Abstract
Various 1,4-benzoxazin-3-one derivatives containing propanolamine groups have been shown to exhibit good antibacterial activity against Pseudomonas syringae pv actinidiae (Psa), X. axonopodis pv citri (Xac) and Xanthomonas oryzae pv oryzae (Xoo). 1,4-benzoxazin-3-one 4n showed the best inhibitory effects against Psa, Xac and Xoo, exhibiting in vitro EC50 values of 4.95, 4.71 and 8.50 μg mL−1, respectively. These potencies were superior to the corresponding EC50 values of the commercial antibiotics bismerthiazol (BT, 89.10, and 116.90 μg mL−1) and thiodiazole copper (TC, 127.30, 82.73 and 87.50 μg mL−1). Treatment on the bacterial leaf blight of rice revealed that compound 4n displayed better curative (51%) and protective (48%) activities for reducing rice BLB than either BT (41%, 39%) or TC (43%, 41%). Scanning electron microscopy (SEM) imaging of Xoo that had been treated with 1,4-benzoxazin-3-one 4n (50–100 μg mL−1) revealed that the bacterial cells had experienced extensive cell wall damage, which is the likely cause of its antimicrobial activity and bacterial death.
Various 1,4-benzoxazin-3-one derivatives containing propanolamine groups have been shown to exhibit good antibacterial activity against Pseudomonas syringae pv actinidiae (Psa), X. axonopodis pv citri (Xac) and Xanthomonas oryzae pv oryzae (Xoo).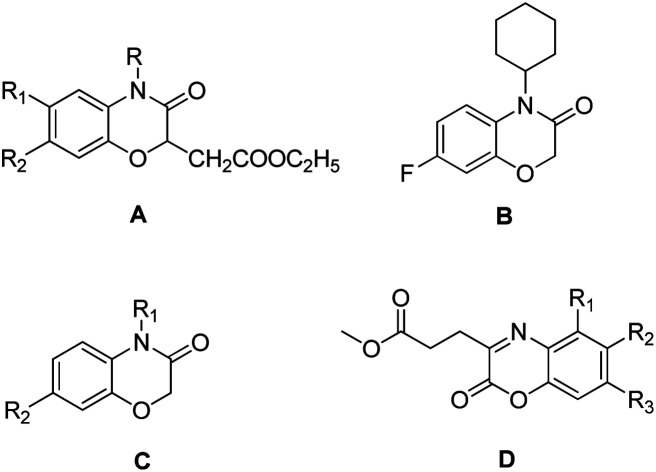
1. Introduction
Plant diseases caused by microbial infections have become one of the world's largest agricultural problems, resulting in serious reductions in crop yields and the production of poor quality agricultural products that can cause significant economic losses.1,2 Amongst these pathogens, Pseudomonas syringae pv actinidiae (Psa; which causes kiwi disease), X. axonopodis pv (Xac; which causes rice bacterial leaf blight [BLB]) and Xanthomonas oryzae pv oryzae (Xoo; which causes citrus canker) are particularly problematic.3–6 Currently, only a few established commercial bactericides, namely, bismerthiazol (BT), etridiazole, benthiazole and thiodiazole copper (TC), are used for the treatment of these plant bacterial diseases. Unfortunately, long-term use of these antimicrobial agents has resulted in the emergence of antimicrobial resistance in many of these pathogenic bacteria,7–9 so the discovery of new antibiotics that elicit their antimicrobial activity through novel modes of action is very important.10,11
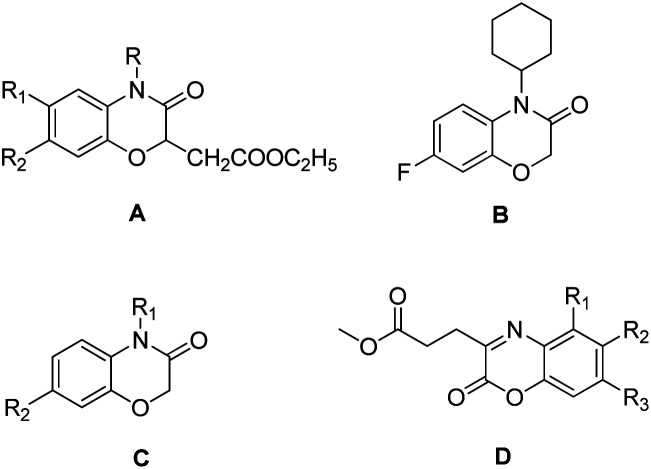
Recently, 1,4-benzoxazin-3-one derivatives have gained increasing attention because of their broad pharmacological activities, including examples that display antibacterial,12,13 antifungal,14,15 herbicidal,16,17 anticancer,18,19 neurodegenerative diseases20 and platelet aggregation inhibitor activities.21 Antibacterial 1,4-benzoxazin-3-one derivatives include ethyl-2-(3-oxo-3,4-dihydro-2H-benzo[b][1,4]oxazin-2-yl)acetate (Fig. 1, A), 4-cyclo-hexyl-7-fluoro-2H-benzo[b][1,4]oxazin-3(4H)-one (Fig. 1, B), 2H-benzo[b] [1,4]oxazin-3(4H)-one (Fig. 1, C), and methyl 3-(2-oxo-2H-benzo[b] [1,4]oxazin-3-yl) propanoate derivatives (Fig. 1, D). Consequently, 2H-1,4-benzoxazin-3-ones can be considered as a potential pharmacophores candidate for developing new agrochemical antibiotics for the treatment of plant bacterial diseases.
