Abstract
Dry age-related macular degeneration (AMD) is a type of progressive blindness that is primarily due to dysfunction and the loss of retinal pigment epithelium (RPE). The accumulation of N-retinylidene-N-retinylethanolamine (A2E), a by-product of the visual cycle, causes RPE and photoreceptor degeneration that impairs vision. Genes associated with dry AMD have been identified using a blue light model of A2E accumulation in the retinal pigment epithelium and transcriptomic studies of retinal tissue from patients with AMD. However, dry macular degeneration progresses slowly, and current approaches cannot reveal changes in gene transcription according to stages of AMD progression. Thus, they are limited in terms of identifying genes responsible for pathogenesis. Here, we created a model of long-term exposure to identify temporally-dependent changes in gene expression induced in human retinal pigment epithelial cells (ARPE-19) exposed to blue light and a non-cytotoxic dose of A2E for 120 days. We identified stage-specific genes at 40, 100, and 120 days, respectively. The expression of genes corresponding to epithelial-mesenchymal transition (EMT) during the early stage, glycolysis and angiogenesis during the middle stage, and apoptosis and inflammation pathways during the late stage was significantly altered by A2E and blue light. Changes in the expression of genes at the late stages of the EMT were similar to those found in human eyes with late-stage AMD. Our results provide further insight into the pathogenesis of dry AMD induced by blue light and a novel model in vitro with which relevant genes can be identified in the future.
Keywords: Dry age-related macular degeneration, Retinal pigment epithelium, Lipofuscin, A2E, Blue light, Transcriptome
INTRODUCTION
Dry age-related macular degeneration (AMD) is a progressive state that causes blindness primarily due to the loss of retinal pigment epithelium (RPE). Damage to RPE cells that function both as a blood-retinal barrier and regulator of the overlying photoreceptor (PR) layer is closely associated with dry AMD. Early-stage dry AMD is characterized by yellowish deposits (drusen) below the Bruch membrane and disordered pigmentation in the macular RPE layers (Jager et al., 2008). The accumulation of lipofuscin in the RPE of the human eye is further accelerated by aging (Yakovleva et al., 2020), particularly in patients with progressive dry AMD (Holz et al., 1999). The advanced form of dry AMD manifests as the irreversible loss of the RPE and PRs that eventually leads to irreversible vision loss. Although antioxidant vitamins and zinc supplements help to slow dry AMD progression, an effective treatment is not presently available.
The etiology of dry AMD is complex and remains unresolved. However, accumulating evidence shows that accelerated aging-like changes in the RPE play a fundamental role in the development of AMD. The lipofuscin fluorophore N-retinylidene-N-retinylethanolamine (A2E) undergoes photo-oxidation and generates oxygen adducts in the presence of blue light (BL) and oxygen; thus, it might contribute to RPE cell degeneration (Wielgus et al., 2010). These adducts subsequently increase oxidative stress and damage proteins and DNA in RPE cells (Ferrington et al., 2016). Apoptosis is induced by A2E in model RPE cells irradiated with low-intensity BL that alone does not induce RPE cell death. The roles of RPE cells include functioning as a blood-retinal barrier and maintaining the viability and function of PRs by supplying nutrients and removing PR by-products generated by the visual cycle.
Degeneration of the RPE induced by light is a popular model for studies of dry AMD in vitro or in vivo. Exposing A2E-loaded RPE cells to BL (400-480 nm) increases mitochondrial and cytoplasmic superoxide dismutase (SOD) activities (Marie et al., 2018), and RPE cell dysfunction (Boutzen et al., 2020), including inflammation, complement, lipid abnormalities, and oxidative stress (Marie et al., 2018; Alaimo et al., 2019). Although A2E itself has a low phototoxic effect, human RPE (ARPE-19) cells treated with A2E and then exposed to BL results in a significant decrease of cell viability in vitro (Jin et al., 2017; Kim et al., 2018a, 2018b). At the same time, the expression of genes in TNFα signaling, UV response, complement, p53, and apoptosis pathways are significantly affected. Additionally, photoactivation of A2E generates reactive oxygen species (ROS) and impairs autophagy in ARPE-19 cells (Jeong et al., 2019). Furthermore, studies in vivo have shown that treatment with A2E and BL irradiation caused cell death of RPE in vitro and retinal damage such as loss of PR nuclear layers and neuronal transmission dysfunction in rat or mouse eyes (Lee et al., 2016; Lin et al., 2019; Fontaine et al., 2020). Most of these studies imply a crucial role of A2E for RPE cell death in dry AMD pathology, but RPE dysfunction and vision loss are chronic and occur over a long period of time. Moreover, a major limitation in studying dry AMD pathology is that it is very difficult to obtain a donor eye from a patient, which makes it virtually impossible to understand the correlation between pathological changes and disease occurrence over time at a clinical level. Thus, additional model systems for studying dry AMD mechanisms and pathobiology are needed.
Here, we investigate the effects of long-term exposure to BL on RPE cells in vitro. We hypothesized that prolonged exposure to sub-lethal doses of A2E and BL induces RPE cell dysfunction that is closely related to the clinical and pathophysiological status at various stages of dry AMD. ARPE-19 cells were incubated with a non-cytotoxic dose of A2E and repeatedly exposed to BL over several months; then, we investigated changes in transcriptomes at each stage of AMD using RNA-seq. Our results suggested that BL induces damage to RPE cells via molecular-level mechanisms that differ according to the stages of pathological damage in dry AMD. Moreover, these results provide further insight into the pathogenesis of dry AMD induced by blue light.
