Abstract
A cyclic analog of natural peptide Yunnanin A was synthesized via photoinduced single electron transfer reaction (SET) in the paper. The resulting compound exhibited potent bioactivity (with IC50 values 29.25 μg mL−1 against HepG-2 cell lines and 65.01 μg mL−1 against HeLa cell lines), but almost have no toxicity to normal cells (with IC50 values 203.25 μg mL−1 against L929 cell lines), which may be served as a potential antitumor drug for medical treatment. The spatial structure was examined by experimental electronic circular dichroism (ECD) and quantum chemistry calculations. Moreover, the theoretical study suggested that special intramolecular hydrogen bonds and γ, β-turn secondary structures may be possible sources affecting cyclic peptide's bioactivity.
The photo-induced synthesis, structure and in vitro bioactivity study of a Yunnanin A cyclopeptide analog was presented.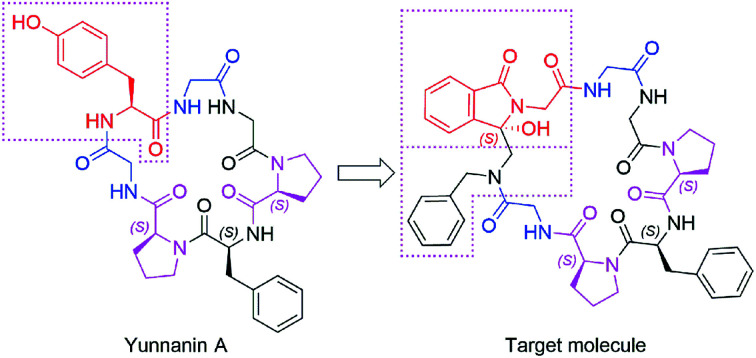
Introduction
It's well known that peptides can broadly affect the physiological and biochemical functions of life. They can bind to specific targets in vivo without significantly interfering with the immune system as their endogenous protein counterparts.1 The low toxicity, good biocompatibility, chemical diversity and protein similarity2 make them promising platforms for drug development and treatment. Consequently, there have been an increasing number of peptide research topics in the past few decades.3,4 However, linear peptides are not very promising therapeutic agents owing to the fact that they are easily cleaved by proteolytic enzymes in vivo, thus losing their biological activity and reducing the availability in the pharmaceutical industry.5 To overcome these obstacles, various peptide modifications have been proposed, including N-methylation (increased membrane permeability), cyclization (increased stability), incorporation of non-standard amino acids (increased specificity and stability, e.g., d-amino acids, ornithine, hydroxyproline, N-methylated residues, thioproline and unnatural amino acids),6,7 PEGylation (reduced clearance)8 and various structural limitations (e.g., introduction of disulfide bonds, ester groups and ether bonds).9 The development of new peptide-based systems obtained by cyclization of linear peptides was recently recognized as one of the most promising methods.
Limiting the peptide into a cyclic structure reduces the conformational freedom of each constituent within the loop and forces the molecule into an ordered secondary structure.10 Compared with the flexible linear analogs, cyclic peptides exhibit higher resistance to endopeptidases,11 less off-target side effects,12 and the elimination of charged terminals in cyclic peptides can effectively improve membrane permeability.13 Furthermore, cyclic peptides are significantly smaller than proteins and consequently more affordable due to reduced manufacturing costs by various chemical methods.14
The active ingredients of cyclic peptides from natural extracts (marine, plants, animals, bacteria and fungi) are widely used in therapeutics, diagnostics, vaccines medicine and agricultural applications.15 Among them, several plant-derived and marine-derived cyclic peptides presented various bioactivities (anticancer, antimicrobial, antioxidant, etc.). However, the content of the well-reacted cyclic peptides extracted from the natural plants and ocean is low and difficult to apply, hence the synthesis and modification of the natural cyclic peptides is called a hot spot.
Among various cyclization methods of linear peptides, intramolecular photo-induced single electron transfer (SET) cyclization reaction is a successful method developed by Yoon and co-workers for synthesis of cyclic peptides.16 During the synthesis process, by using N-terminal phthalimide as an electron acceptor and the C-terminal peptide chain containing trimethylsilyl-benzylamine as an electron donor, an intramolecular donating-accepting electron system was established and their cyclic products were synthesized. The cyclization is affected by the length of the peptide chain and the configuration of the residue, hence not all linear peptides can be cyclized. Our group has successfully synthesized some analogs of natural cyclic peptides Sansalvamide A, Fenestin A and Galaxamide by this method.17–19 We found that the synthesis of cyclopeptides analogs by SET would produce optical pure cyclopeptides in our previous research, which could be regarded as a useful tool to prepare cyclic peptides with specific conformations.
