Abstract
Background
Extended-spectrum β-lactamase-producing Escherichia coli (ESBL-Ec) is a major cause of infections worldwide. An understanding of the reservoirs and modes of transmission of these pathogens is essential, to tackle their increasing frequency.
Objectives
We investigated the contributions of various compartments (humans, animals, environment), to human colonization or infection with ESBL-Ec over a 3 year period, on an island.
Methods
The study was performed on Reunion Island (Southwest Indian Ocean). We collected ESBL-Ec isolates prospectively from humans, wastewater and livestock between April 2015 and December 2018. Human specimens were recovered from a regional surveillance system representative of the island’s health facilities. These isolates were compared with those from livestock and urban/rural wastewater, by whole-genome sequencing.
Results
We collected 410 ESBL-Ec isolates: 161 from humans, 161 from wastewater and 88 from animals. Phylogenomic analysis demonstrated high diversity (100 STs), with different STs predominating among isolates from humans (ST131, ST38, ST10) and animals (ST57, ST156). The large majority (90%) of the STs, including ST131, were principally associated with a single compartment. The CTX-M-15, CTX-M-27 and CTX-M-14 enzymes were most common in humans/human wastewater, whereas CTX-M-1 predominated in animals. Isolates of human and animal origin had different plasmids carrying blaCTX-M genes, with the exception of a conserved IncI1-ST3 blaCTX-M-1 plasmid.
Conclusions
These molecular data suggest that, despite their high level of contamination, animals are not a major source of the ESBL-Ec found in humans living on this densely populated high-income island. Public health policies should therefore focus primarily on human-to-human transmission, to prevent human infections with ESBL-Ec.
Introduction
Infections caused by multidrug-resistant bacteria (MDRB) are a major cause of morbidity and mortality worldwide.1 In this context, the WHO has identified seven specific pathogens in its strategy for fighting antibiotic resistance, including, first and foremost, Escherichia coli resistant to third-generation cephalosporins (3GC) and fluoroquinolones. The main mechanism of 3GC resistance in this species is the production of extended spectrum β-lactamase (ESBL). E. coli is a leading cause of infection in hospitals and in the community.2 If we are to tackle the trend towards an increase in MDRB infections in humans, we will need to use One Health approaches to develop an understanding of the global reservoirs and transmission patterns leading to the acquisition of ESBL-producing Enterobacterales (ESBL-E) in humans.3 Several potential sources of ESBL-producing E. coli (ESBL-Ec) for human infection or colonization have been identified, including food-producing animals, in which the prevalence of ESBL-Ec can be very high.4 ESBL-Ec can also be transmitted via the food chain.5 Moreover, environments contaminated with human and animal faeces can also act as a source of colonization, particularly in developing countries, which tend to have lower health standards than high-income countries.6–9 Studies based on this model addressing this question have yielded conflicting results, leaving the specific roles of these reservoirs unclear.10,11 The evaluation of exchanges of resistance vectors (genes or bacteria) between humans, animals and the environment is challenging and subject to several difficulties: (i) it involves the evaluation of a nearly closed ecosystem weakly influenced by imports from the exterior; (ii) a population representative of the ecosystem must be selected; and (iii) appropriate highly discriminating phylogenetic methods based on both core and accessory genomes, including mobile genetic elements (MGE), must be used.
In this study, we aimed to clarify the contribution of livestock to human colonization or infection with ESBL-Ec, using a delimited high-income area, the French overseas territory of Reunion Island, as a model. We report here the phylogenomic investigation, by WGS, of ESBL-Ec isolated from humans, livestock and wastewater on Reunion Island.
Materials and methods
Study design
Reunion Island is a French overseas territory located close to Madagascar, with 850 000 inhabitants and a surface area of 2500 km2 (340 inhabitants/km2). Together with Mayotte, it forms the French overseas territories of the Southwest Indian Ocean Area (SIOA). Antimicrobial resistance is a major problem on this island, located at the crossroads of Southern Africa and the Indian subcontinent.12 Reunion Island has the same level of healthcare as mainland France, with the University Hospital of Reunion Island (UHRI) as the referral hospital in SIOA, and the same health infrastructure (including access to drinking water, a sewerage system, modern wastewater treatment plants, etc.).
We prospectively collected ESBL-Ec isolates from humans, human and animal wastewater (considered a proxy for polluted human or animal environments) and livestock, between April 2015 and December 2018. All isolates suspected to have originated off the island (travel, medical evacuation, or imported animals) were excluded from the study.
Human specimens
The study was based on the regional antimicrobial resistance surveillance system of Reunion Island, which collects ESBL-Ec from six healthcare facilities: two private laboratory groups and four public hospitals (Figure 1). These four public hospitals reflected a total of 2500 beds and 250 000 hospital admissions accounting for >90% of complete-hospitalization days and 100% of medical, surgical and obstetric (MSO) hospitalizations on Reunion Island. The bacterial isolates were collected between January and December 2018 from clinical or rectal screening samples and from non-duplicate patients. Isolates from patients who had a history of off-island travel within the last 6 months, medical evacuation or a home address (postcode) located off the island were excluded. We then randomly selected three ESBL-Ec isolates per centre and per month for further analysis. Anonymized data were recorded for each isolate: gender, age, origin of patient (community or healthcare facility), specimen source, infection/colonization and postcode.