MATERIALS AND METHODS
Cell culture
Human retinal pigment epithelial cells (ARPE-19; American Type Culture Collection, Manassas, VA, USA) were grown in Dulbecco’s modified Eagle’s medium F-12 (DMEM/F-12; Welgene, Daegu, Korea) containing 10% fetal bovine serum at 37°C under a 5% CO2 atmosphere.
Long-term exposure in ARPE-19 cells
We seeded ARPE-19 cells (5×104 cells/well) in 6-well plates and incubated them with 5 μM A2E (AptaBio, Yongin, Korea) every other day and exposed them to BL (BL; 430 nm, 6,000 lux, 5 min/day) for 0 (D-0), 40 (D-40), 100 (D-100), and 120 (D-120) days. The device for irradiating the ARPE-19 cells with blue light is equipped with an LED on the top of the inside of the device to emit a blue light peak at 430 nm (Four Tech, Gunpo, Korea). The temperature inside the device was kept constant during blue light irradiation (5 min). The intensity of blue light was measured using a display color analyzer (CA-310, Konica Minolta Sensing, Inc., Tokyo, Japan). Total RNA was extracted at each time point using TRIzol (Invitrogen, Carlsbad, CA, USA).
mRNA sequencing
Total RNA was processed to prepare an mRNA sequencing (mRNA-seq) library using TruSeq Stranded mRNA kits (Illumina Inc., San Diego, CA, USA) (Yang et al., 2020). Polyadenylated mRNAs were purified using poly-T oligo-coupled magnetic beads. The mRNAs were fragmented using divalent cations at elevated temperatures. First- and second-strand cDNAs were synthesized from the fragmented RNA by reverse transcriptase with random primers and DNA polymerase I. The cDNA fragments were purified and enriched by the polymerase chain reaction (PCR) to create cDNA libraries that were sequenced using a NextSeq500 instrument (Illumina Inc.). The original image data were converted into sequence data and stored in the FASTQ format. Genes with an absolute fold change (FC) >2 and a false discovery rate (FDR) of <0.05 between groups (n=2) were considered to be differentially expressed. Pathways were analyzed using Enrichr (Kuleshov et al., 2016). Functional enrichment was analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) gene sets in the Molecular Signatures Database (MSigDB; https://www.gsea-msigdb.org/gsea/msigdb).
Real-time quantitative PCR (RT-qPCR)
Total RNA isolated from APRE-19 cells was reverse-transcribed using iScript cDNA synthesis kits (Bio-Rad Laboratories, Hercules, CA, USA) in a total volume of 20 μL containing 2 μL of RT products (Ul-Haq et al., 2019). Quantitative PCR proceeded on a Roche Light Cycler® 480II system with SYBR Green I Master Mix (Roche, Mannheim, Germany) and the primers listed in Table 1. Data are shown as the means ± standard deviation (SD) of triplicate RT-qPCR reactions using the same cDNA samples.
Table 1.
Primer sequences for RT-qPCR
| Gene | Forward | Reverse |
|---|---|---|
| COL1A1 | GTGGCCCAGAAGAACTGGTA | CGCCATACTCGAACTGGAAT |
| CCND1 | GCTGCGAAGTGGAAACCATC | CCTCCTTCTGCACACATTTGAA |
| DUSP5 | GTCCTCACCTCGCTACTC | GGGCTCTCTCACTCTCAAT |
| CXCL8 | CTGGCCGTGGCTCTCTTG | CCTTGGCAAAACTGCACCTT |
| 18S | GAGGATGAGGTGGAACGTGT | TCTTCAGTCGCTCCAGGTCT |
Data collection and analysis of transcriptome datasets
Clinical transcriptome data for comparison with long-term exposure models in vitro were obtained from literature (Radeke et al., 2007). We analyzed the DNA microarray results of retinal tissues donated by three patients (aged 78, 84, and 85 years, AMD) with clinically diagnosed atrophic AMD and normal donors (aged 78 and 84 years, CTR).
Statistical analysis
The RT-qPCR data were statistically analyzed using two-tailed Student t-tests. Values with p<0.05 were considered significantly different.