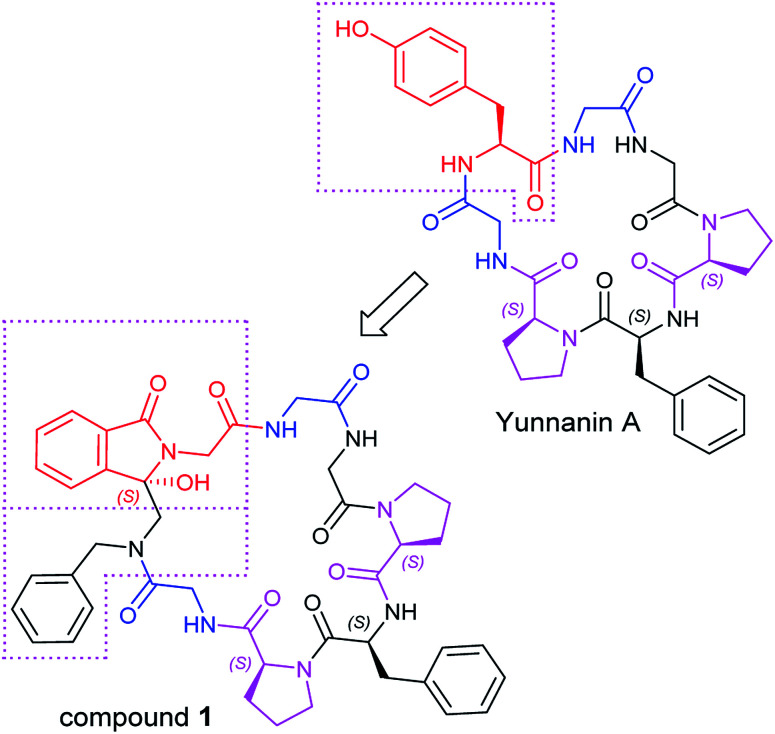
As an ongoing research, in this paper we designed a chiral analogue of Yunnanin A, a proline rich cyclic heptapeptide isolated from the roots of Stellaria yunnanensis franch (Caryophyllaceae),20 and tried to prepare it by photoinduced cyclization to study the its stereochemistry and the bioactivity. Yunnanin A contains two Pro, three Gly, a Phe, and a Tyr residue (Fig. 1), biological evaluation of Yunnanin A showed excellent growth inhibitory activity against murine monocyte/macrophage cells, murine fibrosarcoma cells and human epithelial kidney cells.21 However, there was no research has been reported on the biological action target of Yunnanin A so far, so it was difficult to define which amino acid fragment plays the important role in its bioactivity. On the other hand, it was reported that phthalimide conjugation could efficiently degradation of oncogenic PI3K,22 therefore phthalimide was considered as an essential pharmacophoric section in antitumor agents. It was coincidental that N-terminal phthalimide was an excellent electron acceptor in photo-induced SET cyclization reaction, thus the convenient introduce of phthalimide into natural cyclic peptide Yunnanin A may generate interesting cyclic product with bioactivity. Considering that the introduction of phthalimide into the cyclic skeleton without any modification on other sites would obviously enlarge the ring size of the designed compound and may greatly influence the stereo-structure and binding affinity to the target, the structurally similar Tyr residue in the title compound was legitimately emitted to ensure the skeleton of the ring is not much different from the parent compound. Moreover, the hydroxyl group on isoindolinone (originated from phthalimide during photo-induced SET cyclization reaction) may compensate the loss of hydroxyl group on Tyr residue to some degree, it was also considered to be able to constrain the cyclopeptide conformation due to the H-bond (Fig. 1).23 Due to the crucial role of structure for bioactivity, we also investigated the absolute configuration (AC) by experimental electronic circular dichroism (ECD) combined with quantum chemistry calculations.
Fig. 1. The design of Yunnanin A analog.
