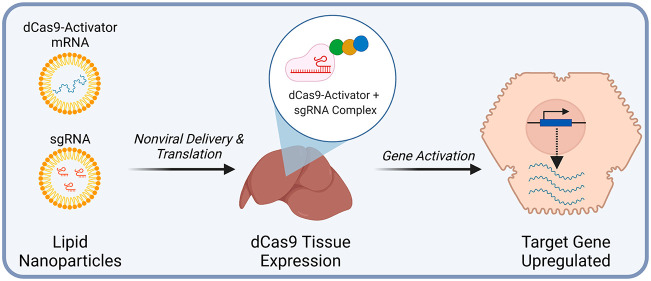
Abstract
Programmable control of gene expression via nuclease-null Cas9 fusion proteins has enabled the engineering of cellular behaviors. Here, both transcriptional and epigenetic gene activation via synthetic mRNA and lipid nanoparticle delivery was demonstrated in vivo. These highly efficient delivery strategies resulted in high levels of activation in multiple tissues. Finally, we demonstrate durable gene activation in vivo via transient delivery of a single dose of a gene activator that combines VP64, p65, and HSF1 with a SWI/SNF chromatin remodeling complex component SS18, representing an important step toward gene-activation-based therapeutics. This induced sustained gene activation could be inhibited via mRNA-encoded AcrIIA4, further improving the safety profile of this approach.
Keywords: CRISPR, dCas9, gene activation, mRNA, lipid nanoparticle, nonviral, in vivo
Genome engineering technologies such as zinc-finger (ZF), transcription activator-like effectors (TALEs), and deactivated Cas9 (dCas9) can be used to manipulate endogenous gene expression through their fusion with both transcriptional activators and epigenetic modifiers.1−5 Early work, primarily demonstrated in vitro, used engineered ZF proteins fused to the herpes simplex virus derived VP16 transactivator or to its tetrameric repeat (VP64) to demonstrate targeted transcriptional gene activation and the benefits of the repeated activator effector. As Cas9-based technology progressed, dCas9 was then utilized to target DNA and demonstrated activation of endogenous genes when fused to VP486 or VP64,7−9 and its performance was improved by the inclusion of p65 and Rta (VPR).10,11 Transcriptional gene activation via CRISPR-targeted epigenetic modifications was later achieved using dCas9 fused to p300, a histone acetyltransferase.5
Recently, there have been demonstrations of these approaches in vivo. The dCas9-based transcriptional gene activation system, using both adeno-associated viruses (AAV) and plasmids, was used to modulate reporter genes and endogenous genes in multiple organs via multiple routes of administration.12 Three additional groups recently utilized AAV in conjunction with VP160,13 VP64,14 and VPR15 to activate three different genes in mouse models: Scn1a to alleviate Dravet Syndrome, Sim1 to rescue obesity in a model of haploinsufficiency, and the cone photoreceptor-specific M-opsin (Opn1mw) in a rhodopsin-deficient mouse model for retinitis pigmentosa. In addition, dCas9-p300 has been implemented in vivo in multiple tissues of transgenic animals.16 Other approaches for delivery in vivo included hydrodynamic delivery of plasmids expressing dCas9 activator systems.17 All of these approaches have advanced the field, but questions remain regarding the long-term safety of AAV18 and other virus-based approaches. Also, there is the issue of inflammation and toxicity associated with DNA electroporation in vivo along with the impractical nature of hydrodynamic delivery. Therefore, there is clearly a need for improved, safer delivery strategies for large dCas9 fusion proteins, especially for their use in relatively healthy individuals. Given the recent successes of synthetic mRNA for vaccines and their inherently improved safety profile,19 we developed synthetic mRNA expressing dCas9 fusion proteins and hypothesized that lipid nanoparticle (LNP) delivery would facilitate mRNA and sgRNA delivery to enable efficient gene activation in vivo. There are many studies demonstrating Cas9 mRNA-based gene editing in different animal models and humans.20−23 However, to our knowledge, mRNA-based gene activation in vivo has not been achieved. As an initial model gene for this work, the glycosyltransferase B4galnt2 was chosen as it is not highly expressed in the liver, muscle, or lymph nodes of C57B/6 mice. Furthermore, the effect of its protein function can easily be detected via lectin-based staining.24 To demonstrate that this approach can be applied to other genes with therapeutic applications, we chose the erythropoietin (Epo) gene as a second target. Epo is a protein hormone secreted by the kidneys that promotes erythropoiesis.25,26 Recombinant forms of this protein are used clinically in scenarios such as chronic renal anemia.27
Results and Discussion
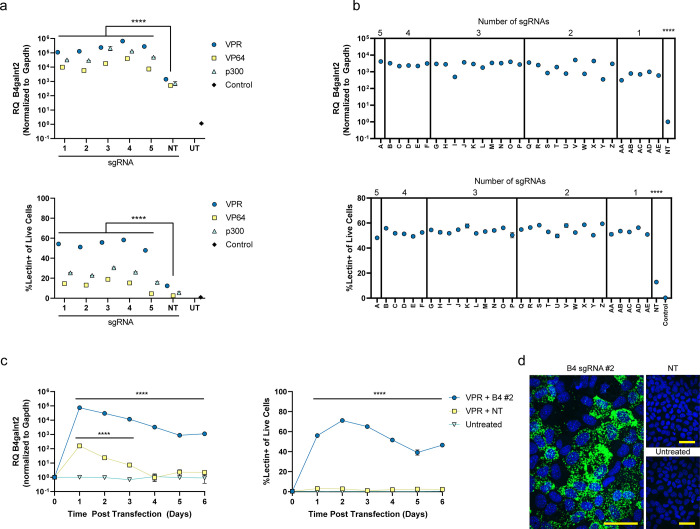
Synthetic mRNAs for dCas9-VP64,7−9 VPR,10 and p300,5 containing a 2A self-cleaving peptide sequence between the activators and mCherry sequences, along with guides previously identified by Sanson et al.28 were screened in AML12 cells for activation of the B4galnt2 gene, identifying five functional sgRNAs (Figure 1a,b and Tables S1 and S2). Dose optimization was performed for viability and percentage of B4galnt2-activated cells using dCas9-VPR mRNA and a single B4galnt2 sgRNA. A dose of 500 ng of mRNA and 500 ng of sgRNA was chosen for in vitro experiments (Figure S1). For simplicity, B4 denotes B4galnt2-targeting guides, and NT denotes nontargeting guides. These sgRNAs were then tested individually with all three activators, demonstrating ∼100–1000-fold activation of the gene by qPCR when compared with nontargeting control sgRNA (Figure 1a, top). Overall, from the RT-qPCR data, VPR was the most efficient among all activators; however, this effect was significantly more pronounced when compared on a single-cell basis via flow cytometry, demonstrating activation in ∼50% of the cells, compared to ∼10 and 20% using VP64 and p300, respectively (Figure 1a, bottom). Comparing mCherry expression levels demonstrates that VPR is a more potent activator, even though the mRNA construct expressed at levels lower than that of p300 or VP64 (Figure S2 and Table S3). All of the possible sgRNA combinations (Table S4) were assessed both by RT-qPCR and flow cytometry for VPR, showing robust activation (∼4000-fold increase) in ∼50% of the cells using all five sgRNAs simultaneously (Figure 1b). Given the transient nature of mRNA expression, time course experiments using VPR were performed in vitro, demonstrating rapid activation in 24 h and durability over 6 days in culture when evaluated both by RT-qPCR and flow cytometry via Dolichos biflorus agglutinin (DBA) lectin staining (Figures 1c and S3). Increased expression of B4galnt2 mRNA was visualized using RNAscope and fluorescence microscopy (Figure 1d). To simultaneously detect B4galnt2 transcripts and lectin staining on a single cell basis, we performed a PrimeFlow assay followed by flow cytometry (Figure S4). When cells were transfected with VPR mRNA and B4galnt2-targeting sgRNAs, ∼60% of the cells were positive for B4galnt2 mRNA and lectin staining, and ∼35% were negative for both staining. Less than 5% of the total cell numbers could be classified as single positive, providing a strong correlation between B4galnt2 gene upregulation and positive lectin staining (Figure S5).
Figure 1.
In vitro gene activation of B4galnt2 in AML12 cells. (a) Single sgRNA screen by qPCR (top) and flow cytometry (bottom) at 24 h post-transfection. (b) Combinatorial screen of five sgRNAs by qPCR (top) and flow cytometry (bottom) at 24 h post-transfection. (c) Time course of gene activation by qPCR (left) and flow cytometry (right). (d) RNAscope assay against B4galnt2 mRNA (green) and DAPI (blue). Scale bar is 50 μm. B4 denotes B4galnt2 sgRNAs. NT denotes nontargeted sgRNAs. UT denotes untreated mice. RQ (relative quantification) denotes the fold change of B4galnt2 mRNA in treated samples relative to untreated samples and normalized to Gapdh mRNA for both. Data are presented as mean ± SEM (n = 3 biological replicates). Statistical significance was assessed using a two-way ANOVA followed by Dunnett’s multiple comparison between the nontargeted condition (****P < 0.0001).