Fig. 1. Selected 1,4-benzoxazin-3-one compounds that exhibit antibacterial activities against plant bacterial pathogens.
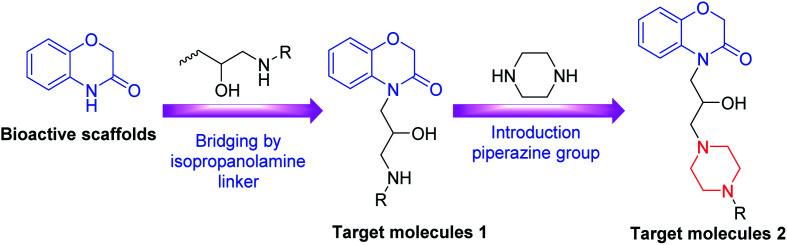
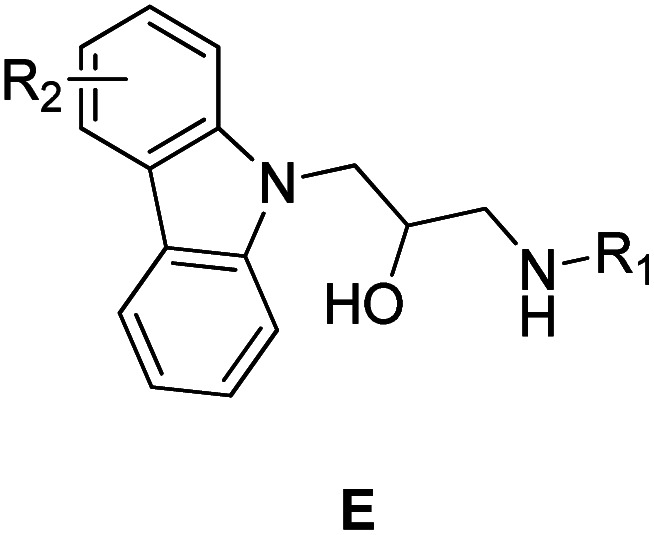
Propanolamine based compounds are also known to exhibit a wide range of medicinally useful properties, including antifungal,22,23 antibacterial,24,25 antimalarial,26,27 antihyperglycemic28 and anticancer drug activities.29,30 For example, Zhao et al. have recently shown that a propanolamine derived carbazole exhibited good antibacterial activities (Fig. 2). Consequently, we decided to investigate whether incorporating a propanolamine group into a 4-benzoxazin-3-one ring system would afford a hybrid compound with good antibiotic activity for the treatment of citrus canker, rice BLB and kiwi disease. These studies reveal that propanolamine 4-benzoxazin-3-ones display good antimicrobial activity against these plant pathogens in vitro, with 1,4-benzoxazin-3-one 4n used as an effective in vivo treatment of BLB in rice. Scanning electron microscopic (SEM) analysis of bacteria treated with 1,4-benzoxazin-3-one 4n revealed significant damage to their cell walls, which is the likely cause of its antibacterial activity.
Fig. 2. Propanolamine containing carbazoles E that exhibit antibiotic activity against plant pathogens.
2. Experimental
2.1. Methods and materials
XT-4 binocular microscope (Beijing Tech Instrument Co., China) was used for melting point determinations with uncorrected documentation. 1H, 13C and 19F NMR spectra were measured using a Bruker Biospin AG-400 (400 MHz) spectrometer in the presence of solvent (CDCl3) solvent and internal standard (TMS). Chemical shifts were recorded in ppm by referring to TMS, while coupling constants were recorded in Hz by referring to the peak multiplicity. SEM images were acquired by Nova NanoSEM 450. TLC with silica gel GF254 as stationary phase was used to monitor the progress of each reaction. All reagents were of analytical grade. All solvents and reagents were in analytically or chemically pure form, and not purified further.
2.2. General procedure for preparing intermediate 1
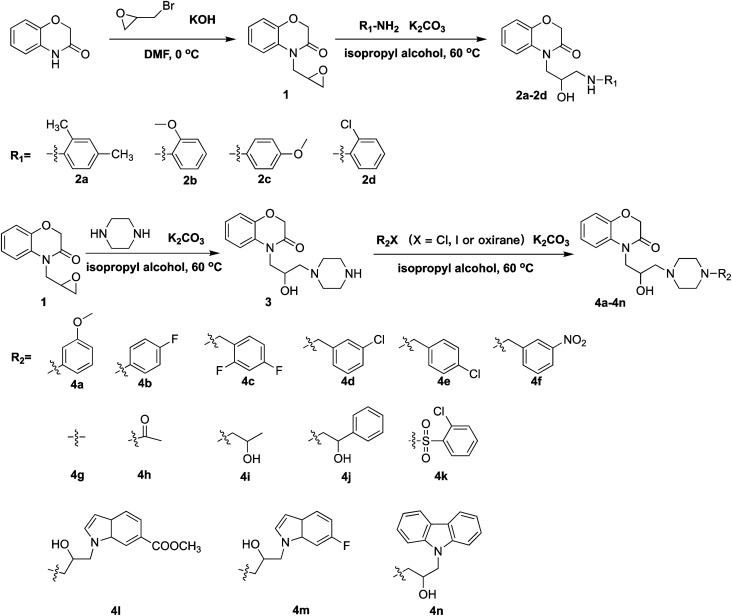
An equimolar amount of KOH (376.17 mg, 6.70 mmol) was mixed with 1,4-benzoxazin-3-one (1 g, 6.70 mmol) dissolved in 5 mL N,N-dimethylformamide. Subsequently, the resultant mixture was stirred at 0 °C for 0.5 hours. Epibromohydrin (1.84 g, 13.41 mmol) was then added portionwise with stirring at 0 °C for 8 hours. After the reaction was completed, the reaction solution was then poured into 60 mL ethyl acetate and rinsed with 3 × 30 mL water, following by drying (Na2SO4), filtering and concentrating in a vacuo. The resultant crude product was then purified with column chromatography or preparative TLC consisting of petroleum ether : ethyl acetate (1 : 8, v/v) mixture as solvent.