Experimental
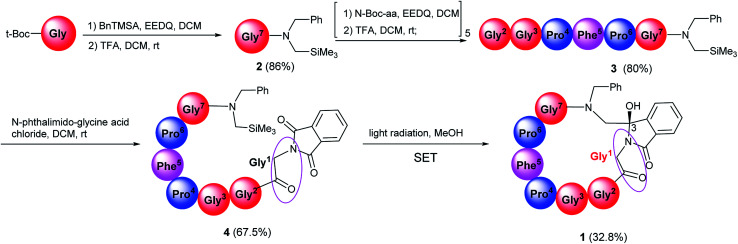
The synthesis of target compound 1 was given in Scheme 1. Firstly, N-Boc-protected amino acids condensed with N-benzyl-1-(trimethylsilyl)-methanamine in the presence of ethoxycarbonyl-ethoxy-dihydroquinoline (EEDQ) to generate the intermediates, which were deprotected by trifluoroacetic acid (TFA) in dichloromethane (DCM) to produce 2. Then 2 condensed with other N-Boc-protected amino acid in sequence in a conventional way. The synthetic intermediate 3 was then acylated by phthaloylglycine chloride to form N-phthaloyl linear peptides 4, subsequently purified by column chromatography (yield: 67.5%), medium-pressure mercury lamp surrounded by a Pyrex glass filter (λ > 290 nm) was used for electronic excitation.24 Specially, nitrogen purged solutions of 4 were irradiated by using a 450 W Hanovia medium-pressure mercury lamp for 40 min. After this, the concentrated product was purified by column chromatography to obtain the pure product 1 (yield: 32.8%). All the reactions were performed under mild conditions at room temperature. The δ 89.17 ppm at 13C-NMR, the molecular weight (M + H)+ 821.3617 in HR-MS have proved the target compound 1 has been successfully synthesized.
Scheme 1. The synthesis of target cyclopeptides. The concentration of photoreaction substrate is 3 × 10−3 mol L−1 in methanol.
The synthesis of Yunnanin A analog began with a series of chiral pure l-amino acids, during which a new C-3 chiral center was formed. In our study, only a chiral pure product was obtained, which may be derived from the chiral starting materials and the greater steric hindrance during cyclization. The absolute configuration (AC) was determined by the electronic circular dichroic (ECD) spectroscopy combined with quantum chemistry calculations. The AC is determined when the simulated spectrum of the selected configuration is consistent with the experimental spectrum. There are two possible ACs for the target compound: C-3R and C-3S isomers. To simulate the ECD, conformational searches by the MMFF method for the two configurations have been performed. The obtained molecules were directly optimized by B3LYP/6-31G(d,p) and subsequently the ECD spectra were simulated using time-dependent density functional method (TDDFT) at the level of B3LYP/6311++(2d,2p).25 Solvent effect was also considered by polarized continuum model (PCM).
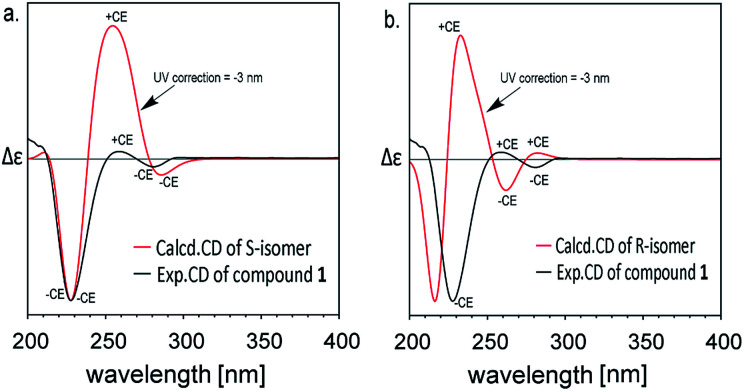
The experimental ECD spectra of compound 1 in CH3OH (2.0 × 10−5 M) in the 200–350 nm range are shown in Fig. 2. The experimental ECD of compound 1 (black line in Fig. 2) showed an obvious negative cotton effect (CE) and a weak negative CE at 228 nm and 281 nm, a weak positive CE at 259 nm. The theoretical ECD spectra of C-3R and C-3S configurations are also depicted in Fig. 2. The simulated UV spectrum of compound 1 (Fig. S4†) showed a slight red shift about 3 nm compared with the experimental spectrum, so here we conducted a ‘UV correction’ of 3 nm to matches well between the simulated spectrum and the experimental ones. The calculated ECD spectra of the S conformer (red line in Fig. 2a) in accord well with the sign of the experimental CEs of compound 1 at 230, 288 and 262 nm. Some discrepancies on the intensity exists, which may be caused by some different conformers in solvents. Nevertheless, the simulated CD of R-configuration (red line in Fig. 2b) showed obvious opposite peak at 221 and 234 nm. This result clearly supports the assignment of a C-3-S configuration to compound 1.
