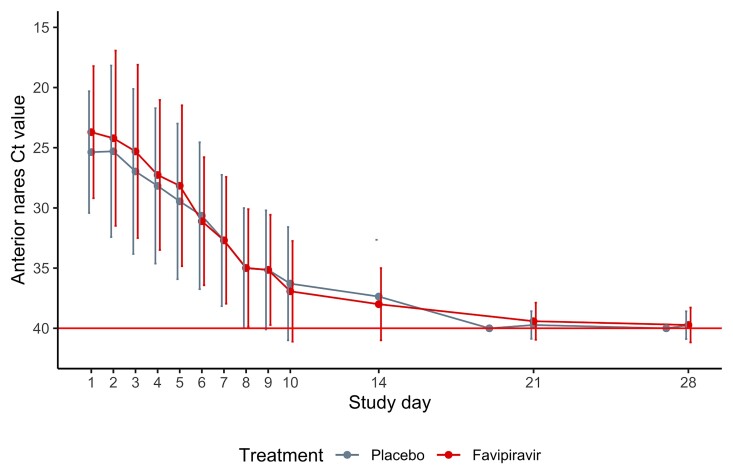
Although the average Ct values increased significantly over time, the magnitude of decline did not differ between treatment arms (Figure 4, Supplementary Figure 2). We found no difference in the proportion of participants in either arm with a negative nasal RT-PCR on days 7 or 10 (Table 2) or a negative oropharyngeal RT-PCR on days 5 and 28 (Table 2, Supplementary Table 2).
Figure 4.
Trajectory of nasal cycle threshold (Ct) in the modified intention-to-treat population. Line plots of nasal Ct values over time by treatment arm. Each dot represents the mean Ct value on that study day by treatment arm; bars represent the standard error around the mean. Lines are slightly jittered to avoid overlap. The red horizontal line at y = 40 represents the limit of detection. The y-axis is reversed so that lower values of Ct represent more virus detected.
Adverse Events
More participants in the favipiravir arm reported adverse events, but this difference was not statistically significant (10/71 [13.5%] for placebo vs 19/75 [25.3%] for favipiravir, P = .11; Table 2). The most common adverse event reported by the favipiravir participants was dizziness. More participants in the favipiravir arm developed hyperuricemia on study day 10 (21/71 [30%] for placebo vs 54/66 [82%] for favipiravir), but only 3 were symptomatic.
Sequencing Analyses
We included 112 PCR-positive nasal samples from 73 study participants (36 placebo, 37 favipiravir) that met our quality and coverage filters, including >1 longitudinal sample from 31 participants (17 placebo, 14 favipiravir). Residual nasal swabs had a mean quantitative PCR (qPCR) CT of 22.3 and a mean depth of coverage of 1738× (95.1% of the genome with depth of coverage >10×). SARS-CoV-2 variation observed within a representative participant is shown in Supplementary Figure 5.
On day 5, we found no difference in mean (standard deviation) low-frequency iSNVs in either arm (favipiravir, 15.7 [11.9] vs placebo, 15.2 [16.5], P = .92; Supplementary Figure 6). After standardizing by sequencing effort (the number of base-pairs sequenced per sample) the mean number of iSNVs was higher in the favipiravir arm, but this difference was not significant (favipiravir mean, 3.09 × 10−8 iSNVs/sequenced base-pairs [SD, 3.24 × 10−8] vs placebo mean, 2.1 × 10−8 iSNVs/sequenced base-pairs [SD, 2.03 × 10−8]; P = .35).
We found no difference in the number of transition iSNVs (P = .76) or the number of transition iSNVs standardized by sequencing effort (P = .17) in the favipiravir arm compared to placebo.
Finally, in linear models, we did not find that treatment arm was significantly associated with within-host SARS-CoV-2 diversity as measured by the raw number of iSNVs, the number of transition iSNVs, or the number of raw or transition iSNVs standardized by sequencing throughput, after controlling for study day.
For 96.7% (30/31) of participants with longitudinal samples available, SARS-CoV-2 exhibited no fixed nucleotide substitutions over time. SARS-CoV-2 consensus genomes obtained from 1 treatment group participant differed by 4 substitutions between day 1 and day 10.
DISCUSSION
In outpatients with asymptomatic or mild COVID-19, we found no difference in time to shedding cessation or symptom resolution between the favipiravir and placebo group.