2.3. General procedure for preparing intermediate 3
A stirred solution of intermediate 1 (1 g, 4.87 mmol), piperazine (2.1 g, 24.36 mmol), K2CO3 (673.47 mg, 4.87 mmol) in isopropyl alcohol (6 mL) was heated at 60 °C for 4 hours. After the reaction was completed, the reaction solution was then poured into 50 mL ethyl acetate and rinsed with 3 × 25 mL water, followed by drying (Na2SO4), filtering and concentrating in a vacuo. The resultant crude product was then purified using column chromatography or preparative TLC consisting of methanol : methylene chloride (1 : 20, v/v) mixture as solvent.
2.4. General procedure for preparing 1,4-benzoxazin-3-one 2a–2d
A stirred solution of intermediate 1 (300 mg, 1.46 mmol), reaction compound (1.46 mmol), and K2CO3 (202.04 mg, 1.46 mmol) dissolved in 3 mL isopropyl alcohol was heated at 60 °C for 6 hours. After the reaction was completed, the reaction solution was then poured into 25 mL ethyl acetate, rinsed with 3 × 15 mL water, followed by drying (Na2SO4), filtering and concentrating in a vacuo. The resultant crude products were then purified using column chromatography or preparative TLC consisting of petroleum ether : ethyl acetate (1 : 8, v/v) mixture as solvent.
2.5. General procedure for preparing 1,4-benzoxazin-3-ones 4a–4n
A stirred solution of intermediate 3 (300 mg, 1.03 mmol), reaction compound (1.03 mmol), and K2CO3 (142.31 mg, 1.03 mmol) dissolved in 3 mL isopropyl alcohol was heated at 60 °C for 8 hours. After the reaction was completed, the reaction solution was then poured into 25 mL ethyl acetate, rinsed with 3 × 15 mL water, followed by drying (Na2SO4) filtering and concentrating in a vacuo. The resultant crude products were then purified using column chromatography or preparative TLC consisting of methanol : CH2Cl2 (1 : 20, v/v) mixture as solvent (Scheme 1).
Scheme 1. Design route of the target compounds.
3. Results and discussion
3.1. Synthesis of propanolamine derived 1,4-benzoxazin-3-one derivatives 2a–2d and 4a–4n
The synthetic approaches used to prepare two different series of novel 1,4-benzoxazin-3-one derivatives consisting of a propanolamine moiety are illustrated in Scheme 2. Intermediates 1 and 3 were prepared using a method described previously.9 Treatment of intermediate 1 (or 3) with different substituted phenylamine nucleophiles resulted in ring-opening reactions to afford the desired 1,4-benzoxazin-3-one derivatives 2a–2d (or 4a–4n). All the new compounds were confirmed by 1H NMR, 13C NMR, 19F NMR and HRMS spectroscopy (ESI).
Scheme 2. Synthesis of propanolamine derived 1,4-benzoxazin-3-one derivatives 4a–4n.
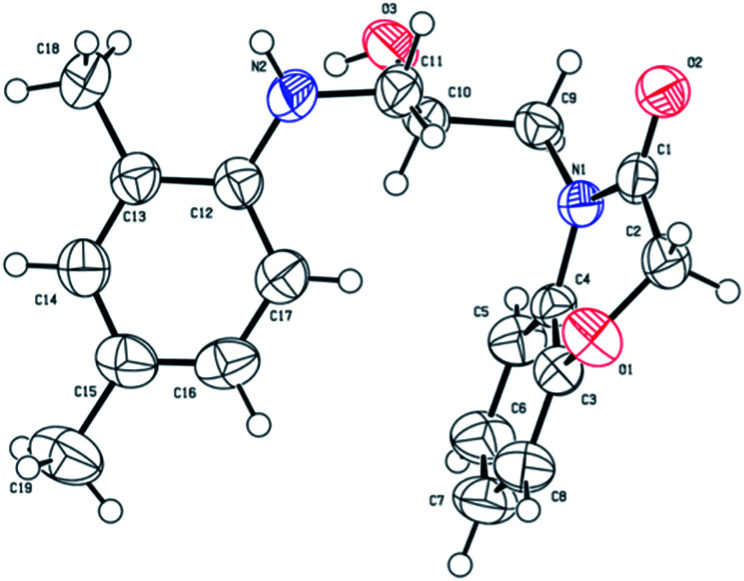
Compound 2a has no intramolecular hydrogen bonds, but a crystal of compound 2a was formed by two intermolecular hydrogen bonds. One intermolecular hydrogen bond was formed by hydroxy oxygen of 2a and the imino hydrogen of the second 2a, the other bond was formed by hydroxyl hydrogen of 2a and the carbonyl oxygen of the third 2a with bond length of 3.012 Å and 2.761 Å respectively. The structural composition of compound 2a was determined using single-crystal X-ray analysis. The deposition number is CCDC 1938560 (see Fig. 3).