Fig. 2. Experimental and calculated (TDDFT/B3LYP/6311++(2d,2p)) ECD spectra of compound 1 after UV correction.
To study the conformational features, we listed the relative free energies and populations of the low-energy conformers significantly populated at room temperature as determined in methanol in Table 1. For C-3S isomers, S-1 is mainly populated at room temperature (96.6%) and the ball-and-stick model was shown in Fig. 3, while other conformers (S-2, S-3, S-4, Fig. S12†) only occupies extremely small percentages (1.6%, 1.3%, 0.5%, respectively). We also performed a variable temperature 1H-NMR experiment for compound 1 to assert the effect of temperature variation on its conformation (Fig. S11†). From the obtained spectrum, when the temperature raised from 25 °C to 80 °C, The 1H-NMR signal peaks were observed with a slight coalesce in some places (e.g., δ 8.2–8.6, δ 7.4–7.9, δ 3.8–4.0, δ 3.0–3.5), which showed mere existence of different conformers besides the predominate conformer, and they exchanged slowly on the NMR timescale. When temperature increases, signals were eventually coalesced when the rate of conformations exchange becomes fast on the NMR timescale (The simulated C-3R isomer was not displayed because its AC didn't accord with the actual structure).
Relative free energies and populations of the conformations as determined in methanol.
| Conf.a | ΔGb | P c% | Conf. | ΔGb | P c% |
|---|---|---|---|---|---|
| S-1a | 0.00 | 96.6 | R-1 | 0.00 | 61.0 |
| S-2a | 2.43 | 1.6 | R-2 | 0.43 | 29.4 |
| S-3a | 2.55 | 1.3 | R-3 | 1.10 | 9.5 |
| S-4a | 3.07 | 0.5 | R-4 | 4.04 | 0.1 |
See Fig. S11 for the structures of the conformers.
The unit for ΔG is kcal mol−1.
Populations based on ΔG values.
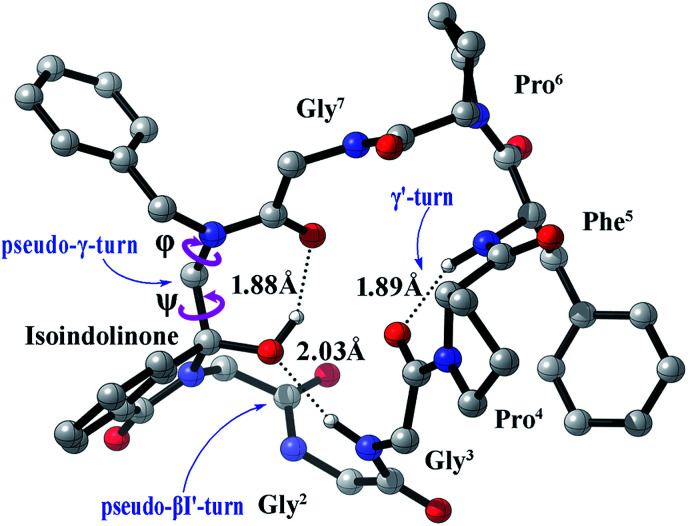
Fig. 3. The most stable conformers S-1 of compound 1 in methanol calculated at the B3LYP/6311++(2d,2p) level of theory.
