Abstract
Background
Neonatal sepsis is a serious bacterial infection of neonates, globally killing up to 680 000 babies annually. It is frequently complicated by antimicrobial resistance, particularly in low- and middle-income country (LMIC) settings with widespread resistance to the WHO’s recommended empirical regimen of ampicillin and gentamicin.
Objectives
We assessed the utility of flomoxef and fosfomycin as a potential alternative empirical regimen for neonatal sepsis in these settings.
Methods
We studied the combination in a 16-arm dose-ranged hollow-fibre infection model (HFIM) experiment and chequerboard assays. We further assessed the combination using clinically relevant regimens in the HFIM with six Enterobacterales strains with a range of flomoxef/fosfomycin MICs.
Results
Pharmacokinetic/pharmacodynamic modelling of the HFIM experimental output, along with data from chequerboard assays, indicated synergy of this regimen in terms of bacterial killing and prevention of emergence of fosfomycin resistance. Flomoxef monotherapy was sufficient to kill 3/3 strains with flomoxef MICs ≤0.5 mg/L to sterility. Three of three strains with flomoxef MICs ≥8 mg/L were not killed by fosfomycin or flomoxef monotherapy; 2/3 of these were killed with the combination of the two agents.
Conclusions
These data suggest that flomoxef/fosfomycin could be an efficacious and synergistic regimen for the empirical treatment of neonatal sepsis in LMIC settings with prevalent antimicrobial resistance. Our HFIM results warrant further assessment of the flomoxef/fosfomycin combination in clinical trials.
Introduction
Neonatal sepsis is a frequent and often fatal infection of newborn infants,1 causing an estimated 430 000–680 000 deaths annually,2,3 with highest mortalities in low- and middle-income countries (LMICs). Leading causative pathogens include Gram-negative (e.g. Escherichia coli, Klebsiella pneumoniae) and Gram-positive bacteria (e.g. Staphylococcus aureus, Streptococcus agalactiae). For the empirical treatment of neonatal sepsis, the WHO currently recommends a narrow-spectrum β-lactam antibiotic (e.g. ampicillin, benzylpenicillin) in combination with gentamicin as first line.4,5 The clinical efficacy of this regimen, however, is increasingly compromised by rising rates of antimicrobial resistance (AMR).
Multiple epidemiological studies have demonstrated significant rates of AMR in bacteria causing neonatal sepsis in a variety of LMIC settings,6–13 including rising rates of ESBLs and aminoglycoside-modifying enzymes (AMEs). Resistance rates to amoxicillin and gentamicin are approximately 80% and 60%, respectively.6–13 Accordingly, alternative therapeutic options that are suitable for use in LMIC settings are needed for the empirical treatment of neonatal sepsis in these LMIC contexts.
Flomoxef is an oxacephem β-lactam with broad activity against Gram-negative and -positive bacteria (including anaerobes, but not pseudomonads).14,15 Of particular interest is its stability to degradation by ESBLs (except for Ambler class C enzymes, i.e. AmpC).16–18 Fosfomycin is a recently revived antibiotic with a unique mechanism of action—inhibition of bacterial cell wall synthesis via MurA inhibition, with broad Gram-negative and -positive activity.15,19 Both agents are off-patent, have a favourable safety profile,14,15,19 have a licence for neonatal use from a stringent regulatory authority, and have activity against the common pathogens that cause neonatal sepsis including those with commonly encountered resistance mechanisms.
Here, we assess the potential utility of the combination of flomoxef and fosfomycin by examining in vitro activity, and the nature and extent of any pharmacodynamic (PD) interaction in both chequerboard assays and hollow-fibre infection models (HFIMs), with a goal of defining a potential candidate regimen suitable for future clinical study.
Materials and methods
Antimicrobial agents
Pure compounds of flomoxef (Shionogi, Osaka) and fosfomycin (Sigma–Aldrich, St Louis) were used for all in vitro experiments, unless otherwise stated. Clinical vials of fosfomycin (Fomicyt, Kent Pharmaceuticals Ltd) were used for the HFIM experiments.
Media and agar
CAMHB (Sigma–Aldrich) was used as the primary medium in all experiments. Where fosfomycin was used, the CAMHB was supplemented with 25 mg/L glucose-6-phosphate (G6P) (Sigma–Aldrich). Mueller–Hinton agar (MHA) was used in all agar plates. For drug-containing plates, MHA was supplemented with antibiotic (with an additional 25 mg/L G6P, if fosfomycin) to a concentration of four times the antibiotic MIC for the specific bacterial strain, prepared within each antibiotic’s stability time limits.
Bacterial isolates
Isolates were supplied by JMI Laboratories, IHMA, PHE, LGC standards, University of Birmingham, University of Oxford and Royal Liverpool University Hospital. For the initial non-dynamic in vitro experiments, a panel of strains was assembled representing a range of possible neonatal sepsis bacterial pathogens and resistance mechanisms in an AMR-prevalent environment consisting of 10 strains of each of the following: S. agalactiae, MRSA, E. coli and K. pneumoniae. All Enterobacterales strains were ESBL (nine E. coli and nine K. pneumoniae strains) or carbapenemase producers (one E. coli and one K. pneumoniae strain) (Table S1, available as Supplementary data at JAC Online). All isolates were stored in glycerol at −80°C and subcultured onto two MHA plates for 18–24 h at 37°C prior to each experiment. In each experiment, colonies were suspended in PBS to McFarland standard 0.5 (1 × 108 cfu/mL) and diluted to the target concentration.
Antimicrobial susceptibility testing
MICs of fosfomycin and flomoxef for a panel of representative neonatal sepsis bacterial pathogens were determined using the EUCAST broth microdilution methodology.20E. coli ATCC 25922 or S. aureus ATCC 29213 were used as controls in all experiments, using quality control MIC values from both EUCAST and the Japanese Society of Chemotherapy.21,22 The antibiotic gradient strip assay method was used for fosfomycin MIC determination from isolates from the HFIM experiment. Briefly, an inoculum of the isolate was made using a suspension of a sweep of colonies into PBS to a McFarland standard of 0.5. A lawn of the inoculum was plated onto an MHA plate and an antibiotic gradient strip (Etest, bioMérieux, Marcy-l’Étoile, France) placed on the plate, which was subsequently incubated for 18–24 h at 37°C before reading.