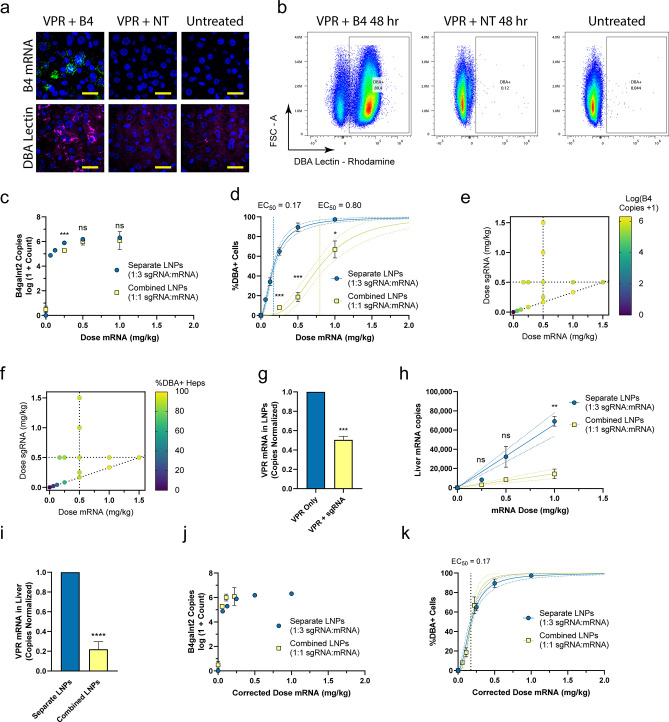
Given the success in vitro, dCas9-VPR mRNA and all five sgRNAs, containing previously identified modifications (Table S1),29 were formulated into a cKK-E12-based LNP together at a 1:1 mass ratio and administered by tail vein injection at 0.5 mg/kg dose of mRNA.29 All five sgRNAs were mixed together in equivalent amounts to form the sgRNA LNPs. The cKK-E12-based LNP was chosen because of its previous proven success of hepatocyte delivery in mice.30 We initially assessed gene activation using lectin staining and a single-molecule-sensitive fluorescent RNAscope assay in liver tissue sections at 1 day postinjection. Fluorescence microscopy revealed abundant activated cells through increased B4galnt2 mRNA puncta and increased lectin staining intracellularly and on the plasma membrane (Figures 2a and S6). With these findings, we repeated the in vivo experiment using three escalating doses of mRNA and sgRNA and assessed activation at 24 and 48 h. For this study, mouse livers were digested into single-cell suspensions in situ based on established protocols followed by processing for RT-qPCR and flow cytometry.31 We gated hepatocytes based on cell size and viability (Figure S7).30 In the B4galnt2-treated groups, we observed a distinct population of lectin positive cells, which constituted up to ∼90% of the viable hepatocytes by 48 h when a 1 mg/kg mRNA dose was used (Figures 2b and S8). By RT-qPCR, we measured increase in B4galnt2 copy number from ∼10 transcripts per 50 ng of isolated RNA in untreated livers to up to ∼106 transcripts in treated livers (Figure S8).
Figure 2.
In vivo dose optimization of B4galnt2 gene activation. (a) Images of liver sections showing RNAscope staining for B4galnt2 mRNA (green), Dolichos biflorus agglutinin (DBA) lectin staining (magenta), and DAPI (blue) at 1 day postinjection. Scale bar is 25 μm. (b) Flow cytometry plots showing DBA lectin staining of hepatocytes at 48 h using 1 mg/kg VPR mRNA and 1 mg/kg sgRNA. B4galnt2 mRNA copy numbers (c) and the percentage of activated hepatocytes (heps) (d) between formulation approaches at varying mRNA doses with constant sgRNA/mRNA mass ratio. Heat maps of B4galnt2 mRNA copies (e) and the percentage of activated hepatocytes (f) with varying mRNA and sgRNA amounts. (g) Direct measurement of LNP encapsulation of mRNA by qPCR. Data were normalized to the separate LNP formulation condition. Data are presented as mean ± SEM (n = 4 technical replicates). (h) VPR mRNA copy numbers from liver samples compared to theoretical doses. Linear regression was performed for each data set (solid line = best fit, dotted lines = 95% CI). (i) Overall delivery of VPR mRNA to livers for each formulation approach is reported as the slope of the linear fits from (h). Data were normalized to the separate LNP formulation condition. Data are presented as mean ±95% CI. (j) Dose-corrected qPCR results from combined and separate formulation experiments. (k) Dose-corrected flow cytometry results from combined and separate formulation experiments. Unless otherwise noted, data represent mean ± SEM (n = 3–4 mice). 4PL curves were fit to data in (d) and (k) (solid lines = best fit curve, dotted lines = 95% CI). An extra sum-of-squares F-test was performed to assess statistical significance between the EC50 values of 4PL fits and slopes of linear fits. When P > 0.05, a combined EC50 value was reported for the curves. Additional statistical significance was assessed using a two-way ANOVA followed by Dunnett’s multiple comparison (c,d,h) and a student’s t test (g,i) between formulation approaches (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
While this formulation approach has been previously used,29 our group has observed reductions in encapsulation of mRNA with increasing length (Figure S9). As a result, we hypothesized that the sgRNA may be outcompeting the mRNA for loading into the LNPs, effectively reducing the in vivo delivery of mRNA. We theorized that an alternative LNP formulation approach in which the sgRNA and mRNA are formulated into separate particles and the final LNPs are mixed prior to injection might result in improved performance. Using the flow cytometry results from the initial dose–response, we chose an mRNA dose of 0.5 mg/kg and a 24 h time point to increase the likelihood of observing an increase in the percentage of activated cells. Compared to the combined formulation approach, we found that a separate formulation approach resulted in a 2-fold increase in the percentage of activated cells in the liver with no significant change in the amount of B4galnt2 mRNA levels in the tissue lysate (Figure S10). Due to improved performance and the convenience of formulation, we elected to use the separate formulation in subsequent experiments. Next, we tested the sensitivity of the system to changes in the amount of mRNA or sgRNA independently. With a fixed mRNA dose at 0.5 mg/kg, changes in the sgRNA dose from 0.167 to 1.5 mg/kg dose resulted in no detectable difference in B4galnt2 mRNA levels or the percentage of activated cells. However, we observed small changes in efficacy when the sgRNA dose was fixed at 0.5 mg/kg, and the mRNA dose was varied from 0.167 to 1.5 mg/kg (Figure S11). Due to the insensitivity of the system to the sgRNA dose, we chose to use a sgRNA/mRNA mass ratio of 1:3 moving forward. Using this ratio, we performed an additional dose–response to characterize the improved performance of the separate formulation approach. We overlaid the results of the separate dose–response at a 1:3 ratio and the results of the combined dose–response at a 1:1 ratio and observed minimal changes in B4galnt2 mRNA transcript levels (Figure 2c). However, we observed a strong leftward shift in the percentage of activated cells; notably, the EC50 dose of a four-parameter logistic (4PL) fit decreased from 0.8 mg/kg in the combined formulation approach to 0.17 mg/kg in the separate formulation approach (Figure 2d). When the results of all three dose–response experiments are combined, we can visualize the response of the system to changes in the dose of mRNA and sgRNA. At doses beyond 0.083 mg/kg sgRNA and 0.25 mg/kg mRNA, we begin to observe diminishing returns; unit increases in sgRNA or mRNA beyond this dose yield smaller increases in the percentage of activated cells or upregulated B4galnt2 transcripts than unit increases below this dose (Figure 2e,f).
The previous results were consistent with our hypothesis that the mRNA was being underdelivered in the combined formulation approach. To directly test this, we quantified the loading of mRNA into the LNPs by performing qPCR on LNP aliquots with and without Triton-X-mediated disruption of the particles. Dilutions were chosen so that if there was no preference for encapsulation of either sgRNA or mRNA over the other, identical copy numbers of encapsulated mRNA should be observed in each condition. In contrast, we observed a 2-fold increase in the amount of VPR mRNA encapsulated using a separate formulation approach compared to the standard combined approach, demonstrating a preference for encapsulation of sgRNA over mRNA (Figure 2g). Similarly, quantification of the copies of VPR mRNA in the livers revealed that the combined approach was underdelivering mRNA at each dose (Figure 2h). The x values of mRNA dose for the combined LNP points in Figure 2h refer to theoretical mRNA doses. Because the RiboGreen assay can only quantify the total mass of encapsulated nucleic acid, it is agnostic to the species of encapsulated nucleic acid (i.e., mRNA or sgRNA). Thus, after formulating combined mRNA and sgRNA at a 1:1 mass ratio input, the theoretical mRNA dose refers to the dose of mRNA that would be delivered if the 1:1 mass ratio was maintained in the encapsulated contents of the LNPs. However, based on the results of the RT-qPCR analysis from liver lysates in (Figure 2i), it is clear that the combined LNP condition is underdelivering mRNA by a factor of approximately 4.5. Thus, the assumption that a 1:1 mass ratio of mRNA to sgRNA is maintained throughout the formulation process is false. Knowing that the actual mRNA delivered is 4.5-fold lower than what we previously assumed, we performed a dose correction on the combined LNP formulation by dividing the theoretical dose of the mRNA from Figure 2c,d by 4.5. This dose correction allowed us to compare the two experiments according to the actual amount of mRNA delivered, which demonstrated strong concordance (Figure 2j,k). These data suggest the primary reason for the lower performance of a combined formulation approach is simply due to a lower amount of mRNA being delivered rather than differential potency of the LNPs.