RESULTS
Identification of differentially expressed genes (DEGs) in long-term exposure model
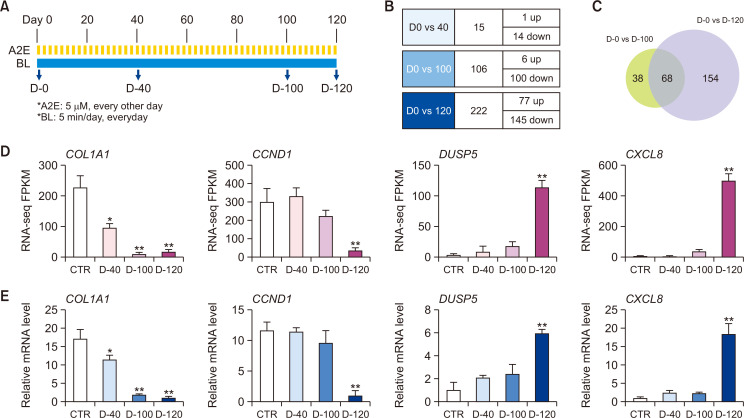
Studies of transcriptomes and related pathways in dry AMD have been limited to RPE cell lines exposed to A2E and BL for short periods or retinal tissues from patients with advanced AMD. However, because dry macular degeneration occurs slowly, current approaches are insufficient to understand changes in gene transcription as the disease progresses and thus have limited ability to identify the genes responsible for AMD pathogenesis. Therefore, we designed a long-term model to overcome these limitations and determine the molecular mechanisms involved in the pathogenesis of dry AMD. We applied a low concentration of A2E (5 μM) to ARPE-19 cells at 2-day intervals and exposed them to BL for 5 min/day for 120 days (Fig. 1A). To identify DEGs in this ARPE-19 model of dry AMD in vitro, mRNA expression was measured on days 0, 40, 100, and 120 (D-0, D-40, D-100, and D-120, respectively). The RNA-seq results showed that 15 (1 upregulated and 14 downregulated), 106 (6 upregulated and 100 downregulated), and 222 (77 up upregulated and 145 downregulated) genes were differentially expressed on D-40, -100, and -120, respectively (Fig. 1B). A few DEGs were found during the early stage (D-40), and then, the numbers of DEGs significantly increased towards the later stage. About 64.2% of genes with altered expression during the middle phase (D-100) were also differentially expressed during the late phase (D-120). Notably, 154 new DEGs in addition to the 68 overlapping with D-100 were identified in the late (D-120) phase, compared with the middle (D-100) phase (Fig. 1C). The RNA-seq results were verified by RT-qPCR. The mRNA levels of downregulated COL1A1 and CCND1 genes and upregulated DUSP5 and CXCL8 genes were similar to the RNA-seq findings (Fig. 1D, 1E).
Fig. 1.
Differential gene expression in ARPE-19 cell models of long-term exposure to BL and A2E. (A) Design of experiment to identify specific expression of genes in ARPE-19 cells exposed to BL (430 nm, 6,000-lux, 5 min/day) and A2E (5 μM every other day) on days 0 (D-0), 40 (D-40), 100 (D-100), and 120 (D-120). (B) Numbers of upregulated or downregulated genes at D-40, D-100, and D-120 compared with D-0. Genes with |FC|>2 and FDR<0.05 between groups were considered to be DEGs. (C) Venn diagram of overlap of D-100 (green) and D-120 (purple) specific genes in ARPE-19 cells induced by BL and A2E. (D) FPKM levels of COL1A1, CCND1, DUSP5, and CXCL8 genes obtained from RNA-seq. (E) Validation of RNA-seq results by RT-qPCR shows mRNA levels of COL1A1, CCND1, DUSP5, and CXCL8 genes in ARPE-19 cells normalized to that of 18S rRNA. Results are shown as means ± SD (n=3). *p<0.05; **p<0.01 (t-tests). A2E, N-retinylidene-N-retinylethanolamine; AMD, age-related macular degeneration; ARPE, human retinal pigment epithelial cells; BL, blue light DEGs, differentially expressed genes.