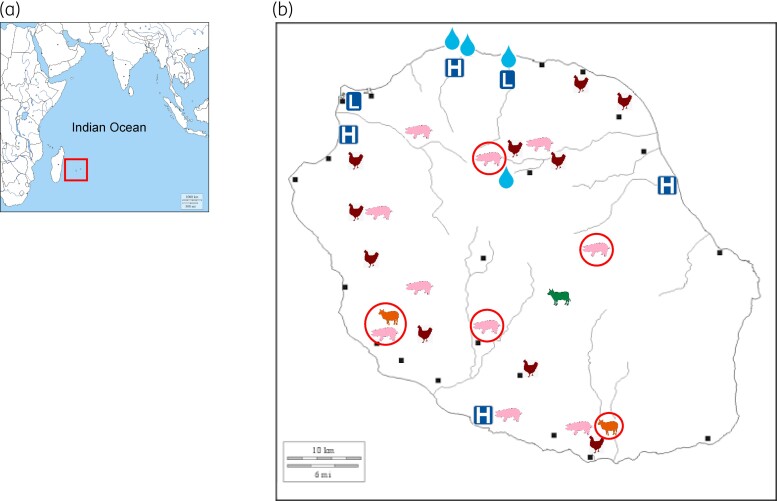
Figure 1.
Map of Reunion Island, showing the locations from which the ESBL-producing E. coli-positive samples were obtained. (a) Map indicating the position of Reunion Island in the Southwest Indian Ocean. (b) Detailed map of Reunion Island showing the locations of the healthcare facilities (L, private laboratories; H, hospitals) at which the human ESBL-Ec isolates were obtained. The wastewater sampling sites are indicated by a teardrop shape; those in the north correspond to the sewage and wastewater treatment plant of Saint-Denis, the largest city on the island. The teardrop in the centre corresponds to the Camp Pierrot animal waste treatment plant in Grand Ilet. The animal symbols indicate the farms of the various livestock sectors. The symbols surrounded by a red circle represent farms with clonally related isolates, belonging to ST156, in common.
Wastewater
Wastewater samples were collected from four sites (Figure 1). The northern sites corresponded to two sewage effluent discharge points (one from a hospital and the other from the community) and one wastewater treatment plant (WWTP) in the major town of the island (separate inflow and outflow sampling). The site in the centre of the island is the only animal waste treatment plant (AWTP) on the island (runoff water and outflow into the lagoon). Each collecting point was sampled (2 × 500 mL) during the first week of each month, between January and December 2018. ESBL-Ec isolates were identified by (i) the serial dilution method for sewage and untreated inflow, and (ii) the filtration method (membranes with 0·45 μm pores) for treated outflow, as previously described.13
Animal specimens
The sampling procedure was adapted from previous studies on the prevalence of ESBL-producing Enterobacterales, to ensure that a collection of samples representative of all the cattle, pig, poultry, rabbit, and small ruminant (sheep and goat) farms on the island was obtained.14 Samples were collected with Sterisox® boot swabs (Sodibox, Nevez, France) or rectal swabs (for cattle). For the boot swabs, the number of samples depended on area considered, with one Sterisox® swab per 100 m2 of building. All samples were maintained at 4°C until analysis, which took place within 48 h of sample reception. In total, 114 farms were sampled, and 566 animal samples (124 swabs from cattle; 177 pig, 176 poultry, 39 rabbit and 50 small-ruminant boot swabs) were obtained between April 2015 and December 2018. Sterisox® boot swabs were introduced into a sterile recipient with 100 mL of saline enriched with brain-heart infusion broth (BHI-T, bioMérieux, Marcy l’Etoile, France). The mixture was incubated overnight at room temperature, and secondly, plated (10 μL) on selective chromogenic agar plates (ChromID-ESBL agar, bioMérieux, Marcy l’Etoile, France) as previously described.15
Collection of ESBL-producing E. coli
The characterization of presumptive ESBL-Ec isolates took place at a single site, at the northern site of the UHRI. Each isolate was identified by MALDI-TOF MS (Bruker Daltonics, Bremen, Germany) and its ESBL production was assessed by the combined disc method, in accordance with the 2019 EUCAST recommendations(https://www.sfm-microbiologie.org/wp-content/uploads/2019/05/CASFM2019_V2.0_MAI.pdf). For the collection of bacteria from animals or environment, only one ESBL-producing isolate for each different resistance profile from each farm or sampling point was retained for further analysis.
Whole-genome sequencing and data analysis
Whole-genome sequencing was performed on an Illumina NextSeq high-output system generating 2 × 150 paired-end reads with a median coverage of 88×. Reads were trimmed with sickle (v1.33) to a minimum length of 50, and were then subsampled to a coverage of at least 80×. Assembly was performed with Spades (v3.13).
STs were determined in silico with pyMLST, using the Enterobase and Achtman schemes. Phylogroup and fimH typing were assigned with ClermonTyping and FimTyper, respectively. We searched for resistance genes with BLAT software and the ResFinder database.
We performed core genome MLST analysis (cgMLST) with pyMLST software and the Enterobase v2 scheme.16 Phylogenetic analysis was performed with bioNJ, using cgMLST distances, and the results were visualized with Itol. Isolates were clustered if their cgMLST differed by fewer than 10 alleles.17 A network was built with the igraph package (v1.2.5), to determine the connections between different sampling sites.