Our results differ from previous open-label studies, possibly due to the added rigor of blinding and robust data collection in our study. In an open-label favipiravir trial, Udwadia et al found no difference in time to viral shedding cessation using both oropharyngeal and nasopharyngeal swabs, but did report a difference in time to clinical cure based on unblinded clinician assessments of fever, oxygen saturation, and cough [2]. Our clinical symptom evaluation was more rigorous, involving daily surveys that included a broader range of COVID-19 symptoms. In an open-label, randomized controlled trial, Doi et al compared early (day 1) and late (day 6) favipiravir initiation and found a difference in fever resolution by day 2, but no difference in time to fever resolution or viral shedding [12]. In another open-label, randomized controlled trial, Ivashchenko et al found a difference in viral clearance by day 5 when they compared 2 favipiravir dosing regimens to standard of care, but this became equivalent by day 10 [3]. Although we used a different primary outcome of time to shedding cessation, we also observed no difference in changes in RT-PCR Ct from day 1 to days 5 and 7.
We used the same favipiravir dosing regimen as other trials investigating favipiravir for COVID-19 [2, 3]. In fact, some trials used the lower dosing regimen approved for patients with pandemic influenza in Japan [13, 14]. However, it is possible that this regimen did not achieve adequate levels to inhibit viral replication. A recent dose-optimizing study of 19 critically ill patients with influenza demonstrated a decrease in plasma trough concentrations (Ctrough) during treatment, estimating that only 42% of patients who received favipiravir 1800 mg BID followed by 800 mg BID achieved the goal Ctrough of ≥20 mg/L for >80% of the treatment duration [15]. Modeling from this work suggested that regimens of ≥3600 mg loading dose followed by 2600 mg might be necessary to achieve target concentrations. Trials investigating favipiravir for Ebola treatment used higher doses of favipiravir (6000 mg/day load, then 2400 mg/day), but also achieved lower drug concentrations than predicted at days 2 and 4 of treatment and did not meet their clinical endpoint [16].
Suboptimal dosing may also explain why we found no evidence of mutagenesis after at least 5 days of favipiravir exposure. Our findings differ from in vitro work demonstrating a 3-fold increase in the number of mutations and a 12-fold increase in C to T or G to A transitions in Vero cells infected with SARS-CoV-2 exposed to favipiravir compared to controls [1]. This is also in contrast to an in vivo study of molnupiravir, a closely related nucleotide analogue, that found a 2-fold increase in SARS-CoV-2 RdRp gene mutations in the treatment group compared to the control group [17]. A study that evaluated favipiravir dosing for Ebola infections in macaques found that viral mutational load was strongly associated with favipiravir dose [9] and that viral mutation accumulation was associated with lower levels of plasma infectious viral particles. Based upon these findings, the authors suggested that an earlier clinical trial in humans may have used suboptimal favipiravir dosing. However, recent in vitro data suggest that even higher favipiravir doses may not be effective against SARS-CoV-2 [18].
In contrast to our findings, a randomized placebo-controlled trial of molnupiravir reported an approximate 30% reduction in mortality and COVID-19–related hospitalizations [19, 20]. The overall hospitalization rates were higher than in our favipiravir study, possibly due to differences in standards of care and the predominance of SARS-CoV-2 B.1.617 (Delta variant) during the molnupiravir study. Of note, in vitro data suggest that molnupiravir may also be mutagenic to mammalian cells [20]. Animal studies suggest that favipiravir administered in combination with molnupiravir may be an effective strategy to allow for lower molnupiravir doses and potentially avoid unintended consequences [21].
Our study has several limitations. Most therapeutic studies for COVID-19, like ours, assess antiviral efficacy by using RT-PCR to detect viral RNA from nasal, nasopharyngeal, or oropharyngeal swabs. However, detectable RNA may not reflect actively replicating virus and individuals can continue to have detectable RNA intermittently and long after illness recovery [22]. Widespread use of cell culture to detect replication-competent virus and to establish viral clearance is limited by feasibility, cost, and safety considerations [22]. Although we use Ct rather than viral load, our analysis was strengthened by serial testing from individuals. Our primary endpoint was based upon participant-collected nasal swabs, which may be less accurate than nasopharyngeal swabs [23]. However, we found similar results from a secondary analysis of study staff–collected oropharyngeal swabs. Our study was powered to detect differences in shedding cessation, not symptom resolution. Although not designed to detect a difference in “long COVID,” we found that nearly half of both groups continued to report symptoms 28 days after enrollment. In addition, we did not limit enrollment to those with very recent symptom onset; this may have impaired our ability to detect a difference in outcomes. Finally, we did not include severely immunocompromised patients in this trial, and we enrolled patients prior to the emergence and dominance of SARS-CoV-2 B.1.617 (Delta) and B.1.1.529 (Omicron) variants in the United States.