In vitro PD assays
Chequerboard assays were used on selected strains (with fosfomycin and flomoxef MICs in the range >0.0625 to ≤32 mg/L) to assess the PD interaction of the flomoxef/fosfomycin combination. An 8 × 8 grid was assembled on a 96-well plate with 100 μL 1:2 serial dilutions of each antibiotic along each axis, with the final row/column having 0 mg/L of the appropriate drug. Each plate was assembled bespoke to each strain, with the maximum concentration of antibiotic being 4× MIC for that strain. An inoculum of 1 × 106 cfu/mL was prepared in CAMHB, with 100 μL added to each well of the prepared plate. The well containing 0 mg/L of each drug acted as the positive control; an additional row of blank CAMHB on the plate acted as negative control. Plates were incubated for 18–24 h at 37°C before being read by an optical densitometer (Varioskan, Thermo Fisher) at 600 nm. Plates were considered valid if the MIC on the monotherapy rows of the chequerboard were within one dilution of the previously determined MIC, the negative controls had no growth, and the prepared inoculum bacterial quantification was within 6–14 × 105 cfu/mL.
Raw OD readings were normalized to the positive control. The readouts were then modelled using Greco’s model of drug synergy23 using ADAPT 5.24 Meta-analysis was performed on the output of the model between individual strains using the R package ‘metafor’.25
HFIM
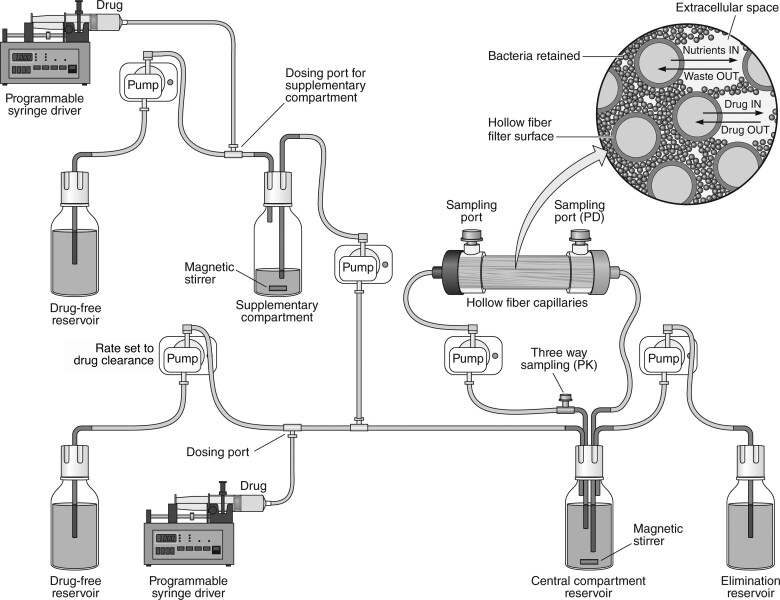
The HFIM is a well-established dynamic model simulating the PD effect of antimicrobials with dynamic concentrations.26 The HFIM method was used largely as described previously.27 Briefly, each arm in the HFIM was set up as demonstrated in Figure 1; monotherapy arms omitted the supplementary compartments. CAMHB was pumped into the central compartment at a rate set to simulate a physiological clearance rate for the drug, with media in the central compartment removed at an equivalent rate. The target-simulated half-lives for fosfomycin and flomoxef were 5.1 and 2.3 h, respectively.28,29 To account for the difference in clearance between flomoxef and fosfomycin, supplementary compartments were set up according the principles laid out by Blaser.30 No drug binding occurred within the model, and drug time–concentration profiles were aimed to replicate that of the in vivo free fraction.
Figure 1.
Schematic configuration of the HFIM with two agents. The supplementary compartments are omitted in arms with monotherapy.
Preliminary monotherapy experiments were performed with the ESBL-producing ST195 E. coli strain (flomoxef MIC = 0.125 mg/L; fosfomycin MIC = 1 mg/L; supplied by the University of Birmingham).31 Pharmacokinetic (PK) and PD outputs of these experiments were modelled using Pmetrics32 and simulated using estimated parameters with ADAPT24 to determine the flomoxef and fosfomycin doses required to achieve EC20, EC50 and EC80 in terms of maximal bactericidal effect within the HFIM. A 16-arm HFIM experiment was performed using a 4 × 4 dosing matrix using these three doses and no dose for both antibiotics in combination. The experiment was run over 96 h, with a target initial inoculum of 1 × 106 cfu/mL of ST195 inoculated into the hollow-fibre cartridges. Flomoxef was administered by bolus every 12 h to the primary central compartment only; fosfomycin was administered by bolus to the primary and supplementary central compartments every 12 h.
PK samples were taken for bioanalysis at four timepoints in dosing windows on Days 1 and 3 of the experiment before storing at −80°C ahead of bioanalysis (see Supplementary Text 1 for bioanalysis method). Samples of inoculum were taken from each hollow-fibre cartridge at four timepoints during the first 24 h, then once daily before administration of antibiotic doses until the 96 h timepoint. These were quantified via the ‘track-dilution’ method33 on drug-free and drug-containing plates. An additional 100 μL of inoculum was plated onto a further drug-free MHA plate to lower the limit of detection for total bacterial quantification (i.e. to 10 cfu/mL). Plates were then incubated at 37°C for 18–24 h for drug-free plates and 42–48 h for drug-containing plates before counting. MICs for any viable colonies from each arm on the final timepoint were determined via antibiotic gradient strip assay for fosfomycin, and broth microdilution method for flomoxef.