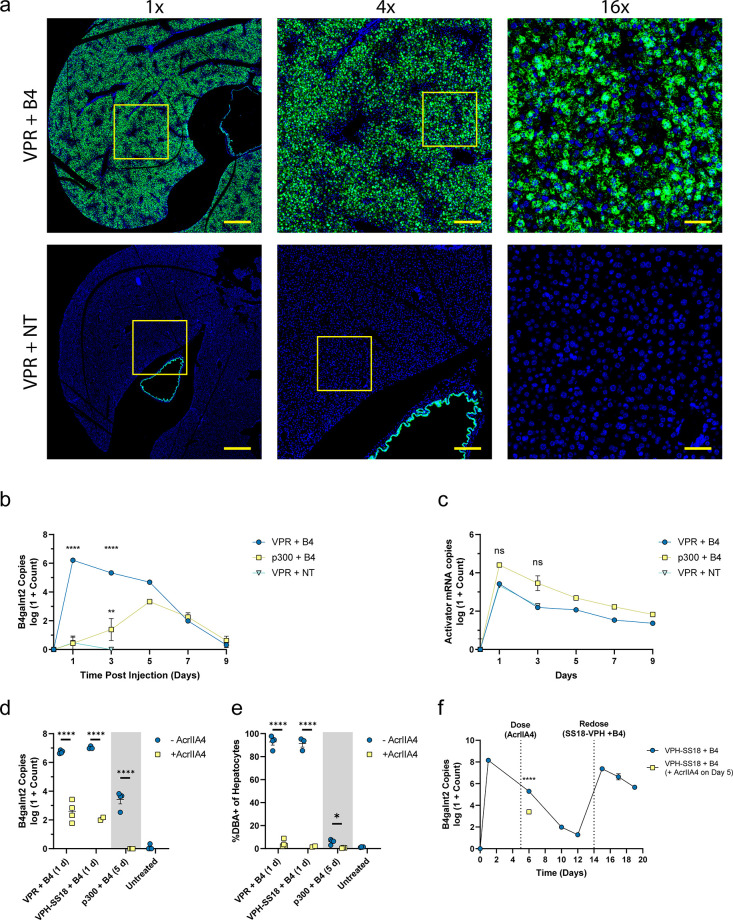
After characterizing doses and formulation approaches in vivo, we chose a model condition of 0.5 mg/kg mRNA and 0.167 mg/kg sgRNA to further investigate the process in the liver. This condition minimized the total amount of nucleic acid delivered to the mice and still resulted in activation of ∼90% of hepatocytes. This result was verified by RNAscope and fluorescence microscopy in whole slide scans (Figure 3a and Figures S12 and S13). To determine if gene activation could be achieved in additional tissue sites, we performed intramuscular injection of LNPs. We observed increased B4galnt2 mRNA levels in multiple cell types within muscle tissue as well as in the periphery of draining lymph nodes; thus, this phenomenon is not unique to hepatocytes (Figures S12–S15).
Figure 3.
In vivo demonstration of optimized B4galnt2 gene activation. (a) Representative slide scan images of liver sections showing RNAscope staining for B4galnt2 mRNA (green) and DAPI (blue). Insets depict the relative locations of 4× and 16× views. Scale bars are 800 μm for 1×, 200 μm for 4×, and 50 μm for 16× images. Time courses of B4galnt2 mRNA (b) and activator mRNA (c) copy numbers from liver tissue over 9 days. B4galnt2 mRNA copy numbers (d) and percentage of activated hepatocytes (e) in mice treated with activator mRNA and B4 sgRNA with or without AcrIIA4 co-delivery. The “–AcrIIA4” groups were dosed with activator mRNA and B4 sgRNA on day 0. The “+AcrIIA4” groups were simultaneously dosed with activator mRNA, B4 sgRNA, and AcrIIA4 mRNA on day 0. All VPR and VPH-SS18-treated mice were euthanized at day 1 postinjection, and all p300-treated mice were euthanized at 5 days postinjection. (f) VPH-SS18 time course with redosing. VPH-SS18 mRNA and B4 sgRNAs were delivered to both groups on day 0. AcrIIA4 treatment was given to one group on day 5, followed by euthanasia on day 6. Remaining mice that did not receive AcrIIA4 mRNA were then redosed with VPH-SS18 mRNA and B4 sgRNAs on day 14. Data represent mean ± SEM (n = 3–4 mice). Statistical significance was assessed using a two-way ANOVA followed by Dunnett’s multiple comparison compared to the NT-treated group (b,c) and a student’s t test (d,e) (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
We next investigated the kinetics of gene activation over time for VPR and p300 gene activator constructs using bulk qPCR. We observed peak B4galnt2 levels of greater than 106 copies per 50 ng of RNA with VPR-mediated gene activation at 24 h. This expression then decreases to control levels by 9 days post-transfection (Figure 3b). The p300 resulted in a peak of ∼103 by day 5 and returned to control levels by 9 days. In addition to RT-qPCR for the B4galnt2 mRNA levels, we measured the copy numbers of delivered activator mRNAs. These mRNAs showed a uniform decay rate after delivery (Figure 3c), indicating that the difference in B4galnt2 levels over time are due to differences in the encoded activator protein and not mRNA half-life. Notably, we observed a ∼100-fold increase in the copy number of B4galnt2 mRNA compared to the copy number of delivered activator mRNA at 24 h. Such high levels of amplification are due to transcriptional activation and likely unachievable with direct delivery of synthetic B4galnt2 encoding mRNA.
As a step toward improved safety of these gene activation approaches, we co-delivered mRNA encoding AcrIIA4 protein which blocks the sgRNA binding site in the Cas9 protein and prevents specific genomic localization of the activator complex.32 In addition to VPR and p300, we tested a third gene activator, VPH-dCas9-SS18 (henceforth abbreviated as VPH-SS18), that combines VP64, p65, and HSF1 with a SWI/SNF chromatin remodeling complex component SS18. The SS18 subunit is sufficient to recruit the full SWI/SNF chromatin remodeling complex.33,34 With simultaneous delivery of LNPs containing AcrIIA4 mRNA along with activator and sgRNA LNPs, we observed a robust difference in bulk B4galnt2 mRNA levels of 4–5 logs in VPR and VPH-SS18 and ∼3 logs in p300 (Figure 3d). Similarly, we observed almost complete inhibition of target activation in hepatocytes by flow cytometry at 24 h for VPR and VPH-SS18 and at 5 days postdelivery for p300 (Figure 3e).