In conclusion, our data do not support favipiravir use at currently recommended doses in outpatients with mild or asymptomatic COVID-19. Dose optimization studies are necessary to elucidate if favipiravir administered at higher doses or delivered in combination with other agents is effective and safe for patients with COVID-19.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Marisa Holubar, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA.
Aruna Subramanian, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA.
Natasha Purington, Quantitative Sciences Unit, Division of Biomedical Informatics Research, Department of Medicine, Stanford University, Palo Alto, California, USA.
Haley Hedlin, Quantitative Sciences Unit, Division of Biomedical Informatics Research, Department of Medicine, Stanford University, Palo Alto, California, USA.
Bryan Bunning, Quantitative Sciences Unit, Division of Biomedical Informatics Research, Department of Medicine, Stanford University, Palo Alto, California, USA.
Katharine S Walter, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA.
Hector Bonilla, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA.
Athanasia Boumis, Stanford Center for Clinical Research, Stanford University, Stanford, California, USA.
Michael Chen, Stanford Solutions, Stanford University School of Medicine, Stanford, California, USA.
Kimberly Clinton, Stanford Center for Clinical Research, Stanford University, Stanford, California, USA.
Liisa Dewhurst, Stanford Center for Clinical Research, Stanford University, Stanford, California, USA.
Carol Epstein, Carol L. Epstein MD Consulting LLC, Wellington, Florida, USA.
Prasanna Jagannathan, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA; Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford, California, USA.
Richard H Kaszynski, Stanford Solutions, Stanford University School of Medicine, Stanford, California, USA.
Lori Panu, Stanford Center for Clinical Research, Stanford University, Stanford, California, USA.
Julie Parsonnet, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA; Department of Epidemiology and Population Health, Stanford University School of Medicine, Stanford, California, USA.
Elizabeth L Ponder, Stanford ChEM-H, Stanford University, Stanford, California, USA.
Orlando Quintero, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA.
Elizabeth Sefton, Stanford ChEM-H, Stanford University, Stanford, California, USA.
Upinder Singh, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA; Department of Microbiology and Immunology, Stanford University School of Medicine, Stanford, California, USA.
Luke Soberanis, Stanford Center for Clinical Research, Stanford University, Stanford, California, USA.
Henry Truong, Mariner Advanced Pharmacy Corporation, San Mateo, California, USA.
Jason R Andrews, Division of Infectious Diseases and Geographic Medicine, Stanford University School of Medicine, Stanford, California, USA.
Manisha Desai, Quantitative Sciences Unit, Division of Biomedical Informatics Research, Department of Medicine, Stanford University, Palo Alto, California, USA.
Chaitan Khosla, Stanford ChEM-H, Stanford University, Stanford, California, USA; Departments of Chemistry and Chemical Engineering, Stanford University, Stanford, California, USA.
Yvonne Maldonado, Department of Epidemiology and Population Health, Stanford University School of Medicine, Stanford, California, USA; Department of Pediatrics, Stanford University School of Medicine, Stanford, California, USA.
Notes
Acknowledgments. The authors sincerely thank all participants for generously volunteering for this study and all study staff for their tireless effort and enthusiasm, including Renu Verma and Eugene Kim. The authors also thank our partners in Stanford Healthcare, Stanford University Occupational Health, and San Mateo Medical Center in assisting in recruitment; Stanford University leadership for providing research space; and the data and safety monitoring board (DSMB) members for their service. The Stanford Research Electronic Data Capture (REDCap) platform (http://redcap.stanford.edu) is developed and operated by the Stanford Medicine Research information technology team. The REDCap platform services at Stanford are subsidized by (1) the Stanford School of Medicine Research Office, and (2) the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health (NIH), through grant number UL1 TR001085. The Quantitative Sciences Unit is partially supported by grant number UL1 TR003142.
Disclaimer. The funders had no role in data collection, analysis, or the decision to submit the manuscript for publication.
Financial support. This work was supported by anonymous donors to Stanford University.