It is widely acknowledged that the secondary structure such as β-turn conformation was a convenient size for potential mimicry by a drug-like molecule,26,27 it was often found on the surface of larger proteins and hence implicated in molecular recognition. Therefore, we tried to inspect the detailed structure of our prepared cyclopeptide. Generally, the β-turn consists of four amino acid residues defined as i, i + 1, i + 2, and i+3, with the distance <7 Å between the Cα1 to Cα4 atom. Different types of β-turns are depending upon the φ and ψ torsion angles of the i + 1 and i + 2 residues.28 The conformation S-1 shown in Fig. 3 suggested that the C-3S conformer presented a weak intramolecular H-bonding (2.03 Å, black dotted line) between the C–3-O and the C O of Gly3, which could be considered as a classical βI′-turn peptidomimetic, here we named it as ‘pseudo-βI′-turn’ according to the φi+1, ψi+1 (73.15°, 36.46°) and φi+2, ψi+2 (104.78°, −1.64°). In this paper, we defined the dihedral angles φi+1, ψi+1 as [C(OH)–N–Cα–Gly1C( O)] and [N–Cα–Gly1C( O)–NHGly2] and φi+2, ψi+2 as [Gly1C( O)–N–Cα–Gly2C( O)] and [N–Cα–Gly2C( O)–NHGly3] (The partial structural parameters are listed in Table 2, Fig. 3). Meanwhile, there exist a weak intramolecular H-bonding (1.88 Å) between the C–3-OH and C O of Gly7, here we name this structural distortion as ‘pseudo-γ-turn’ due to the φ and ψ (85.71°, −52.40°). (The ‘pseudo γ/γ'-turns’ were distinguished referring to the conventional characteristic dihedral angles of γ turns29). Besides, a weak H-bonding (1.89 Å) between the C O of Gly3 and the N–H of Phe5 suggested the existence of classical γ′-turn.
The torsion angles in the secondary structures of the most stable conformer of compound 1 involving H-bondings.
| Compd | Compound 1 (the most stable conformer S-1) | ||
|---|---|---|---|
| H-B | (Gly3) N–H⋯O–C-3 | C–3-O–H⋯O C(Gly7) | (Phe5) N–H⋯O C(Gly3) |
| Type | Pseudo-βI′-turn | Pseudo-γ-turn | γ′-turn |
| Distance Å | 2.03 | 1.88 | 1.89 |
| φ benzylamine-Cα b | — | 85.71 | — |
| ψ benzylamine-Cα b | — | −52.40 | — |
| φGly1 | 73.15 | — | — |
| ψGly1 | 36.46 | — | — |
| φGly2 | 104.78 | — | — |
| ψGly2 | −1.64 | — | — |
| φPro4 | — | — | −81.65 |
| ψPro4 | — | — | 71.99 |
| Referential characteristic torsion in classical βI′-turna | φ i+1 60° ψi+1 30° | ||
| φ i+2 90° ψi+2 0° | |||
| Referential characteristic torsion in classical γ-turna | φ 75° ψ-64° | ||
| Referential characteristic torsion in classical γ′-turna | φ −79° ψ 69° | ||
These data were used to classify the secondary structures of compound 1 according to literature.29 The classification of γ-turn was based on the dihedral angles φ and ψ of the second (i + 1). The characteristic dihedral angles of other types not involving in this paper were not displayed.
The dihedral angles φ and ψ around benzylamine-Cα were shown in Fig. 3.
The cyclic peptide Yunnanin A has been reported to have a very excellent bioactivity against cancer cell lines.21 To verify the antitumor activity of the modified Yunnanin A analog, we performed metabolic viability MTT colorimetric tetrazolium dye assays on compound 1 against HepG-2 and HeLa cell lines, taxol was used as the control group and L929 cell lines used to evaluate the cytotoxicity against normal cells. The HepG-2, HeLa and L929 cell lines were obtained from Harbin Engineering University and maintained in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS), penicillin G (100 U mL−1), and streptomycin (100 g mL−1). The IC50 values of compound 1 against HepG-2 cells, HeLa cells and L929 cell lines were 29.25 μg mL−1, 65.01 μg mL−1 and 203.25 μg mL−1, while the IC50 values of taxol against HepG-2 cells, HeLa cells and L929 cell lines were 100.57 μg mL−1, >400 μg mL−1 and 42.18 μg mL−1, suggesting that the structurally modified compound 1 possessed better anti-tumor growth ability compared with taxol but almost didn't harm for normal cells.