The proximity of mobile genetic elements between strains was evaluated by a method derived from that described by Ludden et al.18 Briefly, mlplasmids (v1.0) was used to classify contigs as potentially plasmid-borne or on the chromosome. Genes were detected with prodigal on contigs that were considered to be plasmid-borne and contained a blaESBL. These genes were then clustered with cd-hit (v4.7) to obtain a non-redundant list of mobile genes associated with blaESBL. We then searched all the genomes for these mobile elements, and their presence/absence was used to build a dendrogram based on a binary distance, with bioNJ.
Long-read sequencing was performed on the PacBio Sequel system (SMRT cell v.2.1 chemistry). The raw data generated were assembled with flye (v2.8.1). Plasmids were extracted and polished with pilon (v1.23). BLAST was used to align complete plasmid sequences with the assembled genomes containing blaCTX-M-1 or blaCTX-M-15.
The sequences of the plasmids and the genomes (raw reads) obtained were deposited on NCBI under the accession numbers OM105937 to OM105940 and BioProject PRJNA795027, respectively.
Results
Collection
In total, we collected 410 ESBL-Ec isolates: 161 from humans, 161 from wastewater and 88 from animals. The mean age of the human patients was 61.1 years (SD 26.2 years) and the sex ratio was 0.96 (Table S1, available as Supplementary data at JAC Online). Most of the isolates (80.7%) were of community origin or were detected in patients during the first 48 h of hospitalization. Most of the isolates were obtained from the urinary tract (59.0%) or rectal colonization (26.1%), whereas bacteraemia and surgical site infections accounted for 6.8% and 5.0% of isolates, respectively. Infections accounted for 67.1% of cases. This bacterial collection included 15% of all the human ESBL-Ec isolates obtained on the island during the study period. For wastewater (Table S2), 70 isolates were obtained from community effluent (n = 29; 18.0%) or hospital effluent (n = 41; 25.4%). For WWTPs, 85 isolates were recovered from inflow (n = 56; 34.8%) and outflow (n = 29; 18.0%). For the AWTP, six isolates were retrieved from runoff water (n = 3; 1.9%) or outflow destined for the lagoon (n = 3, 1.9%).
We collected 88 ESBL-Ec from animals (Table S3): 50 from poultry, 33 from pigs, 2 from cattle, and 3 from small ruminants. No ESBL-Ec strains were detected in rabbits. The rate of ESBL positivity by type of farm was: 70% for poultry farms, 50% for pig farms, 18.5% for small ruminant farms and 8.3% for cattle farms. The overall positivity rate per farm was 36.8%.
Comparative phylogenomics
Whole-genome sequencing was used to type and cluster the isolates. Among the 410 ESBL-Ec, we identified 100 different STs, 42 of which were common to at least two isolates (Figure S1). The top three STs were ST131 (n = 93; 22.7%), ST38 (n = 39; 9.5%) and ST10 (n = 24; 5.9%). We then compared samples from two compartments: those of animal origin (livestock and animal wastewater) and those of human origin (clinical samples and human wastewater). Ten STs were common to these two compartments: ST10, ST38, ST48, ST58, ST88, ST117, ST155, ST162, ST361 and ST3489; however, ST10 was the only ST clustering at least five isolates in both compartments. For isolates of human origin, the top three STs were identical to those for the general population; ST131 was found exclusively in this compartment. Conversely, the predominant animal-related STs (ST57, ST156) were absent from the samples of human origin. Most STs (90%) were associated with only one compartment.
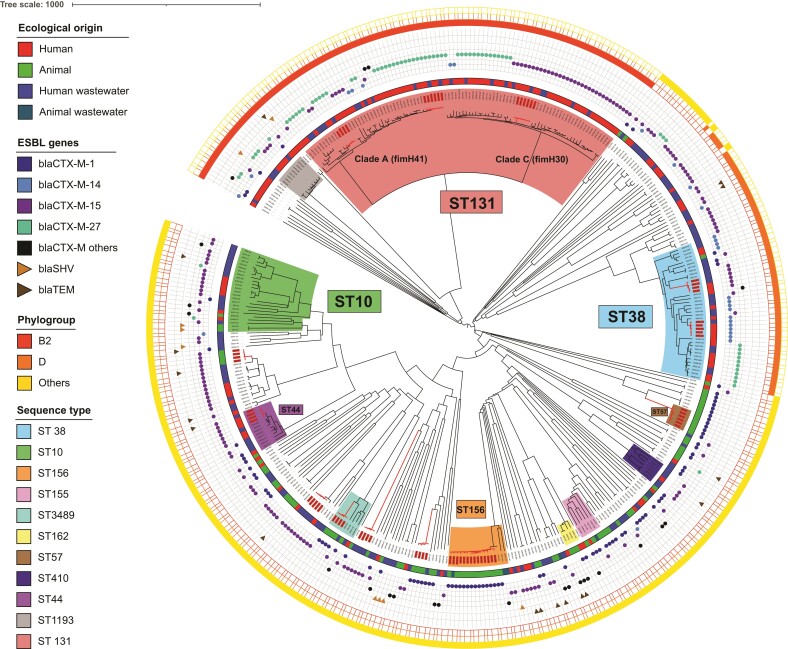
cgMLST analysis demonstrated a high degree of genetic diversity (Figure 2) and showed that the strains belonging to the STs common to human and animal sources, such as ST10, were not clonally related. Multiple clusters were observed (in red, Figure 2), but each corresponded to isolates of animal origin (e.g. ST57 or ST156) or of human origin (e.g. ST38, ST44 or ST131).
Figure 2.