Next, we investigated the kinetics of VPH-SS18-induced gene activation over time. We observed a peak at 24 h of 108 copies that decayed to near baseline levels at 12 days postinjection. At this point, we redosed the mice on day 14 after the initial injection. We saw a similar peak and decay rate in B4galnt2 copies until day 5 after the second injection (Figure 3f). To explore the utility of the AcrIIA4 system in reversing gene activation, we delivered AcrIIA4 mRNA to mice 5 days after activation with VPH-SS18 (Figure 3f). On the following day, we observed a ∼100-fold reduction in B4galnt2 copy number. Because AcrIIA4 protein works by preventing binding of the dCas9 activator protein to genomic DNA, this drop in B4galnt2 copy number indicates that the VPH-dCas9-SS18 protein complex is still interacting with the genomic B4galnt2 promoter by day 5 postinjection. The levels of upregulated B4galnt2 protein were measured by Western blotting (Figure S16). In animals treated with VPR mRNA and B4 sgRNAs, B4galnt2 protein was detectable only on day 1. However, it was detectable on days 1, 5, and 7 in animals treated with VPH-SS18 mRNA and B4 sgRNAs. No detectable increases in B4galnt2 protein were observed in animals treated with p300 mRNA and B4 sgRNAs or with VPR mRNA and nontargeted sgRNAs. The observed molecular weight of B4galnt2 protein was approximately double what is predicted based on sequence, which is likely due to dimerization of the protein, as previously described.25
To demonstrate that this approach can be generalized to other targets, we chose to upregulate the erythropoietin (Epo) gene. The encoded product, Epo, is a 34 kDa serum protein normally secreted by the kidney that regulates erythropoiesis in bone marrow.26,27 Recombinant human Epo protein can be administered as a therapeutic in a variety of clinical scenarios, including chronic renal anemia, HIV infection, and oncologic disorders.35 Epo is also well-established for the evaluation of in vitro-transcribed mRNA performance, and its physiologic effects of increased red blood cell production can be easily measured from blood samples.30,36,37
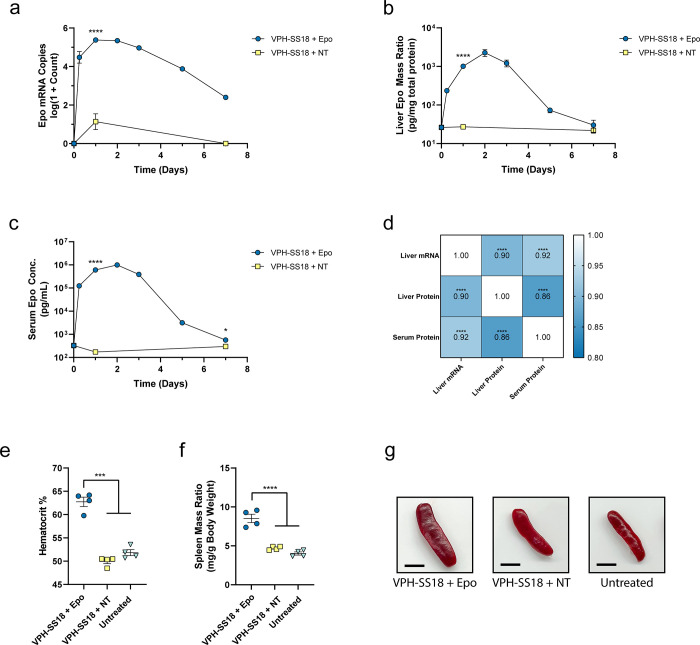
We conducted a time course study to evaluate the potency of VPH-SS18-induced gene activation for the Epo gene using sgRNAs previously identified by Sanson et al.28 (Figure 4). Separate VPH-SS18 LNPs and sgRNA LNPs were delivered to mice, using the previously identified doses of 0.5 mg/kg mRNA and 0.167 mg/kg sgRNA. We observed strong increases in the levels of Epo mRNA in animals treated with targeted sgRNAs to >105 copies per 50 ng RNA, which peaked between 1 and 2 days postinjection (Figure 4a). Similar increases in Epo protein in the liver (Figure 4b) and serum (Figure 4c) peaked on day 2 postinjection. The serum Epo protein concentration reached approximately 1 μg/mL at its maximum, which is approximately 10-fold lower than the peak levels observed by direct mRNA expression of human erythropoietin at a similar mRNA dose of 0.75 mg/kg in cKK-E12 LNPs.30,38 However, while peak Epo levels are lower with our gene activation approach, we observe significant improvement in the duration of elevated Epo protein expression with a broad peak around 2 days. In contrast, direct mRNA approaches demonstrate peak Epo expression at 6 h with rapid decay thereafter.30,38,39 Similarly, rapid kinetics have been observed in a study by Jiang et al., demonstrating a direct mRNA approach for treatment of acute intermittent porphyria.40 These examples highlight the unique ability of gene activation to achieve broad pharmacokinetic profiles from a single dose of LNPs. With broader kinetic profiles, therapeutic levels of a protein of interest could be maintained with a lower frequency of dosing and without sharp spikes in protein amount. This phenomenon is especially important for proteins with short half-lives. Using the matched animal data for liver Epo mRNA, liver Epo protein mass ratio, and serum Epo protein concentration, we performed a Spearman correlation analysis (Figure 4d). The high correlation between the three assays indicates that the upregulated Epo mRNA is readily translated to produce Epo protein, which is freely secreted into the serum.
Figure 4.
In vivo demonstration of optimized erythropoietin gene activation. Time courses of Epo mRNA copy numbers (a) and Epo protein mass ratio (b) from liver tissue over 7 days. (c) Time course of Epo protein concentration in serum. (d) Spearman r correlation matrix of matched animal values for Epo mRNA copy numbers in liver, Epo protein mass ratio in liver, and Epo protein concentration in serum. Values denote the Spearman r coefficient. Stars indicate significance. Hematocrit (e) and spleen mass ratios (f) of animals 7 days postinjection. (g) Representative images of spleens from animals 7 days postinjection. Scale bar is 0.5 cm. Data represent mean ± SEM (n = 4 mice). Statistical significance was assessed using a t test or one-way ANOVA followed by Dunnett’s multiple comparison (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
We next investigated the physiologic response of elevated Epo protein levels to demonstrate that the upregulated protein is functional. On day 7 postinjection, significant increases in hematocrit to approximately 63% were observed in animals treated with Epo-targeting sgRNAs compared to ∼50% in nontargeted sgRNA treated and naïve animals (Figure 4e). Additionally, a significant increase in spleen mass was observed in the targeted group relative to nontargeted and naïve controls (Figures 4f,g and S17). Increased hematocrit demonstrates the signaling function of the upregulated Epo protein is intact, and splenomegaly was expected in these animals due to the increased amount of circulating red blood cells. Similar increases in spleen size can be observed in the setting of polycythemia vera due to red blood cell congestion and extramedullary hematopoiesis.41,42 Overall, this study demonstrates that the gene activation approach can be applied to therapeutically relevant protein targets with clear effects on physiology.
Conclusions
These experiments clearly demonstrate how nonviral, mRNA-expressed dCas9 fusion protein-based gene activation can be utilized in vivo. The data demonstrate the highly efficient nature of nonviral approaches for performing programmable activation of endogenous chromosomal genes. There are a number of important observations and implications of these experiments. First, high levels of activation were achieved in a large majority of the cells in the liver, using low doses of RNA that are well-tolerated in cKK-E12 LNPs without significant elevations in liver enzymes.30 In addition, high levels of activation were achieved in other cell types when administered via the intramuscular route, demonstrating the applicability of the approach to other organ systems.
Second, by formulating the mRNA and sgRNAs separately, encapsulation of mRNA was roughly 4-fold more efficient and enabled significant dose sparing while achieving high levels of activation. This result was not obvious, but after interrogation of the delivered mRNA via qPCR, both in the liver and from the LNPs themselves, it was clear that the discrepancy was due to the preferential encapsulation of guide over mRNA. The majority of the knowledge regarding LNP formulation for mRNA is inherited from studies of siRNA-based formulations, which may be applicable for short mRNAs but has limitations for longer mRNAs, such as those encoding gene activators. We highlight this point in the Figure S9, which shows the encapsulation efficiency of an mRNA decreases with increases in the length. For example, the activator mRNAs used in this study have lengths of approximately 7000 nt, compared to a typical Cas9 mRNA of approximately 4500 nt. The data we presented suggest increases in length would further exacerbate the problem of sgRNA outcompeting mRNA for encapsulation into LNPs. Therefore, formulation of sgRNA and mRNA into separate LNPs is critical to achieving robust performance in vivo. Furthermore, longer mRNAs pose additional challenges during production and formulation due to the risk of degradation during handling.43 Therefore, proper temperature control during formulation and downstream processing followed by frozen storage was essential to maintain the potency of the encapsulated mRNA.
Third, the VPH-SS18 activator containing additional chromatin remodeling factors, SS18, demonstrated ∼100-fold increase in B4galnt2 copies compared to VPR-mediated activation, while maintaining a similar decay rate. The resulting activation and production of B4galnt2 mRNA persisted longer and at higher levels than can likely be achieved using direct expression. Furthermore, similar levels and kinetics were observed after a second dose. This is important because it demonstrates the potential for multiple dosing strategies to be developed for applications where prolonged expression is required.
Fourth, we demonstrated the ability of AcrIIA4 mRNA to inhibit gene activation when given alongside an activator and partially reverse gene activation when given afterward. Anti-CRISPRs are potent inhibitors of Cas9 protein,44,45 and they represent an important step toward improving the safety profile of gene-activation-based therapeutics. We have not come across any previous study that has demonstrated mRNA-based AcrIIA4 to modulate gene activators. Thus, our data represent a proof-of-concept toward the ability to fully reverse transcriptional upregulation induced by gene activators. The combination of transient protein expression of dCas9 activator proteins from mRNA and AcrIIA4-mediated reversal of gene activation creates a safety profile that is highly attractive in the setting of unexpected clinical complications. Furthermore, the ability of AcrIIA4 mRNA to block gene activation when given alongside activator and sgRNA LNPs could be exploited to provide an additional layer of control in gene activation when using LNPs with different tropisms. For example, the delivery of AcrIIA4 mRNA using LNPs with a cell tropism different than that used to deliver activator mRNAs or sgRNAs would provide a method to more precisely control where the gene activation occurs.