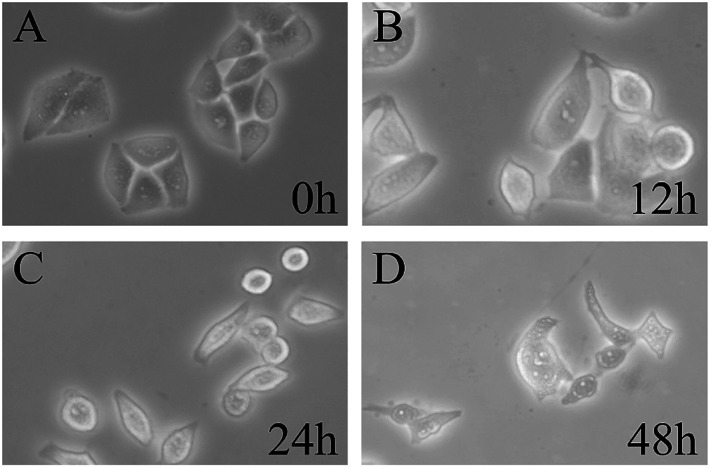
To visually observe the influence of Yunnanin A analog against HepG-2 cells, the cell phenotype in bright field of those HepG-2 cells treated with compound 1 was analysed using a fluorescence inverted microscope (FIM) after incubated with the sample. Fig. 4 showed the morphological changes of HepG-2 cells in bright field at 0, 12, 24 and 48 h (1 mL, 40 μM). As shown in Fig. 4, as time went by, plate-adhered fresh HepG-2 cells with spindle-shape and steady cytoplasm were gradually contracted, the membrane structure damaged and the number of normal cells decreased. During long-playing culture, the number of normal cells became rare and almost all cells extinct. Morphological variations clearly indicated that the Yunnanin A analog could cause cell necrosis and apoptosis in HepG-2 cells.
Fig. 4. Morphological change of HepG-2 cells after incubation by compound 1 in bright field.
Conclusions
Summarily, we have successfully synthesized a natural cyclic peptide Yunnanin A analog by intramolecular photo-induced SET cyclization reaction, which turned out it was a promising method to prepare small ring compounds possessing special constrained conformation due to the rigid isoindolinone moiety and intramolecular hydrogen bonds. The resultant cyclic peptides exhibited effective antitumor activity, which may be attributed to the special γ, β-turn secondary structure, enlightening us to synthesize similar compounds containing such hydrogen bonds and turns to obtain excellent bioactive compounds in future.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
Financial support of this research was provided by Natural Science Foundation of Heilongjiang Province (Key Program) (ZD2018001), University Nursing Program for Young Scholars with Creative Talents in Heilongjiang Province (UNPYSCT-2017182) and the Graduate Innovation Projects of Harbin Normal University (HSDSSCX2019-41). Theoretical calculations were conducted on the ScGrid and Deepcomp7000 at the Supercomputing Center, Computer Network Information Center of the Chinese Academy of Sciences. Also, thanks for High performance computing center of Harbin Normal University.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ra09163g
References
- Barbara C., Margarida B., and Rebeca G. F., Peptide Applications in Biomedicine, Biotechnology and Bioengineering, 2017, p. 87 [Google Scholar]
- Handbook of biologically active peptides, ed. A. Kastin, Academic press, 2013 [Google Scholar]
- Hicks R. P. Bioorg. Med. Chem. 2016;24(18):4056–4065. doi: 10.1016/j.bmc.2016.06.048. [DOI] [PubMed] [Google Scholar]
- Fosgerau K. Hoffmann T. Drug Discovery Today. 2015;20(1):122–128. doi: 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Otvos L., Peptide-based drug design: here and now, Peptide-Based Drug Design, Humana Press, 2008, pp. 1–8 [DOI] [PubMed] [Google Scholar]
- Fernandez-Tejada A. Corzana F. Busto J. H. Aenoza A. Peregrina J. M. J. Org. Chem. 2009;74(24):9305–9313. doi: 10.1021/jo901988w. [DOI] [PubMed] [Google Scholar]
- Chen S. Gfeller D. Buth S. A. Michielin O. Leiman P. G. Heinis C. ChemBioChem. 2013;14(11):1316–1322. doi: 10.1002/cbic.201300228. [DOI] [PubMed] [Google Scholar]