Fig. 3. Crystalline structure of compound 2a.
3.2. Antibacterial bioassay in vitro
The in vitro bioactivities of the 1,4-benzoxazin-3-ones 2a–2d and 4a–4n against Xoo, Xac and Psa were determined using turbidimetric tests, with their bioassay results reported in Tables 1 and 2. Most of the novel 1,4-benzoxazin-3-one derivatives exhibited moderate to excellent antibacterial activities against Psa, Xac and Xoo (see Table 1). Compounds 4b, 4d, 4e, 4f, 4m and 4n exhibited excellent antibacterial activities of ≥62% and ≥51% against Xoo at 100 and 50 μg mL−1, respectively. Notably, these inhibitory values were superior to those of bismerthiazol (BT; 58% and 32%) and thiodiazole copper (TC; 49% and 31%). Similarly, compounds 4b, 4d, 4e, 4m and 4n assayed at 100 and 50 μg mL−1 showed greater antibacterial activities (≥62% and ≥59%, respectively) against Xac compared to BT (59% and 28%, respectively). Finally, the antibacterial activities of compounds 4e, 4m and 4n against Psa were ≥71% and ≥64% at 100 and 50 μg mL−1, respectively, which were better than BT (49% and 21%) and TC (55% and 30%), respectively.
Inhibition effect of 1,4-benzoxazin-3-ones against Xoo, Xac and Psaa.
| Compd | Inhibition (%) | |||||
|---|---|---|---|---|---|---|
| Xoo | Xac | Psa | ||||
| 100 μg mL−1 | 50 μg mL−1 | 100 μg mL−1 | 50 μg mL−1 | 100 μg mL−1 | 50 μg mL−1 | |
| 2a | 27.5 ± 2.0 | 18.1 ± 2.4 | 27.0 ± 1.4 | 15.6 ± 2.3 | 17.7 ± 2.3 | 11.0 ± 1.5 |
| 2b | 12.7 ± 1.1 | 9.1 ± 1.2 | 19.2 ± 1.2 | 5.7 ± 2.1 | 1.4 ± 3.7 | —c |
| 2c | 12.3 ± 1.9 | 8.7 ± 1.2 | 17.5 ± 4.8 | 6.1 ± 1.5 | — | — |
| 2d | 15.7 ± 4.8 | 8.7 ± 5.2 | 12.0 ± 1.2 | 6.7 ± 1.5 | 14.1 ± 2.1 | — |
| 3 | 16.6 ± 4.4 | 14.8 ± 3.5 | 7.9 ± 0.6 | — | — | — |
| 4a | 24.4 ± 4.5 | 14.9 ± 1.0 | 35.4 ± 0.0 | 23.4 ± 1.8 | — | — |
| 4b | 62.4 ± 2.2 | 59.4 ± 0.6 | 62.5 ± 1.2 | 59.1 ± 3.8 | 40.8 ± 0.7 | 24.3 ± 0.9 |
| 4c | 48.6 ± 0.6 | 30.0 ± 3.5 | 56.1 ± 1.8 | 37.2 ± 1.5 | 30.7 ± 1.3 | 26.1 ± 2.6 |
| 4d | 71.7 ± 1.8 | 63.5 ± 1.1 | 90.5 ± 0.7 | 78.7 ± 1.4 | 54.0 ± 1.6 | 43.5 ± 1.4 |
| 4e | 91.6 ± 3.3 | 71.0 ± 2.6 | 96.9 ± 0.5 | 86.0 ± 2.1 | 78.9 ± 0.4 | 64.2 ± 1.9 |
| 4f | 66.9 ± 4.3 | 50.6 ± 1.2 | 57.1 ± 2.1 | 35.5 ± 1.9 | 46.3 ± 3.2 | 27.0 ± 2.1 |
| 4g | 19.4 ± 4.6 | 16.9 ± 3.4 | 22.6 ± 1.6 | 16.2 ± 2.5 | — | — |
| 4h | 17.8 ± 2.6 | 14.3 ± 4.5 | 12.0 ± 4.3 | 8.6 ± 4.2 | 3.1 ± 1.4 | 1.0 ± 4.6 |
| 4i | 18.9 ± 0.9 | 15.0 ± 1.5 | 19.2 ± 3.9 | 12.7 ± 1.6 | 15.0 ± 1.1 | 7.5 ± 1.4 |
| 4j | 27.9 ± 4.4 | 15.9 ± 2.1 | 21.3 ± 0.6 | 16.0 ± 3.7 | 21.5 ± 2.0 | 12.2 ± 2.1 |
| 4k | 15.2 ± 4.8 | 5.6 ± 0.9 | 15.4 ± 3.2 | 8.1 ± 2.8 | 14.2 ± 2.7 | 7.2 ± 1.7 |
| 4l | 34.3 ± 4.5 | 27.9 ± 1.0 | 54.7 ± 1.9 | 33.8 ± 5.0 | 28.8 ± 2.6 | 17.8 ± 3.4 |
| 4m | 80.0 ± 3.8 | 64.9 ± 2.7 | 81.4 ± 2.0 | 77.7 ± 2.3 | 71.4 ± 1.8 | 63.7 ± 2.3 |
| 4n | 100 ± 0.5 | 97.4 ± 1.1 | 100 ± 1.3 | 97.4 ± 1.9 | 83.5 ± 2.1 | 74.0 ± 3.8 |
| BTb | 58.4 ± 3.7 | 31.7 ± 1.5 | — | — | 49.1 ± 1.5 | 21.4 ± 0.8 |
| TCb | 48.7 ± 0.5 | 30.6 ± 1.9 | 58.8 ± 3.5 | 27.5 ± 0.9 | 55.0 ± 2.4 | 30.1 ± 2.7 |
Average of three replicates; BT: bismerthiazol; TC: thiadiazole copper.