Pathway analysis of DEGs in long-term exposure model
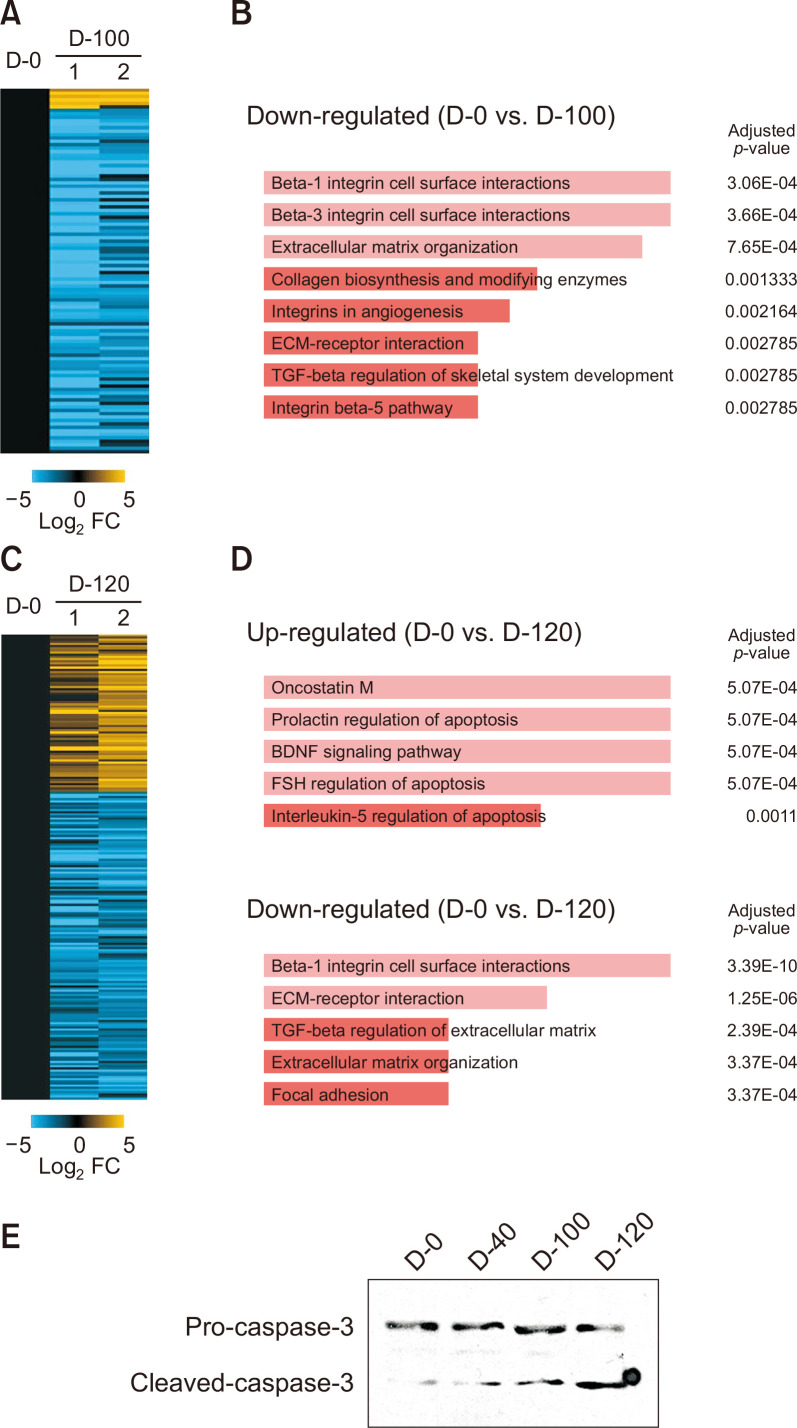
The global expression profiles of A2E-laden RPE cells irradiated with BL for various periods would be valuable for elucidating the mechanism of dry AMD. The RNA-seq data from D-100 revealed 106 DEGs in RPE cells incubated with A2E and exposed to BL compared with control group (D-0). We analyzed pathways based on these findings using Enrichr (BioPlanet 2019) (Kuleshov et al., 2016). At D-100, the three pathways that were most significantly enriched in DEGs were beta-1 integrin cell surface interaction, beta-3 integrin cell surface interactions, and extracellular matrix organization (COL1A1, BMP1, COL5A1, COL7A1, COL5A2, MMP2, THBS1, PLAU, and L1CAM). The expression of genes corresponding to collagen biosynthesis, modifying enzymes, and the ECM-receptor interaction pathway was similarly downregulated (Fig. 2A, 2B, Supplementary Table 1). We identified 222 DEGs in D-120 compared with D-0 (77 upregulated and 145 downregulated, Fig. 2C). Pathway analysis revealed upregulated genes associated with inflammation and apoptosis such as oncostatin (IL11, SOCS3, DYRK3, SERPINB2, MYC, HP, HMOX1, ESR1, and HK2), prolactin regulation of apoptosis (SOCS3, HMOX1, TRIB1, PHLDA1, DUSP6, and MAP3K5), brain-derived neurotrophic factor signaling pathway (DUSP5, IL11, PCLO, MAST4, MYC, TFPI2, UPP1, and GEM), follicle-stimulating hormone regulation of apoptosis (DYRK3, BCL11A, TFPI2, UPP1, ESR1, HK2, GEM, and MAP3K5), and interleukin-5 regulation of apoptosis (DUSP5, TFPI2, UPP1, DUSP6, BIRC3, and RELB) (Fig. 2D, upper panel). Similarly, with D-100, the expression of genes corresponding to pathways associated with cell-cell interaction was also downregulated on D-120 (Fig. 2D, lower panel). These results suggested that BL irradiation initially affects interactions between RPE cells and that prolonged exposure might influence inflammatory and cell cycle processes. Consistent with the pathway analysis results, intracellular caspase-3 was gradually activated as passage increased (Fig. 2E).
Fig. 2.
Pathway analysis of differentially expressed genes in long-term exposure model. (A, C) Heatmap of RNA-seq findings of gene expression at D-100 (A) or D-120 (C) versus control (D-0). Values are log2 fold change (FDR<0.05) relative to control (D-0). (B, D) Pathways associated with DEGs determined by Enrichr at D-100 (B) and D-120 (D) versus control (D-0). (E) The cleaved caspase-3 was determined by Western blotting analysis in ARPE-19 cells (D-0, D-40, D-100, and D-120).