Phylogenetic tree illustrating the relationship between ESBL-producing E. coli isolates from humans, animals and wastewater. Neighbour-joining phylogenetic tree of 410 ESBL-Ec based on cgMLST data. Clonally related isolates are shown in red on the dendrogram. The innermost coloured ring indicates the ecological origin of the isolates, whereas the outermost ring indicates the phylogroup. The symbols located between these two rings (round, triangle) represent the β-lactam resistance-encoding blaBLSE genes. The main STs are represented by a specific colour on the dendrogram, as defined in the legend.
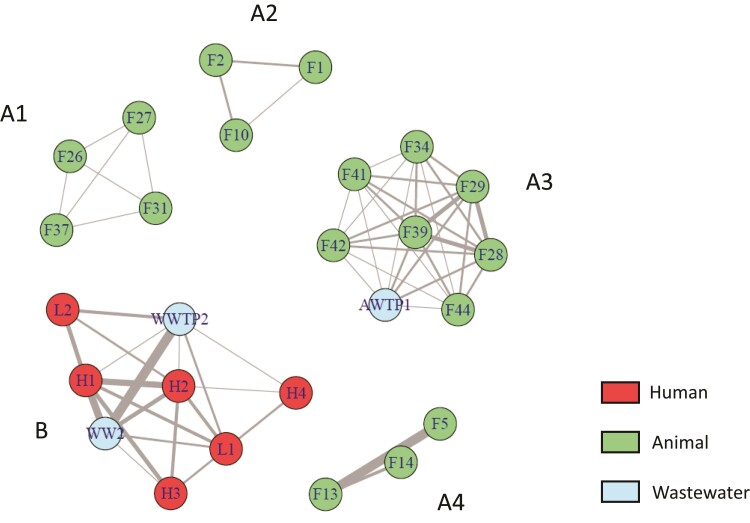
Phylogroups varied with the source, with B2/D predominating among human/human wastewater isolates (B2 was not identified in animal samples and D was identified in only one sample from poultry), whereas B1/A predominated among isolates of animal origin. Phylogroups B2 and D accounted for 40.4% and 9.9% of human isolates, and 60.2% and 14.8% of infection source isolates, respectively. The clonality network (Figure 3) revealed connections between private laboratories and public hospitals, and between these establishments and sewage/WWTPs (cluster B); whereas farms formed isolated networks or were connected to the AWTP (cluster A). Cluster A3 highlighted connections, via ST156 isolates, between pig and small-ruminant farms, which were located in geographically remote sites of the island (symbols surrounded by a red circle in Figure 1).
Figure 3.
Clonality network representing the healthcare facilities, farms and wastewater sources at which clonally related ESBL-producing E. coli isolates were detected. After phylogenetic analysis, each clonally related isolate was assigned to its sampling site. Farms (F), laboratories (L), hospitals (H), wastewater from sewage (WW), wastewater treatment plant (WWTP) and the animal waste treatment plant (AWTP) are symbolized by circles, whereas clonal links are indicated by the lines connecting the circles. The more clonal strains connecting two sites, the wider the line between the two sites.
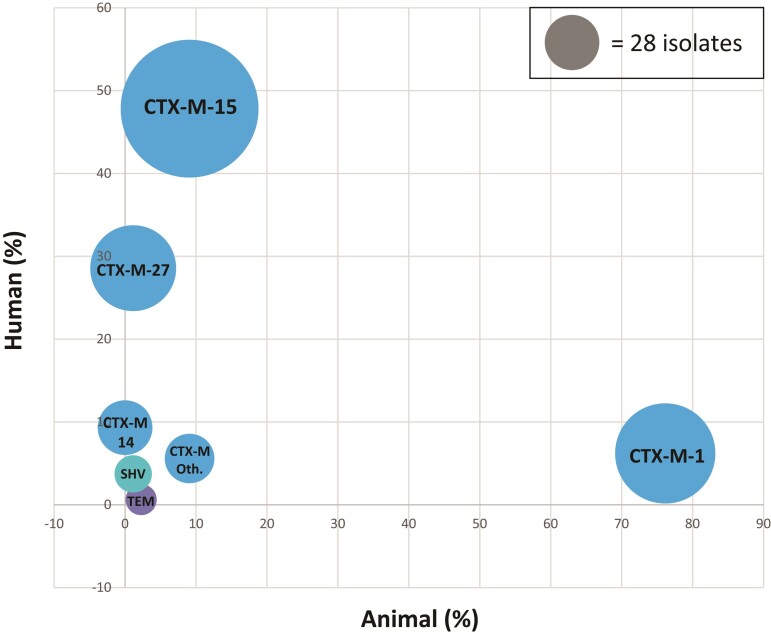
Assessments of the occurrence of genetic determinants of resistance revealed that blaCTX-M-15 was the predominant blaESBL gene (n = 177; 43.2%), followed by blaCTX-M-1 (n = 94; 22.9%), blaCTX-M-27 (n = 69; 16.8%) and blaCTX-M-14 (n = 28; 6.8%) (Figure 4). The CTX-M-15, CTX-M-27 and CTX-M-14 enzymes were most common in humans/human wastewater, whereas CTX-M-1 predominated in ESBL-Ec of animal origin (Figure 4 and Figure S2b). An analysis of ST131 isolates (n = 93) revealed a separation into two clades: (i) a fimH30 clade (n = 49), comprising a C2-M-15 subclade (n = 31) and a C1-M27 subclade (n = 16) (Figure 2); and (ii) a fimH41 clade (n = 37) mostly associated with blaCTX-M-27 (n = 27; 73.0%), corresponding to an ancestral clade A (Figure 2). CTX-M-1 was mostly associated with the animal-related ST57, ST155, ST156 and ST3489. The CTX-M-1 enzyme was found in only 10 (6.2%) of 161 human isolates and CTX-M-15 in only eight (9.1%) of 88 animal isolates. Screening for non-ESBL resistance genes revealed that nine isolates coproduced an acquired cephalosporinase (blaCMY or blaDHA) and five coproduced a carbapenemase (blaNDM-1, blaNDM-5, blaOXA-181 or blaOXA-232) (Figure S2a). The carbapenemase-encoding blaNDM and blaOXA-48-type genes were detected only in isolates from human wastewater.