The antibacterial agents applied to generate comparative antibacterial activities.
“—” indicate as not active.
EC50 values of 1,4-benzoxazin-3-ones against Psa, Xac and Xooa.
| Compd | Xoo | Xac | Psa | |||
|---|---|---|---|---|---|---|
| Regression equation | EC50 (μg mL−1) | Regression equation | EC50 (μg mL−1) | Regression equation | EC50 (μg mL−1) | |
| 4b | y = 0.5434x + 4.1434 | 37.70 ± 1.63 | y = 1.0179x + 3.3998 | 37.33 ± 1.46 | — | — |
| 4d | y = 0.7887x + 3.8552 | 28.28 ± 2.42 | y = 1.2451x + 3.682 | 11.44 ± 1.73 | y = 0.5991x + 3.8205 | 93.07 ± 1.54 |
| 4e | y = 1.2439x + 3.6936 | 11.23 ± 1.16 | y = 1.7851x + 3.0969 | 11.64 ± 1.27 | y = 0.8778x + 3.7733 | 24.97 ± 2.33 |
| 4m | y = 1.1474x + 3.3538 | 27.21 ± 1.61 | y = 2.1693x + 2.57 | 13.19 ± 1.28 | y = 1.0277x + 3.5593 | 25.23 ± 1.32 |
| 4n | y = 1.9139x + 3.6705 | 4.95 ± 0.73 | y = 1.7394x + 3.8296 | 4.71 ± 0.52 | y = 1.1107x + 3.9675 | 8.50 ± 0.83 |
| BTb | y = 1.2472x + 2.5681 | 89.10 ± 1.96 | — | — | y = 1.7949x + 1.2883 | 116.9 ± 1.71 |
| TCb | y = 1.1471x + 2.5854 | 127.3 ± 1.50 | y = 1.6235x + 1.8867 | 82.73 ± 1.45 | y = 1.9802x + 1.1106 | 87.50 ± 1.42 |
Average of three replicates; BT: bismerthiazol; TC: thiadiazole copper.
The antibacterial agents applied to compare antibacterial activities.
The EC50 values of these 1,4-benzoxazin-3-one derivatives against Xoo were found to range from 4.58–37.70 μg mL−1, which were better than BT (89.10 μg mL−1) and TC (127.30 μg mL−1) (see Table 2). These compounds also expressed moderate to good bioactivities (EC50 = 4.57–37.33 μg mL−1) against Xac and excellent antibacterial activities against Psa (EC50 values = 8.08–93.07 μg mL−1), with these inhibitory values superior to BT (116.90 μg mL−1) and TC (87.50 μg mL−1), respectively. Compound 4n exhibited the best overall antibacterial activities against Psa (EC50 = 8.50 μg mL−1), Xac (EC50 = 4.71 μg mL−1) and Xoo (EC50 = 4.95 μg mL−1).
3.3. Structure–activity relationship of antibacterial compounds
A structure–activity relationship (SAR) analysis was carried out according to the antibacterial activities reported in Tables 1 and 2. Analysis of the antibacterial data acquired for compounds 2a–2d revealed that the aryl groups with both electron-donating and-withdrawing groups were well tolerated, which led us to synthesis the second generation 1,4-benzoxazin-3-one derivatives 4a–4n that contained a wider range of substituents. Analysis of the data for 4a–n reveals that the antibacterial activities of 1,4-benzoxazin-3-ones containing electron-withdrawing group on their aryl rings were generally higher than those containing electron-donating groups. Compounds containing a single electron withdrawing group were more active than compounds containing two electron withdrawing groups (cf.4c > 4b). The biological activity of 1,4-benzoxazin-3-one derivatives containing aromatic substituents were much greater than those containing alkyl derivatives (cf.4a, 4b, 4c, 4d, 4e, 4f) > (4g, 4h, 4i). Finally, the best overall antibacterial activity was produced by the carbazole derived 1,4-benzoxazin-3-one derivative 4n, which gave better antibacterial activity than other heterocyclic derivatives (e.g., 4n > 4m > 4j > 4k). Therefore, these novel propanolamine appended this class of 1,4-benzoxazin-3-ones template represent a promising class of lead compound for the discovery potential agrochemicals for the treatment of phytobacterial diseases.