Last, we demonstrated that this approach can be used to upregulate a therapeutically relevant protein that maintains physiologic function within the body. In contrast to direct mRNA approaches for protein replacement therapy, gene activation can exhibit much broader pharmacokinetic curves of therapeutic protein over time from a single LNP dose, provided the protein target of interest is encoded in the genome. Further optimization of dCas9 activator constructs will likely lead to powerful improvements in protein replacement therapies over direct mRNA approaches.
Methods
Cell Culture
AML12 (α mouse liver 12, CRL-2254, ATCC) cells were grown in DMEM/F12 medium supplemented with 10% fetal bovine serum, 10 μg/mL insulin, 5.5 μg/mL transferrin, 5 ng/mL selenium, 40 ng/mL dexamethasone, and 1× penicillin–streptomycin at 5% CO2 at 37 °C. Cells were seeded in 96-well plates at 20,000 cells and transfected the following day with sgRNA and activator mRNA using Lipofectamine MessengerMAX.
mRNA Synthesis
Plasmid DNA constructs coding dCas9-VP64 (Addgene Plasmid #47107), dCas9-VPR (Addgene (Plasmid #63798)), dCas9-p300 (Addgene Plasmid #83889), and VPH-dCas9-SS18 (Charles Gersbach) with cleavable mCherry and AcrIIA4 (Addgene Plasmid #101042) were flanked (3′ and 5′) by UTR regions and synthesized by Genscript Inc. mRNA synthesis was performed as previously described.46 Briefly, in vitro transcription (IVT) of linearized plasmid DNA was done using T7 RNA polymerase by incorporating equimolar ATP, CTP, GTP, and N1-methylpseudouridine. Cap1 and the polyA tail were incorporated to the IVT RNA. mRNA was phosphatase-treated, purified, and stored at −80 °C until use.
sgRNA
The in vitro experiments were performed using sgRNA targeting the mouse B4galnt2 gene.28 Guides were synthesized by Synthego Inc. and contained three 2′-O-methyl RNA bases at the 5′ and 3′ ends with phosphorothioate linkages. For in vivo mouse experiments, the same sgRNA sequences were used except that they were additionally modified (with 2′-O-methyl RNA bases and phosphorothioate linkages) as described previously29 and synthesized by Integrated DNA Technologies.
RNA Extraction, cDNA, and qPCR
Total RNA was extracted from cells using RNeasy Plus kit (Qiagen). For sgRNA and activator screening, TaqMan fast advanced cells-to-CT kit (Thermo Fisher Scientific) was utilized with a 96-well format. Two methods were employed for liver processing, depending on whether or not flow cytometry was to be performed on samples. For PCR analysis only, whole livers were collected in gentleMACS M tubes containing 2.5 mL of DMEM and homogenized using a gentleMACS dissociator (Miltenyi Biotec) RNA_01.01 program. For PCR and flow cytometry analysis, in situ liver perfusion and digestion were performed to obtain a liver cell suspension, as described below (Liver Perfusion). RNA was then extracted from either of the processed liver samples using RNeasy Plus kit (Qiagen). Both methods had similar yields of RNA and downstream performance (data not shown). The cDNA for each sample was prepared from 1 μg of total RNA with a high-capacity cDNA reverse transcription kit (Applied Biosystems) as prescribed by the manufacturer. B4galnt2 gene relative quantification was done by TaqMan primer probe set Mm00484661_m1 (Thermo Fisher Scientific) normalized with the Gapdh gene (Mm99999915_g1) (Table S5). Epo gene quantification was done by a TaqMan primer probe set Mm01202755_m1 (Thermo Fisher Scientific). Each 20 μL PCR reaction mix contained 10 μL of TaqMan fast advanced master mix (Thermo Fisher Scientific), 1 μL of primer-probe set (B4galnt2 and Gapdh), water, and 1 μL of cDNA. PCR was performed using a QuantStudio 7 Flex real-time PCR system (Applied Biosystems). Absolute quantification for B4galnt2 gene and the Epo gene was performed using a standard and the same PCR reaction conditions as described above. Similarly, absolute quantification of activator mRNA was done by designing a primer-probe set that amplified the mCherry sequence from the mRNA (Table S5).
LNP Formulation and RiboGreen Assay
The mRNA and sgRNAs were diluted in 10 mM citrate buffer (pH 3) to create the aqueous phases. When pooling sgRNAs, equivalent amounts of each sgRNA were included in the sgRNA fraction. To prepare the organic phases, cKK-E12 (Organix Inc.), cholesterol (Sigma), C14-PEG 2000-PE (Avanti), and DOPE (Avanti) were added to 100% ethanol at a ratio of 35:46.5:2.5:16. A mass ratio of 20 (lipid/mRNA) was used for all in vivo formulations. The two phases were mixed using a NanoAssemblr benchtop device containing a microfluidic cartridge (Precision NanoSystems Inc.) at an aqueous to organic flow rate ratio of 3:1 at 12 mL/min. The LNPs were next diluted 40× in 10 mM Tris buffer and concentrated using 100 kDa MWCO centrifugal filters (MilliporeSigma). After sterile filtering, particle size was determined by dynamic light scattering on a Zetasizer Nano ZS (Malvern). A RiboGreen assay (Invitrogen) was performed to calculate the encapsulation percentage and concentration of mRNA and sgRNA cargos. After size and concentration were characterized, LNP solutions were mixed with sucrose to 5% (w/v) final concentration, aliquoted, and stored at −80 °C until use.
mRNA LNP Encapsulation Quantification by PCR
To determine differences in the encapsulation of activator mRNA under different formulation conditions, absolute quantification by real-time PCR was used. Stored aliquots of LNPs were used in which particles were formulated with either activator mRNA alone or a mixture of activator mRNA and sgRNA at a 1:1 mass ratio. Using the total encapsulated nucleic acid concentration, as measured by RiboGreen, and the assumption that LNPs formulated with sgRNA and mRNA contain both cargos at a 1:1 mass ratio, LNPs were diluted to theoretically contain the same concentration of encapsulated activator mRNA per milliliter. These dilutions were chosen so that if there was no preference for encapsulation of either sgRNA or mRNA over the other, identical copy numbers of encapsulated mRNA should be observed in each condition. Solutions containing LNPs were mixed with an equal volume of Tris-EDTA buffer containing 2% Triton X-100 and incubated for 10 min at 37 °C to lyse the LNPs. Identical solutions were mixed with an equal volume of Tris-EDTA (without Triton X-100) prior to incubation. Volumes (1 μL) equal to 100 ng of the lysed and mock lysed LNP were added to the cDNA reaction mixture (high-capacity cDNA reverse transcription) so that the total volume is 20 μL. Absolute quantification of activator mRNA was performed (by amplifying mCherry sequence present in the mRNA) using 1 μL of this cDNA. The mRNA copy number of the mock lysed LNP solution (free mRNA) was subtracted from the copy number of the lysed LNP solution (total mRNA) to obtain the copy number of encapsulated activator mRNA.
Animal Studies
All animal experiments were performed in accordance with the approved Institutional Animal Care and Use Committee protocol and Georgia Institute of Technology Physiological Research Laboratory policies. Six to eight week old C57BL/6J mice (Jackson Laboratories) were kept in rooms on a 12 h light/dark cycle with ambient temperature between 22.8 and 23.9 °C with 30–40% relative humidity. Food was provided to mice ad libitum. Animals were acclimatized for at least 6 days before the beginning of experiments. Mice were intravenously injected with LNP via tail vein and sacrificed by CO2 asphyxiation. Injections for separate LNP conditions were prepared as needed by mixing a pure solution of LNPs containing mRNA cargo with another containing sgRNA cargo. Dosing volumes for combined LNP conditions were calculated using the total encapsulated nucleic acid concentration from the RiboGreen assay and assuming that a 1:1 mass ratio of sgRNA to mRNA was maintained inside the LNPs. Based on these assumptions, volumes were modified so that an equivalent dose of activator mRNA was delivered to the mice, regardless of formulation approach. For AcrIIA4 studies, additional LNPs containing AcrIIA4 mRNA cargo were mixed into the injection volumes of activator and sgRNA LNPs. The AcrIIA4 LNPs were dosed at 0.5 mg/kg.
Liver Perfusion
After euthanasia, the livers of the mice were perfused and processed as described previously to achieve single-cell suspensions.31 A 22 gauge catheter was used to cannulate the inferior vena cava and connect the vasculature to a syringe pump with surgical tubing. The portal vein was cut as an outflow tract, and the livers were perfused with 25 mL of EGTA solution followed by 25 mL of enzyme buffer solution (EBS) containing 40 μg/mL Liberase (Sigma). Both solutions were prewarmed to 42 °C and injected at 5 mL/min. The livers were then placed in EBS solution on ice. After all perfusions were completed, livers were gently minced with forceps and filtered through 100 μm cell strainers. Cell suspensions were centrifuged at 30g for 5 min at 4 °C to obtain hepatocyte-enriched pellets. These pellets were washed with 1× PBS containing 1% fetal bovine serum, and their concentration was measured using a Cytek Aurora flow cytometer (Cytek) prior to staining.