Further HFIM experiments were performed assessing the effect of clinically relevant flomoxef and fosfomycin regimens with neonatal-like time–concentration profiles (alone and in combination) against six Enterobacterales strains with a range of different flomoxef and fosfomycin MICs (Table 1). PK profiles of flomoxef and fosfomycin were designed to have the same half-lives as the previous experiments, with Cmax values of 50 and 250 mg/L, respectively.28,29 Each individual experiment consisted of four arms: monotherapy arms for both flomoxef and fosfomycin, a combination therapy arm and an untreated control. Each experiment lasted 7 days to reflect the typical treatment course of neonatal sepsis. Four PK samples were taken for bioanalysis in each of three dose intervals distributed evenly throughout the experiment. Four inoculum samples were taken on Day 1, and once every 24 h thereafter. These samples were quantified as before, with MICs for any viable colonies from each arm on the final timepoint.
Table 1.
Details of strains used in the HFIM experiments
| Strain ID | Species | Resistance mechanisms | MIC (mg/L) | |
|---|---|---|---|---|
| Fosfomycin | Flomoxef | |||
| ST195 | E. coli | CTX-M-14, aph(3′), aac(3), TEM-OSBL, mdf(A) | 1 | 0.125 |
| SPT 719 | E. coli | SHV ESBL, TEM ESBL | 2 | 0.25 |
| BAA2523 | E. coli | OXA-48 | 8 | 0.5 |
| I1025 | E. coli | mdf(A), ampC promoter mutation | 32 | 8 |
| 1216477 | K. pneumoniae | SHV-OSBL, TEM-OSBL, CTX-M-15 | 32 | 0.25 |
| 1280740 | K. pneumoniae | SHV-OSBL, TEM-OSBL, CTX-M-15, DHA-1 | 4 | 32 |
| 1256506 | K. pneumoniae | SHV-OSBL, TEM-OSBL, CTX-M-2, CMY-2 | 32 | 128 |
Modelling
Population PK models were constructed using the PK and PD outputs of the hollow-fibre experiments using the population PK program Pmetrics using a non-parametric adaptive grid (NPAG) estimation routine.32 The structural model was based on Greco’s models of pharmacological synergy.23 For further details, see Supplementary Text 2.
Data availability
The programs ADAPT and Pmetrics are publicly available, with instructions, at https://bmsr.usc.edu/software/adapt/and http://www.lapk.org/pmetrics.php, respectively.
Results
In vitro susceptibility testing
The flomoxef and fosfomycin MIC distributions for the panel of neonatal sepsis bacterial pathogens are shown in Table 2.
Table 2.
Flomoxef (top) and fosfomycin (bottom) MIC distributions for the panel of 40 representative bacterial strains
| Bacterial species | Flomoxef MIC (mg/L) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.0625 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | >32 | |
| E. coli | 1 | 4 | 2 | 1 | — | — | — | 1 | — | — | 1 |
| K. pneumoniae | — | 1 | 4 | 1 | — | — | — | 1 | — | 2 | 1 |
| MRSA | — | — | 1 | 1 | 1 | 1 | 5 | — | 1 | — | — |
| S. agalactiae | 1 | 1 | 7 | 1 | — | — | — | — | — | — | — |
| Fosfomycin MIC (mg/L) | |||||||||||
| ≤0.0625 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | >32 | |
| E. coli | — | — | — | — | — | 5 | 2 | 1 | — | — | 2 |
| K. pneumoniae | — | — | — | — | — | — | 1 | — | — | 1 | 8 |
| MRSA | — | — | 1 | 2 | 2 | 2 | 2 | — | — | — | 1 |
| S. agalactiae | — | — | — | — | — | 2 | 2 | 2 | 1 | 2 | 1 |
The modal flomoxef MIC was 0.25 mg/L. For the Enterobacterales, flomoxef MIC values were either ≤0.5 or ≥8 mg/L. Of the strains inhibited by higher flomoxef MICs, 3/6 carried a plasmid with a gene encoding an AmpC enzyme (e.g. DHA-1 or CMY-II); 1/6 carried an ampC gene promoter mutation causing chromosomal AmpC overexpression; 1/6 was a KPC3 producer; and 1/6 was a CTX-M-3 producer with no identified resistance mechanism for flomoxef. All S. agalactiae strains had flomoxef MICs of ≤0.5 mg/L; 9/10 MRSA strains had flomoxef MICs of ≤4 mg/L. The fosfomycin MIC50 values were 2–4, 1–2 and 8 mg/L for the E. coli, MRSA and S. agalactiae strains, respectively. The K. pneumoniae strains had a fosfomycin MIC50 of >32 mg/L.
In vitro drug–drug interaction modelling
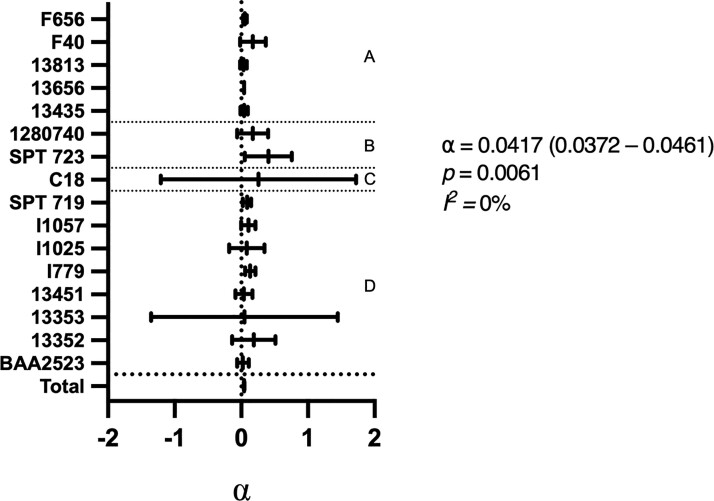
Static chequerboard assays were performed on the strains from the original panel with MICs in the range >0.0625 to ≤32 mg/L for both flomoxef and fosfomycin (n = 16), with the output fitted to the model originally described by Greco23 (Figure 2). The PD interaction was quantified using the point estimate of the interaction parameter α and an assessment of its 95% CI. α is interpreted as follows: a lower CI bound of >0 indicates synergy; an upper CI bound of <0 indicates antagonism; a CI containing 0 indicates additivity only. The magnitude of α is contextual to the experimental conditions and PD measure. Interpretations of the value of α is therefore limited to categorization of interaction and relative comparison with values determined from similar experimental conditions.