Gene enrichment analysis of stage-specific expression changes by long-term exposure
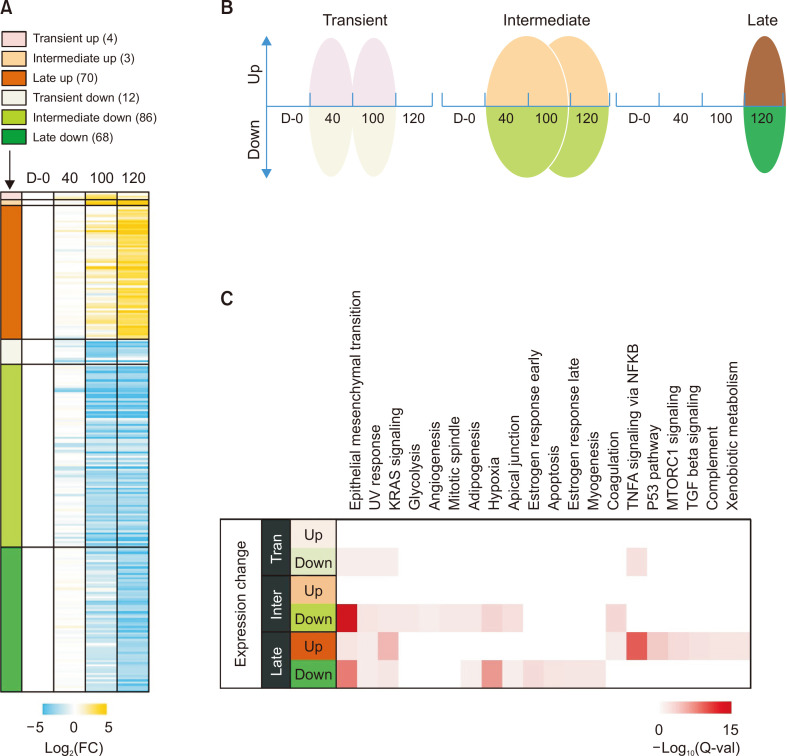
Genes with significant changes in expression at the early stage might affect PRE cell dysfunction and trigger other changes during the middle and late stages. Similarly, functional damage at an early stage induces cellular physiological changes at a later stage and consequently results in AMD lesions. Therefore, to further investigate the relationship between gene expression changes, we separately analyzed DEGs with increased or decreased expression during the early, middle, and late stages. Thus, the DEGs identified by long-term models of dry AMD in vitro were divided into six groups comprising transient increase and decrease, intermediate increase and decrease, and late increase and decrease (Fig. 3A, 3B). Transient increases or decreases included genes that were significantly altered (|FC|>2, FDR<0.05) only at D-40 or D-100. Intermediate increases or decreases included genes that were altered between D-40 and D-100 or between D-100 and D-120. Late increases or decreases involved genes that were altered only at D-120.
Fig. 3.
Gene enrichment analysis of stage-specific changes in expression by long-term exposure to BL and A2E. (A) Heatmap of RNA-seq of gene expression in ARPE-19 cells on D-0, D-40, D-100, and D-120. DEGs were divided into six groups: transiently upregulated (pink), transiently downregulated (light green), intermediate upregulated (orange), intermediate downregulated (green), late upregulated (brown), or late downregulated (dark green). Values are log2 fold change versus control (D-0). Transient increases or decreases included genes that were significantly altered (|FC|>2 and FDR<0.05) only at D-40 or D-100. Intermediate increases or decreases included genes that were altered between D-40 and D-100 or between D-100 and D-120. Late increases or decreases involved genes that were altered only at D-120. (B) Group classification according to differential changes in gene expression at various stages in model of long-term exposure to BL and A2E in vitro. (C) Molecular functions are categorized according to hallmark gene sets in MSigDB (Molecular Signature Database).
We identified cell biological pathways affected by DEGs at each time point using MsigDB (Fig. 3C). The results showed that genes transiently downregulated by BL in RPE cells incubated with A2E were significantly enriched in pathways associated with epithelial-mesenchymal transition (EMT), UV-response, and KRAS signaling. These genes consistently and significantly declined during the middle and late stages. The continuous decrease in the expression of genes involved in the EMT was consistent with the results of the DEG analysis (Fig. 2). Genes involved in glycolysis, angiogenesis, and mitotic spindles were selectively downregulated in the intermediate group. In contrast, genes involved in adipogenesis, hypoxia, apical junctions, and coagulation were affected in the intermediate and late groups. These results suggested that the BL effects on glycolysis, angiogenesis, and mitotic spindle precede those on adipogenesis, hypoxia, apical junctions, and coagulation. Additionally, genes involved in inflammatory pathways such as TNFα signaling through NF-κB, TGF-β signaling, and complement and cell cycle pathways, such as mTORC1 signaling, p53 pathway, and apoptosis, were significantly altered in the late group. Specifically, the genes corresponding to TNFα signaling through NF-κB were downregulated during the early stage (p=1.28E-03) and then remarkably upregulated at the late stage (p=1.10E-10). Taken together, these results indicated that a cascade of molecular biological changes occurs either continuously or sequentially when RPE cells are exposed to BL for long periods.