Cell Staining and Flow Cytometry
Hepatocyte-enriched cell suspensions were adjusted to a concentration of 2 million cells per milliliter and stained using LIVE/DEAD fixable near-IR dye (Invitrogen) at 1:1000 dilution followed by rhodamine-conjugated DBA lectin (Vector Laboratories, RL-1032-2) at a dilution of 1:500. Cells were analyzed using a Cytek Aurora flow cytometer (Cytek). Hepatocytes were gated based on FSC and SSC properties, as previously demonstrated,30 and at least 100,000 live cell events were recorded for analysis of lectin staining.
PrimeFlow and Lectin Staining
At 24 h after transfection, a suspension of AML12 cells was prepared and stained with LIVE/DEAD fixable violet (Invitrogen) at 1:1000 dilution followed by Rhodamine-conjugated DBA lectin (Vector) at a dilution of 1:500. To quantify B4galnt2 and Gapdh transcripts on a single-cell level in treated AML12 cells, a PrimeFlow assay (Invitrogen, 88-18005-204) was performed according to manufacturer’s instructions. Probes against mouse B4galnt2 mRNA (Invitrogen, VB1-3029862-PF), mouse Gapdh mRNA (Invitrogen, VB10-10572-PF), and bacterial dapB (Invitrogen, VF10-10409-PF) were used. After being stained, cells were analyzed by flow cytometry on a Cytek Aurora flow cytometer (Cytek).
Western Blotting
Whole livers were collected in gentleMACS M tubes containing 2.5 mL of DMEM and homogenized using gentleMACS dissociator (Miltenyi Biotec) RNA_01.01 program. Samples were mixed with RIPA buffer (Thermo) supplemented with Halt protease inhibitor cocktail (Thermo) for further lysis. Total protein concentration was determined with by BCA protein assay (Thermo). Samples were mixed with 4× Licor protein loading buffer (LiCor) denatured for 7 min at 95 °C and ran in Bolt 4–12% bis-Tris plus protein gels (Invitrogen). The gels were ran with MES SDS buffer (Thermo) at 200 V for 30 min. Proteins were transferred for 1 h to nitrocellulose membranes at 20 V. Membranes were blocked overnight in 5% bovine serum albumin at 4 °C. Primary antibodies against β-actin (Cell Signaling, 4970S, 1:1000) and B4galnt2 (Novus, NBP1-91229, 1:1000) were incubated overnight at 4 °C in TBST (0.1% Tween 20). Membranes were washed with TBST and incubated with anti-rabbit secondary antibody (LI-COR, 926-68073, 1:5000) in TBST for 1 h at room temperature. Proteins were visualized using an Odyssey infrared imaging system.
Liver Perfusion and Blood Collection for Epo Experiment
After euthanasia, a 22 gauge catheter was used to cannulate the inferior vena cava. Outflow from the catheter was collected and transferred to serum separator tubes (BD 365967) for serum analysis. Heparinized microcapillary tubes (Fisher, 22-362566) were used to collect plasma for hematocrit analysis and sealed with clay sealant. After blood collection, the catheter was connected to a syringe pump with surgical tubing. The portal vein was cut as an outflow tract, and the livers were perfused with 10 mL of 1× PBS. Livers were rinsed in 1× PBS and stored at −80 °C until analysis. Serum samples were allowed to clot for 30 min at room temperature and then centrifuged at 10,000g for 1.5 min. Separated serum was carefully collected from the top of the tube.
Epo ELISA
After mechanical dissociation, liver samples were mixed with cell extraction buffer (Abcam, ab193970) and incubated on ice for 20 min. Samples were centrifuged at 18,000g for 20 min at 4 °C, and supernatants were taken for analysis by ELISA kit (R&D, MEP00B) according to manufacturer’s instructions. Total protein concentration from liver samples was determined by BCA protein assay (Thermo). Serum samples were analyzed directly by the same ELISA kit.
Hematocrit
Microcapillary tubes were centrifuged at 10,000g for 1.5 min to separate out blood components within the tubes. Tubes were imaged on a white background, and the packed red cell height was determined in each image using ImageJ.
Spleen Mass
After euthanasia, the total body mass of each animal was determined on a scale. The spleens were removed and rinsed in 1× PBS. After blotting to remove excess 1× PBS, the mass of each spleen was determined using a microbalance. The spleens were then imaged on a white background.
In Situ Hybridization and Lectin Staining
Liver, muscle, and para-aortic lymph node tissues were collected and fixed in 10% neutral buffered formalin before paraffin-embedded 5 μm sections were prepared by Histowiz services. B4galnt2 mRNA was visualized in tissue sections or 4% paraformaldehyde-fixed AML12 cells using an RNAscope Multiplex fluorescent reagent kit v2 (Advanced Cell Diagnostics, 323136) according to manufacturer’s instructions. Predesigned probes against mouse B4galnt2 mRNA were used (ACD, 529871). Lectin staining was performed by blocking with 5% bovine serum albumin for 1 h and incubating with DBA labeled with rhodamine (Vector Laboratories, RL-1032-2) at 1:100 dilution overnight at 4 °C. After DAPI staining, all samples were mounted with ProLong Gold reagent (Invitrogen) and stored in the dark prior to imaging.
Confocal Microscopy and Slide Scanning
Representative images of AML12 cells and tissue sections were acquired using Plan-Apo 40× 1.3 NA or Plan-Apo 63× 1.4 NA oil objectives on an UltraVIEW spinning disk confocal microscope equipped with a Hamamatsu Flash 4.0v2 CMOS camera. Images were captured and preprocessed using Volocity software (PerkinElmer). Whole slide tissue scans were acquired by the Emory Winship Cancer Tissue and Pathology core on a PerkinElmer Vectra Polaris slide scanner using a 20× 0.5 NA objective. Images were processed and figure images were produced using QuPath software.
Statistical Analysis
The experimental data were analyzed in GraphPad 9 statistical software. Sample size, error bars, and statistical tests are detailed in the figure legends.
Acknowledgments
The authors would like to acknowledge the help of the animal support staff at Georgia Institute of Technology Physiological Research Laboratory (PRL). The authors would also like to acknowledge V. Parihar of the Cancer Tissue and Pathology (CTP) for her help on slide scanning. The table of contents graphic was created with BioRender.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.1c10631.
Supplemental tables and figures referenced in the text (PDF)
Author Contributions
J.P.B., S.B., and P.J.S. conceived the study and designed the experiments. J.P.B., S.B., H.E.P., R.A.H., and J.A.W. performed the experiments. H.E.P. produced the mRNA. J.P.B. and J.A.W. formulated LNPs. N.I. and C.A.G. conceived of, created, and validated the VPH-dCas9-SS18 construct. J.P.B., S.B., and P.J.S. wrote the manuscript. J.P.B., S.B., N.I., C.Z., J.A.W., P.J.S., and C.A.G. edited the manuscript. J.P.B., S.B., and N.I. contributed equally to this work.
This study was supported by the Defense Advanced Research Projects Agency, Grant No. HR00111920008, National Institutes of Health Grant No. U01AI146356, National Science Foundation Grant No. EFMA-1830957, and an Allen Distinguished Investigator Award to C.A.G.
The authors declare the following competing financial interest(s): All authors declare no competing interests related to this work. NI and CAG are inventors on patent applications regarding CRISPR technologies and targeted gene activation. CAG is an advisor to Tune Therapeutics, Sarepta Therapeutics, and Levo Therapeutics, and a co-founder of Tune Therapeutics and Locus Biosciences.