Figure 4.
Bubble graph showing the proportion of blaBLSE resistance genes common to ESBL-producing E. coli from humans and animals. Each blaBLSE resistance gene is positioned according to its prevalence in the human and animal compartments. The size of each bubble represents the number of isolates in which the gene was identified. Bubbles are coloured according to the type of ESBL enzyme: blue for CTX-M, turquoise for SHV and purple for TEM enzymes. With the exception of the blaTEM gene, bubbles are relatively close to the axes, with no bubble in a central position on the bisector. CTX-M Oth, bubble representing the other CTX-M variants.
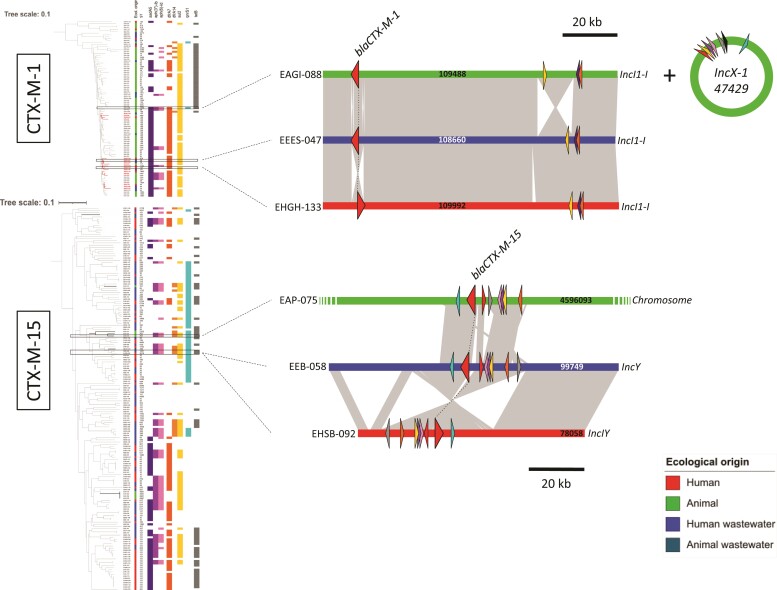
We investigated whether blaCTX-M-1 and blaCTX-M-15, the two major ESBL resistance genes, were carried by the same or different plasmids. For blaCTX-M-1, the analysis identified multiple clusters (with >98% identity), some of which contained isolates of both human and animal origin and from different STs (Figure 5 and Figure S3). For blaCTX-M-15, there was considerable variability between isolates, with no clustering between human and animal isolates (Figure 5 and Figure S4). We then cross-checked this information with MGE data. Two triplets of human-animal-wastewater isolates positive for blaCTX-M-1 and blaCTX-M-15 were selected for long-read sequencing. We searched the complete plasmid sequences (obtained by long-read sequencing) on the assembled genomes. The blaCTX-M-1-positive human isolate carried an IncI1-I blaCTX-M-1 plasmid (109 992 bp) that was highly similar to the plasmids carrying the blaCTX-M-1 gene in the animal and wastewater isolates selected (Figure 5). This conserved IncI1-I (or IncI1-ST3) blaCTX-M-1 plasmid was found in 6 human isolates and 38 animal isolates. In contrast, the blaCTX-M-15 gene in animal isolates was located on the chromosome and probably resulted from the integration of a blaCTX-M-15 IncI/Y plasmid. The region harbouring the blaCTX-M-15 gene in the human, animal and wastewater isolates was less conserved and had undergone multiple recombinations, indicating an absence of recent horizontal transfer (Figure 5).
Figure 5.
Dendrograms of mobile genetic elements identified for blaCTX-M-1 and blaCTX-M-15 genes. Strains containing blaCTX-M-1 and blaCTX-M-15 β-lactam resistance genes were clustered according to the presence/absence of mobile genes associated with these resistance genes. The isolate name is indicated in the first column on the left, the ecological origin of the isolate in the second column, the ST in the third and then the co-resistance genes most frequently shared by the selected isolates. We chose two triplets of isolates of interest on the basis of the inclusion of strains with the same co-resistances but from different STs. The analysis of these six isolates of interest enabled us to reconstitute the plasmids carrying these genes, which are illustrated schematically at the right of the figure. The plasmids carrying the blaCTX-M-1 and blaCTX-M-15 genes are represented in linear form. The resistance genes are indicated by triangles colour-coded as in the dendrogram, with β-lactam resistance genes indicated by red triangles.
Discussion
We compared 410 genomes of ESBL-Ec isolates from humans or human-polluted environments (i.e. human wastewater) with those of isolates from livestock or animal-polluted environments (i.e. animal wastewater) in a circumscribed island area. The main finding of this study was that the populations of these ESBL-Ec isolates from these two reservoirs were genetically different, generally harbouring different blaESBL genes located in different genetic environments. This strongly suggests that livestock have very little effect on the acquisition of ESBL-Ec by humans on Reunion Island, despite all the food produced from the livestock kept on the island being consumed locally.