Figure 2.
Fitted α values from chequerboard assays outputs for 16 strains. The meta-analysis of the 16 strains is shown in the total summary statistic with the α, p and I2 values of this summary statistic alongside. (a) MRSA strains; (b) K. pneumoniae strains; (c) S. agalactiae strain; and (d) E. coli strains.
In this static system, 5/16 strains demonstrated synergy with the combination of flomoxef and fosfomycin (i.e. α was positive with a 95% CI lower bound of >0). In the remaining cases the drug combination was additive. When the outputs were combined using a meta-analysis, the value of α was 0.0417 (95% CI = 0.0372–0.0461) suggesting an overall conclusion of synergy, with low levels of inter-species and inter-strain heterogeneity.
PD interaction of flomoxef and fosfomycin using neonatal PK
The nature and magnitude of the PD interaction between flomoxef and fosfomycin was further explored using an HFIM using neonatal free concentration–time profiles in plasma for both drugs (Figure 1).28,29 The challenge strain was E. coli ST195, which is a CTX-M-14-producing isolate from Laos with flomoxef and fosfomycin MIC values of 0.125 and 1 mg/L, respectively.31 Preliminary dose-finding experiments for each drug as monotherapy defined the dose–exposure response relationships for bactericidal effect and the emergence of resistance. Concentrations corresponding to the EC20, EC50 and EC80 of maximal bactericidal effect were defined. For flomoxef, drug exposures (quantified in terms of the fAUC0–24) of 12, 50 and 90 mg·h/L resulted in the EC20, EC50 and EC80, respectively. Similarly, fAUC0–24 of 60, 250 and 500 mg·h/L resulted in the EC20, EC50 and EC80 for fosfomycin, respectively. The corresponding target Cmax values for these exposures were 2, 8.5 and 15 mg/L for flomoxef and 5, 20 and 40 mg/L for fosfomycin.
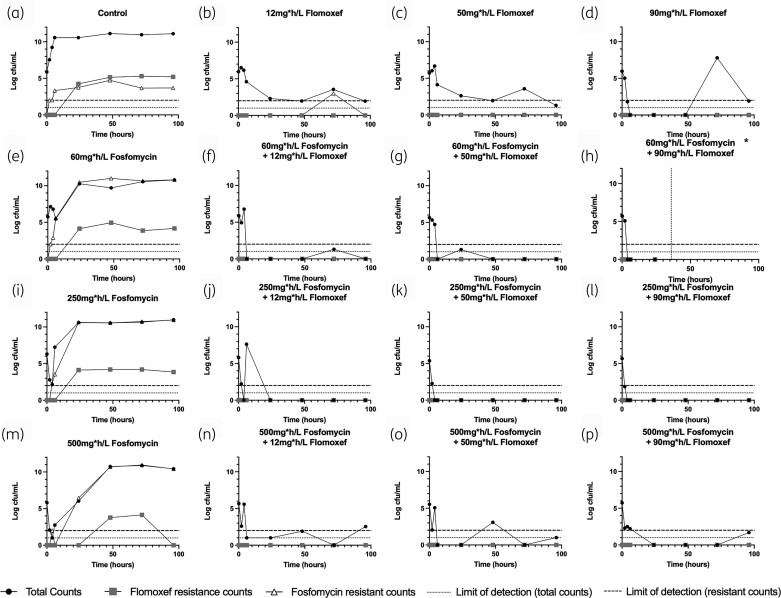
The PD of the combination of flomoxef and fosfomycin were determined in a 16-arm HFIM experiment that consisted of a 4 × 4 experimental design matrix, with a no-treatment control, all EC20, EC50 and EC80 doses supplied as monotherapy, and in all possible two-drug combinations (Figure 3). Target neonatal-like time–concentration profiles were successfully replicated (Figure S1). When administered alone, increasing fosfomycin exposures led to progressively increasing bacterial kill in the first 24 h. However, fosfomycin resistance rapidly emerged, with complete replacement of the WT population with a resistant clone that had fosfomycin MICs of >128 mg/L (Figure 3e, i and m). Flomoxef resistance emergence in these panels was no different to that emerging by mutational frequency in the no-treatment control (Figure 3a).
Figure 3.
PD output of 16-arm experiment. *The arm represented in (h) became contaminated at 36 h, so data after this point (indicated by vertical dashed line) were disregarded.
Progressively increasing flomoxef exposures (as monotherapy) resulted in a dose response in terms of bacterial killing in the first 24 h (i.e. greater magnitude of bacterial killing). However, sterility was not achieved in the monotherapy arms, with bacterial counts static below 4 log cfu/mL or lower for the majority of the experiment, with no observed emergence of resistance to flomoxef. Sustained >3 log kill was observed in all combination arms, with no emergence of resistance to either drug at any point, with greater bacterial kill and suppression of resistance compared with equivalent monotherapy doses.
The nature and magnitude of the PD interaction between flomoxef and fosfomycin was estimated by fitting an interaction model to the PK/PD data (Table 3). The values for the coefficient of determination for the observed-versus-predicted values, using mean parameter values, were 0.987 (flomoxef concentrations), 0.965 (fosfomycin concentrations), 0.815 (total bacterial counts), 0.685 (flomoxef-resistant counts) and 0.987 (fosfomycin-resistant counts) (Figure S2). The model demonstrated synergistic relationships for the antibiotic combination on susceptible and fosfomycin-resistant bacteria, with mean α values of 141.22 (95% credibility interval = 63.98–194.26) and 79.22 (95% credibility interval = 56.22–129.82). Given the lack of emergence of resistance in flomoxef-containing arms, an interpretable estimate of α value for this population is not possible.
Table 3.