3.4. In vivo bioactivity of 1,4-benzoxazin-3-one 4n against rice BLB
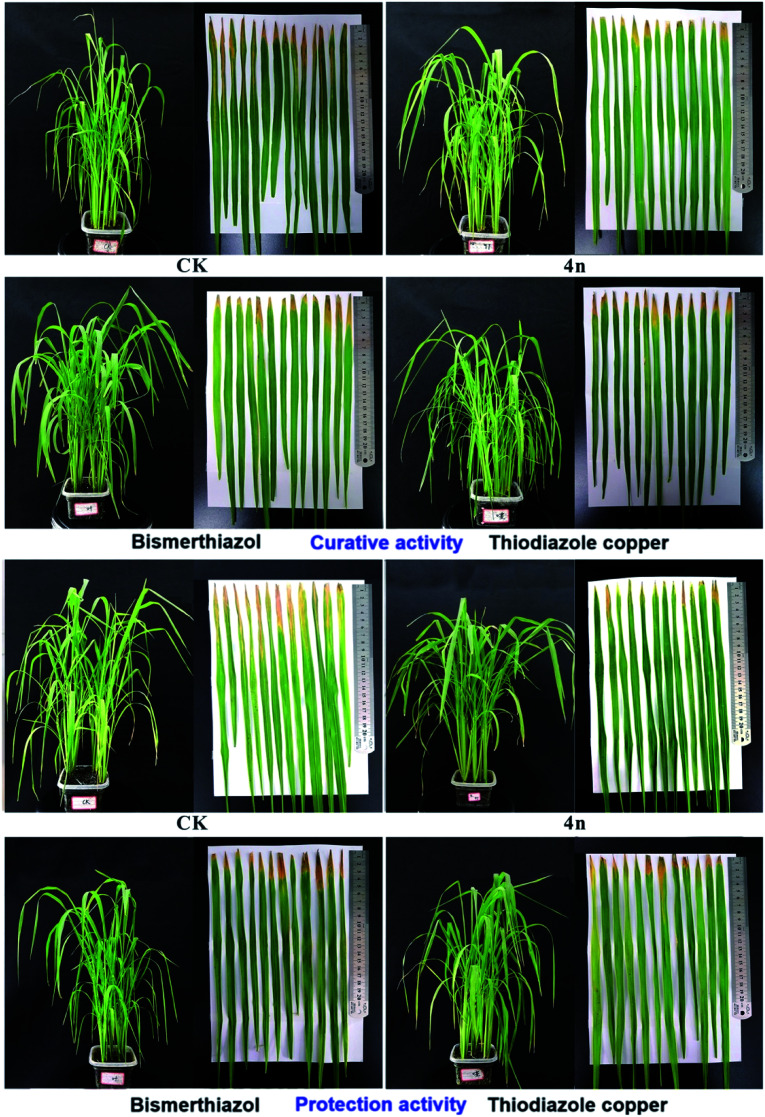
In vivo experiments were carried to determine the potential of 1,4-benzoxazin-3-one as an agrochemical for treating the BLB of rice. As shown in Table 3 and Fig. 4, compound 4n exhibited a greater curative activity (51%) against BLB of rice compared to BT (41%) and TC (42%). Furthermore, compound 4n demonstrated excellent protection activity (48.16%) against rice BLB when compared to BT (38.8%) and TC (41.49%) (see Table 3 and Fig. 4). These results suggests that propanolamine substituted 1,4-benzoxazin-3-ones may be considered as an effective structural template for the development of antibiotic agents to treat phytopathogenic bacterial infections.
Protective and curative effects of compound 4n (200 μg mL−1) on the BLB of rice.
| Treatment | Protection activity (14 days following the last spray application) | Curative activity (14 days following the last spray application) | ||||
|---|---|---|---|---|---|---|
| Morbidity (%) | Disease index (%) | Control efficiencyb (%) | Morbidity (%) | Disease index (%) | Control efficiencyb (%) | |
| 4n | 100 | 40.61 | 48.16 | 100 | 39.48 | 51.08 |
| BT | 100 | 47.92 | 38.82 | 100 | 47.92 | 40.63 |
| TC | 100 | 45.83 | 41.49 | 100 | 46.15 | 42.82 |
| CKa | 100 | 78.33 | — | 100 | 80.71 | — |
Negative control.
Data were analyzed using ANOVA method, and p value less than 0.05 indicated statistically significant different. The same uppercase letters describe the non-significant difference in protective activities between two groups.
Fig. 4. Protective and curative effects of compound 4n (200 μg mL−1) on the BLB of rice. BT and TC were used as positive controls under similar experimental conditions.