References
- 1. Shannon A, Selisko B, Le NT, et al. Rapid incorporation of favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat Commun 2020; 11:4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Udwadia ZF, Singh P, Barkate H, et al. Efficacy and safety of favipiravir, an oral RNA-dependent RNA polymerase inhibitor, in mild-to-moderate COVID-19: a randomized, comparative, open-label, multicenter, phase 3 clinical trial. Int J Infect Dis 2021; 103:62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ivashchenko AA, Dmitriev KA, Vostokova NV, et al. AVIFAVIR for treatment of patients with moderate coronavirus disease 2019 (COVID-19): interim results of a phase II/III multicenter randomized clinical trial. Clin Infect Dis 2021; 73:531–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harris PA, Taylor R, Minor BL, et al. The REDCap consortium: building an international community of software platform partners. J Biomed Inform 2019; 95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. United States Food and Drug Administration . Assessing COVID-19-related symptoms in outpatient adult and adolescent subjects in clinical trials of drugs and biological products for COVID-19 prevention or treatment: guidance for industry. Silver Spring, MD: FDA, 2020.
- 7. Trotti A, Colevas A, Setser A, et al. CTCAE v3.0: development of a comprehensive grading system for the adverse effects of cancer treatment. Semin Radiat Oncol 2003; 13:176–81. [DOI] [PubMed] [Google Scholar]
- 8. Ewels PA, Peltzer A, Fillinger S, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 2020; 38:276–8. [DOI] [PubMed] [Google Scholar]
- 9. Guedj J, Piorkowski G, Jacquot F, et al. Antiviral efficacy of favipiravir against Ebola virus: a translational study in cynomolgus macaques. PLoS Med 2018; 15:e1002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kassambara A. rstatix: pipe-friendly framework for basic statistical tests. R package version 0.6.0. Available at:https://CRAN.R-project.org/package=rstatix. Accessed September 2021.
- 11. R Core Team . R: A language and environment for statistical computing. Vienna, Austria, 2019. [Google Scholar]
- 12. Doi Y, Hibino M, Hase R, et al. A prospective, randomized, open-label trial of early versus late favipiravir therapy in hospitalized patients with COVID-19. Antimicrob Agents Chemother 2020; 64:e01897-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lou Y, Liu L, Yao H, et al. Clinical outcomes and plasma concentrations of baloxavir marboxil and favipiravir in COVID-19 patients: an exploratory randomized, controlled trial. Eur J Pharm Sci 2021; 157:105631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cai Q, Yang M, Liu D, et al. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering (Beijing) 2020; 6:1192–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang Y, Zhong W, Salam A, et al. Phase 2a, open-label, dose-escalating, multi-center pharmacokinetic study of favipiravir (T-705) in combination with oseltamivir in patients with severe influenza. EBioMedicine 2020; 62:103125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sissoko D, Laouenan C, Folkesson E, et al. Experimental treatment with favipiravir for ebola virus disease (the JIKI Trial): a historically controlled, single-arm proof-of-concept trial in Guinea. PLoS Med 2016; 13:e1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fischer W, Eron JJ, Holman W, et al. Molnupiravir, an oral antiviral treatment for COVID-19. medRxiv [Preprint]. Posted online 17 June 2021. doi:10.1101/2021.06.17.21258639. [Google Scholar]
- 18. Tomita Y, Takeda M, Matsuyama S. The anti-influenza virus drug favipiravir has little effect on replication of SARS-CoV-2 in cultured cells. Antimicrob Agents Chemother 2021; 65:e00020-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jayk Bernal A, Gomes da Silva MM, Musungaie DB, et al. Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N Engl J Med 2022; 386:509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou S, Hill CS, Sarkar S, et al. β-D-N4-hydroxycytidine inhibits SARS-CoV-2 through lethal mutagenesis but is also mutagenic to mammalian cells. J Infect Dis 2021; 224:415–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abdelnabi R, Foo CS, Kaptein SJF, et al. The combined treatment of molnupiravir and favipiravir results in a potentiation of antiviral efficacy in a SARS-CoV-2 hamster infection model. EBioMedicine 2021; 72:103595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jefferson T, Spencer EA, Brassey J, Heneghan C. Viral cultures for COVID-19 infectious potential assessment—a systematic review. Clin Infect Dis 2020; 73:e3884–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee RA, Herigon JC, Benedetti A, Pollock NR, Denkinger CM. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS-CoV-2 molecular detection: a systematic review and meta-analysis. J Clin Microbiol 2021; 59:e02881-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.