Phylogenomic and resistance gene analyses revealed the existence of very little connection between the human and animal compartments, which had different major STs and predominant ESBL-encoding genes. In contrast, human and human wastewater isolates were much more strongly connected, indicating that wastewater can be used as a proxy for human gut colonization.19 As expected, phylogroups B2 and D were associated with most of the human infections (75.0%).20 For the predominant ST131 clone, the high proportion of H41-M27 isolates within the ancestral clade A (already described in Thailand and Cambodia) suggests an influence of South-East Asia on the epidemiology of ESBL-Ec on Reunion Island.5,21 ST10 was the only top-three ST common to both compartments, but phylogenomic analysis nevertheless highlighted the presence of different subclones that appeared to be adapted to human or animal hosts.18,22 The clonality networks established between the different structures (farms or hospitals) revealed the partitioning between the human and animal compartments, each of which was connected to its own wastewater treatment circuit (Figure 3). This finding supports the hypothesis that human-to-human transmission accounts for most cases of infection or colonization in humans. This clonality network connected previously described spatially remote care facilities and farms on the island.23 In animals, this clonal relationship between farms remained unclear and could be explained by the exchange of some animals (especially male breeding stock) between farms. The finding of separate compartments was confirmed by MGE analysis, as the animal compartment was not identified as a progenitor of MGEs for humans; with the exception of the blaCTX-M-1 gene carried by IncI1-ST3 plasmids, which may represent a minor source of horizontal transfer between compartments.24 CTX-M-15 was the main ESBL found in humans (47.8%), whereas CTX-M-1 accounted for only 6.2% of human ESBL isolates. We can therefore estimate that <5% of human isolates carry antimicrobial resistance genes closely related to those found in livestock, consistent with previous findings.18 A limitation of this analysis is that only two triplets of isolates could be sequenced in long reads, which could potentially cause us to underestimate this result.
The partitioning of ESBL-Ec between the human and animal compartments observed here goes against the findings of several previous studies.25,26 However, these previous studies often involved less-thorough analyses or focused on sporadic transmission in specific exposed populations (farmers, in particular), and the results obtained were sometimes overturned by subsequent WGS analyses.27 In contrast, our results are consistent with two models that have recently emerged from major studies published on the subject: a One Health model in low-income countries, in which people are much more closely linked to their environment,5,8,28 and a partitioned model in high-income countries, in which the infrastructure in place probably significantly limits these interactions.18,22–23,29 These findings highlight the importance of the human-to-human oral–faecal route as the probable most frequent route of transmission for adapted ESBL-Ec, although transmission may also occur via other minority routes. This implies that improvements in community infrastructure (e.g. sanitation or housing) and farming practices (biosecurity and hygiene) would probably help to prevent the development of antimicrobial resistance.30 As recently pointed out by Mughini-Gras et al.31, other models based not only on the prevalence of ESBL-producing Enterobacterales but also on the probability of exposure in humans, should be explored further.
The key strengths of our study are: (i) the representativeness of the human ESBL-Ec isolate collection with respect to the population of strains in the community; (ii) the prospective sampling of these strains over a period of 3.5 years; and (iii) the optimal geographic context of collection in a well-defined, densely populated island environment. The large collection of human isolates (n = 161, 39.3% of all the isolates) originated mostly from the community (80.7%) and corresponded to both infections (two-thirds) and colonization (one-third). This collection was complemented by 155 urban wastewater isolates (sewage and WWTP), which can be considered a proxy for the gut flora of the population living in the community.13,18,22 The animal isolates were also highly representative, with recruitment covering all the livestock sectors present on the island. Samples were also taken from an AWTP, although few isolates were obtained from this source. A recent study on the island of Zanzibar (Tanzania) also highlighted the benefits of One Health studies of the interconnection between different compartments in a circumscribed territory.28 One potential limitation of this study is the lack of exploration of the food chain, but several studies have shown that the ESBL-Ec populations from livestock and the food chain are genetically similar.22,28,32,33 This pathway should nevertheless be considered as a potential source of direct transmission to humans.
ESBL-Ec was isolated from 36.8% of livestock farms, with particularly high rates of isolation for poultry (70.0%) and pig (50.0%) farms. These rates are higher than those for mainland France during the same period but consistent with data for other European countries, such as Germany, the Netherlands or the United Kingdom.18,34–37
As a consequence, national and European public health policies aiming to reduce antibiotic use in livestock are likely to have only a limited and delayed impact on human infections over time.38 In contrast with the rapid decline in mcr-1-linked colistin resistance in China, limiting the prevalence of ESBL-Ec in livestock will have only a limited impact on the rate of blaESBL transmission for human-adapted E. coli clones.39 The priority should therefore be to limit the selection pressure imposed by antibiotic use in humans, to maximize the impact of measures.
This study on the exchange of ESBL-Ec between human and animal compartments is the first to demonstrate a clear sectorization of animal and human isolates on a densely populated, high-income island with a high prevalence of ESBL-Ec in livestock. Our findings suggest that livestock is not a major source of ESBL-Ec acquisition in humans, either through direct contact with animals or indirectly, via the food chain.