3.5. Scanning electron microscopy (SEM) of Xoo bacteria treated with 1,4-benzoxazin-3-one 4n
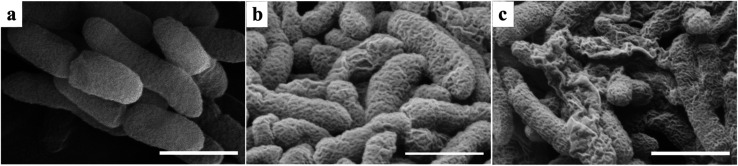
SEM images of Xoo cells were acquired to determine whether any morphological changes could be detected after treatment with different dosages of compound 4n. Treatment of Xoo with compound 4n at a dose of 50 μg mL−1 resulted in the morphologies of the surfaces of the cells being changed from being well-rounded (Fig. 5a) to being partially corrugated with a number of pore-like shapes visible (Fig. 5b). Furthermore, the extent of cell wall damage was increased significantly when bacteria were exposed to a higher 100 μg mL−1 concentration of 4n, with numerous corrugated and fractured bacterial cells being observed (Fig. 5c). This suggests that the antibiotic 4n causes physiological changes within the bacteria that result in cell-wall damage that results in lysis and bacterial cell death.
Fig. 5. SEM images of Xoo treated with different amounts of compound 4n. Images of Xoo treated with 0 (a), 50 (b), and 100 (c) μg mL−1 of 4n. Scale bars are 1 μm.
4. Conclusions
Novel 1,4-benzoxazin-3-one derivatives containing propanolamine groups were prepared, and their antibacterial activities were evaluated against three phytopathogenic bacteria. Bioassay results showed that most of these 1,4-benzoxazin-3-ones possess outstanding antibacterial activities against Psa, Xac and Xoo in vitro, with the best compound 4n exhibiting EC50 values for Xoo, Xac and Psa of 4.95, 4.71 and 8.50 μg mL−1, respectively. 1,4-benzoxazin-3-one 4n also displayed good in vivo antibacterial activities against the BLB of rice (curative activity 51% and protective activity 48%) which was better than the curative and protection activities of BT (41% and 39%) and TC (43%, 41%), respectively. SEM images of Xoo incubated with compound 4n revealed the presence of corrugated and broken cells, consistent with bacterial cell wall damage having occurred that results in bacterial death. These results demonstrate that 1,4-benzoxazin-3-one derivatives containing propanolamine moieties can effectively control the growth of Xoo, Xac and Psa bacteria, thus providing new structural templates for the development of promising antibiotic agents against phytobacterial pathogens.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
We acknowledge the financial support of National Natural Science Foundation of China (21877021, 31860516, 21662009).
Electronic supplementary information (ESI) available. CCDC 1938560. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9ra09639f
Notes and references
- Song X. P. Li P. Li M. W. Yang A. Yu L. Luo L. Z. Hu D. Y. Song B. A. Pestic. Biochem. Physiol. 2018;147:11–19. doi: 10.1016/j.pestbp.2017.10.011. [DOI] [PubMed] [Google Scholar]
- Li B. Liu B. P. Shan C. L. Lbrahim M. Lou Y. H. Wang Y. L. Xie G. L. Li H. Y. Sun G. C. Pest Manage. Sci. 2013;69:312–320. doi: 10.1002/ps.3399. [DOI] [PubMed] [Google Scholar]
- Dai P. Luo K. Yu X. Yang W.-C. Wu L. Zhang W.-H. Adv. Synth. Catal. 2018;360:468–473. doi: 10.1002/adsc.201701364. [DOI] [Google Scholar]
- Lorenzo F. D. Palmigiano A. Silipo A. Desaki Y. Garozzo D. Lanzetta R. Shibuya N. Molinaro A. Carbohydr. Res. 2016;427:38–43. doi: 10.1016/j.carres.2016.03.026. [DOI] [PubMed] [Google Scholar]
- Li Z. Chen S. Zhu S. Luo J. Zhang Y. Weng Q. Molecules. 2015;20:13941–13957. doi: 10.3390/molecules200813941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. F. Wu S. L. Bai X. F. Liu Y. Lu J. F. Liu Y. Xiao B. G. Lu X. P. Fan L. J. J. Bacteriol. 2011;193:6088–6089. doi: 10.1128/JB.06009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo H. P. T. Ho T. H. Lee I. Tran H. T. Sur B. Kim S. Kim J. G. Ahn Y. J. Cha S. S. Kang L. W. J. Agric. Food Chem. 2016;64:7307–7314. doi: 10.1021/acs.jafc.6b02976. [DOI] [PubMed] [Google Scholar]