Correlation between long-term exposure model in vitro and dry AMD
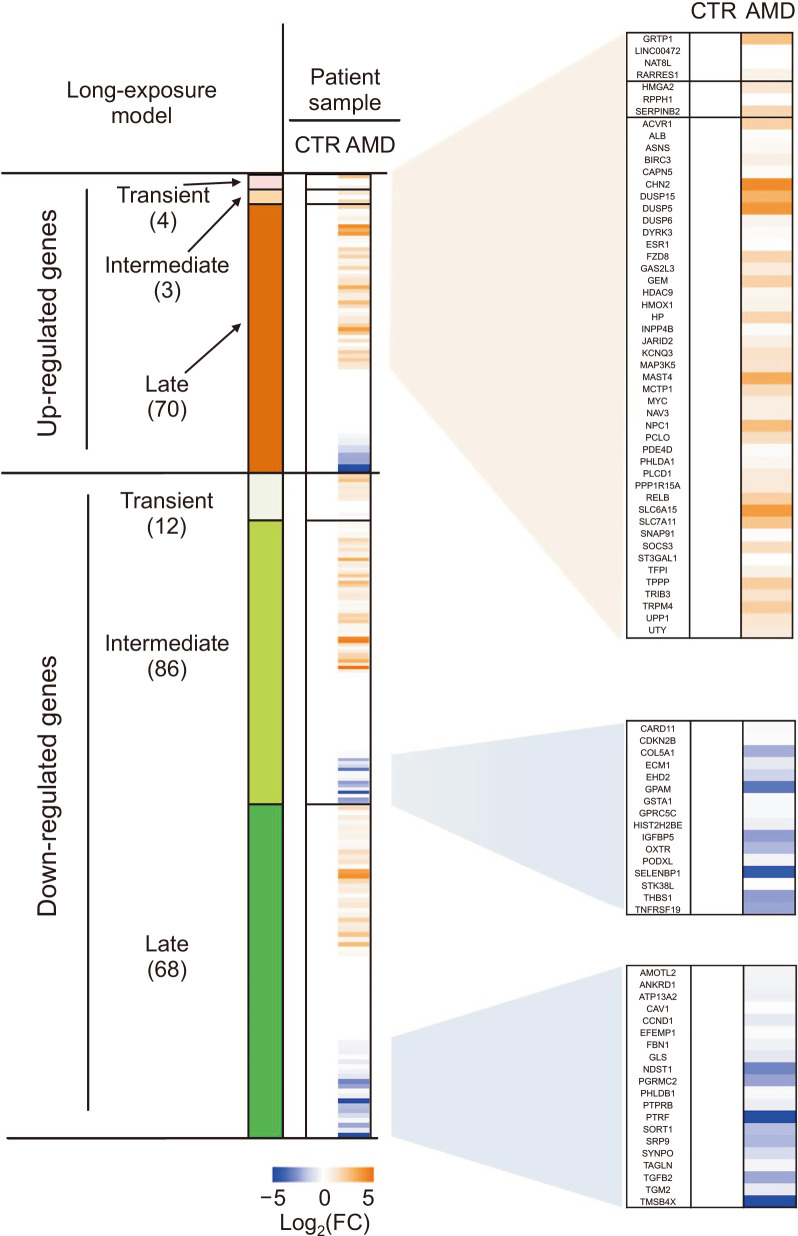
We compared our RNA-seq results with gene expression profiles in retinas from AMD patients to validate the clinical relevance of our model. We analyzed retinal tissues donated by three patients (aged 78, 84, and 85 years, AMD) with a clinical diagnosis of atrophic AMD and by two normal donors (aged 78, 84 years, CTR) using a DNA microarray (Radeke et al., 2007). The DEGs that increased late, decreased intermediate, or decreased late in our RNA-seq were similarly increased or decreased in samples from patients with atrophic AMD (Fig. 4). In particular, the genes that increased late in our model also increased in samples from the patients (|FC|>2) at a higher rate than those in the transient or intermediate groups. These results indicated that our RPE cell model of long-term exposure in vitro is clinically relevant to dry AMD.
Fig. 4.
Correlation between long-term exposure model in vitro and dry AMD. Comparison of differentially expressed genes between ARPE-19 cells exposed to BL+A2E and eye samples from three patients with dry AMD (aged 78, 84, 85 years, AMD) and two normal donors (aged 78 and 84 years, CTR). Heatmap shows that the genes identified long-term exposure model in vitro overlap with DEGs in patients with dry AMD. In clinical transcriptome data analysis, genes with |FC|>2 and FDR<0.05 were considered differentially expressed.
DISCUSSION
Clinically, dry AMD including early and geographic atrophy (GA), accounts for approximately 90% of all patients with AMD (Holz et al., 2014). The early stage of dry AMD is when yellowish drusen (hyperpigmentation, a normal feature of aging) is deposited in the macula. Later, pigment changes and RPE disturbances are visible and visual function is often affected (Ferris et al., 2013). During the advanced dry AMD, also known as geographic atrophy, RPE cells are lost and the loss of central vision progresses (Rickman et al., 2013). These pathological features show that RPE dysfunction plays a central role in the development of PR loss in dry AMD.
The combination of BL illumination and A2E is toxic to RPE, ARPE-19, and primary porcine PRE cells in vitro (Marie et al., 2018). The mechanism of RPE phototoxicity has been more widely studied in APRE-19 cells, suggesting that ARPE-19 could serve as a model system for studies of dry AMD in vitro. However, from a clinical perspective, RPE cells are gradually exposed to relatively small amounts of A2E and BL throughout their lifetime, and the current model system does not precisely reflect this environment. Here, we showed that progressive RPE dysfunction due to long-term exposure to BL and A2E in ARPE-19 cells is similar to the stages of dry AMD from the viewpoint of gene expression. Genes corresponding to extracellular matrix proteins were downregulated in ARPE-19 cells exposed to BL and A2E for 40 days, resulting in phenotypic changes resembling those of the early stage of dry AMD. Changes in extracellular matrix components lead to increased deposition of lipids and protein components in the Bruch membrane, which changes during the early stages of AMD (Al Gwairi et al., 2016).