Supplementary Material
Acknowledgements
This study is based on independent research co-ordinated by the University of Reunion Island (UMR PIMIT), the University Hospital of Reunion Island and the University of Bourgogne Franche-Comté (UMR Chrono-Environnement). The research team thank all the staff who participated in specimen collection at the hospitals, laboratory group and agencies. We warmly thank Olivier Pince, Kevin Ronceray, Serge Hoarau of the CISE Réunion and Laurent Lai-Kan-Thon, Jean-Louis Duchemann, Anthony Gardebien of Runéo for their help with environmental sampling. We thank Morgan Laval and Maël Jégo for help for animal sampling. We also thank Pablo Tortosa and Jérôme Allyn, who participated in the steering committee for this work.
Funding
This study was supported by Reunion Regional Health Agency (ARS Réunion), Indian Ocean Commission (COI), French Agricultural Research Centre for International Development (CIRAD), UMR PIMIT, through its national institutions (Reunion University, CNRS, INSERM, IRD) and the Biological Resource Centre of Besançon University Hospital. This research was also partially funded by the grants from POE FEDER 2014/2020 of the Conseil Régional de La Réunion (RESISTORUN program).
Transparency declarations
None to declare.
Supplementary data
Information on the whole genome sequencing and data analysis, plus Tables S1 to S3 and Figures S1 to S4 are available as Supplementary data at JAC Online.
References
- 1. Cassini A, Högberg LD, Plachouras Det al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect Dis 2019; 19: 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pitout JDD, Laupland KB. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 2008; 8: 159–66. [DOI] [PubMed] [Google Scholar]
- 3. Walsh TR. A one-health approach to antimicrobial resistance . Nat Microbiol 2018; 3: 854–5. [DOI] [PubMed] [Google Scholar]
- 4. Thanner S, Drissner D, Walsh F. Antimicrobial resistance in agriculture. mBio 2016; 7: e02227-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nadimpalli M, Vuthy Y, de Lauzanne Aet al. Meat and fish as sources of extended-spectrum β-lactamase-producing Escherichia coli, Cambodia. Emerg Infect Dis 2019; 25: 126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van Hoek AHAM, Dierikx C, Bosch Tet al. Transmission of ESBL-producing Escherichia coli between broilers and humans on broiler farms. J Antimicrob Chemother 2020; 75: 543–9. [DOI] [PubMed] [Google Scholar]
- 7. Leonard AFC, Zhang L, Balfour AJet al. Human recreational exposure to antibiotic resistant bacteria in coastal bathing waters. Environ Int 2015; 82: 92–100. [DOI] [PubMed] [Google Scholar]
- 8. Purohit MR, Chandran S, Shah Het al. Antibiotic resistance in an Indian rural community: a ‘One-Health’ observational study on commensal coliform from humans, animals, and water. Int J Environ Res Public Health 2017; 14: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seni J, Moremi N, Matee Met al. Preliminary insights into the occurrence of similar clones of extended-spectrum β-lactamase-producing bacteria in humans, animals and the environment in Tanzania: a systematic review and meta-analysis between 2005 and 2016. Zoonoses Public Health 2018; 65: 1–10. [DOI] [PubMed] [Google Scholar]
- 10. Lazarus B, Paterson DL, Mollinger JLet al. Do human extraintestinal Escherichia coli infections resistant to expanded-spectrum cephalosporins originate from food-producing animals? A systematic review. Clin Infect Dis 2015; 60: 439–52. [DOI] [PubMed] [Google Scholar]
- 11. Madec J-Y, Haenni M, Nordmann Pet al. Extended-spectrum β-lactamase/AmpC- and carbapenemase-producing Enterobacteriaceae in animals: a threat for humans? Clin Microbiol Infect 2017; 23: 826–33. [DOI] [PubMed] [Google Scholar]
- 12. Miltgen G, Cholley P, Martak Det al. Carbapenemase-producing Enterobacteriaceae circulating in the Reunion Island, a French territory in the Southwest Indian Ocean. Antimicrob Resist Infect Control 2020; 9: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bréchet C, Plantin J, Sauget Met al. Wastewater treatment plants release large amounts of extended-spectrum β-lactamase-producing Escherichia coli into the environment. Clin Infect Dis 2014; 58: 1658–65. [DOI] [PubMed] [Google Scholar]
- 14. Gay N, Belmonte O, Collard J-Met al. Review of antibiotic resistance in the Indian Ocean commission: a Human and animal health issue. Front Public Health 2017; 5: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gay N, Lugagne N, Miltgen Get al. Reunion, a sentinel island for multidrug-resistant bacteria surveillance in South-western Indian ocean: a retrospective survey using hospitalised patients screening, 2015-2017. BMC Public Health 2020; 20: 1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pietsch M, Irrgang A, Roschanski Net al. Whole genome analyses of CMY-2-producing Escherichia coli isolates from humans, animals and food in Germany. BMC Genomics 2018; 19: 601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schürch AC, Arredondo-Alonso S, Willems RJLet al. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect 2018; 24: 350–4. [DOI] [PubMed] [Google Scholar]