Parameter value estimates with 95% credibility interval from HFIM PK/PD interaction model
| Parameter | Mean | Median | 95% credibility interval |
|---|---|---|---|
| V1 (L) | 0.391 | 0.400 | 0.368–0.419 |
| V2 (L) | 0.307 | 0.280 | 0.252–0.335 |
| CL1 (L/h) | 0.094 | 0.096 | 0.090–0.097 |
| CL2 (L/h) | 0.031 | 0.031 | 0.026–0.036 |
| Kgs | 1.08 | 1.02 | 0.916–1.180 |
| Kks | 3.71 | 3.53 | 2.88–4.75 |
| EC501s (mg/L) | 2.74 | 1.52 | 0.34–5.33 |
| EC502s (mg/L) | 11.20 | 10.92 | 4.26–17.63 |
| αs | 123.35 | 141.22 | 63.98–194.26 |
| Kgr1 | 0.80 | 0.61 | 0.34–0.78 |
| Kkr1 | 3.41 | 3.69 | 2.28–4.52 |
| EC501r1 (mg/L) | 3.32 | 1.86 | 0.51–5.54 |
| αr1a | 27.34 | 27.11 | 13.48–51.96 |
| Kgr2 | 1.50 | 0.88 | 0.52–2.64 |
| Kkr2 | 3.13 | 3.22 | 1.63–4.64 |
| EC502r2 (mg/L) | 30.08 | 30.59 | 25.10–33.12 |
| αr2 | 82.48 | 79.23 | 56.23–129.82 |
| H1s | 2.08 | 1.36 | 1.14–4.31 |
| H2s | 2.85 | 3.11 | 1.02–4.68 |
| H1r1 | 1.72 | 1.51 | 0.12–3.03 |
| H2r2 | 2.92 | 3.50 | 1.35–4.72 |
Kg, bacterial growth constant; Kk, bacterial kill constant; α, interaction parameter; H, Hill constant. Parameter suffices are defined as follows: 1 = relating to flomoxef; 2 = relating to fosfomycin; s = relating to WT bacterial population; r1 = relating to ‘flomoxef-resistant’ bacterial population; r2 = relating to ‘fosfomycin-resistant’ bacterial population.
αr1 is shown here for completeness, but given the lack of flomoxef resistance emerging in flomoxef-containing arms, this cannot be reliably interpreted.
Assessment of a candidate regimen of flomoxef and fosfomycin
The PD of the combination of flomoxef and fosfomycin were further assessed using neonate-like concentration–time profiles in the HFIM over a 7 day period. For fosfomycin, a dose equivalent to 100 mg/kg q12h (fAUC0–24 = 2400 mg·h/L) in humans was used, with a half-life of 5.2 h, replicating PK data from the NeoFosfo trial.28 For flomoxef, a dose equivalent to 20 mg/kg q12h (fAUC0–24 = 360 mg·h/L) in humans was used, with a half-life of 2.3 h.29 Six Enterobacterales strains with a range of resistance mechanisms and inhibited by a range of flomoxef and fosfomycin MICs (Table 1) were selected to be assessed in these HFIM experiments.
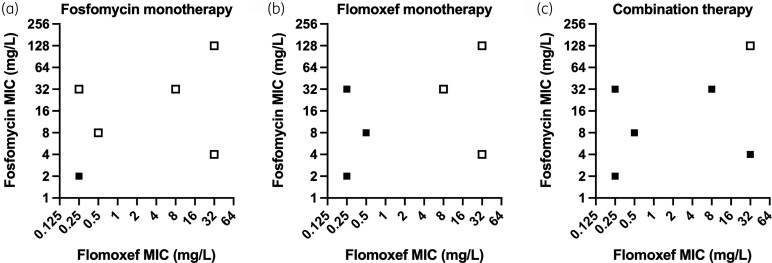
A summary of the PD outputs of these experiments is shown in Figure 4 (full PD outputs are shown in Figures S3–8). Successful replication of neonatal-like time–concentration profiles for both drugs was attained in all experiments (Figures S9–10). Fosfomycin monotherapy failed to achieve sustained kill in 5/6 strains inhibited by fosfomycin MICs of ≥4 mg/L, in keeping with previous studies.34 Flomoxef monotherapy resulted in sustained kill in the 3/6 strains with flomoxef MICs of ≤0.5 mg/L whilst it failed to achieve sustained kill in the remaining three strains with flomoxef MICs of ≥8 mg/L. Two of three strains with flomoxef MICs of ≥8 mg/L had identified production of plasmid-mediated AmpC enzymes, with the third having AmpC overexpression due to a mutation in the gene promoter. When combined, fosfomycin and flomoxef were able to kill 5/6 strains; the single strain that failed to be killed had flomoxef and fosfomycin MICs of 32 and 128 mg/L, respectively.
Figure 4.
Summary of the PD endpoints of the HFIM experiments replicating neonatal regimens of (a) fosfomycin monotherapy; (b) flomoxef monotherapy; and (c) flomoxef and fosfomycin in combination. Filled squares represent bacteria successfully killed to sterility; open squares represent failure to kill bacteria to sterility.
As with the 16-arm experiment, strains that survived fosfomycin-containing regimens (i.e. both monotherapy and combination arms) developed fosfomycin resistance (MICs of >128 mg/L). For all strains that survived a flomoxef-containing regimen, the flomoxef MICs increased by ≥2–4-fold. We saw no evidence of cross-resistance between the tested antibiotics.
Discussion
The combination of flomoxef and fosfomycin is potentially useful for the treatment of neonatal sepsis caused by ESBL-producing Enterobacterales. Both agents have well-established efficacy and safety, are licensed for use in neonates, and are off-patent, allowing potential affordability in LMIC settings. There is evidence of synergy in both bacterial killing and the prevention of emergence of fosfomycin resistance. Collectively, these observations provide the rationale for considering further clinical studies with this combination in neonates for clinically diagnosed neonatal sepsis that could be caused by a range of bacterial pathogens (including Enterobacterales, S. aureus and S. agalactiae).