- Nordström R. Nyström L. Ilyas H. Atreya H. S. Borro B. C. Bhunia A. Malmsten M. Colloids Surf., A. 2019;565:8–15. doi: 10.1016/j.colsurfa.2018.12.049. [DOI] [Google Scholar]
- Denisov S. S. Ippel J. H. Mans B. J. Dijkgraaf I. Hackeng T. M. Chem. Commun. 2019;55(10):1374–1377. doi: 10.1039/C8CC08777F. [DOI] [PubMed] [Google Scholar]
- Guharoy M. Chakrabarti P. Bioinformatics. 2007;23(15):1909–1918. doi: 10.1093/bioinformatics/btm274. [DOI] [PubMed] [Google Scholar]
- Shibata K. Suzawa T. Soga S. Mizukami T. Yamada K. Hanai N. et al. . Bioorg. Med. Chem. Lett. 2003;13(15):2583–2586. doi: 10.1016/S0960-894X(03)00476-1. [DOI] [PubMed] [Google Scholar]
- Rezai T. Yu B. Millhauser G. L. Jacobson M. P. Lokey R. S. J. Am. Chem. Soc. 2006;128(8):2510–2511. doi: 10.1021/ja0563455. [DOI] [PubMed] [Google Scholar]
- Lindgren M. Hallbrink M. Prochiantz A. Langel U. Trends Pharmacol. Sci. 2000;21(3):99–103. doi: 10.1016/S0165-6147(00)01447-4. [DOI] [PubMed] [Google Scholar]
- Terrett N. RSC Med. Chem. 2013;4(3):474–475. doi: 10.1039/C2MD90062A. [DOI] [Google Scholar]
- Thell K. Hellinger R. Sahin E. Michenthaler P. Gold-Binder M. Haider T. et al. . Proc. Natl. Acad. Sci. U. S. A. 2016;113(15):3960–3965. doi: 10.1073/pnas.1519960113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon U. C. Jin Y. X. Oh S. W. et al. . J. Am. Chem. Soc. 2003;125(35):10664–10671. doi: 10.1021/ja030297b. [DOI] [PubMed] [Google Scholar]
- Zhao L. S. Zhang H. Y. Tan G. H. Wang Z. Q. Jin Y. X. Tetrahedron Lett. 2017;58(16):1669. doi: 10.1016/j.tetlet.2017.03.042. [DOI] [Google Scholar]
- Zhao L. S. Zhang H. Y. Cui J. N. Zhao M. Q. Wang Z. Q. Yue Q. F. Jin Y. X. New J. Chem. 2017;41(23):14044. doi: 10.1039/C7NJ03363J. [DOI] [Google Scholar]
- Wu J. W. Zhao L. S. Wang Z. Q. Jin Y. X. New J. Chem. 2018;42(24):19779–19784. doi: 10.1039/C8NJ04978E. [DOI] [Google Scholar]
- Morita H. Shishido A. Kayashita T. Shimamura M. Takeya K. Itokawa H. Chem. Lett. 1994;23(12):2415. doi: 10.1246/cl.1994.2415. [DOI] [Google Scholar]
- Napolitano A. et al. . Tetrahedron. 2003;59(51):10203–10211. doi: 10.1016/j.tet.2003.10.073. [DOI] [Google Scholar]
- Li W. Gao C. Zhao L. Yuan Z. Chen Y. Z. Jiang Y. Eur. J. Med. Chem. 2018;151:237–247. doi: 10.1016/j.ejmech.2018.03.066. [DOI] [PubMed] [Google Scholar]
- De L. R. Luis M. Elyse T. W. Margaret A. B. Chem.–Eur. J. 2018;24(68):17869–17880. doi: 10.1002/chem.201802533. [DOI] [PubMed] [Google Scholar]
- Lee Y. J. Ling R. Mariano P. S. J. Org. Chem. 1996;61(10):3304. doi: 10.1021/jo9522623. [DOI] [PubMed] [Google Scholar]
- Ding S. Jia L. Durandin A. Crean C. Kolbanovskiy A. Shafirovich V. Geacintov N. E. Chem. Res. Toxicol. 2009;22(6):1189. doi: 10.1021/tx900107q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N., Blaney F. E., Garland S., Tehan B., and Wall I., Comprehensive Medicinal Chemistry II, 2007, pp. 669–701 [Google Scholar]
- Tyndall J. D. Pfeiffer B. Abbenante G. Fairlie D. P. Chem. Rev. 2005;105(3):793–826. doi: 10.1021/cr040689g. [DOI] [PubMed] [Google Scholar]
- Suat K. Jois S. Curr. Pharm. Des. 2003;9(15):1209–1224. doi: 10.2174/1381612033454900. [DOI] [PubMed] [Google Scholar]
- Sewald N., and Jakubke H. D., Peptides: chemistry and biology, John Wiley & Sons, 2015 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.