- Piazza A. Zimaro T. Garavaglia B. S. A Ficarra F. Thomas L. Marondedze C. Feil R. Lunn J. E. Gehring C. Ottado J. Gottig N. Exp. Bot. 2015;66:2795–2811. doi: 10.1093/jxb/erv095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. L. Huang X. Liu L. W. Wang P. Y. Long Q. S. Tao Q. Q. Li Z. Yang S. J. Agric. Food Chem. 2019;67:7512–7525. doi: 10.1021/acs.jafc.9b02036. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Pan X. Duan Y. B. Zhu X. F. Ma X. W. Huang T. T. Gao T. C. Zhou M. G. Australas. Plant Pathol. 2015;44:541–543. doi: 10.1007/s13313-015-0370-7. [DOI] [Google Scholar]
- Li P. Shi L. Gao M. N. Yang X. Xue W. Jin L. H. Hu D. Y. Song B. A. Bioorg. Med. Chem. Lett. 2015;25:481–484. doi: 10.1016/j.bmcl.2014.12.038. [DOI] [PubMed] [Google Scholar]
- Gleńsk M. Gajda B. Franiczek R. Krzyżanowska B. Biskup I. Włodarczyk M. Nat. Prod. Res. 2015;30:1–4. doi: 10.1080/14786419.2015.1054284. [DOI] [PubMed] [Google Scholar]
- Bollu R. Banu S. Bantu R. Reddy A. G. Nagarapu L. Sirisha K. Kumar C. G. Gunda S. K. Shaik K. Bioorg. Med. Chem. Lett. 2017;27:5158–5162. doi: 10.1016/j.bmcl.2017.10.061. [DOI] [PubMed] [Google Scholar]
- Jiang Y. Q. Mao L. F. Zhu L. Ren B. Q. Li W. Xu G. Q. Z. Naturforsch. B Chem. Sci. 2014;69b:103–108. [Google Scholar]
- Śmist M. Kwiecień H. Krawczyk M. J. Environ. Sci. Health, Part B. 2016;51:1–9. doi: 10.1080/03601234.2016.1142744. [DOI] [PubMed] [Google Scholar]
- Calle M. E. d. l. Cabrera G. Cantero D. Valle A. Bolivar J. New Biotechnol. 2019;50:9–19. doi: 10.1016/j.nbt.2019.01.002. [DOI] [PubMed] [Google Scholar]
- Li H. B. Li L. Li J. X. Han T. F. He J. L. Zhu Y. Q. Pest Manage. Sci. 2017;74:579–589. doi: 10.1002/ps.4739. [DOI] [PubMed] [Google Scholar]
- Su C. L. Tseng C. L. Ramesh C. Liu H. S. Huang C. Y. F. Yao C. F. Eur. J. Med. Chem. 2017;132:90–107. doi: 10.1016/j.ejmech.2017.03.034. [DOI] [PubMed] [Google Scholar]
- Wang P. Liu F. Zhong Q. Zheng S. L. Chen Y. Wang G. D. He L. Chin. Chem. Lett. 2017;28:1243–1247. doi: 10.1016/j.cclet.2016.12.028. [DOI] [Google Scholar]
- Wang L. L. Ankati H. Akubathini S. K. Balderamos M. Storey C. A. Patel A. V. Price V. Kretzschmar D. Biehl E. R. D'Mello S. R. J. Neurosci. Res. 2010;88:1970–1984. doi: 10.1002/jnr.22352. [DOI] [PubMed] [Google Scholar]
- Xia S. Liu J. Q. Wang X. H. Tian Y. Wang Y. Wang J. H. Fang L. Zuo H. Bioorg. Med. Chem. Lett. 2014;24:1479–1483. doi: 10.1016/j.bmcl.2014.02.014. [DOI] [PubMed] [Google Scholar]
- Zhu S. P. Wang W. Y. Fang K. Li Z. G. Dong G. Q. Miao Z. Y. Yao J. Z. Zhang W. N. Sheng C. Q. Chin. Chem. Lett. 2014;25:229–233. doi: 10.1016/j.cclet.2013.10.022. [DOI] [Google Scholar]
- Zhang Y. Tangadanchu V. K. R. Bheemanaboina R. R. Y. Cheng Y. Zhou C. H. Eur. J. Med. Chem. 2018;155:579–589. doi: 10.1016/j.ejmech.2018.06.022. [DOI] [PubMed] [Google Scholar]
- Cheng C. Y. Chang C. P. Lauderdale T. L. Y. Yu G. Y. Lee J. C. Jhang Y. W. Wu C. H. Ke Y. Y. Sadani A. A. Yeh C. F. Huang I. W. Kuo Y. P. Tsai D. J. Yeh T. K. Tseng C. T. Song J. S. Liu Y. W. Tsou L. K. Shia K. S. ACS Med. Chem. Lett. 2016;7:1191–1196. doi: 10.1021/acsmedchemlett.6b00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Tangadanchu V. K. R. Cheng Y. Yang R. G. Lin J. M. Zhou C. H. ACS Med. Chem. Lett. 2018;9:244–249. doi: 10.1021/acsmedchemlett.7b00514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin A. Brown F. Bahamontes-Rosa N. Wu B. H. Beitz E. Kun J. F. J. Flitsch S. L. J. Med. Chem. 2007;50:4243–4249. doi: 10.1021/jm070553l. [DOI] [PubMed] [Google Scholar]
- Molette J. Routier J. Abla N. Besson D. Bombrun A. Brun R. Burt H. Georgi K. Kaiser M. Nwaka S. Muzerelle M. Scheer A. ACS Med. Chem. Lett. 2013;4:1037–1041. doi: 10.1021/ml400015f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries P. S. Bersot R. Kincaid J. Mabery E. McCluskie K. Park T. Renner T. Riegler E. Steinfeld T. Turtle E. D. Wei Z. L. Willis E. Bioorg. Med. Chem. Lett. 2018;28:293–297. doi: 10.1016/j.bmcl.2017.12.051. [DOI] [PubMed] [Google Scholar]
- Jin J. Y. Miao C. X. Wang Z. L. Zhang W. L. Zhang X. W. Xie X. Lu W. Eur. J. Med. Chem. 2018;150:757–770. doi: 10.1016/j.ejmech.2018.03.032. [DOI] [PubMed] [Google Scholar]
- Chen Z. Yang T. Y. Wang W. S. Yao J. M. Han S. M. Tao Y. Wang R. Duan L. P. ChemistrySelect. 2018;3:12630–12638. doi: 10.1002/slct.201803029. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.