Flomoxef exhibits a bimodal MIC distribution when assessed against Enterobacterales species.18,35–39 WT isolates have flomoxef MICs of ≤1 mg/L, whereas isolates with flomoxef MICs of ≥8 mg/L predominantly produce either various Ambler class A or B carbapenemases (that are also cephalosporinases) or AmpC, usually encoded by plasmids.37 Our HFIM experiments suggest that when flomoxef MICs are ≤1 mg/L, monotherapy has potent activity against WT Enterobacterales, even if the strain is producing an Ambler Class A ESBL. This observation is consistent with clinical data from adults where flomoxef had similar treatment outcomes to carbapenem treatment when flomoxef MICs were ≤1 mg/L for the infecting ESBL-producing Enterobacterales.40
Our data further show that flomoxef and fosfomycin in combination have synergy in both direct bactericidal effect and prevention of emergence of resistance to fosfomycin, allowing the combination to kill bacteria that neither flomoxef nor fosfomycin monotherapy could kill alone. The benefit of the combination is particularly relevant when flomoxef MICs are ≥8 mg/L for the infecting bacterium. Therefore, as an empirical treatment, the benefit of flomoxef and fosfomycin in combination over flomoxef monotherapy will depend on the prevalence of such ‘flomoxef-resistant’ bacteria balanced against the potential toxicity caused by the additional administration of fosfomycin. The epidemiological benefit of this combination may be better defined by the determination of a treatment success threshold, defined by the MIC values of both drugs, as suggested with other combinations.34 However, whilst a success threshold is suggested by our data, further work will be needed to define it in detail.
The predominant prevalent mechanism of resistance to flomoxef is degradation by AmpC β-lactamase production.37 The ampC gene is present in the chromosomes of many Gram-negative bacteria, including some Enterobacterales.41 AmpC is only usually produced at high levels by induction or stable derepression in the ESCPM group of organisms (i.e. Enterobacter spp., Serratia spp., Citrobacter freundii, Providencia spp. and Morganella spp.);41,42 these species are typically relatively infrequent causes of neonatal sepsis.1 Chromosomal ampC can be present in other Enterobacterales (e.g. E. coli) but AmpC is usually produced at low levels due to lacking the component required for induction and derepression and are so considered clinically insignificant. However, mutations in the ampC promoter region in these species can lead to clinically significant AmpC production.41 Additionally, non-AmpC-producing Enterobacterales can acquire plasmid-mediated ampC (e.g. blaCMY or blaDHA) and produce AmpC in clinically significant quantities.41
The BARNARDS study of neonatal sepsis in several LMIC settings demonstrated that infecting species with inducible AmpC accounted for 247/1038 (23.8%) Gram-negative and 247/2483 (9.9%) total identified blood culture isolates in neonatal sepsis.13,43 However, it is noted that many of these isolates came from a small proportion of study sites that may have biased the dataset to suggest a higher prevalence of infecting bacteria with inducible AmpC in neonates than the true prevalence across the geographical areas in the study. The study also demonstrated that 17/258 (6.6%) K. pneumoniae and 2/75 (2.7%) E. coli isolates contained a plasmid-mediated ampC gene. Preliminary data from the GARDP-commissioned NeoOBS study suggest similar rates: 5.6% and 2.5% plasmid-borne ampC carriage rates for K. pneumoniae and E. coli, respectively (S. Ellis, personal communication).
Flomoxef resistance may be mediated by porin modifications and increased production of efflux pumps too, as with other β-lactams, but the effect of these has largely not been characterized for flomoxef. Where specific associations have been characterized (e.g. loss of the OmpF porin leading to flomoxef resistance) the effect on the flomoxef MIC has been small.16
While the HFIM is increasingly used to establish neonatal antimicrobial PD,27,34,44 there are several limitations. The HFIM can only simulate drug concentrations in one PK compartment at a time. Examination of the PD in specific compartments (e.g. CSF, lung) is possible with a modified experimental set-up, but this requires data informing the PK distribution to these compartments. In our experiments we examined the systemic circulation compartment only. Whilst this is the relevant compartment of distribution for most cases of neonatal sepsis, these experiments did not model the CSF penetration of either drug and we cannot therefore comment on the suitability of these agents for treating neonatal meningitis. The neonatal CSF/plasma ratio is estimated to be 0.32 for fosfomycin28 and 0.05 for flomoxef.45
A further limitation is that the HFIM lacks the immune effectors present in neonates (even if immature), which will have an impact on the in vivo PD outcome. Similarly, the size of the inoculum used in these HFIM experiments was 105–106 cfu/mL—several orders of magnitude higher than the 100–103 cfu/mL estimated in the blood of neonates with sepsis.46,47 However, the impact of both of these factors only potentially worsens the in vitro performance of these agents in the HFIM compared with in vivo performance; the conclusions drawn are therefore conservative.
We propose that fosfomycin and flomoxef will be an efficacious and suitable regimen for the treatment of neonatal sepsis in LMIC settings. Our work establishes that this combination of agents is potentially useful by extending the spectrum of either agent as monotherapy, enabling enhanced bacterial killing and preventing the emergence of fosfomycin resistance. The regimens used in this study are therefore suitable for further assessment in clinical settings.
Supplementary Material
Acknowledgements
We thank Jonathan Folb for supplying S. agalactiae strains from the Royal Liverpool Hospital, and JMI and IMHA for gifting their strains for this work. We also thank Shionogi for gifting flomoxef material for this project and Yoshinori Yamano (Shionogi) for his comments on the manuscript. This work was presented, in part, at the European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) 2021 (abstract number 842).