A genome-wide meta-analysis of AMD associations identified extracellular matrix tissue (VTN, COL15A1, COL8A1, and MMP9) and collagen fibrillar (COL15A1, COL8A1, and MMP9) pathways during early and intermediate AMD. The complement system (C3, CFH, C9, CFI, and CFB) and lipoprotein metabolism (ABCA1, CETP, LIPC, and APOE) pathways are specifically changed in patients with advanced AMD (Winkler et al., 2020). We found altered transcription of genes associated with cell-cell interaction, extracellular matrix organization, ECM-receptor interaction, and collagen biosynthesis in ARPE-19 cells exposed to BL and incubated with a non-lethal dose of A2E for 100 days in vitro. Such exposure for 120 days resulted in the upregulation of genes enriched in oncostatin and apoptosis and the prolonged downregulation of genes enriched in cell-cell interaction, extracellular matrix organization, ECM-receptor interaction, and collagen biosynthesis. These results are similar to those in patients with AMD. Specifically, the genes corresponding to TNFα signaling through NF-κB were upregulated during the late stage, which differed from previous findings of TNFα signaling, being a major pathway that is rapidly affected by a single exposure of high-dose A2E and BL (Pham et al., 2021). Notably, the changes in late gene (e.g., CHN2, DUSP5, and SLC6A15) expression in our model correlated with the changes in expression that are characteristic of patients with advanced dry AMD dry AMD (Radeke et al., 2007). Thus, our model of long-term exposure in vitro is clinically relevant to dry AMD.
To our knowledge, this is the first study to identify genome-wide changes in gene expression in ARPE-19 cells after prolonged exposure to low doses of A2E and BL. Our findings not only provide a new perspective on the pathogenesis of dry AMD, but also provide a useful model for studying the mechanism of dysfunction induced by BL in RPE cells. The novel information about the different stages of the molecular pathogenesis of dry AMD provides new insights into identifying novel therapeutic targets to treat dry AMD by reducing lasting damage or by replacing, repairing, or regenerating damaged cells.
SUPPLEMENTARY MATERIALS
ACKNOWLEDGMENTS
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2021R1A2C1011132) to K.W.J. and National Natural Science Foundation of China (No. 81960667) to H.L.J.
Footnotes
CONFLICT OF INTEREST
The authors have declared that there is no conflict of interest.
REFERENCES
- Al Gwairi O., Thach L., Zheng W., Osman N., Little P. J. Cellular and molecular pathology of age-related macular degeneration: potential role for proteoglycans. J. Ophthalmol. 2016;2016:2913612. doi: 10.1155/2016/2913612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaimo A., Liñares G. G., Bujjamer J. M., Gorojod R. M., Alcon S. P., Martínez J. H., Baldessari A., Grecco H. E., Kotler M. L. Toxicity of blue led light and A2E is associated to mitochondrial dynamics impairment in ARPE-19 cells: implications for age-related macular degeneration. Arch. Toxicol. 2019;93:1401–1415. doi: 10.1007/s00204-019-02409-6. [DOI] [PubMed] [Google Scholar]
- Boutzen J., Valet M., Alviset A., Fradot V., Rousseau L., Français O., Picaud S., Lissorgues G. Impedance spectroscopy study of the retinal pigment epithelium: application to the monitoring of blue light exposure effect on A2E-loaded in-vitro cell cultures. Biosens. Bioelectron. 2020;161:112180. doi: 10.1016/j.bios.2020.112180. [DOI] [PubMed] [Google Scholar]
- Ferrington D. A., Sinha D., Kaarniranta K. Defects in retinal pigment epithelial cell proteolysis and the pathology associated with age-related macular degeneration. Prog. Retin. Eye Res. 2016;51:69–89. doi: 10.1016/j.preteyeres.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris F. L., Wilkinson C. P., Bird A., Chakravarthy U., Chew E., Csaky K., Sadda S. R. Beckman Initiative for Macular Research Classification Committee, author. Clinical classification of age-related macular degeneration. Ophthalmology. 2013;120:844–851. doi: 10.1016/j.ophtha.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine V., Monteiro E., Fournié M., Brazhnikova E., Boumedine T., Vidal C., Balducci C., Guibout L., Latil M., Dilda P. J., Veillet S., Sahel J. A., Lafont R., Camelo S. Systemic administration of the di-apocarotenoid norbixin (BIO201) is neuroprotective, preserves photoreceptor function and inhibits A2E and lipofuscin accumulation in animal models of age-related macular degeneration and Stargardt disease. Aging (Albany N.Y.) 2020;12:6151–6171. doi: 10.18632/aging.103014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz F. G., Bellmann C., Margaritidis M., Schütt F., Otto T. P., Völcker H. E. Patterns of increased in vivo fundus autofluorescence in the junctional zone of geographic atrophy of the retinal pigment epithelium associated with age-related macular degeneration. Graefes Arch. Clin. Exp. Ophthalmol. 1999;237:145–152. doi: 10.1007/s004170050209. [DOI] [PubMed] [Google Scholar]
- Holz F. G., Strauss E. C., Schmitz-Valckenberg S., van Lookeren Campagne M. Geographic atrophy: clinical features and potential therapeutic approaches. Ophthalmology. 2014;121:1079–1091. doi: 10.1016/j.ophtha.2013.11.023. [DOI] [PubMed] [Google Scholar]