Supplementary Material
References
- Beerli R. R.; Segal D. J.; Dreier B.; Barbas C. F. Toward Controlling Gene Expression at Will: Specific Regulation of the ErbB-2/HER-2 Promoter by Using Polydactyl Zinc Finger Proteins Constructed from Modular Building Blocks. Proc. Natl. Acad. Sci. U. S. A. 1998, 95 (25), 14628–14633. 10.1073/pnas.95.25.14628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerli R. R.; Dreier B.; Barbas C. F. Positive and Negative Regulation of Endogenous Genes by Designed Transcription Factors. Proc. Natl. Acad. Sci. U. S. A. 2000, 97 (4), 1495–1500. 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.; Cong L.; Lodato S.; Kosuri S.; Church G. M.; Arlotta P. Efficient Construction of Sequence-Specific TAL Effectors for Modulating Mammalian Transcription. Nat. Biotechnol. 2011, 29 (2), 149–153. 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. C.; Tan S.; Qiao G.; Barlow K. A.; Wang J.; Xia D. F.; Meng X.; Paschon D. E.; Leung E.; Hinkley S. J.; Dulay G. P.; Hua K. L.; Ankoudinova I.; Cost G. J.; Urnov F. D.; Zhang H. S.; Holmes M. C.; Zhang L.; Gregory P. D.; Rebar E. J. A TALE Nuclease Architecture for Efficient Genome Editing. Nat. Biotechnol. 2011, 29 (2), 143–148. 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Hilton I. B.; D’Ippolito A. M.; Vockley C. M.; Thakore P. I.; Crawford G. E.; Reddy T. E.; Gersbach C. A. Epigenome Editing by a CRISPR-Cas9-Based Acetyltransferase Activates Genes from Promoters and Enhancers. Nat. Biotechnol. 2015, 33 (5), 510–517. 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A. W.; Wang H.; Yang H.; Shi L.; Katz Y.; Theunissen T. W.; Rangarajan S.; Shivalila C. S.; Dadon D. B.; Jaenisch R. Multiplexed Activation of Endogenous Genes by CRISPR-on, an RNA-Guided Transcriptional Activator System. Cell Res. 2013, 23 (10), 1163–1171. 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P.; Kocak D. D.; Vockley C. M.; Adler A. F.; Kabadi A. M.; Polstein L. R.; Thakore P. I.; Glass K. A.; Ousterout D. G.; Leong K. W.; Guilak F.; Crawford G. E.; Reddy T. E.; Gersbach C. A. RNA-Guided Gene Activation by CRISPR-Cas9–Based Transcription Factors. Nat. Methods 2013, 10 (10), 973–976. 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. A.; Horlbeck M. A.; Adamson B.; Villalta J. E.; Chen Y.; Whitehead E. H.; Guimaraes C.; Panning B.; Ploegh H. L.; Bassik M. C.; Qi L. S.; Kampmann M.; Weissman J. S. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159 (3), 647–661. 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder M. L.; Linder S. J.; Cascio V. M.; Fu Y.; Ho Q. H.; Joung J. K. CRISPR RNA-Guided Activation of Endogenous Human Genes. Nat. Methods 2013, 10 (10), 977–979. 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A.; Scheiman J.; Vora S.; Pruitt B. W.; Tuttle M.; Iyer E. P. R.; Lin S.; Kiani S.; Guzman C. D.; Wiegand D. J.; Ter-Ovanesyan D.; Braff J. L.; Davidsohn N.; Housden B. E.; Perrimon N.; Weiss R.; Aach J.; Collins J. J.; Church G. M. Highly Efficient Cas9-Mediated Transcriptional Programming. Nat. Methods 2015, 12 (4), 326–328. 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A.; Tuttle M.; Pruitt B. W.; Ewen-Campen B.; Chari R.; Ter-Ovanesyan D.; Haque S. J.; Cecchi R. J.; Kowal E. J. K.; Buchthal J.; Housden B. E.; Perrimon N.; Collins J. J.; Church G. Comparison of Cas9 Activators in Multiple Species. Nat. Methods 2016, 13 (7), 563–567. 10.1038/nmeth.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H.-K.; Hatanaka F.; Araoka T.; Reddy P.; Wu M.-Z.; Sui Y.; Yamauchi T.; Sakurai M.; O’Keefe D. D.; Núñez-Delicado E.; Guillen P.; Campistol J. M.; Wu C.-J.; Lu L.-F.; Esteban C. R.; Izpisua Belmonte J. C. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-Epigenetic Modulation. Cell 2017, 171 (7), 1495–1507. 10.1016/j.cell.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasante G.; Lignani G.; Brusco S.; Di Berardino C.; Carpenter J.; Giannelli S.; Valassina N.; Bido S.; Ricci R.; Castoldi V.; Marenna S.; Church T.; Massimino L.; Morabito G.; Benfenati F.; Schorge S.; Leocani L.; Kullmann D. M.; Broccoli V. DCas9-Based Scn1a Gene Activation Restores Inhibitory Interneuron Excitability and Attenuates Seizures in Dravet Syndrome Mice. Mol. Ther. 2020, 28 (1), 235–253. 10.1016/j.ymthe.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matharu N.; Rattanasopha S.; Tamura S.; Maliskova L.; Wang Y.; Bernard A.; Hardin A.; Eckalbar W. L.; Vaisse C.; Ahituv N. CRISPR-Mediated Activation of a Promoter or Enhancer Rescues Obesity Caused by Haploinsufficiency. Science 2019, 363 (6424), eaau0629. 10.1126/science.aau0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard K.; Rengstl B.; Oehm P.; Michel K.; Billmeier A.; Hayduk N.; Klein O.; Kuna K.; Ouchan Y.; Wöll S.; Christ E.; Weber D.; Suchan M.; Bukur T.; Birtel M.; Jahndel V.; Mroz K.; Hobohm K.; Kranz L.; Diken M.; Kühlcke K.; Türeci Ö.; Sahin U. An RNA Vaccine Drives Expansion and Efficacy of Claudin-CAR-T Cells against Solid Tumors. Science 2020, eaay5967. 10.1126/science.aay5967. [DOI] [PubMed] [Google Scholar]
- Gemberling M. P.; Siklenka K.; Rodriguez E.; Tonn-Eisinger K. R.; Barrera A.; Liu F.; Kantor A.; Li L.; Cigliola V.; Hazlett M. F.; Williams C. A.; Bartelt L. C.; Madigan V. J.; Bodle J. C.; Daniels H.; Rouse D. C.; Hilton I. B.; Asokan A.; Ciofani M.; Poss K. D.; Reddy T. E.; West A. E.; Gersbach C. A. Transgenic Mice for in Vivo Epigenome Editing with CRISPR-Based Systems. Nat. Methods 2021, 18 (8), 965–974. 10.1038/s41592-021-01207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nihongaki Y.; Otabe T.; Ueda Y.; Sato M. A Split CRISPR–Cpf1 Platform for Inducible Genome Editing and Gene Activation. Nat. Chem. Biol. 2019, 15 (9), 882–888. 10.1038/s41589-019-0338-y. [DOI] [PubMed] [Google Scholar]
- Nguyen G. N.; Everett J. K.; Kafle S.; Roche A. M.; Raymond H. E.; Leiby J.; Wood C.; Assenmacher C.-A.; Merricks E. P.; Long C. T.; Kazazian H. H.; Nichols T. C.; Bushman F. D.; Sabatino D. E. A Long-Term Study of AAV Gene Therapy in Dogs with Hemophilia A Identifies Clonal Expansions of Transduced Liver Cells. Nat. Biotechnol. 2021, 39 (1), 47–55. 10.1038/s41587-020-0741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N.; Hogan M. J.; Porter F. W.; Weissman D. MRNA Vaccines — a New Era in Vaccinology. Nat. Rev. Drug Discovery 2018, 17 (4), 261–279. 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonov D. R.; Gowen B. G.; Boontanrart M.; Roth T. L.; Gagnon J. D.; Mumbach M. R.; Satpathy A. T.; Lee Y.; Bray N. L.; Chan A. Y.; Lituiev D. S.; Nguyen M. L.; Gate R. E.; Subramaniam M.; Li Z.; Woo J. M.; Mitros T.; Ray G. J.; Curie G. L.; Naddaf N.; Chu J. S.; Ma H.; Boyer E.; Van Gool F.; Huang H.; Liu R.; Tobin V. R.; Schumann K.; Daly M. J.; Farh K. K.; Ansel K. M.; Ye C. J.; Greenleaf W. J.; Anderson M. S.; Bluestone J. A.; Chang H. Y.; Corn J. E.; Marson A. Discovery of Stimulation-Responsive Immune Enhancers with CRISPR Activation. Nature 2017, 549 (7670), 111–115. 10.1038/nature23875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M.; Glass Z.; Chen J.; Haas M.; Jin X.; Zhao X.; Rui X.; Ye Z.; Li Y.; Zhang F.; Xu Q. Lipid Nanoparticle-Mediated Codelivery of Cas9MRNA and Single-Guide RNA Achieves Liver-Specific in Vivo Genome Editing of Angptl3. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (10), e2020401118. 10.1073/pnas.2020401118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothgangl T.; Dennis M. K.; Lin P. J. C.; Oka R.; Witzigmann D.; Villiger L.; Qi W.; Hruzova M.; Kissling L.; Lenggenhager D.; Borrelli C.; Egli S.; Frey N.; Bakker N.; Walker J. A.; Kadina A. P.; Victorov D. V.; Pacesa M.; Kreutzer S.; Kontarakis Z.; Moor A.; Jinek M.; Weissman D.; Stoffel M.; van Boxtel R.; Holden K.; Pardi N.; Thöny B.; Häberle J.; Tam Y. K.; Semple S. C.; Schwank G. In Vivo Adenine Base Editing of PCSK9 in Macaques Reduces LDL Cholesterol Levels. Nat. Biotechnol. 2021, 1–9. 10.1038/s41587-021-00933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillmore J. D.; Gane E.; Taubel J.; Kao J.; Fontana M.; Maitland M. L.; Seitzer J.; O’Connell D.; Walsh K. R.; Wood K.; Phillips J.; Xu Y.; Amaral A.; Boyd A. P.; Cehelsky J. E.; McKee M. D.; Schiermeier A.; Harari O.; Murphy A.; Kyratsous C. A.; Zambrowicz B.; Soltys R.; Gutstein D. E.; Leonard J.; Sepp-Lorenzino L.; Lebwohl D. CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N. Engl. J. Med. 2021, NEJMoa2107454. 10.1056/NEJMoa2107454. [DOI] [PubMed] [Google Scholar]