Funding
This work was funded the Global Antibiotic Research and Development Partnership (GARDP). GARDP was funded by the German Federal Ministry of Education and Research; German Federal Ministry of Health; Médecins Sans Frontières; Netherlands Ministry of Health, Welfare and Sport; United Kingdom Department for International Development; and the United Kingdom National Institute of Health Research. C.D. is an MRC Clinical Training Fellow based at the University of Liverpool supported by the North West England Medical Research Council Fellowship Scheme in Clinical Pharmacology and Therapeutics, which is funded by the Medical Research Council (Award Ref. MR/N025989/1), Roche Pharma, Eli Lilly and Company Limited, UCB Pharma, Novartis, the University of Liverpool and the University of Manchester. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Transparency declarations
Shionogi provided the flomoxef material free of charge, but provided no additional funding. W.H. holds or has recently held research grants with UKRI, National Institutes of Health, National Institute of Health Research, F2G, Spero Therapeutics, Antabio, Pfizer, Bugworks, Phico Therapeutics, BioVersys, GARDP and NAEJA-RGM. He is (or has recently been) a consultant for Appili Therapeutics, F2G, Spero Therapeutics, NAEJA-RGM, Centauri, Pfizer, Phico Therapeutics and Venatorx. He is a member of the Specialist Advisory Committee for GARDP and the Specialty National Co-lead for Infectious Diseases for the National Institute of Health Research (NIHR). There is nothing further to declare for all other authors.
Supplementary data
Supplementary Text 1 and Text 2, Table S1 and Figures S1 to S10 are available as Supplementary data at JAC Online.
References
- 1. Shane AL, Sánchez PJ, Stoll BJ. Neonatal sepsis. Lancet 2017; 390: 1770–80. [DOI] [PubMed] [Google Scholar]
- 2. Oza S, Lawn JE, Hogan DRet al. Neonatal cause-of-death estimates for the early and late neonatal periods for 194 countries: 2000-2013. Bull World Health Organ 2015; 93: 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seale AC, Blencowe H, Manu AAet al. Estimates of possible severe bacterial infection in neonates in sub-Saharan Africa, south Asia, and Latin America for 2012: a systematic review and meta-analysis. Lancet Infect Dis 2014; 14: 731–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fuchs A, Bielicki J, Mathur Set al. Antibiotic use for sepsis in neonates and children: 2016 evidence update. WHO-Reviews 2016: 7: http://www.who.int/selection_medicines/committees/expert/21/applications/s6_paed_antibiotics_appendix4_sepsis.pdf?ua=1. [Google Scholar]
- 5. WHO . Pocket book of hospital care for children. Second edn. 2013. https://www.who.int/publications/i/item/978-92-4-154837-3.
- 6. Investigators of the Delhi Neonatal Infection Study (DeNIS) collaboration . Characterisation and antimicrobial resistance of sepsis pathogens in neonates born in tertiary care centres in Delhi, India: a cohort study. Lancet Glob Heal 2016; 4: e752–60. [DOI] [PubMed] [Google Scholar]
- 7. Labi A-K, Obeng-Nkrumah N, Bjerrum Set al. Neonatal bloodstream infections in a Ghanaian tertiary hospital: are the current antibiotic recommendations adequate? BMC Infect Dis 2016; 16: 598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bandyopadhyay T, Kumar A, Saili Aet al. Distribution, antimicrobial resistance and predictors of mortality in neonatal sepsis. J Neonatal Perinatal Med 2018; 11: 145–53. [DOI] [PubMed] [Google Scholar]
- 9. Jajoo M, Manchanda V, Chaurasia Set al. Alarming rates of antimicrobial resistance and fungal sepsis in outborn neonates in North India. PLoS One 2018; 13: e0180705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yadav NS, Sharma S, Chaudhary DKet al. Bacteriological profile of neonatal sepsis and antibiotic susceptibility pattern of isolates admitted at Kanti Children’s Hospital, Kathmandu, Nepal. BMC Res Notes 2018; 11: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pokhrel B, Koirala T, Shah Get al. Bacteriological profile and antibiotic susceptibility of neonatal sepsis in neonatal intensive care unit of a tertiary hospital in Nepal. BMC Pediatr 2018; 18: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chaurasia S, Sivanandan S, Agarwal Ret al. Neonatal sepsis in South Asia: huge burden and spiralling antimicrobial resistance. BMJ 2019; 364: k5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sands K, Carvalho MJ, Portal Eet al. Characterization of antimicrobial-resistant Gram-negative bacteria that cause neonatal sepsis in seven low- and middle-income countries. Nat Microbiol 2021; 6: 512–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ito M, Ishigami T. The meaning of the development of flomoxef and clinical experience in Japan. Infection 1991; 19: 253–7. [DOI] [PubMed] [Google Scholar]
- 15. Darlow CA, da Costa RMA, Ellis Set al. Potential antibiotics for the treatment of neonatal sepsis caused by multidrug-resistant bacteria. Pediatr Drugs 2021; 23: 465–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jacoby GA, Carreras I. Activities of β-lactam antibiotics against Escherichia coli strains producing extended-spectrum β-lactamases. Antimicrob Agents Chemother 1990; 34: 858–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Neu HC, Aswapokee N. Antibacterial activity of a new 1-oxa cephalosporin compared with that of other β-lactam compounds. Antimicrob Agents Chemother 1979; 16: 141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang Q, Zhang H, Cheng Jet al. In vitro activity of flomoxef and comparators against Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis producing extended-spectrum β-lactamases in China. Int J Antimicrob Agents 2015; 45: 485–90. [DOI] [PubMed] [Google Scholar]
- 19. Silver LL. Fosfomycin: mechanism and resistance. Cold Spring Harb Perspect Med 2017; 7: a025262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) . Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin Microbiol Infect 2003; 9: ix–xv. [DOI] [PubMed] [Google Scholar]
- 21. EUCAST . Routine and extended internal quality control for MIC determination and disk diffusion as recommended by EUCAST. Version 11.0. 2021. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/QC/v_11.0_EUCAST_QC_tables_routine_and_extended_QC_pdf.pdf.