- Jager R. D., Mieler W. F., Miller J. W. Age-related macular degeneration. N. Engl. J. Med. 2008;358:2606–2617. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- Jeong S. Y., Gu X., Jeong K. W. Photoactivation of N-retinylidene-N-retinylethanolamine compromises autophagy in retinal pigmented epithelial cells. Food Chem. Toxicol. 2019;131:110555. doi: 10.1016/j.fct.2019.06.002. [DOI] [PubMed] [Google Scholar]
- Jin H. L., Choung S. Y., Jeong K. W. Protective mechanisms of polyphenol-enriched fraction of Vaccinium uliginosum L. Against blue light-induced cell death of human retinal pigmented epithelial cells. J. Funct. Foods. 2017;39:28–36. doi: 10.1016/j.jff.2017.10.009. [DOI] [Google Scholar]
- Kim J., Jin H. L., Jang D. S., Jeong K. W., Choung S. Y. Hyperoside (quercetin-3-O-β-D-galactopyranoside) protects A2E-laden retinal pigmented epithelium cells against UVA and blue light-induced apoptosis in vitro and in vivo. J. Funct. Foods. 2018a;40:426–437. doi: 10.1016/j.jff.2017.11.021. [DOI] [Google Scholar]
- Kim J., Jin H. L., Jang D. S., Jeong K. W., Choung S. Y. Quercetin-3-O-alpha-l-arabinopyranoside protects against retinal cell death via blue light-induced damage in human RPE cells and Balb-c mice. Food Funct. 2018b;9:2171–2183. doi: 10.1039/C7FO01958K. [DOI] [PubMed] [Google Scholar]
- Kuleshov M. V., Jones M. R., Rouillard A. D., Fernandez N. F., Duan Q., Wang Z., Koplev S., Jenkins S. L., Jagodnik K. M., Lachmann A., McDermott M. G., Monteiro C. D., Gundersen G. W., Ma'ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. L., Kang J. H., Kim H. M., Jeong S. H., Jang D. S., Jang Y. P., Choung S. Y. Polyphenol-enriched Vaccinium uliginosum L. fractions reduce retinal damage induced by blue light in A2E-laden ARPE19 cell cultures and mice. Nutr. Res. 2016;36:1402–1414. doi: 10.1016/j.nutres.2016.11.008. [DOI] [PubMed] [Google Scholar]
- Lin C. H., Wu M. R., Huang W. J., Chow D. S., Hsiao G., Cheng Y. W. Low-luminance blue light-enhanced phototoxicity in A2E-laden RPE cell cultures and rats. Int. J. Mol. Sci. 2019;20:1799. doi: 10.3390/ijms20071799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie M., Bigot K., Angebault C., Barrau C., Gondouin P., Pagan D., Fouquet S., Villette T., Sahel J. A., Lenaers G., Picaud S. Light action spectrum on oxidative stress and mitochondrial damage in A2E-loaded retinal pigment epithelium cells. Cell Death Dis. 2018;9:287. doi: 10.1038/s41419-018-0331-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T. N. M., Shin C. Y., Park S. H., Lee T. H., Ryu H. Y., Kim S. B., Auh K., Jeong K. W. Solanum melongena L. extract protects retinal pigment epithelial cells from blue light-induced phototoxicity in in vitro and in vivo models. Nutrients. 2021;13:359. doi: 10.3390/nu13020359.8e3cdbaf95044e9e8d461c594ec07736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeke M. J., Peterson K. E., Johnson L. V., Anderson D. H. Disease susceptibility of the human macula: differential gene transcription in the retinal pigmented epithelium/choroid. Exp. Eye Res. 2007;85:366–380. doi: 10.1016/j.exer.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Rickman C. B., Farsiu S., Toth C. A., Klingeborn M. Dry age-related macular degeneration: mechanisms, therapeutic targets, and imaging. Invest. Ophthalmol. Vis. Sci. 2013;54:ORSF68–ORSF80. doi: 10.1167/iovs.13-12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ul-Haq A., Jin M. L., Jeong K. W., Kim H. M., Chun K. H. Isolation of MLL1 inhibitory RNA aptamers. Biomol. Ther. (Seoul) 2019;27:201–209. doi: 10.4062/biomolther.2018.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielgus A., Collier R., Martin E., Lih F., Tomer K., Chignell C., Roberts J. Blue light induced A2E oxidation in rat eyes - experimental animal model of dry AMD. Photochem. Photobiol. Sci. 2010;9:1505–1512. doi: 10.1039/c0pp00133c. [DOI] [PubMed] [Google Scholar]
- Winkler T. W., Grassmann F., Brandl C., Kiel C., Günther F., Strunz T., Weidner L., Zimmermann M. E., Korb C. A., Poplawski A., Schuster A. K., Müller-Nurasyid M., Peters A., Rauscher F. G., Elze T., Horn K., Scholz M., Cañadas-Garre M., McKnight A. J., Quinn N., Hogg R. E., Küchenhoff H., Heid I. M., Stark K. J., Weber B. H. F. Genome-wide association meta-analysis for early age-related macular degeneration highlights novel loci and insights for advanced disease. BMC Med. Genomics. 2020;13:120. doi: 10.1186/s12920-020-00760-7.24f90290407540b99d3e0817a15540eb [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovleva M. A., Radchenko A. S., Feldman T. B., Kostyukov A. A., Arbukhanova P. M., Borzenok S. A., Kuzmin V. A., Ostrovsky M. A. Fluorescence characteristics of lipofuscin fluorophores from human retinal pigment epithelium. Photochem. Photobiol. Sci. 2020;19:920–930. doi: 10.1039/C9PP00406H. [DOI] [PubMed] [Google Scholar]
- Yang L., Jin M., Park S. J., Seo S. Y., Jeong K. W. SETD1A promotes proliferation of castration-resistant prostate cancer cells via FOXM1 transcription. Cancers (Basel) 2020;12:1736. doi: 10.3390/cancers12071736.b3f25ce3657c42d58714838fc6e30391 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.