- 18. Ludden C, Raven KE, Jamrozy Det al. One health genomic surveillance of Escherichia coli demonstrates distinct lineages and mobile genetic elements in isolates from humans versus livestock. mBio 2019; 10: e02693-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zarfel G, Galler H, Feierl Get al. Comparison of extended-spectrum-β-lactamase (ESBL) carrying Escherichia coli from sewage sludge and human urinary tract infection. Environ Pollut 2013; 173: 192–9. [DOI] [PubMed] [Google Scholar]
- 20. Johnson JR, Delavari P, Kuskowski Met al. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J Infect Dis 2001; 183: 78–88. [DOI] [PubMed] [Google Scholar]
- 21. Runcharoen C, Raven KE, Reuter Set al. Whole genome sequencing of ESBL-producing Escherichia coli isolated from patients, farm waste and canals in Thailand. Genome Med 2017; 9: 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Day MJ, Hopkins KL, Wareham DWet al. Extended-spectrum β-lactamase-producing Escherichia coli in human-derived and foodchain-derived samples from England, Wales, and Scotland: an epidemiological surveillance and typing study. Lancet Infect Dis 2019; 19: 1325–35. [DOI] [PubMed] [Google Scholar]
- 23. Alzayn M, Findlay J, Schubert Het al. Characterization of AmpC-hyperproducing Escherichia coli from humans and dairy farms collected in parallel in the same geographical region. J Antimicrob Chemother 2020; 75: 2471–9. [DOI] [PubMed] [Google Scholar]
- 24. Valcek A, Roer L, Overballe-Petersen Set al. IncI1 ST3 and IncI1 ST7 plasmids from CTX-M-1-producing Escherichia coli obtained from patients with bloodstream infections are closely related to plasmids from E. coli of animal origin. J Antimicrob Chemother 2019; 74: 2171–5. [DOI] [PubMed] [Google Scholar]
- 25. Leverstein-van Hall MA, Dierikx CM, Cohen Stuart Jet al. Dutch patients, retail chicken meat and poultry share the same ESBL genes, plasmids and strains. Clin Microbiol Infect 2011; 17: 873–80. [DOI] [PubMed] [Google Scholar]
- 26. Tansawai U, Walsh TR, Niumsup PR. Extended spectrum β-lactamase-producing Escherichia coli among backyard poultry farms, farmers, and environments in Thailand. Poult Sci 2019; 98: 2622–31. [DOI] [PubMed] [Google Scholar]
- 27. de Been M, Lanza VF, de Toro Met al. Dissemination of cephalosporin resistance genes between Escherichia coli strains from farm animals and humans by specific plasmid lineages. PLoS Genet 2014; 10: e1004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Büdel T, Kuenzli E, Campos-Madueno EIet al. On the island of Zanzibar people in the community are frequently colonized with the same MDR Enterobacterales found in poultry and retailed chicken meat. J Antimicrob Chemother 2020; 75: 2432–41. [DOI] [PubMed] [Google Scholar]
- 29. Börjesson S, Ny S, Egervärn Met al. Limited dissemination of extended-spectrum β-lactamase- and plasmid-encoded AmpC-producing Escherichia coli from food and farm animals, Sweden. Emerg Infect Dis 2016; 22: 634–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Collignon P, Beggs JJ, Walsh TRet al. Anthropological and socioeconomic factors contributing to global antimicrobial resistance: a univariate and multivariable analysis. Lancet Planet Health 2018; 2: e398–405. [DOI] [PubMed] [Google Scholar]
- 31. Mughini-Gras L, Dorado-García A, van Duijkeren Eet al. Attributable sources of community-acquired carriage of Escherichia coli containing β-lactam antibiotic resistance genes: a population-based modelling study. Lancet Planet Health 2019; 3: e357–69. [DOI] [PubMed] [Google Scholar]
- 32. Dorado-García A, Smid JH, van Pelt Wet al. Molecular relatedness of ESBL/AmpC-producing Escherichia coli from humans, animals, food and the environment: a pooled analysis. J Antimicrob Chemother 2018; 73: 339–47. [DOI] [PubMed] [Google Scholar]
- 33. Nguyen MN, Hoang HTT, Xavier BBet al. Prospective One Health genetic surveillance in Vietnam identifies distinct blaCTX-M-harbouring Escherichia coli in food-chain and human-derived samples. Clin Microbiol Infect 2021; 27: 1515.e1–e8. [DOI] [PubMed] [Google Scholar]
- 34. Hille K, Felski M, Ruddat Iet al. Association of farm-related factors with characteristics profiles of extended-spectrum β-lactamase-/plasmid-mediated AmpC β-lactamase-producing Escherichia coli isolates from German livestock farms. Vet Microbiol 2018; 223: 93–9. [DOI] [PubMed] [Google Scholar]
- 35. Dierikx C, van der Goot J, Fabri Tet al. Extended-spectrum-β-lactamase- and AmpC-β-lactamase-producing Escherichia coli in Dutch broilers and broiler farmers. J Antimicrob Chemother 2013; 68: 60–7. [DOI] [PubMed] [Google Scholar]
- 36. Blaak H, van Hoek AHAM, Hamidjaja RAet al. Distribution, numbers, and diversity of ESBL-producing E. coli in the poultry farm environment. PLoS One 2015; 10: e0135402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bourély C, Chauvin C, Jouy Éet al. Comparative epidemiology of E. coli resistance to third-generation cephalosporins in diseased food-producing animals. Vet Microbiol 2018; 223: 72–8. [DOI] [PubMed] [Google Scholar]
- 38. Bourély C, Coeffic T, Caillon Jet al. Trends in antimicrobial resistance among Escherichia coli from defined infections in humans and animals. J Antimicrob Chemother 2020; 75: 1525–9. [DOI] [PubMed] [Google Scholar]
- 39. Wang Y, Xu C, Zhang Ret al. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect Dis 2020; 20: 1161–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.