- 22. Nagayama A, Yamaguchi K, Watanabe Ket al. Final report from the Committee on Antimicrobial Susceptibility Testing, Japanese Society of Chemotherapy, on the agar dilution method (2007). J Infect Chemother 2008; 14: 383–92. [DOI] [PubMed] [Google Scholar]
- 23. Greco WR, Park HS, Rustum YM. Application of a new approach for the quantitation of drug synergism to the combination of cis-diamminedichloroplatinum and 1-β-d-arabinofuranosylcytosine. Cancer Res 1990; 50: 5318–27. [PubMed] [Google Scholar]
- 24. D’Argenio DZ, Schumitzky A, Wang X. ADAPT 5 User’s Guide: Pharmacokinetic/Pharmacodynamic Systems Analysis Software. Biomedical Simulations Resource; 2009. https://bmsr.usc.edu/files/2013/02/ADAPT5-User-Guide.pdf. [Google Scholar]
- 25. Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw 2010; 36. 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- 26. Cadwell JJS. The hollow fiber infection model for antimicrobial pharmacodynamics and pharmacokinetics. Adv Pharmacoepidemiol Drug Saf 2012; Special Issue: 10.4172/2167-1052.S1-007. [DOI] [Google Scholar]
- 27. Ramos-Martín V, Johnson A, Livermore Jet al. Pharmacodynamics of vancomycin for CoNS infection: experimental basis for optimal use of vancomycin in neonates. J Antimicrob Chemother 2016; 71: 992–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kane Z, Gastine S, Obiero Cet al. IV and oral fosfomycin pharmacokinetics in neonates with suspected clinical sepsis. J Antimicrob Chemother 2021; 76: 1855–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shionogi . Flumarin for intravenous injection SPC. 2009.
- 30. Blaser J. In-vitro model for simultaneous simulation of the serum kinetics of two drugs with different half-lives. J Antimicrob Chemother 1985; 15: 125–30. [DOI] [PubMed] [Google Scholar]
- 31. Anu K, Esther K, Dunn Steven Jet al. Real-time sampling of travelers shows intestinal colonization by multidrug-resistant bacteria to be a dynamic process with multiple transient acquisitions. bioRxiv 2019; 827915. [Google Scholar]
- 32. Neely MN, Van Guilder MG, Yamada WMet al. Accurate detection of outliers and subpopulations with Pmetrics, a nonparametric and parametric pharmacometric modeling and simulation package for R. Ther Drug Monit 2012; 34: 467–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jett BD, Hatter KL, Huycke MMet al. Simplified agar plate method for quantifying viable bacteria. Biotechniques 1997; 23: 648–50. [DOI] [PubMed] [Google Scholar]
- 34. Darlow CA, Docobo-Perez F, Farrington Net al. Amikacin combined with fosfomycin for treatment of neonatal sepsis in the setting of highly prevalent antimicrobial resistance. Antimicrob Agents Chemother 2021; 65: e0029321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee C-H, Liu J-W, Li C-Cet al. Spread of ISCR1 elements containing blaDHA-1 and multiple antimicrobial resistance genes leading to increase of flomoxef resistance in extended-spectrum-β-lactamase-producing Klebsiella pneumoniae. Antimicrob Agents Chemother 2011; 55: 4058–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang Q, Zhang H, Cheng Jet al. Flomoxef showed excellent in vitro activity against clinically important gram-positive and gram-negative pathogens causing community- and hospital-associated infections. Diagn Microbiol Infect Dis 2015; 81: 269–74. [DOI] [PubMed] [Google Scholar]
- 37. Matsumura Y, Yamamoto M, Nagao Met al. In vitro activities and detection performances of cefmetazole and flomoxef for extended-spectrum β-lactamase and plasmid-mediated AmpC β-lactamase-producing Enterobacteriaceae. Diagn Microbiol Infect Dis 2016; 84: 322–7. [DOI] [PubMed] [Google Scholar]
- 38. Lee C-H, Su L-H, Tang Y-Fet al. Treatment of ESBL-producing Klebsiella pneumoniae bacteraemia with carbapenems or flomoxef: a retrospective study and laboratory analysis of the isolates. J Antimicrob Chemother 2006; 58: 1074–7. [DOI] [PubMed] [Google Scholar]
- 39. Matsumura Y, Yamamoto M, Nagao Met al. Multicenter retrospective study of cefmetazole and flomoxef for treatment of extended-spectrum-β-lactamase-producing Escherichia coli bacteraemia. Antimicrob Agents Chemother 2015; 59: 5107–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee CH, Su LH, Chen FJet al. Comparative effectiveness of flomoxef versus carbapenems in the treatment of bacteraemia due to extended-spectrum β-lactamase-producing Escherichia coli or Klebsiella pneumoniae with emphasis on minimum inhibitory concentration of flomoxef: a retrospective study. Int J Antimicrob Agents 2015; 46: 610–5. [DOI] [PubMed] [Google Scholar]
- 41. Jacoby GA. AmpC β-lactamases. Clin Microbiol Rev 2009; 22: 161–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meini S, Tascini C, Cei Met al. AmpC β-lactamase-producing Enterobacterales: what a clinician should know. Infection 2019; 47: 363–75. [DOI] [PubMed] [Google Scholar]
- 43. Thomson KM, Dyer C, Liu Fet al. Effects of antibiotic resistance, drug target attainment, bacterial pathogenicity and virulence, and antibiotic access and affordability on outcomes in neonatal sepsis: an international microbiology and drug evaluation prospective substudy (BARNARDS). Lancet Infect Dis 2021; 21: 1677–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramos-Martín V, Johnson A, McEntee Let al. Pharmacodynamics of teicoplanin against MRSA. J Antimicrob Chemother 2017; 72: 3382–9. [DOI] [PubMed] [Google Scholar]
- 45. Okada T, Furukawa S. Clinical evaluation of flomoxef in pediatrics and a study on the penetration into cerebrospinal fluid. Jpn J Antibiot 1987; 40: 1477–85. [PubMed] [Google Scholar]
- 46. Kellogg JA, Ferrentino FL, Goodstein MHet al. Frequency of low level bacteremia in infants from birth to two months of age. Pediatr Infect Dis J 1997; 16: 381–5. [DOI] [PubMed] [Google Scholar]
- 47. Dietzman DE, Fischer GW, Schoenknecht FD. Neonatal Escherichia coli septicemia—bacterial counts in blood. J Pediatr 1974; 85: 128–30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The programs ADAPT and Pmetrics are publicly available, with instructions, at https://bmsr.usc.edu/software/adapt/and http://www.lapk.org/pmetrics.php, respectively.