- Heaton B. E.; Kennedy E. M.; Dumm R. E.; Harding A. T.; Sacco M. T.; Sachs D.; Heaton N. S. A CRISPR Activation Screen Identifies a Pan-Avian Influenza Virus Inhibitory Host Factor. Cell Rep. 2017, 20 (7), 1503–1512. 10.1016/j.celrep.2017.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskiewicz E.; Zhu G.; Bassi R.; Darling D. S.; Young W. W. B1,4-N-Acetylgalactosaminyltransferase (GM2 Synthase) Is Released from Golgi Membranes as a Neuraminidase-Sensitive, Disulfide-Bonded Dimer by a Cathepsin D-like Protease. J. Biol. Chem. 1996, 271 (42), 26395–26403. 10.1074/jbc.271.42.26395. [DOI] [PubMed] [Google Scholar]
- Dey S.; Lee J.; Noguchi C. T. Erythropoietin Non-Hematopoietic Tissue Response and Regulation of Metabolism During Diet Induced Obesity. Front. Pharmacol. 2021, 12, 725734. 10.3389/fphar.2021.725734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares F.; Havron A.; Fima E. Designing a Long Acting Erythropoietin by Fusing Three Carboxyl-Terminal Peptides of Human Chorionic Gonadotropin Subunit to the N-Terminal and C-Terminal Coding Sequence. Int. J. Cell Biol. 2011, 2011, e275063. 10.1155/2011/275063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson K. R.; Hanna R. E.; Hegde M.; Donovan K. F.; Strand C.; Sullender M. E.; Vaimberg E. W.; Goodale A.; Root D. E.; Piccioni F.; Doench J. G. Optimized Libraries for CRISPR-Cas9 Genetic Screens with Multiple Modalities. Nat. Commun. 2018, 9 (1), 5416. 10.1038/s41467-018-07901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn J. D.; Smith A. R.; Patel M. C.; Shaw L.; Youniss M. R.; van Heteren J.; Dirstine T.; Ciullo C.; Lescarbeau R.; Seitzer J.; Shah R. R.; Shah A.; Ling D.; Growe J.; Pink M.; Rohde E.; Wood K. M.; Salomon W. E.; Harrington W. F.; Dombrowski C.; Strapps W. R.; Chang Y.; Morrissey D. V. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018, 22 (9), 2227–2235. 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- Miao L.; Lin J.; Huang Y.; Li L.; Delcassian D.; Ge Y.; Shi Y.; Anderson D. G. Synergistic Lipid Compositions for Albumin Receptor Mediated Delivery of MRNA to the Liver. Nat. Commun. 2020, 11 (1), 2424. 10.1038/s41467-020-16248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mederacke I.; Dapito D. H.; Affò S.; Uchinami H.; Schwabe R. F. High-Yield and High-Purity Isolation of Hepatic Stellate Cells from Normal and Fibrotic Mouse Livers. Nat. Protoc. 2015, 10 (2), 305–315. 10.1038/nprot.2015.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.; Jiang F.; Liu J.-J.; Bray N. L.; Rauch B. J.; Baik S. H.; Nogales E.; Bondy-Denomy J.; Corn J. E.; Doudna J. A. Disabling Cas9 by an Anti-CRISPR DNA Mimic. Sci. Adv. 1620, 3 (7), e170. 10.1126/sciadv.1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C.; Williams R. T.; Calarco J. P.; Miller E. L.; Weber C. M.; Braun S. M. G.; Pulice J. L.; Chory E. J.; Crabtree G. R. Dynamics of BAF–Polycomb Complex Opposition on Heterochromatin in Normal and Oncogenic States. Nat. Genet. 2017, 49 (2), 213–222. 10.1038/ng.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S. M. G.; Kirkland J. G.; Chory E. J.; Husmann D.; Calarco J. P.; Crabtree G. R. Rapid and Reversible Epigenome Editing by Endogenous Chromatin Regulators. Nat. Commun. 2017, 8 (1), 560. 10.1038/s41467-017-00644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajpoot K.; Tekade M.; Sharma M. C.; Sreeharsha N.; Sharma A.; Tekade R. K.. Recombinant Blood Products and Therapeutic Enzymes: An Update. In The Future of Pharmaceutical Product Development and Research; Tekade R. K., Ed.; Advances in Pharmaceutical Product Development and Research; Academic Press, 2020; pp 447–482. [Google Scholar]
- Pardi N.; Tuyishime S.; Muramatsu H.; Kariko K.; Mui B. L.; Tam Y. K.; Madden T. D.; Hope M. J.; Weissman D. Expression Kinetics of Nucleoside-Modified MRNA Delivered in Lipid Nanoparticles to Mice by Various Routes. J. Controlled Release 2015, 217, 345–351. 10.1016/j.jconrel.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabnis S.; Kumarasinghe E. S.; Salerno T.; Mihai C.; Ketova T.; Senn J. J.; Lynn A.; Bulychev A.; McFadyen I.; Chan J.; Almarsson Ö.; Stanton M. G.; Benenato K. E. A Novel Amino Lipid Series for MRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-Human Primates. Mol. Ther. 2018, 26 (6), 1509–1519. 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton O. S.; Kauffman K. J.; McClellan R. L.; Appel E. A.; Dorkin J. R.; Tibbitt M. W.; Heartlein M. W.; DeRosa F.; Langer R.; Anderson D. G. Bioinspired Alkenyl Amino Alcohol Ionizable Lipid Materials for Highly Potent In Vivo MRNA Delivery. Adv. Mater. 2016, 28 (15), 2939–2943. 10.1002/adma.201505822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K.; Muramatsu H.; Keller J. M.; Weissman D. Increased Erythropoiesis in Mice Injected With Submicrogram Quantities of Pseudouridine-Containing MRNA Encoding Erythropoietin. Mol. Ther. 2012, 20 (5), 948–953. 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L.; Berraondo P.; Jericó D.; Guey L. T.; Sampedro A.; Frassetto A.; Benenato K. E.; Burke K.; Santamaría E.; Alegre M.; Pejenaute Á.; Kalariya M.; Butcher W.; Park J.-S.; Zhu X.; Sabnis S.; Kumarasinghe E. S.; Salerno T.; Kenney M.; Lukacs C. M.; Ávila M. A.; Martini P. G. V.; Fontanellas A. Systemic Messenger RNA as an Etiological Treatment for Acute Intermittent Porphyria. Nat. Med. 2018, 24 (12), 1899–1909. 10.1038/s41591-018-0199-z. [DOI] [PubMed] [Google Scholar]
- Zaleskas V. M.; Krause D. S.; Lazarides K.; Patel N.; Hu Y.; Li S.; Etten R. A. V. Molecular Pathogenesis and Therapy of Polycythemia Induced in Mice by JAK2 V617F. PLoS One 2006, 1 (1), e18. 10.1371/journal.pone.0000018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak J. L. Polycythemia Vera: Myths, Mechanisms, and Management. Blood 2002, 100 (13), 4272–4290. 10.1182/blood-2001-12-0349. [DOI] [PubMed] [Google Scholar]
- Wayment-Steele H. K.; Kim D. S.; Choe C. A.; Nicol J. J.; Wellington-Oguri R.; Watkins A. M.; Sperberg R. A. P.; Huang P.-S.; Participants E.; Das R.. Theoretical Basis for Stabilizing Messenger RNA through Secondary Structure Design. bioRxiv 2021, 10.1101/2020.08.22.262931 (accessed March 22, 2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters K. E.; Shivram H.; Fellmann C.; Lew R. J.; McMahon B.; Doudna J. A. Potent CRISPR-Cas9 Inhibitors from Staphylococcus Genomes. Proc. Natl. Acad. Sci. U. S. A. 2020, 117 (12), 6531–6539. 10.1073/pnas.1917668117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J.; Pawluk A.; Maxwell K. L.; Davidson A. R. Bacteriophage Genes That Inactivate the CRISPR/Cas Bacterial Immune System. Nature 2013, 493 (7432), 429–432. 10.1038/nature11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard E. L.; Vanover D.; Bawage S. S.; Tiwari P. M.; Rotolo L.; Beyersdorf J.; Peck H. E.; Bruno N. C.; Hincapie R.; Michel F.; Murray J.; Sadhwani H.; Vanderheyden B.; Finn M. G.; Brinton M. A.; Lafontaine E. R.; Hogan R. J.; Zurla C.; Santangelo P. J. Treatment of Influenza and SARS-CoV-2 Infections via MRNA-Encoded Cas13a in Rodents. Nat. Biotechnol. 2021, 717–726. 10.1038/s41587-021-00822-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.